In this study, Pontes et al. show that impaired protein synthesis alone triggers a Pi starvation response even when Pi is plentiful in the extracellular milieu in yeast and bacteria. Their findings identify a regulatory connection between protein synthesis and Pi homeostasis that is widespread in nature.
Keywords: ATP, Mg2+, Pi, PhoB/PhoR, PhoP/PhoQ, translation
Abstract
Phosphorus is an essential element assimilated largely as orthophosphate (Pi). Cells respond to Pi starvation by importing Pi from their surroundings. We now report that impaired protein synthesis alone triggers a Pi starvation response even when Pi is plentiful in the extracellular milieu. In the bacterium Salmonella enterica serovar Typhimurium, this response entails phosphorylation of the regulatory protein PhoB and transcription of PhoB-dependent Pi transporter genes and is eliminated upon stimulation of adenosine triphosphate (ATP) hydrolysis. When protein synthesis is impaired due to low cytoplasmic magnesium (Mg2+), Salmonella triggers the Pi starvation response because ribosomes are destabilized, which reduces ATP consumption and thus free cytoplasmic Pi. This response is transient because low cytoplasmic Mg2+ promotes an uptake in Mg2+ and a decrease in ATP levels, which stabilizes ribosomes, resulting in ATP consumption and Pi increase, thus ending the response. Notably, pharmacological inhibition of protein synthesis also elicited a Pi starvation response in the bacterium Escherichia coli and the yeast Saccharomyces cerevisiae. Our findings identify a regulatory connection between protein synthesis and Pi homeostasis that is widespread in nature.
Phosphorus is an integral component of a large repertoire of biological molecules ranging from nucleotides and nucleic acids to lipids and sugar phosphates (Pis). Phosphorus is assimilated and used in the form of Pis (Wanner 1996; Kroger and Fuchs 1999) during the synthesis of adenosine triphosphate (ATP). Because Pi groups carry negative charges, they exist as salts with positively charged ionic species, prominently magnesium (Mg2+), the most abundant divalent cation in living cells (Maguire and Cowan 2002; Wolf and Cittadini 2003; Pontes et al. 2015b). For example, Mg2+ neutralizes negative charges in ATP (Storer and Cornish-Bowden 1976; Maguire and Cowan 2002; Wolf and Cittadini 2003) and ribosomal RNAs (rRNAs) (Wolf and Cittadini 2003; Klein et al. 2004; Pontes et al. 2016). Given that protein synthesis is the activity that consumes the majority of cellular ATP (Stouthamer 1973) and that Mg2+ is required for the assembly of functional ribosomes (Wolf and Cittadini 2003; Klein et al. 2004; Pontes et al. 2016), we reasoned that cells coordinate protein synthesis with the metabolisms of Pi and Mg2+.
Enteric bacteria harbor distinct regulatory systems that maintain the cytoplasmic levels of Pi and Mg2+ within the physiological range. On the one hand, the PhoB/PhoR system consists of the membrane-bound sensor PhoR, which responds to low Pi by activating PhoB (Horiuchi et al. 1959; Makino et al. 1988, 1989; Yamada et al. 1990; Gao and Stock 2013b), a transcriptional regulator that promotes its own expression as well as that of many genes, including those specifying Pi transporters (Rosenberg et al. 1977; Cox et al. 1981; Shinagawa et al. 1983; Makino et al. 1988, 1989; Gao and Stock 2013a, 2015) and enzymes that scavenge Pi from extracytoplasmic organic molecules (Fig. 1; Horiuchi et al. 1959; Makino et al. 1988, 1989; Hsieh and Wanner 2010).
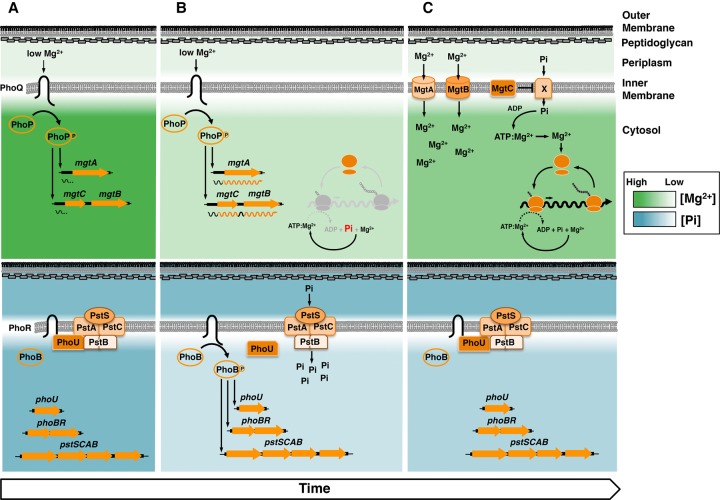
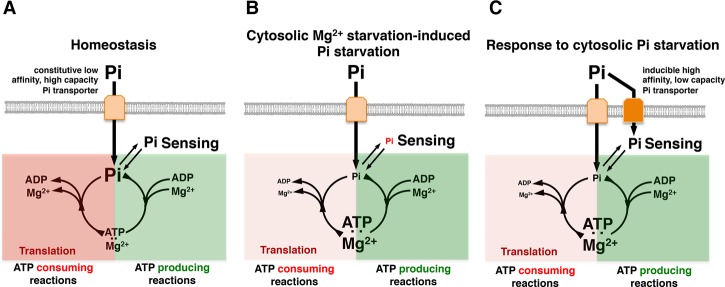
Figure 1.
Cartoon representation of the PhoP/PhoQ and PhoB/PhoR systems, regulated targets, and cellular behavior during Pi starvation induced in low cytoplasmic Mg2+. (A, top) Low extracytoplasmic Mg2+ activates PhoQ, which promotes the phosphorylated state of PhoP (PhoP-P), which in turn promotes transcription initiation from the mgtA and mgtCB promoters. Transcription stops before RNA polymerase (RNAP) reaches the mgtA- and mgtCB-coding regions. (Bottom) High extracellular and intracellular free Pi promotes an interaction between the sensor kinase PhoR and the PstB component of the high-affinity Pi transporter via the regulatory protein PhoU, which inhibits the activity of the PhoB/PhoR system. (B, top) As free cytoplasmic Mg2+ decreases, ribosome subunits are unable to associate efficiently, translation slows down, ATP consumption decreases, and transcription elongation into mgtA- and mgtCB-coding regions takes place. (Bottom) The decrease in ATP consumption prevents the liberation of Pi into the cytosol, leading to a decrease in free cytoplasmic Pi. The low concentration of free cytoplasmic Pi disrupts the inhibitory activity of PhoU, resulting in PhoB/PhoR activation even when extracytoplasmic Pi is high. (C, top) Restoration of free cytoplasmic Mg2+ by the MgtA and MgtB proteins importing Mg2+ into the cytoplasm and the MgtC protein reducing the importation of Pi (via an unknown mechanism/Pi transporter X), which feeds into the synthesis of the Mg2+-chelating ATP. The increase in free cytoplasmic Mg2+ restores translation, which results in ATP hydrolysis and recycling of intracellular Pi. (Bottom) Elevated ATP hydrolysis increases free cytoplasmic Pi levels, restoring the PhoU-mediated inhibition of PhoB/PhoR activity.
On the other hand, the PhoP/PhoQ system consists of the sensor PhoQ, which responds to low extracytoplasmic Mg2+ by activating PhoP (Soncini et al. 1996; Véscovi et al. 1996), a regulatory protein that promotes its own transcription (Soncini et al. 1995) as well as that of dozens of genes (Zwir et al. 2012; Groisman et al. 2013). In Salmonella enterica serovar Typhimurium, where the PhoP/PhoQ system is best understood (Groisman et al. 2013), PhoP promotes transcription of genes that mediate the chemical modification of Pis in the bacterial outer membrane (Guo et al. 1997; Bishop et al. 2000; Shi et al. 2004), Mg2+ uptake into the cytoplasm (Soncini et al. 1996; Véscovi et al. 1996), and inhibition of the ATP-generating F1Fo ATPase (Fig. 1; Lee et al. 2013).
We now report that the metabolisms of Pi and Mg2+ are linked to each other and to the translation status of the cell. We establish that cytoplasmic Mg2+ limitation activates the Salmonella PhoB/PhoR system even when Pi levels in the bacterium's surroundings are high. We determine that protein synthesis inhibitors activate the PhoB/PhoR system in both Salmonella and Escherichia coli and trigger a Pi starvation response in Saccharomyces cerevisiae. Moreover, genomic analysis reveals that genes involved in Pi metabolism are often adjacent to those responsible for Mg2+ homeostasis in the chromosomes of divergent bacterial species. Our findings demonstrate that the coordination of Pi and Mg2+ metabolisms with ribosome activity exhibits a broad phylogenetic distribution. This is true even though the particular gene products responsible for this coordination vary across species. The identified coordination of Pi and Mg2+ metabolisms with ribosome activity likely reflects the universality of phosphorus as a cellular building block, Mg2+ as a biological Pi counter ion, and both ions as central players in protein synthesis.
