Abstract
A small subset of HIV-1 infected individuals, the “Elite Controllers” (EC), can control viral replication and restrain progression to immunodeficiency without antiretroviral therapy (ART). In this study, a cross-sectional transcriptomics and targeted proteomics analysis were performed in a well-defined Swedish cohort of untreated EC (n = 19), treatment naïve patients with viremia (VP, n = 32) and HIV-1-negative healthy controls (HC, n = 23). The blood transcriptome identified 151 protein-coding genes that were differentially expressed (DE) in VP compared to EC. Genes like CXCR6 and SIGLEC1 were downregulated in EC compared to VP. A definite distinction in gene expression between males and females among all patient-groups were observed. The gene expression profile between female EC and the healthy females was similar but did differ between male EC and healthy males. At targeted proteomics analysis, 90% (29/32) of VPs clustered together while EC and HC clustered separately from VP. Among the soluble factors, 33 were distinctive to be statistically significant (False discovery rate = 0.02). Cell surface receptor signaling pathway, programmed cell death, response to cytokine and cytokine-mediated signaling seem to synergistically play an essential role in HIV-1 control in EC.
Keywords: HIV-1 Elite Controllers, Transcriptome, Proteome
Graphical Abstract
Highlights
-
•
The AIDS restriction genes do not have any role in immune control mechanism in EC.
-
•
Strong distinction in gene expression between males and females among all patient groups studied.
-
•
Multiple pathways play a synergistic role in controlling the viral replication control in EC.
-
•
Pathways involving TNFSF/TNFRSF and the immune checkpoint are of importance for immune control in HIV-infected patients.
A group of HIV-1 infected individuals termed Elite Controllers (EC) controls viral replication naturally without antiretroviral therapy. In this study, we used an explorative molecular data-first approach using high-throughput transcriptomics and targeted proteomics analysis to understand mechanisms of viral replication control in EC. We observed that EC are more similar to HIV-1 negative healthy controls but a robust gender-specific differentiation was seen in blood transcriptomics profile. Further, combining the gene expression and plasma proteomics profile analysis clearly implicated that several immunological pathways play an important role in HIV-1 control in EC, synergistically through interconnecting molecules.
1. Introduction
A small subset of human immunodeficiency virus type 1 (HIV-1) infected individuals, the “Elite Controllers” (EC), can control the virus and restrain progression to immunodeficiency without antiretroviral therapy (ART). It has been suggested that EC can hold the key to how a functional HIV-cure can be reached. Strict criteria have been used for defining EC and only < 0.5% of the HIV-infected population fulfill such criteria, e.g., known HIV-1 positivity for ≥ 10 year with none or very few episodes of viremia and stable CD4+ T cell counts (Olson et al., 2014). However, we hypothesize that EC still is a heterogeneous group as a result of that several definitions exists (Olson et al., 2014) which is illustrated by the claim that EC also can develop acquired immunodeficiency syndrome (Deeks and Walker, 2007).Therefore, we suggest that an even more strict definition of EC should be applied to minimize the bias of false phenotypic classification (Zhang et al., 2017).
Genome-wide association studies (GWAS) have tried to identify the background for HIV control. Multiple independent polymorphisms have been defined within the HLA and CCR5-CCR2 locus that together explains ~ 25% of the observed variability in viral load (McLaren et al., 2012, Pereyra et al., 2010, McLaren et al., 2015). Apart from the HLAs, the AIDS restriction genes (ARGs) like CCR5-Δ32 (rs333), CCR5 59029G (rs1799987), and SDF1-3′A (rs1801157) that have been reported to have a strong association with disease control (Poropatich and Sullivan Jr., 2011, O'Brien and Nelson, 2004). Though understanding about HIV-1 disease progression was generated, the studies were not designed to assess the impact of the full spectrum of functional variants within coding regions. Recent research has shown that several GWAS hits have no specific biological relevance to disease (Boyle et al., 2017). Most of the earlier studies trying to elicit the mechanism of disease progression and control in HIV have focused on specific predefined molecules or pathways, thereby ignoring the systemic, interconnected, immunological programs that are associated with individual immune defense mechanisms.
Gene expression data from EC are limited, mainly derived from the CD4+ T cells. Using low-throughput TaqMan® Low-Density Arrays (TILDA) assay, elevated levels of Schlafen-11 were identified as a signature in T cells of the EC, as one of only 34 genes tested (Abdel-Mohsen et al., 2013). Another study using high throughput RNA sequencing on CD4+ T cells from EC did not reveal an entirely distinctive mRNA expression pattern (Rotger et al., 2010). Further transcriptome analysis of EC in a third recent study reported that seven genes were differentially expressed in EC compared to healthy controls. Notably, the gene encoding for macrophage inflammatory protein 1α (MIP-1α), a natural ligand for CCR5, was found to be upregulated (Walker et al., 2015). However, as the immune system is a complex, adaptable system and specific immune cells are dependent upon each other for stimulations and inhibition (Kaczorowski et al., 2017), analysis of single cell population may not reveal the complete picture.
Therefore, to understand the underlying mechanisms, which ensure that disease progression is prevented in EC, a comprehensive analysis of clinical phenotypes coupled to genetics and biomolecular mechanisms is required. The rapidly increasing accessibility of genetic and biomolecular expression data from new high-throughput technologies is the foundation to shift the traditional phenotype-first approach to explorative genetic or molecular data-first approaches. In this study, we aimed to explore a comprehensive analysis of host transcriptomics and proteomics data coupled to clinical phenotypes in a well-defined Swedish EC cohort with up to 20 years of clinical follow-up data.
2. Materials and Methods
2.1. Patients
Whole peripheral blood was obtained between 2014 and 2016 from three categories of individuals; an unbiased cohort of untreated HIV-1-infected EC (n = 19), treatment naïve patients with viremia (VP, n = 8), and HIV-1-negative persons (HC, n = 14). Viral load was below detection limit (< 20 copies/mL) in all EC at the time of sample collection. Peripheral blood mononuclear cells (PBMCs) were isolated using density gradient centrifugation. For the proteomics profiling, additional plasma samples were obtained from therapy naïve patients with viremia (n = 24) and HIV-1-negative persons (n = 9) from the InfCareHIV cohort at the Karolinska University Hospital, Huddinge collected between 2010 and 2016. The EC was defined as known HIV-positivity more than a year and ≥ 3 consecutive viral load < 75 copies/ml over one year (and all previous VLs < 1000 cp/ml) and known HIV-1 positivity ≥ 10 years and minimum two VL-measurements (≥ 90% of all VLs < 400 copies/ml). Group characteristics of the patient-categories are presented in Table 1. The individual characteristics of ECs have been given elsewhere (Zhang et al., 2017).
Table 1.
Cohort characteristics.
| Elite controllers | Viremic progressors | Healthy controls | p-Value | |
|---|---|---|---|---|
| n | 19 | 32 | 23 | |
| Age in years, median (IQR) | 46 (40–52) | 39 (32–52) | 45 (39–52) | 0.36a |
| Male, n (%) | 10 (53) | 15 (47) | 13 (57) | 0.77a |
| Ethnicity, n (%) | ||||
| Caucasian | 8 (42) | 20 (63) | 18 (78) | 0.19 |
| Black | 10 (53) | 11 (34) | 4 (17) | |
| Other | 1 (5) | 1 (3) | 1 (4) | |
| Mode of transmission, n (%) | ||||
| Heterosexual | 10 (53) | 23 (72) | NA | 0.11 |
| MSM | 4 (22) | 8 (25) | ||
| PWID | 2 (10) | 0 | ||
| Blood product | 2 (10) | 0 | ||
| Unknown | 1 (5) | 1 (3) | ||
| Time since diagnosis, years- median (IQR) | 9.0 (5.0–14.0) | 1.3 (0.1–3.6) | NA | < 0.001b |
| CD4 count, cells/mm3, median (IQR) | 950 (695–1160) | 410 (302–527) | NA | < 0.001b |
| CD8 count, cells/mm3, median (IQR) | 670 (570–900) | 975 (735–1215) | NA | 0.005b |
| CD4/CD8 ratio | 1.52 (0.82–1.74) | 0.46 (0.29–0.57) | NA | < 0.001b |
| Viral load, Log10 copies/mL, median (IQR) | < 1.27 | 4.60 (3.85–5.03) | NA | < 0.001b |
| HIV-1 subtype, n (%) | ||||
| A1 | 0 | 6 (19) | NA | 0.001 |
| B | 3 (16) | 11 (34) | ||
| C | 5 (26) | 5 (16) | ||
| Recombinant forms | 2 (11) | 10 (31) | ||
| Unknown | 9 (47) | 0 |
NA: Not available, IQR: Interquartile range, MSM: Men who sex with men, PWID: People who inject drugs.
The Kruskal–Wallis test.
Mann-Whitney U test.
