Significance
Capicua (Cic) is a conserved transcriptional repressor involved in receptor tyrosine kinase (RTK)-dependent gene regulation. Cic controls numerous physiological and pathological processes in flies and mammals, but whether all Cic functions are connected to RTK signaling remains unclear. Here, we show that Cic represses Toll/IL-1 signaling targets in Drosophila embryos independently of RTK control. In this context, Cic functions via suboptimal DNA sites that it cannot recognize on its own. Instead, Cic binding requires the Dorsal/NF-kB protein, which enters the nucleus following Toll activation and binds next to the Cic sites. Thus, Cic exerts more diverse mechanisms of control than previously realized, raising the possibility that RTK-independent mechanisms and/or suboptimal DNA binding may also underlie Cic function in other contexts.
Keywords: transcriptional repression, Dorsal, Groucho, low-affinity binding sites, ChIP-nexus
Abstract
The HMG-box protein Capicua (Cic) is a conserved transcriptional repressor that functions downstream of receptor tyrosine kinase (RTK) signaling pathways in a relatively simple switch: In the absence of signaling, Cic represses RTK-responsive genes by binding to nearly invariant sites in DNA, whereas activation of RTK signaling down-regulates Cic activity, leading to derepression of its targets. This mechanism controls gene expression in both Drosophila and mammals, but whether Cic can also function via other regulatory mechanisms remains unknown. Here, we characterize an RTK-independent role of Cic in regulating spatially restricted expression of Toll/IL-1 signaling targets in Drosophila embryogenesis. We show that Cic represses those targets by binding to suboptimal DNA sites of lower affinity than its known consensus sites. This binding depends on Dorsal/NF-κB, which translocates into the nucleus upon Toll activation and binds next to the Cic sites. As a result, Cic binds to and represses Toll targets only in regions with nuclear Dorsal. These results reveal a mode of Cic regulation unrelated to the well-established RTK/Cic depression axis and implicate cooperative binding in conjunction with low-affinity binding sites as an important mechanism of enhancer regulation. Given that Cic plays a role in many developmental and pathological processes in mammals, our results raise the possibility that some of these Cic functions are independent of RTK regulation and may depend on cofactor-assisted DNA binding.
The HMG-box protein Capicua (Cic) is a conserved transcriptional repressor with key functions downstream of receptor tyrosine kinase (RTK) signaling (1). In both Drosophila and mammals, Cic acts antagonistically to the RTK pathway: Cic represses RTK-responsive genes in unstimulated cells and tissues, whereas RTK signaling via mitogen-activated protein kinase (MAPK) activation inhibits Cic activity, and thus leads to derepression of its targets. This molecular switch has been well studied during Drosophila development (1, 2). For example, during early embryogenesis, Cic represses two zygotic genes, tailless (tll) and huckebein (hkb), in the central region of the embryo, and this repression is relieved by Torso RTK signaling at the anterior and posterior embryonic poles (3–6). In mammals, Cic also controls multiple processes and is implicated in neurodegeneration and tumorigenesis, where it behaves as a tumor and metastasis suppressor (1, 7–10). However, it is not yet clear whether Cic functions are always under RTK regulation or can also involve other types of responses.
Here, we define a mechanism of Cic transcriptional regulation distinct from the RTK switch. This mechanism operates during dorsoventral (DV) patterning of the Drosophila embryo, which is controlled by the localized activation of the Toll/IL-1 receptor on the ventral surface of the embryo (11, 12) (Fig. 1A). During Toll signaling, the phosphorylation-dependent disassembly of cytoplasmic Cactus (Cact)–Dorsal complexes allows Dorsal (an NF-κB–related transcription factor) to enter the nucleus (13). This produces a nuclear gradient of Dorsal with maximal levels in ventral regions and progressively lower levels in lateral and dorsal regions. Dorsal controls DV patterning by mediating either activation or repression of multiple targets (11, 12) (Fig. 1A). Although this network is largely independent of RTK regulation, we now demonstrate that Cic plays an essential role in Dorsal-mediated repression.
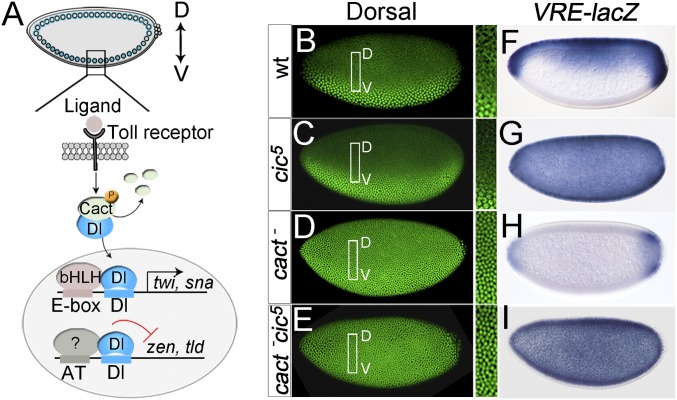
Fig. 1.
Cic acts downstream of Cact to repress the zen VRE enhancer. (A) Toll- and Dorsal-dependent transcriptional control in the ventral region of the embryo. Toll activation leads to the nuclear translocation of Dl, which then activates zygotic genes such as twi and snail (sna), while repressing others such as zen and tld. These differential outcomes depend on the presence of additional transcription factor binding sites in target enhancers, which are recognized by basic helix–loop–helix (bHLH) activator factors and at least one unidentified repressor (question mark). D, dorsal; V, ventral. (B–E, Left) Early stage 5 embryos of the indicated maternal genotypes stained for Dorsal protein. (B–E, Right) Boxed areas are shown in higher magnification. Note the presence of Dorsal in dorsal nuclei of cact− and cact− cic5 mutant embryos. (F–I) Patterns of VRE-lacZ reporter expression in the indicated maternal genotypes. In I, the reporter is derepressed in cic5 mutant embryos, even when Dorsal is constitutively nuclear. Photographs in B–I were taken at 200× magnification.
