Abstract
Early diagnosis and appropriate treatment for carbapenem-resistant Klebsiella pneumoniae (CR-Kp) infection is a big challenge for clinicians due to its high mortality. Every effort has been made to improve its clinical outcomes. However, treatment according to synergy susceptibility testing has never been reported in the literature. We reported a 29-year-old systemic lupus erythematosus female with CR-Kp blood stream infection. We highlighted the identification by next generation sequencing and treatment according to synergy susceptibility testing in the case.
1. Introduction
Systemic lupus erythematosus (SLE) is one of the most common autoimmune disease in Chinese population [1]. Chronic microbial colonization of the intestine, leading to exacerbations of infection when patients with immunosuppression, is the major cause of illness in SLE patients [2]. Klebsiella pneumoniae carbapenemases (KPCs) are plasmid-encoded carbapenem-hydrolyzing enzymes which have the potential to spread widely through gene transfer [3]. Treatment of carbapenem-resistant Klebsiella pneumoniae (CR-Kp) is difficult due to the limitation of antibiotics choice. Although next generation sequencing (NGS) is becoming increasingly important in clinical microbiology, only sporadic reports of NGS-analyzed clinical specimens have been published to date. Furthermore, synergy susceptibility testing as a new attempt to treat CR-Kp has never been reported in the literature.
2. Case Presentation
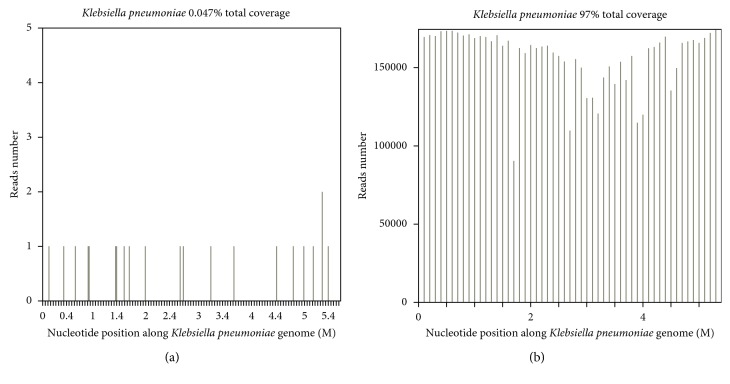
A 29-year-old female was admitted to our intensive care unit (ICU) from the rescue room because of dyspnea for one week. Her medical history included systemic lupus erythematosus (SLE) for one year which was treated with daily oral prednisone 60 mg. Prednisone was discontinued by herself last month because she was preparing for pregnancy. On physical examination, she appeared cyanotic with fever up to 38.5°C, and moist rale could be clearly heard on the lower lobe of both sides. Chest CT scan showed diffuse interstitial pulmonary fibrosis. Acute respiratory failure was managed with protective mechanical ventilation (tidal volume 360 mL, plateau pressure 25 cm H2O, and positive end-expiratory pressure 15 cm H2O) [4]. Laboratory evaluation at the time of ICU admission revealed negative blood cultures and normal procalcitonin (0.08 ng/mL). Systemic lupus erythematosus disease activity index (SLEDAI) was 5 points which indicated that SLE was flare. Next generation sequencing (NGS) was performed to exclude infectious diseases which revealed negative in bronchoalveolar lavage fluid sample. However, Klebsiella pneumoniae was detected in blood sample with low coverage (0.047%) and depth (Figure 1(a)). Klebsiella pneumoniae genome concordance was 97% (Figure 1(b)).
Figure 1.
The results of next generation sequencing in blood sample. (a) Total coverage of Klebsiella pneumoniae in blood sample. (b) Klebsiella pneumoniae genome concordance.
Methylprednisone (40 mg q12h) was initiated prescribed to prevent progression of SLE and pulmonary fibrosis. Meropenem (1 g q8h) was instituted to treat the probable blood stream infection (BSI). Three days after admission to ICU, fever of the patient was up to 40.1°C, and procalcitonin increased to 43.7 ng/mL. Blood cultures were positive for carbapenem-resistant Klebsiella pneumoniae (CR-Kp) according to bioMérieux Vitek-2 automated system. Antimicrobial susceptibility testing was performed, and the breakpoint (susceptible, intermediate, or resistant) was determined according to Enterobacteriaceae M100-S27 provided by the Clinical and Laboratory Standards Institute (CLSI) standards. The minimal inhibition concentration (MIC) of meropenem was ≥16 mg/L. The antibiotic susceptibility profiling revealed that this strain of CR-Kp was susceptible only to tigecycline (MIC = 0.75 mg/L) and colistin (MIC = 0.25 mg/L). However, colistin was not available in the mainland China.
