Significance
The repair of DNA double-strand breaks (DSBs) entails complex and highly coordinated machinery, the detailed molecular organization of which remains to be fully understood. In this study, the cold-inducible RNA-binding protein (CIRBP) was identified as an active contributor during DSB repair. On DNA damage, CIRBP was found to temporarily accumulate at DNA damage sites through an interaction with poly(ADP ribose) polymerase-1 (PARP-1)-generated poly(ADP-ribose). CIRBP was also shown to modulate association of the MRN (Mre11, Rad50, and NBS1) complex and ATM kinase with chromatin and to reduce the activation of downstream signaling. The complex interactions among CIRBP, PARP-1, ATM kinase, and MRN provide compelling evidence supporting a role for CIRBP in the regulation of DSB repair and genome stability.
Keywords: poly(ADP-ribose) polymerase-1, cold-inducible RNA-binding protein, genome stability, double-strand break responses, homologous recombination
Abstract
Maintenance of genome integrity is critical for both faithful propagation of genetic information and prevention of mutagenesis induced by various DNA damage events. Here we report cold-inducible RNA-binding protein (CIRBP) as a newly identified key regulator in DNA double-strand break (DSB) repair. On DNA damage, CIRBP temporarily accumulates at the damaged regions and is poly(ADP ribosyl)ated by poly(ADP ribose) polymerase-1 (PARP-1). Its dissociation from the sites of damage may depend on its phosphorylation status as mediated by phosphatidylinositol 3-kinase-related kinases. In the absence of CIRBP, cells showed reduced γH2AX, Rad51, and 53BP1 foci formation. Moreover, CIRBP-depleted cells exhibited impaired homologous recombination, impaired nonhomologous end-joining, increased micronuclei formation, and higher sensitivity to gamma irradiation, demonstrating the active involvement of CIRBP in DSB repair. Furthermore, CIRBP depleted cells exhibited defects in DNA damage-induced chromatin association of the MRN complex (Mre11, Rad50, and NBS1) and ATM kinase. CIRBP depletion also reduced phosphorylation of a variety of ATM substrate proteins and thus impaired the DNA damage response. Taken together, these results reveal a previously unrecognized role for CIRBP in DSB repair.
Genome stability is susceptible to continuous challenge by various intrinsic replication errors and extrinsic genotoxic mutagens. Double-strand breaks (DSBs) represent the most deleterious type of DNA damage. If left unattended, DSBs will cause mutations, chromosome aberrations, and genome instability, which could eventually lead to cell death and severe physiological disorders, such as neurodegeneration, immunodeficiency, and cancer (1, 2).
To counteract the detrimental effects of DSBs, mammalian cells have evolved at least two sophisticated genome surveillance mechanisms to recognize, signal, and resolve DNA DSBs (3). Nonhomologous end-joining (NHEJ), which comprises both canonical and alternative pathways, promotes direct ligation of DSB-damaged DNA and is the major, yet more error-prone, DSB repair pathway in the mammalian cell cycle (4, 5). In contrast, homologous recombination (HR), which occurs in the S or G2 phase of the cell cycle, uses a homologous DNA sequence as a template to guide the faithful repair of a DSB (6). The recognition of DSBs by the MRN complex (Mre11, Rad50, and NBS1) results in activation of ATM kinase (7–9), which then initiates DSB signaling by phosphorylating multiple protein targets, such as the histone variant H2AX, checkpoint kinase CHK2, and the chromatin structure regulator KAP-1 (10). In addition, the MRN complex cooperates with several nucleases and helicases to generate a 3′ single-strand DNA (ssDNA) overhang that will pair with the homologous template to restore the damaged DNA (6).
Poly(ADP ribose) polymerase-1 (PARP-1) participates in the repair response to both single-strand breaks and DSBs. Activated PARP-1 synthesizes poly(ADP ribose) (PAR) polymers to modify itself and other protein targets during the early stages of DNA damage responses (DDRs), such as local chromatin relaxation, transcription regulation, and recruitment of DNA repair machinery (11, 12). Emerging evidence indicates that PARP-1 and PAR polymer play active roles in the localization of RNA-binding proteins at the site of DNA damage (13–18). In particular, PARP-1 recruits FUS protein to damaged chromatin, which is a prerequisite for both HR and NHEJ pathways (15, 16). PAR polymer-dependent recruitment of RNA-Binding Motif Protein, X-linked (RBMX) is also necessary for HR, and RMBX may mediate DSB repair by regulating the expression of BRCA2 (19).
In an in-house genome-wide screening effort to find PARP-1 protein targets, the cold-inducible RNA-binding protein (CIRBP; hnRNP A18) was identified as a likely substrate candidate for PARP-1 (20). Recently, several mass spectrometry-based screens also revealed an association between PARP-1 and CIRBP under genotoxic stressed and unstressed conditions (21–24). CIRBP is a stress-responsive protein that is up-regulated on cold treatment or UV irradiation (25, 26). Previous studies have shown that CIRBP plays a protective role during genotoxic stress by binding to the 3′-UTR of stress-responsive transcripts, such as RPA2, TRX, and ATR (27–29). CIRBP also accumulates in cytosolic stress granules, where it participates in repressing the translation of messenger RNA (mRNA) (30). Additional biological activities of CIRBP, including roles in circadian gene regulation (31), spermatogenesis (32), the inflammatory response during hemorrhagic shock and sepsis (33, 34), alternative polyadenylation (35), inhibition of DNA damage-induced apoptosis (36), regulation of telomerase activity (37), and tumorigenesis (38) have also been reported. These findings imply a diverse set of functions for CIRBP under various conditions of cellular stress. This prompted us to further investigate the possible link between CIRBP and PARP-1 activities in the response to DNA damage. Herein we report CIRBP as a previously uncharacterized protein that participates in DSB repair though its interaction with PARP-1.
Results
CIRBP Promotes Genome Stability and DSB Repair.
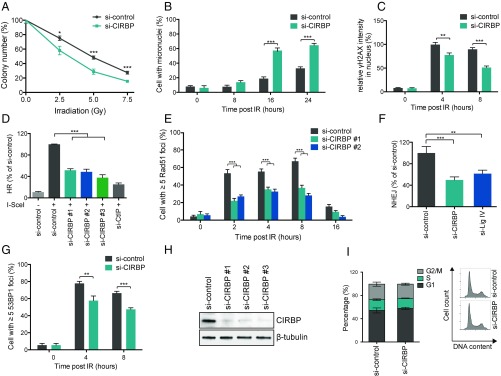
CIRBP is a member of the RNA-binding protein (RBP) family. Many proteins of this family are known to serve different functions in the DDR (39). Therefore, CIRBP was examined to determine whether it also plays a role in maintaining genome stability and modulating cellular sensitivity toward exogenous ionizing radiation (IR). To investigate this hypothesis, U2OS cells were depleted of CIRBP using siRNA and then treated with IR. The effect of CIRBP depletion was assessed based on a colony-formation assay. A control was run in parallel using nontargeting siRNA-treated cells. CIRBP-depleted cells were more sensitive to IR, growing 20% fewer colonies compared with the control (Fig. 1A and SI Appendix, Fig. S1A). Furthermore, CIRBP-depleted cells exhibited a threefold increase in micronuclei formation after IR exposure (Fig. 1B and SI Appendix, Fig. S1B). The appearance of micronuclei has been related to genome instability, because it typically occurs when acentric chromosome fragments or whole chromosomes are not properly segregated to the daughter nuclei during cell division.
Fig. 1.
CIRBP promotes genome stability and DSB repair. (A) CIRBP depletion increases IR sensitivity. U2OS cells were treated with control (nontargeting siRNA) or CIRBP siRNA, followed by exposure to 0, 2.5, 5, and 7.5 Gy of IR. The surviving colonies were visualized with crystal violet, and colony counts were standardized vs. non–IR-treated cells. Data are presented as mean ± SEM from two independent experiments. (B) CIRBP depletion increases IR-induced micronuclei formation. Control or CIRBP siRNA-transfected U2OS cells were treated with 5 Gy of IR and then fixed at the indicated time points. The cells were then stained with DAPI, and the percentage of cells with associated micronuclei were determined. (C) CIRBP depletion decreases nuclear γH2AX intensity. Control or CIRBP siRNA-transfected U2OS cells were treated with 5 Gy of IR, fixed at the indicated time points, and stained with γH2AX antibody. Cells were then counterstained with DAPI, and the nuclear γH2AX intensity was analyzed with Fiji ImageJ. (D) CIRBP knockdown reduces HR efficiency. HR efficiency was determined by monitoring GFP-positive cells by FACS and normalized to nontargeting siRNA-treated cells. (E) CIRBP depletion reduces IR-induced Rad51 foci formation. U2OS cells were treated as in C, and the percentage of cells with five or more Rad51 foci was calculated. (F) CIRBP knockdown reduces NHEJ. NHEJ efficiency was determined by random plasmid incorporation assay. Data collected from CIRBP- or Lig IV-depleted cells were normalized to control siRNA treated cells. (G) CIRBP depletion reduces IR-induced 53BP1 foci formation. U2OS cells were treated as C, and the percentage of cells with five or more 53BP1 foci was calculated. (H) CIRBP knockdown efficiency using three different siRNAs. (I) FACS analysis showing that CIRBP knockdown has no significant effect on cell cycle distribution. Control or CIRBP siRNA-transfected U2OS cells were fixed and stained with propidium iodide and analyzed with FlowJo software. Data are presented as mean ± SD from experiments performed in triplicate. In B–G, data are presented as mean ± SEM from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001, unpaired t test. For micronuclei, γH2AX, Rad51, and 53BP1 foci counting, approximately 400 cells were collected from randomly selected fields.