The mechanisms responsible for Dorsal-mediated repression have proven difficult to unravel (11, 12). This repression is known to require conserved A/T-rich (hereafter AT) sites located close to Dorsal binding sites in target enhancers (14, 15), as well as auxiliary factors such as the Groucho (Gro) corepressor (16, 17). However, the identity of the factors that recognize the AT sites and how they function together with Dorsal and Gro remain unclear. In addition, we previously proposed that Cic could be involved in repression of Dorsal-dependent targets (3), but this potential role has been difficult to study for two reasons. First, Cic has an earlier role in regulating DV patterning during oogenesis (4, 18, 19), which is critical for the formation of the Dorsal gradient and complicates the study of Cic activity in Dorsal-dependent controls. Second, the analysis of Cic DNA binding has been problematic, and it only recently came to light that Cic requires not only its HMG-box domain but an additional conserved domain, referred to as C1, for DNA binding (20).
In this study, we have overcome these limitations and analyzed the roles of Cic, Dorsal, and Gro in repression of Dorsal targets such as zerknullt (zen), tolloid (tld), and decapentaplegic (dpp). We report that Cic directly represses these genes in ventral regions of the embryo by binding to AT sites of lower affinity than Cic sites present in its RTK-regulated targets. We show that Dorsal facilitates, and thus spatially controls, the binding of Cic to these suboptimal sites in ventral nuclei, and that binding does not occur in dorsal nuclei lacking Dorsal protein. We also show that Cic represses Dorsal targets via Gro, and that Dorsal is dispensable for repression per se, indicating that Dorsal functions in this context mainly as a DNA binding cofactor for Cic.
Results
Cic Functions in Dorsal-Mediated Repression in the Embryo.
To further characterize the role of Cic in DV regulation, we searched for new cic mutant alleles that might uncouple Cic functions in oogenesis and embryogenesis. In both Drosophila and mammals, Cic is expressed as short (Cic-S) and long (Cic-L) isoforms (21, 22). Although Drosophila Cic-S (isoform A in FlyBase) is believed to function in both the early embryo and the ovarian follicle cells (3, 4), we used CRISPR-Cas9–mediated mutagenesis to generate specific mutations affecting only this isoform. One resulting mutation, cic5, is a frameshift allele that truncates Cic-S after the initiator methionine residue (Fig. S1A and Table S1), thereby inactivating this isoform completely without affecting Cic-L. Surprisingly, cic5 did not affect Cic function in the follicle cells as judged by normal expression of Cic targets in those cells (Fig. S1 B–D), indicating that the described Cic-S isoform is dispensable in that context (also Fig. S2). Accordingly, the embryos laid by cic5 females exhibit normal expression of twist (twi), a gene activated by Dorsal in ventral regions (Fig. S1 E–G). However, these embryos show full derepression of zen up to the ventral midline (Fig. S1 E–G). A similar result was obtained by analyzing a reporter driven by the ventral repression element (VRE) cis-regulatory module (CRM) of zen, which controls its early embryonic expression (refs. 14 and 15 and references therein) (Fig. 1 B, C, F, and G). These findings strongly indicate that Cic acts in the early embryo to repress the zen VRE.
To confirm this idea, we reasoned that Cic should be required for repression of the VRE even when Dorsal is constitutively present in embryonic nuclei independently of prior patterning events in the follicle cells. To this end, we used embryos devoid of Cact protein, in which Dorsal is present in all embryonic nuclei, thus leading to repression of the VRE in dorsal regions (23) (Fig. 1 A, D, and H). We found that such repression is fully abolished along the entire DV axis by the additional loss of Cic function (Fig. 1 E and I), showing that Cic is indeed required for VRE repression in the embryo.
Cic Represses zen and tld by Directly Binding to Their AT Sites.
We wondered whether Cic might act through the AT sites present in the VRE (Fig. 2A). Indeed, these sites resemble optimal Cic binding sites [CBSs; T(C/G)AATGAA] except for a single mismatch (Fig. 2B). When tested in electrophoretic mobility shift assays (EMSAs), these AT sites were detectably bound by Cic, albeit with much lower affinity (between 7.5- and 12.5-fold less) than a regular CBS (Fig. 2 C and D). This weaker binding was specific, as it was abolished by point mutations in the AT sites, as well as in the HMG-box or C1 domain of Cic, which are both required for binding to regular CBSs (20) (Fig. 2 C and D). Conversely, converting an AT site into an optimal CBS (AT2opt) enhanced Cic binding considerably (Fig. 2 B and D). Together, these findings suggested that Cic could repress zen by binding to the AT sites via its HMG-box and C1 domains.
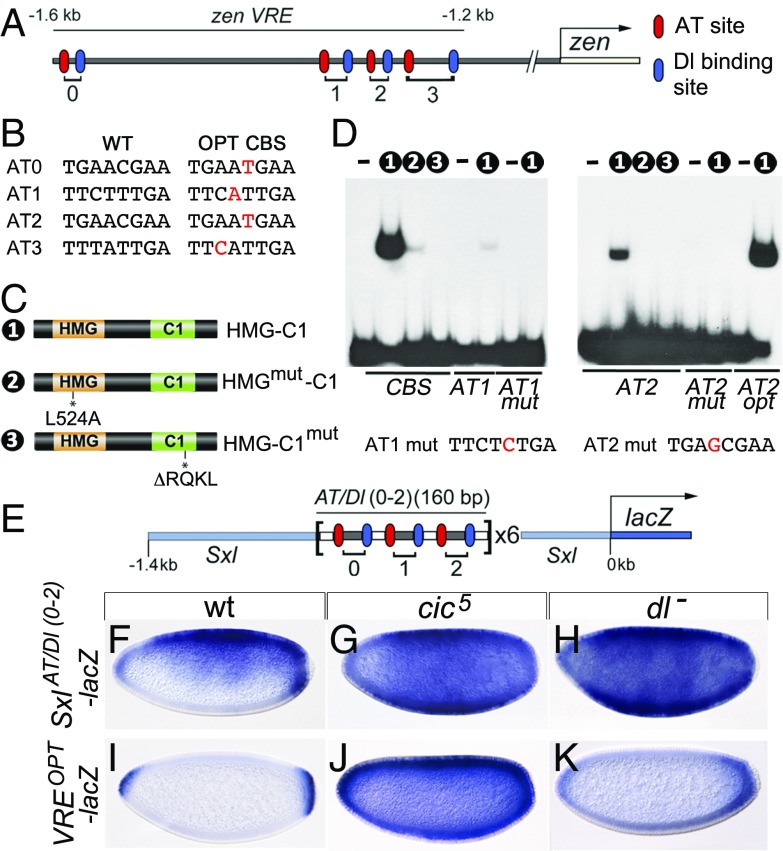
Fig. 2.
