A comprehensive overview of published associations between genetic variants and human neuropathic pain conditions with validation of reported findings in the U.K. Biobank.
Keywords: Neuropathic pain, Genetic association studies, Genetic variants, Single nucleotide polymorphisms, U.K. Biobank
Abstract
Numerous studies have shown associations between genetic variants and neuropathic pain disorders. Rare monogenic disorders are caused by mutations of substantial effect size in a single gene, whereas common disorders are likely to have a contribution from multiple genetic variants of mild effect size, representing different biological pathways. In this review, we survey the reported genetic contributors to neuropathic pain and submit them for validation in a 150,000-participant sample of the U.K. Biobank cohort. Successfully replicated association with a neuropathic pain construct for 2 variants in IL10 underscores the importance of neuroimmune interactions, whereas genome-wide significant association with low back pain (P = 1.3e-8) and false discovery rate 5% significant associations with hip, knee, and neck pain for variant rs7734804 upstream of the MAT2B gene provide evidence of shared contributing mechanisms to overlapping pain conditions at the molecular genetic level.
1. Introduction
Neuropathic pain arises from a lesion or disease of the somatosensory system.66 Although some conditions have a known genetic cause, others develop as part of disease sequelae or posttraumatic complications.
The defining feature is an aberrant nociceptive network manifesting as pain occurring spontaneously or without adequate stimulation.6,21 In the case of rare familial disorders, abnormal nociceptive signalling is genetically encoded, and many causal variants are known. Acquired neuropathic pain may develop secondarily to another condition, such as diabetes or cancer, in which nerve damage is often a consequence of disease progression. Alternatively, nerve damage or lesion may occur during physical trauma or surgery and result in a neuropathic pain condition. In all these cases, susceptibility to chronic pain varies beyond what environmental factors can explain. Although twin studies for common neuropathic pain conditions have not been done, substantial heritability has been reported in multiple other chronic pain conditions103—including back and neck pain, which often have a neuropathic component. Animal models of neuropathic pain have likewise revealed a significant genetic contribution.32,101
During the past decade, the number of studies aiming to identify genetic factors in neuropathic pain conditions has grown in the hope of elucidating the molecular risk factors and identifying treatment targets. In this review, we summarise these studies and present the current landscape of neuropathic pain molecular pathophysiology as it has been informed by them.
2. Methods
To obtain a list of original studies reporting genetic association or linkage analysis with neuropathic pain conditions, we used the search method described in Ref. 158. Briefly, after drawing the initial list from the Human Pain Genetics Database (https://humanpaingenetics.org/hpgdb), a search was conducted in Google Scholar using the name of each disorder and one of the following terms: “genetic association,” “variant,” or “polymorphism.” In addition, we performed a search in PubMed, using the string: (((gene OR variant OR polymorphism) AND neuropath*) AND pain*) under the category “Text Word.” Last, recent reviews of neuropathic pain genetics were perused for studies overlooked using the above methods. Studies reporting association with a multi-symptom condition were included if neuropathic pain was one of the described symptoms, using the rationale that pleiotropic genetic loci should be considered neuropathic pain modulators whether or not they affect other clinical phenotypes. Although publications were primarily screened by title and abstract, the text and relevant tables were perused if clarifications were necessary.
After compiling the list of genes containing variants with reported associations in common neuropathic pain conditions, we took all variants with a minimum minor allele frequency of 1% (except human leukocyte antigen [HLA]-region variants because of their complex haplotypic structure) and checked them for validation in the U.K. Biobank, UKBB (application no. 20802). This public repository of genotypic and phenotypic data for 500,000 U.K. individuals, aged 40 to 69 years, is a data set of unprecedented proportions in human genetic association studies.131 To construct a neuropathic pain phenotype, we grouped the following conditions (self-reported or determined by interview with a clinical nurse, UKBB Data Field, hereafter DF, 20002): peripheral neuropathy (code 1255), diabetic neuropathy and ulcers (1468), shingles (1573), trigeminal neuralgia (1523), sciatica (1476), spinal stenosis (1536), peripheral nerve injury (1394), trapped or compressed nerve (1257), prolapsed or slipped disk (1312), varicella zoster virus (1674), and peripheral nerve disorder (1254). Individuals self-reporting one or more of these conditions were classified as cases and those reporting no pain conditions (DF 6159) as controls. In addition to neuropathic pain, we checked the list of variants in Table 1 for association with 4 site-specific pain conditions (>3 months' duration) in the UKBB: back (DF 3571), hip (DF 3414), knee (DF 3773), and neck (DF 3404). Controls were the same as in the neuropathic pain group (no pain under DF 6159). To test for association with these phenotypes, we ran regression analyses on the available sample of 150,000 individuals of predominantly Caucasian ancestry using SNPTEST,92 version 2.5.2.
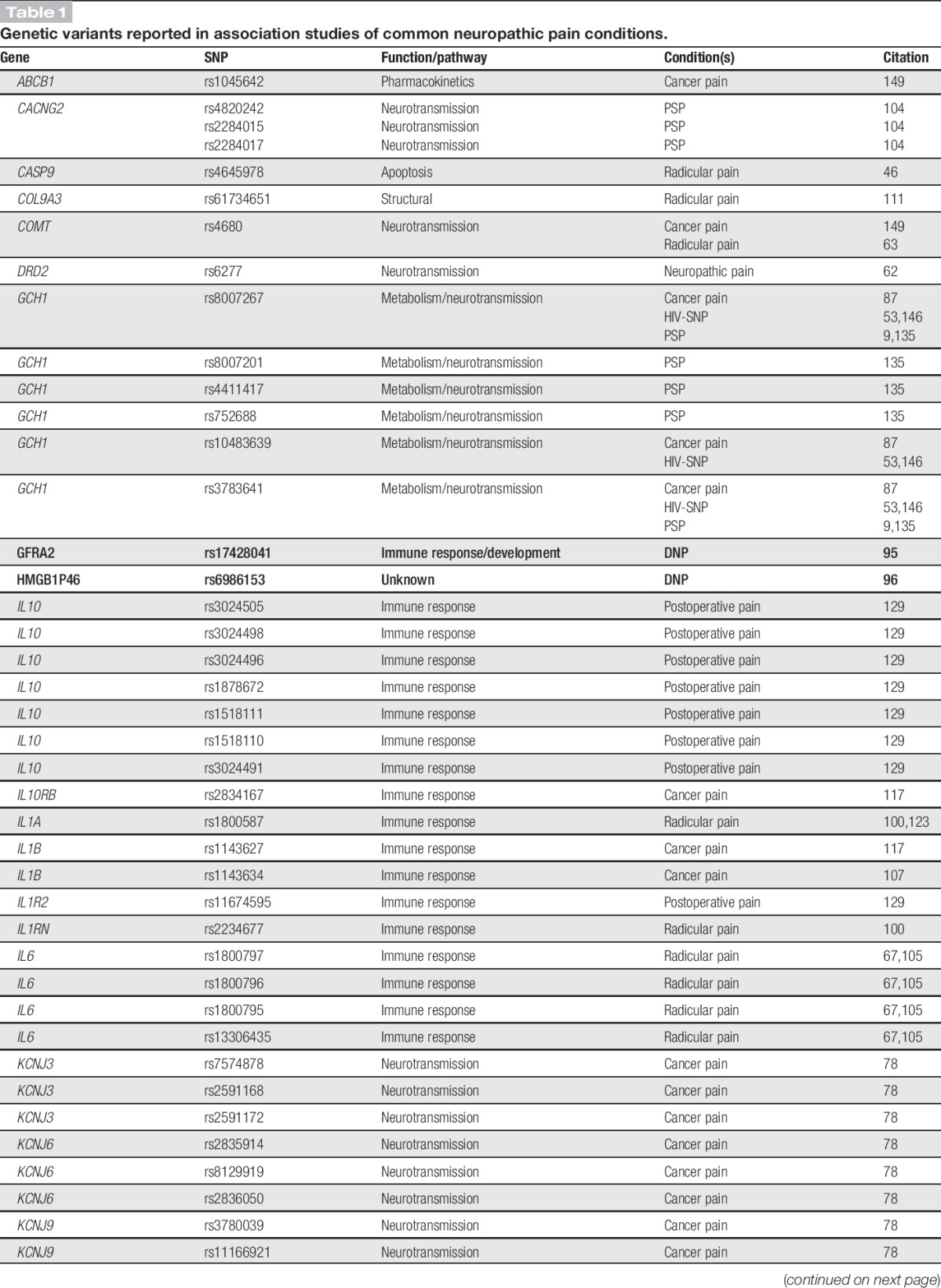
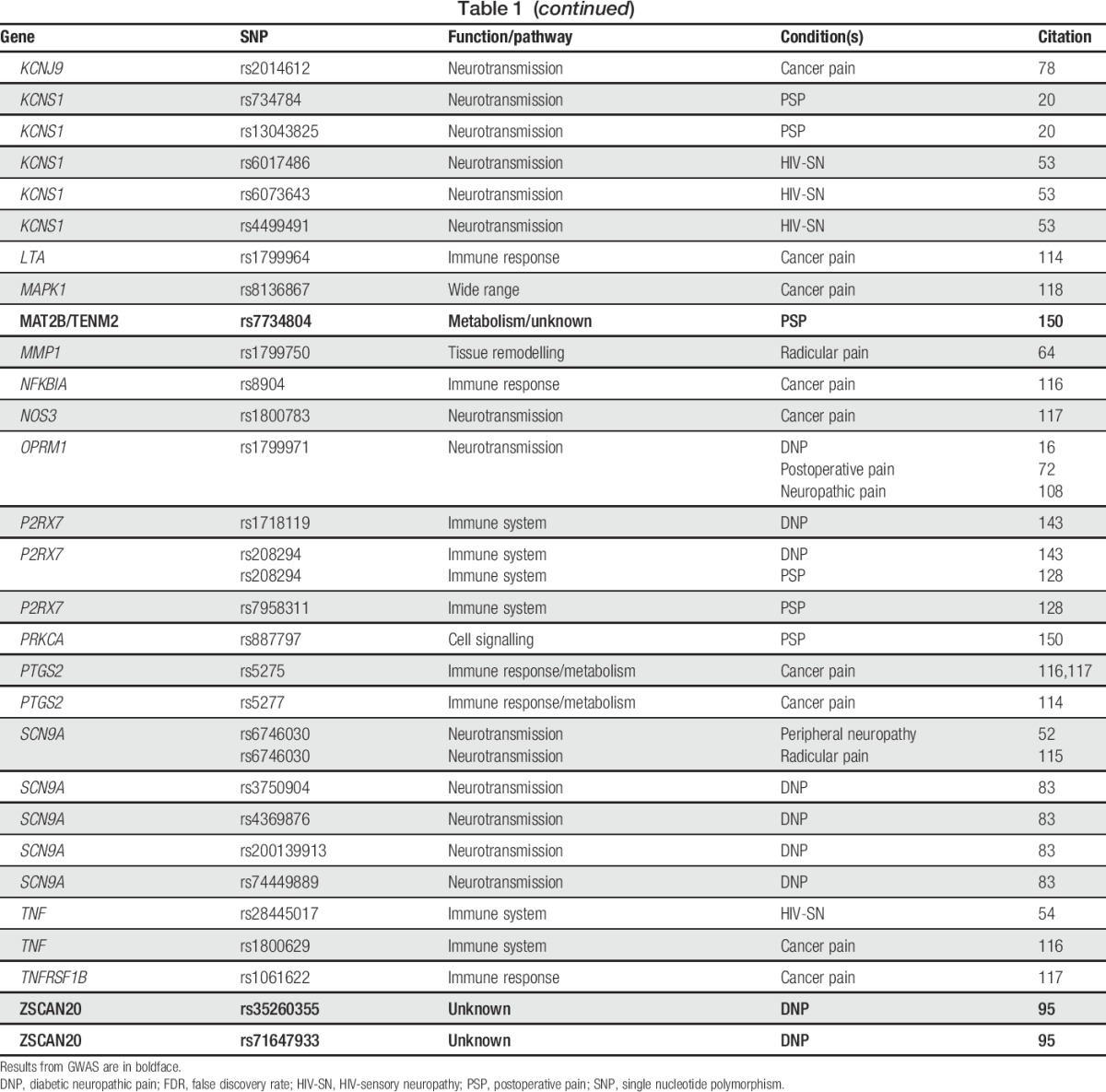
Table 1.