Because the formation of aberrant chromosomes can result from defects in the repair of DSBs (40, 41), the possibility that CIRBP plays a role in this process was investigated. Localized accumulation of γH2AX (i.e., member X of the phosphorylated H2A histone family) is one of the earliest steps during assembly of the DNA repair machinery at sites of DSB damage and thus is a known biomarker for DSBs (42). CIRBP-depleted cells displayed up to a 50% decrease in the intensity of nuclear γH2AX foci following gamma irradiation (Fig. 1C and SI Appendix, Fig. S1C).
The effect of CIRBP on the efficiency of HR was investigated using a U2OS direct-repeat green fluorescence protein (DR-GFP) cell line containing two nonfunctional GFP genes. The first GFP gene harbors an I-SceI endonuclease cleavage site, whereas the second GFP gene is truncated at both ends. Cleavage of the first gene via I-SceI and subsequent repair via HR using the second gene results in production of GFP that can be monitored by flow cytometry (43). Cells depleted of CtBP-interacting protein (CtIP) using siRNA were used as a positive control to reduce HR efficiency (44), because CtIP is a key component in the HR pathway. As shown in Fig. 1 D and H, knockdown of CIRBP by three different siRNA constructs all led to a 50–60% drop in HR repair.
The involvement of CIRBP in HR repair was further examined by monitoring the formation of Rad51 foci. Rad51 foci reflect the accumulation of recombinase at sites of DNA damage, where it facilitates the pairing of homologous DNA sequences and strand exchange (45). The results shown in Fig. 1E and SI Appendix, Fig. S1D indicate that CIRBP-depleted U2OS cells exhibit an ∼50% reduction in formation of Rad51 foci following IR exposure.
A random plasmid integration assay was next used to assess the effect of CIRBP on DSB repair mediated by NHEJ. A linearized pEGFP-C1 vector was incorporated into the genome of the U2OS cell line to provide resistance to the aminoglycoside antibiotic G418, and colonies grown in the presence of G418 were counted to assess NHEJ efficiency. It was found that CIRBP depletion significantly reduced NHEJ efficiency to a level comparable to that of ligase IV depletion, a known component in the NHEJ pathway. It is also possible that CIRBP depletion affected the noncanonical end-joining pathway (5) (Fig. 1F and SI Appendix, Fig. S1E). In addition, CIRBP depletion also caused a 20% reduction in DNA damage-induced 53BP1 foci formation, which facilitates NHEJ repair of DSBs (46) (Fig. 1G and SI Appendix, Fig. S1F). Importantly, the CIRBP depletion-induced reductions in HR and NHEJ efficiency were not due to perturbation of the cell cycle (Fig. 1I). Taken together, these results demonstrate the importance of CIRBP in the maintenance of genome integrity and strongly suggest an active role for CIRBP in repairing DNA DSBs.
Transient Localization of CIRBP at Regions of Damaged DNA.
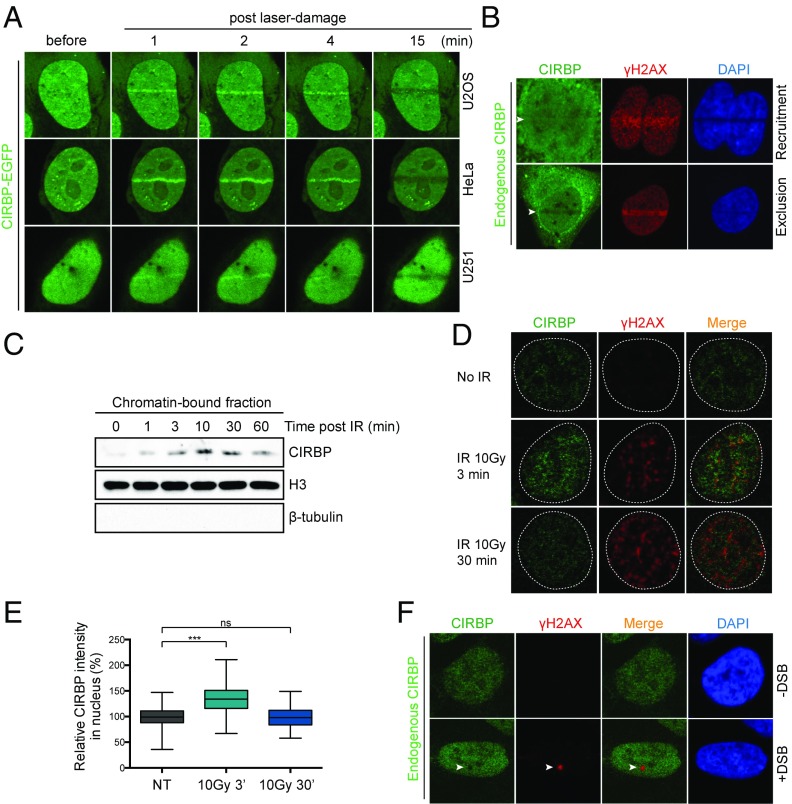
To gain additional insight into how CIRBP contributes to the DDR, experiments were performed to characterize the cellular distribution of CIRBP following DNA damage. Laser microirradiation was carried out to create localized DNA damage. Live-cell imaging revealed rapid accumulation of ectopically expressed EGFP-tagged CIRBP at the sites of laser-induced DNA damage within 1 min, followed by prolonged exclusion. Localization of CIRBP at DNA lesions was not specific to the cell type, with similar phenomena observed in several different cancer cell lines, including U2OS, HeLa, and U251 (Fig. 2A). Immunofluorescence analysis indicated that the endogenous CIRBP accumulated at sites overlapping foci of γH2AX, a marker of DNA damage (Fig. 2B). Consistent with the results of live-cell imaging, an increase in the level of chromatin-bound CIRBP was also noted following treatment of cells with IR (Fig. 2C). This association was time-dependent, showing an initial, transient increase followed by a decrease in total chromatin-bound CIRBP within 1 h of IR treatment. Similarly, when cells were preextracted with CSK buffer containing 0.2% Triton X-100 before immunofluorescence staining, there was a transient enrichment of CIRBP at the detergent-resistant chromatin-bound regions following exposure to IR (Fig. 2 D and E).
Fig. 2.
Transient accumulation and exclusion of CIRBP at DNA damage regions. (A) Transient relocation of CIRBP to DNA damage sites in various cells. U2OS, HeLa, and U251 cells were overexpressed with EGFP-tagged CIRBP and subjected to laser microirradiation assay. (B) Recruitment and exclusion of endogenous CIRBP at laser-microirradiated regions. U2OS cells were laser microirradiated and then immediately fixed and immunoblotted with CIRBP- and γH2AX-specific antibodies. DAPI staining shows the nucleus. (C) Transient accumulation of endogenous CIRBP on chromatin-bound fractions after irradiation. U2OS cells were treated with 5 Gy of IR and harvested at the indicated time points. The chromatin-containing fractionation was isolated and subjected to immunoblotting against indicated antibodies. (D) Representative pictures showing transient accumulation of CIRBP on CSK nonextractable fractions. U2OS cells were untreated or irradiated with 10 Gy of IR, postincubated for the indicated times, and fixed with CSK preextraction. (E) Quantification of the results shown in D. The data are presented as median, interquartile range (boxes), and minimum to maximum percentile range (whiskers) from two independent experiments. ns, no significant difference; ***P < 0.001, Mann–Whitney U test. (F) Representative pictures showing the exclusion of CIRBP from the DSB region. The U2OS-DSB reporter cell line was treated with Shield-1 plus 4-OHT to induce DSBs. The control was treated only with ethanol solvent. At 4 h after induction, cells were fixed and immunoblotted with CIRBP and γH2AX antibodies. DAPI staining shows the nucleus. Results represent at least 20 cells from two independent experiments.
Whether CIRBP exhibits similar behavior at sites of DSBs was probed with a DSB reporter system in which DSBs in an integrated transgene in U2OS cells was created using an inducible mCherry-LacI-FokI endonuclease (47) (Fig. 7A). In this experiment, CIRBP exclusion from DSBs generated by site-specific FokI endonuclease was very apparent (Fig. 2F). Collectively, these results demonstrate that CIRBP localizes briefly to the sites of DNA damage before being excluded from the damaged sites.