Results
Cytoplasmic Mg2+ limitation induces a Pi starvation response
When bacteria are placed in low-Mg2+ medium, low extracellular Mg2+ activates the PhoP/PhoQ system (Fig. 1A; Véscovi et al. 1996). Initially, bacteria grow and consume the limited Mg2+ present in the medium via the housekeeping Mg2+ transporter CorA (Snavely et al. 1989b). Eventually, as CorA is unable to maintain an adequate supply of Mg2+, the concentration of cytoplasmic Mg2+ drops to levels that compromise ribosome assembly (Pontes et al. 2016). Impaired translation and/or direct detection of low cytoplasmic Mg2+ promotes transcription of the mgtA-, mgtB-, and mgtC-coding regions via the formation of secondary structures in the leader regions of their respective mRNAs that hinder transcription termination within the leader region (Cromie et al. 2006; Spinelli et al. 2008; Park et al. 2010; Zhao et al. 2011; Lee and Groisman 2012a,b). The mgtA and mgtB genes specify two distinct Mg2+ transporters (Snavely et al. 1989a,b), and the mgtC gene specifies an inhibitor of the F1Fo ATPase (Lee et al. 2013). Production of the MgtA, MgtB, and MgtC proteins increases the concentration of free cytosolic Mg2+, inhibits ribosome production, and stabilizes the remaining ribosomes (Pontes et al. 2016).
To identify additional genes differentially transcribed in response to a decrease in cytoplasmic Mg2+ concentration, we assessed RNA polymerase (RNAP) occupancy of the Salmonella genome by chromatin immunoprecipitation and high-throughput sequencing (ChIP-seq) following growth in defined liquid medium with two different Mg2+ concentrations (10 or 50 µM). Growth in either Mg2+ concentration activates the PhoP/PhoQ system (Véscovi et al. 1996; Cromie et al. 2006) and results in a similar optical density (OD600) of the bacterial cultures by 4 h (Supplemental Fig. S1A). However, the threshold of cytoplasmic Mg2+ concentration that triggers transcription of the mgtA-, mgtB-, and mgtC-coding regions takes place following 4 h of growth in 10 µM Mg2+ but not in 50 µM Mg2+ (Cromie et al. 2006; Spinelli et al. 2008).
RNAP occupancy was similar under 10 and 50 µM Mg2+ for the vast majority of genes, including the PhoP-activated phoPQ, slyA, and slyB genes (Fig. 2A) and those not regulated by PhoP, such as the ompA, ybdQ, and his genes (Supplemental Fig. S1B). In contrast, RNAP occupancy of the mgtA-, mgtB-, and mgtC-coding regions was much higher following growth in 10 µM Mg2+ than in 50 µM Mg2+ (Fig. 2A), in agreement with previous mRNA and transcriptional reporter data (Cromie et al. 2006; Spinelli et al. 2008).
Figure 2.
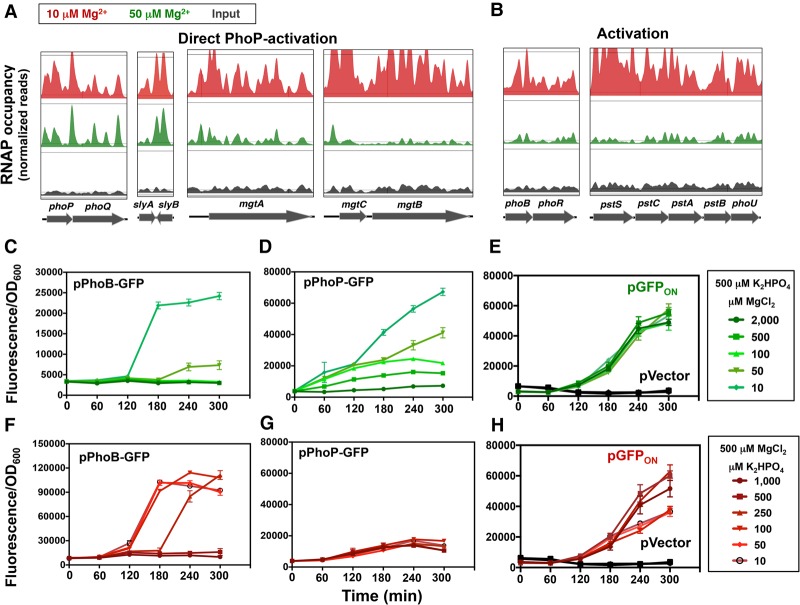
Low cytoplasmic Mg2+ induces a transcriptional signature of Pi starvation. (A,B) RNAP ChIP-seq results of selected genes in wild-type Salmonella (14028s) grown in N-minimal medium containing 10 µM Mg2+ (red) or 50 µM MgCl2 (green) to an OD600 of ∼0.3. Input (nonimmunoprecipitated DNA) is also shown (black). Peak heights were normalized to the total number of assembled nucleotides. (C–E) Fluorescence from wild-type Salmonella (14028s) harboring pPhoB-GFP (C), pPhoP-GFP (D), and the promoterless GFP vector pVector and inducible GFP vector pGFPON (pUH-GFP; 325 µM IPTG) (E) grown in MOPS medium containing 500 µM K2HPO4 and the indicated concentrations of MgCl2. (F–H) Fluorescence from wild-type Salmonella (14028s) harboring pPhoB-GFP (F), pPhoP-GFP (G), and the promoterless GFP vector pVector and constitutive GFP vector pGFPON (pUH-GFP; 100 µM IPTG) (H) grown in MOPS medium containing 500 µM MgCl2 and the indicated concentrations of K2HPO4. Error bars represent the standard deviations. Graphs are representative of at least three independent experiments with a total of at least six biological replicates. See also Supplemental Figure S1.
Surprisingly, RNAP occupancies of the PhoB-activated phoBR, pstSCAB, and phoU genes were higher during growth in 10 µM Mg2+ than in 50 µM Mg2+ (Fig. 2B). This result was unexpected for two reasons. First, Pi was abundant in the growth medium (∼1.16 mM), implying that PhoR should be promoting the inactive state of the PhoB protein, resulting in low levels of PhoB-activated mRNAs (Gao and Stock 2013a,b). Second, the promoters of the phoBR, pstSAB, and phoU genes lack sequences resembling PhoP-binding sites (Kato et al. 1999; Zwir et al. 2012), suggesting that PhoP does not directly activate these promoters.
In agreement with the RNAP ChIP-seq results (Fig. 2B), quantitative PCR (qPCR) revealed that the mRNA levels of the phoB, pstB, pstC, and pstS genes were fivefold to 25-fold higher following growth in 10 µM Mg2+ than in 50 µM Mg2+ (Supplemental Fig. S1C). In contrast, no differences in mRNA amounts were observed for the slyB, ompA, and ybdQ genes (Supplemental Fig. S1C). In sum, low cytoplasmic Mg2+ promotes transcription of both PhoB-activated genes and PhoP-activated genes that maintain protein synthesis (Pontes et al. 2016).
Delayed activation of PhoB in bacteria experiencing low Mg2+ or low Pi
To examine the expression behavior of PhoB-dependent genes resulting from changes in cytoplasmic Mg2+, we monitored the fluorescence of wild-type Salmonella harboring a medium copy number plasmid with a transcriptional fusion between the PhoB-activated phoB promoter and a promoterless gfp gene. This approach enabled a temporal resolution of the response to cytoplasmic Mg2+ concentration in an organism with an otherwise wild-type genome.
Fluorescence was low in the first 2 h of growth in 10 µM Mg2+, increased dramatically by 3 h, and remained at the same high levels for the following 2 h (Fig. 2C). When grown in 50 µM Mg2+, fluorescence increased only at 4 h and reached values less than one-fifth of those observed during growth in 10 µM Mg2+ (Fig. 2C). Organisms experiencing ≥100 µM Mg2+ displayed no change in fluorescence (Fig. 2C). Thus, activation of the phoB promoter requires growth in <100 µM Mg2+ for >2 h.
Transcription from the phoB promoter is in contrast to that from the PhoP-activated phoP promoter because the fluorescence of a strain harboring a transcriptional fusion between the phoP promoter and a promoterless gfp gene increased in medium with ≤500 µM Mg2+ in the first hour and continued to increase over the course of the experiment (Fig. 2D). As expected, fluorescence remained low in wild-type Salmonella carrying the vector control harboring a promoterless gfp gene (Fig. 2E, pVector) and was not impacted by the Mg2+ concentration in the medium in a strain in which the lac promoter transcribes a promoterless gfp gene (Fig. 2E, pGFPON).
In contrast, low Pi does not activate the PhoP/PhoQ system: Fluorescence of the strain harboring the phoP-gfp fusion was the same regardless of the Pi concentration in the medium (Fig. 2G). Control experiments revealed that fluorescence of wild-type Salmonella harboring the phoB-gfp fusion increased dramatically at 3 h in medium with ≤100 µM Pi and at 4 h in medium with 250 µM Pi (Fig. 2F). No increase in fluorescence was observed in organisms grown in ≥500 µM Pi (Fig. 2F) or in the strains harboring the vector control (Fig. 2H, pVector). Additionally, when gfp was transcribed from the lac promoter, fluorescence was only 50% lower in cells grown in low Pi relative to those grown in high-Pi medium (Fig. 2H), indicating that low Pi per se does not lead to increased GFP fluorescence. Cumulatively, the results presented above establish that both low Mg2+ and low Pi activate the PhoB/PhoR system after a delay of several hours.