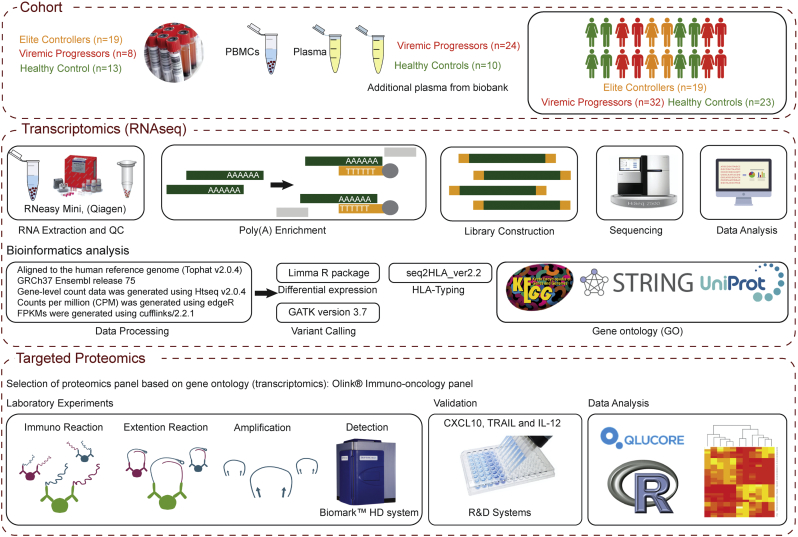
The study was approved by regional ethics committees of Stockholm (2013/1944–31/4). All participants gave informed consent. The patient identity was anonymized and delinked before analysis. The complete study design is illustrated in Fig. 1.
Fig. 1.
Flow diagram of study designing and analysis plan. Cross-sectional blood samples from HIV-1-infected EC (n = 19), treatment naïve patients with viremia (VP, n = 8), and HIV-1-negative persons (HC, n = 13) were obtained for transcriptomics study. Additional plasma samples were obtained from therapy naïve patients with viremia (n = 24) and HIV-1-negative persons (n = 10) for the proteomics profiling. The data analysis plan for transcriptomics and proteomics study is shown. The figure for the PEA was kindly provided by Olink AB, Sweden.
2.2. RNA Sequencing (RNA-Seq)
Total RNA was extracted from PBMCs using RNeasy Mini (Qiagen, Hilden, Germany) according to the manufacturer's protocol and the RNA quality was determined by Agilent RNA 6000 p kit on an Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). The mean and SD RIN value was 8.85 (0.94). The library preparation, and RNA-Seq, were performed at the National Genomics Infrastructure, Science for Life Laboratory, Stockholm, Sweden. The libraries were prepared from 100 ng of total RNA samples using Illumina TruSeq® Stranded mRNA Library Prep Kit with poly-A selection following manufacturer's guidelines. The clonal cluster amplification was carried out with Illumina cBOT System. The pooled libraries were sequenced on HiSeq2500 (HiSeq Control Software 2.2.58/RTA 1.18.64) with a 2 × 126 setup using ‘HiSeq SBS Kit v4’ chemistry. The Base call (Bcl) files to FastQ conversion was performed using bcl2fastq from the Illumina's Consensus Assessment of Sequence and Variation (CASAVA) software suite (Illumina, US).
2.3. Data Processing
The quality control of raw sequencing reads was performed with FastQC, (www.bioinformatics.babraham.ac.uk/projects/fastqc/). The reads were then aligned to the human reference genome GRCh37 Ensembl release 75 using Tophat v2.0.4. Gene-level count data was generated using Htseq v2.0.4 to summarize read counts for each gene. Counts per million (CPM) for protein-coding genes and non-protein coding transcripts were generated using edgeR (Robinson et al., 2010), and the CPM were log transformed to reduce the sequencing depth differences among the libraries. Transcripts with CPM value < 0.5 in the whole group of minimal sample size were discarded to remove transcripts that were not expressed at a biologically meaningful level in any condition. Fragments per kilobase of transcript per Million mapped reads (FPKMs) were generated using cufflinks/2.2.1 (http://cufflinks.cbcb.umd.edu/).
2.4. Clustering Analysis
To analyze the similarities and dissimilarities between the samples, unsupervised principal component analysis and clustering analysis were performed using PlotMDS function in the Limma (linear models for microarray data) R package (Ritchie et al., 2015) to generate Multi-Dimensional Scaling plot (MDS). Heatmap of hierarchical clustering of the samples was generated by calculating the matrix of Euclidean distances from the logCPM to examine the relationships between samples.
2.5. Differential Expression (DE)
The data were normalized to eliminate composition biases between libraries. The calcNormFactors function in edgeR, which uses Trimmed Mean of M-values (TMM) method, was executed for the normalization (Robinson and Oshlack, 2010). The normalized CPM was transformed to logCPMs using voom function in Limma package while taking into account the mean-variance relationship in the data (Law et al., 2014). The voom transformation data was used for differential expression (DE) analysis using Limma R package. Transcripts with adjusted P-value (FDR) < 0.05 were selected as significantly differentially expressed. Transcripts with a log2-fold-change value greater than two were considered as upregulated and those with less than minus two as downregulated.
2.6. Variant Calling and HLA Typing
Variant calling was performed using the uniquely mapped duplicate-removed data to obtain unbiased variant frequency calls using Genome Analysis Tool Kit (GATK version 3.7). The duplicates are defined as the paired-end reads where both mates map to the same genomic positions. First, the local realignment was performed around insertion-deletions (indels) to correct mapping-related artifacts to minimize the chances of false positive calling to obtain the bam files. Corrected bam files were further used for Base Quality Score Recalibration (BQSR) using BaseRecalibrator module, which resulted in more accurate base qualities, to improve the accuracy of the variant calls. The report on callable loci was considered for the variant discovery. Finally, variant discovery was performed by UnifiedGenotyper module. The module used Bayesian genotype likelihood model to call the variants. HLA-typing was performed using seq2HLA_ver2.2 (Boegel et al., 2015).
2.7. Gene Ontology (GO) Annotation
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (http://string.embl.de/) was used to identify known and predicted interactions among the differential expressed genes and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Gene ontology (GO) term biological process was only considered with FDR < 0.05. Further gene functions were cross-checked with Uniprot (Pundir et al., 2017) by using the in-house script.
2.8. Targeted Proteomic Profiling of the Plasma Soluble Factors
Plasma samples were analyzed using the proximity extension assay (PEA). Based on the GO annotation of the transcriptomics data we have selected the Olink® Immuno-oncology panel (Olink Bioscience AB, Uppsala, Sweden) (Assarsson et al., 2014). This panel includes 92 proteins. The protein analysis is reported as normalized protein expression levels (NPX), which are Ct values normalized by the subtraction of values for extension control, as well as an inter-plate control. The scale is shifted using a correction factor (normal background noise) and reported in Log2 scale (Berggrund et al., 2016). We further validated the co-relation between protein concentrations in log2 picogram/mL with NPX, using ELISA targeted for three soluble factors CXCL10, TRAIL and IL-12 (R&D Systems, US) (Supplementary Fig. S1).
2.9. Statistical Analysis
Demographics and the characteristics of patients were compiled with descriptive statistical methods. We used the χ2 test to compare groups' categorical data. The Mann-Whitney U test (between two groups) and the Kruskal–Wallis test (for more than two groups) were used to compare continuous data. The protein levels in the proteomics study were compared using the Mann-Whitney U test between two groups. P values < 0.05 and < 0.001 were shown and considered significant.
2.10. Data Visualization
Venn diagram was created using InteractiVenn (Heberle et al., 2015). The differential profile (heatmap) of the soluble factors (proteome) was analyzed using Qlucore Omics Explorer version 3.2. CIRCOS plot was used to visualize the circular plot of up and downregulated genes, SNPs and HLA typing (Krzywinski et al., 2009).
2.11. Data Availability
RNAseq raw sequencing data along with sample level metadata have been deposited in the NCBI/SRA Web service. Data can be accessed using BioProject accession number PRJNA420459.
3. Results
3.1. Class I HLA Typing and AIDS Restriction Genes
We first characterized EC concerning class I HLA typing and variant calling in the AIDS restriction genes (ARGs) that have been reported to have a strong association with disease control (Poropatich and Sullivan Jr., 2011, McLaren et al., 2012, Pereyra et al., 2010). Several protective HLA I alleles were identified in EC (Supplementary Fig. S2). Within the EC, 63% (12/19) had at least one protective allele (A*25:01, A*74:01, B*14:02, B*27:05, B*42:01, B*51:01, B*57:01, B*57:03, B*58:01, and B*81:01), of whom four had two alleles. However, some EC also had disease-susceptible HLA alleles (B*07:01, B*08:0, B*35:03, and B*53:01). The single nucleotide polymorphism (SNP) in the HLA C region rs9264608, which has been shown to have an independent protective effect, was observed in 21% (4/19) of EC and 12% (1/8) of VP, but not in HC.
While analyzing ARGs, CCR5-Δ32 (rs333), CCR5 59029G (rs1799987), and SDF1-3′A (rs1801157) polymorphisms were not identified in any of the samples. However, HCP5 rs2395029, an SNP in the non-protein coding region, was observed in three of the EC. The Caucasian who was positive for the HCP5 rs2395029 was also positive for HLA B*57:01.
