Significance
Histidine biosynthesis is a target for herbicide and antibacterial agents, with imidazoleglycerol-phosphate dehydratase (IGPD) a key enzyme within this pathway. As a result, IGPD is the focus of inhibitor design programs, with several potent herbicides in development. Interestingly, the lead inhibitor is more potent against yeast (Saccharomyces) compared with plant (Arabidopsis) IGPD. To understand this change, we have determined their structure by electron microscopy to reveal a possible mechanism behind differences in inhibitor potency, with Saccharomyces IGPD containing a 24-amino acid insert that forms an extended surface loop that stabilizes an inhibitor binding loop. This study provides insights into the IGPD family and demonstrates the power of using an electron microcopy approach to study inhibitor binding.
Keywords: electron microscopy, structure-based drug design, histidine biosynthesis, IGPD, enzyme inhibition
Abstract
Histidine biosynthesis is an essential process in plants and microorganisms, making it an attractive target for the development of herbicides and antibacterial agents. Imidazoleglycerol-phosphate dehydratase (IGPD), a key enzyme within this pathway, has been biochemically characterized in both Saccharomyces cerevisiae (Sc_IGPD) and Arabidopsis thaliana (At_IGPD). The plant enzyme, having been the focus of in-depth structural analysis as part of an inhibitor development program, has revealed details about the reaction mechanism of IGPD, whereas the yeast enzyme has proven intractable to crystallography studies. The structure–activity relationship of potent triazole-phosphonate inhibitors of IGPD has been determined in both homologs, revealing that the lead inhibitor (C348) is an order of magnitude more potent against Sc_IGPD than At_IGPD; however, the molecular basis of this difference has not been established. Here we have used single-particle electron microscopy (EM) to study structural differences between the At and Sc_IGPD homologs, which could influence the difference in inhibitor potency. The resulting EM maps at ∼3 Å are sufficient to de novo build the protein structure and identify the inhibitor binding site, which has been validated against the crystal structure of the At_IGPD/C348 complex. The structure of Sc_IGPD reveals that a 24-amino acid insertion forms an extended loop region on the enzyme surface that lies adjacent to the active site, forming interactions with the substrate/inhibitor binding loop that may influence inhibitor potency. Overall, this study provides insights into the IGPD family and demonstrates the power of using an EM approach to study inhibitor binding.
Imidazoleglycerol-phosphate dehydratase (IGPD) is an essential enzyme in plants and microorganisms, and thus it is an attractive target for the development of herbicides and antibacterial agents, for which there are presently a limited number of biological targets. IGPD catalyzes the sixth step in histidine biosynthesis, where manganese(II)-dependent dehydration of imidazoleglycerol-phosphate occurs to form imidazoleacetol-phosphate and a concomitant water (1–3). The structure of IGPD has been well studied by X-ray crystallography in several organisms, including the plant Arabidopsis thaliana (At_IGPD) (3), the bacterial and archaeal species including Mycobacterium tuberculosis and Pyrococcus furiosus (4, 5), and the fungus Cryptococcus neoformans (6). These studies show that the core IGPD structure is conserved, comprising a 24-mer with 432 symmetry with two octahedrally coordinated Mn2+ ions in each active site. The manganese ions have been shown to have an important role in binding the substrate and coordinating turnover of the intermediates during the reaction (3, 7–9). Although the majority of the early biochemical studies (8) and various herbicide development programs have focused on IGPD from Saccharomyces cerevisiae as a model system (Sc_IGPD), diffraction from crystals of the enzyme has never been obtained to better than ∼6 Å resolution (10), and the structure has not been determined. Although there is generally high sequence homology between IGPD homologs from different species, especially surrounding the active site, Sc_IGPD has an intriguing sequence difference arising from a 24-amino acid insertion between β2 and β3 that is only found in budding yeast. Interestingly, the most potent lead compound that has emerged from herbicide development, the triazole-phosphonate compound 2-hydroxy-3-(1,2,4-triazol-1-yl) (C348), is an order of magnitude more potent against Sc_IGPD (0.6 nM Ki) (2) than At_IGPD (∼25 nM Ki) (5). Although the mode of binding of C348 has been well studied by X-ray crystallography in At_IGPD, the molecular basis of this difference in potency is unknown. Understanding differences in potency across different species is important if we are to develop more specific and potent inhibitors of IGPD against bacteria, fungi, and plants for future structure-based inhibitor development programs.
The technical advances occurring in the field of cryo-electron microcopy (EM) has resulted in the resolution expectations for single-particle EM reconstructions to be significantly improved with, for example, the number of sub 4 Å structures in the Electron Microscopy Data Bank (EMDB) rising from 32 to 171 between 2012 and 2014 (11). This improvement in resolution has opened the door to a host of research directions gained by single-particle cryo-EM experiments. One such example is rational inhibitor design (12, 13), a field previously driven by X-ray crystallography and NMR data, but that more recently has seen cryo-EM used to determine structures of the TRPA1 ion channel, the proteasome, p97, the 80S ribosome, and β-galactosidase in complex with their respective inhibitors (14–18).