Cic represses VRE activity by binding to low-affinity DNA sites. (A) Diagram of the VRE indicating the positions of AT and Dl binding sites labeled according to refs. 14 and 15. In these and similar drawings, the transcription start site is indicated by an arrow. (B) Sequences of native (WT) AT sites and the corresponding optimal (OPT) CBSs. (C) Cic protein constructs used in EMSAs; the HMG-box and C1 domains are placed next to each other (20). HMG-C1mut contains the cic4 mutation (20). (D) EMSAs of Cic derivatives binding to WT or mutant probes. Numbers indicate the constructs used in the binding reactions; unlabeled lanes do not contain protein. Probes are indicated below the gels. CBS carries a control, high-affinity CBS. The sequence of the mutated AT1 mut and AT2 mut sites is also shown, with their respective substitutions indicated in red. Note the weak binding of HMG-C1 to the intact AT1 and AT2 probes. (E) Structure of the SxlAT/Dl(0-2)-lacZ reporter (not drawn to scale) (also Fig. S3). (F–K) Expression of SxlAT/Dl(0-2)-lacZ and VREOPT-lacZ reporters in the indicated backgrounds. Repression of SxlAT/Dl(0-2)-lacZ depends on both Cic and Dorsal, whereas Cic represses VREOPT-lacZ independently of Dorsal. Embryos in F–K were photographed at 200× magnification.
To evaluate this further, we dissected out three AT/Dorsal (Dl) binding site pairs from the VRE and inserted them as a tandem array in the upstream region of the Sex-lethal (Sxl) gene, a regulator of sex determination that is expressed uniformly in female blastoderm embryos (24) (Fig. 2E and Fig. S3). Therefore, the final reporter construct [SxlAT/Dl(0-2)-lacZ] contains multimerized AT/Dl binding motifs but lacks other intervening and flanking sequences of the VRE. When assayed in otherwise wild-type embryos, SxlAT/Dl(0-2)-lacZ was repressed in ventral regions (Fig. 2F), much like the normal VRE-lacZ reporter (Fig. 1F). In contrast, repression was abolished in both cic5 and dorsal (dl) mutant embryos (Fig. 2 G and H), indicating that Cic acts through the AT sites in a Dorsal-dependent fashion. Furthermore, in a set of analogous experiments, we found that Cic represses another Dorsal target, tld, through conserved AT sites linked to Dorsal sites (Fig. S4; also discussed below).
Cic is present in ventral and dorsal embryonic nuclei, yet zen and tld are repressed only ventrally where Dorsal is also present. This suggests that the in vivo binding of Cic to the AT sites might depend on the binding of Dorsal to neighboring sites. Alternatively, Cic could bind the AT sites in dorsal regions but be unable to repress their targets without Dorsal. To help distinguish between these possibilities, we converted each AT site in the VRE to an optimal CBS (Fig. 2B). If Dorsal is critical for Cic’s ability to repress but not for Cic binding, repression in this reporter should still be Dorsal-dependent. However, the resulting reporter, VREOPT-lacZ, is repressed along the entire DV axis except at the embryo poles (Fig. 2I). This expression actually resembles that of hkb, a target of Cic regulated by Torso signaling (6). The VREOPT-lacZ reporter was strongly derepressed in cic5 embryos (Fig. 2J), suggesting that this effect is mediated by Cic and not by other repressors binding to these sites. Importantly, the expression was unaltered in embryos devoid of Dorsal (Fig. 2K), suggesting that Cic binding to these sites no longer required Dorsal. Thus, when Cic binds the VRE through optimal sites, it readily represses it in either ventral or dorsal regions independently of Dorsal, but when Cic binds through suboptimal AT sites, it can only repress in ventral nuclei, where Dorsal is also present. Together, these results indicate that Dorsal facilitates the binding of Cic to its low-affinity sites in ventral regions.
Dorsal Promotes Binding of Cic to Suboptimal AT Sites.
To directly test the requirement of Dorsal for Cic binding to low-affinity sites, we used chromatin immunoprecipitation with nucleotide resolution through exonuclease, unique barcode and single ligation (ChIP-nexus), a ChIP-exo–based protocol for high-resolution detection of binding sites in vivo (25). We obtained ChIP-nexus signals for Cic and Dorsal in two genetic backgrounds: embryos with constitutive Toll signaling, and hence uniform Dorsal nuclear localization (derived from Toll10b females), and embryos without Toll activity, and therefore lacking Dorsal protein in nuclei [derived from gastrulation defective mutant (gd7) females]. In Toll10b embryos, Dorsal produced footprints in zen and tld, including those corresponding to Dorsal sites 0–3 within the zen VRE and site 1 within the tld CRM (Fig. 3A and Fig. S5). Cic, on the other hand, produced specific peaks in close proximity to the AT sites flanking the Dorsal sites. Notably, the Dorsal profile also showed binding around the AT sites, which indicates a dependency between Dorsal and Cic binding, perhaps through direct physical interactions (Fig. 3A, close-up of the zen VRE). Most importantly, Cic did not bind its zen and tld sites in gd7 embryos devoid of nuclear Dorsal protein (Fig. 3A and Fig. S5). In contrast, Cic binding was not reduced in gd7 versus Toll10b mutants at control enhancers such as those of hkb and tll (Fig. 3A), which are repressed by Cic via optimal CBSs along the entire DV axis (6). This further supports our conclusion that Cic binding to suboptimal AT sites depends on Dorsal, while Cic binding to optimal CBSs does not.
