Significance
The brassinosteroid (BR) receptor BRI1 provides a paradigm for understanding receptor-mediated signaling in plants. Different posttranslational modifications have been implicated in the regulation of BRI1 activity. Here, we show that BR perception promotes BRI1 association with plant U-box E3 ubiquitin ligases PUB12 and PUB13, which in turn directly ubiquitinate BRI1. Importantly, the BRI1 protein abundance and plasma membrane-residence time are increased while the endosomal pool of BRI1 is reduced in the pub12pub13 mutant, indicating that PUB12/PUB13-mediated ubiquitination regulates BRI1 endocytosis and degradation. BRI1 phosphorylates PUB13 on a specific residue to enhance its association with BRI1, suggesting a unique regulatory circuit of phosphorylation-regulated E3 ligase–substrate association. Our study elucidates a mechanism of BRI1 internalization through E3 ubiquitin ligase-mediated ubiquitination.
Keywords: Arabidopsis, BRI1, ubiquitination, E3 ligase, endocytosis
Abstract
Plants largely rely on plasma membrane (PM)-resident receptor-like kinases (RLKs) to sense extracellular and intracellular stimuli and coordinate cell differentiation, growth, and immunity. Several RLKs have been shown to undergo internalization through the endocytic pathway with a poorly understood mechanism. Here, we show that endocytosis and protein abundance of the Arabidopsis brassinosteroid (BR) receptor, BR INSENSITIVE1 (BRI1), are regulated by plant U-box (PUB) E3 ubiquitin ligase PUB12- and PUB13-mediated ubiquitination. BR perception promotes BRI1 ubiquitination and association with PUB12 and PUB13 through phosphorylation at serine 344 residue. Loss of PUB12 and PUB13 results in reduced BRI1 ubiquitination and internalization accompanied with a prolonged BRI1 PM-residence time, indicating that ubiquitination of BRI1 by PUB12 and PUB13 is a key step in BRI1 endocytosis. Our studies provide a molecular link between BRI1 ubiquitination and internalization and reveal a unique mechanism of E3 ligase–substrate association regulated by phosphorylation.
Being sessile and autotrophic organisms, plants live in a relatively constrained niche with constant challenges from environmental stresses while coordinating growth and developmental processes. Plants have evolved a largely expanded collection of plasma membrane (PM)-resident receptor-like kinases (RLKs), many of which have been implicated in sensing external or internal signals and relaying the signaling cascades to various downstream outputs that are central to plant growth, development, and immunity (1, 2). For instance, BRASSINOSTEROID INSENSITIVE1 (BRI1) perceives the polyhydroxylated steroid hormone brassinosteroids (BRs) in regulating growth and development (3), and FLAGELLIN-SENSING2 (FLS2) perceives bacterial flagellin or its conserved 22-aa peptide (flg22) in regulating plant pattern-triggered immunity (PTI) (4). Both BRI1 and FLS2 belong to the leucine-rich repeat (LRR) domain-containing RLK family with more than 200 members in Arabidopsis (1). Despite distinct signaling outputs in growth and immunity, both BRI1 and FLS2 heterodimerize with another LRR RLK, BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1), also known as SOMATIC EMBRYOGENESIS RECEPTOR KINASE3 (SERK3), and other SERK members upon the cognate ligand perception (5–9). BAK1/SERKs are the shared coreceptors of FLS2, BRI1, and several other LRR RLK receptors, including ELONGATION FACTOR-TU (EF-Tu) RECEPTOR (EFR) regulating immunity and ERECTA regulating stomatal patterning (10). FLS2 and BRI1 reside in PM nanoclusters, the majority of which are spatiotemporally separated at the steady state (11).
The activity of LRR RLK receptor complexes is under multilayered positive and negative regulations to fine-tune signaling outputs (1, 12). Protein posttranslational modifications, such as phosphorylation and ubiquitination, play key roles in the activation and attenuation of LRR RLK complexes. BAK1-mediated transphosphorylation with its associated LRR RLK receptors is essential to activate or amplify intracellular signaling (13). FLS2 is ubiquitinated by two closely related plant U-box (PUB) E3 ubiquitin ligases PUB12 and PUB13. Upon flg22 perception, FLS2 associates with PUB12 and PUB13, resulting in ligand-induced FLS2 ubiquitination and degradation to down-regulate FLS2 signaling (14, 15). In addition, PUB13 ubiquitinates LYSIN MOTIF RECEPTOR KINASE 5 (LYK5), an RLK perceiving the fungal cell wall component chitin, and leads to LYK5 degradation and down-regulation of chitin-triggered immune responses (16). BRI1 also undergoes polyubiquitination in planta and it appears that ubiquitination regulates BRI1 internalization and vacuolar targeting (17). However, the E3 ubiquitin ligase that mediates BRI1 ubiquitination remains elusive.
PM-resident receptors often undergo dynamic trafficking between the PM and the endosomes, which is critical for signaling attenuation or activation (18). BRI1 constitutively cycles between the PM and the trans-Golgi network/early endosome (TGN/EE) and is targeted for degradation in the vacuole in a ligand-independent manner (19, 20). In contrast, flg22 induces FLS2 translocation into intracellular mobile vesicles, followed by intracellular degradation (21). It has also been shown that the nonactivated FLS2 constitutively cycles between the PM and the TGN/EE, which is proposed to regulate the abundance of receptor in the PM and maintain a constant pool of signaling receptor (22). Clathrin-mediated endocytosis is the major internalization route of many plant PM proteins including BRI1, FLS2, and the pattern recognition LRR RLK PEP RECEPTOR1 (PEPR1) (23–25). In addition, the internalization of BRI1 requires the functional adaptors AP-2 (26) and TPLATE (27) and the guanine nucleotide exchange factor for ARF GTPase (ARF GEFs) GNOM (19, 23). In contrast to FLS2 and PEPR1 where a functional endocytic machinery is important for downstream signaling activation (24, 25), endocytosis of BRI1 is mainly needed for signaling attenuation as blocking BRI1 internalization resulted in enhanced BR responses (23). Besides endocytosis, secretion and recycling also contribute to maintaining the PM pool of BRI1 required for signaling (28).
Here we demonstrate that E3 ubiquitin ligases PUB12 and PUB13 mediate BRI1 polyubiquitination in vitro and in vivo. BR perception stimulates BRI1–PUB13 association and PUB13-dependent BRI1 ubiquitination. In contrast to BAK1-mediated association and phosphorylation of FLS2 and PUB13 in PTI signaling, BRI1 directly phosphorylates PUB13 at serine 344 residue. BRI1-mediated phosphorylation further regulates the association between PUB13 and BRI1, suggesting an intertwined regulation of protein phosphorylation and ubiquitination in the BRI1–PUB13 complex. Consequently, the BRI1 protein levels were increased in the pub12pub13 mutant, suggesting that loss of ubiquitination interferes with the degradation of BRI1. Consistently, the endocytosis of BRI1 in the pub12pub13 mutant was impaired, which results in a prolonged residence time of BRI1 proteins in PM and BR hypersensitivity. Thus, PUB12- and PUB13-mediated BRI1 ubiquitination is a key step for its subsequent internalization and intracellular degradation.
Results
PUB12 and PUB13 Mediate BRI1 Ubiquitination.