3.2. Differential Expression (DE) Analysis
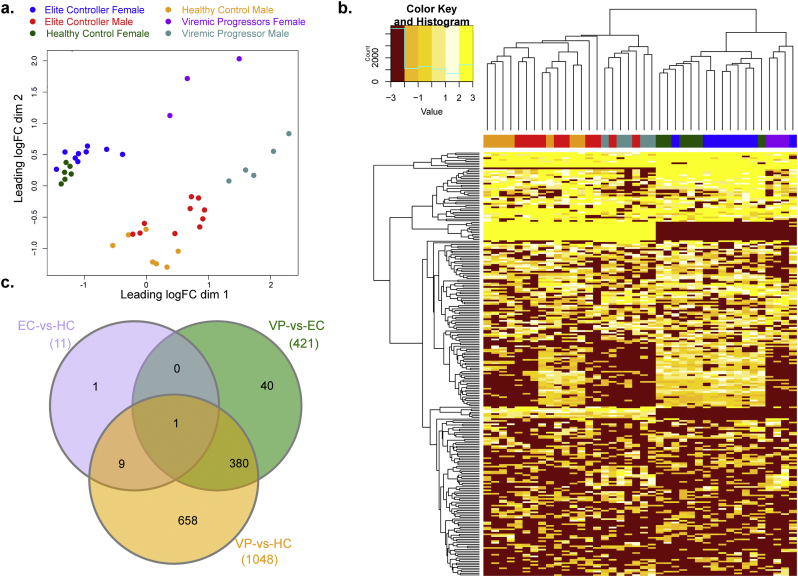
DE analysis was carried out to study the transcript (protein-coding and non-protein coding) expression profile of PBMCs from the HIV-1 infected patients compared to the HC. Bias in the gender of the patients was observed in the unsupervised principal component analysis (Fig. 2a). The heatmap of the top 500 significant differentially expressed transcripts showed similar clustering of the samples (Fig. 2b). DE analysis was performed in two ways; one with gender differentiation and another one without. DE without gender differentiation gave rise to 11 differentially expressed transcripts in EC compared to HC, 421 in VP compared to EC, and 1048 in VP compared to HC (Fig. 2c). Out of the 11 differentially expressed transcripts in EC compared to HC, eight were protein-coding genes. Similarly, there were 270 protein-coding genes among the 421 differentially expressed transcripts in VP compared to EC, and 688 protein-coding genes among the 1048 transcripts in VP compared to HC. The corresponding numbers of non-protein coding transcripts were thus three in EC compared to HC, 151 in VP compared to EC, and 360 in VP compared to HC. This data indicates that EC is more similar to HC than to VP regarding the host gene expression profile. Among the unique transcripts (n = 40) (Fig. 2c) in the Venn diagram, 22 were protein-coding genes that were differentially expressed between EC and VP only. Among them, CXCR6 and SIGLEC1 were downregulated in EC. The differentially expressed transcripts between EC and VP and the Venn annotation are given in Supplementary data 1.
Fig. 2.
Differential expression (DE) of the transcript (protein-coding and non-protein coding) expression profile of cells from HIV-1 infected patients compared to healthy individuals' cells. (a) The unsupervised principal component analysis identified clustering of different diseases states in a gender-specific manner. The color codes for each group were mentioned above the plot. (b) Unsupervised hierarchical clustering analysis is showing the expression signature of the top 500 significant differentially expressed transcripts. Log2 fold change (− 3 to 3) values are represented in six colors shade. (c) Venn diagram of DE transcripts. The sum of the numbers in each large circle represents the total number of differentially expressed genes among various combinations (EC vs HC, VP vs EC and VP vs HC). The overlapping part of the circles represents common DEGs between combinations.
3.3. Gender-specific DE Analysis
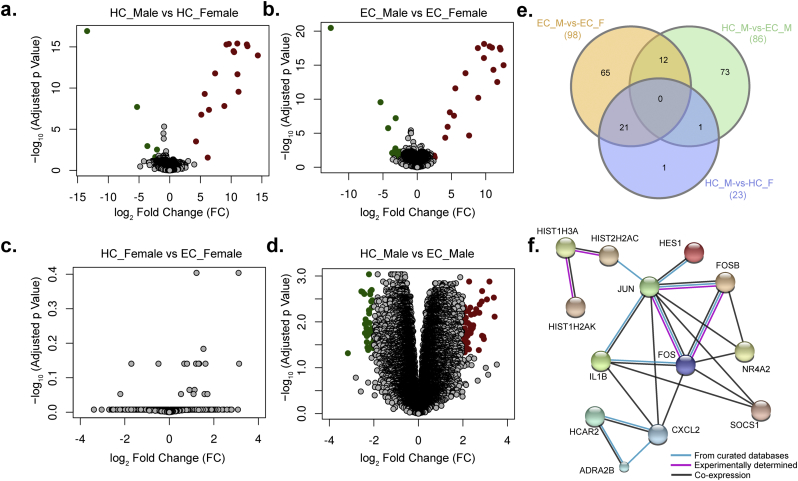
A separate DE analysis was performed in response to the gender-specific difference observed. Samples of the EC and the HC were categorized into males (EC_M: n = 10; HC_M: n = 7) and females (EC_F: n = 9; HC_F: n = 6) prior to analysis. The results of the DE are represented as volcano plots (Fig. 3a–d). There were 17 up-regulated and six downregulated transcripts in male HC when compared with female HC. However larger differences were observed in male EC over female EC with 24 upregulated and 74 downregulated transcripts. Interestingly, no DE transcripts were found in female EC compared to female HC. In contrast, 53 upregulated and 33 downregulated transcripts were identified in male HC when compared with male EC. A total number of differentially expressed transcripts in all comparisons is represented in Venn diagram (Fig. 3e).
Fig. 3.
Differential expression (DE) of the transcript (protein-coding and non-protein coding) expression profile in a gender-specific manner. (a–d) DE are represented as volcano plots in different categories of the HC and EC groups were categorized into males (HC_M: n = 7; EC_M: n = 10) and females (HC_F: n = 6; EC_F: n = 9) before analysis. Transcripts with log2-fold-change value greater than two and adjusted p-value (FDR) < 0.05 (filled red circle), were considered as upregulated and those with less than minus two and adjusted p-value (FDR) < 0.05 (filled green circle) were considered as downregulated. (e) Venn diagram of DE transcripts. The sum of the numbers in each large circle represents the total number of differentially expressed genes among various combinations (EC_M vs. EC_F, HC_M vs. EC_M and HC_M vs. HC_F). The overlapping part of the circles represents common DEGs between combinations. (f) The network analysis in STRING with 38 proteins that were upregulated in female EC compared to the male EC.
Further, the transcripts were classified into protein-coding and non-protein coding. Out of 98 differentially expressed transcripts in female EC compared to male EC, 51 transcripts were protein-coding, and 47 transcripts were non-protein coding. Similarly, in male EC compared to male HC, 29 differentially expressed transcripts were protein-coding, and 57 transcripts were non-protein coding. Also in female HC compared to male HC, 12 protein-coding transcripts and 11 non-protein coding transcripts were differentially expressed. The differential expression data for all the comparisons is given in supplementary data 2. The network analysis in STRING with 38 proteins that were upregulated in female EC compared to the male EC identified a prominent protein-protein interaction (PPI) with 13 proteins (p = 0.0002) including JUN, FOS, FOSB, and SOCS1 (Fig. 3f) in nine pathways that were shown to be significantly enriched. The functional enrichments in the network identified by KEGG are given as supplementary data 3. Among the nine pathways osteoclast differentiation (no of genes = 5; FDR = 0.001), TNF signaling pathway (no. of genes = 4; FDR = 0.004) and Toll-like receptor signaling pathway (no. of genes: 3; FDR = 0.026) were significantly enriched.