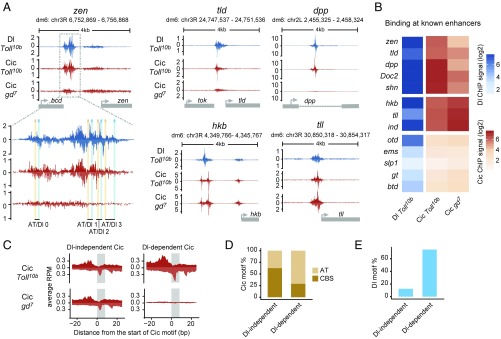
Fig. 3.
Dorsal mediates binding of Cic to low-affinity AT sites. (A) ChIP-nexus profiles of Dl and Cic at the indicated genes in Toll10b or gd7 mutant background; a zoom-in view of the zen VRE enhancer is included. Peaks on the upper (positive) and lower (negative) strands represent pile-ups of 5′ read ends after digestion with exonuclease, which stops at sites of protein–DNA cross-linking. Cic binding to zen, tld, and dpp (but not to hkb and tll) is strongly reduced in gd7 embryos lacking nuclear Dorsal protein. (B) Heat maps of Dorsal and Cic binding at known enhancers. ChIP-nexus signal was calculated within a 200-bp region centered on the highest Dorsal summit. (C) Average ChIP-nexus signals for Dorsal-independent (n = 56) and Dorsal-dependent (n = 59) Cic binding motifs across the genome. RPM, reads per million. (D) Type of Cic motif found in the Dorsal-independent and Dorsal-dependent sets: CBS motifs are an exact match to TSAATGAA, while AT motifs contain one mismatch relative to this consensus site. (E) Fraction of Cic binding motifs flanked by Dorsal binding motifs (GGRWWTTCC with up to one mismatch) within less than 50 bp.
Similar Dorsal-dependent binding of Cic was also observed at other enhancers driving dorsal-specific expression in the early embryo. For example, the dpp enhancer, which, like the zen and tld CRMs, was known to contain ventral-specific repression sequences (26), has a conserved AT/Dl pair with Dorsal-dependent Cic binding (Fig. 3A and Figs. S5 and S6). Furthermore, more recently identified dorsal ectodermal enhancers such as those of schnurri (shn) and Dorsocross2 (Doc2) (27), but not the intermediate neuroblasts defective (ind) enhancer, which is regulated by Cic via optimal CBSs and RTK-mediated derepression (6, 28), show Dorsal-dependent binding of Cic (Fig. 3B and Fig. S5). We also noted that anterior-posterior (AP) genes previously found to exhibit altered expression in cic mutants, including orthodenticle (otd) and giant (gt) (5, 29), did not show Cic ChIP-nexus footprints, suggesting that this effect is indirect (Fig. 3B).
We next tested more systematically whether Dorsal-dependent binding of Cic to the genome tends to be mediated through low-affinity Cic sites next to Dorsal sites. We first identified two sets of Cic-bound regions containing a Cic motif, one where Cic binds in both Toll10b and gd7 embryos (Dorsal-independent Cic binding, n = 56) and one where Cic binds in Toll10b but not gd7 samples (Dorsal-dependent Cic binding, n = 59). The average Dorsal-dependent Cic footprint is remarkably strong in signal and looks very similar in shape to the Dorsal-independent footprint (Fig. 3C). However, the motif content shows striking differences between both sets. In the first set, most Cic sites are high-affinity CBSs, while the second set has a much larger fraction of suboptimal sites with mismatches (71% AT sites versus 37.5% in the first set; P < 0.0004, Fisher test) (Fig. 3D). At the same time, the second set has a much higher fraction of Dorsal binding sites in the vicinity of the Cic motif (75% Dorsal sites versus 12.5% in the first set; P < 10−11, Fisher test) (Fig. 3E). Together, these results provide genome-wide evidence that Dorsal-dependent binding of Cic to DNA depends on low-affinity sites linked to nearby Dorsal sites.
Cic Represses zen via the Gro Corepressor.
The preceding results indicate that Dorsal mediates repression of its DV targets by facilitating Cic binding to low-affinity sites, rather than enabling Cic’s ability to repress. To further test this idea, we analyzed the relationship between Dorsal, Cic, and Gro, the latter of which is a corepressor required for Dorsal-mediated repression (16, 17). Gro is a WD-repeat factor that is recruited to target enhancers via interactions with short peptide motifs present in transcriptional repressors (30, 31). Both Cic and Dorsal contain eh1-like motifs that have been implicated in Gro-mediated repression (22, 32), but their relative importance in Dorsal-mediated repression has not been assessed. We have therefore used CRISPR-Cas9 to create cic and dl mutants that lack the eh1-like motif (Fig. S7 and Table S1). We found that the eh-1–like motif of Cic (referred to as N2; Fig. 4A) plays a more important role in zen repression (Fig. S7 A–H). Consistent with this result, we also found that N2 has stronger intrinsic repressor activity than the Dorsal eh-1–like motif in an in vivo repressor assay (Fig. S7 I–O).
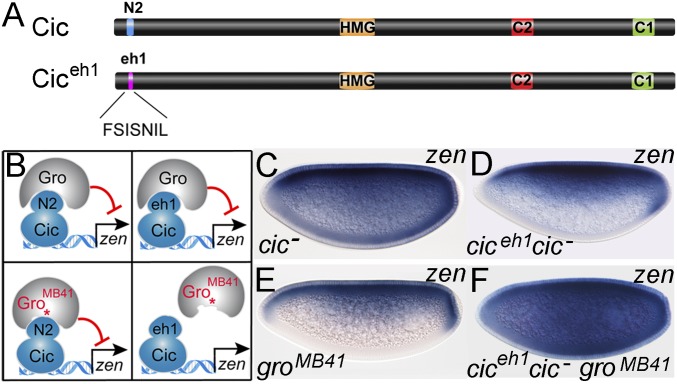
Fig. 4.