Genetic variants reported in association studies of common neuropathic pain conditions.
3. Results
The literature review findings are divided into rare monogenic disorders and common disorders with multiple associated risk loci and a complex etiology. The 2 classes of disorders are studied using different approaches: while rare conditions require linkage analysis of multi-generation pedigrees, common conditions require association studies of large cohorts of unrelated individuals. Section 3.1 is devoted to findings from familial rare variant studies, and section 3.2 focuses on common disorders with neuropathic pain as a secondary attribute.
3.1. Monogenic disorders
As the name suggests, monogenic disorders are often caused by variants in a single gene. The effect of such a variant can either exacerbate nociception or annul it. Although not painful, the latter class of disorders is tied to genetic loci that directly participate in pain signalling or contribute to the vitality of sensory neurons and are therefore equally important to our understanding of pain processing. In fact, some of the same genes that harbour painful neuropathic variants also carry mutations leading to painless states.
3.1.1. Painful rare monogenic disorders
Erythromelalgia is marked by redness and painful swelling of hands and feet, symptoms that have been attributed to C-fibre hypersensitivity.109 Its hereditary form, primary erythromelalgia, has shown linkage to rare hyperfunctional variants in sodium channel NaV1.7 (SCN9A), discovered as causal153 and repeatedly replicated in Chinese49,79,84,85,157 and Caucasian33,35,36,97,121 individuals.
The implicated SCN9A variants have been reported to change the electrophysiological properties of dorsal root ganglion neurons, thereby affecting nociceptive signalling.26,33,39 The magnitude of effect on these functions seems to modulate the timing of disease onset. Thus, a variant with a smaller effect on hyperpolarization has been reported to be associated with later onset of erythromelalgia.47 An alternative theory posits that SCN9A variants affecting different electrophysiological properties translate to different neuropathic conditions.38 In cellular assays, this group has demonstrated that alleles responsible for erythromelalgia disrupted fast inactivation in nociceptors, whereas alleles that lower firing thresholds, slow deactivation, and potentiate currents result in paroxysmal extreme pain disorder.38 The latter is also a rare neuropathic disorder, which manifests as rectal, periocular, and perimandibular pain, and affected individuals have been reported to carry gain-of-function variants in SCN9A.38,42,94 The genetic contribution of SCN9A variants to paroxysmal extreme pain disorder and their cellular phenotype has been also reported in Ref. 34. This gene's variants have likewise been reported to be associated with unexplained chronic neuropathic pain.28
Sodium channels NaV1.7, NaV1.8, and NaV1.9 (SCN9A, SCN10A, and SCN11A, respectively), have been found to harbour variants involved in a set of conditions collectively known as idiopathic painful small fibre neuropathies. These conditions affect small-diameter A-delta and C fibers, and their clinical manifestations include sudden bouts of pain propagating inward from the extremities. Associations with SCN9A,28,40,48 SCN10A,27,41,57 and SCN11A50,56,81 are supported by their cellular phenotype—hyperexcitability in dorsal root ganglion neurons.40,41,48,50,56,57
Aside from sodium channels, variants in 4 other genes have been reported in connection with painful peripheral neuropathies. α-galactosidase A, GLA, has been reported in a small fibre neuropathy patient.31 Myelin protein zero, MPZ, has been reported in a family with debilitating neuropathic pain and demyelination. A variant in a subunit of kinesin, KIF5A, a protein involved in intracellular motility, has been reported to cause a form of hereditary spastic paraplegia with axonal neuropathy and pain.119 Individuals with a rare case of late-onset hereditary peripheral neuropathy have been reported to carry a variant in α-N-acetyl-glucosaminidase, NAGLU.138
3.1.2. Painless rare monogenic disorders
Among rare congenital sensory disorders, a class of conditions defined by insensitivity to pain has been linked to hypofunctional variants in SCN9A22,23,70,74,90,112,126 and hyperfunctional variants in SCN11A.80,113 These findings are consistent with the electrophysiological properties of the 2 sodium channels. Increased activity in NaV1.9 leads to longer neuronal depolarisation, which inhibits NaV1.7 and NaV1.8 activity in nociceptors, effectively shutting down pain signal transmission.
In addition, insensitivity to pain is one of the defining symptoms in a group of disorders known collectively as hereditary sensory and autonomic neuropathies (HSANs). Primarily, variations in autonomic symptoms and genetic causes segregate these disorders into 8 major subtypes, 7 of which include insensitivity to pain. The different pathways leading to pain insensitivity are tagged by the 11 operative genes whose variants have been found in affected individuals: SPTLC1, DMNT1, WNK1, KIF1A, RETREG1, SCN9A, ELP1, NTRK1, NGF, SCN11A, and PRDM12. SPTLC1 encodes a subunit of serine palmitoyltransferase, whose variant-compromised activity contributes to neuronal toxicity and death. DNMT1 encodes a DNA methyltransferase, whose impaired function disrupts neuronal maintenance. Variants in these 2 genes have been reported as causal in HSAN, type I.3,5,8,30,55,69,155 WNK1 encodes WNK lysine deficient protein kinase 1, whose variants lead to a reduced number of sensory neurons, although the exact mechanism for this is unknown. KIF1A encodes kinesin family member 1A, an axonal transporter of synaptic vescicles, and variants in this gene lead to impaired neuronal function. RETREG1 encodes reticulophagy regulator 1, whose variants disrupt its physiological function in autophagy leading to neuronal toxicity and death. WNK1, KIF1A, and RETREG1 variants lead to different subtypes of HSAN, type II.29,76,77,102,120,125 Variants in SCN9A have also been implicated in HSAN, type II.156 ELP1 encodes elongator complex protein 1, a scaffolding protein whose variants have been found in patients with HSAN, type III.2,127 HSAN, type IV, as its alternative name—congenital insensitivity to pain with anhidrosis—suggests, is defined by insensitivity to pain as its primary symptom. Variants in NTRK1, which encodes neurotrophic receptor tyrosine kinase 1, disrupt its role in neuronal cell maintenance, thereby leading to congenital insensitivity to pain with anhidrosis.1,12,13,45,58,60,61,75,82,86,88,98,124,134,141,147,148,154 NGF encodes nerve growth factor beta, the binding partner of NTRK1. Its variants lead to HSAN, type V.14,37 HSAN subtypes VII and VIII were characterised more recently, and their genetic causes have been determined to be variants in SCN11A80,113,152 and PRDM12,15 respectively. A recent study showed variants in FLVCR1, a heme transporter, in a patient with an unclassified HSAN.17 All of these genes, in their pathological variant form, abolish pain sensitivity by depleting the number of viable sensory neurons.
3.2. Common disorders
Genetic studies of common neuropathic pain conditions suffer from a lack of clearly defined phenotyping.144 Despite the recently updated definition and grading system published by the neuropathic pain task force,43,66,140 this type of pain remains resistant to accurate diagnosis. Although diabetic neuropathy, radicular pain, trigeminal neuralgia, and viral infection-related sensory neuropathies are among the more clearly defined neuropathic pain conditions, cancer pain and postoperative pain display mixed phenotypes of neuropathic and nociceptive pain. Many genetic reports do not adhere to standardised terminology and diagnostic procedures,10 and some studies report an association with chronic pain in cancer or postsurgery patients without characterising it. Nevertheless, given the substantial neuropathic component of these conditions, we include these studies in our review.
Common neuropathic pain conditions, in which suspected genetic contribution derives from many different loci, benefit from studies in large cohorts with hypothesis-free scans of the entire genome. Thus, the publication of 3 genome-wide association studies (GWAS) in neuropathic pain during the past 3 years is an exciting recent development, even if the top findings in these studies are just below the threshold of genome-wide significance.95,96,150 Aside from these GWAS, studies done in patients genotyped or sequenced at targeted gene panels have been informative about loci that may modulate susceptibility to developing neuropathic pain after a traumatic event, physical injury, or the onset of another disease. Variants identified in this disease category are listed in Table 1. The most frequently investigated gene with variants reported to be associated with common neuropathic pain is GCH1.
3.2.1. Diabetic neuropathy
Diabetic neuropathy is a condition of polar extremes, characterised by pain at one extreme and insensitivity at the other, and its prevalence is up to 50% in diabetes patients.136 Prolonged glycemic mismanagement and disruption of nerve microvasculature are the putative disease-associated risk factors.136,137 However, given the incomplete penetrance of neuropathic pain in diabetic patients, the search for genetic contributors is ongoing. The first GWAS in the domain of painful neuropathies to be published was on diabetic neuropathic pain.95 Closely following came a second report from the same group.96 Both studies were conducted with the same cohort of almost 7000 genotyped diabetic patients. In the first study, cases of neuropathic pain were defined as individuals with a history of at least 1 specified diabetic peripheral neuropathy drug prescription and a positive monofilament test indicative of sensory neuropathy. In the second study, the monofilament test requirement was dropped, but cases had to have at least 2 prescriptions. The results in the first study showed a nearly genome-wide significant association for glial cell-derived neurotrophic factor family receptor alpha 2, encoded by GFRA2. In the second study, the same level of significance was reported for a wide region in women on chr1p35.1, gated by zinc-finger and SCAN domain-encoding ZSCAN20 on one end and toll-like receptor 12 pseudogene TLR12P on the other, and in men a high-mobility group box 1 pseudogene 46, HMGB1P46, on chr8p23.1.