3.4. Pathway Analysis of Protein-coding Genes
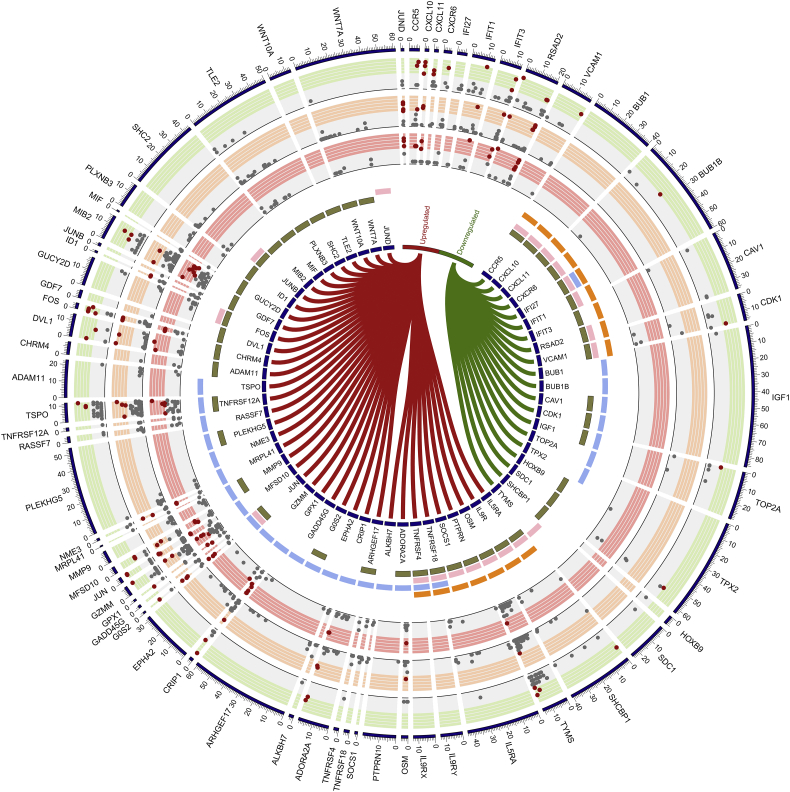
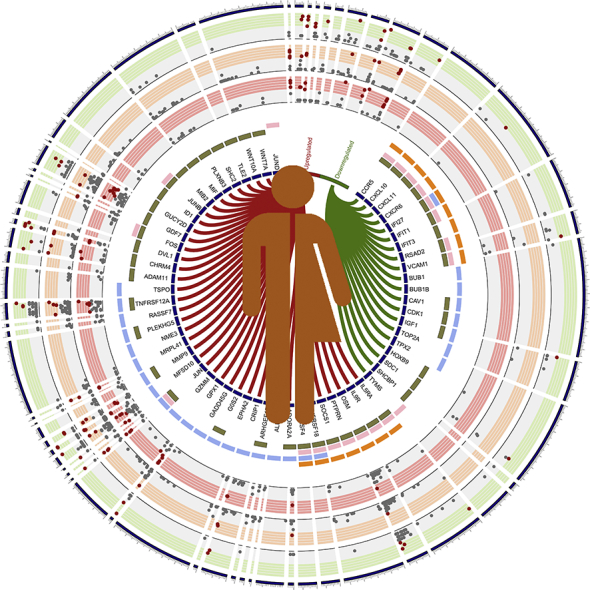
As EC and HC showed a similar transcriptomic profile, we restricted our pathway analysis to protein-coding genes (n = 270) that were differentially expressed in EC compared to VP to understand the potential biological pathways associated with those genes. The functional enrichments identified by gene ontology (GO) revealed that 37 biological processes were significantly associated with the gene sets (Supplementary data 4). Based on the biological significance, cell surface receptor signaling pathway (GO.0007166) had the maximum number of genes (n = 44; FDR = 0.046) (Fig. 4). There were several genes identified in this process that was also present in the pathways related to programmed cell death (GO.0012501; FDR = 0.029), response to cytokine (GO.0034097; FDR = 0.046), or cytokine-mediated signaling (GO.0019221; FDR = 0.017) (Fig. 4). From these genes, we further compared the SNPs among the three groups of individuals EC, HC, and VP. Several SNPs in genes like CCR5 (rs1800874 and rs746492), CXCL10 (rs8878), and CXCL11 (rs13130018, rs10003382, and rs13130221) were present at low frequency in EC, but not in VP. Given the small number of identified SNPs, we did not perform any statistical analysis. The rs2234358-T allele in CXCR6 has been significantly associated with long-term nonprogression (LTNP), but not with EC status (Limou et al., 2010). In our cohort, we identified a higher proportion of this allele in VP (75%, 6/8) compared to EC (27%, 5/19).
Fig. 4.
DE of the protein-coding region between VP and EC, SNPs within the differently expressed genes (DEGs) and GO analysis: Outer three rings are the variant calling results of VP (green), EC (orange) and HC (pink) in DEGs. Variants present in ≥ 50% of the samples in each group are denoted by red dots and others by grey dots in an x-y scatter plot manner. Inner CIRCOS plot visualizes differential expression and GO annotation where each arc label indicates DEGs. Up-regulated genes linked by red ribbon and down-regulated genes by a green ribbon. Outer four circles indicate GOs identified, cytokine-mediated signaling pathway (orange), programmed cell death (blue), response to cytokine (pink), cell surface receptor signaling pathway (khaki).
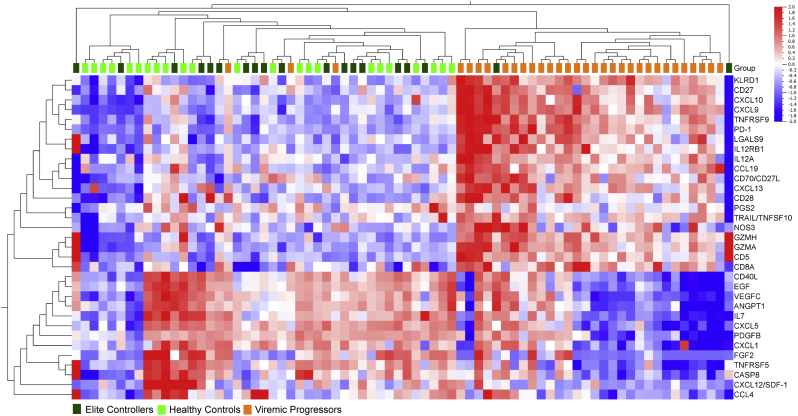
3.5. Proteomic Profiling of the Plasma Soluble Factors
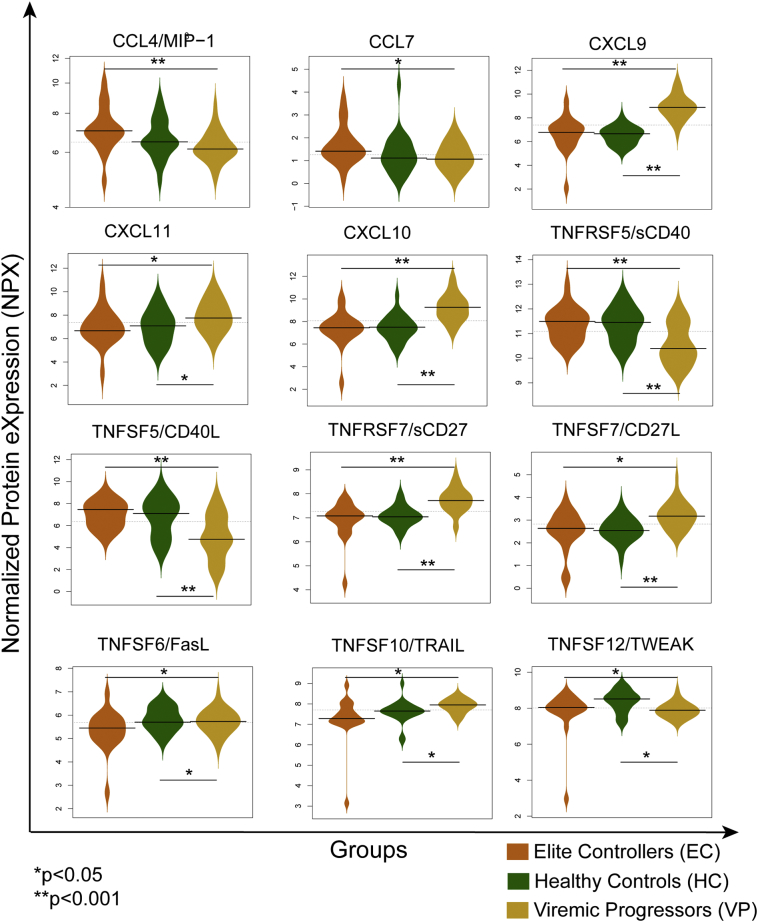
Based on the GO annotation of the transcriptome, we selected 92 soluble factors belonging to the Olink® Immuno-oncology panel. The panel constitutes 52 proteins involved in cell surface receptor signaling pathway (GO.0007166), 27 in programmed cell death (GO:0012501), 24 in response to cytokine (GO.0034097) and 20 in cytokine-mediated signaling (GO.0019221) (Supplementary data 5). Among the 92 proteins, 82 were detectable in > 50% of the samples and were further used for the analysis. Multi-group analysis by ANOVA identified clustering of 90% (29/32) of VPs together while EC and HC clustered separately from VP, but intermingled with each other (Fig. 5). Among the factors, 33 were distinctive to be statistically significant at a false discovery rate (FDR) adjusted p (q) = 0.02. When proteins were compared individually between groups, a less stringent cut-off of p < 0.05 was applied as significantly different. Protein levels of 61 out of 82 analyzed soluble factors were significantly different between at least two of the patient groups. In Fig. 6, Normalized Protein Expression (NPX) in VP, EC, and HC is shown for 12 selected soluble factors of which five belong to the chemokine family. Chemokine ligands that share the same receptor were concurrently increased or decreased when compared between the three groups. CCL4 and CCL7 that both binds to CCR5 showed significantly higher levels in EC compared to VP. CXCL9, CXCL10, and CXCL11 are ligands of CXCR3 and significantly decreased levels were observed for all ligands in EC compared to VP. Among the 61 significantly different soluble factors, there were six members of the Tumor Necrosis Factor Super Family (TNFSF) and five members of the Tumor Necrosis Factor Receptor Super Family (TNFRSF). These protein families are involved in pathways regulating apoptosis, inflammation, cellular differentiation, and antiviral state (Kumar et al., 2013). The soluble protein levels of TNFSF5 and TNFRSF5 (CD40L/CD40) were both significantly increased in EC while CD27 and CD70 were decreased, suggesting that downstream pathways could also be affected. Interestingly, ligands for death receptors TNFSF6 (FasL), TNFSF10 (TRAIL), and TNFSF12 (TWEAK) did not show concomitant expression patterns between the groups. While EC had lower levels of FasL and TRAIL compared to VP (significantly) and HC (significantly for FasL, but not for TRAIL), TWEAK was significantly decreased in VP and EC compared to HC, but no significant difference was observed between the EC and VP. The cell surface receptor PD-1, an immune checkpoint that also promotes apoptosis in immune cells (Bardhan et al., 2016), but does not belong to the TNFSF family, showed a similar pattern as TRAIL; levels were significantly increased in VP compared to both EC and HC.