N2 motif of Cic mediates recruitment of Gro during zen repression. (A) Diagram of Cic and Ciceh1 proteins. (B) Model of interactions between Cic and Ciceh1 with intact and mutant Gro proteins. Cic can bind to both Gro and GroMB41, whereas the Ciceh1 chimera can only associate with intact Gro. GroMB41 carries the amino acid substitution R483H (asterisk) in the WD region (33). (C–F) Expression of zen in the indicated backgrounds. The cic mutant genotypes are cic1/cicQ474X (C and D) and cic1 (F). Embryos in D and F express maternally contributed Ciceh1 product. Images in C–F were obtained at 200× magnification.
To confirm a critical role of N2 in the repression of DV targets, we used a gro allele (groMB41), which prevents Gro from binding to canonical eh1 motifs of proteins such as Engrailed (En), but does not affect Cic-mediated repression via the N2 motif in AP patterning (22, 33) (Fig. 4B). Consistent with the above results, groMB41 embryos (laid by females carrying groMB41 germline clones) show normal zen expression (Fig. 4E), indicating that GroMB41 is functional in DV repression. We then reasoned that if N2 is the main element mediating Gro-dependent repression of zen, replacing this motif with a canonical eh1 peptide should render zen repression sensitive to the groMB41 mutation (Fig. 4 A and B). Indeed, replacing N2 by the eh1 peptide of En produced a functional chimera (Ciceh1; Fig. 4A) that was capable of repressing zen (Fig. 4 C and D). However, Ciceh1 was unable to repress zen in groMB41 embryos (Fig. 4F), indicating that N2 is essential for GroMB41 repressor activity in this context. This strongly suggests that binding of the Cic N2 motif to Gro is critical for zen repression in vivo, while the eh1-like motif of Dorsal is dispensable. This further supports our conclusion that the main repressive function of Dorsal is to facilitate DNA binding of Cic, which, in turn, recruits Gro.
Cic Mediates Cross-Regulation Between Torso and Toll Signaling.
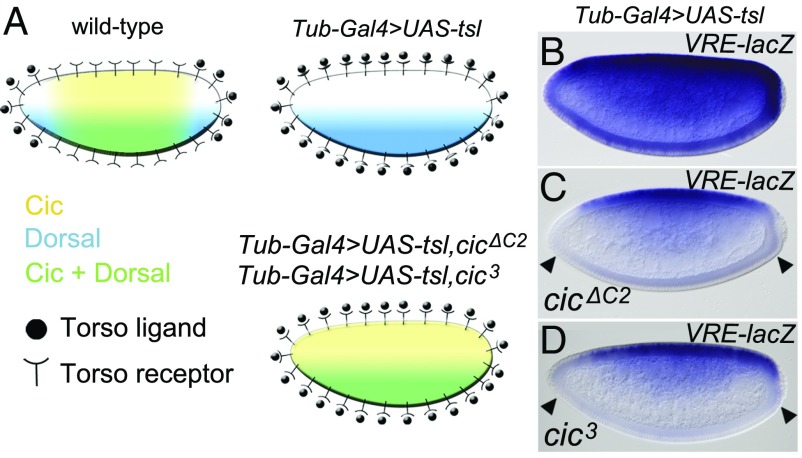
The above results identify a role for Cic in DV patterning that is distinct from its usual mode of regulation by RTK signaling. RTK signaling by Torso (and MAPK activity) is localized at the embryo poles at this stage and cannot account for the differential regulation of Cic targets across the DV axis. However, VRE- and VREOPT-lacZ reporter expression is notably derepressed at the poles, suggesting that Torso signaling might limit the repression of DV targets by MAPK-mediated phosphorylation and down-regulation of Cic. Indeed, this would explain why constitutive Torso activity throughout the embryo leads to zen derepression in ventral regions (34). To test if Cic mediates the cross-regulation between Torso and Toll signaling (Fig. 5A), we ectopically activated Torso via ubiquitous expression of the Torso-like (Tsl) determinant in follicle cells (35). As expected, this caused clear derepression of the VRE-lacZ reporter (Fig. 5B). If this derepression is mediated through MAPK-mediated down-regulation of Cic, we reasoned that a MAPK-insensitive form of Cic should restore VRE-lacZ repression on the ventral side. Indeed, when crossed with cic alleles in which the C2 MAPK-docking motif was deleted [cicΔC2 (4)] or mutated [cic3 (36)], VRE-lacZ expression was almost normal again (Fig. 5 C and D). The only difference was that repression was now also observed at the poles (Fig. 5 C and D), consistent with Torso signaling being no longer able to antagonize VRE-lacZ repression by down-regulating Cic. This demonstrates that Cic is sensitive to RTK down-regulation during the repression of Dorsal targets but that its principle mode of operation in this context, selective repression in the presence of Dorsal, occurs independently of RTK signaling. Similarly, while Cic also responds to EGFR RTK signaling at a slightly later stage of DV patterning (6), this mechanism is again distinct from the activities discussed here.
Fig. 5.
Cic is the key component of zen repression targeted by Torso signaling. (A) Diagram illustrating the spatial relationship between Cic, Dorsal, and the Torso pathway in wild-type embryos, embryos derived from mothers with uniform Tsl expression in the follicle cells (Tub-Gal4 > UAS-tsl), and embryos from Tub-Gal4 > UAS-tsl mothers that express MAPK-insensitive forms of Cic. (B–D) Effects of uniform Torso activation on VRE-lacZ expression. (B) Control embryo expressing intact Cic. (C and D) Embryos expressing MAPK-insensitive forms of Cic produced by either a cicΔC2 transgene (4) or the cic3 allele (36), respectively. Note that both cic mutations lead to VRE-lacZ repression at the poles, since Cic is no longer down-regulated at those positions (arrowheads; compare with Fig. 1F). Photographs in B–D were taken at 200× magnification.