Here we present single-particle cryo-EM structures of At_IGPD and Sc_IGPD in complex with C348 to global resolutions of 3.1 and 3.2 Å, respectively. The core of each complex displays higher local resolution, permitting de novo building and the identification of both the bound inhibitor and coordinating metal ions, with the At structure validated through a corresponding X-ray structure (PDB ID: 5EKW) (5). Importantly, clear structural differences could be seen between the At and Sc IGPD homologs that may account for the significant differences seen between the binding affinities of potent inhibitors.
Results and Discussion
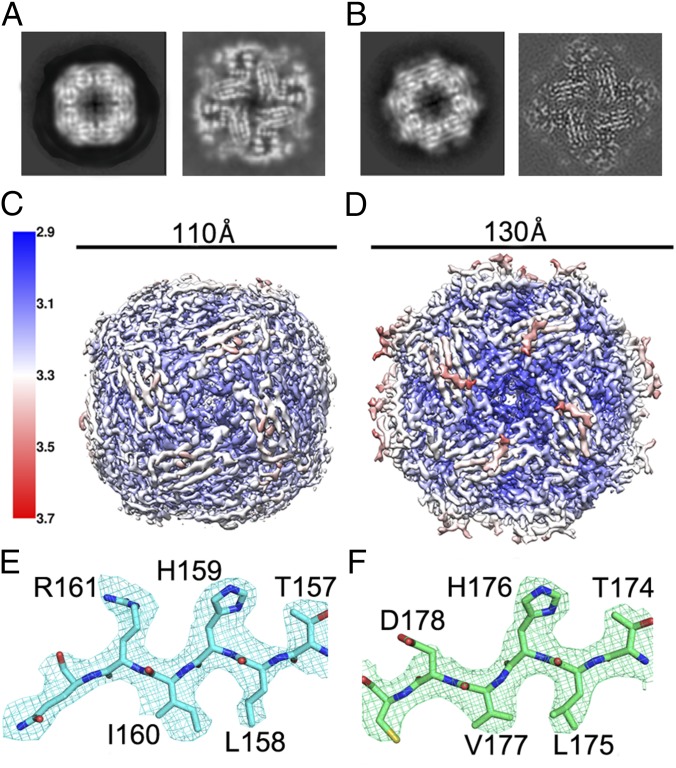
Grids of both Sc_IGPD and At_IGPD in complex with an excess of the racemate of C348 displayed good ice thickness, with the particles having a distinct square shape. For At_IGPD, swarm picking was performed in EMAN2 (19), resulting in 110,977 particles. For Sc_IGPD, autopicking was carried out using RELION, resulting in 365,498 particles. For both enzymes, the resulting 2D classes in RELION provided clear structural detail, indicative of a high-resolution data set (Fig. 1 A and B). 3D refinement was performed on particles belonging to the best 2D classes in RELION (20), with the resolution further improved through the removal of bad particles through iterative 2D and 3D classification and also cutting back the dose to ∼20 e−/Å2 in the case of At_IGPD. The final EM maps have a global resolution of 3.1 and 3.2 Å (Fig. S1), with a higher resolution within the core of the map of 2.9 and 3.1 Å, respectively, for At_IGPD and Sc_IGPD (Fig. 1 C and D). In both structures, the resulting maps showed clear and continuous density for the protein backbone and side chains (Fig. 1 E and F). For the At_IGPD/C348 complex, the EM-derived density map was of sufficient quality to carry out de novo model building in Coot (21, 22), with the resulting model clearly identifying four helical components and eight β-strands within each subunit, which were arranged in a 24-mer with 432 symmetry (Fig. 2 A and B). For Sc_IGPD, the structure was built by docking a homology model based on At_IGPD into the EM map before rebuilding any unconserved amino acid sidechains and fitting the loop regions. The quality of the EM map for both structures permitted the identification of side chains and the ability to trace the sequence with confidence (Fig. 1 E and F).
Fig. 1.
The cryo-EM structure of both At_IGPD and Sc_IGPD. Representative 2D class average (Left) and a slice through from the final 3D reconstruction showing the internal β-strands (Right) for At_IGPD (A) and Sc_IGPD (B). 3D reconstruction colored by local resolution (determined within RELION) for At_IGPD (C) and Sc_IGPD (D) (color key is shown on the right and approximate dimensions above). Example of the density in the EM maps for the equivalent region in At_IGPD (E; blue carbon atoms) and Sc_IGPD (F; green carbon atoms); note the different shape of the density for the Ile (At_IGPD) and Val (Sc_IGPD) residue. In E and F, nitrogen, oxygen, and sulfur atoms are colored blue, red, and yellow, respectively.