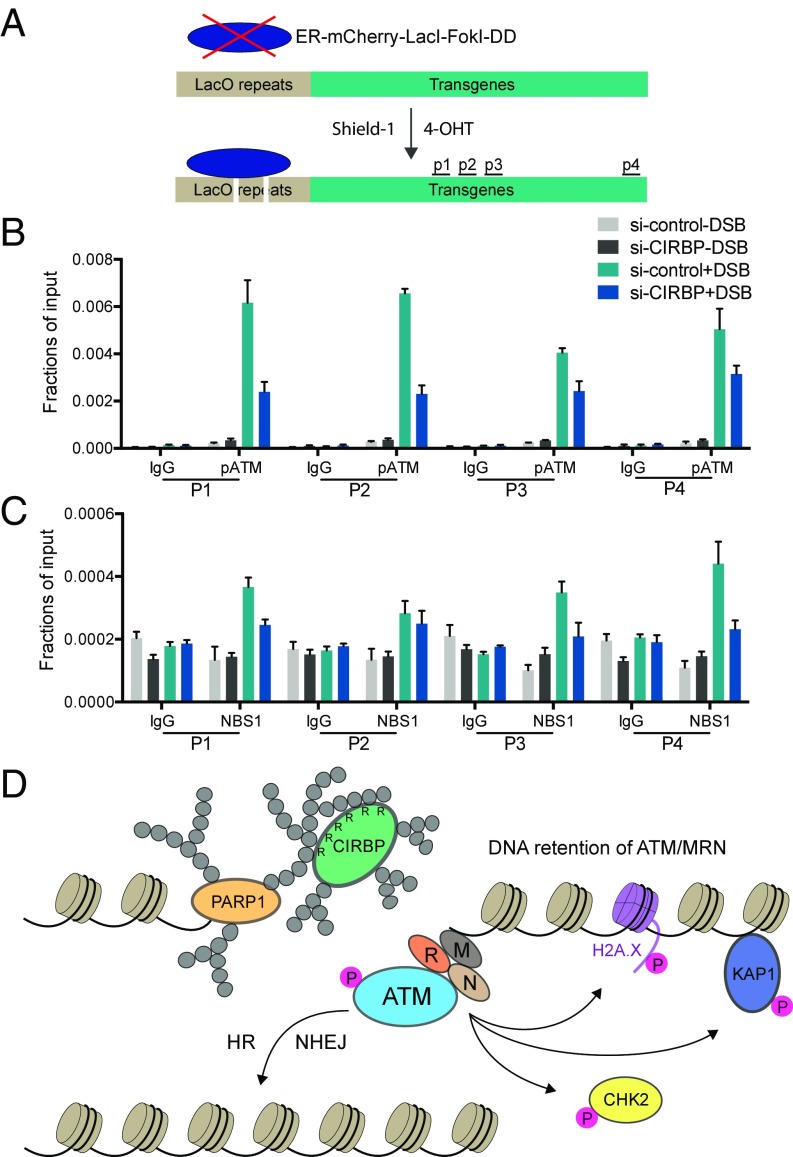
Fig. 7.
CIRBP regulates enrichment of phospho-ATM and NBS1 at DSBs. (A) Scheme adapted from ref. 47 showing inducible DSB generated by mCherry-LacI-FokI at an integrated transgene in U2OS cells. DD, destabilization domain; ER, estrogen receptor; p1–p4, ChIP-qPCR primer targeting sites. (B and C) CIRBP depletion affects the enrichment of phospho-ATM and NBS1 at DSB regions. In the U2OS mCherry-LacI-FokI reporter cell line, ChIP was performed using antibodies against ATM-p1981 and NBS1 in the presence or absence of FokI endonuclease-induced DSBs in cells transfected with CIRBP or control siRNA. The immunoprecipitated DNA was separated from the protein-antibody complex and subjected to qPCR analysis against p1–p4 primers. IgG was used as a negative control. The data are presented as mean ± SD from triplicate experiments. (D) Proposed model for PARP-1–dependent recruitment of CIRBP after induction of DSBs. CIRBP deficiency is proposed to cause defective chromatin binding of ATM, which impairs downstream signaling and DSB repair, eventually leading to genome instability.
PARP-1–Dependent Accumulation and Phosphatidylinositol 3-Kinase–Related Kinase–Dependent Exclusion of CIRBP at DNA-Damaged Regions.
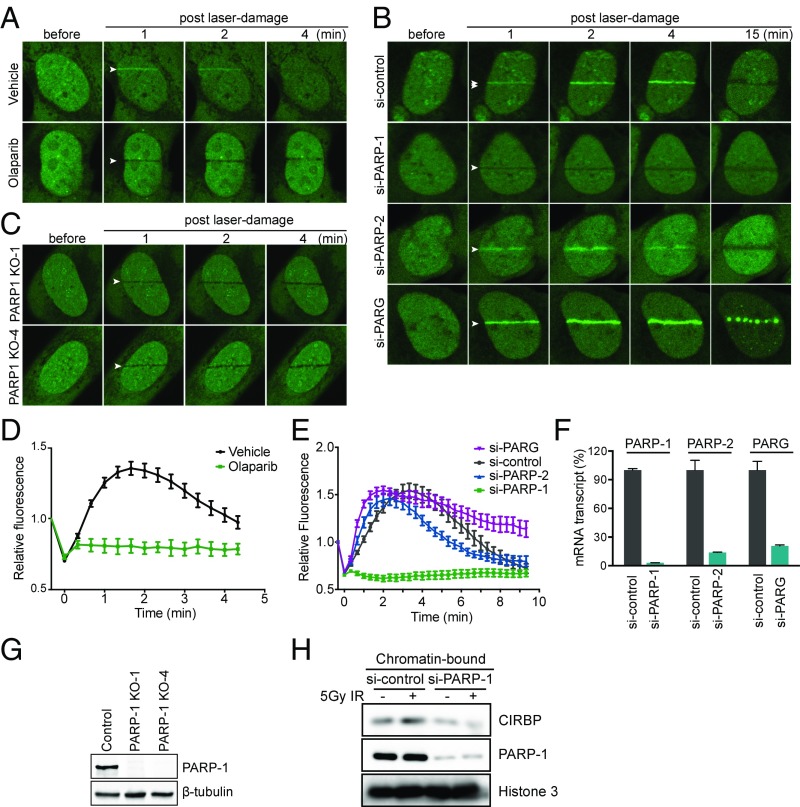
On DNA damage, PARP-1 catalyzes the polymerization of NAD+ to modify itself and other acceptor proteins with long, branched PAR polymers. It has been reported that PAR can serve as a scaffold to help localize DDR proteins at the sites of DNA damage to facilitate break repair (12, 48). Since CIRBP was implicated to interact with PAR polymer and/or to be a potential PARP-1 substrate in several mass spectroscopy-based screening efforts (21–24), the role of PAR polymer in the localization of CIRBP at sites of DNA damage and in DSB repair were examined. As shown in Fig. 3 A and D, when cells were pretreated with the PARP-1 inhibitor olaparib, accumulation of CIRBP at DNA damage sites was prevented, suggesting that PAR polymer is a necessary component of this process.
Fig. 3.
PARP-1–dependent recruitment of CIRBP to DNA damage sites. (A) Inhibition of CIRBP recruitment by PARP-1 inhibitor. U2OS cells overexpressed with EGFP-CIRBP were pretreated with or without 1 μM olaparib for 1 h, followed by laser microirradiation to induce localized DNA damage. The fluorescent intensity at the site of damage was normalized to the intensity before laser microirradiation. (B) PARP-1 and PAR polymer-dependent recruitment of CIRBP. EGFP-CIRBP–overexpressed U2OS cells were transfected separately with nontargeting PARP-1, PARP-2, and PARG siRNA and then subjected to laser microirradiation assay. (C) No CIRBP recruitment in PARP-1 knockout cells. CIRBP-EGFP was overexpressed in PARP-1 knockout U2OS cells and subjected to laser microirradiation assay. (D) Quantification of results in A. The data are presented as mean ± SEM. (E) Quantification of the results in B. (F) Knockdown efficiency of B. Data show individual mRNA levels normalized to nontargeting siRNA-transfected cells. (G) Immunoblotting to check PARP-1 knockout efficiency. (H) PARP-1 dependency of binding of CIRBP to chromatin. U2OS cells transfected with nontargeting or PARP-1 siRNA were treated with or without 5 Gy of IR. Chromatin fractions were isolated and subjected to immunoblotting against antibodies as indicated.
To further characterize the dependence of CIRBP localization on PARP-1 activity, the redistribution of CIRBP in PARP-1, PARP-2, and PAR glycohydrolase (PARG) knockdown cells was investigated. Notably, depletion of PARP-1, but not of PARP-2, significantly reduced CIRBP accumulation at regions of laser-irradiated chromatin, and also led to earlier exclusion of the protein from the irradiated regions. In contrast, depletion of PARG, which is known to degrade PAR polymer, prolonged the localization of CIRBP at the DNA damage sites (Fig. 3 B and F, quantified in Fig. 3E). Two PARP-1 knockout U2OS cells were also generated using the CRISPR-Cas9 system (Fig. 3G). Similar to cells depleted of PARP-1 using siRNA, no accumulation of CIRBP at sites of damaged chromatin was observed in the PARP-1 knockout cells (Fig. 3C). In addition, PARP-1 depletion eliminated the enrichment of endogenous CIRBP with chromatin after IR (Fig. 3H). Taken together, these results demonstrate that CIRBP accumulation at damaged sites depends on the presence of PARP-1 and PAR polymer.