Discussion
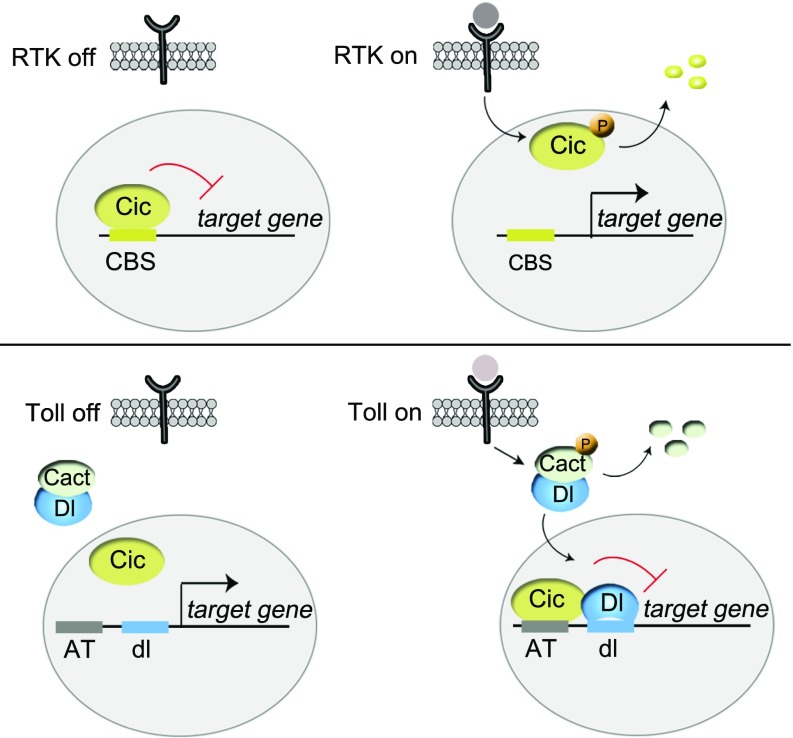
Cic is a conserved regulator of RTK responses, but it has been unclear whether Cic always functions in connection with RTK signaling. This study reveals an RTK-independent role of Cic downstream of Toll signaling, which is based on a different regulatory principle: Whereas RTK signaling down-regulates Cic, causing derepression of its targets, Toll signaling promotes Cic-mediated repression (Fig. 6). It does so by inducing the nuclear translocation of Dorsal, which, in turn, is required for Cic binding to low-affinity AT sites in target genes. In this manner, Cic recognizes sites of different affinities to orchestrate transcriptional responses along the AP and DV embryonic axes (Fig. S8). Recently, it has been shown that some ubiquitously expressed transcriptional repressors may play a role across both the Drosophila AP and DV embryonic axes by coordinating the timing of gene expression (37). Our work, on the other hand, shows how a single ubiquitously expressed transcriptional repressor can control the simultaneous spatial subdivision of two orthogonal axes by receiving input from two different signaling pathways and by binding to either high- or low-affinity sites.
Fig. 6.
Different modes of Cic regulation in RTK- and Toll-dependent transcriptional control. (Upper) As a default repressor of RTK target genes, Cic binds to optimal CBSs in those genes and represses them in the absence of signaling. Following RTK activation, Cic is down-regulated via MAPK-mediated phosphorylation (P), which leads to derepression of its targets. (Lower) In contrast, in the context of Toll signaling, Cic functions via suboptimal AT sites that are not recognized by Cic when the signal is off. Upon Toll activation, Dorsal (Dl/dl) translocates into the nucleus and facilitates Cic binding to the AT sites.
Our findings significantly extend previous work highlighting the importance of suboptimal DNA binding sites in conferring specificity to developmental enhancers (38–40). Suboptimal sites can help increase the binding specificity for different members of the Hox proteins and their cofactors (38). They may also increase the specificity for combinatorial input, as suboptimal binding sites in the Ciona Otx-a enhancer are important to ensure that activation only occurs in tissues where GATA factor is expressed and FGF signaling is active (39). However, it has been unclear how such combinatorial control occurs at the molecular level in this paradigm of enhancer regulation. Our results suggest that combinatorial control occurs at the level of binding and that it extends to repression.
Our data provide direct in vivo evidence that low-affinity sites make binding dependent on another transcription factor. Despite being low-affinity sites in vitro, the suboptimal AT sites show strong in vivo occupancy by Cic in the presence of Dorsal and very low occupancy in tissues where Dorsal is not nuclear. Likewise, changing the AT sites to optimal CBSs did not uniformly increase repression in the embryo but caused a dramatic increase in areas without Dorsal. This suggests that Cic and Dorsal cooperate at the level of binding. Based on the ChIP-nexus profile, we favor a model in which Dorsal physically stabilizes the binding of Cic. Alternatively, it is also possible that binding of Dorsal produces local changes in chromatin that facilitate the subsequent binding of Cic to the AT sites. Future studies will be aimed at further dissecting how Cic and Dorsal cooperate in binding at the molecular level.
Another important insight is that suboptimal binding sites can be critical for regulating repression. This helps explain how transcription factors such as Dorsal can have dual functions in activation and repression. Dorsal is inherently a weak activator that associates with basic helix–loop–helix factors for robust DNA binding and activation (25, 41–43). Similarly, our data suggest that Dorsal lacks intrinsic repressor activity and that its main repressive function is to facilitate the binding of repressors such as Cic. Therefore, by acting as a DNA binding cofactor for either activator or repressor proteins, Dorsal exerts opposite modes of transcriptional control to pattern the entire DV embryonic axis.
These insights have implications for studying enhancer function in development and disease. If relevant binding sequences in enhancers can be of low affinity, they can be hard to find despite exerting critical functions in vivo. For example, Cic has important roles in tumor formation (reviewed in refs. 1, 10), yet target genes are often scored based on the presence of canonical CBSs (7, 9, 44). In light of our data, we suggest that suboptimal “cryptic” sites linked to cofactor sites may also control a subset of mammalian Cic targets, thereby expanding the regulatory potential of this repressor.