High-quality sequencing data were generated by removing low-quality reads, adapter contamination, and reads with a certain proportion of Ns' bases. Then, the remaining reads were aligned to Bacterial Virulence Factor Database (VFDB) using the Burrows Wheeler Alignment tool (BWA, Version 0.7.10). The coverage rate and depth of every bacterial virulence gene were calculated by Short Oligonucleotide Analysis Package (SOAP, http://soap.genomics.org.cn, Version 2.7.7). Drug resistance and virulence genes were detected, and Klebsiella pneumoniae carbapenemases-2 (KPC-2) was confirmed as the main (Table 1).
Table 1.
The top five drug resistant and virulence genes of this strain Klebsiella pneumoniae.
| Gene names | Protein | Percentage | Depth |
|---|---|---|---|
| Drug resistance gene | |||
| KPC-2 | Carbapenem-hydrolyzing class A beta-lactamase, KPC-2 | 100 | 230 |
| aadA2 | ANT(3″)-Ia family aminoglycoside nucleotidyltransferase | 100 | 220 |
| CTX-M-24 | Class A extended-spectrum beta-lactamase, CTX-M-24 | 100 | 180 |
| QnrS1 | Quinolone resistance pentapeptide repeat protein, QnrS1 | 100 | 180 |
| dfrA12 | Trimethoprim-resistant dihydrofolate reductase, DfrA12 | 100 | 180 |
| Virulence gene | |||
| bla | Methylmalonic aciduria type A protein | 100 | 1400 |
| fepA | Ferrienterobactin receptor | 100 | 810 |
| mrkD | Fimbria adhesin protein | 100 | 810 |
| iutA | Putative ferric siderophore receptor | 100 | 650 |
| manB | Mannosidase, beta A, lysosomal-like | 100 | 590 |
Annotation of drug resistance gene: KPC-2, carbapenem-hydrolyzing class A; aadA2, aminoglycoside resistance; CTX-M-24, beta-lactamase class A; QnrS1, quinolone resistance pentapeptide repeat protein; dfrA12, dihydrofolate reductase (trimethoprim resistance protein).
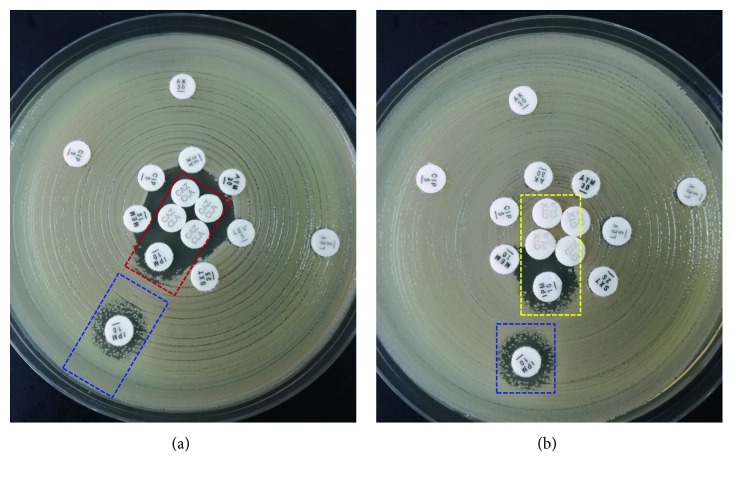
A systematic review of the literature showed that tazobactam and clavulanic acid are partial inhibitors of KPCs and the hydrolysis of the third generation cephalosporin is relatively weak in KPCs [5]. We performed a synergy susceptibility testing based on large doses of ceftazidime-clavaminic acid and found that it represented synergism between imipenem and ceftazidime (Figures 2(a) and 2(b)). Although it was just in vitro, it might be the best therapeutic option at that time. Imipenem-cilastatin (1 g q6h) and ceftazidime (2 g q8h) combined with tigecycline (100 mg q12h) were started. Three days later, blood cultures turned negative, procalcitonin level dropped to 0.23 ng/mL, and the fever disappeared. One week later, NGS of blood sample revealed negative, and antibiotics were discontinued due to kidney injury and digestive side effects. The patient was discharged with a good health condition 10 days later, and there was no recurrence of BSI.