The Mg2+ transporters MgtA and MgtB and the ATPase inhibitor MgtC regulate PhoB activity in opposite ways
To explore whether the PhoB activation (Fig. 2B) and the transcription of the mgtA-, mgtB-, and mgtC-coding regions taking place in low cytoplasmic Mg2+ (Figs. 1B, 2A; Cromie et al. 2006; Spinelli et al. 2008) are physiologically linked, we examined fluorescence over time in isogenic strains harboring gfp transcriptional fusions to the phoB promoter, the mgtA promoter and leader region, and the mgtC promoter and leader region (mgtB follows mgtC in the mgtCBR operon) (Fig. 1).
Transcription from the phoB-gfp fusion preceded that of the mgtA-gfp and mgtC-gfp fusions following a shift from 10 mM to 10 µM Mg2+ (Fig. 3A); that is, fluorescence from the phoB-gfp fusion began at 120 min, remained stable for 80 min, and decreased during the following 16.6 h (Fig. 3A). In contrast, fluorescence from the mgtA-gfp and mgtC-gfp fusions began at 135 min and continued to increase throughout the course of the experiment (Fig. 3A). As expected, the strain with the vector control displayed no major changes in fluorescence (Fig. 3A). Thus, the phoB-gfp fusion differs from the mgtA-gfp and mgtC-gfp fusions in both the time of induction and the behavior in the subsequent 17 h.
Figure 3.
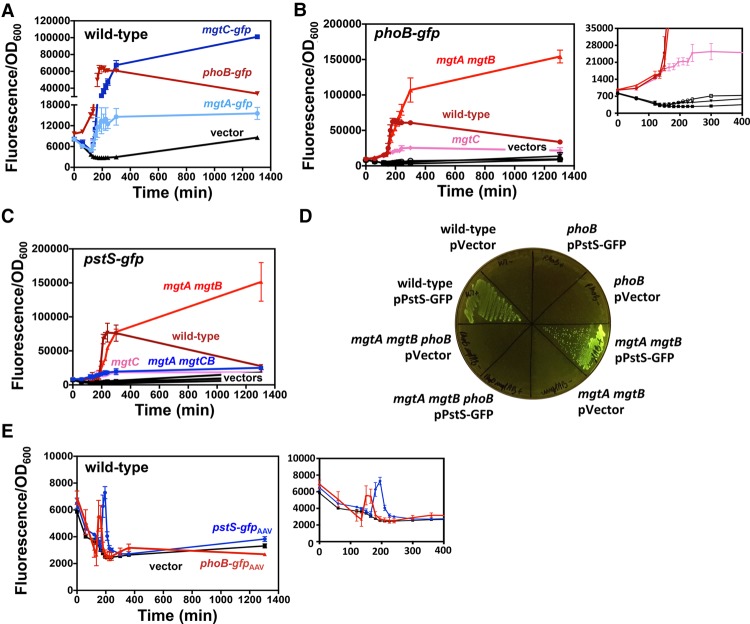
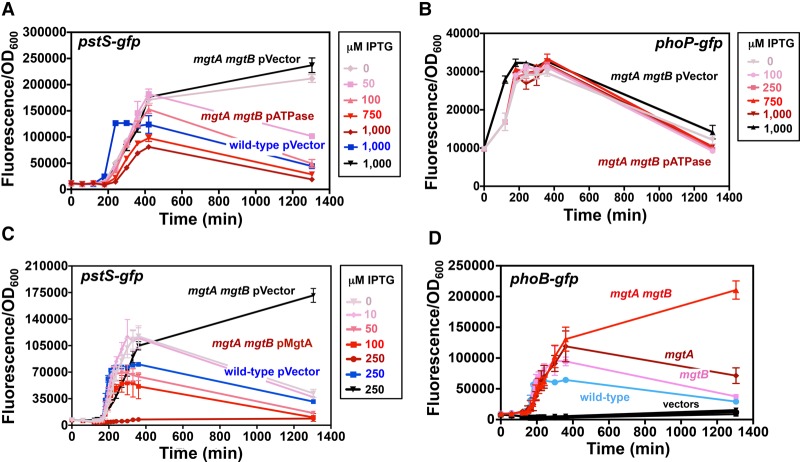
The Mg2+ transporters MgtA and MgtB and the ATPase inhibitor MgtC regulate PhoB activity in opposite ways. (A) Fluorescence from wild-type Salmonella (14028s) harboring pPhoB-GFP (phoB-gfp), pMgtA-leader-GFP (mgtA-gfp), pMgtC-leader-GFP (mgtC-gfp), or the promoterless GFP vector pVector plasmid. (B) Fluorescence from wild-type (14028s), mgtA mgtB (EG17048), and mgtC (EL4) Salmonella harboring pPhoB-GFP or pVector. (C) Fluorescence from wild-type (14028s), mgtA mgtB (EG17048), mgtC (EL4), and mgtA mgtCB (MP363) Salmonella harboring pPstS-GFP or pVector. (D) Fluorescence from wild-type (14028s), phoB (EG9054), mgtA mgtB (EG17048), and mgtA mgtB (EG17048) and phoB mgtA mgtB (MP1184) Salmonella following 24 h of growth on solid medium. (E) Fluorescence from wild-type Salmonella (14028s) harboring pPhoB-GFPAAV, pPstS-GFPAAV, or pVectorAAV. For all experiments, cells were grown in MOPS liquid or on solid (1% agarose) medium containing 10 µM MgCl2 and 500 µM K2HPO4. Error bars represent the standard deviations. Graphs are representative of at least three independent experiments with a total of at least six biological replicates. See also Supplemental Figure S2.
We reasoned that PhoB activation results from disturbances in free cytoplasmic Mg2+ occurring prior to the production of the MgtA, MgtB, and MgtC proteins, which maintain physiological levels of free cytoplasmic Mg2+ by importing Mg2+ ions and reducing the levels of Mg2+-chelating ATP molecules (Pontes et al. 2016). If so, the decrease in fluorescence of the PhoB-activated phoB-gfp fusion happening after transcription of the mgtA- and mgtCB-coding regions (Fig. 3A) may reflect changes brought about by the MgtA, MgtB, and/or MgtC proteins. This hypothesis predicts that inactivation of the mgtA, mgtB, and/or mgtC genes should abolish the decrease in fluorescence of the phoB-gfp fusion observed in wild-type cells at 200 min (Fig. 3A) but not alter the time at which fluorescence starts.
As hypothesized, fluorescence from the phoB-gfp fusion began at ∼120 min in wild-type, mgtC, and mgtA mgtB Salmonella (Fig. 3B). However, whereas fluorescence decreased in the wild-type strain after 200 min (Fig. 3B), it rose throughout the course of the experiment in the mgtA mgtB double mutant (Fig. 3B). Unexpectedly, fluorescence was lower in the mgtC mutant than in wild-type Salmonella (Fig. 3B). The behavior of the strain harboring the phoB-gfp fusion reflects activation of the PhoB protein (as opposed to being confined to the phoB promoter) because the wild-type, mgtC, and mgtA mgtB strains displayed analogous fluorescence patterns when harboring a gfp fusion to the pstS promoter (Fig. 3C), which is also activated by PhoB (Makino et al. 1988). The mgtC mutation is epistatic to the mgtA and mgtB mutations because fluorescence was similarly low in the mgtC single mutant and mgtA mgtB mgtC triple mutant (Fig. 3C). Control experiments showed unchanged low fluorescence in strains harboring the vector control (Fig. 3B,C).
The PhoB protein is responsible for the transcriptional induction of the phoB-gfp and pstS-gfp fusions taking place in low Mg2+ because deletion of the phoB gene eliminated the high fluorescence exhibited by wild-type and mgtA mgtB strains harboring the pstS-gfp fusion on solid medium containing 10 µM Mg2+ (Fig. 3D). In agreement with the results described above (Fig. 3B,C), the mgtA mgtB double mutant and the mgtC mutant displayed more and less fluorescence than the wild-type strain, respectively, when harboring either the phoB-gfp (Supplemental Fig. S2A) or pstS-gfp (Supplemental Fig. S2B) fusion. There was no fluorescence in strains harboring the vector control (Supplemental Fig. S2A,B).
Taken together, the results presented in this section indicate that low cytoplasmic Mg2+ activates the PhoB protein and that the Mg2+ transporters MgtA and MgtB and the ATPase inhibitor MgtC regulate PhoB activity in opposite ways.
MgtC activates PhoB by decreasing cytoplasmic Pi levels
To understand how MgtC activates the PhoB protein, we probed the effects of time and levels of mgtC induction and whether Mg2+ controls PhoB activity independently of its effect on MgtC expression (Spinelli et al. 2008). Fluorescence from the pstS-gfp fusion increased sharply in an mgtC mutant harboring a plasmid with a copy of the mgtC-coding region under the control of a derivative of the lac promoter (pMgtC) following addition of isopropyl β-D-1-thiogalactopyranoside (IPTG), the gratuitous inducer of the lac promoter (Fig. 4A). MgtC acts in a dose-dependent manner because fluorescence progressively rose as the IPTG concentration increased (Fig. 4A) and surpassed that of the wild-type strain carrying the plasmid vector at ≥250 µM IPTG (Fig. 4A). As expected, fluorescence from the mgtC mutant harboring the vector control remained low and unaffected by the presence of IPTG (Fig. 4A); the same was true when no IPTG was added to the mgtC/pMgtC strain (Fig. 4A).
Figure 4.