Fig. 5.
The plasma soluble factor profiles using hierarchical clustering analysis. The number of samples included in the analysis were VP (Orange, n = 32), EC, (light green, n = 19) and HC (dark greenn = 23). The analysis was performed at a stringent false discovery rate (FDR) adjusted p (q) < 0.001 using ANOVA using the normalized protein expression levels (NPX). In the heatmap, Red: high NPX, blue: low NPX.
Fig. 6.
The normalized expression profiling of the 12 selected proteins. These proteins belong to the chemokine family, Tumor Necrosis Factor Super Family (TNFSF), Tumor Necrosis Factor Receptor Super Family (TNFRSF) and ligands for death receptors. * and ** indicate p < 0.05 and p < 0.001 respectively (Mann-Whitney U test).
4. Discussion
In this study, the high-throughput proteo-transcriptomics analysis was performed using a well-defined Swedish HIV-1 EC cohort. More than half of the EC had a previously reported protective allele, including B*57:01 and B*57:03. In contrast, SNPs in AIDS restriction genes (ARGs) were not detected which thus may not play any significant role in the viral control in EC. Overall the EC and HC possessed similar gene expression profiles with only 11 differentially expressed transcripts (protein-coding and non-protein coding). However, sharp gender-specific differences were observed. In our GO analysis of EC compared to VP, 37 biological processes were identified that were significantly associated with gene sets and same genes are shared by multiple candidate pathways forming interactome among the pathways.
More than half of our EC (n = 12) had an HLA-allele that is considered protective in HIV-1 disease, most frequently HLA-B alleles (9/12). This is not surprising since HLA-B is known to be strongly associated with delayed disease progression (Goulder and Walker, 2012). In contrast, other known protective SNPs in ARGs (O'Brien and Nelson, 2004) (CCR5-Δ32, CCR5 59029G, SDF1-3′A) were not detected implicating that their impact on viral control might not be as crucial as observed with HLA alleles.
Clustering analysis of our transcriptomics data revealed a definite distinction in gene expression between males and females among all patient groups. Surprisingly, the transcriptomic profile between female EC and healthy females were similar, whereas gene expression did differ between male EC and healthy males. Differences between healthy males and females could mostly be explained by the X- and Y-linked genes. However, differentially expressed genes between male and female EC were extended outside sex-linked genes, indicating that after HIV-1 infection sex-specific processes occur that could play a part in different mechanisms of HIV-1 control. Among the unique differentially expressed genes between male and female EC were JUN, FOS, FOSB, and SOCS1. The former three genes encode for transcription factors that can dimerize with each other or with other members of the FOS and JUN families to form the AP-1 transcription factor complex. This complex can in a dynamic, multifaceted network be activated by a variety of stimuli and regulate transcription of a plethora of genes involved in various biological processes, such as immune activation in leukocytes (Chinenov and Kerppola, 2001, Hess et al., 2004). SOCS1 is a negative immune regulator that offers negative feedback to the JAK/STAT pathway, which regulates cytokine signaling (Starr et al., 1997). Dysregulation of SOCS1 is associated with immune exhaustion in chronic infections, like hepatitis C virus and HIV-1 (Frazier et al., 2010, Zhang et al., 2011, Miller et al., 2011, Sachdeva et al., 2017). Since persistent immune activation is considered a driver for HIV-1 disease progression (Lawn et al., 2001, Khaitan and Unutmaz, 2011), changes in SOCS1 expression levels could, therefore, be involved in establishing EC phenotype. Gender-specific differences in HIV-1 infection and systemic acute innate responses could shape the dimension of persistent immune activation in the chronic stage of infection (Markle and Fish, 2014, vom Steeg and Klein, 2016, Griesbeck et al., 2016). We speculate that these gender-specific differences are one of the determining factors for whether an individual becomes EC or not. Indeed, the proportion of females within EC is higher compared to other groups of HIV-infected individuals (Crowell et al., 2015, de Azevedo et al., 2017). Despite extensive data on the male and female difference on disease outcome, research does not sufficiently take gender into account. Altogether, our study showed that it would be important to carefully consider gender in cohort design for future transcriptomic and intervention studies on HIV-1 patients.
Previous studies have suggested numerous correlates associated with HIV disease progression including the impaired response of innate and adaptive immune functions, increased plasma/serum levels of pro-inflammatory cytokines and chemokines (Peretz et al., 2012). The transcriptomic analyses revealed that the differentially expressed gene (DEG) profile of EC closely resembles that of HC. However, there were unique differences in gene expression between EC and VP. Interestingly, both CXCR6 and SIGLEC1 were among the 22 protein-coding DEGs. The chemokine receptor CXCR6 is expressed by various subsets of leukocytes and mediates leukocyte adhesion and migration through its exclusive ligand CXCL16 (Wilbanks et al., 2001). Interestingly, CXCR6 is considered as a minor co-receptor for HIV-1 entry into T lymphocytes and a polymorphism within CXCR6 (rs2234358) correlates to HIV-1 control in long-term non-progressors (Liao et al., 1997, Limou et al., 2010). Although this SNP was absent in our EC cohort, CXCR6 gene expression was significantly downregulated in EC compared to VP. SIGLEC1 is expressed by myeloid cells and encodes for the sialic acid binding cell surface protein sialoadhesin, which is involved in the regulation of several immune responses (Hartnell et al., 2001, Pulliam et al., 2004, Crocker and Redelinghuys, 2008, Martinez-Pomares and Gordon, 2012). Furthermore, macrophages and dendritic cells (DCs) can bind pathogens such as HIV-1 through sialoadhesin, internalize virions into virus-containing compartments and mediate cell-to-cell transmission (Pino et al., 2015, Hammonds et al., 2017). Increased SIGLEC1 expression in PBMCs, which is most likely restricted to the myeloid population, could, therefore, contribute to increased HIV-1 transmission and larger HIV-1 latency reservoir size. Since both CXCR6 and SIGLEC1 were uniquely differentially expressed between VP and EC and not between VP and HC, downregulation of both genes in EC could point to a mechanism of increased HIV-1 control in EC caused by decreased susceptibility of T lymphocytes for HIV-1 entry and declined HIV-1 cell-to-cell transmission mediated by myeloid cells.
The proteomic profile of plasma strongly co-related with the transcriptome profile. Among the factors tested was the chemokine CCL4 (MIP-1β), which is an HIV-suppressive factor produced by CD8+ T cells (Cocchi et al., 1995). It acts as a competitor for the viral binding site (Blanpain et al., 1999). The CCL4 levels were increased in EC compared to VP in our cohort as observed earlier (Walker et al., 2015). CCL7 (MCP-3) also binds to CCR5, albeit with low affinity, but has not been shown to have anti-HIV-1 activities (Blanpain et al., 1999). However, CCL7 levels were also increased in EC compared to VP in our cohort. The levels of three other chemokines CXCL9, CXCL10, and CXCL11 which all binds CXCR3, were increased in VP compared to both EC and HC. Our finding concerning CXCL10 is consistent with earlier studies (Jacobs et al., 2017). This inflammatory factor is considered to be a biomarker of chronic immune activation and as a marker for disease progression during HIV infection. It has recently been reported to suppress T cell and NK cell function in HIV-infected patients (Mhandire et al., 2017, Wang et al., 2017).
Immune checkpoints molecules regulate the responses to self-proteins upon invading infection. The PD1-inhibitory pathway limits the activity of T cells and other immune cells. Blocking this pathway is, therefore, a strong candidate not only in cancer treatment but also in HIV infection where clinical trials have been conducted (Bardhan et al., 2016, Gay et al., 2017, Pardoll, 2012, Velu et al., 2015). Interestingly, the levels of both PD-1 and its ligand PD1-L2 were significantly lower in EC compared to VP assuming that downstream signaling is also decreased in this study group. Levels of both CD27 and CD27L (CD70), were increased in VP, but not in EC, compared to HC. This has been claimed to result in impaired IgG responses and exhaustion of T cells, which further need experimental validation (Beishuizen et al., 2009, Tesselaar et al., 2003). Also, in our cohort CD40 signaling seemed to be downregulated in VP compared to EC and HC as levels for both CD40 and CD40L were low in VP compared to HC and EC though it contradicts earlier studies (Jenabian et al., 2014, Kaltenmeier et al., 2015). This reduced CD40L and increased CD70 signaling might contribute to immune dysregulation in HIV-infected individuals.