Other groups have conducted association studies to examine the effects of a priori determined genetic variants, based on their roles in other related diseases. Among them, 1 has reported an association for the well-known A188G hypofunctional variant in µ-opioid receptor, OPRM1, with foot ulcer pain in diabetic patients.16 Another reported 2 hyperfunctional variants in purinergic receptor 7, P2RX7, to be associated with higher pain in women diagnosed with diabetic neuropathy.143 Interleukin-4 receptor, encoded by IL4R, has been reported to have its variable number of tandem repeats associated with diabetic neuropathy.7 Last, several hyperfunctional variants in sodium channel NaV1.7 (SCN9A), were found in a cohort of nearly 1000 individuals with diabetes to be associated with neuropathic pain.83
3.2.2. Radicular pain
Spinal disk herniation or prolapse leads to neuropathic pain through a combination of inflammation and nerve compression.59 Accompanying pain intensity and duration vary, and several studies have reported genetic variants as risk modifiers. Inflammatory mediators have shown association with herniated disk-related pain intensity, namely IL1A100,123; IL1RN100; and IL6.67,105 In addition, associations have been published for variants in OPRM1,108 COMT,63 COL9A3 (encoding a chain of type IX collagen),111 MMP1 (encoding matrix metalloproteinase 1),64 and CASP9 (encoding caspase-9).64
3.2.3. Trigeminal neuralgia
Trigeminal neuralgia manifests as paroxysmal bursts of pain along the innervation pathway of the trigeminal nerve.73 According to recently proposed diagnostic criteria, its onset may be: (1) idiopathic, (2) caused by an underlying condition, or (3) accompanying pressure exerted on the trigeminal nerve root by the surrounding blood vessels.24 Genetic studies of trigeminal neuralgia have been scarce, with 1 report suggesting a variant in serotonin transporter, SLC6A4,25 and a recent study suggesting sodium channel NaV1.6, encoded by SCN8A.133 Both proteins are involved in neurotransmission and are suggestive of the nociceptive pathway. The finding of NaV1.6 is unique, because this is the first report of this channel's involvement in pain. Given its distribution in high-frequency firing neurons, it had previously been studied for its role in epilepsy.133
3.2.4. Viral infection–related sensory neuropathies
Painful neuropathy as a sequela of HIV infection is common and has been investigated by several groups in the recent years. An excellent review of the genetics of HIV-associated painful neuropathy on the African continent has just been published.93 Two genes harbouring variants associated with pain intensity in HIV-infected Southern Africans are KCNS153 and TNF.54,145 Two groups have also investigated the involvement of GCH1 in HIV-associated neuropathic pain in Africans but found no association.53,146
Postherpetic neuralgia is a condition characterised by persistent spontaneous or innocuous stimulus-evoked pain. Several studies in Japanese patients have examined the role of genetic variants in the HLA region.18,110,122,132 Associations have been found for both class I molecules: HLA-A, HLA-B, and HLA-C18,110,122,132; and class II HLA-DRB1.122,132 The proposed mechanism whereby HLA-complex variants contribute to postherpetic neuropathic pain is nerve damage permitted by inadequate immune system response to the initial viral infection.132
3.2.5. Cancer pain
Up to 40% of all cancer pain has a neuropathic component.10 Aside from cancer-related surgery and chemotherapy, the cancer itself leads to neuropathic pain either by tumour invasion of nociceptors or by inflammatory cytokine leakage from cancerous cells.142 Given the protracted inflammation, a sustained level of nociceptor activation could lead to persistent changes in neuronal connectivity, changing the response thresholds and intensities and transmitting innocuous stimuli as painful.116
Variants in prostaglandin-endoperoxide synthase 2, PTGS2, tumour necrosis factor, TNF, and NFκB inhibitor-α, NFKBIA, genotyped in Ref. 116, and tumour necrosis factor-β, LTA, genotyped in Ref. 114, have been reported to be associated with severe cancer pain. In another study, an aggregate of phenotypes that includes high pain intensity has been reported to be associated with the cumulative effect of variants in nitric oxide synthase-3, NOS3; interleukin-1β, IL1B; tumor necrosis factor receptor superfamily member 1B, TNFRSF1B; PTGS2; and interleukin-10 receptor-β, IL10RB.117 In addition, pain-protective variants in GCH1-encoded GTP cyclohydrolase, also involved in nitric oxide production, have been reported in patients with advanced cancer.87 These studies converge on a suggestive role for the immune system in modulating the extent of neuropathic pain accompanying cancer.
On the other hand, several studies have shown association with mediators of neurotransmission and even members of the pain-inhibition pathway. Variants in voltage-gated potassium ion channel encoding KCNS1, KCNJ3, KCNJ6, and KCNK9 have been reported in women with breast cancer pain before surgery,78 and variants in catechol-O-methyltransferase (COMT) and membrane-bounded P-glycoprotein (ABCB1) in charge of clearing exogenous opioids have also been reported to be associated with pain in cancer patients.149
Last, mitogen-activated protein kinase 1 (MAPK1)—a broad-spectrum regulator—has also been implicated in cancer pain.118
3.2.6. Postoperative pain
Persistent postoperative pain is generally defined by the lower duration boundary of 2 to 6 months.89 Among putative causal mechanisms are nerve damage, which recruits immune cells and a prolonged state of inflammation during the acute period.89,151 In either case, inflammatory cytokine barrage leads to sustained nociceptor activity, which may result in a rewired pain transmission system.19,65,68,99
Two studies of postmastectomy patients, in whom neuropathic pain prevalence has been estimated to be up to 68%,51 reported associations between persistent breast pain and variants in interleukin-1 receptor type 2, IL1R2, interleukin-10, IL10,129 and purinergic receptor 7, P2RX7.128
Several variants in genes directly involved in neurotransmission have also been reported as risk modifiers in persistent postoperative pain, specifically µ-opioid receptor (OPRM1) in a postabdominal surgery cohort,72 voltage-gated potassium channel subunit (KCNS1) in 2 limb amputation cohorts and 1 postmastectomy cohort,20 and stargazin (CACNG2)—involved in the trafficking of AMPA receptors—in a postmastectomy cohort.104 Variants in GTP hydrolase, encoded by GCH1, have been reported to modulate postoperative pain in 2 studies.9,135
The first genome-wide scan in a postoperative pain cohort (individuals with knee and hip replacement surgery) was published this year.150 The strongest associated variant, just shy of genome-wide significance (rs887797, P = 1.29 × 10e-7), lies in PRKCA, which encodes protein kinase C alpha, and the next best association is for a variant in MAT2B (rs7734804, P = 5.25 × 10e-6). Both of these associations were confirmed in one of their 2 replication joint-related neuropathic pain cohorts.
3.2.7. Other conditions
In several studies, neuropathic pain conditions were grouped into 1 phenotype, such that an association would indicate a link to condition-agnostic neuropathic pain. In 1 such study, a variant in dopamine receptor DRD2 has been shown to be associated with susceptibility to pain, given one of the following primary conditions: nerve injury, atypical facial pain burning mouth syndrome, and trigeminal neuropathy.62 In addition, transient receptor potential channels, TRPA1 and TRPV1, have been reported to affect somatosensory sensitivity in patients with neuropathic pain who had a variety of neuropathic conditions.11 A variant in SCN9A has also been reported to have association with pain by a group that examined 5 different cohorts with neuropathic pain.115
3.3. Replication in UKBB
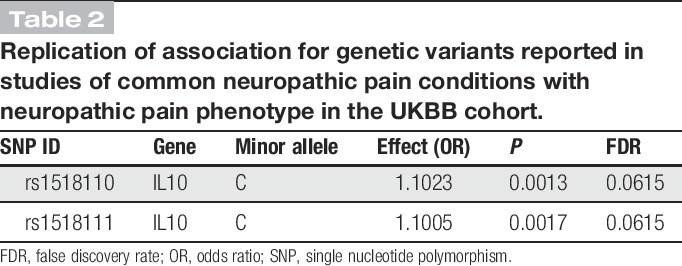
We analysed the list of variants reported in association studies of common neuropathic pain conditions (Table 1) for replication with neuropathic pain in UKBB. Associations for 2 single nucleotide polymorphisms (SNPs) on 1 haploblock of IL10 pass correction for multiple testing at false discovery rate 20% (Table 2). Previously reported in a postoperative pain cohort,129 these replicated associations, albeit nominal, give us increased confidence in the contribution of inflammatory mediators to neuropathic pain in a condition-agnostic manner, underscoring the importance of neuroimmune interactions already suspected to contribute to neuropathic pain.4,91
Table 2.
Replication of association for genetic variants reported in studies of common neuropathic pain conditions with neuropathic pain phenotype in the UKBB cohort.
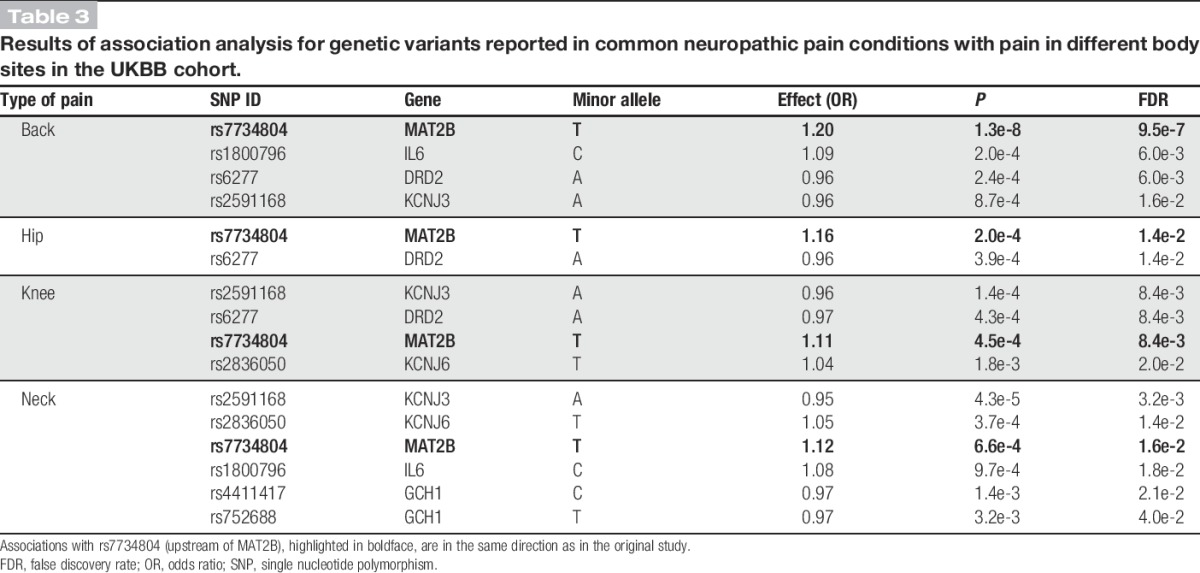
In addition, we tested the list of variants in Table 1 for association with pain in 4 body sites—back, hip, knee, and neck—in all of which chronic pain may indicate neuropathy.44,71,106,130,139 Results of these association analyses are shown in quantile-quantile plots (Fig. 1), which are a statistical tool to visualise the deviation of the observed distribution of association P values (log transformed) from the one expected by chance, given uniform sampling in the P-value space. Notably, although in all 4 sites several SNPs are associated with P values passing the false discovery rate 5% threshold, SNP rs7734804, whose minor allele was originally reported as risk-conferring in a postoperative pain GWAS,150 is associated with the same direction of effect in all 4 sites and with back pain with genome-wide significance (P = 1.3e-8, odds ratio = 1.2; Table 3).