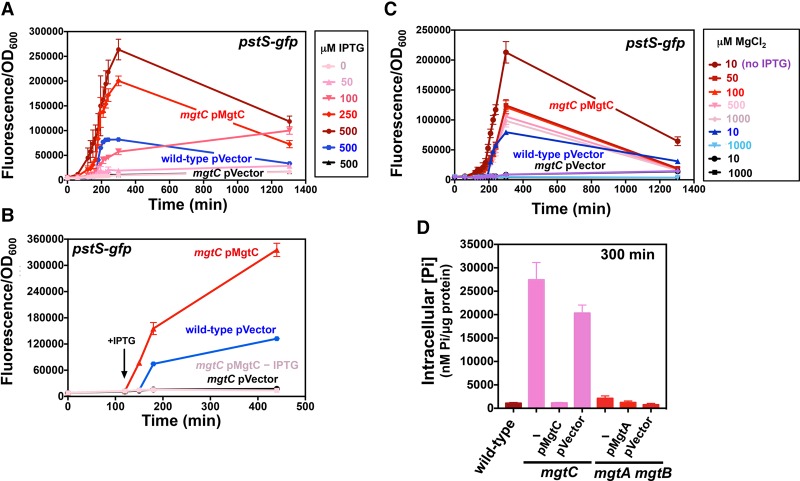
MgtC reduces cytoplasmic Pi during cytoplasmic Mg2+ starvation. (A) Fluorescence from wild-type (14028s) and mgtC (EL4) Salmonella carrying pPstS-GFPc and either pUHE-21 (pVector) or pUHE-MgtC (pMgtC) grown in the presence of different concentrations of IPTG. The fluorescence of mgtC (EL4)/pMgtC cultures lacking IPTG is also shown. (B) Fluorescence from wild-type (14028s) and mgtC (EL4) strains carrying pPstS-GFPc and pVector in the presence of 500 µM IPTG and mgtC (EL4)/pMgtC at the indicated IPTG concentrations. The arrow indicates the time when 250 µM IPTG was added to the cultures. (C) Fluorescence from wild-type (14028s) and mgtC (EL4) Salmonella carrying pPstS-GFPc and pVector during growth in 10 or 1000 µM MgCl2 and of mgtC (EL4)/pMgtC Salmonella in the presence of the indicated MgCl2 concentrations. Unless indicated in the figure, bacteria were grown in the presence of 250 µM IPTG. (D) Total intracellular Pi in wild type (14028s), mgtC (EL4), or mgtC (EL4)/pMgtC or pVector, mgtA mgtB (EG17048) Salmonella, and mgtA mgtB (EG17048)/pMgtA or pVector following 300 min of growth. All experiments were carried out in MOPS medium containing 500 µM K2HPO4 and either 10 µM MgCl2 or the specified MgCl2 concentration. Error bars represent the standard deviations. Graphs are representative of at least three independent experiments with a total of at least six biological replicates. See also Supplemental Figure S2.
The wild-type strain harboring the vector control instead of pMgtC fluoresced much later than the mgtC/pMgtC strain treated with IPTG (Fig. 4B). This result reflects normal transcriptional control of the mgtC-coding region by the wild-type promoter (Soncini et al. 1996) and leader (Cromie et al. 2006; Spinelli et al. 2008) regions.
MgtC activates PhoB independently of the Mg2+ concentration in the medium because IPTG induced fluorescence from phoB-gfp even when the mgtC/pMgtC strain was grown in 1 mM Mg2+ (Fig. 4C), a condition in which the mgtC-coding region is not transcribed in wild-type Salmonella (Spinelli et al. 2008). In contrast, wild-type Salmonella harboring the vector control fluoresced following growth in 10 µM Mg2+ but not in 1 mM Mg2+ (Fig. 4C). The fluorescence of the mgtC mutant harboring the vector control remained low in all tested conditions (Fig. 4C).
We reasoned that MgtC activates PhoB by decreasing cytoplasmic Pi levels because low Pi medium activates PhoB (Horiuchi et al. 1959; Makino et al. 1988, 1989) and because the sensor PhoR has a small periplasmic region. In agreement with this notion, Pi levels were 18-fold to 25-fold lower in wild-type Salmonella than in the mgtC mutant following 5 h of growth in 10 µM Mg2+ (Fig. 4D). (This time was chosen because wild-type Salmonella exhibited low steady-state levels of phoB and pstS transcription [Fig. 3A–C,E], suggesting that intracellular Pi levels are stable.) Moreover, mgtC expression from a heterologous promoter lowered intracellular Pi levels in the mgtC mutant, whereas the vector control had no effect (Fig. 4D). In sum, MgtC activates PhoB by reducing cytoplasmic Pi levels.
The MgtA and MgtB proteins prevent a sustained Pi starvation response when Pi is plentiful
The PhoB protein is hyperactivated in the mgtA mgtB double mutant following growth in 10 µM Mg2+ for 120 min (Figs. 3B,C, 5A,C,D; Supplemental Fig. S2). Thus, we anticipated the mgtA mgtB mutant to have low cytoplasmic Pi. Surprisingly, the mgtA mgtB double mutant exhibited wild-type levels of cytoplasmic Pi (Fig. 4D). What, then, activates PhoB in the mgtA mgtB double mutant or in the wild-type strain before the MgtA and MgtB proteins are expressed?
We hypothesized that PhoB activation results from Pi being sequestered in ATP molecules that are not effectively recycled when translation is compromised. In agreement with this notion, translation consumes the majority of cellular ATP (Stouthamer 1973), and the mgtA mgtB double mutant, unable to efficiently assemble ribosomes and carry out translation (Pontes et al. 2016), displays higher ATP levels than the wild-type strain (Supplemental Fig. S3A; Pontes et al. 2016). If this hypothesis is correct, releasing Pi trapped in ATP molecules should decrease PhoB activity.
As hypothesized, fluorescence from the pstS-gfp fusion was inversely proportional to the IPTG concentration in a culture of the mgtA mgtB double mutant carrying plasmid pATPase (Fig. 5A), which promotes IPTG-inducible ATP hydrolysis (Supplemental Fig. S3B). In contrast, there was no change in fluorescence in the isogenic strain with the vector control (Fig. 5A). Fluorescence of an isogenic strain with the PhoP-activated phoP-gfp fusion was unaffected by the presence of pATPase (Fig. 5B), indicating that the pATPase plasmid does not cause a systemic decrease in transcription. Taken together, these results indicate that MgtA and MgtB inhibit PhoB activation by maintaining the integrity of ATP-hydrolyzing reactions.
Figure 5.
Free cytoplasmic Mg2+ inhibits Pi starvation response. (A) Fluorescence from wild-type (14028s), mgtA (EG16735), mgtB (EL5), and mgtA mgtB (EG17048) Salmonella harboring pPhoB-GFP. (B) Fluorescence from wild-type Salmonella (14028s) carrying pPstS-GFPc and pUHE-21 (pVector) and from mgtA mgtB mutant (EG17048) harboring pPstS-GFPc and either pUHE-21 (pVector) or pUHE-MgtA (pMgtA) in the presence of the indicated IPTG concentrations. (C) Fluorescence from wild-type Salmonella (14028s) carrying pPstS-GFPc and pVector and from mgtA mgtB mutant Salmonella (EG17048) harboring pPstS-GFPc and either pVector or pUHE-ATPase (pATPase) in the presence of the indicated IPTG concentrations. (D) Fluorescence from mgtA mgtB (EG17048) Salmonella harboring pPhoP-GFP and either pATPase or pVector in the presence of various IPTG concentrations. For all experiments, cells were grown in MOPS liquid medium containing 10 µM MgCl2 and 500 µM K2HPO4. Error bars represent the standard deviations. Graphs are representative of at least three independent experiments with a total of at least six biological replicates. See also Supplemental Figures S2 and S3.
The Mg2+ transporters MgtA and MgtB are ∼50% identical at the amino acid level but display different specificities and modes of regulation (Snavely et al. 1991; Tao et al. 1995; Groisman et al. 2013). We determined that either Mg2+ transporter is sufficient for the decrease in PhoB activation after it reaches a peak; that is, fluorescence from the pstS-gfp fusion in the mgtA mgtB double mutant carrying the IPTG-inducible pMgtA decreased as the IPTG concentration increased (Fig. 5C), but no decrease in fluorescence was observed in the isogenic strain harboring the vector control (Fig. 5C). In contrast to the sustained high fluorescence displayed by the mgtA mgtB double mutant (Figs. 3B,C, 5A,C,D), fluorescence from the phoB-gfp fusion increased and subsequently decreased in the mgtA and mgtB single mutants (Fig. 5D), like in the wild-type strain (Fig. 5D). However, the mgtA and mgtB single mutants achieved higher fluorescence peak values than the wild-type strain (Fig. 5D). As expected, the fluorescence of strains carrying the vector control remained low throughout the course of the experiment (Fig. 5D). Thus, the MgtA and MgtB proteins prevent sustained activation of the PhoB protein.
The data presented above argue that PhoB activation is transient in wild-type Salmonella experiencing low cytoplasmic Mg2+, lasting only until the MgtA and MgtB proteins normalize Mg2+ concentration to levels that support ribosome assembly (Pontes et al. 2016). To test this possibility, we examined the behavior of a set of isogenic strains harboring plasmid-borne transcriptional fusions of the PhoB-activated phoB and pstS promoters to a gfp gene specifying an unstable GFP variant (gfpAAV) (Andersen et al. 1998), which allowed examination of short-lived changes in transcriptional activity.