Phosphatidylinositol 3-kinase-related kinases (PIKKs), including ATM, ATR, and DNA-PK, are transducers that coordinate the cellular responses to DNA damage via the phosphorylation of a wide variety of cellular substrates (10). For example, the exclusion of several RNA-binding proteins, such as SAF-A and THRAP3, from microirradiated regions is believed to be mediated by PIKKs (14, 18). This activity appears to be affected by the PIKK phosphorylation of the TQ/SQ motif of the protein substrates (10). A similar result was thus observed with CIRBP when its retention at sites of laser-damaged chromatin appeared to be prolonged from 4 min to 10 min in the presence of KU55933, VE-821, and NU7441, known inhibitors of PIKKs (SI Appendix, Fig. S2 A, B, and D). Furthermore, a T43A/S146A mutant of CIRBP, which targets the likely TQ/SQ equivalent residues, also exhibited prolonged accumulation at sites of laser-damaged chromatin in the absence of PIKK inhibitors compared with the wild-type (WT) CIRBP (SI Appendix, Fig. S2 C and D). Subsequent experiments using U2OS cells overexpressing EGFP-tagged WT CIRBP and the T43A/S146A mutant showed that only the WT was phosphorylated following irradiation (SI Appendix, Fig. S2E). These results support the hypothesis that CIRBP can be phosphorylated by PIKKS likely at T43 and S146, and that such modifications may be the cause of its exclusion from the DNA-damaged regions.
Characterization of the PAR Polymer-Binding Domain in CIRBP.
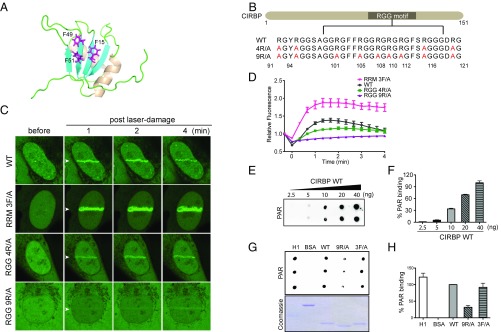
CIRBP contains an Arg-Gly-Gly (RGG)-rich motif at the C-terminus as well as an RNA-recognition motif (RRM) at the N-terminus that is conserved among RNA-binding proteins. Hydrophobic interactions between three highly conserved aromatic residues in the RRM domain and nucleotide bases in RNA are known to be important for the affinity of RNA-binding proteins to RNA (49) (Fig. 4A). Given the structural resemblance between PAR polymer and RNA, the RRM domain in CIRBP may also act as a PAR-binding domain. Moreover, the positively charged arginine residues in the C-terminal RGG motif may interact with negatively charged PAR polymer, further contributing to PAR binding (Fig. 4B).
Fig. 4.
The RGG motif of CIRBP is involved in PAR polymer binding. (A) Key residues in RRM domain that are responsible for RNA binding. The structure was adapted from Protein Data Bank ID code 1X5S. The key RNA-interacting residues, including F15, F49, and F51, are labeled in magenta. In 3F/A mutant, the three F residues were mutated to A. (B) Sequences of CIRBP WT and R to A mutants. In 4R/A mutant, R91, R94, R116, and R121 were mutated to A. In 9R/A mutant, R91, R94, R101, R105, R 108, R110, R112, R116, and R121 were mutated to A. (C) Arginines in RGG motifs mediate the recruitment of CIRBP to laser-microirradiated regions. EGFP-tagged WT, 3F/A, 4R/A, and 9R/A CIRBP were overexpressed in U2OS cells, and the fluorescent signals at laser microirradiation sites were recorded. (D) Quantification of the results in C. (E) Dose-dependent PAR binding of WT CIRBP. Increasing amounts of recombinant His-tagged WT CIRBP (2.5, 5, 10, 20, and 40 ng) were dot-blotted on nitrocellulose membrane and then incubated with PAR polymers. Binding of CIRBP to PAR polymers was visualized by anti-PAR antibody. (F) Quantification of the results in E. The signal intensity of PAR binding of each dot was analyzed using Fuji ImageJ software and normalized to that of 40 ng of membrane-bound CIRBP. Data are presented as mean ± SD from duplicate experiments. (G) In vitro PAR binding of CIRBP WT and mutants. His-tagged CIRBP WT, 3F/A, and 9R/A mutants (8 ng) were used to perform an in vitro PAR-binding assay. Histone 1 (H1) and BSA served as positive and negative controls, respectively. (H) Quantification of the results in G. The intensities of PAR-binding signals of different proteins were normalized to the WT CIRBP. Data are presented as mean ± SD from experiments done in triplicate.
To test which domain is required for PAR polymer binding by CIRBP, several CIRBP mutants targeting the RRM and RGG motifs were constructed. When the three key phenylalanine residues (F15, F49, and F51) in the RRM domain of CIRBP were mutated to alanine, the resulting 3F/A mutant displayed earlier and greater accumulation at loci of DNA damage following laser irradiation compared with the WT. Cells pretreated with the transcription inhibitor 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole to obstruct mRNA synthesis also showed enhanced WT CIRBP accumulation (SI Appendix, Fig. S3). In contrast, Arg-to-Ala mutations in the RGG motif of CIRBP to produce either a 4R/A mutant (i.e., R91, R94, R116, and R121 to A) or a 9R/A mutant (i.e., R91, R94, R101, R105, R108, R110, R112, R116, and R121 to A) exhibited little to no accumulation at the sites of DNA damage (Fig. 4 C and D).
To further establish the ability of the RGG motif to contribute to PAR polymer binding, WT CIRBP as well as the 3F/A and 9R/A mutants were purified and subjected to an in vitro PAR-binding assay. Consistent with the in vivo results, WT CIRBP bound to PAR polymer in a dose-dependent manner (Fig. 4 E and F), whereas the 9R/A mutant showed an around 70% reduction of PAR polymer binding (Fig. 4 G and H). Thus, both the in vitro and in vivo results support the hypothesis that CIRBP binds PAR polymer, and that the RGG motif is important for this binding.
Characterization of the PARylation Sites in CIRBP.
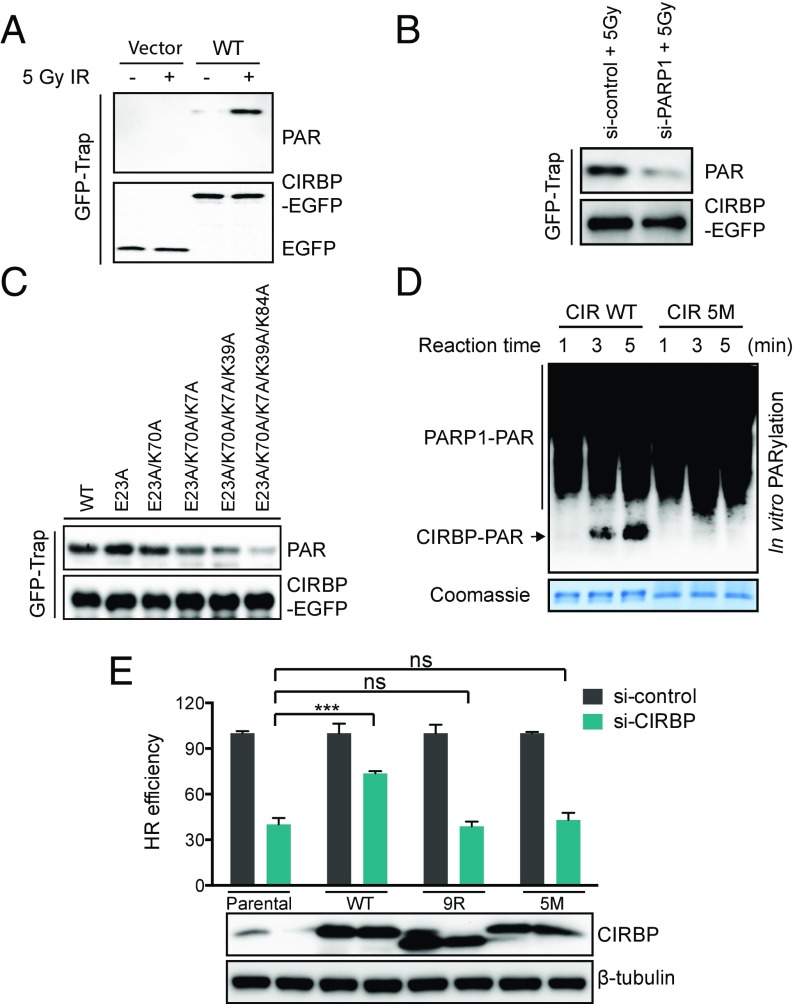
Having demonstrated the ability of CIRBP to bind PAR polymer, an effort was made to determine whether CIRBP is PARylated under conditions of DNA damage. U2OS cells expressing EGFP-tagged CIRBP were prepared, and the tagged protein was pulled down from U2OS cells using GFP trap beads following treatment with IR. As shown in Fig. 5A, cellular irradiation results in covalent modification of CIRBP with PAR polymer. Furthermore, prior knockdown of PARP-1 using siRNA resulted in a near-complete loss of CIRBP PARylation following gamma irradiation (Fig. 5B). These results imply that CIRBP is indeed a protein substrate for PARP-1 and can be PARylated in vivo under DNA damage conditions.
Fig. 5.