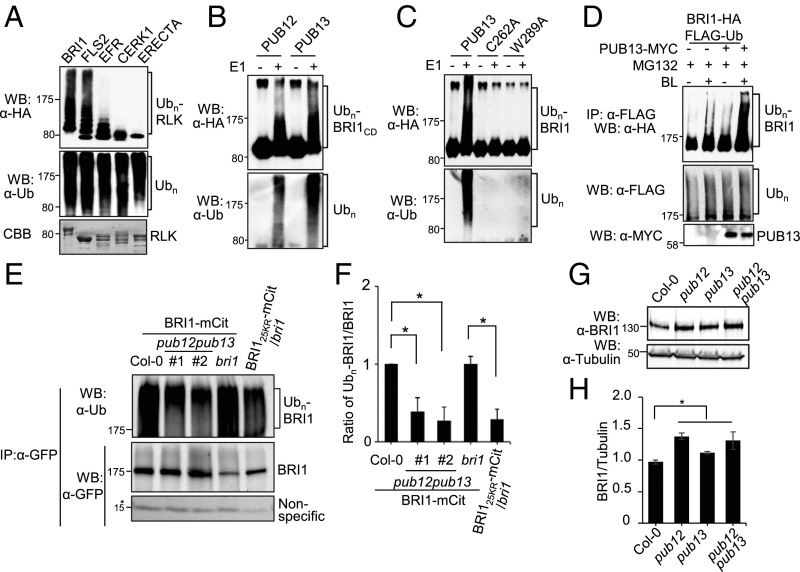
We previously identified PUB13 and its closest homolog, PUB12, as functional E3 ubiquitin ligases mediating RLK FLS2 ubiquitination and subsequent degradation (14, 15). To test whether PUB12 and PUB13 ubiquitinate other RLKs, including BRI1, EFR, ERECTA, and CHITIN ELICITOR RECEPTOR KINASE1 (CERK1), we performed an in vitro ubiquitination assay of maltose binding protein (MBP)-fused cytosolic domain (CD) of RLKs by GST-fused PUB13 in the presence of recombinant proteins AtUBA1 (E1), AtUBC8 (E2), ubiquitin, and ATP. The MBP fusion proteins were tagged with an HA epitope at the carboxyl (C) terminus. Similar to FLS2CD, BRI1CD was strongly ubiquitinated by PUB13 as evidenced by the detection of a ladder-like smear with high-molecular-weight proteins in a Western blot (WB) using an α-HA antibody (Fig. 1A). PUB12 also ubiquitinated BRI1CD in vitro (Fig. 1B). The absence of E1 abolished BRI1CD ubiquitination (Fig. 1B). Mutation of the conserved E2-binding cysteine or tryptophan residue to alanine (C262A or W289A) in the U-box motif of PUB13 diminished its ubiquitination on BRI1CD (Fig. 1C). We further examined whether PUB13 modulated BRI1 ubiquitination in vivo. The in vivo ubiquitination assay was performed using Arabidopsis protoplasts coexpressing C-terminal HA-tagged BRI1 (BRI1-HA), MYC-tagged PUB13 (PUB13-MYC), and N-terminal FLAG-tagged ubiquitin (FLAG-Ub) (29). The cells were treated with the 26S proteasome inhibitor MG132 (Fig. 1D), vacuolar-type H+-ATPase inhibitor bafilomycin A1 (Baf-A1) or vacuolar degradation inhibitor wortmannin (Fig. S1A) to inhibit the potential degradation after ubiquitination. Ubiquitination of BRI1 was detected as a ladder-like smear using an α-HA antibody after immunoprecipitation (IP) with an α-FLAG antibody (Fig. 1D). Apparently, treatment with 1 μM brassinolide (BL), the most active form of BRs, stimulated the formation of ubiquitinated BRI1 in vivo (Fig. 1D). Furthermore, coexpression of PUB13 with BRI1 enhanced BRI1 ubiquitination, in particular after BL treatment (Fig. 1D and Fig. S1A), suggesting that PUB13 mediates BL-stimulated BRI1 ubiquitination. To examine the requirement of PUB12/PUB13 for BRI1 ubiquitination in Arabidopsis seedlings we expressed the functional BRI1-mCitrine fusion under the control of the endogenous BRI1 promoter in the pub12pub13 mutant (Fig. S2 A and B). As a control, we used the previously characterized BRI1-mCitrine–expressing wild-type (Col-0) plants (17). Two independent transgenic lines with BRI1 transcript levels comparable to those of the control BRI1-mCitrine–expressing plants (Fig. S2) were selected. The BRI1-mCitrine/pub12pub13 transgenic plants were phenotypically indistinguishable from BRI1-mCitrine/Col-0 transgenic plants (Fig. S2 A and B). BRI1 was immunoprecipitated using α-GFP antibodies from the BRI1-mCitrine–expressing plants following isolation of microsomal proteins after MG132 treatment and the immunoprecipitates were probed with the P4D1 α-ubiquitin antibodies (Fig. 1E). A high-molecular-weight smear above 170 kDa was detected in immunoprecipitated samples, corresponding to the ubiquitinated BRI1 proteins (Fig. 1E). Significantly, BRI1 was less ubiquitinated in pub12pub13 compared with the wild-type plants measured as relative signal intensity ratio between ubiquitinated BRI1 (Ubn-BRI1) detected by P4D1 α-ubiquitin antibodies and immunoprecipitated BRI1 detected by α-GFP antibodies (Fig. 1F). Notably, the observed reduction in BRI1 ubiquitination in the pub12pub13 mutant resembled the BRI125KR-mCitrine/bri1 plants, in which BRI1 bears 25 lysine (K)-to-arginine (R) mutations, compared with BRI1-mCitrine/bri1 plants (17) (Fig. 1 E and F). However, PUB13 ubiquitinated BRI1K866RCD, an arginine mutation of an in vivo BRI1 ubiquitination site (17), to a level similar to the wild-type BRI1CD, consistent with the notion that multiple sites in BRI1 are ubiquitinated (Fig. S3). In addition, we detected more BRI1 in total protein extracts of each single pub12 and pub13 as well as pub12pub13 double mutant using α-BRI1 antibodies (Fig. 1 G and H). Altogether, our results show that PUB12 and PUB13 mediate BRI1 ubiquitination in vitro and in vivo and regulate BRI1 protein levels.
Fig. 1.
PUB12 and PUB13 ubiquitinate BRI1. (A) Ubiquitination of different RLKs by PUB13 in vitro. The CDs of RLKs, including BRI1, FLS2, EFR, CERK1, and ERECTA, were purified as the MBP-fusion proteins with an HA tag at the C terminus. The ubiquitination of RLKs by GST-fused PUB13 was detected by a WB with an α-HA antibody after an in vitro ubiquitination reaction (Top). The total ubiquitinated proteins, including both RLK and PUB13, were detected by a WB with an α-ubiquitin (α-Ub) antibody (Middle). RLK inputs were indicated by Coomassie Brilliant Blue (CBB) staining. (B) PUB12 and PUB13 ubiquitinate BRI1 in vitro. The ubiquitination of MBP-BRI1CD-HA was carried out by using GST-fused PUB12 or PUB13 as the E3 ligase. The E1 enzyme AtUBA1 was excluded (−) or included (+) in the ubiquitination reaction. (C) The PUB13 residues C262 and W289 are essential for its autoubiquitination and ubiquitination on BRI1. The in vitro ubiquitination of MBP-BRI1CD-HA was performed using GST-PUB13 wild type or mutants as the E3 ligase. (D) PUB13 mediates BRI1 ubiquitination in vivo. Arabidopsis protoplasts were cotransfected with FLAG-tagged ubiquitin (FLAG-Ub), HA-tagged BRI1 (BRI1-HA) and together with a control vector or MYC-tagged PUB13 (PUB13-MYC) and incubated for 10 h followed by treatment with 1 μM BL for 3 h in the presence of 2 μM MG132. The ubiquitinated BRI1 was detected with an α-HA WB after IP with α-FLAG antibody (Top). The total ubiquitinated proteins were detected by an α-FLAG WB (Middle) and PUB13 proteins were detected by an α-MYC WB (Bottom). (E) Reduced BRI1 ubiquitination in pub12pub13. IP was performed using α-GFP antibodies on solubilized microsomal fraction protein extracts from homozygous plants expressing BRI1-mCitrine (mCit) in either wild-type (Col-0) or pub12pub13 mutant background, and from plants expressing BRI1-mCit and BRI125KR-mCit in bri1, and subjected to immunoblotting with α-Ub (P4D1) (Top) or α-GFP (Middle). The asterisk indicates nonspecific signals from the same blot as a loading control. (F) Quantification of BRI1 ubiquitination profiles. Error bars represent SD (n = 3). The asterisks indicate statistical significance by using t test (*P < 0.05). (G) BRI1 protein accumulation in pub12, pub13, and pub12pub13. Total proteins were isolated from 5-d-old seedlings and detected by WB using α-BRI1 antibodies. The protein inputs were equilibrated using α-Tubulin antibodies. (H) Quantification of BRI1 abundance (BRI1/Tubulin) (n = 3 biological replicates).
BR Perception Promotes BRI1 and PUB13 Association.
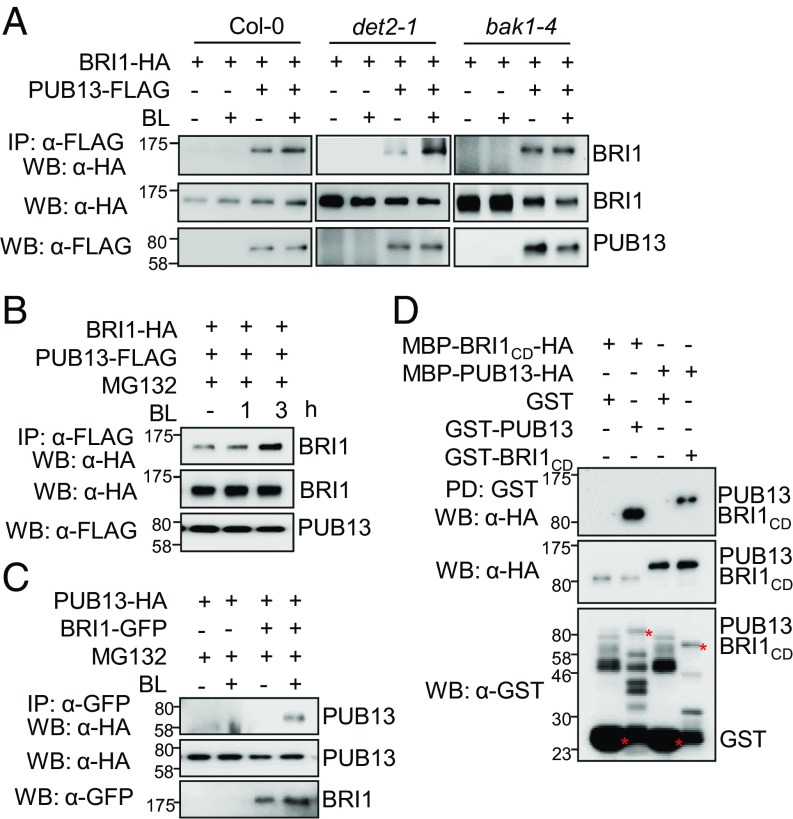
To test the association between BRI1 and PUB13 we performed a co-IP assay in Arabidopsis wild-type protoplasts coexpressing PUB13-FLAG and BRI1-HA. BRI1-HA coimmunoprecipitated with PUB13-FLAG and the BL treatment slightly enhanced BRI1–PUB13 association (Fig. 2A). To avoid the complication of endogenous BRs we tested the BRI1–PUB13 association in the det2-1 mutant, a BR biosynthesis mutant with a reduced level of endogenous BRs (30). The association between BRI1 and PUB13 was markedly increased upon BL treatment in the det2-1 mutant, indicating that BL stimulates BRI1–PUB13 complex formation (Fig. 2A). In addition, the BL-induced BRI1–PUB13 association was not evident in bak1-4, a mutant of the coreceptor BAK1 for BRI1 (Fig. 2A). The BL-induced BRI1–PUB13 association was more apparent in the presence of proteasome inhibitor MG132, vacuolar-type H+-ATPase inhibitor Baf-A1, or vacuolar degradation inhibitor wortmannin in the wild-type plants (Fig. 2B and Fig. S1B), suggesting that BRI1 might be degraded upon association with E3 ligase PUB13. The association of BRI1–PUB13 was further confirmed by co-IP assay using transgenic plants carrying GFP-tagged BRI1 under the control of its endogenous promoter (pBRI1::BRI1-GFP) and HA-tagged PUB13 under the control of the 35S promoter (35S::PUB13-HA) in the presence of MG132. Consistently, BL induced BRI1–PUB13 association in the transgenic plants (Fig. 2C). To examine whether BRI1 directly interacts with PUB13 we performed an in vitro pull-down assay. GST-PUB13 immobilized on glutathione beads specifically pulled down MBP-BRI1CD with a C-terminal HA tag (MBP-BRI1CD-HA) (Fig. 2D). Similarly, GST-BRI1CD pulled down MBP-PUB13-HA (Fig. 2D), suggesting a direct interaction between BRI1CD and PUB13. Thus, the data reveal that BRI1 directly interacts with PUB13, which could be promoted upon BR perception.