Figure 2.
The results of synergy susceptibility testing. (a) Synergy susceptibility testing represented synergism between imipenem and ceftazidime (red square). CR-Kp was resistant to imipenem (blue square). (b) No synergism between imipenem and clavulanic acid (yellow square). CR-Kp was resistant to imipenem (blue square). CAZ, ceftazidime; IPM, imipenem; CLA, clavulanic acid.
3. Discussion
KPCs are plasmid-encoded carbapenem hydrolyzing enzymes which have the potential to spread widely through gene transfer. Notable among the drugs under development against KPC are mostly derivatives of polymixin, β-lactamase inhibitor NXL104 with combination of oxyimino cephalosporin as well as with ceftazidime [6]. The KPCs have become endemic, and treatment options for these infections are limited because colistin and ceftazidime–avibactam are not available in the mainland China. In these patients, treating with tigecycline alone showed a higher mortality despite the in vitro susceptibility [7]. Therefore, new methods should be sought to improve the clinical outcome.
Susceptibility methods vary in terms of choice of media, inoculum preparation, antimicrobial disk content, breakpoints, and interpretation of those breakpoints around the world [8]. Even when these variables are taken into consideration, susceptibility testing of Klebsiella pneumoniae remains challenging given the multiple mechanisms of resistance, both intrinsic and acquired, which are frequently expressed concurrently, often at low levels [9]. The hydrolysis of the third generation cephalosporin is relatively weak in KPCs; thus, we took ceftazidime as the foundation of the synergy susceptibility testing. The in vivo experiment demonstrated synergism between imipenem and ceftazidime and achieved a better therapeutic effect in this patient. However, disk diffusion testing does not correlate well with MIC results, and no cutoff value was defined about synergy susceptibility testing. Maybe it is just a new attempt and exploration, and we still need more studies with large samples to prove the real effect.
BSI remains one of the major challenges in critical patients, which, if not treated promptly, will lead to septic shock and multiple organ failure [10]. Due to the lack of timely diagnostic approaches with sufficient sensitivity, mortality of these patients is still unacceptably high. Blood culture-based diagnostic procedures represent the standard of care, although they are associated with relevant limitations [11]. Two caveats should be born in mind in interpreting the blood culture results: (1) culture-based diagnostic procedures often reveal false negative results due to the administration of an empiric antibiotic therapy, and (2) growth of causative pathogen often takes up to 5 days for results to become available [12]. Therefore, an empirical diagnosis of the causative microorganism is critical to improve outcome of BSI.
NGS-based diagnostic testing might offer several advantages over blood culture. First, it is an open platform, providing the opportunity to detect bacterial, fungal, and viral pathogens in a single assay. Second, it is quantitative through counting of sequence reads and calculation of statistical significance. Third, it is unbiased and untargeted, so it benefits from any DNA sequence information within patient specimens, potentially delivering higher sensitivity and specificity [13, 14]. However, although NGS is becoming increasingly important in clinical microbiology, only sporadic reports of NGS-analyzed clinical specimens have been published to date. Furthermore, it remains largely unknown whether treatment strategy based on NGS is able to shorten the course of antibiotics which is worth further discussing.
In summary, the KPCs have become endemic, and treatment options for these infections are limited. Synergy susceptibility testing is a new attempt and exploration, which still needs more studies with large samples to prove the true effect. NGS analyzes the circulating cell-free DNA (cfDNA) from plasma samples of septic patients and has higher sensitivity and specificity. Combining NGS with synergy susceptibility testing may improve the outcome of CR-Kp BSI patients.
Acknowledgments
The authors appreciate Honglong Wu from BGI-Tianjin for detecting the drug resistance and virulence genes. The study was supported by the National Key Research and Development Program of China (2017YFC0909002).
Abbreviations
- CR-Kp:
Carbapenem-resistant Klebsiella pneumoniae
- NGS:
Next generation sequencing
- MIC:
Minimal inhibition concentration
- KPCs:
Klebsiella pneumoniae carbapenemases.