PARylation of CIRBP at K7/E23/K39/K70/K84 by PARP-1. (A) CIRBP is PARylated after gamma irradiation. U2OS cells overexpressing EGFP-CIRBP were treated with 5 Gy of gamma IR and then lysed in RIPA buffer. EGFP-tagged CIRBPs were pulled down by GFP-trap beads and subjected to immunoblotting against PAR and GFP antibodies. (B) PARP-1–dependent PARylation of CIRBP. EGFP-tagged CIRBP was overexpressed in U2OS cells subjected to knockdown with different siRNAs, as indicated. After 48 h, cells were treated with 5 Gy of IR and then lysed in RIPA buffer 10 min after IR. Subsequent analysis followed the same procedures as described in A. (C) CIRBP is PARylated by PARP-1 at multiple residues. EGFP-tagged WT and several mutants were overexpressed in 293T cells. The same procedures described in A were used to study their reactions with PARP-1. (D) In vitro PARylation assays show stronger PARylation on WT CIRBP than on the 5M (K7/E23/K39/K70/K84 to A) mutant. The reaction mixtures were either stained with Coomassie blue (Bottom) or immunoblotted with anti-PAR antibody (Top). (E) WT, but not PAR-binding (9R/A) or PARylation (5M) mutant, rescues HR deficiency in CIRBP-depleted cells. U2OS DR-GFP cells stably expressing siRNA-resistant WT or mutant CIRBP were knocked down with control or CIRBP siRNA. The GFP-positive cells in CIRBP-depleted cells were normalized to nontargeting siRNA-treated cells. Data are presented as mean ± SEM from three independent experiments. ns, no significant difference; ***P < 0.001, unpaired t test.
To identify the site of CIRBP PARylation, hydroxylamine was used as a nucleophile to cleave the ester linkage between the modified protein residues, which are likely to be glutamates or aspartates (24), and the PAR polymer. Formation of a hydroxamic acid moiety instead of a carboxylate group at the cleavage site would thus result in a 15-Da mass increase associated with the originally PARylated residue (24). Subsequent liquid chromatography-tandem mass spectrometry analysis revealed Glu23 as the sole site of PARylation (SI Appendix, Fig. S4A); however, a CIRBP E23A mutant could still be PARylated (SI Appendix, Fig. S4B), indicating modification at sites other than Glu and Asp residues. Besides Glu and Asp, Lys is another residue known to be modified by PARP-1 (50). Alignment of CIRBP from several species led to the identification of a number of highly conserved lysine residues (K7, K28, K39, K61, K70, and K84) in CIRBP as potential PARylation sites (SI Appendix, Fig. S4C). These lysine residues were mutated to Ala, and the PARylation levels of various combination mutants were visualized by Western blot analysis. Mutation of five candidate residues—K7A, E23A, K39A, K70A and K84A—to produce a CIRBP 5M mutant resulted in the near-complete loss of PARylation compared with the WT protein (Fig. 5C). Consistent with the in vivo results, an in vitro PARylation assay also supported K7, E23, K39, K70, and K84 as residues important for PARylation (Fig. 5D).
The impact of these Lys-to-Ala mutations on the accumulation of CIRBP at the laser-microirradiated regions was also examined. The CIRBP 5M construct was found to behave similarly to the WT protein (SI Appendix, Fig. S4 D and E). This observation suggested that impaired PARylation of CIRBP 5M does not affect its ability to accumulate at the laser-microirradiated regions. To gain more information about the effect of CIRBP binding to PAR polymer and PARylation of CIRBP on DSB repair, siRNA-resistant WT, 5M mutant, and 9R/A mutant CIRBP were stably transduced separately into endogenous CIRBP-depleted HR reporter cells using siRNA. While the WT CIRBP rescued the HR repair defects caused by CIRBP depletion, the PARylation mutant (5M) and recruitment mutant (9R/A) did not (Fig. 5E). These results indicate that both PARP-1–dependent PARylation of CIRBP and binding of CIRBP to PAR polymer are critical to promoting HR.
Effect of CIRBP on the Cellular Level of the R-Loop.
Some RNA processing factors have been identified to facilitate the resolution of dexterous DNA/RNA hybrids (R-loop) during transcription and RNA processing and promote genome stability (39). To investigate whether CIRBP has anything to do with formation or removal of the R-loop, we knocked down RNase H1, which specifically degrades the RNA component of the DNA/RNA hybrids to facilitate R-loop formation. The results showed that RNase H1 depletion does not affect the accumulation of CIRBP at the regions of microirradiated chromatin (SI Appendix, Fig. S5 A and B), where R-loop formation is elevated (14). In addition, depletion of CIRBP did not affect the level of R-loop formation (SI Appendix, Fig. S5 C and D), whereas depletion of RNase H1 increased R-loop formation in the nucleolus by twofold.
CIRBP Promotes DNA Damage-Induced Chromatin Association of MRN and ATM.
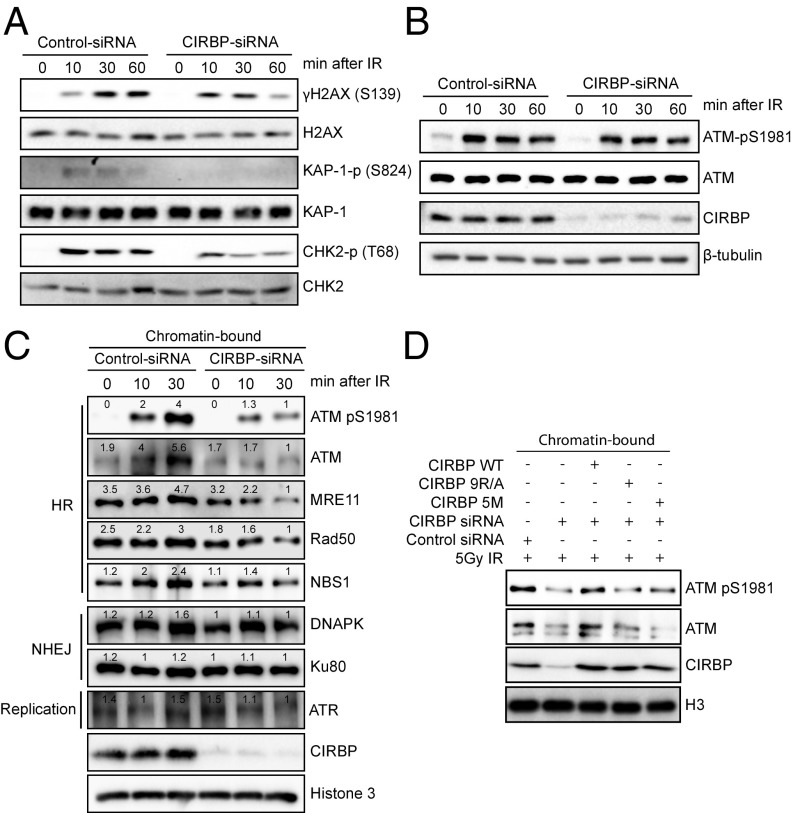
It has been reported that on DNA damage involving DSBs, the MRN complex will bind ATM and trigger its autophosphorylation (8). The activated ATM will subsequently catalyze the phosphorylation of a subset of downstream substrates to initiate the early DSB repair response (51). In contrast, ATR (ATM- and Rad3-related kinase) signaling can be activated by a broad spectrum of DNA damage in addition to DSBs (52). As shown in Fig. 6A, CIRBP knockdown resulted in decreased phosphorylation of the ATM substrates CHK2, KAP1, and H2AX, whereas phosphorylation of ATR downstream substrates, such as CHK1 and RPA, was not affected (SI Appendix, Fig. S6A). These results provide evidence that CIRBP is involved in ATM-dependent, but not ATR-dependent, signaling.
Fig. 6.
CIRBP promotes DNA damage-induced chromatin-association of MRN and ATM. (A) CIRBP knockdown reduces phosphorylation of ATM downstream substrates. Control or CIRBP siRNA-treated U2OS cells were either unexposed or exposed to 5 Gy IR. The total cell lysates were prepared using Laemmli buffer and subjected to immunoblotting against the antibodies indicated. (B) CIRBP knockdown does not affect total protein expression and autophosphorylation of ATM. Control or CIRBP siRNA-treated U2OS cells were exposed to 5 Gy IR. The total cell lysates were prepared using RIPA buffer and subjected to immunoblotting against the indicated antibodies. (C) CIRBP knockdown decreases the amount of chromatin-bound ATM and MRN complex. Control and CIRBP siRNA-treated U2OS cells were untreated or treated with 5 Gy IR. The chromatin-bound protein fraction was extracted at the indicated time points using CSK buffer and then subjected to immunoblotting against the antibodies indicated. The band intensity, quantified with ImageJ, is shown on top of each lane. (D) CIRBP mediates chromatin binding of ATM in a PAR polymer-dependent manner. siRNA-resistant WT, 5M, and 9R/A CIRBPs were transduced into U2OS cells. After 24 h, cells were treated with control or CIRBP siRNA. The chromatin-bound protein fraction was extracted using CSK buffer and subjected to immunoblotting against the indicated antibodies. All results in A–D are representative of at least two independent experiments.
Additional experiments were then performed to test whether CIRBP knockdown affects the activation of ATM kinase. It was found that CIRBP knockdown did not significantly change the total phosphorylation level of ATM suggesting that CIRBP is not required for ATM activation (Fig. 6B). However, there was a 75% decrease in the level of DNA damage-induced association of ATM-pSer1981 with chromatin in cells lacking CIRBP (Fig. 6C). Similarly, the amount of chromatin-bound MRN complex was also reduced by 40∼80%, while the binding of DNA-PK and Ku80 to chromatin, which is associated with the NHEJ repair pathway (4), was not significantly affected (Fig. 6C).