Fig. 2.
BRI1 interacts with PUB13. (A) BL application stimulates BRI1–PUB13 association in Arabidopsis protoplasts. Arabidopsis protoplasts from wild-type (Col-0), det2-1, or bak1-4 mutant plants were cotransfected with BRI1-HA and PUB13-FLAG or a control vector and incubated for 10 h followed by 1 μM BL treatment for 3 h. The association of BRI1–PUB13 was detected by an α-HA WB after α-FLAG IP (Top). The protein levels of BRI1 and PUB13 before IP were detected by α-HA (Middle) or α-FLAG (Bottom) WB, respectively. (B) BL-induced BRI1–PUB13 association in the presence of the proteasome inhibitor MG132 in protoplasts. Arabidopsis Col-0 protoplasts were cotransfected with BRI1-HA and PUB13-FLAG and incubated for 10 h. Protoplasts were pretreated with 2 μM MG132 for 2 h before 1 μM BL treatment for 1 or 3 h. The association of BRI1–PUB13 was detected by an α-HA WB after α-FLAG IP. (C) BL treatment induces BRI1–PUB13 association in Arabidopsis plants. Arabidopsis transgenic seedlings carrying 35S::PUB13-HA alone or with pBRI1::BRI1-GFP were used for Co-IP assay. Fourteen-day-old seedlings were pretreated with 50 µM MG132 for 5 h before treatment with 1 µM BL for 3 h. (D) BRI1 interacts with PUB13 in an in vitro pull-down assay. HA-tagged MBP-fusion proteins (MBP-PUB13-HA or MBP-BRI1CD-HA) were incubated with glutathione beads coupled with GST, GST-PUB13, or GST-BRI1CD and the beads were collected and washed for α-HA WB (Top). The protein inputs were determined by α-HA (Middle) or α-GST (Bottom) WB. The position of the corresponding proteins is labeled with an asterisk in α-GST WB.
BRI1 Interacts with and Phosphorylates the ARM Domain of PUB13.
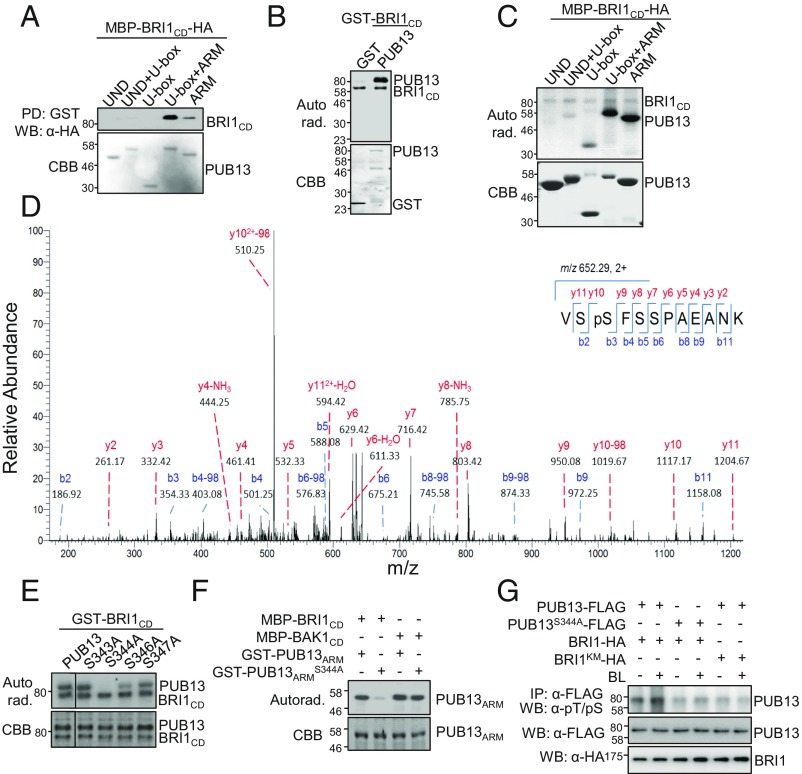
PUB13 contains a U-box N-terminal domain (UND), a U-box domain, and a C-terminal ARM repeat domain (15) (Fig. S4A). It has been suggested that the ARM domain mediates protein–protein interaction whereas the U-box domain confers E3 ubiquitin ligase activity (31). To examine whether the ARM domain, including six ARM repeats and the linker between U-box and ARM repeats, is necessary and sufficient to mediate PUB13 interaction with BRI1, we performed a pull-down assay using GST-fused different truncations of PUB13 with MBP-BRI1CD-HA. As shown in Fig. 3A, MBP-BRI1CD-HA was pulled down by the ARM domain or the U-box-ARM truncation of PUB13, indicating that the ARM domain mediates a direct interaction with BRI1CD. We observed that the interaction of BRI1CD with the U-box-ARM truncation appeared to be stronger than the ARM domain only (Fig. 3A). It is possible that the U-box domain of PUB13 is also involved in the interaction with BRI1 or stabilizes the interaction.
Fig. 3.
BRI1 interacts with and phosphorylates the ARM domain of PUB13. (A) BRI1 interacts with the PUB13 ARM domain in an in vitro pull-down assay. MBP-BRI1CD-HA fusion proteins were incubated with glutathione beads coupled with GST or GST-fused various PUB13 truncated proteins and the beads were collected and washed for an α-HA WB (Top). The protein inputs are indicated by CBB staining (Bottom). (B) GST-BRI1CD phosphorylates GST-PUB13 in vitro. GST-PUB13 proteins were used as substrates and GST-BRI1CD proteins were used as the kinase in an in vitro kinase assay. Phosphorylation was detected by autoradiography (Top), and the protein loading is shown by CBB staining (Bottom). (C) MBP-BRI1CD-HA phosphorylates the ARM domain of PUB13. GST-fused various PUB13 truncated proteins were used as substrates and MBP-BRI1CD-HA as the kinase in an in vitro kinase assay. (D) PUB13S344 is phosphorylated by BRI1. MS analysis identified the phosphorylated S344 residue of PUB13 after an in vitro kinase assay using GST-BRI1CD as the kinase. The graph indicates MS/MS spectrum of a doubly charged peptide of m/z 652.29. The 2+ indicates doubly charged and −98 indicates neutral loss of the phosphate group. The MS/MS spectrum was exported from the raw MS/MS file using Xcalibur and annotated manually. (E) PUB13S344 is required for BRI1-mediated phosphorylation. The serine residues around PUB13S344 (S343S344FS346S347) were individually mutated to alanine (A). GST-tagged wild-type or various mutated PUB13 proteins were subjected to an in vitro kinase assay using GST-BRI1CD as the kinase. (F) PUB13S344 is not required for BAK1-mediated phosphorylation. GST-fused ARM domains of PUB13 or PUB13S344A proteins were subjected to an in vitro kinase assay with MBP-BRI1CD or MBP-BAK1CD as the kinases. (G) BL-induced phosphorylation of PUB13 in vivo. Arabidopsis protoplasts were cotransfected with BRI1-HA and PUB13-FLAG and incubated for 10 h followed by 1 μM BL treatment for 3 h. BRI1KM is a kinase-dead mutant of BRI1. The phosphorylation of PUB13 was detected by WB using α-phospho-threonine/serine (α-pT/pS) after α-FLAG IP (Top). PUB13 and BRI1 were detected by WB using α-FLAG (Middle) or α-HA (Bottom) antibodies, respectively.