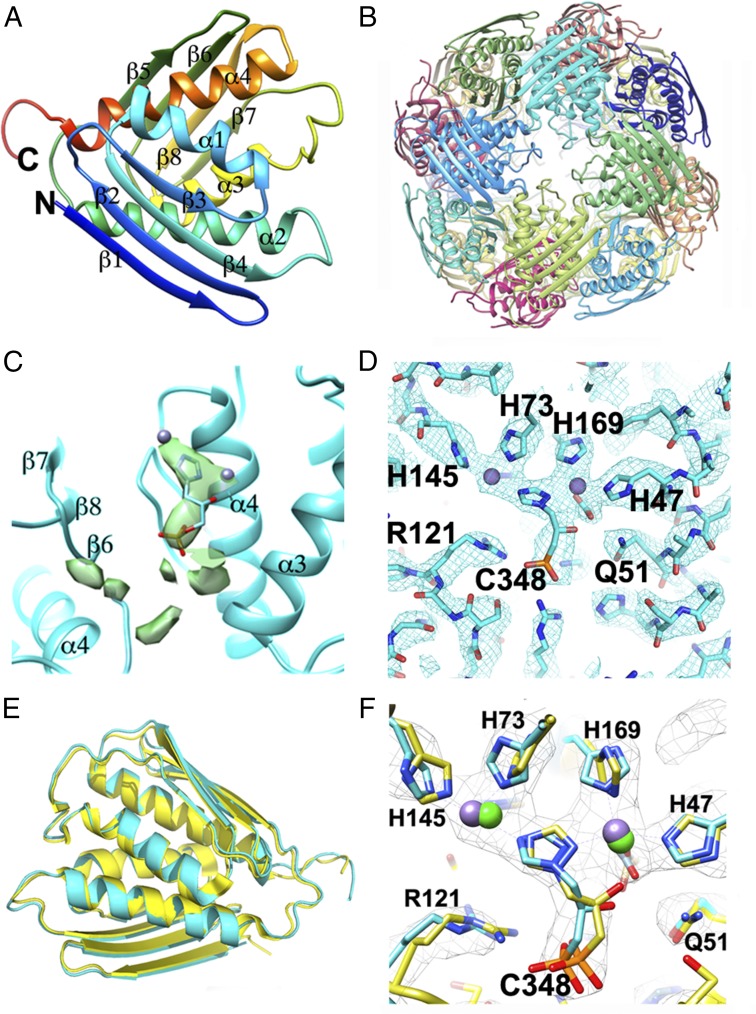
Fig. 2.
Analysis of the inhibitor binding pocket and comparison with a representative crystal structure of At_IGPD. (A) Ribbon diagram showing the final model of the At_IGPD monomer colored blue to red from N to C termini, with secondary structure labeled. (B) Ribbon diagram of the full At_IGPD 24-mer, with each subunit colored differently. (C) Positive density within the difference map (green surface) generated by subtracting the map of the de novo built At_IGPD structure from the At_IGPD EM map. This resulted in strong density only within the active site and flanking region, with no other area of significant density not accounted for by the de novo built model. The inhibitor, C348, is shown as sticks, with blue carbon atoms within the difference map. Note that C348 sits within a subunit interface, with the pocket partly formed by α4 from two different subunits. (D) Fitting of C348 within the active site of IGPD, showing strong density that accommodates both the inhibitor and the two Mn2+ ions (purple spheres). (E) A superposition of the At_IGPD structure as derived from EM (blue) and X-ray crystallography [yellow; PDB ID: 5EKW (5)], shown in ribbon format. The conformation of the secondary structure elements within monomer is largely identical, with only minor conformational differences observed within the loop regions. (F) The inhibitor binding pocket of the EM and X-ray crystallography-derived models, colored as in E with key residues and inhibitor shown in stick format. Mn2+ ions are shown as purple (EM) or green (X-ray) spheres. In all parts, nitrogen, oxygen, and phosphorous are colored blue, red, and orange, respectively.
To identify any regions of unexplained density within the At_IGPD map, which would account for any intrinsically bound ligands, the protein model was filtered to the same resolution as the EM map, and a difference map between the EM map and the At_IGPD model was generated. Two significant areas of positive density were present in the difference map located within the active site of the enzyme, which, based on evidence from previous studies (3, 5, 23), account for the inhibitor and a pair of manganese ions (Fig. 2C). The metal ions were modeled into the two strongest peaks in positive difference density within two clusters of histidine and glutamate residues (His47, His73, His74, His145, His169, His170, Glu173, and Glu77) that octahedrally coordinate each metal ion with positions ∼2.2 Å away. The remaining density accounted for the inhibitor, and although the resolution of the EM map was not sufficient to assign the pose or stereochemistry, the approximate orientation and location of the inhibitor could be modeled (Fig. 2D). The negatively charged phosphonate group of C348 is located within a cavity surrounded by a cluster of positively charged residues: Arg99, Arg121, and Lys177. The lone pairs of the 1,2,4-triazole ring nitrogen atoms of the inhibitor occupy a position close (∼2.6 Å) to both Mn2+ ions, suggesting they could complete the coordination sphere of each metal ion. The docked inhibitor and Mn2+ ions were then subject to refinement in PHENIX (24), using the symmetrized 24-mer.