To gain more insight into how CIRBP depletion impairs DNA damage-induced association of ATM with chromatin, the siRNA-resistant WT CIRBP, the PARylation mutant (5M), and the PAR polymer-binding mutant (9R/A) were overexpressed in CIRBP-depleted U2OS cells, and the levels of chromatin-bound ATM following exposure to IR was measured. Notably, only WT CIRBP successfully restored the level of chromatin-associated ATM-pSer1981 in the CIRBP-depleted cells, whereas the 5M and 9R/A mutants were unable to do so (Fig. 6D). These results indicate that PAR polymer-mediated localization of CIRBP and PARylation of CIRBP at sites of DNA damage both contribute to facilitating the interaction between ATM-pSer1981 and chromatin. FokI endonuclease induced site-specific DSBs followed by chromatin immunoprecipitation (ChIP) and quantitative real-time PCR was also carried out to investigate the impact of CIRBP depletion on the enrichment of phospho-ATM and NBS1 at DSBs (Fig. 7A). This assay demonstrated that the enrichment of phosphorylated ATM and NBS1 at DSBs was decreased by around 67% and 50%, respectively, on knockdown of CIRBP (Fig. 7 B and C). Overall, these results support the hypothesis that CIRBP plays a crucial role in mediating the associations of MRN and ATM with chromatin.
Discussion
In this study, a previously unrecognized function for CIRBP during the DDR has been identified and characterized. This function is distinct from the established roles of CIRBP in RNA metabolism, circadian gene regulation, and the inflammatory response. Specifically, CIRBP was found to temporarily accumulate at regions of microirradiated chromatin in a PARP-1 and PAR polymer-dependent manner (Figs. 2 and 3). On DNA damage, activated PARP-1 can catalyze PARylation of many proteins involved in DNA repair. The early localization of CIRBP to sites of DNA damage can be attributed to its association with the PAR polymers generated by PARP-1 (Fig. 3 A–C). Binding of CIRBP with PAR polymer is likely mediated by the RGG motif (Fig. 4G), a common structural feature shared by proteins that bind PAR polymer. CIRBP was also found to be a substrate for PARP-1, with PARylation occurring at multiple sites, including Glu23 and several lysine residues (Fig. 5 C and D). Although the specific function of PARylated CIRBP remains to be further elucidated, it does appear to be important for promoting DSB repair (Figs. 5E and 6D). Since PAR polymers are highly negatively charged (12), PARylation of CIRBP may weaken its interaction with RNA. In addition, PARylation of CIRBP can also facilitate its interaction with other PAR polymer-binding DDR proteins.
Notably, an extended exclusion of CIRBP from microirradiated regions (Fig. 2 A and B) and DSB sites (Fig. 2F) was observed after the PARP-1–mediated recruitment. This exclusion cannot be ascribed to PARylation of CIRBP, because the same phenomenon could still be seen in PARP-1–depleted cells (Fig. 3 A–C). Instead, evidence suggests that CIRBP dissociation from damaged DNA may be due to phosphorylation mediated by PIKKs, ATM, ATR and DNA-PK, which phosphorylate many DDR proteins to coordinate the DDR (10). While it is unclear whether the exclusion of CIRBP is a prerequisite for proper DNA repair or merely a readout of reduced RNA concentration due to local transcription inhibition induced by DNA damage, similar behavior has been reported for several other RNA-binding proteins actively involved in DDR (39). Thus, the foregoing results strongly support an active role of CIRBP in DNA repair.
Interestingly, cells that were subjected to transcriptional inhibition to impede mRNA synthesis (SI Appendix, Fig. S3A) or carry CIRBP with the key RNA-binding residues mutated to alanine (Fig. 4C) showed earlier and enhanced accumulation of CIRBP at sites of microirradiation. This implies that free CIRBP, as opposed to RNA-bound CIRBP, accumulates at the sites of DNA damage. This preference may simply be due to the repulsive charge–charge interaction between the CIRBP-bound RNA and the PAR polymers carried by other DDR proteins gathered at the DNA damage sites. Importantly, CIRBP also mediates chromatin association with ATM induced by DNA damage and thereby impacts the downstream ATM signaling cascades, as well as the formation of the γH2AX, Rad51, and 53BP1 foci required for efficient DSB repair and genome stability (Fig. 7D).
Of particular interest is how CIRBP promotes and stabilizes the binding of ATM with damaged DNA. Since both ATM and the MRN complex bind PAR polymer (53, 54), the PAR polymer on PARylated CIRBP may interact with ATM and the MRN complex and thereby facilitate their association with chromatin. Indeed, PARylation of CIRBP was shown to be required for the binding of ATM to chromatin as well as the homology-directed repair. Recently, a model was proposed in which PAR polymer may seed intrinsically disordered proteins containing an RGG motif to form aggregates (17). PAR-initiated aggregation of RNA-binding proteins might create a microenvironment to control and filter DNA repair proteins at damaged chromatin. In this study, we found that PAR polymer was effective to polymerize CIRBP (SI Appendix, Fig. S7A) and accelerate the formation of protein aggregates (SI Appendix, Fig. S7B). Although the detailed mechanism remains to be elucidated, the PAR polymer-seeded CIRBP aggregates may form a temporary complex along with other RNA-binding proteins around ATM and facilitate its association with chromatin. Finally, PARylated CIRBP may also function as an adaptor that forms bound complexes with other DDR proteins to retain ATM at sites of DNA damage.
RNA-binding proteins have been found to directly interact with damaged DNA and mediate the DDR at different stages (39). Most of them display an early localization to damaged DNA through their interaction with PARP-1 generated PAR. For instance, hnRNPUL1 & 2 play an active role in HR through direct interaction with MRN and CtIP, and contribute to DNA end resection by facilitating the recruitment of BLM helicase (55). FUS protein also interacts with HDAC1 to mediate DSB repair (15, 16, 56). In addition, RBMX is necessary for DSB repair through regulating the expression of BRCA2 (19). Here, CIRBP is identified as a novel DDR-regulating protein that interacts with damaged DNA and promotes DSB repair by mediating the association of ATM with chromatin. This new function is directly related to its PARylation catalyzed by PARP-1 and its ability to bind PAR polymer. Moreover, the above-mentioned RNA-binding proteins which are related to DSB repair were also identified to be CIRBP-interacting proteins in our preliminary mass spectrometry screening results. Further characterization of the crosstalk between CIRBP and other RNA-binding proteins with damaged DNA is underway and may provide more insight into the crucial role of RNA-binding proteins in DDRs.
Materials and Methods
Antibodies, Plasmid Constructions, and Chemicals.
Antibodies used in this study were purchased from commercial sources and are summarized in SI Appendix, Table S1. Plasmid constructs are listed in SI Appendix, Table S2. Chemicals are listed in SI Appendix, Table S3. Primers were synthesized by Integrated DNA Technologies.
Abbreviation Information.
The abbreviations and full names are provided in SI Appendix, Table S5.
Cell Culture, Overexpression, and siRNA Silencing.
U251MG cells and HeLa cells were kindly provided by Andy Ellington and Robert Krug, The University of Texas at Austin, respectively. The U2OS FokI DSB reporter cell was a generous gift from Roger A. Greenberg, University of Pennsylvania, Philadelphia. The 293T, U2OS, and U2OS DR-GFP cells were kindly provided by Kyle Miller, The University of Texas at Austin. The cells were cultured in DMEM (Sigma-Aldrich) supplemented with 10% FBS (HyClone) at 37 °C. Overexpression was carried out using HilyMax reagent (Dojindo). siRNA knockdown was carried out with RNAiMax (Invitrogen) according to the manufacturer’s instructions. The siRNA sequences used in this work are listed in SI Appendix, Table S4, and these siRNAs were customarily synthesized by Sigma-Aldrich.
Genome Editing by CRISPR-Cas9.
PARP-1 knockout U2OS cells were generated by CRISPR-Cas9 technology as described previously (57). Two individual guide RNAs —5′-tgggttctctgagcttcggt-3′ and 5′-gcaccctgacgttgaggtgg-3′—were designed and cloned into a pSpCas9(BB)-2A-Puro (PX459) vector (Addgene, plasmid 48139), a gift from Feng Zhang. U2OS cells were transfected with an expression vector containing PARP-1 single-guide RNA using HilyMax reagent (Dojindo). The cells were then cultured for 48 h in the presence of 2 μg/mL puromycin, which was used as selection agent. After serial dilution into 96-well plates, the resulting single clones were subjected to Western blot and immunofluorescence analysis to check PARP-1 knockout efficiency.
Protein Expression and Purification.
His-tagged recombinant proteins were expressed using the Baculovirus-expressing system (Invitrogen). In brief, bacmid containing a gene of interest and genes required for virus production were cloned and expressed in Spodoptera frugiperda (Sf21) insect cells. At 4 d after transfection, the supernatant containing the virus particles (P1 virus) was used to infect more Sf21 insect cells for protein expression. The cell pellets were harvested at 3 d after P1 virus infection and lysed with lysis buffer (20 mM Hepes, 300 mM NaCl, 10 mM imidazole, 10% glycerol, 1% Triton X-100, 1 μM β-mercaptoethanol, 1 mM PMSF, and 1 mM benzamidine-HCl). The lysates were sonicated and then treated with 2 μg/mL DNase I and 0.5 μg/mL RNase A for 1 h at 4 °C. The lysates were spun down at 16,000 × g for 30 min, and the supernatants were pooled and incubated with Ni-NTA resin for 1 h at 4 °C. The resin-bound, pulled-down proteins were washed four times with wash buffer (20 mM Hepes, 1 M NaCl, 10 mM imidazole, and 10% glycerol) and then eluted with buffer containing 20 mM Hepes, 300 mM NaCl, 250 mM imidazole, and 10% glycerol.