Figure 1.
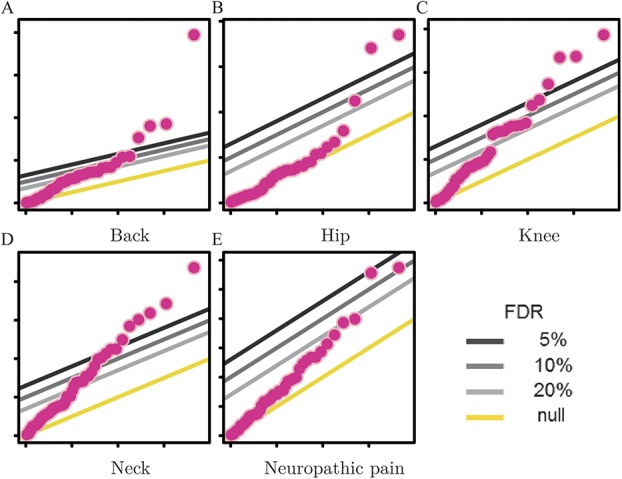
Results of association analysis for genetic variants reported for common neuropathic pain conditions (Table 1) with pain in different body sites (A-D) and neuropathic pain (E) in the UKBB cohort. In each quantile-quantile plot, the P values smaller than expected by chance surpass at least 3 of the fixed thresholds of statistical significance (false discovery rate = 5%, 10%, and 20%).
Table 3.
Results of association analysis for genetic variants reported in common neuropathic pain conditions with pain in different body sites in the UKBB cohort.
4. Conclusion
This survey of literature provides an overview of genetic variants implicated in a variety of neuropathic pain conditions. Rare monogenic painful conditions are firmly rooted in the ion channel—specifically sodium channel—mutations, underscoring the critical role of these channels in pain processing. Among painless monogenic conditions, mutations disrupting nociceptive neuron maintenance are overrepresented. In common nonfamilial neuropathic pain conditions, the landscape of implicated molecules is more varied; the effect of genetic variants is considerably smaller and often harder to demonstrate. Nevertheless, neuroimmune interactions have emerged with a central role in neuropathic pain pathophysiology, supported by additional evidence from the UKBB study. Although further studies are needed, this evidence supports the hypothesis that timely treatment targeting the immune system could be helpful in mitigating neuropathic pain. In addition, the involvement of neuropathic pain genetic variants in other pain conditions with a neuropathic pain component—in particular, a variant upstream of MAT2B whose association is prominent in back, hip, knee, and neck pain—provides preliminary evidence of shared contributing mechanisms at the genetic-molecular level.
Diagnosing pain and confirming it as neuropathic in origin remains a challenge. The difficulty of identifying a nerve lesion or disease is exacerbated by other pain comorbidities and by the fact that diagnosis relies heavily on verbal interpretation of pain, far removed from the pathophysiological mechanisms that engender it. Thus, it is our hope that genetic studies will enable a more comprehensive assessment of patients presenting with painful conditions and become a powerful tool in diagnosing and treating these conditions with requisite specificity.
Conflict of interest statement
L. Diatchenko declares a potential conflict of interest as a coinventor of the patent-pending application on genetic variants of the COMT enzyme contributing to pain phenotypes. L. Diatchenko is also a board member, consultant, and shareholder of Algynomics, Inc and Proove Biosciences. The other authors have no conflict of interest to declare.
This work is supported by the Canadian Excellence Research Chairs (CERC) Program (http://www.cerc.gc.ca/home-accueil-eng.aspx), grant CERC09 to L. Diatchenko.
Acknowledgements
The authors acknowledge the contribution of research clinician Dr Rodrigo Benavides, who identified neuropathic pain-relevant conditions in the UKBB.
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
References
- [1].Altassan R, Saud HA, Masoodi TA, Dosssari HA, Khalifa O, Al-Zaidan H, Sakati N, Rhabeeni Z, AlHassnan Z, Binamer Y, Alhashemi N, Wade W, Al-Zayed Z, Al-Sayed M, Al-Muhaizea M, Meyer B, Al-Owain M, Wakil SM. Exome sequencing identifies novel NTRK1 mutations in patients with HSAN-IV phenotype. Am J Med Genet A 2017;173:1009–16. [DOI] [PubMed] [Google Scholar]
- [2].Anderson SL, Coli R, Daly IW, Kichula EA, Rork MJ, Volpi SA, Ekstein J, Rubin BY. Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet 2001;68:753–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Auer-Grumbach M, Bode H, Pieber TR, Schabhüttl M, Fischer D, Seidl R, Graf E, Wieland T, Schuh R, Vacariu G, Grill F, Timmerman V, Strom TM, Hornemann T. Mutations at Ser331 in the HSN, type I gene SPTLC1 are associated with a distinct syndromic phenotype. Eur J Med Genet 2013;56:266–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Austin PJ, Moalem-Taylor G. The neuro-immune balance in neuropathic pain: involvement of inflammatory immune cells, immune-like glial cells and cytokines. J Neuroimmunol 2010;229:26–50. [DOI] [PubMed] [Google Scholar]
- [5].Baets J, Duan X, Wu Y, Smith G, Seeley WW, Mademan I, McGrath NM, Beadell NC, Khoury J, Botuyan MV, Mer G, Worrell GA, Hojo K, DeLeon J, Laura M, Liu YT, Senderek J, Weis J, van den Bergh P, Merrill SL, Reilly MM, Houlden H, Grossman M, Scherer SS, de Jonghe P, Dyck PJ, Klein CJ. Defects of mutant DNMT1 are linked to a spectrum of neurological disorders. Brain 2015;138:845–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Baron R. Neuropathic pain: a clinical perspective. In: Sensory nerves. Berlin: Springer, 2009. p. 3–30. [DOI] [PubMed] [Google Scholar]
- [7].Basol N, Inanir A, Yigit S, Karakus N, Kaya SU. High association of IL-4 gene intron 3 VNTR polymorphism with diabetic peripheral neuropathy. J Mol Neurosci 2013;51:437–41. [DOI] [PubMed] [Google Scholar]
- [8].Bejaoui K, Wu C, Scheffler MD, Haan G, Ashby P, Wu L, de Jong P, Brown RH. SPTLC1 is mutated in hereditary sensory neuropathy, type 1. Nat Genet 2001;27:261. [DOI] [PubMed] [Google Scholar]
- [9].Belfer I, Dai F, Kehlet H, Finelli P, Qin L, Bittner R, Aasvang EK. Association of functional variations in COMT and GCH1 genes with postherniotomy pain and related impairment. PAIN 2015;156:273–9. [DOI] [PubMed] [Google Scholar]
- [10].Bennett MI, Rayment C, Hjermstad M, Aass N, Caraceni A, Kaasa S. Prevalence and aetiology of neuropathic pain in cancer patients: a systematic review. PAIN 2012;153:359–65. [DOI] [PubMed] [Google Scholar]
- [11].Binder A, May D, Baron R, Maier C, Tölle TR, Treede RD, Berthele A, Faltraco F, Flor H, Gierthmühlen J, Haenisch S, Huge V, Magerl W, Maihöfner C, Richter H, Rolke R, Scherens A, Üçeyler N, Ufer M, Wasner G, Zhu J, Cascorbi I. Transient receptor potential channel polymorphisms are associated with the somatosensory function in neuropathic pain patients. PLoS One 2012;6:e17387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bodzioch M, Lapicka K, Aslanidis C, Kacinski M, Schmitz G. Two novel mutant alleles of the gene encoding neurotrophic tyrosine kinase receptor type 1 (NTRK 1) in a patient with congenital insensitivity to pain with anhidrosis: a splice junction mutation in intron 5 and cluster of four mutations in exon 15. Hum Mutat 2001;17:72. [DOI] [PubMed] [Google Scholar]
- [13].Bonkowsky JL, Johnson J, Carey JC, Smith AG, Swoboda KJ. An infant with primary tooth loss and palmar hyperkeratosis: a novel mutation in the NTRK1 gene causing congenital insensitivity to pain with anhidrosis. Pediatrics 2003;112:e237–41. [DOI] [PubMed] [Google Scholar]
- [14].Capsoni S, Covaceuszach S, Marinelli S, Ceci M, Bernardo A, Minghetti L, Ugolini G, Pavone F, Cattaneo A. Taking pain out of NGF: a “painless” NGF mutant, linked to hereditary sensory autonomic neuropathy type V, with full neurotrophic activity. PLoS One 2011;6:e17321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chen YC, Auer-Grumbach M, Matsukawa S, Zitzelsberger M, Themistocleous AC, Strom TM, Samara C, Moore AW, Cho LTY, Young GT, Weiss C, Schabhüttl M, Stucka R, Schmid AB, Parman Y, Graul-Neumann L, Heinritz W, Passarge E, Watson RM, Hertz JM, Moog U, Baumgartner M, Valente EM, Pereira D, Restrepo CM, Katona I, Dusl M, Stendel C, Wieland T, Stafford F, Reimann F, von Au K, Finke C, Willems PJ, Nahorski MS, Shaikh SS, Carvalho OP, Nicholas AK, Karbani G, McAleer MA, Cilio MR, McHugh JC, Murphy SM, Irvine AD, Jensen UB, Windhager R, Weis J, Bergmann C, Rautenstrauss B, Baets J, de Jonghe P, Reilly MM, Kropatsch R, Kurth I, Chrast R, Michiue T, Bennett DL, Woods CG, Senderek J. Transcriptional regulator PRDM12 is essential for human pain perception. Nat Genet 2015;47:803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cheng KI, Lin SR, Chang LL, Wang JY, Lai CS. Association of the functional A118G polymorphism of OPRM1 in diabetic patients with foot ulcer pain. J Diabetes Complications 2010;24:102–8. [DOI] [PubMed] [Google Scholar]
- [17].Chiabrando D, Castori M, di Rocco M, Ungelenk M, Gießelmann S, Di Capua M, Madeo A, Grammatico P, Bartsch S, Hübner CA, Altruda F, Silengo L, Tolosano E, Kurth I. Mutations in the heme exporter FLVCR1 cause sensory neurodegeneration with loss of pain perception. PLoS Genet 2016;12:e1006461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chung HY, Song EY, Yoon JA, Suh DH, Lee SC, Kim YC, Park MH. Association of human leukocyte antigen with postherpetic neuralgia in Koreans. APMIS 2016;124:865–71. [DOI] [PubMed] [Google Scholar]
- [19].Coderre TJ, Katz J, Vaccarino AL, Melzack R. Contribution of central neuroplasticity to pathological pain: review of clinical and experimental evidence. PAIN 1993;52:259–85. [DOI] [PubMed] [Google Scholar]