Death receptor ligands FasL (TNFSF6), TRAIL (TNFSF10), and TWEAK (TNFSF12) were also included in our analysis. Regarding the fact that they regulate similar signaling pathways, it is interesting that their level between study groups differs. Fas-signaling is assumed to be one of the reasons for depletion of infected and uninfected lymphocytes during infection (Kumar et al., 2013, Cummins and Badley, 2010). It is somewhat surprising that we could not confirm this higher level of FasL in VP compared to HC in our cohort. However, FasL was decreased in EC compared to VP and even compared to HC, suggesting that the virus might not induce Fas/FasL signaling and lymphocytes might be less sensitive to this signaling as observed earlier (Dianzani et al., 2003). Our findings for TRAIL were also in agreement with earlier studies where elevated levels are seen in VP compared to HC (Herbeuval et al., 2005). Interestingly, TRAIL-induced apoptosis has also been found in infected and uninfected bystander T cells, offering another possible explanation for T cell depletion in HIV-infected patients (Herbeuval et al., 2005, Kumar et al., 2013, Cummins and Badley, 2010). As for FasL, TRAIL levels in EC were significantly lower compared to VP. Taking together our observations in EC and findings in previous studies, downregulation of Fas and/or TRAIL signaling mediated by FasL and/or TRAIL blocking antibodies could be promising in HIV treatment in order to maintain number and function of immune cells (Lichtner et al., 2004, Herbeuval et al., 2005, Poonia et al., 2009). In contrast to those two death receptor ligands, TWEAK levels were found to be significantly lower in VP compared to HC, which has been reported earlier (Beltran et al., 2014), and levels in EC were in between those of VP and HC. Reduced soluble TWEAK plasma levels have been described in other inflammatory diseases like rheumatoid arthritis or atherosclerosis, but so far it is unknown which mechanisms are responsible for this decrease in HIV-infected patients.
Our study has a few limitations that merit comments. First, although our research included the highest number of EC as compared to earlier reports (Abdel-Mohsen et al., 2013, Rotger et al., 2010, Walker et al., 2015) despite using the strict criteria, the number of samples used for the transcriptomics study were relatively low. Second, a targeted approach was employed for the proteomics studies, which does not provide a complete picture of the proteomic profile. Finally, this is a hypothesis-generating explorative study. Future detailed mechanistic studies of individual pathways will dissect the actual role of each pathway in the immune control of HIV-1.
In conclusion, our data point to one of the potential mechanisms of HIV-1 control in EC; decreased susceptibility of T-lymphocytes for HIV-1 entry and decreased HIV-1 cell-to-cell transmission mediated by myeloid cells. Further, combining the gene expression and plasma proteomics profile of the soluble factors strongly implicate that cell surface receptor signaling pathway, programmed cell death, response to cytokine and cytokine-mediated signaling play synergistically an essential role in HIV-1 control in EC. After carefully considering gender in the cohort analysis, future detailed trans-omics studies could potentially indicate a precise biomarker profile of natural HIV-1 control that could be of clinical benefit or target for therapy.
Acknowledgments
Acknowledgements
The authors would like to acknowledge support from Science for Life Laboratory, the National Genomics Infrastructure, NGI, and Uppmax for assisting in massive parallel sequencing and computational infrastructure. We would also like to thank Olink AB for assistance in plasma proteome profiling. WZ acknowledge the support from China Scholarship Council (CSC). The authors would like to thank all the patients, nurses and clinicians who supported the InfCare system.
Funding Sources
The study is funded by grants from Swedish Research Council (2016–01675), Stockholm County Council (ALF 20160074) and Jeanssons Stiftelser (JS2016–0185). UN acknowledges the support received from Jonas Söderquist's Stipendium for Experimental Virology and Immunology Research-2016. PN is a clinical research fellow supported by Stockholm County Council – Karolinska Institute grant. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interests
None to declare.
Author Contributions
UN conceived and designed the studies and analysis plan. WZ, MS and KN perform the laboratory analysis. ATA and UN performed the bioinformatics analysis. AS and KN have identified and designed the EC cohort. KN, PN and UN performed the data collection. UN, WZ, MS, RvD and ATA wrote the respective portion of first draft of the manuscript reviewed by PN, AR and AS. PN and AS interpreted clinical data. All the authors approved the final version of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2017.11.031.
Appendix A. Supplementary Data
Supplementary Figure S1: The co-relation between protein concentrations in log2 picogram/mL with NPX, using ELISA targeted for three soluble factors CXCL10, TRAIL and IL-12 (R&D Systems, US).
Supplementary Figure S2: HLA class I alleles in EC.
Supplementary Data 1: The differentially expressed transcripts between EC and VP and the Venn annotations.
Supplementary Data 2: The differential expression data for all the comparisons.
Supplementary Data 3: The functional enrichments in the network identified by KEGG.
Supplementary Data 4: The functional enrichments identified by gene ontology (GO).
Supplementary Data 5: GO annotation of the Olink® Immuno-oncology panel.
References
- Abdel-Mohsen M., Raposo R.A., Deng X., Li M., Liegler T., Sinclair E., Salama M.S., Ghanem Hel D., Hoh R., Wong J.K., David M., Nixon D.F., Deeks S.G., Pillai S.K. Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology. 2013;10:106. doi: 10.1186/1742-4690-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assarsson E., Lundberg M., Holmquist G., Bjorkesten J., Thorsen S.B., Ekman D., Eriksson A., Rennel Dickens E., Ohlsson S., Edfeldt G., Andersson A.C., Lindstedt P., Stenvang J., Gullberg M., Fredriksson S. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9:e95192. doi: 10.1371/journal.pone.0095192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo S.S.D., Caetano D.G., Cortes F.H., Teixeira S.L.M., Dos Santos Silva K., Hoagland B., Grinsztejn B., Veloso V.G., Morgado M.G., Bello G. Highly divergent patterns of genetic diversity and evolution in proviral quasispecies from HIV controllers. Retrovirology. 2017;14:29. doi: 10.1186/s12977-017-0354-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardhan K., Anagnostou T., Boussiotis V.A. The PD1:PD-L1/2 pathway from discovery to clinical implementation. Front. Immunol. 2016;7:550. doi: 10.3389/fimmu.2016.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beishuizen C.R., Kragten N.A., Boon L., Nolte M.A., van Lier R.A., van Gisbergen K.P. Chronic CD70-driven costimulation impairs IgG responses by instructing T cells to inhibit germinal center B cell formation through FasL-Fas interactions. J. Immunol. 2009;183:6442–6451. doi: 10.4049/jimmunol.0901565. [DOI] [PubMed] [Google Scholar]
- Beltran L.M., Munoz Hernandez R., de Pablo Bernal R.S., Garcia Morillo J.S., Egido J., Noval M.L., Ferrando-Martinez S., Blanco-Colio L.M., Genebat M., Villar J.R., Moreno-Luna R., Moreno J.A. Reduced sTWEAK and increased sCD163 levels in HIV-infected patients: modulation by antiretroviral treatment, HIV replication and HCV co-infection. PLoS One. 2014;9:e90541. doi: 10.1371/journal.pone.0090541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berggrund M., Ekman D., Gustavsson I., Sundfeldt K., Olovsson M., Enroth S., Gyllensten U. Protein detection using the multiplexed proximity extension assay (PEA) from plasma and vaginal fluid applied to the indicating FTA elute micro card. J. Circ. Biomark. 2016;5:9. doi: 10.5772/64000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C., Migeotte I., Lee B., Vakili J., Doranz B.J., Govaerts C., Vassart G., Doms R.W., Parmentier M. CCR5 binds multiple CC-chemokines: MCP-3 acts as a natural antagonist. Blood. 1999;94:1899–1905. [PubMed] [Google Scholar]
- Boegel S., Scholtalbers J., Lower M., Sahin U., Castle J.C. In silico HLA typing using standard RNA-Seq sequence reads. Methods Mol. Biol. 2015;1310:247–258. doi: 10.1007/978-1-4939-2690-9_20. [DOI] [PubMed] [Google Scholar]