Wild-type Salmonella harboring the phoB-gfpAAV or pstS-gfpAAV fusion displayed a sharp increase in fluorescence that lasted 45 min when shifted to 10 µM Mg2+ (Fig. 3E). In contrast, no changes in fluorescence were observed in the isogenic strain carrying the vector control (Fig. 3E). In agreement with these results, wild-type Salmonella harboring the phoB-gfp or pstS-gfp fusion were fluorescent following overnight growth on 10 µM Mg2+ solid medium (Supplemental Fig. S2A–C) but not when harboring fusions of the same two promoters to the gfpAAV variant (Supplemental Fig. S2D–F). In contrast, the mgtA mgtB double mutant with either the phoB-gfpAAV or pstS-gfpAAV fusion was fluorescent after overnight growth on 10 µM Mg2+ (Supplemental Fig. S2E,F). Thus, by promoting Pi recycling, the MgtA and MgtB proteins avoid a Pi starvation response.
A physiological connection between protein synthesis and PhoB activation
Bacteria link metabolic energy with translation potential (Gaal et al. 1997). For example, the translation inhibitor chloramphenicol promotes an increase in cytoplasmic ATP levels (Bagnara and Finch 1973). Under normal physiological conditions (i.e., in the absence of translation inhibitors), an increase in ATP levels stimulates rRNA transcription (Gaal et al. 1997; Schneider et al. 2002; Murray et al. 2003), which derepresses expression of ribosomal proteins (Nomura et al. 1984; Zengel and Lindahl 1994), resulting in a larger number of translating ribosomes and boosting translation rates (Bremer and Dennis 1996; Murray et al. 2003).
We reasoned that the translation inhibitor chloramphenicol activates the PhoB/PhoR system because Pi is largely incorporated through assimilation into ATP molecules (Wanner 1996; Kroger and Fuchs 1999) and because translation is the activity that uses most cellular ATP (Stouthamer 1973). Therefore, if translation is inhibited, Pi would remain trapped in ATP, which bacteria might experience as low cytoplasmic Pi even if the Pi concentration in their surroundings is high.
As proposed, addition of chloramphenicol to wild-type Salmonella increased both cytoplasmic ATP levels (Fig. 6A) and the amounts of the phoB (∼16-fold) and pstS (∼40-fold) mRNAs (Fig. 6B). Notably, these increases took place in the presence of 2 mM Pi, a concentration that normally hinders activation of the PhoB/PhoR system (Horiuchi et al. 1959; Makino et al. 1988, 1989). The large increases in phoB and pstS mRNA amounts appear specific to PhoB-activated genes because they were not observed in a phoB mutant (Fig. 6C) and because the mRNA level of the PhoB-independent thrS gene was not altered (Fig. 6B).
Figure 6.
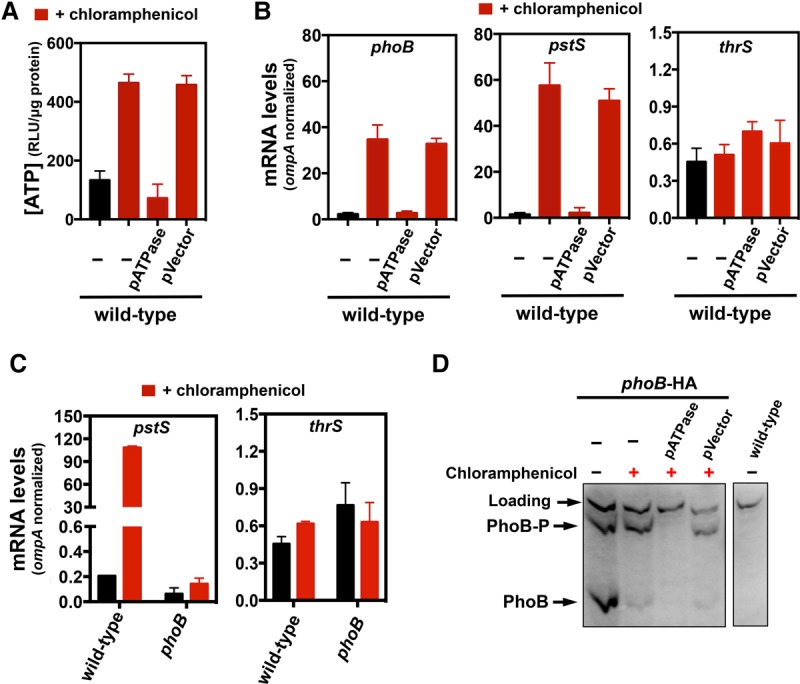
Translation recycles intracellular Pi from ATP, preventing a Pi starvation response. (A) Intracellular ATP levels of wild-type Salmonella (14028s) lacking or harboring either pUHE-ATPase (pATPase) or pUHE-21 (pVector). (B) mRNA amounts of the phoB, pstS, and thrS genes produced by wild-type Salmonella (14028s) lacking or harboring either pATPase or pVector. (C) mRNA amounts of the pstS and thrS genes produced by wild-type (14028s) and phoB (EG9054) Salmonella. For A–C, cells were grown in MOPS containing 10 mM MgCl2 and 2 mM K2HPO4 for 2 h followed by 30 min of treatment with 1 mM IPTG (when harboring pATPase or pVector) followed by 30 min of treatment with 25 µg/mL chloramphenicol. mRNA amounts were normalized to those of the ompA gene. (D) Western blot analysis of extracts prepared from wild-type (14028s) and phoB-HA (MP1429) Salmonella and phoB-HA (MP1429) Salmonella harboring either pATPase or pVector following separation on Phos-tag SDS-PAGE to detect PhoB-HA and PhoB-HA-P. Cells were grown in MOPS containing 10 mM MgCl2 and 200 µM K2HPO4 for 2 h and treated for 35 min with 1 mM IPTG followed by chloramphenicol as in A–C. Error bars represent the standard deviations. Graphs are representative of at least three independent experiments with a total of at least six biological replicates. See also Supplemental Figures S3 and S4.
If excessive ATP resulting from the inhibition of translation promotes PhoB activation, then chloramphenicol should increase the fraction of phosphorylated relative to unphosphorylated PhoB protein. As predicted, we established that when Salmonella was grown in an intermediate Pi concentration to promote PhoB activation, the addition of chloramphenicol increased the ratio PhoB-P to PhoB (Fig. 6D). Liberating Pi from ATP by expressing pATPase prior to chloramphenicol addition prevented PhoB activation, whereas the vector control had no effect (Fig. 6D). Because PhoB positively regulates its own transcription, cells experiencing pATPase prior to chloramphenicol addition had undetectable levels of PhoB protein (Fig. 6D).
The increases in ATP levels and transcription of PhoB-dependent genes taking place upon chloramphenicol treatment result from inhibition of protein synthesis (rather than from a different action of chloramphenicol) because similar increases in ATP levels (Supplemental Fig. S4A) and phoB and pstS mRNA amounts (Supplemental Fig. S4B) were observed when translation was inhibited with spectinomycin. Furthermore, they result from Pi being trapped in ATP because the pATPase plasmid prevented rises in ATP (Fig. 6A), pstS, and phoB mRNA amounts (Fig. 6B) and PhoB-P levels (Fig. 6D) caused by chloramphenicol. This is in contrast to cultures harboring the vector control, which failed to overcome the effect of chloramphenicol (Fig. 6A,B,D). Taken together, these results indicate that ATP consumption by active ribosomes increases the intracellular pools of free Pi, thereby reducing PhoB activity.
Translation inhibitors promote Pi starvation responses in E. coli and S. cerevisiae
We hypothesized that inhibition of protein synthesis triggers a Pi starvation response in other organisms because all living cells use ATP to synthesize and power ribosomes and because Pi is a precursor of ATP. Chloramphenicol treatment increased the mRNA levels of the phoB and pstS genes in E. coli (Supplemental Fig. S4D), just as it did in Salmonella (Fig. 6B). The latter increase is caused by Pi being trapped in ATP because the pATPase plasmid normalized ATP levels (Supplemental Fig. S4C) and prevented the increase in the levels of phoB and pstS mRNAs (Supplemental Fig. S4D), whereas the vector control had no effect (Supplemental Fig. S4C,D).
The Pi starvation response triggered by protein synthesis inhibitors extends beyond the bacterial domain; that is, the inhibitor of eukaryotic translation cycloheximide increased the mRNA levels of the PHO89 and PHO84 genes in S. cerevisiae (Supplemental Fig. S4E), which specify high-affinity Pi transporters normally induced when Pi is limited (Willsky and Malamy 1980; Bun-Ya et al. 1991; Auesukaree et al. 2003). In contrast, the mRNA levels of MAG1, which is induced by DNA damage (Chen and Samson 1991), remained unaltered (Supplemental Fig. S4E). These results argue that ATP hydrolysis resulting from translation maintains Pi homeostasis by replenishing free Pi to the cytosol.
Discussion
We uncovered a fundamental relationship between protein synthesis and the cytoplasmic levels of Pi and Mg2+. This relationship enables cells to coordinate Mg2+ availability with Pi acquisition. Moreover, it protects the integrity of critical processes, such as translation, that require the stabilization of negative charges of Pi-containing biomolecules by Mg2+ ions (Pontes et al. 2016). Specifically, a decrease in free cytoplasmic Mg2+ triggers a Pi starvation response that lasts until cells normalize the pools of free cytoplasmic Mg2+ by promoting Mg2+ transport into the cytosol and inhibiting the acquisition of Pi. The resulting physiological changes reduce growth rate (Pontes et al. 2016), enabling cells to maintain viability (Soncini et al. 1996) and achieve higher growth yields (Blanc-Potard and Groisman 1997; Pontes et al. 2015a).