- [20].Costigan M, Belfer I, Griffin RS, Dai F, Barrett LB, Coppola G, Wu T, Kiselycznyk C, Poddar M, Lu Y, Diatchenko L, Smith S, Cobos EJ, Zaykin D, Allchorne A, Shen PH, Nikolajsen L, Karppinen J, Männikkö M, Kelempisioti A, Goldman D, Maixner W, Geschwind DH, Max MB, Seltzer Z, Woolf CJ. Multiple chronic pain states are associated with a common amino acid—changing allele in KCNS1. Brain 2010;133:2519–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Costigan M, Scholz J, Woolf CJ. Neuropathic pain: a maladaptive response of the nervous system to damage. Annu Rev Neurosci 2009;32:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cox JJ, Reimann F, Nicholas AK, Thornton G, Roberts E, Springell K, Karbani G, Jafri H, Mannan J, Raashid Y, Al-Gazali L, Hamamy H, Valente EM, Gorman S, Williams R, McHale DP, Wood JN, Gribble FM, Woods CG. An SCN9A channelopathy causes congenital inability to experience pain. Nature 2006;444:894–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cox JJ, Sheynin J, Shorer Z, Reimann F, Nicholas AK, Zubovic L, Baralle M, Wraige E, Manor E, Levy J, Woods CG, Parvari R. Congenital insensitivity to pain: novel SCN9A missense and in-frame deletion mutations. Hum Mutat 2010;31:E1670–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cruccu G, Finnerup NB, Jensen TS, Scholz J, Sindou M, Svensson P, Treede RD, Zakrzewska JM, Nurmikko T. Trigeminal neuralgia new classification and diagnostic grading for practice and research. Neurology 2016;87:220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cui W, Yu X, Zhang H. The serotonin transporter gene polymorphism is associated with the susceptibility and the pain severity in idiopathic trigeminal neuralgia patients. J Headache Pain 2014;15:42–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cummins TR, Dib-Hajj SD, Waxman SG. Electrophysiological properties of mutant Nav1.7 sodium channels in a painful inherited neuropathy. J Neurosci 2004;24:8232–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dabby R, Sadeh M, Broitman Y, Yosovich K, Dickman R, Leshinsky Silver E. Painful small fiber neuropathy with gastroparesis: a new phenotype with a novel mutation in the SCN10A gene. J Clin Neurosci 2016;26:84–8. [DOI] [PubMed] [Google Scholar]
- [28].Dabby R, Sadeh M, Gilad R, Lampl Y, Cohen S, Inbar S, Leshinsky-Silver E. Chronic non-paroxysmal neuropathic pain – novel phenotype of mutation in the sodium channel SCN9A gene. J Neurol Sci 2011;301:90–2. [DOI] [PubMed] [Google Scholar]
- [29].Davidson G, Murphy S, Polke J, Laura M, Salih M, Muntoni F, Blake J, Brandner S, Davies N, Horvath R, Price S, Donaghy M, Roberts M, Foulds N, Ramdharry G, Soler D, Lunn M, Manji H, Davis M, Houlden H, Reilly M. Frequency of mutations in the genes associated with hereditary sensory and autonomic neuropathy in a UK cohort. J Neurol 2012;259:1673–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dawkins JL, Hulme DJ, Brahmbhatt SB, Auer-Grumbach M, Nicholson GA. Mutations in SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, cause hereditary sensory neuropathy type I. Nat Genet 2001;27:309. [DOI] [PubMed] [Google Scholar]
- [31].de Greef BT, Hoeijmakers JG, Wolters EE, Smeets HJ, van den Wijngaard A, Merkies IS, Faber CG, Gerrits MM. No Fabry disease in patients presenting with isolated small fiber neuropathy. PLoS One 2016;11:e0148316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Devor M, Raber P. Heritability of symptoms in an experimental model of neuropathic pain. PAIN 1990;42:51–67. [DOI] [PubMed] [Google Scholar]
- [33].Dib-Hajj S, Rush A, Cummins T, Hisama F, Novella S, Tyrrell L, Marshall L, Waxman S. Gain-of-function mutation in Nav1.7 in familial erythromelalgia induces bursting of sensory neurons. Brain 2005;128:1847–54. [DOI] [PubMed] [Google Scholar]
- [34].Dib-Hajj SD, Estacion M, Jarecki BW, Tyrrell L, Fischer TZ, Lawden M, Cummins TR, Waxman SG. Paroxysmal extreme pain disorder M1627K mutation in human Nav1.7 renders DRG neurons hyperexcitable. Mol Pain 2008;4:37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Drenth JP, te Morsche RH, Guillet G, Taieb A, Kirby RL, Jansen JB. SCN9A mutations define primary erythermalgia as a neuropathic disorder of voltage gated sodium channels. J Invest Dermatol 2005;124:1333–8. [DOI] [PubMed] [Google Scholar]
- [36].Drenth JP, te Morsche RH, Mansour S, Mortimer PS. Primary erythermalgia as a sodium channelopathy: screening for SCN9A mutations: exclusion of a causal role of SCN10A and SCN11A. Arch Dermatol 2008;144:320–4. [DOI] [PubMed] [Google Scholar]
- [37].Einarsdottir E, Carlsson A, Minde J, Toolanen G, Svensson O, Solders G, Holmgren G, Holmberg D, Holmberg M. A mutation in the nerve growth factor beta gene (NGFB) causes loss of pain perception. Hum Mol Genet 2004;13:799–805. [DOI] [PubMed] [Google Scholar]
- [38].Estacion M, Dib-Hajj S, Benke P, te Morsche RH, Eastman E, Macala L, Drenth J, Waxman S. Nav1.7 gain-of-function mutations as a continuum: a1632E displays physiological changes associated with erythromelalgia and paroxysmal extreme pain disorder mutations and produces symptoms of both disorders. J Neurosci 2008;28:11079–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Estacion M, Harty TP, Choi JS, Tyrrell L, DibHajj SD, Waxman SG. A sodium channel gene SCN9A polymorphism that increases nociceptor excitability. Ann Neurol 2009;66:862–6. [DOI] [PubMed] [Google Scholar]
- [40].Faber CG, Hoeijmakers JG, Ahn HS, Cheng X, Han C, Choi JS, Estacion M, Lauria G, Vanhoutte EK, Gerrits MM, Dib-Hajj S, Drenth JP, Waxman SG, Merkies IS. Gain of function Nav1.7 mutations in idiopathic small fiber neuropathy. Ann Neurol 2012;71:26–39. [DOI] [PubMed] [Google Scholar]
- [41].Faber CG, Lauria G, Merkies IS, Cheng X, Han C, Ahn HS, Persson AK, Hoeijmakers JG, Gerrits MM, Pierro T, Lombardi R, Kapetis D, Dib-Hajj SD, Waxman SG. Gain-of-function Nav1.8 mutations in painful neuropathy. Proc Natl Acad Sci USA 2012;109:19444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fertleman CR, Baker MD, Parker KA, Moffatt S, Elmslie FV, Abrahamsen B, Ostman J, Klugbauer N, Wood JN, Gardiner RM, Rees M. SCN9A mutations in paroxysmal extreme pain disorder: allelic variants underlie distinct channel defects and phenotypes. Neuron 2006;52:767–74. [DOI] [PubMed] [Google Scholar]
- [43].Finnerup NB, Haroutounian S, Kamerman P, Baron R, Bennett DL, Bouhassira D, Cruccu G, Freeman R, Hansson P, Nurmikko T, Raja SN, Rice AS, Serra J, Smith BH, Treede RD, Jensen TS. Neuropathic pain: an updated grading system for research and clinical practice. PAIN 2016;157:1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Freynhagen R, Baron R. The evaluation of neuropathic components in low back pain. Curr Pain Headache Rep 2009;13:185–90. [DOI] [PubMed] [Google Scholar]
- [45].Gao L, Guo H, Ye N, Bai Y, Liu X, Yu P, Xue Y, Ma S, Wei K, Jin Y, Wen L, Xuan K. Oral and craniofacial manifestations and two novel missense mutations of the NTRK1 gene identified in the patient with congenital insensitivity to pain with anhidrosis. PLoS One 2013;8:e66863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Guo TM, Liu M, Zhang YG, Guo WT, Wu SX. Association between caspase-9 promoter region polymorphisms and discogenic low back pain. Connect Tissue Res 2011;52:133–8. [DOI] [PubMed] [Google Scholar]
- [47].Han C, Dib-Hajj SD, Lin Z, Li Y, Eastman EM, Tyrrell L, Cao X, Yang Y, Waxman SG. Early- and late-onset inherited erythromelalgia: genotype-phenotype correlation. Brain 2009;132:1711–22. [DOI] [PubMed] [Google Scholar]
- [48].Han C, Hoeijmakers JG, Liu S, Gerrits MM, te Morsche RH, Lauria G, Dib-Hajj SD, Drenth JP, Faber CG, Merkies IS, Waxman SG. Functional profiles of SCN9A variants in dorsal root ganglion neurons and superior cervical ganglion neurons correlate with autonomic symptoms in small fibre neuropathy. Brain 2012;135:2613–28. [DOI] [PubMed] [Google Scholar]
- [49].Han C, Rush AM, Dib-Hajj SD, Li S, Xu Z, Wang Y, Tyrrell L, Wang X, Yang Y, Waxman SG. Sporadic onset of erythermalgia: a gain-of-function mutation in Nav1.7. Ann Neurol 2006;59:553–8. [DOI] [PubMed] [Google Scholar]
- [50].Han C, Yang Y, de Greef BT, Hoeijmakers JG, Gerrits MM, Verhamme C, Qu J, Lauria G, Merkies IS, Faber CG, Dib-Hajj SD, Waxman SG. The domain II S4-S5 linker in Nav1.9: a missense mutation enhances activation, impairs fast inactivation, and produces human painful neuropathy. Neuromolecular Med 2015;17:158–69. [DOI] [PubMed] [Google Scholar]
- [51].Haroutiunian S, Nikolajsen L, Finnerup NB, Jensen TS. The neuropathic component in persistent postsurgical pain: a systematic literature review. PAIN 2013;154:95–102. [DOI] [PubMed] [Google Scholar]
- [52].Harrer JU, Üçeyler N, Doppler K, Fischer TZ, Dib-Hajj SD, Waxman SG, Sommer C. Neuropathic pain in two-generation twins carrying the sodium channel Nav1.7 functional variant R1150W. PAIN 2014;155:2199–203. [DOI] [PubMed] [Google Scholar]
- [53].Hendry L, Lombard Z, Wadley A, Kamerman P. KCNS1, but not GCH1, is associated with pain intensity in a black Southern African population with HIV-associated sensory neuropathy: a genetic association study. J Acquir Immune Defic Syndr 2013;63:27–30. [DOI] [PubMed] [Google Scholar]
- [54].Hendry LM, Wadley AL, Cherry CL, Price P, Lombard Z, Kamerman PR. TNF block gene variants associate with pain intensity in black Southern Africans with HIV-associated sensory neuropathy. Clin J Pain 2016;32:45–50. [DOI] [PubMed] [Google Scholar]
- [55].Houlden H, King R, Blake J, Groves M, Love S, Woodward C, Hammans S, Nicoll J, Lennox G, O'Donovan DG, Gabriel C, Thomas PK, Reilly MM. Clinical, pathological and genetic characterization of hereditary sensory and autonomic neuropathy type 1 (HSAN I). Brain 2005;129:411–25. [DOI] [PubMed] [Google Scholar]