As BRI1 is a functional kinase we examined whether it can phosphorylate PUB13 using an in vitro kinase assay with GST-BRI1CD as a kinase and GST-PUB13 as a substrate. In the presence of [32P]-γ-ATP, GST-BRI1CD autophosphorylated and phosphorylated GST-PUB13 (Fig. 3B). We next determined which domain(s) of PUB13 was phosphorylated by BRI1CD. Apparently, BRI1CD strongly phosphorylated the ARM or U-box-ARM truncation yet exhibited little phosphorylation activity toward those truncations lacking the ARM domain, including UND, UND-U-box, or U-box truncation (Fig. 3C). The data indicate that BRI1 mainly phosphorylates the ARM domain of PUB13, consistent with the observation that BRI1 interacts with the PUB13 ARM domain (Fig. 3A). To identify the PUB13 residue(s) phosphorylated by BRI1 we performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis after an in vitro phosphorylation reaction using GST-BRI1CD as a kinase and GST-PUB13ARM as a substrate. The serine 344 (S344) residue, located between the U-box domain and the first ARM repeat of PUB13, was identified as the primary BRI1CD phosphorylation site (Fig. 3D). To reveal the importance of this site in PUB13 phosphorylation by BRI1 we mutagenized S344 to alanine (S344A) and found that GST-BRI1CD was no longer able to phosphorylate GST-PUB13S344A (Fig. 3E). S344 lies in a region with multiple adjacent serine residues, including S343, S346, and S347. However, mutation of these residues in PUB13 to alanine had little effects on PUB13 phosphorylation status by BRI1CD (Fig. 3E), suggesting that S344 is a major BRI1 phosphorylation site in PUB13. S344 is conserved between PUB13 and PUB12 (Fig. S4B) and lies in a loosely conserved region before the first ARM repeat among homologs from different plant species (Fig. S4C). BAK1 also phosphorylates the ARM domain of PUB13 (15). Interestingly, PUB13ARMS344A was able to be phosphorylated by BAK1CD to a level similar to wild-type PUB13ARM protein (Fig. 3F), indicating that BRI1 and BAK1 mediate PUB13 phosphorylation at different residues. Apparently, BAK1 phosphorylates PUB13 at multiple sites in the ARM domain since four different truncations of PUB13ARM were all able to be phosphorylated by BAK1CD (Fig. S5). To detect the BRI1-mediated PUB13 phosphorylation in vivo we coexpressed PUB13-FLAG with BRI1-HA in Arabidopsis protoplasts and examined PUB13 phosphorylation by immunoblotting using α-phosphor-serine or threonine antibody upon IP of PUB13 proteins (Fig. 3G). Phosphorylation of PUB13 was enhanced upon BL treatment, while the S344A mutation blocked BL-induced PUB13 phosphorylation (Fig. 3G). In addition, coexpression of BRI1KM, a kinase-dead mutant bearing a mutation in the ATP-binding lysine residue K911, did not lead to BL-induced PUB13 phosphorylation (Fig. 3G), indicating BL-induced and BRI1 kinase-dependent phosphorylation of the S344 residue in PUB13. To further uncover the role of S344 in PUB13-mediated ubiquitination of BRI1 we introduced the 35S promoter-driven wild-type PUB13 and the phosphorylation mutant PUB13S344A into BRI1-mCitrine/bri1 plants (Fig. S6A) (17). Overexpression of PUB13, but not PUB13S344A, increased the ubiquitination of BRI1 (Fig. S6 B and C). Seemingly, BRI1 ubiquitination was reduced in 35S::PUB13S344A-HA/BRI1-mCitrine/bri1 plants compared with 35S::PUB13-HA/BRI1-mCitrine/bri1 or BRI1-mCitrine/bri1 plants (Fig. S6 B and C). Collectively, the data indicate that PUB13S344 is a major site specifically phosphorylated by BRI1 and this phosphorylation might be involved in PUB13-mediated ubiquitination of BRI1.
BRI1 Phosphorylation Is Required for PUB13-Mediated Ubiquitination and Interaction.
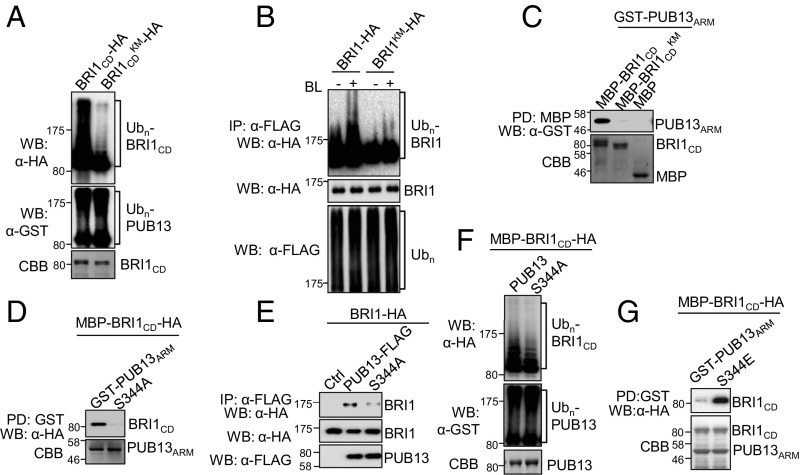
Ubiquitination of receptor tyrosine kinases (RTKs) often depends on their activation through phosphorylation (32, 33). However, ubiquitination of FLS2 by PUB13 is independent of FLS2 phosphorylation (14, 15). We determined whether PUB13-mediated BRI1 ubiquitination requires BRI1 phosphorylation by using a kinase-dead mutant, BRI1KM. An in vitro ubiquitination assay indicated that ubiquitination of MBP-BRI1CDKM-HA by GST-PUB13 was much weaker than wild-type MBP-BRI1CD-HA (Fig. 4A). Consistently, an in vivo ubiquitination assay with the full-length BRI1-HA expressed in protoplasts indicated that BRI1KM was less ubiquitinated after BL treatment than the wild-type BRI1 (Fig. 4B), suggesting that BRI1 phosphorylation is required for PUB13-mediated BRI1 ubiquitination. Notably, the K911 residue in BRI1 is not surface-exposed (34), and it is unlikely to be ubiquitinated by PUB13. Furthermore, BRI1 phosphorylation was essential for the interaction between BRI1 and PUB13, as GST-PUB13ARM was no longer pulled down with MBP-BRI1CDKM (Fig. 4C) and a treatment with the kinase inhibitor K252a reduced both the basal and BL-stimulated BRI1–PUB13 association in Arabidopsis det2-1 protoplasts (Fig. S7). Altogether, our data indicate that BRI1 phosphorylation is required for its interaction with PUB13 and subsequent ubiquitination by PUB13.
Fig. 4.
BRI1 kinase activity is required for PUB13-mediated ubiquitination and interaction. (A) The BRI1 kinase-dead mutant (BRI1KM) blocks ubiquitination mediated by PUB13. The ubiquitination of MBP-BRI1CD-HA or MBP-BRI1CDKM-HA by GST-PUB13 was detected by an α-HA WB after an in vitro ubiquitination reaction. The autoubiquitination of GST-PUB13 was detected by an α-GST WB. The protein loading is indicated by CBB staining. (B) BRI1KM blocks ubiquitination in vivo. Arabidopsis protoplasts were cotransfected with FLAG-Ub and BRI1-HA or BRI1KM-HA and incubated for 10 h followed by treatment with 1 μM BL for 3 h with 2 μM MG132. The ubiquitinated BRI1 was detected with an α-HA WB after an α-FLAG IP (Top). The total ubiquitinated proteins were detected by an α-FLAG WB (Bottom). The BRI1 proteins are shown by an α-HA WB (Middle). (C) BRI1KM blocks interaction with PUB13. GST-PUB13ARM proteins were incubated with amylose beads coupled with MBP, MBP-BRI1CD, or MBP-BRI1CDKM and the beads were collected and washed for an α-GST WB. The protein inputs are shown by CBB staining. (D) PUB13S344A blocks interaction with BRI1. MBP-BRI1CD-HA was incubated with glutathione beads coupled with GST-PUB13ARM or GST-PUB13ARMS344A (S344A) mutant proteins and the beads were collected and washed for an α-HA WB. (E) Reduced interaction between BRI1 and PUB13S344A in Arabidopsis protoplasts. The Co-IP was performed in Col-0 protoplasts expressing BRI1-HA and PUB13-FLAG or PUB13S344A-FLAG. (F) PUB13S344A shows reduced ubiquitination on BRI1. The ubiquitination of MBP-BRI1CD-HA by GST-PUB13 or GST-PUB13S344A was detected by an α-HA WB after an in vitro ubiquitination assay. The protein inputs are shown by CBB staining. (G) PUB13S344E enhances interaction with BRI1. MBP-BRI1CD-HA was incubated with glutathione beads coupled with GST-PUB13ARM or GST-PUB13ARMS344E (S344E) and the beads were collected and washed for an α-HA WB.