We determined that conditions that compromise translation trigger a Pi starvation response even when Pi is plentiful in a cell's surroundings (Fig. 7). This is because translation is the cellular activity that consumes most cellular ATP (Stouthamer 1973), and Pi incorporation into ATP enables its assimilation into cells through cycles of ATP synthesis and hydrolysis (Wanner 1996; Kroger and Fuchs 1999). For instance, a decrease in free cytoplasmic Mg2+ reduces the number of active ribosomes by compromising ribosome assembly, thereby drastically lowering the fraction of ATP-consuming reactions (Fig. 7A; Pontes et al. 2016). As the available Pi transporters are unable to maintain a steady supply of Pi relative to the fraction being incorporated into ATP molecules, the levels of free cytoplasmic Pi decrease (Fig. 7B). This decrease activates the PhoB/PhoR system, promoting the phosphorylated state of PhoB (Fig. 6D) and transcription of a high-affinity Pi transporter (Figs. 6B, 7C). That translation inhibitors activate the PhoB/PhoR system (Fig. 6B–D; Supplemental Fig. S4B,D) by “trapping” cytoplasmic Pi in ATP molecules is supported by the prevention of PhoB/PhoR activation resulting from stimulation of ATP hydrolysis using the pATPase plasmid (Fig. 6A,B; Supplemental Fig. S4A–D).
Figure 7.
Free cytoplasmic Mg2+ inhibits Pi starvation response. (A) When neither Pi nor Mg2+ are limiting in the cytoplasm, extracellular Pi is imported via constitutive low-affinity high-capacity Pi transporters. Free intracellular Pi is sensed by components of the cellular machinery and incorporated into ATP. Pi from ATP is transferred to other biomolecules and assimilated into the cell or liberated back into the cytosol through ATP hydrolysis (e.g., the result of active ribosomes). (B) During Pi starvation, the intracellular concentration of free Pi decreases either because Pi is limited in the extracellular environment or because low free cytoplasmic Mg2+ levels compromise the function of cellular activities that recycle Pi from ATP (i.e., translation). (C) The cellular machinery senses a drop in free intracellular Pi, eliciting a Pi starvation response that is characterized, among other things, by the expression of high-affinity Pi transporters. See also Supplemental Figure S5.
Our results provide a physiological explanation for the finding that mutations conditionally expanding cytoplasmic pools of adenine nucleotides promote expression of PhoB-dependent genes even when Pi is abundant in the growth medium (Wilkins 1972); that is, when the adenine pool increases, cells experience low cytoplasmic Pi because Pi is trapped as ATP. Moreover, our results elucidate a reciprocal regulatory relationship between translation and Pi homeostasis: Pi acquisition allows the synthesis and operation of ribosomes, which in turn control Pi acquisition. Notably, the majority of Pi in living cells is present in ATP and rRNA (Bremer and Dennis 1996; Elser et al. 2003; Gillooly et al. 2005).
The intimate relationship between translation and the PhoB/PhoR system is further supported by the bacterial behavior elicited when protein synthesis is compromised due to an increase in the levels of the alarmone (p)ppGpp: Transcription of PhoB-dependent genes is inhibited even when environmental Pi is limited (Spira et al. 1995; Spira and Yagil 1998). Thus, just as an increase in ATP (Fig. 6A) drives expression of Pi acquisition genes (Fig. 6B), (p)ppGpp-mediated inhibition of rRNA transcription (Sarmientos et al. 1983) and adenine biosynthesis (Pao and Dyess 1981; Hou et al. 1999) alleviates the consumption of free cytoplasmic Pi pools, preventing PhoB/PhoR activation even when cells experience low environmental Pi.
Our findings indicate that the PhoB/PhoR system is activated by conditions promoting a decrease in free cytoplasmic Pi (Scholten and Tommassen 1993; Hoffer and Tommassen 2001) even in organisms experiencing high Pi in their surroundings (Fig. 1). Furthermore, they argue against the prevailing notion that the sensor PhoR responds to extracytoplasmic Pi (Hsieh and Wanner 2010). Moreover, they are in agreement with the proposal that induction of the PHO genes in S. cerevisiae is triggered by low cytoplasmic (as opposed to extracytoplasmic) Pi (Auesukaree et al. 2004).
The PhoU protein inhibits PhoB activation through an interaction with PhoR and the PstB high-affinity Pi transporter component (Fig. 1; Gardner et al. 2014). In E. coli, inactivation of phoU results in the hyperactivation of PhoB (Steed and Wanner 1993; Rice et al. 2009). Notably, mutations in phoU result in severe growth defects that can be suppressed by inactivation of either phoB or the PhoB-activated high-affinity Pi transporter encoded by the pstSCBA operon (Steed and Wanner 1993) or ameliorated by decreasing the Pi concentration in the growth medium (Rice et al. 2009). This indicates that excessive cytoplamic Pi inhibits E. coli growth. The cytosolic activation of the PhoB/PhoR system constitutes a simple mechanism to avoid the deleterious effects resulting from high cytoplasmic Pi that would inevitably arise from premature pstSCBA expression.
In prokaryotes, natural selection often favors close genomic linkage among genes participating in related or complementary cellular processes, presumably because acquisition of certain adjacent gene pairs by horizontal gene transfer is more beneficial than the acquisition of either gene alone (Lawrence 1997, 2002). In support of the physiological interplay between Pi and Mg2+, genomic analysis reveals that genes involved in the acquisition and/or metabolism of Pi are often found in close proximity to those responsible for the maintenance of free cytoplasmic Mg2+ (Supplemental Fig. S5). For instance, the mgtC gene of the gram-negative bacterium Polynucleobacter asymbioticus is adjacent to phoBR, phoU, and pstSCAB homologs; the corA Mg2+ transporter gene of the gram-positive Bacillus cereus is located near one specifying a phosphatase (Supplemental Fig. S5). Notably, close linkage between these classes of genes is also observed in archaeal species such as Halorhabdus utahensis, Natrialba magadii, and Haloarcula spp. (Supplemental Fig. S5).
Finally, cells typically rely on Mg2+ to neutralize the negative charge of Pi-containing molecules. These include the rRNA Pi backbone, which stabilizes the ribosome structure, enabling translation (Gesteland 1966; Klein et al. 2004; Pontes et al. 2016); the DNA Pi backbone, which stabilizes base pairing and base stacking in chromosomes (Shaw and Wang 1993); and ATP and other (deoxy)nucleotide triphosphates [(d)NTPs], enabling catalysis of most (d)NTP-dependent reactions, such as those carried out by DNA polymerase and RNAP (Kornberg and Gefter 1972; Zaychikov et al. 1996; Cowan 2002). Given that these fundamental cellular processes require Mg2+, we postulate that inhibition of Pi acquisition during cytoplasmic Mg2+ starvation is widespread in nature.
Materials and methods
Microbial strains, plasmids, and growth conditions
Microbial strains and plasmids used in this study are listed in Supplemental Table S1. All S. enterica serovar Typhimurium strains were derived from strain 14028s (Fields et al. 1986) and constructed by λ-red-mediated recombination (Datta et al. 2006) followed by phage P22-mediated transductions as described (Davis et al. 1980). E. coli MG1655 (Blattner et al. 1997) and S. cerevisiae DY1457 (Askwith et al. 1994) were used in physiological experiments. S. cerevisiae was grown in YPD medium supplemented with 5 mM MgSO4 at 250 rpm and 30°C; when necessary, cultures were supplemented with 250 µg/mL cycloheximide. Bacterial strains used in transformation or recombination were grown in LB medium at 30°C or 37°C (Datta et al. 2006; Khetrapal et al. 2015). When required, LB medium was supplemented with 100 µg/mL ampicillin, 20 µg/mL chloramphenicol, 50 µg/mL kanamycin, and/or 30 mM glucose or rhamnose.
In ChIP-seq experiments, S. enterica was grown in N-minimal medium (Véscovi et al. 1996) containing 10 or 50 µM MgCl2. Unless stated otherwise, physiological experiments with bacteria were carried out at 37°C with shaking at 250 rpm in MOPS medium (Neidhardt et al. 1974) lacking CaCl2 (to avoid repression of the PhoP/PhoQ system) (Véscovi et al. 1996) and supplemented with 0.1% (w/v) bacto casamino acids (BD Difco) and 30 mM glucose. Experiments were conducted as follows: After overnight (∼16- to 20-h) growth in MOPS containing high Mg2+ (10 mM MgCl2) and Pi (2 mM K2HPO4), cells washed twice in medium lacking Mg2+ and Pi and inoculated (1:100) in fresh medium containing 10 mM Mg2+ and 2 mM Pi, 10 mM Mg2+ and 250 µM Pi, or 10 µM Mg2+ and 500 µM Pi and propagated at 37°C with shaking at 250 rpm. (It should be noted that at a concentration of 0.1% [w/v] bacto casamino acids [BD Difco], the medium already contained ∼163 µM Pi.) During physiological experiments, selection of plasmids was accomplished by the addition of ampicillin at 35 µg/mL (overnight growth) or 15 µg/mL (experimental condition) and/or chloramphenicol at 20 µg/mL (overnight growth) or 8 µg/mL (experimental condition). Unless specified otherwise, heterologous expression of proteins was achieved by treatment of cultures with 100 µM (pMgtA), 250 µM (pMgtC), or 1 mM (pATPase) IPTG. In experiments involving pharmacological inhibition of translation, bacteria were treated with approximately three times their minimal inhibitory concentration: either 25 µg/mL chloramphenicol or 500 µg/mL spectinomycin.