- [56].Huang J, Han C, Estacion M, Vasylyev D, Hoeijmakers JG, Gerrits MM, Tyrrell L, Lauria G, Faber CG, Dib-Hajj SD, Merkies IS, Waxman SG; PROPANE Study Group. Gain-of-function mutations in sodium channel Nav1. 9 in painful neuropathy. Brain 2014;137:1627–42. [DOI] [PubMed] [Google Scholar]
- [57].Huang J, Yang Y, Zhao P, Gerrits MM, Hoeijmakers G, Bekelaar K, Merkies IS, Faber CG, Dib-Hajj SD, Waxman SG. Small-fiber neuropathy Nav1. 8 mutation shifts activation to hyperpolarized potentials and increases excitability of dorsal root ganglion neurons. J Neurosci 2013;33:14087–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Huehne K, Zweier C, Raab K, Odent S, BonnaureMallet M, Sixou JL, Landrieu P, Goizet C, Sarlangue J, Baumann M, Eggermann T, Rauch A, Ruppert S, Stettner GM, Rautenstrauss B. Novel missense, insertion and deletion mutations in the neurotrophic tyrosine kinase receptor type 1 gene (NTRK1) associated with congenital insensitivity to pain with anhidrosis. Neuromuscul Disord 2008;18:159–66. [DOI] [PubMed] [Google Scholar]
- [59].Hurri H, Karppinen J. Discogenic pain. PAIN 2004;112:225–8. [DOI] [PubMed] [Google Scholar]
- [60].Indo Y. Molecular basis of congenital insensitivity to pain with anhidrosis (CIPA): mutations and polymorphisms in TRKA (NTRK1) gene encoding the receptor tyrosine kinase for nerve growth factor. Hum Mutat 2001;18:462–71. [DOI] [PubMed] [Google Scholar]
- [61].Indo Y, Mardy S, Miura Y, Moosa A, Ismail EA, Toscano E, Andria G, Pavone V, Brown DL, Brooks A, Endo F, Matsuda I. Congenital insensitivity to pain with anhidrosis (CIPA): novel mutations of the TRKA (NTRK1) gene, a putative uniparental disomy, and a linkage of the mutant TRKA and PKLR genes in a family with CIPA and pyruvate kinase deficiency. Hum Mutat 2001;18:308–18. [DOI] [PubMed] [Google Scholar]
- [62].Jääskeläinen SK, Lindholm P, Valmunen T, Pesonen U, Taiminen T, Virtanen A, Lamusuo S, Forssell H, Hagelberg N, Hietala J, Pertovaara A. Variation in the dopamine D2 receptor gene plays a key role in human pain and its modulation by transcranial magnetic stimulation. PAIN 2014;155:2180–7. [DOI] [PubMed] [Google Scholar]
- [63].Jacobsen L, Schistad E, Storesund A, Pedersen L, Rygh L, Røe C, Gjerstad J. The COMT rs4680 met allele contributes to long-lasting low back pain, sciatica and disability after lumbar disc herniation. Eur J Pain 2012;16:1064–9. [DOI] [PubMed] [Google Scholar]
- [64].Jacobsen LM, Schistad EI, Storesund A, Pedersen LM, Espeland A, Rygh LJ, Røe C, Gjerstad J. The MMP1 rs1799750 2G allele is associated with increased low back pain, sciatica, and disability after lumbar disk herniation. Clin J Pain 2013;29:967–71. [DOI] [PubMed] [Google Scholar]
- [65].Jänig W, Levine J, Michaelis M. Interactions of sympathetic and primary afferent neurons following nerve injury and tissue trauma. Prog Brain Res 1996;113:161–84. [DOI] [PubMed] [Google Scholar]
- [66].Jensen TS, Baron R, Haanpää M, Kalso E, Loeser JD, Rice AS, Treede RD. A new definition of neuropathic pain. PAIN 2011;152:2204–5. [DOI] [PubMed] [Google Scholar]
- [67].Karppinen J, Daavittila I, Noponen N, Haapea M, Taimela S, Vanharanta H, Ala-Kokko L, Männikkö M. Is the interleukin-6 haplotype a prognostic factor for sciatica? Eur J Pain 2008;12:1018–25. [DOI] [PubMed] [Google Scholar]
- [68].Kehlet H, Jensen TS, Woolf CJ. Persistent postsurgical pain: risk factors and prevention. Lancet 2006;367:1618–25. [DOI] [PubMed] [Google Scholar]
- [69].Klein CJ, Botuyan MV, Wu Y, Ward CJ, Nicholson GA, Hammans S, Hojo K, Yamanishi H, Karpf AR, Wallace DC, Simon M, Lander C, Boardman LA, Cunningham JM, Smith GE, Litchy WJ, Boes B, Atkinson EJ, Middha S, Dyck PJB, Parisi JE, Mer G, Smith DI, Dyck PJ. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet 2011;43:595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Klein CJ, Wu Y, Kilfoyle DH, Sandroni P, Davis MD, Gavrilova RH, Low PA, Dyck PJ. Infrequent SCN9A mutations in congenital insensitivity to pain and erythromelalgia. J Neurol Neurosurg Psychiatry 2013;84:386–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kleinman N, Patel AA, Benson C, Macario A, Kim M, Biondi DM. Economic burden of back and neck pain: effect of a neuropathic component. Popul Health Manag 2014;17:224–32. [DOI] [PubMed] [Google Scholar]
- [72].Kolesnikov Y, Gabovits B, Levin A, Veske A, Qin L, Dai F, Belfer I. Chronic pain after lower abdominal surgery: do catechol-O-methyl transferase/opioid receptor µ-1 polymorphisms contribute? Mol Pain 2013;9:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Krafft RM. Trigeminal neuralgia. Am Fam Physician 2008;77:1291–6. [PubMed] [Google Scholar]
- [74].Kurban M, Wajid M, Shimomura Y, Christiano AM. A nonsense mutation in the SCN9A gene in congenital insensitivity to pain. Dermatology 2010;221:179–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kurth I, Baumgartner M, Schabhüttl M, Tomni C, Windhager R, Strom TM, Wieland T, Gremel K, Auer-Grumbach M. Whole exome sequencing in congenital pain insensitivity identifies a novel causative intronic NTRK1-mutation due to uniparental disomy. Am J Med Genet B Neuropsychiatr Genet 2016;171:875–8. [DOI] [PubMed] [Google Scholar]
- [76].Kurth I, Pamminger T, Hennings JC, Soehendra D, Huebner AK, Rotthier A, Baets J, Senderek J, Topaloglu H, Farrell SA, Nürnberg G, Nürnberg P, de Jonghe P, Gal A, Kaether C, Timmerman V, Hübner CA. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat Genet 2009;41:1179–81. [DOI] [PubMed] [Google Scholar]
- [77].Lafrenière RG, MacDonald ML, Dubé MP, MacFarlane J, O'Driscoll M, Brais B, Meilleur S, Brinkman RR, Dadivas O, Pape T, Platon C, Radomski C, Risler J, Thompson J, Guerra-Escobio AM, Davar G, Breakefield XO, Pimstone SN, Green R, Pryse-Phillips W, Goldberg YP, Younghusband HB, Hayden MR, Sherrington R, Rouleau GA. Identification of a novel gene (HSN2) causing hereditary sensory and autonomic neuropathy type II through the study of Canadian genetic isolates. Am J Hum Genet 2004;74:1064–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Langford DJ, West C, Elboim C, Cooper BA, Abrams G, Paul SM, Schmidt BL, Levine JD, Merriman JD, Dhruva A, Neuhaus J, Leutwyler H, Baggott C, Sullivan CW, Aouizerat BE, Miaskowski C. Variations in potassium channel genes are associated with breast pain in women prior to breast cancer surgery. J Neurogenet 2014;28:122–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lee MJ, Yu HS, Hsieh ST, Stephenson DA, Lu CJ, Yang CC. Characterization of a familial case with primary erythromelalgia from Taiwan. J Neurol 2007;254:210–14. [DOI] [PubMed] [Google Scholar]
- [80].Leipold E, Liebmann L, Korenke GC, Heinrich T, Gießelmann S, Baets J, Ebbinghaus M, Goral RO, Stödberg T, Hennings JC, Bergmann M, Altmüller J, Thiele H, Wetzel A, Nürnberg P, Timmerman V, de Jonghe P, Blum R, Schaible HG, Weis J, Heinemann SH, Hübner CA, Kurth I. A de novo gain-of-function mutation in SCN11A causes loss of pain perception. Nat Genet 2013;45:1399–404. [DOI] [PubMed] [Google Scholar]
- [81].Leng XR, Qi XH, Zhou YT, Wang YP. Gain-of-function mutation p. Arg225Cys in SCN11A causes familial episodic pain and contributes to essential tremor. J Hum Genet 2017;62:641–6. [DOI] [PubMed] [Google Scholar]
- [82].Li M, Liang J, Sun Z, Zhang H, Yao Z. Novel nonsense and frameshift NTRK1 gene mutations in Chinese patients with congenital insensitivity to pain with anhidrosis. Genet Mol Res 2012;11:2056–162. [DOI] [PubMed] [Google Scholar]
- [83].Li QS, Cheng P, Favis R, Wickenden A, Romano G, Wang H. Scn9a variants may be implicated in neuropathic pain associated with diabetic peripheral neuropathy and pain severity. Clin J Pain 2015;31:976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Li S, Yang Y, Wang Y, Xu Z, Bu DF, Tu P, Zhu XJ. SCN9A gene mutation in a primary erythermalgia pedigree. J Clin Dermatol 2005;2:002. [Google Scholar]
- [85].Lin ZM, Li Y, Zhang LL, Cao XW, Zhang YL, Yang Y. Mutations of SCN9A gene in five patients with primary erythermalgia. J Clin Dermatol 2008;8:016. [Google Scholar]
- [86].Liu S, Wu N, Liu J, Ming X, Chen J, Pavelec D, Su X, Qiu G, Tian Y, Giampietro P, Wu Z. Novel NTRK1 frameshift mutation in congenital insensitivity to pain with anhidrosis. J Child Neurol 2015;30:1357–61. [DOI] [PubMed] [Google Scholar]
- [87].Lötsch J, Klepstad P, Doehring A, Dale O. A GTP cyclohydrolase 1 genetic variant delays cancer pain. PAIN 2010;148:103–6. [DOI] [PubMed] [Google Scholar]
- [88].Lv F, Xu XJ, Song YW, Li LJ, Wang O, Jiang Y, Xia WB, Xing XP, Gao P, Li M. Recurrent and novel mutations in the NTRK1 gene lead to rare congenital insensitivity to pain with anhidrosis in two Chinese patients. Clinica Chim Acta 2017;468:39–45. [DOI] [PubMed] [Google Scholar]
- [89].Macrae W. Chronic post-surgical pain: 10 years on. Br J Anaesth 2008;101:77–86. [DOI] [PubMed] [Google Scholar]
- [90].Mansouri M, Elalaoui SC, Bencheikh BOA, El Alloussi M, Dion PA, Sefiani A, Rouleau GA. A novel nonsense mutation in SCN9A in a Moroccan child with congenital insensitivity to pain. Pediatr Neurol 2014;51:741–4. [DOI] [PubMed] [Google Scholar]
- [91].Marchand F, Perretti M, McMahon S. Role of the immune system in chronic pain. Nat Rev Neurosci 2005;6:521–32. [DOI] [PubMed] [Google Scholar]