Materials and Methods
Drosophila Genetics.
The cic5, cic6, and dl28 alleles were generated as described (36) via CRISPR-Cas9–induced nonhomologous end joining. Target sites of guide RNA sequences and information on other alleles, transgenes, and genetic procedures used in this study are provided in SI Materials and Methods.
In Situ Hybridization and Immunostaining.
Embryos were collected, dechorionated, and fixed in 4% formaldehyde-PBS-heptane using standard procedures. In situ hybridizations were performed using digoxigenin-UTP–labeled antisense RNA probes. Dorsal protein was detected using monoclonal antibody 7A4-c (Developmental Studies Hybridoma Bank). Additional information is provided in SI Materials and Methods.
EMSAs.
The HMG-C1 and HMG-C1mut constructs have been described by Forés et al. (20) and correspond to Dm CIC HMG-C1-His and Dm CIC HMG-C1ΔRQKL-His, respectively, with Dm standing for Drosophila melanogaster. HMGmut-C1 was made similarly in pET-29b vector. EMSAs were carried out using standard protocols. Details of probes can be found in SI Materials and Methods.
ChIP-nexus.
Embryos derived from heterozygous Toll10b or homozygous gd7 females were collected on apple juice plates at 25 °C for 2–4 h after egg deposition as done previously (27). Immunoprecipitations were carried out using previously published antibodies against Dorsal (25) and Cic (45). ChIP-nexus experiments and data processing were performed as previously described (25). Further details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Ainoa Olza for Drosophila injections; Marco Milán, Albert Courey, and Mike Levine for discussions; and Barbara Jennings, Iswar Hariharan, Mike Levine, Laura Nilson, David Stein, and the Bloomington Drosophila Stock Center for fly strains. N.K.’s contribution was part of her PhD thesis with the Open University, Milton Keynes, United Kingdom. This work was funded by grants from the Spanish Government (Grants BFU2011-23611 and BFU2014-52863-P), Fundació La Marató de TV3 (Grant 20131730), and Stowers Institute for Medical Research. Z.P. is an incumbent of the Lady Davis Chair in Experimental Medicine and Cancer Research, and is supported by the Jan M. and Eugenia Król Charitable Foundation.
Footnotes
Conflict of interest statement: J.Z. owns a patent on ChIP-nexus.
This article is a PNAS Direct Submission.
Data deposition: The raw and processed ChIP data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE104839).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713930115/-/DCSupplemental.
References
- 1.Jiménez G, Shvartsman SY, Paroush Z. The Capicua repressor–A general sensor of RTK signaling in development and disease. J Cell Sci. 2012;125:1383–1391. doi: 10.1242/jcs.092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin Y, et al. EGFR/Ras signaling controls Drosophila intestinal stem cell proliferation via Capicua-regulated genes. PLoS Genet. 2015;11:e1005634. doi: 10.1371/journal.pgen.1005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiménez G, Guichet A, Ephrussi A, Casanova J. Relief of gene repression by torso RTK signaling: Role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 2000;14:224–231. [PMC free article] [PubMed] [Google Scholar]
- 4.Astigarraga S, et al. A MAPK docking site is critical for downregulation of Capicua by torso and EGFR RTK signaling. EMBO J. 2007;26:668–677. doi: 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Löhr U, Chung HR, Beller M, Jäckle H. Antagonistic action of Bicoid and the repressor Capicua determines the spatial limits of Drosophila head gene expression domains. Proc Natl Acad Sci USA. 2009;106:21695–21700. doi: 10.1073/pnas.0910225106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ajuria L, et al. Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila. Development. 2011;138:915–924. doi: 10.1242/dev.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okimoto RA, et al. Inactivation of Capicua drives cancer metastasis. Nat Genet. 2017;49:87–96. doi: 10.1038/ng.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu HC, et al. Disruption of the ATXN1-CIC complex causes a spectrum of neurobehavioral phenotypes in mice and humans. Nat Genet. 2017;49:527–536. doi: 10.1038/ng.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simón-Carrasco L, et al. Inactivation of Capicua in adult mice causes T-cell lymphoblastic lymphoma. Genes Dev. 2017;31:1456–1468. doi: 10.1101/gad.300244.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka M, Yoshimoto T, Nakamura T. A double-edged sword: The world according to Capicua in cancer. Cancer Sci. 2017;108:2319–2325. doi: 10.1111/cas.13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeves GT, Stathopoulos A. Graded Dorsal and differential gene regulation in the Drosophila embryo. Cold Spring Harb Perspect Biol. 2009;1:a000836. doi: 10.1101/cshperspect.a000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stein DS, Stevens LM. Maternal control of the Drosophila dorsal-ventral body axis. Wiley Interdiscip Rev Dev Biol. 2014;3:301–330. doi: 10.1002/wdev.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belvin MP, Jin Y, Anderson KV. Cactus protein degradation mediates Drosophila dorsal-ventral signaling. Genes Dev. 1995;9:783–793. doi: 10.1101/gad.9.7.783. [DOI] [PubMed] [Google Scholar]
- 14.Jiang J, Cai H, Zhou Q, Levine M. Conversion of a dorsal-dependent silencer into an enhancer: Evidence for dorsal corepressors. EMBO J. 1993;12:3201–3209. doi: 10.1002/j.1460-2075.1993.tb05989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirov N, Zhelnin L, Shah J, Rushlow C. Conversion of a silencer into an enhancer: Evidence for a co-repressor in dorsal-mediated repression in Drosophila. EMBO J. 1993;12:3193–3199. doi: 10.1002/j.1460-2075.1993.tb05988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubnicoff T, et al. Conversion of Dorsal from an activator to a repressor by the global corepressor Groucho. Genes Dev. 1997;11:2952–2957. doi: 10.1101/gad.11.22.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valentine SA, et al. Dorsal-mediated repression requires the formation of a multiprotein repression complex at the ventral silencer. Mol Cell Biol. 1998;18:6584–6594. doi: 10.1128/mcb.18.11.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goff DJ, Nilson LA, Morisato D. Establishment of dorsal-ventral polarity of the Drosophila egg requires capicua action in ovarian follicle cells. Development. 2001;128:4553–4562. doi: 10.1242/dev.128.22.4553. [DOI] [PubMed] [Google Scholar]