As PUB13S344 is the primary phosphorylation site by BRI1, next we investigated whether the phosphorylation status of PUB13 affected its interaction with BRI1. As shown in Fig. 4D, GST-PUB13ARMS344A no longer interacted with MBP-BRI1CD in an in vitro pull-down assay, consistent with the fact that BRI1 kinase activity is required for its interaction with PUB13 (Fig. 4C). Similarly, the association between BRI1-HA and PUB13S344A-FLAG was markedly reduced in Arabidopsis protoplasts compared with BRI1-HA and PUB13-FLAG association (Fig. 4E). PUB13-GFP could be detected in the PM in the transgenic plants under the PUB13 native promotor (Fig. S8A) or with protoplast transient assay (Fig. S8B). Moreover, GST-PUB13S344A had reduced ubiquitination on MBP-BRI1CD-HA as evidenced by the reduced ladder-like smear formation in an in vitro ubiquitination assay (Fig. 4F), consistent with the reduced BRI1 ubiquitination in vivo in 35S::PUB13S344A-HA/BRI1-mCitrine/bri1 plants (Fig. S6). In contrast to PUB13S344A, the phosphomimetic mutant PUB13S344E had a stronger interaction with BRI1CD than the wild-type PUB13 (Fig. 4G). However, PUB13S344E ubiquitinated BRI1CD at a level similar to PUB13 (Fig. S9A), likely due to BRI1CD-mediated phosphorylation of PUB13 in the in vitro ubiquitination assay (Fig. S9B). Thus, BRI1 is both the substrate and activator of E3 ligase PUB13. Taken together, the data suggest a model in which BL-activated BRI1 interacts with and phosphorylates PUB13, which in turn enhances PUB13 activity to ubiquitinate BRI1.
PUB12 and PUB13 Contribute to BR Sensitivity via Controlling BRI1 Endocytosis and Degradation.
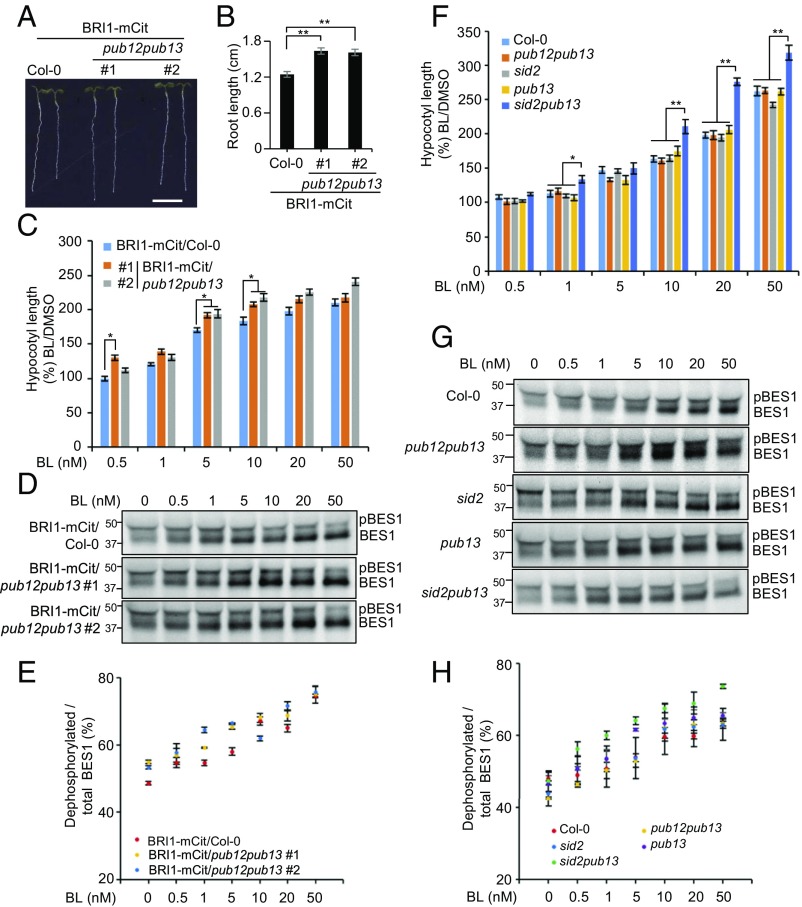
BR signaling intensity is largely determined by the abundance of BRI1 proteins in the PM (23, 28). Given that pub12pub13 plants displayed subsided BRI1 ubiquitination but increased BRI1 protein levels (Fig. 1 E–G), we hypothesize that pub12pub13 plants would lead to BR hypersensitivity, resembling plants overexpressing either wild-type BRI1 (35) or the BRI125KR mutant (17). As expected, BRI1-mCitrine/pub12pub13 plants displayed longer primary roots than the respective control BRI1-mCitrine/Col-0 when grown under light (Fig. 5 A and B and Fig. S2) and they were hypersensitive to exogenous BRs when grown in the presence of increasing concentrations of BL (0.5–50 nM) (Fig. 5C). The activated BR signaling results in dephosphorylation of the transcription factor bri1-Ems-Suppressor1 (BES1), which can be detected as mobility shifts of proteins on immunoblots using a specific α-BES1 antibody (36). Compared with the BRI1-mCitrine/Col-0 plants, BRI1-mCitrine/pub12pub13 showed increased accumulation of dephosphorylated BES1 upon exogenous BL treatment and the difference appeared even without BL treatment (Fig. 5 D and E). We did not observe notable differences in terms of hypocotyl length when pub12pub13 seedlings were grown in the absence or in the presence of BL (Fig. 5F). The pub13 mutant displayed an elevated level of salicylic acid (SA) when grown under certain conditions (37, 38), which may interfere with BR responses. When the SA biosynthesis-deficient mutant sid2 was introduced into pub13 we observed hypersensitivity to BLs in the sid2pub13 mutant, while neither pub13 nor sid2 exhibited difference compared with wild type (Fig. 5F). A further increased accumulation of dephosphorylated BES1 was observed in sid2pub13 compared with sid2, pub13, or wild-type plants (Fig. 5 G and H). Altogether, the pub12pub13 mutation exaggerated the sensitivity of these plants to BRs, indicating the genetic involvement of PUB12 and PUB13 in BR responses.
Fig. 5.
Loss of PUB12 and PUB13 causes BR hypersensitivity. (A and B) Seedlings expressing BRI1-mCitrine (mCit) in wild type (Col-0) and in pub12pub13 (two independent lines) were grown for 5 d and used for quantification of the root length. The representative picture is shown in A and quantification is shown in B (n > 20 seedlings). (Scale bar, 5 mm.) (C and F) Averaged hypocotyl length of 5-d-old seedlings grown in the presence of increasing concentrations of BL. Experiments were done in triplicate. Error bars indicate SE (n = 20). The asterisks indicate statistical significance by using t test (*P < 0.05, **P < 0.01). DMSO is the control for BL. Seedlings used in C and F were subjected to WB analysis of BES1 dephosphorylation using an α-BES1 antibody (D and G). The percentage of dephosphorylated BES1 relative to the total BES1 from three biological repeats is shown in E and H.