Protein Extraction.
Total protein lysates were prepared with RIPA buffer (50 mM Tris-Cl pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitor mixture). For chromatin fractionation, cells were lysed in CSK buffer (10 mM Pipes pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, and 0.2% Triton X-100) on ice for 30 min and centrifuged for 10 min at 10,000 × g at 4 °C to remove chromatin unbound proteins. The pellets were washed twice with CSK buffer, resuspended in Laemmli buffer (4% SDS, 20% glycerol, 120 mM Tris⋅HCl pH 6.8, and 0.02% bromophenol), and subjected to sonication, followed by boiling for 10 min to denature proteins. The resulting solution containing proteins associated with chromatin was collected as the chromatin-bound fractionation and subjected to standard Western blot analysis.
Purification of GFP-Tagged Proteins Using GFP-Trap Beads.
GFP-tagged proteins were overexpressed in U2OS or 293T cells. At 2 d later, cells were lysed in RIPA buffer (50 mM Tris-Cl pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitor mixture) supplemented with 1 mM PMSF (AMRESCO), 1 mM benzamidine-HCl (Sigma-Aldrich), and 10 μM tannic acid (Sigma-Aldrich). The lysates were centrifuged at 16,000 × g for 10 min, and the supernatants were incubated with GFP-trap MA beads overnight. The pulled-down protein-beads complex was washed with RIPA buffer four times, then resuspended in Laemmli buffer and boiled for 10 min. The samples were analyzed by Western blot against various antibodies.
RNA Isolation and cDNA Synthesis.
Total RNA was extracted using the QIAGEN RNeasy Mini Kit. RNA concentration was determined using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies). RNA (1 μg) was reversely transcribed to cDNA with SuperScript III Reverse Transcriptase (Invitrogen) using Oligo-dT (Invitrogen) as the primer.
ChIP.
The ChIP assay was carried out with a Simple ChIP Enzymatic Chromatin IP kit (9003; Cell Signaling Technology) following the manufacturer’s protocol. In brief, U2OS DSB reporter cells with or without Shield-1 [(1R)-3-(3,4-dimethoxyphenyl)-1-[3-(2-morpholin-4-ylethoxy)phenyl]propyl)(2S)-1-[(2S)-2-(3,4,5-trimethoxyphenyl)butanoyl]piperidine-2-carboxylate; Clontech] and 4-hydroxytamoxifen (4-OHT; Sigma-Aldrich) to induce DSBs were cross-linked with 1% formaldehyde and neutralized with 125 mM glycine. Cells were lysed, and the cross-linked nucleus lysates were digested with micrococcal nuclease and then sonicated with a Model 100 Sonic Dismembrator (Thermo Fisher Scientific) to yield genomic DNA fragments between 150 and 900 bp. The digested chromatin (5 μg) was immunoprecipitated with indicated primary antibody overnight at 4 °C. Normal rabbit antibody was used in parallel as a control. The immunocomplexes were pulled down with magnetic beads, reversely cross-linked at 65 °C for 30 min, and digested with proteinase K overnight. The DNA samples were purified using QIAprep Spin Miniprep Columns (QIAGEN). Real-time PCR was performed with the ViiA 7 system (Applied Biosystems) using SYBR Select Master Mix (Applied Biosystems). The ChIP-qPCR primers used are listed in SI Appendix, Table S4. The qPCR conditions were as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. This was followed by melting curve analysis to confirm singly amplified product: 95 °C for 15 s, 60 °C for 1 min, 0.05 °C/s increments from 60 °C to 95 °C, and holding at 95 °C for 15 s. The results were analyzed with QuantStudio real-time PCR software.
Immunofluorescence.
Cells seeded on poly-l-lysine coverslips (BD Biosciences) were fixed with 2% formaldehyde solution (Sigma-Aldrich) for 15 min and permeabilized with 0.1% Triton X-100 or CSK buffer for 10 min at room temperature. After blocking with 5% BSA (Sigma-Aldrich), samples were incubated with indicated primary antibodies overnight at 4 °C. Samples were then washed and incubated with secondary antibodies plus DAPI for 1 h at room temperature. Samples were mounted onto glass slides with VECTASHIELD anti-fade mounting medium (Vector Laboratories) and visualized with a Zeiss LSM 710 confocal microscope. For CSK preextraction, cells were preincubated with CSK buffer containing 10 μM tannic acid for 3 min at room temperature and then subjected to standard immunofluorescence procedures. For immunostaining with monoclonal S9.6 antibody, cells were specifically fixed and permeabilized with ice-cold methanol at −20 °C for 15 min before blocking. Z-stack images for each cell were recorded and the nuclear S9.6 intensity was analyzed using Fiji software.
Live-Cell Laser Microirradiation.
Cells seeded on 35-mm glass-bottom dishes (WillCo Wells) were presensitized with 0.5 μM Hoechst 33342 for 1 h at 37 °C. Laser microirradiation was carried out using a Zeiss LSM 710 confocal microscope equipped with a 37 °C chamber and CO2 module. For endogenous protein, a selected region was microirradiated for 20 iterations by a 405-nM laser with 100% power to induce localized DNA breaks (56). The treated cells were immediately fixed, followed by detection based on the protocol described above. For EGFP-tagged protein, time-lapse images were acquired at 20-s time intervals after laser microirradiation. The GFP intensities were recorded with Zen 2012 software (Zeiss) and analyzed with Fiji ImageJ software. The fluorescence values of more than 10 cells from three independent experiments were normalized to the original signal and plotted as a fluorescence-vs.-time graph using GraphPad Prism software. The error bars represent SEM.
In Vitro PAR-Binding Assay.
Nitrocellulose membrane was equilibrated with TBS-T (20 mM Tris⋅HCl pH 7.5, 150 mM NaCl, and 0.05% Tween 20) and air-dried. Recombinant proteins (0.25 μg each) were dot-blotted on the nitrocellulose membrane and then air-dried. The membrane was then incubated with purified PAR polymer (200 nM, determined based on OD260) for 1 h with gentle agitation followed by extensive washing with TBS-T. After blocking with 5% milk, the membrane was washed with TBS-T and then subjected to immunoblotting with PAR antibody. Histone 1 and BSA served as positive and negative controls, respectively.
In Vitro PARylation Assay.
Purified recombinant proteins were incubated with purified PARP-1 protein in the PARylation buffer (50 μM Tris⋅HCl pH 8.0, 4 mM MgCl2, 250 μM DTT, 20 mM NaCl, 400 μM NAD+, and nicked calf thymus DNA) for 1∼5 min at 37 °C. The reactions were quenched with Laemmli buffer and subjected to immunoblotting against anti-PAR antibody.
Colony-Formation Assay.
U2OS cells treated with different siRNA were counted and plated in a six-well plate. Cells were exposed to different dosages of IR and then incubated at 37 °C for 10–14 d until colonies could be visualized under a light microscope. The colonies were stained with 20% ethanol solution containing 0.5% crystal violet. After gentle washing, the colonies were counted and normalized to control siRNA-treated cells. The plotted graph is based on the average of two independent experiments, and error bars represent mean ± SEM.
Cell Cycle Analysis.
Ethanol-fixed cells were stained with propidium iodide (Life Technologies) solution with 3.8 μM sodium citrate, 50 μg/mL propidium iodide, and 5 μg/mL RNase A at 4 °C overnight. DNA contents were measured using a BD Accuri Flow Cytometer (BD Biosciences) and analyzed with FlowJo software.
Homologous Recombination Assay.
The U2OS DR-GFP cells were treated with siRNA of the control or targeted genes. After 24 h, control pCAG vector or pCAG-I-SceI vector was overexpressed in the siRNA-treated cells. The cells were grown for an additional 48 h, then trypsinized, washed with PBS, and resuspended in PBS. The GFP-positive cells were counted using a BD Accuri Flow Cytometer (BD Biosciences) and normalized to control siRNA-treated cells with I-SceI expression.
Random Plasmid Integration Assay.
U2OS cells were treated with various siRNAs, and BamHI-XhoI linearized pEGFP-C1 vector was introduced into the siRNA-treated cells 24 h later. After another 48 h of incubation, the overexpression efficiency of the linearized pEGFP-C1 was determined by flow cytometry. The cells were also counted and plated in two six-well plates. Two duplicated plates with (for random plasmid integration) or without (for plating efficiency) G418 (0.5 mg/mL) were incubated at 37 °C for approximately 10 d to monitor the formation of colonies. The colonies were stained with a 20% ethanol solution containing 0.5% crystal violet. Random plasmid integration events were normalized to plating efficiency and pEGFP-C1 overexpression efficiency, and were compared with control siRNA-treated cells. The average of three independent experiments are graphed; the error bars indicate mean ± SEM.
PAR Polymer Synthesis.