- 19.Andreu MJ, et al. Mirror represses pipe expression in follicle cells to initiate dorsoventral axis formation in Drosophila. Development. 2012;139:1110–1114. doi: 10.1242/dev.076562. [DOI] [PubMed] [Google Scholar]
- 20.Forés M, et al. A new mode of DNA binding distinguishes Capicua from other HMG-box factors and explains its mutation patterns in cancer. PLoS Genet. 2017;13:e1006622. doi: 10.1371/journal.pgen.1006622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam YC, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 22.Forés M, et al. Origins of context-dependent gene repression by Capicua. PLoS Genet. 2015;11:e1004902. doi: 10.1371/journal.pgen.1004902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth S, Hiromi Y, Godt D, Nüsslein-Volhard C. cactus, a maternal gene required for proper formation of the dorsoventral morphogen gradient in Drosophila embryos. Development. 1991;112:371–388. doi: 10.1242/dev.112.2.371. [DOI] [PubMed] [Google Scholar]
- 24.Cline TW, Meyer BJ. Vive la différence: Males vs females in flies vs worms. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- 25.He Q, Johnston J, Zeitlinger J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat Biotechnol. 2015;33:395–401. doi: 10.1038/nbt.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang JD, Schwyter DH, Shirokawa JM, Courey AJ. The interplay between multiple enhancer and silencer elements defines the pattern of decapentaplegic expression. Genes Dev. 1993;7:694–704. doi: 10.1101/gad.7.4.694. [DOI] [PubMed] [Google Scholar]
- 27.Koenecke N, Johnston J, Gaertner B, Natarajan M, Zeitlinger J. Genome-wide identification of Drosophila dorso-ventral enhancers by differential histone acetylation analysis. Genome Biol. 2016;17:196. doi: 10.1186/s13059-016-1057-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim B, et al. Kinetics of gene derepression by ERK signaling. Proc Natl Acad Sci USA. 2013;110:10330–10335. doi: 10.1073/pnas.1303635110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Xu Z, Mei C, Yu D, Small S. A system of repressor gradients spatially organizes the boundaries of Bicoid-dependent target genes. Cell. 2012;149:618–629. doi: 10.1016/j.cell.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jennings BH, Ish-Horowicz D. The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biol. 2008;9:205. doi: 10.1186/gb-2008-9-1-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turki-Judeh W, Courey AJ. Groucho: A corepressor with instructive roles in development. Curr Top Dev Biol. 2012;98:65–96. doi: 10.1016/B978-0-12-386499-4.00003-3. [DOI] [PubMed] [Google Scholar]
- 32.Flores-Saaib RD, Jia S, Courey AJ. Activation and repression by the C-terminal domain of Dorsal. Development. 2001;128:1869–1879. doi: 10.1242/dev.128.10.1869. [DOI] [PubMed] [Google Scholar]
- 33.Jennings BH, et al. Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol Cell. 2006;22:645–655. doi: 10.1016/j.molcel.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 34.Rusch J, Levine M. Regulation of the dorsal morphogen by the Toll and torso signaling pathways: A receptor tyrosine kinase selectively masks transcriptional repression. Genes Dev. 1994;8:1247–1257. doi: 10.1101/gad.8.11.1247. [DOI] [PubMed] [Google Scholar]
- 35.Stevens LM, Beuchle D, Jurcsak J, Tong X, Stein D. The Drosophila embryonic patterning determinant Torsolike is a component of the eggshell. Curr Biol. 2003;13:1058–1063. doi: 10.1016/s0960-9822(03)00379-8. [DOI] [PubMed] [Google Scholar]
- 36.Forés M, Papagianni A, Rodríguez-Muñoz L, Jiménez G. Using CRISPR-Cas9 to study ERK signaling in Drosophila. In: Jimenez G, editor. ERK Signaling: Methods and Protocols. Springer; New York: 2017. pp. 353–365. [DOI] [PubMed] [Google Scholar]
- 37.Koromila T, Stathopoulos A. Broadly expressed repressors integrate patterning across orthogonal axes in embryos. Proc Natl Acad Sci USA. 2017;114:8295–8300. doi: 10.1073/pnas.1703001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crocker J, et al. Low affinity binding site clusters confer Hox specificity and regulatory robustness. Cell. 2015;160:191–203. doi: 10.1016/j.cell.2014.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farley EK, et al. Suboptimization of developmental enhancers. Science. 2015;350:325–328. doi: 10.1126/science.aac6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crocker J, Noon EP, Stern DL. The soft touch: Low-affinity transcription factor binding sites in development and evolution. Curr Top Dev Biol. 2016;117:455–469. doi: 10.1016/bs.ctdb.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 41.Szymanski P, Levine M. Multiple modes of dorsal-bHLH transcriptional synergy in the Drosophila embryo. EMBO J. 1995;14:2229–2238. doi: 10.1002/j.1460-2075.1995.tb07217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72:741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- 43.González-Crespo S, Levine M. Interactions between dorsal and helix-loop-helix proteins initiate the differentiation of the embryonic mesoderm and neuroectoderm in Drosophila. Genes Dev. 1993;7:1703–1713. doi: 10.1101/gad.7.9.1703. [DOI] [PubMed] [Google Scholar]
- 44.Kawamura-Saito M, et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15:2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- 45.Pascual J, et al. Hippo reprograms the transcriptional response to Ras signaling. Dev Cell. 2017;42:667–680.e4. doi: 10.1016/j.devcel.2017.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.