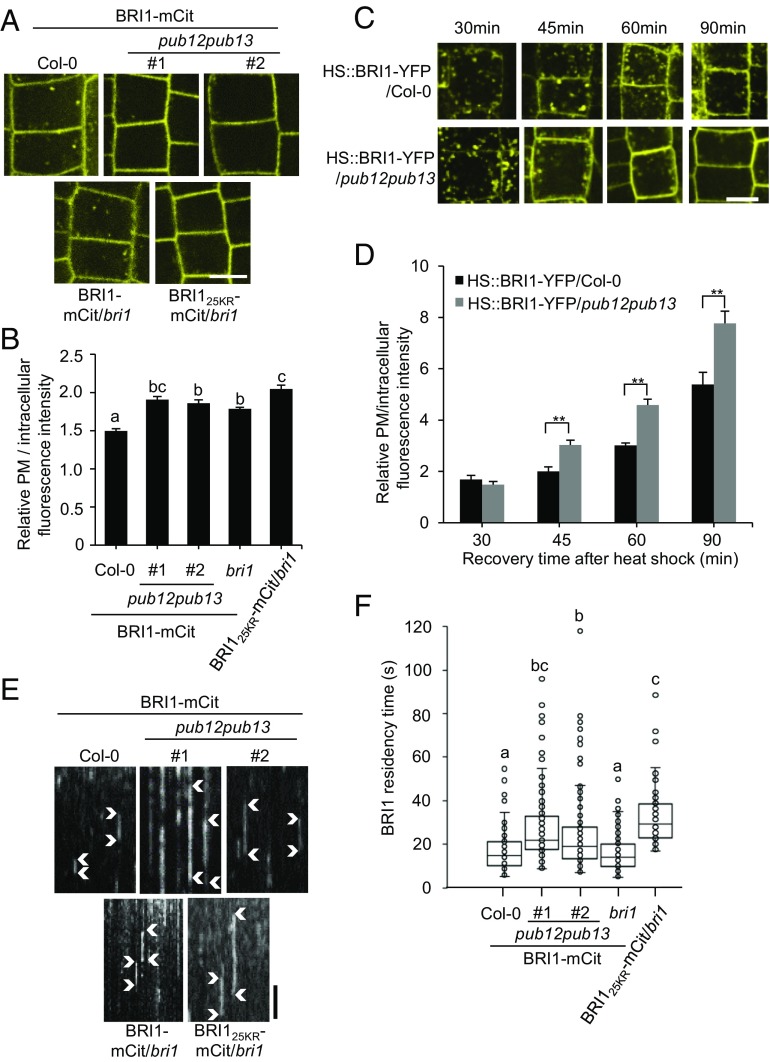
It was previously shown that decreased ubiquitination contributes to the accumulation of BRI1 in the PM because of defective endocytosis and degradation (17). We next examined if the loss of PUB12 and PUB13 would impair BRI1 internalization. Using quantitative microscopy (39), we evaluated the PM pool of BRI1 in the root meristem of 5-d-old BRI1-mCitrine/pub12pub13 seedlings after treatment with cycloheximide (CHX) to inhibit de novo protein synthesis. We observed a significant increase in the relative ratio of the PM to intracellular fluorescence intensity in BRI1-mCitrine/pub12pub13 plants compared with the BRI1-mCitrine/Col-0 control, similarly to the differences observed between BRI125KR-mCitrine/bri1 and BRI1-mCitrine/bri1 plants (Fig. 6 A and B), suggesting a decreased internalization of BRI1 from the PM in pub12pub13. Comparable results were obtained when we crossed the heat shock-inducible BRI1-YFP line (HS::BRI1-YFP) (19) into the pub12pub13 mutant and monitored the YFP fluorescence signal in root meristem epidermal cells in a time course (recovery time) after 1-h heat shock treatment. Recovery of 30 min was sufficient to drive visibly the same amount of newly synthesized BRI1-YFP to the PM in both wild-type and pub12pub13 plants (Fig. 6 C and D). However, the PM/intracellular fluorescence intensity ratio increased more dramatically in HS::BRI1-YFP/pub12pub13 plants than in HS::BRI1-YFP/Col-0 plants after 45-min recovery (Fig. 6 C and D), implying impaired internalization of BRI1 in pub12pub13 plants. We next examined the dynamic localization of BRI1-mCitrine in the PM of the pub12pub13 mutant using variable-angle epifluorescence microscopy (VAEM)/spinning-disk confocal microscopy. Because the root meristem area is uneven, making it difficult to bring the root sufficiently close to the coverslip for imaging, we imaged hypocotyl cells of etiolated seedlings taking advantage of the flat cells. We detected numerous bright dynamic foci when focusing in the focal plane of the PM (Movies S1–S4). The movement of the foci was largely in the z axis, which represented the fusion and fission events of BRI1-mCitrine in the PM. By using kymographs analysis (27, 40) we measured the dwell time of BRI1-mCitrine–labeled foci in the PM in different backgrounds (Fig. 6 E and F). The residence time of BRI1 in the PM of the control wild-type or bri1 plants was on average 16.54 s. We observed a significant increase in the lifetime of BRI1-mCitrine in pub12pub13 (between 24–27 s for different transgenic lines), indicating that endocytosis of BRI1 is significantly inhibited in the pub12pub13 mutant. The residence time of BRI125KR-mCitrine (32.15 s) was also significantly longer than the control (Fig. 6 E and F). In summary, our data suggest that PUB12 and PUB13 are required for BRI1 internalization through direct ubiquitination.
Fig. 6.
PUB12 and PUB13 regulate BRI1 endocytosis. (A) Analysis of BRI1 internalization in the pub12pub13 mutant. BRI1-mCitrine (mCit) and BRI125KR-mCit expressed in either Col-0 or bri1 background were used as controls. Five-day-old seedlings were pretreated with CHX (50 μM) for 1.5 h and root epidermal cells were imaged. (Scale bar, 5 μm.) (B) Relative PM to intracellular fluorescence intensity values were derived from the images shown in A using ImageJ. At least 15 cells from five roots were measured for each line. (C) Analysis of BRI1 internalization by using heat shock-induced BRI1-YFP in Col-0 or in pub12pub13 background. YFP intensity in 5-d-old HS::BRI1-YFP/Col-0 and HS::BRI1-YFP/pub12pub13 root epidermal cells chased for 30, 45, 60, and 90 min after 1 h 37 °C induction was analyzed. (Scale bar, 5 μm.) (D) Relative PM to intracellular fluorescence intensity values were determined from images shown in C with ImageJ. At least 15 cells from five roots were measured for each line. (E) Analysis of BRI1 dwell time in the PM in kymographs obtained from spinning-disk movies of BRI1-mCitrine/Col-0, BRI1-mCit/pub12pub13 (#1 and #2), and BRI125KR-mCit/bri1. Arrowheads mark the dwell-time tracks of the proteins at the PM. Timescale, 20 s. (F) Time of residency in the PM of BRI1-mCit in Col-0 and in pub12pub13. The plot graph was based on kymograph analyses of at least 150 tracks from 10 cells of at least five seedlings. The asterisks in D indicate statistical significance by using t test (**P < 0.01). The different letters in B and F indicate statistically significant difference analyzed with one-way ANOVA followed by Tukey’s test (P < 0.05).
Discussion
The BR receptor BRI1 provides a paradigm to understand RLK-mediated signaling in plants (41). The activity of BRI1 and its interacting partners is regulated by different posttranslational modifications. Phosphorylation between BRI1, coreceptor SERKs, and different downstream receptor-like cytoplasmic kinases plays a key role in the activation of BR signaling (41), whereas ubiquitination has been implicated in BRI1 degradation and signaling attenuation (17). We show here that the PUB E3 ubiquitin ligases PUB12 and PUB13 interact with and ubiquitinate BRI1 directly. BRI1 phosphorylates PUB13, which likely enhances its ubiquitination activity on BRI1. BR perception promotes BRI1 and PUB13 interaction, which also depends on BRI1 and PUB13 phosphorylation (Fig. S10). Importantly, the BRI1 protein abundance and PM-residence time are enhanced and the amount of BRI1 proteins associated with endosomes is reduced in pub12pub13, indicating that PUB12/PUB13-mediated ubiquitination regulates BRI1 endocytosis and intracellular degradation (Fig. S10). Thus, our studies provide a missing link between BRI1 ubiquitination and internalization and reveal a mechanism of E3 ligase–substrate association regulated by phosphorylation.
Plant RLKs are functional counterparts of mammalian RTKs, ubiquitination of which is often regulated by ligand-induced RTK phosphorylation (32, 33). Similarly, the BRI1 kinase-dead mutant is no longer ubiquitinated by PUB13, which is consistent with the previous findings that the ubiquitination of BRI1 requires its kinase activity (17). However, phosphorylation of BRI1 may not have a direct effect on its ubiquitination; instead it may promote or stabilize its interaction with PUB13 since the BRI1 kinase-dead mutant failed to interact with PUB13. Thus, the mode-of-action of BRI1 ubiquitination by PUB13 is different from RTK ubiquitination. The mechanism of PUB13 ubiquitination on BRI1 is also different from its ubiquitination on FLS2. In the case of FLS2, the kinase-dead mutant of FLS2 is normally ubiquitinated by PUB13, which is consistent with the negligible kinase activity of FLS2 (14). However, BAK1-mediated phosphorylation is important for FLS2-PUB13 association and ubiquitination (14, 15). Unlike FLS2, BRI1 is able to directly phosphorylate PUB13 at serine 344 and this phosphorylation mediates the interaction between PUB13 and BRI1. Interestingly, PUB13S344 is not an important phosphorylation residue by BAK1, suggesting the differential phosphorylation of PUB13 may stimulate its association with different RLKs and result in distinct signaling outputs.
In addition, several interacting pairs of PUB and kinase have been implicated in plant development and immunity (14, 16, 42–44). The Brassica ARC1, the homolog of Arabidopsis PUB4, interacts with SRK910, an S-domain-containing RLK, in a phosphorylation-dependent manner and functions in self-incompatibility response (45). PUB1, a negative regulator of infection and nodulation in Medicago truncatula with Sinorhizobium meliloti, interacts with and is phosphorylated by the LYK3 symbiotic receptor, a lysin motif RLK (46). The rice B-lectin domain RLK PID2-mediated phosphorylation on PUB15 and Arabidopsis MPK3-mediated phosphorylation on PUB22 are critical for their E3 ligase activities (47, 48). Interestingly, MPK3 phosphorylates PUB22 on two sites, threonine 62 (T62) located in the U-box domain adjacent to the E2 docking domain and T88 located in a predicated disordered region between the U-box and the first ARM repeat (48). T62 phosphorylation inhibits oligomerization and autoubiquitination of PUB22, thereby stabilizing PUB22, whereas T88 phosphorylation additively stabilizes PUB22 through an unclear mechanism (48). Despite not being conserved, considering the fact that both T88 in PUB22 and S344 in PUB13 are located between the U-box and the first ARM repeat, T88 phosphorylation may also affect PUB22 interaction with its substrate, as the case in which S344 phosphorylation promotes PUB13 association with BRI1. It is possible that a common mechanism may exist in terms of regulating the PUB activities or dimerization through substrate-mediated phosphorylation.