To assess the validity of the At_IGPD EM model, it was compared with a previously determined 1.1 Å crystal structure of the same complex (PDB ID: 5EKW) (5). The structures were superimposed using GESAMT superpose in CCP4 (25) with a rmsd value of 0.7 Å (Fig. 2E). Conformational differences between the two structures are concentrated within the loop regions (residues Pro131 to Asp139 and Asp190 to Lys201), which could reflect differences in the model building resulting from the lower resolution of the EM map. Comparisons of the inhibitor binding region showed similarities with regard to the positioning of the bound inhibitor and the pattern of coordination around the metal ions (Fig. 2F), with small differences in the side chain position of the coordinating histidine residues (Fig. 2F) accounted for by the lower resolution of the EM structure. In the At_IGPD/C348 crystal structure, the C-terminal 15 residues of each subunit form interactions with the phosphonate group of the inhibitor via the side chains of Ser199 and Lys201. This interaction is also conserved in the complexes of an inactive mutant (E21Q) of At_IGPD2 in complex with substrate (PDB ID: 4MU4) (3), where the position of the loop buries the active site from solvent. Mutations that truncate IGPD at the C terminus, removing the loop, render the enzyme inactive, suggesting the interaction of the C-terminal loop with the active site is critical for activity (5). The C-terminal loop can be seen as a weak feature in the At_IGPD EM map and was not of sufficient quality to permit the unambiguous modeling of this loop region. The weak density likely represents the averaging of multiple positions of the loop on each enzyme subunit on freezing the sample.
Both the crystal structure and EM structure were determined in the presence of a mix of R and S forms of the inhibitor. In the crystal structure, both enantiomers could be interpreted within the electron density map at the resolution obtained, which showed that they bind with mirror-image packing (5). However, the corresponding EM map does not offer this level of detail because of the lower resolution, and therefore we cannot assess the ratio of R and S isomer binding.
To assess the possible role of radiation damage on the pose of the bound inhibitor, the At_IGPD map was generated with a significantly reduced electron dose. This was achieved by reducing the number of frames used for the reconstruction, with the particle number remaining constant. The lowest dose that was useable before significant loss of resolution was ∼12 e−/Å2, which produced an equivalent map of 3.25 Å global resolution. Although the lower dose map had stronger features for some of the negative side chains (Fig. S2), the resulting density about the inhibitor binding domain did not show any significant difference between the high- and low-dose map, with a local resolution in this area of 2.9 Å.
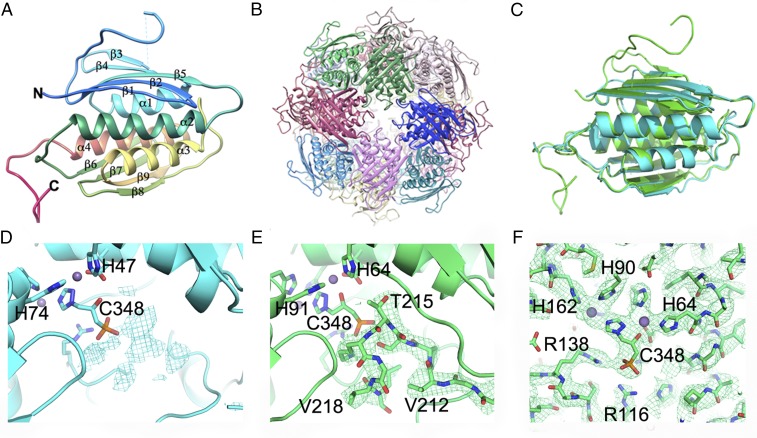
After the successful structure determination of At_IGPD, we also studied Sc_IGPD by single-particle cryo-EM, which has thus far been intractable to structure determination by crystallography because of the diffraction limit of the crystals being ∼6 Å (9). The sequence identity between At_IGPD and Sc_IGPD is 42%, with 54% of residues being similar or identical (Fig. S3). For the 16 residues that make up the inhibitor binding site, 14 are fully conserved with changes in Gln51Ala and Ser54Lys (At_IGPD numbering). The most significant difference between the two homologs is a 24-amino acid insertion between β2 and β3 in Sc_IGPD, which seems to be a unique feature of IGPD homologs from budding yeast. The single-particle EM map of Sc_IGPD at 3.2 Å resolution showed that the enzyme has the same overall architecture made up of a four-α-helical bundle flanked by a four-stranded β-sheet on either side (Fig. 3A). Each monomer is arranged in a 432 symmetrical 24-mer, consistent with other members of the IGPD family (4, 23), with a larger overall size compared with At_IGPD resulting from the additional 24-amino acid insert on the surface of Sc_IGPD situated between β2 and β3 (Fig. 3B). Comparison of the overall fold for both Sc_IGPD and At_IGPD shows close similarity with an rmsd of 0.9 Å (Fig. 3C). In addition to the region between β2 and β3, the C-terminal substrate binding loop, which is also involved in inhibitor binding, is better resolved within the Sc_IGPD structure compared with At_IGPD, permitting all but the final Met residue in the C-terminal loop to be built (Fig. 3 D and E). The inhibitor binding site, as with At_IGPD, shows clear density for the bound inhibitor, surrounding side chain residues and coordinating metal ions, allowing it to be unambiguously placed within the binding site (Fig. 3F).