- Boyle E.A., Li Y.I., Pritchard J.K. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y., Kerppola T.K. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- Cocchi F., DeVico A.L., Garzino-Demo A., Arya S.K., Gallo R.C., Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8 + T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- Crocker P.R., Redelinghuys P. Siglecs as positive and negative regulators of the immune system. Biochem. Soc. Trans. 2008;36:1467–1471. doi: 10.1042/BST0361467. [DOI] [PubMed] [Google Scholar]
- Crowell T.A., Gebo K.A., Blankson J.N., Korthuis P.T., Yehia B.R., Rutstein R.M., Moore R.D., Sharp V., Nijhawan A.E., Mathews W.C., Hanau L.H., Corales R.B., Beil R., Somboonwit C., Edelstein H., Allen S.L., Berry S.A. Hospitalization rates and reasons among HIV elite controllers and persons with medically controlled HIV infection. J. Infect. Dis. 2015;211:1692–1702. doi: 10.1093/infdis/jiu809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins N.W., Badley A.D. Mechanisms of HIV-associated lymphocyte apoptosis: 2010. Cell Death Dis. 2010;1:e99. doi: 10.1038/cddis.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks S.G., Walker B.D. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Dianzani U., Bensi T., Savarino A., Sametti S., Indelicato M., Mesturini R., Chiocchetti A. Role of FAS in HIV infection. Curr. HIV Res. 2003;1:405–417. doi: 10.2174/1570162033485131. [DOI] [PubMed] [Google Scholar]
- Frazier A.D., Zhang C.L., Ni L., Ma C.J., Zhang Y., Wu X.Y., Atia A.N., Yao Z.Q., Moorman J.P. Programmed death-1 affects suppressor of cytokine signaling-1 expression in T cells during hepatitis C infection. Viral Immunol. 2010;23:487–495. doi: 10.1089/vim.2010.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay C.L., Bosch R.J., Ritz J., Hataye J.M., Aga E., Tressler R.L., Mason S.W., Hwang C.K., Grasela D.M., Ray N., Cyktor J.C., Coffin J.M., Acosta E.P., Koup R.A., Mellors J.W., Eron J.J., Team, A. C. T. S Clinical trial of the anti-PD-L1 antibody BMS-936559 in HIV-1 infected participants on suppressive antiretroviral therapy. J. Infect. Dis. 2017;215:1725–1733. doi: 10.1093/infdis/jix191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder P.J., Walker B.D. HIV and HLA class I: an evolving relationship. Immunity. 2012;37:426–440. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck M., Scully E., Altfeld M. Sex and gender differences in HIV-1 infection. Clin. Sci. (Lond.) 2016;130:1435–1451. doi: 10.1042/CS20160112. [DOI] [PubMed] [Google Scholar]
- Hammonds J.E., Beeman N., Ding L., Takushi S., Francis A.C., Wang J.J., Melikyan G.B., Spearman P. Siglec-1 initiates formation of the virus-containing compartment and enhances macrophage-to-T cell transmission of HIV-1. PLoS Pathog. 2017;13:e1006181. doi: 10.1371/journal.ppat.1006181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartnell A., Steel J., Turley H., Jones M., Jackson D.G., Crocker P.R. Characterization of human sialoadhesin, a sialic acid binding receptor expressed by resident and inflammatory macrophage populations. Blood. 2001;97:288–296. doi: 10.1182/blood.v97.1.288. [DOI] [PubMed] [Google Scholar]
- Heberle H., Meirelles G.V., da Silva F.R., Telles G.P., Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinforma. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbeuval J.P., Boasso A., Grivel J.C., Hardy A.W., Anderson S.A., Dolan M.J., Chougnet C., Lifson J.D., Shearer G.M. TNF-related apoptosis-inducing ligand (TRAIL) in HIV-1-infected patients and its in vitro production by antigen-presenting cells. Blood. 2005;105:2458–2464. doi: 10.1182/blood-2004-08-3058. [DOI] [PubMed] [Google Scholar]
- Hess J., Angel P., Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- Jacobs E.S., Keating S.M., Abdel-Mohsen M., Gibb S.L., Heitman J.W., Inglis H.C., Martin J.N., Zhang J., Kaidarova Z., Deng X., Wu S., Anastos K., Crystal H., Villacres M.C., Young M., Greenblatt R.M., Landay A.L., Gange S.J., Deeks S.G., Golub E.T., Pillai S.K., Norris P.J. Cytokines elevated in HIV elite controllers reduce HIV replication in vitro and modulate HIV restriction factor expression. J. Virol. 2017;91 doi: 10.1128/JVI.02051-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenabian M.A., Patel M., Kema I., Vyboh K., Kanagaratham C., Radzioch D., Thebault P., Lapointe R., Gilmore N., Ancuta P., Tremblay C., Routy J.P. Soluble CD40-ligand (sCD40L, sCD154) plays an immunosuppressive role via regulatory T cell expansion in HIV infection. Clin. Exp. Immunol. 2014;178:102–111. doi: 10.1111/cei.12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczorowski K.J., Shekhar K., Nkulikiyimfura D., Dekker C.L., Maecker H., Davis M.M., Chakraborty A.K., Brodin P. Continuous immunotypes describe human immune variation and predict diverse responses. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E6097–E6106. doi: 10.1073/pnas.1705065114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenmeier C., Gawanbacht A., Beyer T., Lindner S., Trzaska T., van der Merwe J.A., Harter G., Gruner B., Fabricius D., Lotfi R., Schwarz K., Schutz C., Honig M., Schulz A., Kern P., Bommer M., Schrezenmeier H., Kirchhoff F., Jahrsdorfer B. CD4 + T cell-derived IL-21 and deprivation of CD40 signaling favor the in vivo development of granzyme B-expressing regulatory B cells in HIV patients. J. Immunol. 2015;194:3768–3777. doi: 10.4049/jimmunol.1402568. [DOI] [PubMed] [Google Scholar]
- Khaitan A., Unutmaz D. Revisiting immune exhaustion during HIV infection. Curr. HIV/AIDS Rep. 2011;8:4–11. doi: 10.1007/s11904-010-0066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Abbas W., Herbein G. TNF and TNF receptor superfamily members in HIV infection: new cellular targets for therapy? Mediat. Inflamm. 2013;2013:484378. doi: 10.1155/2013/484378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C.W., Chen Y., Shi W., Smyth G.K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawn S.D., Butera S.T., Folks T.M. Contribution of immune activation to the pathogenesis and transmission of human immunodeficiency virus type 1 infection. Clin. Microbiol. Rev. 2001;14:753–777. doi: 10.1128/CMR.14.4.753-777.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao F., Alkhatib G., Peden K.W., Sharma G., Berger E.A., Farber J.M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J. Exp. Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtner M., Maranon C., Vidalain P.O., Azocar O., Hanau D., Lebon P., Burgard M., Rouzioux C., Vullo V., Yagita H., Rabourdin-Combe C., Servet C., Hosmalin A. HIV type 1-infected dendritic cells induce apoptotic death in infected and uninfected primary CD4 T lymphocytes. AIDS Res. Hum. Retrovir. 2004;20:175–182. doi: 10.1089/088922204773004897. [DOI] [PubMed] [Google Scholar]
- Limou S., Coulonges C., Herbeck J.T., van Manen D., An P., Le Clerc S., Delaneau O., Diop G., Taing L., Montes M., van't Wout A.B., Gottlieb G.S., Therwath A., Rouzioux C., Delfraissy J.F., Lelievre J.D., Levy Y., Hercberg S., Dina C., Phair J., Donfield S., Goedert J.J., Buchbinder S., Estaquier J., Schachter F., Gut I., Froguel P., Mullins J.I., Schuitemaker H., Winkler C., Zagury J.F. Multiple-cohort genetic association study reveals CXCR6 as a new chemokine receptor involved in long-term nonprogression to AIDS. J. Infect. Dis. 2010;202:908–915. doi: 10.1086/655782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markle J.G., Fish E.N. SeXX matters in immunity. Trends Immunol. 2014;35:97–104. doi: 10.1016/j.it.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Martinez-Pomares L., Gordon S. CD169 + macrophages at the crossroads of antigen presentation. Trends Immunol. 2012;33:66–70. doi: 10.1016/j.it.2011.11.001. [DOI] [PubMed] [Google Scholar]