- [92].Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007;39:906–13. [DOI] [PubMed] [Google Scholar]
- [93].Mbenda HGN, Wadley A, Lombard Z, Cherry C, Price P, Kamerman P. Genetics of HIV-associated sensory neuropathy and related pain in Africans. J Neurovirol 2017;23:511–9. [DOI] [PubMed] [Google Scholar]
- [94].Meglič A, Perkovič-Benedik M, Podkrajšek KT, Bertok S. Painful micturition in a small child: an unusual clinical picture of paroxysmal extreme pain disorder. Pediatr Nephrol 2014;29:1643–6. [DOI] [PubMed] [Google Scholar]
- [95].Meng W, Deshmukh H, Zuydam N, Liu Y, Donnelly L, Zhou K, Morris A, Colhoun H, Palmer C, Smith B. A genome-wide association study suggests an association of Chr8p21.3 (GFRA2) with diabetic neuropathic pain. Eur J Pain 2015;19:392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Meng W, Deshmukh HA, Donnelly LA; Wellcome Trust Case Control Consortium 2 (WTCCC2), Surrogate markers for Micro- and Macro-vascular hard endpoints for Innovative diabetes Tools (SUMMIT) study group, , Torrance N, Colhoun HM, Palmer CN, Smith BH. A genome-wide association study provides evidence of sex-specific involvement of Chr1p35.1 (ZSCAN20-TLR12P) and Chr8p23.1 (HMGB1P46) with diabetic neuropathic pain. EBioMedicine 2015;2:1386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Michiels JJ, te Morsche RH, Jansen JB, Drenth JP. Autosomal dominant erythermalgia associated with a novel mutation in the voltage-gated sodium channel α subunit Nav1.7. Arch Neurol 2005;62:1587–90. [DOI] [PubMed] [Google Scholar]
- [98].Miura Y, Mardy S, Awaya Y, Nihei K, Endo F, Matsuda I, Indo Y. Mutation and polymorphism analysis of the TRKA (NTRK1) gene encoding a high-affinity receptor for nerve growth factor in congenital insensitivity to pain with anhidrosis (CIPA) families. Hum Genet 2000;106:116–24. [DOI] [PubMed] [Google Scholar]
- [99].Moalem G, Tracey DJ. Immune and inflammatory mechanisms in neuropathic pain. Brain Res Rev 2006;51:240–64. [DOI] [PubMed] [Google Scholar]
- [100].Moen A, Schistad EI, Rygh LJ, Røe C, Gjerstad J. Role of IL1A rs1800587, IL1B rs1143627 and IL1RN rs2234677 genotype regarding development of chronic lumbar radicular pain; a prospective one-year study. PLoS One 2014;9:e107301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Mogil JS, Wilson SG, Bon K, Lee SE, Chung K, Raber P, Pieper JO, Hain HS, Belknap JK, Hubert L, Elmer GI, Chung JM, Devor M. Heritability of nociception I: responses of 11 inbred mouse strains on 12 measures of nociception. PAIN 1999;80:67–82. [DOI] [PubMed] [Google Scholar]
- [102].Murphy SM, Davidson GL, Brandner S, Houlden H, Reilly MM. Mutation in FAM134B causing severe hereditary sensory neuropathy. J Neurol Neurosurg Psychiatry 2012;83:119–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Nielsen C, Knudsen G, Steingrímsdóttir Ó. Twin studies of pain. Clin Genet 2012;82:331–40. [DOI] [PubMed] [Google Scholar]
- [104].Nissenbaum J, Devor M, Seltzer Z, Gebauer M, Michaelis M, Tal M, Dorfman R, Abitbul-Yarkoni M, Lu Y, Elahipanah T, del Canho S, Minert A, Fried K, Persson AK, Shpigler H, Shabo E, Yakir B, Pisanté A, Darvasi A. Susceptibility to chronic pain following nerve injury is genetically affected by CACNG2. Genome Res 2010;20:1180–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Noponen-Hietala N, Virtanen I, Karttunen R, Schwenke S, Jakkula E, Li H, Merikivi R, Barral S, Ott J, Karppinen J, Ala-Kokko L. Genetic variations in IL6 associate with intervertebral disc disease characterized by sciatica. PAIN 2005;114:186–94. [DOI] [PubMed] [Google Scholar]
- [106].Ohtori S, Orita S, Yamashita M, Ishikawa T, Ito T, Shigemura T, Nishiyama H, Konno S, Ohta H, Takaso M, Inoue G, Eguchi Y, Ochiai N, Kishida S, Kuniyoshi K, Aoki Y, Arai G, Miyagi M, Kamoda H, Suzkuki M, Nakamura J, Furuya T, Kubota G, Sakuma Y, Oikawa Y, Suzuki M, Sasho T, Nakagawa K, Toyone T, Takahashi K. Existence of a neuropathic pain component in patients with osteoarthritis of the knee. Yonsei Med J 2012;53:801–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Oliveira A, Dinis-Oliveira RJ, Nogueira A, Gonçalves F, Silva P, Vieira C, Silvestre R, Carvalho F, Medeiros R. Interleukin-1β genotype and circulating levels in cancer patients: metastatic status and pain perception. Clin Biochem 2014;47:1209–13. [DOI] [PubMed] [Google Scholar]
- [108].Olsen MB, Jacobsen LM, Schistad EI, Pedersen LM, Rygh LJ, Røe C, Gjerstad J. Pain intensity the first year after lumbar disc herniation is associated with the A118G polymorphism in the opioid receptor mu 1 gene: evidence of a sex and genotype interaction. J Neurosci 2012;32:9831–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Ørstavik K, Weidner C, Schmidt R, Schmelz M, Hilliges M, Jørum E, Handwerker H, Torebjörk E. Pathological c-fibres in patients with a chronic painful condition. Brain 2003;126:567–78. [DOI] [PubMed] [Google Scholar]
- [110].Ozawa A, Sasao Y, Iwashita K, Miyahara M, Sugai J, Iizuka M, Kawakubo Y, Ohkido M, Naruse T, Anzai T, Takashige N, Ando A, Inoko H. HLA-A33 and -B44 and susceptibility to postherpetic neuralgia (PHN). HLA 1999;53:263–8. [DOI] [PubMed] [Google Scholar]
- [111].Paassilta P, Lohiniva J, Göring HH, Perälä M, Räinä SS, Karppinen J, Hakala M, Palm T, Kröger H, Kaitila I, Vanharanta H, Ott J, Ala-Kokko L. Identification of a novel common genetic risk factor for lumbar disk disease. JAMA 2001;285:1843–9. [DOI] [PubMed] [Google Scholar]
- [112].Peddareddygari LR, Oberoi K, Grewal RP. Congenital insensitivity to pain: a case report and review of the literature. Case Rep Neurol Med 2014;2014:141953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Phatarakijnirund V, Mumm S, McAlister WH, Novack DV, Wenkert D, Clements KL, Whyte MP. Congenital insensitivity to pain: Fracturing without apparent skeletal pathobiology caused by an autosomal dominant, second mutation in SCN11A encoding voltage-gated sodium channel 1.9. Bone 2016;84:289–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Rausch SM, Gonzalez BD, Clark MM, Patten C, Felten S, Liu H, Li Y, Sloan J, Yang P. SNPs in PTGS2 and LTA predict pain and quality of life in long term lung cancer survivors. Lung Cancer 2012;77:217–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Reimann F, Cox JJ, Belfer I, Diatchenko L, Zaykin DV, McHale DP, Drenth JP, Dai F, Wheeler J, Sanders F, Wood L, Wu TX, Karppinen J, Nikolajsen L, Männikkö M, Max MB, Kiselycznyk C, Poddar M, te Morsche RH, Smith S, Gibson D, Kelempisioti A, Maixner W, Gribble FM, Woods CG. Pain perception is altered by a nucleotide polymorphism in SCN9A. Proc Natl Acad Sci USA 2010;107:5148–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Reyes-Gibby CC, Spitz MR, Yennurajalingam S, Swartz M, Gu J, Wu X, Bruera E, Shete S. Role of inflammation gene polymorphisms on pain severity in lung cancer patients. Cancer Epidemiol Biomarkers Prev 2009;18:2636–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Reyes-Gibby CC, Swartz MD, Yu X, Wu X, Yennurajalingam S, Anderson KO, Spitz MR, Shete S. Symptom clusters of pain, depressed mood, and fatigue in lung cancer: assessing the role of cytokine genes. Support Care Cancer 2013;21:3117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Reyes-Gibby CC, Wang J, Silvas MRT, Yu R, Yeung SCJ, Shete S. MAPK1/ERK2 as novel target genes for pain in head and neck cancer patients. BMC Genet 2016;17:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Rinaldi F, Bassi MT, Todeschini A, Rota S, Arnoldi A, Padovani A, Filosto M. A novel mutation in motor domain of KIF5A associated with an HSP/axonal neuropathy phenotype. J Clin Neuromuscul Dis 2015;16:153–8. [DOI] [PubMed] [Google Scholar]
- [120].Rivière JB, Ramalingam S, Lavastre V, Shekarabi M, Holbert S, Lafontaine J, Srour M, Merner N, Rochefort D, Hince P, Gaudet R, Mes-Masson AM, Baets J, Houlden H, Brais B, Nicholson GA, van Esch H, Nafissi S, de Jonghe P, Reilly MM, Timmerman V, Dion PA, Rouleau GA. KIF1A, an axonal transporter of synaptic vesicles, is mutated in hereditary sensory and autonomic neuropathy type II. Am J Hum Genet 2011;89:219–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Samuels ME, Te Morsche R, Lynch ME, Drenth P. Compound heterozygosity in sodium channel Nav1.7 in a family with hereditary erythermalgia. Mol Pain 2008;4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Sato M, Ohashi J, Tsuchiya N, Kashiwase K, Ishikawa Y, Arita H, Hanaoka K, Tokunaga K, Yabe T. Association of HLA-A* 3303-B* 4403-DRB1* 1302 haplotype, but not of TNFA promoter and NKp30 polymorphism, with postherpetic neuralgia (PHN) in the Japanese population. Genes Immun 2002;3:477. [DOI] [PubMed] [Google Scholar]
- [123].Schistad EI, Jacobsen LM, Røe C, Gjerstad J. The interleukin-1α gene C>T polymorphism rs1800587 is associated with increased pain intensity and decreased pressure pain thresholds in patients with lumbar radicular pain. Clin J Pain 2014;30:869–74. [DOI] [PubMed] [Google Scholar]
- [124].Shatzky S, Moses S, Levy J, Pinsk V, Hershkovitz E, Herzog L, Shorer Z, Luder A, Parvari R. Congenital insensitivity to pain with anhidrosis (CIPA) in Israeli-Bedouins: genetic heterogeneity, novel mutations in the TrkA/NGF receptor gene, clinical findings, and results of nerve conduction studies. Am J Med Genet 2000;92:353–60. [DOI] [PubMed] [Google Scholar]