Fig. 3.
Analysis of the Sc_IGPD structure and comparisons to At_IGPD. (A) A ribbon diagram of the Sc_IGPD monomer colored blue to red from the N to C termini and with secondary structure labeled. (B) Ribbon diagram of the full At_IGPD 24-mer, with each subunit colored differently. (C) Superposition of the At_IGPD (blue) and Sc_IGPD (green) structures, shown in ribbon format, with the additional 24-residue loop region in the later shown above the N-terminal β-sheet. (D) Density within the At_IGPD EM map around the C-terminal domain showing weak density for this region. (E) The C-terminal loop region of Sc_IGPD, which can be unambiguously assigned within the EM map. Both D and E are shown in an equivalent view with the same map contouring level. (F) Fitting of C348 within the active site of IGPD, showing strong density that accommodates the inhibitor, surrounding residues, and the two Mn2+ ions (purple spheres).
Intriguingly, enzyme assays have shown that C348 is a more potent inhibitor of Sc_IGPD (0.6 nM Ki) (2) than of At_IGPD (∼25 nM Ki) (5), but a structural basis for this difference has not been determined. To better understand this difference, the position of the residues that flank the inhibitor biding site were compared in each structure, showing no significant difference (Figs. 2D and 3F and Fig. S4A). All but two residues that form the inhibitor binding site are fully conserved between Sc and At_IGPD, with the changes in Gln51Ala and Ser54Lys being distant from the bound inhibitor (Fig. S4 B and C). The quality of the map density for the inhibitor and the metal ions is also consistent between the two structures. Therefore, with the sequence and structural conservation within the binding site between the At_IGPD and Sc_IGPD, the difference in inhibitor potency does not seem to be directly related to those residues that make up the inhibitor binding pocket. It is interesting to note that the replacement of Ala and Ile for Cys at position 81 and 96, respectively, within Sc_IGPD places two cysteine residues within a distance compatible for a disulphide bond. This is consistent with studies that have shown β-mercaptoethanol to be required for activity of Sc_IGPD (8). However, there is no evidence within the EM-derived map to show the presence of a disulphide bond (Fig. S5).
The EM map clearly indicates the position of the 24-amino acid insertion between β2 and β3 in Sc_IGPD, which lies on the outside face of each subunit, surrounding the spherical particle (Fig. 3 A and B). The EM map for this loop region permits 19 of the 24 amino acids to be modeled (Fig. 4A). However, there is significant ambiguity in the location of Ser39 to Val43, where the EM map density is at its weakest and does not enable the full path of the main chain to be determined. To better define this poorly resolved region, comparative modeling was conducted with Rosetta (26, 27), which showed a consistent positioning of Ser39 to Val43 within the map (Fig. S6). The surface loop emerges from β2 (Gly-25 to Ile-34) and folds back over the surface of the external β-sheet, forming a strip of hydrophobic packing interactions with the surface residues on β2 and β3 (Figs. 3A and 4A). The loop then becomes less defined, with residues Pro36 to Val43 unresolved within the map. The loop then forms a β-strand adjacent to β3 on the external β-sheet before rejoining the structure at β4 (Fig. 4A). This structural arrangement presents a highly charged –EKEAEAVAEQ–motif on the surface loop, the role for which remains to be determined. This additional loop region is a feature specific to the fungal IGPD family, and multiple sequence analysis shows it can vary from 24 (Saccharomyces) to 42 amino acids in, for example, Penicillium and Aspergillus species (Fig. S7). Overall, there is poor conservation across the different homologs, but it’s interesting to note that strong sequence conservation is observed around the negatively charged region that forms the additional β-strand, suggesting this may be a conserved feature across the different fungal species.
Fig. 4.
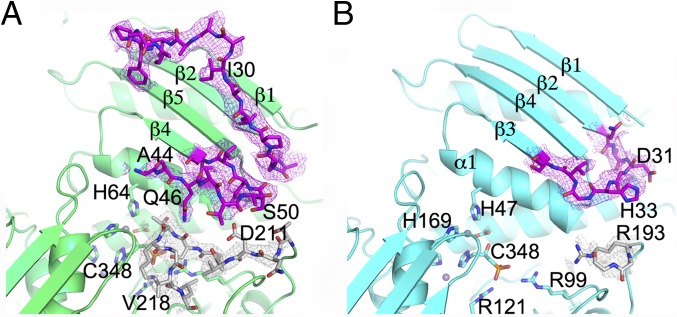
Insertion loop in Sc_IGPD. (A) The 24-residue insertion loop in Sc_IGPD, situated between β2 and β4 colored in purple and the C-terminal inhibitor/substrate binding loop is shown in gray. The insertion (purple) within Sc_IGPD lies next to the C-terminal substrate/inhibitor binding loop (gray), possibly stabilizing the interaction. The 24-residue insert in Sc_IGPD compared with At_IGPD forms an additional β-strand (β3) between A44 and Q46, and therefore there is one extra strand within its external β-sheet. Residues between K38 and A44 are not well resolved within the EM map, and have not been modeled, and some residues in the loop region have been truncated because of poor side chain density. (B) The equivalent view in At_IGPD with a much smaller loop between β2 and β3, which is well defined in the EM map (shown in purple). The C-terminal loop (gray) is more poorly defined and makes no interactions with the loop region between β2 and β3.