Recombineering and qPCR primers
Generation of chromosomal mutations was carried out using PCR products generated with primers listed in Supplemental Table S2, as described (Datta et al. 2006). The counterselection method developed by Khetrapal et al. (2015) was used to construct strain MP1429.
Construction of plasmids
Phusion high-fidelity DNA polymerase (New England BioLabs) was used in reactions with primers listed in Supplemental Table S2. PCR products were assembled into pFPV25 or pFPV25AAV (digested with restriction enzymes BamHI and EcoRI), pUHE-21-2-lacIq (digested with BamHI and HinDIII), or pACYC184 (digested with SalI and HinDIII) using NEBuilder HiFi DNA assembly cloning kit (New England BioLabs). The integrity of constructs was verified by DNA sequencing and/or functional assays.
RNAP ChIP-seq
ChIP assays were performed as described (Shin and Groisman 2005) with the modifications described in the Supplemental Material.
RNA extraction, cDNA synthesis, and qPCR
For all experiments involving bacteria, RNA extraction was carried out using RNeasy (Qiagen) according to the manufacturer's instructions. For S. cerevisiae, RNA extraction was carried as described (Aiba et al. 1981) with the minor modifications described in the Supplemental Material. cDNA was synthesized from RNA samples using the SuperScript VILO cDNA synthesis lit (Thermo Fisher Scientific) according to the manufacturer's instructions. Relative amounts of cDNA were determined using standard curves obtained from qPCR with serially diluted wild-type E. coli, Salmonella, or S. cerevisiae genomic DNA. Quantification of cDNA was performed by real-time PCR using Fast SYBR Green master mix (Thermo Fisher Scientific) in an ABI 7500 sequence detection system (Thermo Fisher Scientific).
Estimation of intracellular ATP
Intracellular ATP was estimated as described (Pontes et al. 2015a) with the minor modifications described in the Supplemental Material.
Estimation of intracellular Pi
Total Pi in the samples was estimated from crude cell extracts using the molybdenum blue method as described (Kanno et al. 2016). Total Pi levels in samples were normalized by the amount of protein in the samples. Detailed sample extraction and Pi and protein measurements are described in the Supplemental Material.
Monitoring gene expression via fluorescence
Fluorescence derived from reporter constructs was measured into clear-bottomed 96-well black plates (Corning) using a Synergy H1 plate reader (BioTek). Green fluorescence was measured with excitation 485 nm and emission 535 nm, and the absorbance in each well was measured at 600 nm. Detailed sample extraction and Pi and protein measurements are described in the Supplemental Material.
Phos-tag SDS-PAGE and Western blotting
Phos-tag SDS-PAGE and Western blotting were carried out as described (Wayne et al. 2012) with the modifications described in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank Dr. Colin MacDiarmid (University of Wisconsin-Madison) for kindly providing S. cerevisiae strain DY1457, and Dr. Andrew Goodman, Dr. Patricia Sanchez-Vazquez, and Dr. Ann M. Stock for critical reading of this manuscript. We also thank the reviewers of this submission for their comments. This research was supported by National Institutes of Health grant AI49561 to E.A.G.
Author contributions: M.H.P. and E.A.G. conceived the study and designed the research, M.H.P. performed the research, and M.H.P. and E.A.G. analyzed the data and wrote the paper.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.309245.117.
References
- Aiba H, Adhya S, de Crombrugghe B. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem 256: 11905–11910. [PubMed] [Google Scholar]
- Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol 64: 2240–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askwith C, Eide D, Van Ho A, Bernard PS, Li L, Davis-Kaplan S, Sipe DM, Kaplan J. 1994. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76: 403–410. [DOI] [PubMed] [Google Scholar]
- Auesukaree C, Homma T, Kaneko Y, Harashima S. 2003. Transcriptional regulation of phosphate-responsive genes in low-affinity phosphate-transporter-defective mutants in Saccharomyces cerevisiae. Biochem Biophys Res Commun 306: 843–850. [DOI] [PubMed] [Google Scholar]
- Auesukaree C, Homma T, Tochio H, Shirakawa M, Kaneko Y, Harashima S. 2004. Intracellular phosphate serves as a signal for the regulation of the PHO pathway in Saccharomyces cerevisiae. J Biol Chem 279: 17289–17294. [DOI] [PubMed] [Google Scholar]
- Bagnara AS, Finch LR. 1973. Relationships between intracellular contents of nucleotides and 5-phosphoribosyl 1-pyrophosphate in Escherichia coli. Eur J Biochem 36: 422–427. [DOI] [PubMed] [Google Scholar]
- Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. 2000. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J 19: 5071–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc-Potard AB, Groisman EA. 1997. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J 16: 5376–5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462. [DOI] [PubMed] [Google Scholar]
- Bremer H, Dennis PP. 1996. Modulation of chemical composition and other parameters of the cell by growth rate. In Escherichia coli and Salmonella. (ed. Neidhardt FC), pp. 1553–1569. ASM Press, Washington, DC. [Google Scholar]
- Bun-Ya M, Nishimura M, Harashima S, Oshima Y. 1991. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol 11: 3229–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Samson L. 1991. Induction of S. cerevisiae MAG 3-methyladenine DNA glycosylase transcript levels in response to DNA damage. Nucleic Acids Res 19: 6427–6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan JA. 2002. Structural and catalytic chemistry of magnesium-dependent enzymes. Biometals 15: 225–235. [DOI] [PubMed] [Google Scholar]
- Cox GB, Rosenberg H, Downie JA, Silver S. 1981. Genetic analysis of mutants affected in the Pst inorganic phosphate transport system. J Bacteriol 148: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie MJ, Shi Y, Latifi T, Groisman EA. 2006. An RNA sensor for intracellular Mg2+. Cell 125: 71–84. [DOI] [PubMed] [Google Scholar]
- Datta S, Costantino N, Court DL. 2006. A set of recombineering plasmids for gram-negative bacteria. Gene 379: 109–115. [DOI] [PubMed] [Google Scholar]
- Davis RW, Botstein D, Roth JR. 1980. A manual for genetic engineering advanced bacterial genetics. Cold Springer Harbor, New York. [Google Scholar]
- Elser JJ, Nagy JD, Kuang Y. 2003. Biological stoichiometry: an ecological perspective on tumor dynamics. Bioscience 53: 1112–1120. [Google Scholar]
- Fields PI, Swanson RV, Haidaris CG, Heffron F. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci 83: 5189–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal T, Bartlett MS, Ross W, Turnbough CL Jr, Gourse RL. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278: 2092–2097. [DOI] [PubMed] [Google Scholar]
- Gao R, Stock AM. 2013a. Evolutionary tuning of protein expression levels of a positively autoregulated two-component system. PLoS Genet 9: e1003927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Stock AM. 2013b. Probing kinase and phosphatase activities of two-component systems in vivo with concentration-dependent phosphorylation profiling. Proc Natl Acad Sci 110: 672–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Stock AM. 2015. Temporal hierarchy of gene expression mediated by transcription factor binding affinity and activation dynamics. MBio 6: e00686-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner SG, Johns KD, Tanner R, McCleary WR. 2014. The PhoU protein from Escherichia coli interacts with PhoR, PstB, and metals to form a phosphate-signaling complex at the membrane. J Bacteriol 196: 1741–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland RF. 1966. Unfolding of Escherichia coli ribosomes by removal of magnesium. J Mol Biol 18: 356–371. [DOI] [PubMed] [Google Scholar]
- Gillooly JF, Allen AP, Brown JH, Elser JJ, Martinez del Rio C, Savage VM, West GB, Woodruff WH, Woods HA. 2005. The metabolic basis of whole-organism RNA and phosphorus content. Proc Natl Acad Sci 102: 11923–11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA, Hollands K, Kriner MA, Lee EJ, Park SY, Pontes MH. 2013. Bacterial Mg2+ homeostasis, transport, and virulence. Annu Rev Genet 47: 625–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP–phoQ. Science 276: 250–253. [DOI] [PubMed] [Google Scholar]
- Hoffer SM, Tommassen J. 2001. The phosphate-binding protein of Escherichia coli is not essential for P(i)-regulated expression of the pho regulon. J Bacteriol 183: 5768–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T, Horiuchi S, Mizuno D. 1959. A possible negative feedback phenomenon controlling formation of alkaline phosphomonoesterase in Escherichia coli. Nature 183: 1529–1530. [DOI] [PubMed] [Google Scholar]
- Hou Z, Cashel M, Fromm HJ, Honzatko RB. 1999. Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase. J Biol Chem 274: 17505–17510. [DOI] [PubMed] [Google Scholar]
- Hsieh YJ, Wanner BL. 2010. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol 13: 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno S, Cuyas L, Javot H, Bligny R, Gout E, Dartevelle T, Hanchi M, Nakanishi TM, Thibaud MC, Nussaume L. 2016. Performance and limitations of phosphate quantification: guidelines for plant biologists. Plant Cell Physiol 57: 690–706. [DOI] [PubMed] [Google Scholar]