- McLaren P.J., Ripke S., Pelak K., Weintrob A.C., Patsopoulos N.A., Jia X., Erlich R.L., Lennon N.J., Kadie C.M., Heckerman D., Gupta N., Haas D.W., Deeks S.G., Pereyra F., Walker B.D., de Bakker P.I., International, H. I. V. C. S Fine-mapping classical HLA variation associated with durable host control of HIV-1 infection in African Americans. Hum. Mol. Genet. 2012;21:4334–4347. doi: 10.1093/hmg/dds226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren P.J., Coulonges C., Bartha I., Lenz T.L., Deutsch A.J., Bashirova A., Buchbinder S., Carrington M.N., Cossarizza A., Dalmau J., De Luca A., Goedert J.J., Gurdasani D., Haas D.W., Herbeck J.T., Johnson E.O., Kirk G.D., Lambotte O., Luo M., Mallal S., van Manen D., Martinez-Picado J., Meyer L., Miro J.M., Mullins J.I., Obel N., Poli G., Sandhu M.S., Schuitemaker H., Shea P.R., Theodorou I., Walker B.D., Weintrob A.C., Winkler C.A., Wolinsky S.M., Raychaudhuri S., Goldstein D.B., Telenti A., de Bakker P.I., Zagury J.F., Fellay J. Polymorphisms of large effect explain the majority of the host genetic contribution to variation of HIV-1 virus load. Proc. Natl. Acad. Sci. U. S. A. 2015;112:14658–14663. doi: 10.1073/pnas.1514867112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhandire K., Mlambo T., Zijenah L.S., Duri K., Mateveke K., Tshabalala M., Mhandire D.Z., Musarurwa C., Wekare P.T., Mazengera L.R., Matarira H.T., Stray-Pedersen B. Plasma IP-10 concentrations correlate positively with Viraemia and inversely with CD4 counts in untreated HIV infection. Open AIDS J. 2017;11:24–31. doi: 10.2174/1874613601711010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.C., Schlaepfer E., Baenziger S., Crameri R., Zeller S., Byland R., Audige A., Nadal D., Speck R.F. HIV interferes with SOCS-1 and -3 expression levels driving immune activation. Eur. J. Immunol. 2011;41:1058–1069. doi: 10.1002/eji.201041198. [DOI] [PubMed] [Google Scholar]
- O'Brien S.J., Nelson G.W. Human genes that limit AIDS. Nat. Genet. 2004;36:565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- Olson A.D., Meyer L., Prins M., Thiebaut R., Gurdasani D., Guiguet M., Chaix M.L., Amornkul P., Babiker A., Sandhu M.S., Porter K. An evaluation of HIV elite controller definitions within a large seroconverter cohort collaboration. PLoS One. 2014;9:e86719. doi: 10.1371/journal.pone.0086719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretz Y., Cameron C., Sekaly R.P. Dissecting the HIV-specific immune response: a systems biology approach. Curr. Opin. HIV AIDS. 2012;7:17–23. doi: 10.1097/COH.0b013e32834ddb0e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereyra F., Jia X., McLaren P.J., Telenti A., de Bakker P.I., Walker B.D., Ripke S., Brumme C.J., Pulit S.L., Carrington M., Kadie C.M., Carlson J.M., Heckerman D., Graham R.R., Plenge R.M., Deeks S.G., Gianniny L., Crawford G., Sullivan J., Gonzalez E., Davies L., Camargo A., Moore J.M., Beattie N., Gupta S., Crenshaw A., Burtt N.P., Guiducci C., Gupta N., Gao X., Qi Y., Yuki Y., Piechocka-Trocha A., Cutrell E., Rosenberg R., Moss K.L., Lemay P., O'Leary J., Schaefer T., Verma P., Toth I., Block B., Baker B., Rothchild A., Lian J., Proudfoot J., Alvino D.M., Vine S., Addo M.M., Allen T.M., Altfeld M., Henn M.R., Le Gall S., Streeck H., Haas D.W., Kuritzkes D.R., Robbins G.K., Shafer R.W., Gulick R.M., Shikuma C.M., Haubrich R., Riddler S., Sax P.E., Daar E.S., Ribaudo H.J., Agan B., Agarwal S., Ahern R.L., Allen B.L., Altidor S., Altschuler E.L., Ambardar S., Anastos K., Anderson B., Anderson V., Andrady U., Antoniskis D., Bangsberg D., Barbaro D., Barrie W., Bartczak J., Barton S., Basden P., Basgoz N., Bazner S., Bellos N.C., Benson A.M., Berger J., Bernard N.F., Bernard A.M., Birch C., Bodner S.J., Bolan R.K., Boudreaux E.T., Bradley M., Braun J.F., Brndjar J.E., Brown S.J., Brown K., Brown S.T. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino M., Erkizia I., Benet S., Erikson E., Fernandez-Figueras M.T., Guerrero D., Dalmau J., Ouchi D., Rausell A., Ciuffi A., Keppler O.T., Telenti A., Krausslich H.G., Martinez-Picado J., Izquierdo-Useros N. HIV-1 immune activation induces Siglec-1 expression and enhances viral trans-infection in blood and tissue myeloid cells. Retrovirology. 2015;12:37. doi: 10.1186/s12977-015-0160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poonia B., Pauza C.D., Salvato M.S. Role of the Fas/FasL pathway in HIV or SIV disease. Retrovirology. 2009;6:91. doi: 10.1186/1742-4690-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poropatich K., Sullivan D.J., Jr. Human immunodeficiency virus type 1 long-term non-progressors: the viral, genetic and immunological basis for disease non-progression. J. Gen. Virol. 2011;92:247–268. doi: 10.1099/vir.0.027102-0. [DOI] [PubMed] [Google Scholar]
- Pulliam L., Sun B., Rempel H. Invasive chronic inflammatory monocyte phenotype in subjects with high HIV-1 viral load. J. Neuroimmunol. 2004;157:93–98. doi: 10.1016/j.jneuroim.2004.08.039. [DOI] [PubMed] [Google Scholar]
- Pundir S., Martin M.J., O'Donovan C. UniProt protein knowledgebase. Methods Mol. Biol. 2017;1558:41–55. doi: 10.1007/978-1-4939-6783-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotger M., Dang K.K., Fellay J., Heinzen E.L., Feng S., Descombes P., Shianna K.V., Ge D., Gunthard H.F., Goldstein D.B., Telenti A. Genome-wide mRNA expression correlates of viral control in CD4 + T-cells from HIV-1-infected individuals. PLoS Pathog. 2010;6:e1000781. doi: 10.1371/journal.ppat.1000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M., Sharma A., Arora S.K. Increased expression of negative regulators of cytokine signaling during chronic HIV disease cause functionally exhausted state of dendritic cells. Cytokine. 2017;91:118–123. doi: 10.1016/j.cyto.2016.08.010. [DOI] [PubMed] [Google Scholar]
- Starr R., Willson T.A., Viney E.M., Murray L.J., Rayner J.R., Jenkins B.J., Gonda T.J., Alexander W.S., Metcalf D., Nicola N.A., Hilton D.J. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- vom Steeg L.G., Klein S.L. SeXX matters in infectious disease pathogenesis. PLoS Pathog. 2016;12:e1005374. doi: 10.1371/journal.ppat.1005374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesselaar K., Arens R., van Schijndel G.M., Baars P.A., van der Valk M.A., Borst J., van Oers M.H., van Lier R.A. Lethal T cell immunodeficiency induced by chronic costimulation via CD27-CD70 interactions. Nat. Immunol. 2003;4:49–54. doi: 10.1038/ni869. [DOI] [PubMed] [Google Scholar]
- Velu V., Shetty R.D., Larsson M., Shankar E.M. Role of PD-1 co-inhibitory pathway in HIV infection and potential therapeutic options. Retrovirology. 2015;12:14. doi: 10.1186/s12977-015-0144-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker W.E., Kurscheid S., Joshi S., Lopez C.A., Goh G., Choi M., Barakat L., Francis J., Fisher A., Kozal M., Zapata H., Shaw A., Lifton R., Sutton R.E., Fikrig E. Increased levels of macrophage inflammatory proteins result in resistance to R5-tropic HIV-1 in a subset of elite controllers. J. Virol. 2015;89:5502–5514. doi: 10.1128/JVI.00118-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wu T., Ma M., Zhang Z., Fu Y., Liu J., Xu J., Ding H., Han X., Chu Z., Wu Y., Shang H., Jiang Y. Elevated interferon-gamma-induced protein 10 and its receptor CXCR3 impair NK cell function during HIV infection. J. Leukoc. Biol. 2017;102:163–170. doi: 10.1189/jlb.5A1016-444R. [DOI] [PubMed] [Google Scholar]
- Wilbanks A., Zondlo S.C., Murphy K., Mak S., Soler D., Langdon P., Andrew D.P., Wu L., Briskin M. Expression cloning of the STRL33/BONZO/TYMSTRligand reveals elements of CC, CXC, and CX3C chemokines. J. Immunol. 2001;166:5145–5154. doi: 10.4049/jimmunol.166.8.5145. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ma C.J., Ni L., Zhang C.L., Wu X.Y., Kumaraguru U., Li C.F., Moorman J.P., Yao Z.Q. Cross-talk between programmed death-1 and suppressor of cytokine signaling-1 in inhibition of IL-12 production by monocytes/macrophages in hepatitis C virus infection. J. Immunol. 2011;186:3093–3103. doi: 10.4049/jimmunol.1002006. [DOI] [PubMed] [Google Scholar]
- Zhang W., Morshed M.M., Noyan K., Russom A., Sonnerborg A., Neogi U. Quantitative humoral profiling of the HIV-1 proteome in elite controllers and patients with very long-term efficient antiretroviral therapy. Sci. Rep. 2017;7:666. doi: 10.1038/s41598-017-00759-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: The co-relation between protein concentrations in log2 picogram/mL with NPX, using ELISA targeted for three soluble factors CXCL10, TRAIL and IL-12 (R&D Systems, US).
Supplementary Figure S2: HLA class I alleles in EC.
Supplementary Data 1: The differentially expressed transcripts between EC and VP and the Venn annotations.
Supplementary Data 2: The differential expression data for all the comparisons.
Supplementary Data 3: The functional enrichments in the network identified by KEGG.
Supplementary Data 4: The functional enrichments identified by gene ontology (GO).
Supplementary Data 5: GO annotation of the Olink® Immuno-oncology panel.
Data Availability Statement
RNAseq raw sequencing data along with sample level metadata have been deposited in the NCBI/SRA Web service. Data can be accessed using BioProject accession number PRJNA420459.