In addition to the loop insert between β2 and β3 in Sc_IGPD, a further difference is seen for the C-terminal loop, which has been shown to be important for substrate and inhibitor binding. The C-terminal loop is poorly defined in the At_IGPD EM map, but in Sc_IGPD, the position of the C-terminal loop is better resolved and could be modeled, despite both structures being at equivalent resolutions (Figs. 3 D and E and 4B). Analysis of the position and conformation of the C-terminal loop in Sc_IGPD shows that it sits adjacent to the part of the extended surface loop that forms the additional β-strand on the external β-sheet. Moreover, Gln46, which is at the end of the additional β-strand, is positioned such that it could form a hydrogen bond with the backbone carbonyl of Thr215. Additional interactions could be made between Ser50 and Asp211. Importantly, Gln46 is fully conserved between the different fungal species, and position 50 varies between a Ser and Thr residue, both of which could make hydrogen bond interactions. Therefore, the extended surface loop may act as a “latch” to the active site, stabilizing the C-terminal substrate/inhibitor binding loop; this is why it is better resolved in the Sc_IGPD EM map and perhaps goes some way to explaining the enzyme’s increased binding affinity for C348. The sequence conservation between the different fungal species may point toward this being a conserved aspect of the fungal IGPD family.
Conclusions
IGPD catalyzes the sixth step in histidine biosynthesis and is essential in plants and microorganisms, making it an attractive target for antibacterial and herbicide development. Although X-ray crystallography has been a powerful approach in determining the structure of several homologs of IGPD from, for example, A. thaliana, M. tuberculosis, and P. furiosus, it has been unsuccessful in determining the S. cerevisiae IGPD structure. This is crucial because the yeast homolog contains a significant insert between β2 and β3 and is more sensitive to IGPD inhibitors (2, 5). Here we have used an EM approach to determine the 3D structures of both At_IGPD and Sc_IGPD to investigate the binding of potent inhibitors. We have shown that the difference in inhibitor potency against the different homologs is unlikely to be through direct changes in the inhibitor binding region. Instead, the position of a 24-amino acid insertion in Sc_IGPD shows that it can form an additional β-strand and potentially make hydrogen bonding interactions to the C-terminal loop that would stabilize the binding of the inhibitor and the substrate and intermediates generated during catalysis. The presence of this β-strand in Sc_IGPD and its absence in At_IGPD may explain why the lead compounds are more potent inhibitors of IGPD from S. cerevisiae and why diffraction quality crystals of Sc_IGPD were difficult to obtain. Moreover, through sequence comparisons, this region has been shown to be conserved in fungal species, and although variable in length, those residues that interact with the C-terminal loop and form the additional β-strand are highly conserved, suggesting this feature is common within fungi. This is important both in the design of antifungal therapies and in understanding the basic biology of this important class of proteins.
The use of cryo-EM as a high-resolution structural technique has been shown during the last 5 years with a diverse range of proteins and protein complexes determined to sub 4 Å, with some now breaking the 2 Å barrier (28). Therefore, EM can now be used to directly visualize inhibitor binding and is a powerful approach for those systems intractable to other techniques; for example X-ray crystallography (17). Moreover, by being able to de novo build the structure and identify the bound inhibitor, the EM studies are not dependant on other structural techniques to provide initial models. Structural biology can underpin therapeutic design, and it is becoming clear that with advances in EM, this can also become a complementary approach to developing a toolkit for structure-based drug design.
Methods
Protein Production.
At_IGPD was produced and purified using procedures previously published (3, 5). The gene encoding for S. cerevisiae IGPD (HIS3) was cloned into a pET vector and transformed into a BL21 (DE3) expression strain of Escherichia coli cells (Novagen). Protein expression was performed at 37 °C by induction with 1 mM isopropyl-b-d-thiogalactopyranoside in Luria Broth supplemented with 5 mM MnCl2 at the point of induction. Cells were harvested by centrifugation (5,000 × g), and pellets were frozen at −80 °C. For purification, 2 g cell paste was defrosted in buffer A (50 mM Tris⋅HCl at pH 8.0) and disrupted by ultrasonication (3 × 20 s bursts). Cell debris was removed by centrifugation at 70,000 × g for 10 min. Cell-free extract was applied on a 5-mL DEAE Fast Flow cartridge (GE Healthcare). Elution was performed by a 50-mL gradient of NaCl from 0 to 0.5 M concentration in buffer A. Fractions containing Sc_IGPD were combined, and the enzyme was precipitated by addition of 1.7 M ammonium sulfate (0.75 mL of 4 M ammonium sulfate was added per milliliter of protein solution). The pellet was collected by centrifugation (5 min at 45,000 × g) and then dissolved in 2 mL buffer A and applied on a gel filtration column 16 × 600 HiLoad Superdex200 that had been equilibrated with buffer A + 0.5 M NaCl. Gel filtration was performed at flow rate 1.5 mL/min and 2-mL fractions of the 24-mer of Sc_IGPD (∼500 kDa MW) were combined. Purity of the preparation was about 90%, as estimated by SDS/PAGE, and yield was about 5 mg/g cells. The protein was concentrated in elution buffer to 9 mg/mL and later diluted to 0.5 mg/mL with 50 mM Tris at pH 8 before the inhibitor, C348 (pH adjusted to 7.5 in water), was added to a final concentration of 5 mM.