The application of proteasome or vacuolar degradation inhibitors facilitated us to detect a BR-stimulated BRI1–PUB13 interaction and BRI1 ubiquitination. A previous study suggested a ligand-independent ubiquitination of BRI1 in the absence of inhibitors (17). These results suggest that the ubiquitinated BRI1 might be quickly degraded upon BL perception. Consistent with the observations that BRI1 ubiquitination negatively regulates BRI1 function and signaling (17), we revealed that the ubiquitination levels of BRI1 in the pub12pub13 mutant were largely reduced compared with the wild-type plants, leading to the accumulation of BRI1 proteins. Thus, we concluded that PUB12/PUB13-mediated ubiquitination of BRI1 controls the receptor levels. Consistently, we observed a BR hypersensitivity in the BRI1-mCitrine/pub12pub13 plants compared with the BRI1-mCitrine–expressing wild-type plants, which is likely linked to the elevated BRI1 levels in pub12pub13. However, compared with wild type, the pub12pub13 mutant had little altered BR sensitivity, except for the slightly increased levels of dephosphorylated BES1 upon short-term BL application. One explanation is that additional PUB members may play a redundant function with PUB12 and PUB13 in regulating BRI1 signaling. The other explanation could be the pleiotropic phenotype of the pub12pub13 mutant, which may mask the BR sensitivity upon long-term BR application. PUB12 and PUB13 were initially identified as BAK1-interacting E3 ligases that regulate FLS2 stability in flg22 signaling (14, 15). Recently, PUB12 and PUB13 were reported to regulate the abscisic acid (ABA) signaling and the chitin-induced immune responses by modulating the ubiquitination and protein abundance of the key ABA coreceptor ABA-INSENSITIVE1 (ABI1) and the chitin receptor LYK5, respectively (16, 49). In addition, the pub13 mutant displayed certain autoimmune responses, including spontaneous cell death and accumulation of hydrogen peroxide and plant defense hormone SA when grown under certain conditions (37, 38). Interestingly, the sid2pub13 mutant, but not the sid2 or pub13 mutants, displayed hypersensitivity to BRs. Thus, the elevated SA in the pub13 mutant may mask the responses of mutants to BRs. PUB13 also negatively regulates Arabidopsis flowering time and leaf senescence (15, 37), suggesting broad roles of PUB13 in controlling plant immunity, hormone signaling, and growth. BRs are known to cross-talk with the ABA signaling pathways (50–52) and plant immunity (53), thus complicating the interpretation of the pub12pub13 growth phenotypes.
In addition to its well-known role in recognition by the proteasome, ubiquitination is also involved in down-regulation of PM proteins through their internalization followed by targeting to the lysosome/vacuole for degradation (54). It is likely that these processes are also conserved in plants, where many PM transporters and receptors are found ubiquitinated (55), consistent with the characterized role of ubiquitin-mediated endocytosis in the functions of the iron and the boron transporters (56, 57) and the efflux auxin carrier component PIN2 (58, 59). Ubiquitination was also demonstrated for BRI1 and shown to play dual roles in receptor internalization and vacuolar degradation (17). A ubiquitination-compromised BRI125KR mutant accumulated in the PM and displayed reduced endosomal localization (17). Similarly, BRI1 showed an increased PM localization and reduced internalization in the pub12pub13 mutant. By using VAEM and spinning-disk confocal microscopy we also observed a significant increase in the residence time of BRI1 proteins in the PM of pub12pub13 mutant, comparable to the BRI125KR mutant, indicating that endocytosis of BRI1, if not completely abolished, was significantly reduced by the loss of function of PUB12 and PUB13. The effect of BRI125KR mutant on BRI1 ubiquitination was, however, stronger than the effect of pub12pub13 mutant, possibly due to redundancy of the E3 ligases as besides PUB13 and PUB12, a RING finger E3 ligase was identified to interact with BRI1 (60). As BRI1 is endocytosed constitutively both in a ligand-bound and a ligand-free form (19, 23) it remains to be determined if the ubiquitination of the ligand-bound and active BRI1 can specify the interaction with the endocytic machinery and thus distinguish between ligand-dependent and ligand-independent internalization. Although the endocytosis of BRI1 is mediated by the clathrin adaptor AP-2, the internalization of the ligand-bound BRI1 was not completely abolished in the loss of function AP-2 mutants (26). This implies that the internalization of ubiquitinated BRI1 might require different endocytic machinery, similarly to what was shown for the G protein-coupled receptor Protease-activated receptor-1 (PAR1), where AP-2 was required for constitutive but not for agonist-induced PAR1 internalization (61). In yeast and mammals, posttranslational modification by ubiquitin is recognized by the ubiquitin-binding adaptor proteins Epsin/Eps15-like with a function in endocytosis of different PM cargoes (54, 62, 63). These adaptors are able to associate with clathrin, AP-2, and phosphatidylinositol 4,5-bisphosphate via their epsin N-terminal homology (ENTH) domain (56). The same scenario might also be relevant for BRI1, although the plant ENTH-like proteins do not contain conserved ubiquitin-binding motifs (64), suggesting that ubiquitinated BRI1 is recognized in the PM by plant-specific and not-yet-characterized adaptors. Therefore, it remains to be determined if the ubiquitinated BRI1 binds the TARGET OF MYB1 (TOM1)-LIKE family of ubiquitin-binding proteins shown to function in the endocytosis of ubiquitinated plant PM proteins (65).
Materials and Methods
Plant Materials and Growth Conditions.
The following mutant and transgenic Arabidopsis thaliana lines have been described previously: pub12, pub13, pub12pub13, det2-1, bak1-4, sid2, sid2pub13 mutants in Col-0 background (14, 15, 37, 66), pBRI1::BRI1-GFP/Col-0 (67), pBRI1::BRI1-mCitrine/Col-0; pBRI1::BRI1-mCitrine/bri1 and pBRI1::BRI125KR-mCitrine/bri1 (17), and HS::BRI1-YFP/Col-0 (19). Media and growth conditions are described in SI Materials and Methods.
Plasmid Construction and Generation of Transgenic Plants and Crosses.
The constructs of BRI1, BAK1, full-length or truncated PUB13 in the plant expression vector (pHBT) or protein expression vector (pMAL or pGST) were reported previously (14, 15, 66). PUB13 or BRI1 point mutation variants were generated by site-specific mutagenesis with primers listed in Table S1. To construct pCB302-35S::PUB13-HA binary vector for Agrobacterium-mediated transformation in Arabidopsis, PUB13 was subcloned into a modified plant transformation binary vector pCB302 derivative under the control of the 35S promoter with an HA-epitope tag at its C terminus. The pCB302-PUB13::PUB13-GFP transgenic plants were generated by Agrobacterium-mediated transformation and screened with the herbicide BASTA (resistance conferred by the pCB302 binary vector). PUB13 expression was detected by WB with α-HA or GFP antibodies. The homozygous lines were selected based on the survival ratio of T2 and T3 generation plants after BASTA spray. The entry clones pEN-P1P2-BRI1 (no-stop), pEN-P2P3-promoterBRI1, pEN-P4P1-mCitrine were used together with pH7m34GW by multisite Gateway reactions (Life Technologies) to generate pH7m34GW-pBRI1::BRI1-mCitrine bearing a resistance to Hygromycin to allow transformation of the pub12pub13 mutant. The pBRI1::BRI1-mCitrine/pub12pub13 transgenic lines were generated by Agrobacterium-mediated transformation of pub12pub13 with pH7m34GW-pBRI1::BRI1-mCitrine. The 35S::PUB13-HA/BRI1-mCitrine/bri1 lines were generated by Agrobacterium-mediated transformation of homozygous BRI1-mCitrine/bri1 (17) plants with 35S::PUB13-HA and 35S::PUB13S344A-HA constructs. The HS::BRI1-YFP/pub12pub13 plants were generated by crossing HS::BRI1-YFP/Col-0 (19) into the pub12pub13 mutant.
Used primers (Tables S1 and S2), ubiquitination, co-IP, pull-down, phosphorylation, RT-PCR, mass spectrometry, confocal spinning-disk microscopy, and image analyses are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Yanhai Yin, Gregory Vert, Michael Hothorn, and Guo-Liang Wang for providing materials and Daniël Van Damme and Evelien Mylle for help with imaging and data analysis. This work was supported by NSF Grant IOS-1252539 (to P.H.), NIH Grant R01GM097247 and Robert A. Welch Foundation Grant A-1795 (to L.S.), Young Eastern Scholar Grant QD2016035 and Shanghai Sailing Program Grant 17YF1406400 (to J.Z.), Special Research Fund Grant BOF15/24J/048 (to E.R.), and Research Foundation-Flanders joint project with Bulgarian Academy of Sciences Grant VS.025.13N (to E.R. and K.M.). R.K. received a postdoctoral fellowship from the Belgian Science Policy Office.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.Z. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712251115/-/DCSupplemental.