- [125].Shekarabi M, Girard N, Rivière JB, Dion P, Houle M, Toulouse A, Lafrenière RG, Vercauteren F, Hince P, Laganière J, Vercauteren F, Hince P, Laganière J, Rocheford D, Faivre L, Samuels M, Rouleau G. Mutations in the nervous system—specific HSN2 exon of WNK1 cause hereditary sensory neuropathy type II. J Clin Invest 2008;118:2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Shorer Z, Wajsbrot E, Liran TH, Levy J, Parvari R. A novel mutation in SCN9A in a child with congenital insensitivity to pain. Pediatr Neurol 2014;50:73–6. [DOI] [PubMed] [Google Scholar]
- [127].Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, Cuajungco MP, Liebert CB, Chadwick B, Idelson M, Reznik L, Robbins C, Makalowska I, Brownstein M, Krappmann D, Scheidereit C, Maayan C, Axelrod FB, Gusella JF. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet 2001;68:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Sorge RE, Trang T, Dorfman R, Smith SB, Beggs S, Ritchie J, Austin JS, Zaykin DV, Vander Meulen H, Costigan M, Herbert TA, Yarkoni-Abitbul M, Tichauer D, Livneh J, Gershon E, Zheng M, Tan K, John SL, Slade GD, Jordan J, Woolf CJ, Peltz G, Maixner W, Diatchenko L, Seltzer Z, Salter MW, Mogil JS. Genetically determined P2X7 receptor pore formation regulates variability in chronic pain sensitivity. Nat Med 2012;18:595–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Stephens K, Cooper BA, West C, Paul SM, Baggott CR, Merriman JD, Dhruva A, Kober KM, Langford DJ, Leutwyler H, Luce JA, Schmidt BL, Abrams GM, Elboim C, Hamolsky D, Levine JD, Miaskowski C, Aouizerat BE. Associations between cytokine gene variations and severe persistent breast pain in women following breast cancer surgery. J Pain 2014;15:169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Sterling M, Pedler A. A neuropathic pain component is common in acute whiplash and associated with a more complex clinical presentation. Man Ther 2009;14:173–9. [DOI] [PubMed] [Google Scholar]
- [131].Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R. UK BioBank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Sumiyama D, Kikkawa EF, Kita Y, Shinagawa H, Mabuchi T, Ozawa A, Inoko H. HLA alleles are associated with postherpetic neuralgia but not with herpes zoster. Tokai J Exp Clin Med 2008;33:150–3. [PubMed] [Google Scholar]
- [133].Tanaka BS, Zhao P, Dib-Hajj FB, Morisset V, Tate S, Waxman SG, Dib-Hajj SD. A gain-of-function mutation in NaV1.6 in a case of trigeminal neuralgia. Mol Med 2016;22:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Tang Y, Zheng D, Li Q, Wang Z, Lin Y, Lan F. A novel mutation of NTRK1 gene in a family with congenital insensitivity to pain with anhidrosis. Chin J Med Genet 2014;31:574–7. [DOI] [PubMed] [Google Scholar]
- [135].Tegeder I, Costigan M, Griffin RS, Abele A, Belfer I, Schmidt H, Ehnert C, Nejim J, Marian C, Scholz J, Wu T, Allchorne A, Diatchenko L, Binshtok AM, Goldman D, Adolph J, Sama S, Atlas SJ, Carlezon WA, Parsegian A, Lötsch J, Fillingim R, Maixner W, Geisslinger G, Max MB, Woolf CJ. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nat Med 2006;12:1269–77. [DOI] [PubMed] [Google Scholar]
- [136].Tesfaye S, Selvarajah D. Advances in the epidemiology, pathogenesis and management of diabetic peripheral neuropathy. Diabetes Metab Res Rev 2012;28(suppl 1):8–14. [DOI] [PubMed] [Google Scholar]
- [137].Tesfaye S, Stevens L, Stephenson J, Fuller J, Plater M, Ionescu-Tirgoviste C, Nuber A, Pozza G, Ward J. Prevalence of diabetic peripheral neuropathy and its relation to glycaemic control and potential risk factors: the EURODIAB IDDM Complications Study. Diabetologia 1996;39:1377–84. [DOI] [PubMed] [Google Scholar]
- [138].Tétreault M, Gonzalez M, Dicaire MJ, Allard P, Gehring K, Leblanc D, Leclerc N, Schondorf R, Mathieu J, Zuchner S, Brais B. Adult-onset painful axonal polyneuropathy caused by a dominant NAGLU mutation. Brain 2015;138:1477–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Thakur M, Dickenson AH, Baron R. Osteoarthritis pain: nociceptive or neuropathic? Nat Rev Rheumatol 2014;10:374–80. [DOI] [PubMed] [Google Scholar]
- [140].Treede RD, Jensen TS, Campbell J, Cruccu G, Dostrovsky J, Griffin J, Hansson P, Hughes R, Nurmikko T, Serra J. Neuropathic pain redefinition and a grading system for clinical and research purposes. Neurology 2008;70:1630–5. [DOI] [PubMed] [Google Scholar]
- [141].Tüysüz B, Bayrakli F, DiLuna ML, Bilguvar K, Bayri Y, Yalcinkaya C, Bursali A, Ozdamar E, Korkmaz B, Mason CE, Ozturk AK, Lifton RP, State MW, Gunel M. Novel NTRK1 mutations cause hereditary sensory and autonomic neuropathy type IV: demonstration of a founder mutation in the Turkish population. Neurogenetics 2008;9:119–25. [DOI] [PubMed] [Google Scholar]
- [142].Urch CE, Suzuki R, Higginson IJ, Hearn J, Murtagh F, Twycross R, Bennett M, El Osta B, Bruera E, Monroe B, Miaskowski C. Pathophysiology of somatic, visceral, and neuropathic cancer pain. In: Clinical pain management second edition: cancer pain. Boca Raton, FL: CRC Press, 2008;3:13. [Google Scholar]
- [143].Ursu D, Ebert P, Langron E, Ruble C, Munsie L, Zou W, Fijal B, Qian YW, McNearney TA, Mogg A, Grubisha O, Mechant K, Sher E. Gain and loss of function of P2X7 receptors: mechanisms, pharmacology and relevance to diabetic neuropathic pain. Mol Pain 2014;10:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Van Hecke O, Kamerman PR, Attal N, Baron R, Bjornsdottir G, Bennett DL, Bennett MI, Bouhassira D, Diatchenko L, Freeman R, Freynhagen R, Haanppää M, Jensen TS, Raja SN, Rice ASC, Seltzer Z, Thorgeirsson TE, Yarnitsky D, Smith BH. Neuropathic pain phenotyping by international consensus (NeuroPPIC) for genetic studies: a NeuPSIG systematic review, Delphi survey, and expert panel recommendations. PAIN 2015;156:2337–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Wadley AL, Hendry LM, Kamerman PR, Chew CS, Price P, Cherry CL, Lombard Z. Role of TNF block genetic variants in HIV-associated sensory neuropathy in black Southern Africans. Eur J Hum Genet 2015;23:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Wadley AL, Lombard Z, Cherry CL, Price P, Kamerman PR. Analysis of a previously identified “pain-protective” haplotype and individual polymorphisms in the GCH1 gene in Africans with HIV-associated sensory neuropathy: a genetic association study. J Acquir Immune Defic Syndr 2012;60:20–3. [DOI] [PubMed] [Google Scholar]
- [147].Wang Q, Guo S, Duan G, Xiang G, Ying Y, Zhang Y, Zhang X. Novel and novel de novo mutations in NTRK1 associated with congenital insensitivity to pain with anhidrosis: a case report. Medicine 2015;94:e871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Wang T, Li H, Xiang J, Wei B, Zhang Q, Zhu Q, Liu M, Sun M, Li H. Identification of a novel nonsense mutation of the neurotrophic tyrosine kinase receptor type 1 gene in two siblings with congenital insensitivity to pain with anhidrosis. J Int Med Res 2017;45:549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Wang XS, Song HB, Chen S, Zhang W, Liu JQ, Huang C, Wang HR, Chen Y, Chu Q. Association of single nucleotide polymorphisms of ABCB1, OPRM1 and COMT with pain perception in cancer patients. J Huazhong Univ Sci Technolog Med Sci 2015;35:752–8. [DOI] [PubMed] [Google Scholar]
- [150].Warner SC, van Meurs JB, Schiphof D, BiermaZeinstra SM, Hofman A, Uitterlinden AG, Richardson H, Jenkins W, Doherty M, Valdes AM. Genome-wide association scan of neuropathic pain symptoms post total joint replacement highlights a variant in the protein-kinase C gene. Eur J Hum Genet 2017;25:446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Watkins LR, Milligan ED, Maier SF. Glial activation: a driving force for pathological pain. Trends Neurosci 2001;24:450–5. [DOI] [PubMed] [Google Scholar]
- [152].Woods CG, Babiker MOE, Horrocks I, Tolmie J, Kurth I. The phenotype of congenital insensitivity to pain due to the NaV1. 9 variant p. L811P. Eur J Hum Genet 2015;23:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Yang Y, Wang Y, Li S, Xu Z, Li H, Ma L, Fan J, Bu D, Liu B, Fan Z, Wu G, Jin J, Ding B, Zhu X, Shen Y. Mutations in SCN9A, encoding a sodium channel alpha subunit, in patients with primary erythermalgia. J Med Genet 2004;41:171–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Yis U, Mademan I, Kavukçu S, Baets J. A novel NTRK1 mutation in a patient with congenital insensitivity to pain with anhidrosis. Acta Neurol Belg 2015;115:509–11. [DOI] [PubMed] [Google Scholar]
- [155].Yuan J, Higuchi Y, Nagado T, Nozuma S, Nakamura T, Matsuura E, Hashiguchi A, Sakiyama Y, Yoshimura A, Takashima H. Novel mutation in the replication focus targeting sequence domain of DNMT1 causes hereditary sensory and autonomic neuropathy IE. J Peripher Nervous Syst 2013;18:89–93. [DOI] [PubMed] [Google Scholar]
- [156].Yuan J, Matsuura E, Higuchi Y, Hashiguchi A, Nakamura T, Nozuma S, Sakiyama Y, Yoshimura A, Izumo S, Takashima H. Hereditary sensory and autonomic neuropathy type IID caused by an SCN9A mutation. Neurology 2013;80:1641–9. [DOI] [PubMed] [Google Scholar]
- [157].Zhang LL, Lin ZM, Ma ZH, Xu Z, Yang YL, Yang Y. Mutation hotspots of SCN9A in primary erythermalgia. Br J Dermatol 2007;156:767–9. [DOI] [PubMed] [Google Scholar]
- [158].Zorina-Lichtenwalter K, Meloto C, Khoury S, Diatchenko L. Genetic predictors of human chronic pain conditions. Neuroscience 2016;338:36–62. [DOI] [PubMed] [Google Scholar]