EM.
Grids of both At_IGPD and Sc_IGPD were prepared by adding 0.5 mg/mL IGPD/inhibitor complex to a 2:2 Quantifoil grid that had been glow discharged for 20 s before use. Grids were blotted and frozen using a Vitrobot Mark IV with 6-s blot time and a force of 6. Data were collected on an FEI Titan Krios microscope operating at 300 kV.
At_IGPD data were collected at 75,000× magnification, corresponding to 1.075 Å/pix sampling on a Falcon II 4 k × 4 k direct detector with 32 frames per micrograph. Data were automatically collected within a defocus range of 1.0 and 3.5 μm, using SerialEM (29). Six hundred and eighty-two micrographs were collected, with those displaying a high degree of astigmatism or defocus removed from data processing. Data were drift corrected in MotionCor (30) (Motioncor2 was not available at the time of data processing) before defocus calculations in CTFFIND4 (31). Particles were picked using the swarm function in EMAN2, with a box size of 180 resulting in 110,977 particles (19). 2D classification within RELION was used to sort the data, with 83,446 particles belonging to well-defined classes, which displayed clear secondary structure (20). Subsequent 3D classification into two classes gave one good-quality class, with the other resulting in a poorer reconstruction with no signs of an alternative conformational state. 3D classification resulted in a final dataset of 55,481 particles.
Sc_IGPD data were collected automatically, using EPU with a Falcon III detector operating in linear mode at a magnification corresponding to 1.065 Å/pix, with a total dose of ∼50 e−/Å2. Then, 2,827 micrographs were collected and drift correction and dose weighting carried out in MotionCor2 and defocus estimation in Gctf (32, 33). Automated particle picking was carried out in RELION 2.0, resulting in 365,498 particles. Particles from the center of 2D lattices were removed via 2D classification, leaving 61,480 particles. Subsequent 3D classification into three classes gave one-good quality class, with the other two resulting in poorer reconstructions with no signs of an alternative conformational state. 3D classification resulted in a final dataset of 11,560 particles.
Model Building, Refinement, and Validation.
For At_IGPD, secondary structure assignment was initially conducted in silico, using Buccaneer (21) using a segmented map that represented only one of the 24 subunits to reduce computation time. Visual inspection of this model in Coot was used to correct for any clear errors in connectivity, assign unbuilt residues, and alter incorrectly built rotamers (22). Independently for additional validation, de novo model building was also carried out within Rosetta3 (26, 27), producing a consistent fold with all core helices well modeled. On completion of the model, clear density could be identified, which was not part of the peptide chain and was designated as inhibitor and metal density. The inhibitor and metals were subsequently fitted by hand, assuming standard bond length and geometry for the metals and bound inhibitor. The completed model containing the inhibitor was then symmetrized into the map, using UCSF Chimera (34), and refined against the map, using PHENIX real space refinement (24, 35). Maps were locally filtered within RELION for model building and map analysis.
For Sc_IGPD, a homology model was generated using Phyre2 1:1 threading based on the EM-derived At_IGPD structure (36). The model was corrected by hand in COOT, and the additional loop region was added, before real space refinement within PHENIX (for refinement statistics, refer to Tables S1 and S2). Missing residues Ser39 to Val43 were modeled using the comparative modeling tool in Rosetta (26, 27) to generate an ensemble of 100 models that were then scored and sorted.
Supplementary Material
Acknowledgments
We thank Syngenta for kindly gifting the C348 compound. This work was supported by an MRC career development grant (G100567 to S.P.M.), and S.R. and D.L.H. are Wellcome Trust-supported PhD students (009752/Z/12/Z) and (102572/B/13/Z). At_IGPD EM data were collected at the Chinese Academy of Sciences (Beijing). This work was also supported by a Biotechnology and Biological Sciences Research Council (BBSRC) Grant (BB/I003703/1 to P.J.B. and D.W.R.). Sc_IGPD EM data were collected at the Astbury Biostructure Laboratory with the assistance of Rebecca Thompson (Leeds) and supported through the Wellcome Trust (108466/Z/15/Z).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: EM-derived maps in this paper have been deposited in the Electron Microscopy Data Bank (EMDB) www.ebi.ac.uk/pdbe/emdb/, (codes EMD-3999 At_IGPD and EMD-4160 Sc_IGPD). Models produced from the EM map in this paper have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6EZJ At_IGPD and 6EZM Sc_IGPD).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708839115/-/DCSupplemental.