Consent
Written informed consent was obtained from the patient and her family for publication of their individual details and accompanying images in this manuscript.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Yuetian Yu and Cheng Zhu both helped to draft and edit the article. Fupin Hu did the synergy susceptibility testing. Yuetian Yu, Cheng Zhu, and Erzhen Chen collected the laboratory analysis reports of the patient. Yuan Gao and Liangjing Lu revised and approved the final manuscript. Yuetian Yu and Fupin Hu contributed equally to this work.
References
- 1.Chen D., Xie J., Chen H., et al. Infection in southern chinese patients with systemic lupus erythematosus: spectrum, drug resistance, outcomes, and risk factors. Journal of Rheumatology. 2016;43(9):1650–1656. doi: 10.3899/jrheum.151523. [DOI] [PubMed] [Google Scholar]
- 2.Sciascia S., Cuadrado M. J., Karim M. Y. Management of infection in systemic lupus erythematosus. Best Practice & Research Clinical Rheumatology. 2013;27(3):377–389. doi: 10.1016/j.berh.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Mathlouthi N., Al-Bayssari C., Bakour S., Rolain J. M., Chouchani C. Prevalence and emergence of carbapenemases-producing Gram-negative bacteria in Mediterranean basin. Critical Reviews in Microbiology. 2017;43(1):43–61. doi: 10.3109/1040841x.2016.1160867. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z. Protective ventilation for patients without acute respiratory distress syndrome. JAMA. 2013;309(7):p. 654. doi: 10.1001/jama.2012.197014. [DOI] [PubMed] [Google Scholar]
- 5.Mandrawa C. L., Cronin K., Buising K. L., Lorenzo Y. S. P., Waters M. J., Jeremiah C. J. Carbapenemase-producing Klebsiella pneumoniae: a major clinical challenge. Medical Journal of Australia. 2016;204(7):277–278. doi: 10.5694/mja15.00885. [DOI] [PubMed] [Google Scholar]
- 6.Zheng B., Dai Y., Liu Y., et al. Molecular epidemiology and risk factors of carbapenem-resistant Klebsiella pneumoniae infections in Eastern China. Frontiers in Microbiology. 2017;8:p. 1061. doi: 10.3389/fmicb.2017.01061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J., Pan Y., Shen J., Xu Y. The efficacy and safety of tigecycline for the treatment of bloodstream infections: a systematic review and meta-analysis. Annals of Clinical Microbiology and Antimicrobials. 2017;16(1):p. 24. doi: 10.1186/s12941-017-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnham C. D., Leeds J., Nordmann P., O’Grady J., Patel J. Diagnosing antimicrobial resistance. Nature Reviews Microbiology. 2017;15(11):697–703. doi: 10.1038/nrmicro.2017.103. [DOI] [PubMed] [Google Scholar]
- 9.Waters V., Ratjen F. Standard versus biofilm antimicrobial susceptibility testing to guide antibiotic therapy in cystic fibrosis. Cochrane Database of Systematic Reviews. 2017;10:p. CD009528. doi: 10.1002/14651858.CD009528.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z., Smischney N. J., Zhang H., et al. AME evidence series 001–the society for translational medicine: clinical practice guidelines for diagnosis and early identification of sepsis in the hospital. Journal of Thoracic Disease. 2016;8(9):2654–2665. doi: 10.21037/jtd.2016.08.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Opota O., Croxatto A., Prod’hom G., Greub G. Blood culture-based diagnosis of bacteraemia: state of the art. Clinical Microbiology and Infection. 2015;21(4):313–322. doi: 10.1016/j.cmi.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Opota O., Jaton K., Greub G. Microbial diagnosis of bloodstream infection: towards molecular diagnosis directly from blood. Clinical Microbiology and Infection. 2015;21(4):323–331. doi: 10.1016/j.cmi.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Quainoo S., Coolen J. P. M., van Hijum S. A. F. T., et al. Whole-Genome Sequencing of Bacterial Pathogens: the Future of Nosocomial Outbreak Analysis. Clinical Microbiology Reviews. 2017;30(4):1015–1063. doi: 10.1128/cmr.00016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shelburne S. A., Kim J., Munita J. M., et al. Whole-Genome Sequencing Accurately Identifies Resistance to Extended-Spectrum beta-Lactams for Major Gram-Negative Bacterial Pathogens. Clinical Infectious Diseases. 2017;65(5):738–745. doi: 10.1093/cid/cix417. [DOI] [PMC free article] [PubMed] [Google Scholar]