- Kato A, Tanabe H, Utsumi R. 1999. Molecular characterization of the PhoP–PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J Bacteriol 181: 5516–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetrapal V, Mehershahi K, Rafee S, Chen S, Lim CL, Chen SL. 2015. A set of powerful negative selection systems for unmodified Enterobacteriaceae. Nucleic Acids Res 43: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA. 2004. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 10: 1366–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg T, Gefter ML. 1972. Deoxyribonucleic acid synthesis in cell-free extracts. IV. Purification and catalytic properties of deoxyribonucleic acid polymerase III. J Biol Chem 247: 5369–5375. [PubMed] [Google Scholar]
- Kroger A, Fuchs G. 1999. Basic prerequisites for cellular life: growth and nutrition. In Biology of the prokaryotes (ed. Lengeler JW, et al. ), pp. 88–109. Georg Thieme Verlag, Stuttgart, Germany. [Google Scholar]
- Lawrence JG. 1997. Selfish operons and speciation by gene transfer. Trends Microbiol 5: 355–359. [DOI] [PubMed] [Google Scholar]
- Lawrence JG. 2002. Shared strategies in gene organization among prokaryotes and eukaryotes. Cell 110: 407–413. [DOI] [PubMed] [Google Scholar]
- Lee EJ, Groisman EA. 2012a. Control of a Salmonella virulence locus by an ATP-sensing leader messenger RNA. Nature 486: 271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Groisman EA. 2012b. Tandem attenuators control expression of the Salmonella mgtCBR virulence operon. Mol Microbiol 86: 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Pontes MH, Groisman EA. 2013. A bacterial virulence protein promotes pathogenicity by inhibiting the bacterium's own F1Fo ATP synthase. Cell 154: 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire ME, Cowan JA. 2002. Magnesium chemistry and biochemistry. Biometals 15: 203–210. [DOI] [PubMed] [Google Scholar]
- Makino K, Shinagawa H, Amemura M, Kimura S, Nakata A, Ishihama A. 1988. Regulation of the phosphate regulon of Escherichia coli. Activation of pstS transcription by PhoB protein in vitro. J Mol Biol 203: 85–95. [DOI] [PubMed] [Google Scholar]
- Makino K, Shinagawa H, Amemura M, Kawamoto T, Yamada M, Nakata A. 1989. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J Mol Biol 210: 551–559. [DOI] [PubMed] [Google Scholar]
- Murray HD, Schneider DA, Gourse RL. 2003. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell 12: 125–134. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Gourse R, Baughman G. 1984. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem 53: 75–117. [DOI] [PubMed] [Google Scholar]
- Pao CC, Dyess BT. 1981. Effect of unusual guanosine nucleotides on the activities of some Escherichia coli cellular enzymes. Biochim Biophys Acta 677: 358–362. [DOI] [PubMed] [Google Scholar]
- Park SY, Cromie MJ, Lee EJ, Groisman EA. 2010. A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell 142: 737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes MH, Lee EJ, Choi J, Groisman EA. 2015a. Salmonella promotes virulence by repressing cellulose production. Proc Natl Acad Sci 112: 5183–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes MH, Sevostyanova A, Groisman EA. 2015b. When too much ATP is bad for protein synthesis. J Mol Biol 427: 2586–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes MH, Yeom J, Groisman EA. 2016. Reducing ribosome biosynthesis promotes translation during low Mg2+ stress. Mol Cell 64: 480–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice CD, Pollard JE, Lewis ZT, McCleary WR. 2009. Employment of a promoter-swapping technique shows that PhoU modulates the activity of the PstSCAB2 ABC transporter in Escherichia coli. Appl Environ Microbiol 75: 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg H, Gerdes RG, Chegwidden K. 1977. Two systems for the uptake of phosphate in Escherichia coli. J Bacteriol 131: 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmientos P, Sylvester JE, Contente S, Cashel M. 1983. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell 32: 1337–1346. [DOI] [PubMed] [Google Scholar]
- Schneider DA, Gaal T, Gourse RL. 2002. NTP-sensing by rRNA promoters in Escherichia coli is direct. Proc Natl Acad Sci 99: 8602–8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholten M, Tommassen J. 1993. Topology of the PhoR protein of Escherichia coli and functional analysis of internal deletion mutants. Mol Microbiol 8: 269–275. [DOI] [PubMed] [Google Scholar]
- Shaw SY, Wang JC. 1993. Knotting of a DNA chain during ring closure. Science 260: 533–536. [DOI] [PubMed] [Google Scholar]
- Shi Y, Cromie MJ, Hsu FF, Turk J, Groisman EA. 2004. PhoP-regulated Salmonella resistance to the antimicrobial peptides magainin 2 and polymyxin B. Mol Microbiol 53: 229–241. [DOI] [PubMed] [Google Scholar]
- Shin D, Groisman EA. 2005. Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in vivo. J Biol Chem 280: 4089–4094. [DOI] [PubMed] [Google Scholar]
- Shinagawa H, Makino K, Nakata A. 1983. Regulation of the pho regulon in Escherichia coli K-12. Genetic and physiological regulation of the positive regulatory gene phoB. J Mol Biol 168: 477–488. [DOI] [PubMed] [Google Scholar]
- Snavely MD, Florer JB, Miller CG, Maguire ME. 1989a. Magnesium transport in Salmonella typhimurium: expression of cloned genes for three distinct Mg2+ transport systems. J Bacteriol 171: 4752–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snavely MD, Florer JB, Miller CG, Maguire ME. 1989b. Magnesium transport in Salmonella typhimurium: 28Mg2+ transport by the CorA, MgtA, and MgtB systems. J Bacteriol 171: 4761–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snavely MD, Miller CG, Maguire ME. 1991. The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J Biol Chem 266: 815–823. [PubMed] [Google Scholar]
- Soncini FC, Véscovi EG, Groisman EA. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J Bacteriol 177: 4364–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncini FC, Véscovi EG, Solomon F, Groisman EA. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol 178: 5092–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli SV, Pontel LB, Véscovi EG, Soncini FC. 2008. Regulation of magnesium homeostasis in Salmonella: Mg2+ targets the mgtA transcript for degradation by RNase E. FEMS Microbiol Lett 280: 226–234. [DOI] [PubMed] [Google Scholar]
- Spira B, Yagil E. 1998. The relation between ppGpp and the pho regulon in Escherichia coli. Mol Gen Genet 257: 469–477. [DOI] [PubMed] [Google Scholar]
- Spira B, Silberstein N, Yagil E. 1995. Guanosine 3′,5′-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J Bacteriol 177: 4053–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed PM, Wanner BL. 1993. Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in the phosphate regulon. J Bacteriol 175: 6797–6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storer AC, Cornish-Bowden A. 1976. Concentration of MgATP2− and other ions in solution: calculation of the true concentrations of species present in mixtures of associating ions. Biochem J 159: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouthamer AH. 1973. A theoretical study on the amount of ATP required for synthesis of microbial cell material. Antonie Van Leeuwenhoek 39: 545–565. [DOI] [PubMed] [Google Scholar]
- Tao T, Snavely MD, Farr SG, Maguire ME. 1995. Magnesium transport in Salmonella typhimurium: mgtA encodes a P-type ATPase and is regulated by Mg2+ in a manner similar to that of the mgtB P-type ATPase. J Bacteriol 177: 2654–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Véscovi EG, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84: 165–174. [DOI] [PubMed] [Google Scholar]
- Wanner BL. 1996. Phosphorus assimilation and control of the phosphate regulon. In Escherichia coli and Salmonella. (ed. Neidhardt FC), pp. 1357–1381. ASM Press, Washington, DC. [Google Scholar]
- Wayne KJ, Li S, Kazmierczak KM, Tsui HC, Winkler ME. 2012. Involvement of WalK (VicK) phosphatase activity in setting WalR (VicR) response regulator phosphorylation level and limiting cross-talk in Streptococcus pneumoniae D39 cells. Mol Microbiol 86: 645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins AS. 1972. Physiological factors in the regulation of alkaline phosphatase synthesis in Escherichia coli. J Bacteriol 110: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsky GR, Malamy MH. 1980. Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J Bacteriol 144: 356–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FI, Cittadini A. 2003. Chemistry and biochemistry of magnesium. Mol Aspects Med 24: 3–9. [DOI] [PubMed] [Google Scholar]
- Yamada M, Makino K, Shinagawa H, Nakata A. 1990. Regulation of the phosphate regulon of Escherichia coli: properties of phoR deletion mutants and subcellular localization of PhoR protein. Mol Gen Genet 220: 366–372. [DOI] [PubMed] [Google Scholar]
- Zaychikov E, Martin E, Denissova L, Kozlov M, Markovtsov V, Kashlev M, Heumann H, Nikiforov V, Goldfarb A, Mustaev A. 1996. Mapping of catalytic residues in the RNA polymerase active center. Science 273: 107–109. [DOI] [PubMed] [Google Scholar]
- Zengel JM, Lindahl L. 1994. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog Nucleic Acid Res 47: 331–370. [DOI] [PubMed] [Google Scholar]
- Zhao G, Kong W, Weatherspoon-Griffin N, Clark-Curtiss J, Shi Y. 2011. Mg2+ facilitates leader peptide translation to induce riboswitch-mediated transcription termination. EMBO J 30: 1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwir I, Latifi T, Perez JC, Huang H, Groisman EA. 2012. The promoter architectural landscape of the Salmonella PhoP regulon. Mol Microbiol 84: 463–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.