References
- 1.Gohda K, Kimura Y, Mori I, Ohta D, Kikuchi T. Theoretical evidence of the existence of a diazafulvene intermediate in the reaction pathway of imidazoleglycerol phosphate dehydratase: Design of a novel and potent heterocycle structure for the inhibitor on the basis of the electronic structure-activity relationship study. Biochim Biophys Acta. 1998;1385:107–114. doi: 10.1016/s0167-4838(98)00049-1. [DOI] [PubMed] [Google Scholar]
- 2.Hawkes TR, et al. Proceedings of the Brighton Crop Protection Conference. Vol 2. British Crop Protection Council; Farnham, UK: 1993. Imidazole glycerol phosphate dehydratase: A herbicide target; pp. 739–744. [Google Scholar]
- 3.Bisson C, et al. Crystal structures reveal that the reaction mechanism of imidazoleglycerol-phosphate dehydratase is controlled by switching Mn(II) coordination. Structure. 2015;23:1236–1245. doi: 10.1016/j.str.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahangar MS, Vyas R, Nasir N, Biswal BK. Structures of native, substrate-bound and inhibited forms of Mycobacterium tuberculosis imidazoleglycerol-phosphate dehydratase. Acta Crystallogr D Biol Crystallogr. 2013;69:2461–2467. doi: 10.1107/S0907444913022579. [DOI] [PubMed] [Google Scholar]
- 5.Bisson C, et al. Mirror-image packing provides a molecular basis for the nanomolar equipotency of enantiomers of an experimental herbicide. Angew Chem Int Ed Engl. 2016;55:13485–13489. doi: 10.1002/anie.201607185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinha SC, et al. Crystal structure of imidazole glycerol-phosphate dehydratase: Duplication of an unusual fold. J Biol Chem. 2004;279:15491–15498. doi: 10.1074/jbc.M312733200. [DOI] [PubMed] [Google Scholar]
- 7.Petersen J, Hawkes TR, Lowe DJ. The metal-binding site of imidazole glycerol phosphate dehydratase; EPR and ENDOR studies of the oxo-vanadyl enzyme. J Biol Inorg Chem. 1997;2:308–319. [Google Scholar]
- 8.Hawkes TR, et al. Purification and characterization of the imidazoleglycerol-phosphate dehydratase of Saccharomyces cerevisiae from recombinant Escherichia coli. Biochem J. 1995;306:385–397. doi: 10.1042/bj3060385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tada S, et al. Insect cell expression of recombinant imidazoleglycerolphosphate dehydratase of Arabidopsis and wheat and inhibition by triazole herbicides. Plant Physiol. 1995;109:153–159. doi: 10.1104/pp.109.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkinson KW, et al. Crystallization and analysis of the subunit assembly and quaternary structure of imidazoleglycerol phosphate dehydratase from Saccharomyces cerevisiae. Acta Crystallogr D Biol Crystallogr. 1995;51:845–847. doi: 10.1107/S0907444995001569. [DOI] [PubMed] [Google Scholar]
- 11.Kühlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 12.Subramaniam S, Earl LA, Falconieri V, Milne JL, Egelman EH. Resolution advances in cryo-EM enable application to drug discovery. Curr Opin Struct Biol. 2016;41:194–202. doi: 10.1016/j.sbi.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rawson S, McPhillie MJ, Johnson RM, Fishwick CWG, Muench SP. The potential use of single-particle electron microscopy as a tool for structure-based inhibitor design. Acta Crystallogr D Struct Biol. 2017;73:534–540. doi: 10.1107/S2059798317004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang F, et al. Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat Chem Biol. 2015;11:518–524. doi: 10.1038/nchembio.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, et al. Structure- and function-based design of Plasmodium-selective proteasome inhibitors. Nature. 2016;530:233–236. doi: 10.1038/nature16936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banerjee S, et al. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong W, et al. Mefloquine targets the Plasmodium falciparum 80S ribosome to inhibit protein synthesis. Nat Microbiol. 2017;2:17031. doi: 10.1038/nmicrobiol.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartesaghi A, et al. 2.2 Å resolution cryo-EM structure of β-galactosidase in complex with a cell-permeant inhibitor. Science. 2015;348:1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glynn SE, et al. Structure and mechanism of imidazoleglycerol-phosphate dehydratase. Structure. 2005;13:1809–1817. doi: 10.1016/j.str.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krissinel E. Enhanced fold recognition using efficient short fragment clustering. J Mol Biochem. 2012;1:76–85. [PMC free article] [PubMed] [Google Scholar]
- 26.DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using rosetta. J Mol Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang RY, et al. De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nat Methods. 2015;12:335–338. doi: 10.1038/nmeth.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merk A, et al. Breaking cryo-EM resolution barriers to facilitate drug discovery. Cell. 2016;165:1698–1707. doi: 10.1016/j.cell.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen EF, et al. UCSF ChimeraA visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Headd JJ, et al. Use of knowledge-based restraints in phenix.refine to improve macromolecular refinement at low resolution. Acta Crystallogr D Biol Crystallogr. 2012;68:381–390. doi: 10.1107/S0907444911047834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.