PAR polymer was enzymatically synthesized as described previously (58) with some modifications. In brief, PAR polymer was produced in 20 mL of incubation mixture containing 50 mM Tris⋅HCl pH 8, 4 mM MgCl2, 250 μM DTT, 20 mM NaCl, 5 mM NAD+, 10 mM DTT, 50 μg/mL histone H1, 10 μg/mL 8-mer oligonucleotide (GGAATTCC), and 150 nM human PARP-1 at 37 °C for 90 min. The reaction was stopped by adding 20 mL of cold 20% (wt/vol) TCA and incubating on ice for 15 min. After precipitation and washing with 100% ethanol, PAR polymer was detached from the modified proteins using 0.5 M KOH/50 mM EDTA and purified as described previously (59). The PAR concentration (based on mono-ADP ribose equivalent) was determined with a Nanodrop ND1000 spectrophotometer based on the measured absorption at 258 nm and an extinction coefficient of 13,500 M−1cm−1.
Formaldehyde Cross-Linking Assay.
His-tagged recombinant CIRBP protein was diluted in PBS (pH 7.4) to a final concentration of 2 μM and then incubated with or without purified PAR polymer (50–200 μM based on mono ADP ribose equivalent) for 15 min at 37 °C. The incubation was continued at –20 °C for 1 h to accelerate protein assembly, and then cross-linked with 0.4% formaldehyde at room temperature for 15 min. Cross-linking was stopped by adding glycine to a final concentration of 100 mM. The cross-linked samples were boiled at 95 °C in 2× sample buffer for 5 min and then separated by SDS PAGE. The SDS gel was stained with SYPRO Ruby protein gel stain solution (Thermo Fisher Scientific) and visualized by UV.
TEM.
Recombinant His-tagged CIRBP was diluted in the TEM sample buffer containing 40 mM Hepes-KOH and 150 mM KCl (pH 7.4) to a final concentration of 50 nM and the incubated with substoichiometric amounts of PAR (1 nM). The CIRBP-PAR mixtures were agitated at 1,200 rpm in an Eppendorf ThermoMixer at 37 °C for 20 h, after which 2 μL of the CIRBP-PAR mixture was spread onto Formar carbon films (FCF400-Cu; EMS), followed by staining with 2% phosphotunastic acid, pH 7.4 (Sigma-Aldrich). Images were acquired using an FEI Tecnai transmission electron microscope.
Supplementary Material
Acknowledgments
We thank Professors Andy Ellington, Roger Greenberg, Robert Krug, and Kyle Miller for their generous gifts of cell lines; Li-Ya Chiu for technical support; Kyle Miller for use of the Faxitron X-ray generator; and Kyle Miller and Tanya Paull for valuable comments on the paper. This work was supported in part by a grant from the Welch Foundation (F-1511).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713912115/-/DCSupplemental.
References
- 1.Khanna KK, Jackson SP. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 2.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 3.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zelensky AN, Schimmel J, Kool H, Kanaar R, Tijsterman M. Inactivation of Pol θ and C-NHEJ eliminates off-target integration of exogenous DNA. Nat Commun. 2017;8:66. doi: 10.1038/s41467-017-00124-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 7.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 8.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 9.Uziel T, et al. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 11.Ko HL, Ren EC. Functional aspects of PARP1 in DNA repair and transcription. Biomolecules. 2012;2:524–548. doi: 10.3390/biom2040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 13.Krietsch J, et al. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012;40:10287–10301. doi: 10.1093/nar/gks798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Britton S, et al. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 2014;42:9047–9062. doi: 10.1093/nar/gku601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem. 2013;288:24731–24741. doi: 10.1074/jbc.M113.497974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rulten SL, et al. PARP-1–dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014;42:307–314. doi: 10.1093/nar/gkt835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altmeyer M, et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose) Nat Commun. 2015;6:8088. doi: 10.1038/ncomms9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beli P, et al. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell. 2012;46:212–225. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao P. 2007. Studies of activation, automodification and transmodification of poly(ADP-ribose) polymerase-1. PhD thesis (University of Texas at Austin)
- 21.Wright RH, et al. ADP-ribose-derived nuclear ATP synthesis by NUDIX5 is required for chromatin remodeling. Science. 2016;352:1221–1225. doi: 10.1126/science.aad9335. [DOI] [PubMed] [Google Scholar]
- 22.Gibson BA, et al. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science. 2016;353:45–50. doi: 10.1126/science.aaf7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jungmichel S, et al. Proteome-wide identification of poly(ADP-ribosyl)ation targets in different genotoxic stress responses. Mol Cell. 2013;52:272–285. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Wang J, Ding M, Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat Methods. 2013;10:981–984. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- 25.Nishiyama H, et al. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J Cell Biol. 1997;137:899–908. doi: 10.1083/jcb.137.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheikh MS, et al. Identification of several human homologs of hamster DNA damage-inducible transcripts: Cloning and characterization of a novel UV-inducible cDNA that codes for a putative RNA-binding protein. J Biol Chem. 1997;272:26720–26726. doi: 10.1074/jbc.272.42.26720. [DOI] [PubMed] [Google Scholar]
- 27.Yang C, Carrier F. The UV-inducible RNA-binding protein A18 (A18 hnRNP) plays a protective role in the genotoxic stress response. J Biol Chem. 2001;276:47277–47284. doi: 10.1074/jbc.M105396200. [DOI] [PubMed] [Google Scholar]
- 28.Yang R, Weber DJ, Carrier F. Post-transcriptional regulation of thioredoxin by the stress-inducible heterogenous ribonucleoprotein A18. Nucleic Acids Res. 2006;34:1224–1236. doi: 10.1093/nar/gkj519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang R, et al. Functional significance for a heterogenous ribonucleoprotein A18 signature RNA motif in the 3′-untranslated region of ataxia telangiectasia mutated and Rad3-related (ATR) transcript. J Biol Chem. 2010;285:8887–8893. doi: 10.1074/jbc.M109.013128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Leeuw F, et al. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp Cell Res. 2007;313:4130–4144. doi: 10.1016/j.yexcr.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 31.Morf J, et al. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 2012;338:379–383. doi: 10.1126/science.1217726. [DOI] [PubMed] [Google Scholar]
- 32.Masuda T, et al. Cold-inducible RNA-binding protein (Cirp) interacts with Dyrk1b/Mirk and promotes proliferation of immature male germ cells in mice. Proc Natl Acad Sci USA. 2012;109:10885–10890. doi: 10.1073/pnas.1121524109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiang X, et al. Cold-inducible RNA-binding protein (CIRP) triggers inflammatory responses in hemorrhagic shock and sepsis. Nat Med. 2013;19:1489–1495. doi: 10.1038/nm.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan MM, Yang WL, Brenner M, Bolognese AC, Wang P. Cold-inducible RNA-binding protein (CIRP) causes sepsis-associated acute lung injury via induction of endoplasmic reticulum stress. Sci Rep. 2017;7:41363. doi: 10.1038/srep41363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, et al. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci Rep. 2013;3:2054. doi: 10.1038/srep02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee HN, Ahn SM, Jang HH. Cold-inducible RNA-binding protein, CIRP, inhibits DNA damage-induced apoptosis by regulating p53. Biochem Biophys Res Commun. 2015;464:916–921. doi: 10.1016/j.bbrc.2015.07.066. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, et al. Cold-inducible RNA-binding protein CIRP/hnRNP A18 regulates telomerase activity in a temperature-dependent manner. Nucleic Acids Res. 2016;44:761–775. doi: 10.1093/nar/gkv1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang ET, Parekh PR, Yang Q, Nguyen DM, Carrier F. Heterogenous ribonucleoprotein A18 (hnRNP A18) promotes tumor growth by increasing protein translation of selected transcripts in cancer cells. Oncotarget. 2016;7:10578–10593. doi: 10.18632/oncotarget.7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wickramasinghe VO, Venkitaraman AR. RNA processing and genome stability: Cause and consequence. Mol Cell. 2016;61:496–505. doi: 10.1016/j.molcel.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fenech M, et al. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 2011;26:125–132. doi: 10.1093/mutage/geq052. [DOI] [PubMed] [Google Scholar]
- 41.Zhang CZ, et al. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonner WM, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chowdhury D, Choi YE, Brault ME. Charity begins at home: Non-coding RNA functions in DNA repair. Nat Rev Mol Cell Biol. 2013;14:181–189. doi: 10.1038/nrm3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lord CJ, Ashworth A. RAD51, BRCA2 and DNA repair: A partial resolution. Nat Struct Mol Biol. 2007;14:461–462. doi: 10.1038/nsmb0607-461. [DOI] [PubMed] [Google Scholar]
- 46.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 47.Tang J, et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: Where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cléry A, Blatter M, Allain FH. RNA recognition motifs: Boring? Not quite. Curr Opin Struct Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 50.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiloh Y, Ziv Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 52.Bekker-Jensen S, et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haince JF, et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 54.Haince JF, et al. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J Biol Chem. 2007;282:16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- 55.Polo SE, et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol Cell. 2012;45:505–516. doi: 10.1016/j.molcel.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang WY, et al. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci. 2013;16:1383–1391. doi: 10.1038/nn.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kiehlbauch CC, Aboul-Ela N, Jacobson EL, Ringer DP, Jacobson MK. High-resolution fractionation and characterization of ADP-ribose polymers. Anal Biochem. 1993;208:26–34. doi: 10.1006/abio.1993.1004. [DOI] [PubMed] [Google Scholar]
- 59.Malanga M, Bachmann S, Panzeter PL, Zweifel B, Althaus FR. Poly(ADP-ribose) quantification at the femtomole level in mammalian cells. Anal Biochem. 1995;228:245–251. doi: 10.1006/abio.1995.1346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.