References
- 1.Belkhadir Y, Yang L, Hetzel J, Dangl JL, Chory J. The growth-defense pivot: Crisis management in plants mediated by LRR-RK surface receptors. Trends Biochem Sci. 2014;39:447–456. doi: 10.1016/j.tibs.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shiu SH, Bleecker AB. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 4.Gómez-Gómez L, Boller T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 5.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 6.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 7.Chinchilla D, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 8.Karlova R, et al. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell. 2006;18:626–638. doi: 10.1105/tpc.105.039412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heese A, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma X, Xu G, He P, Shan L. SERKing coreceptors for receptors. Trends Plant Sci. 2016;21:1017–1033. doi: 10.1016/j.tplants.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Bücherl CA, et al. Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. Elife. 2017;6:e25114. doi: 10.7554/eLife.25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol. 2016;16:537–552. doi: 10.1038/nri.2016.77. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 14.Lu D, et al. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science. 2011;332:1439–1442. doi: 10.1126/science.1204903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, et al. The dominant negative ARM domain uncovers multiple functions of PUB13 in Arabidopsis immunity, flowering, and senescence. J Exp Bot. 2015;66:3353–3366. doi: 10.1093/jxb/erv148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao D, et al. Arabidopsis E3 ubiquitin ligase PLANT U-BOX13 (PUB13) regulates chitin receptor LYSIN MOTIF RECEPTOR KINASE5 (LYK5) protein abundance. New Phytol. 2017;214:1646–1656. doi: 10.1111/nph.14472. [DOI] [PubMed] [Google Scholar]
- 17.Martins S, et al. Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat Commun. 2015;6:6151. doi: 10.1038/ncomms7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geldner N, Robatzek S. Plant receptors go endosomal: A moving view on signal transduction. Plant Physiol. 2008;147:1565–1574. doi: 10.1104/pp.108.120287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geldner N, Hyman DL, Wang X, Schumacher K, Chory J. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 2007;21:1598–1602. doi: 10.1101/gad.1561307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russinova E, et al. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1) Plant Cell. 2004;16:3216–3229. doi: 10.1105/tpc.104.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robatzek S, Chinchilla D, Boller T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 2006;20:537–542. doi: 10.1101/gad.366506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beck M, Zhou J, Faulkner C, MacLean D, Robatzek S. Spatio-temporal cellular dynamics of the Arabidopsis flagellin receptor reveal activation status-dependent endosomal sorting. Plant Cell. 2012;24:4205–4219. doi: 10.1105/tpc.112.100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irani NG, et al. Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat Chem Biol. 2012;8:583–589. doi: 10.1038/nchembio.958. [DOI] [PubMed] [Google Scholar]
- 24.Mbengue M, et al. Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc Natl Acad Sci USA. 2016;113:11034–11039. doi: 10.1073/pnas.1606004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortiz-Morea FA, et al. Danger-associated peptide signaling in Arabidopsis requires clathrin. Proc Natl Acad Sci USA. 2016;113:11028–11033. doi: 10.1073/pnas.1605588113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Rubbo S, et al. The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell. 2013;25:2986–2997. doi: 10.1105/tpc.113.114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gadeyne A, et al. The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell. 2014;156:691–704. doi: 10.1016/j.cell.2014.01.039. [DOI] [PubMed] [Google Scholar]
- 28.Luo Y, et al. V-ATPase activity in the TGN/EE is required for exocytosis and recycling in Arabidopsis. Nat Plants. 2015;1:15094. doi: 10.1038/nplants.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou J, He P, Shan L. Ubiquitination of plant immune receptors. Methods Mol Biol. 2014;1209:219–231. doi: 10.1007/978-1-4939-1420-3_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujioka S, et al. The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell. 1997;9:1951–1962. doi: 10.1105/tpc.9.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samuel MA, Salt JN, Shiu SH, Goring DR. Multifunctional arm repeat domains in plants. Int Rev Cytol. 2006;253:1–26. doi: 10.1016/S0074-7696(06)53001-3. [DOI] [PubMed] [Google Scholar]
- 32.Hunter T. The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 33.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bojar D, et al. Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 2014;78:31–43. doi: 10.1111/tpj.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 36.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 37.Li W, et al. The U-Box/ARM E3 ligase PUB13 regulates cell death, defense, and flowering time in Arabidopsis. Plant Physiol. 2012;159:239–250. doi: 10.1104/pp.111.192617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antignani V, et al. Recruitment of PLANT U-BOX13 and the PI4Kβ1/β2 phosphatidylinositol-4 kinases by the small GTPase RabA4B plays important roles during salicylic acid-mediated plant defense signaling in Arabidopsis. Plant Cell. 2015;27:243–261. doi: 10.1105/tpc.114.134262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo Y, Russinova E. Quantitative microscopic analysis of plasma membrane receptor dynamics in living plant cells. Methods Mol Biol. 2017;1564:121–132. doi: 10.1007/978-1-4939-6813-8_10. [DOI] [PubMed] [Google Scholar]
- 40.Dejonghe W, et al. Mitochondrial uncouplers inhibit clathrin-mediated endocytosis largely through cytoplasmic acidification. Nat Commun. 2016;7:11710. doi: 10.1038/ncomms11710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Belkhadir Y, Jaillais Y. The molecular circuitry of brassinosteroid signaling. New Phytol. 2015;206:522–540. doi: 10.1111/nph.13269. [DOI] [PubMed] [Google Scholar]
- 42.Ding X, et al. A rice kinase-protein interaction map. Plant Physiol. 2009;149:1478–1492. doi: 10.1104/pp.108.128298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim M, Cho HS, Kim DM, Lee JH, Pai HS. CHRK1, a chitinase-related receptor-like kinase, interacts with NtPUB4, an armadillo repeat protein, in tobacco. Biochim Biophys Acta. 2003;1651:50–59. doi: 10.1016/s1570-9639(03)00234-6. [DOI] [PubMed] [Google Scholar]
- 44.Samuel MA, et al. Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 2008;147:2084–2095. doi: 10.1104/pp.108.123380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu T, Mazzurco M, Sulaman W, Matias DD, Goring DR. Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc Natl Acad Sci USA. 1998;95:382–387. doi: 10.1073/pnas.95.1.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mbengue M, et al. The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell. 2010;22:3474–3488. doi: 10.1105/tpc.110.075861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, et al. The E3 ligase OsPUB15 interacts with the receptor-like kinase PID2 and regulates plant cell death and innate immunity. BMC Plant Biol. 2015;15:49. doi: 10.1186/s12870-015-0442-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furlan G, et al. Changes in PUB22 ubiquitination modes triggered by MITOGEN-ACTIVATED PROTEIN KINASE3 Dampen the immune response. Plant Cell. 2017;29:726–745. doi: 10.1105/tpc.16.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong L, et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat Commun. 2015;6:8630. doi: 10.1038/ncomms9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Northey JG, et al. Farnesylation mediates brassinosteroid biosynthesis to regulate abscisic acid responses. Nat Plants. 2016;2:16114. doi: 10.1038/nplants.2016.114. [DOI] [PubMed] [Google Scholar]
- 51.Hu Y, Yu D. BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell. 2014;26:4394–4408. doi: 10.1105/tpc.114.130849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai Z, et al. GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:9651–9656. doi: 10.1073/pnas.1316717111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lozano-Durán R, Zipfel C. Trade-off between growth and immunity: Role of brassinosteroids. Trends Plant Sci. 2015;20:12–19. doi: 10.1016/j.tplants.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 54.Piper RC, Dikic I, Lukacs GL. Ubiquitin-dependent sorting in endocytosis. Cold Spring Harb Perspect Biol. 2014;6:a016808. doi: 10.1101/cshperspect.a016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson A, Vert G. Unraveling K63 polyubiquitination networks by sensor-based proteomics. Plant Physiol. 2016;171:1808–1820. doi: 10.1104/pp.16.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barberon M, et al. Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci USA. 2011;108:E450–E458. doi: 10.1073/pnas.1100659108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kasai K, Takano J, Miwa K, Toyoda A, Fujiwara T. High boron-induced ubiquitination regulates vacuolar sorting of the BOR1 borate transporter in Arabidopsis thaliana. J Biol Chem. 2011;286:6175–6183. doi: 10.1074/jbc.M110.184929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leitner J, et al. Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc Natl Acad Sci USA. 2012;109:8322–8327. doi: 10.1073/pnas.1200824109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leitner J, Retzer K, Korbei B, Luschnig C. Dynamics in PIN2 auxin carrier ubiquitylation in gravity-responding Arabidopsis roots. Plant Signal Behav. 2012;7:1271–1273. doi: 10.4161/psb.21715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones AM, et al. Border control–A membrane-linked interactome of Arabidopsis. Science. 2014;344:711–716. doi: 10.1126/science.1251358. [DOI] [PubMed] [Google Scholar]
- 61.Wolfe BL, Marchese A, Trejo J. Ubiquitination differentially regulates clathrin-dependent internalization of protease-activated receptor-1. J Cell Biol. 2007;177:905–916. doi: 10.1083/jcb.200610154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Traub LM. Tickets to ride: Selecting cargo for clathrin-regulated internalization. Nat Rev Mol Cell Biol. 2009;10:583–596. doi: 10.1038/nrm2751. [DOI] [PubMed] [Google Scholar]
- 63.Fortian A, et al. Endocytosis of ubiquitylation-deficient EGFR mutants via clathrin-coated pits is mediated by ubiquitylation. Traffic. 2015;16:1137–1154. doi: 10.1111/tra.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song K, et al. An A/ENTH domain-containing protein functions as an adaptor for clathrin-coated vesicles on the growing cell plate in Arabidopsis root cells. Plant Physiol. 2012;159:1013–1025. doi: 10.1104/pp.112.199380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Korbei B, et al. Arabidopsis TOL proteins act as gatekeepers for vacuolar sorting of PIN2 plasma membrane protein. Curr Biol. 2013;23:2500–2505. doi: 10.1016/j.cub.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 66.Lin W, et al. Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc Natl Acad Sci USA. 2013;110:12114–12119. doi: 10.1073/pnas.1302154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.