Significance
Identifying and explaining regional differences in tropical forest dynamics, structure, diversity, and composition are critical for anticipating region-specific responses to global environmental change. Floristic classifications are of fundamental importance for these efforts. Here we provide a global tropical forest classification that is explicitly based on community evolutionary similarity, resulting in identification of five major tropical forest regions and their relationships: (i) Indo-Pacific, (ii) Subtropical, (iii) African, (iv) American, and (v) Dry forests. African and American forests are grouped, reflecting their former western Gondwanan connection, while Indo-Pacific forests range from eastern Africa and Madagascar to Australia and the Pacific. The connection between northern-hemisphere Asian and American forests is confirmed, while Dry forests are identified as a single tropical biome.
Keywords: biogeographic legacies, forest classification, forest functional similarity, phylogenetic community distance, tropical forests
Abstract
Knowledge about the biogeographic affinities of the world’s tropical forests helps to better understand regional differences in forest structure, diversity, composition, and dynamics. Such understanding will enable anticipation of region-specific responses to global environmental change. Modern phylogenies, in combination with broad coverage of species inventory data, now allow for global biogeographic analyses that take species evolutionary distance into account. Here we present a classification of the world’s tropical forests based on their phylogenetic similarity. We identify five principal floristic regions and their floristic relationships: (i) Indo-Pacific, (ii) Subtropical, (iii) African, (iv) American, and (v) Dry forests. Our results do not support the traditional neo- versus paleotropical forest division but instead separate the combined American and African forests from their Indo-Pacific counterparts. We also find indications for the existence of a global dry forest region, with representatives in America, Africa, Madagascar, and India. Additionally, a northern-hemisphere Subtropical forest region was identified with representatives in Asia and America, providing support for a link between Asian and American northern-hemisphere forests.
The biogeographic origin of species, in combination with dispersal limitation and environmental filtering, are the principal determinants of spatial variation in the species composition of tropical forests (1, 2). Despite evidence of long-distance dispersal (1, 3–5), tropical forests maintain conspicuous regional differences in species composition. For example, only ∼4% of tropical tree species are shared among Africa, America, and Asia (6). The lack of species overlap among continents makes global inference of relationships among tropical forests problematic, because such classifications depend on comparison of the amount of shared species. Therefore, pan-tropical biogeographic analyses have been based on comparison of compositional patterns at higher taxonomic levels, namely genus or family (6–8). However, such analyses treat taxa as independent units, while in reality taxa vary in their degree of phylogenetic relatedness and, as a consequence, their morphological and ecological similarity (1, 2). Taking phylogenetic relatedness into consideration enhances our ability to delimit phytogeographical boundaries that characterize functional and biogeographic affinities among forest regions (1, 2, 9, 10). Here we include phylogenetic relationships in a floristic analysis to provide such insight.
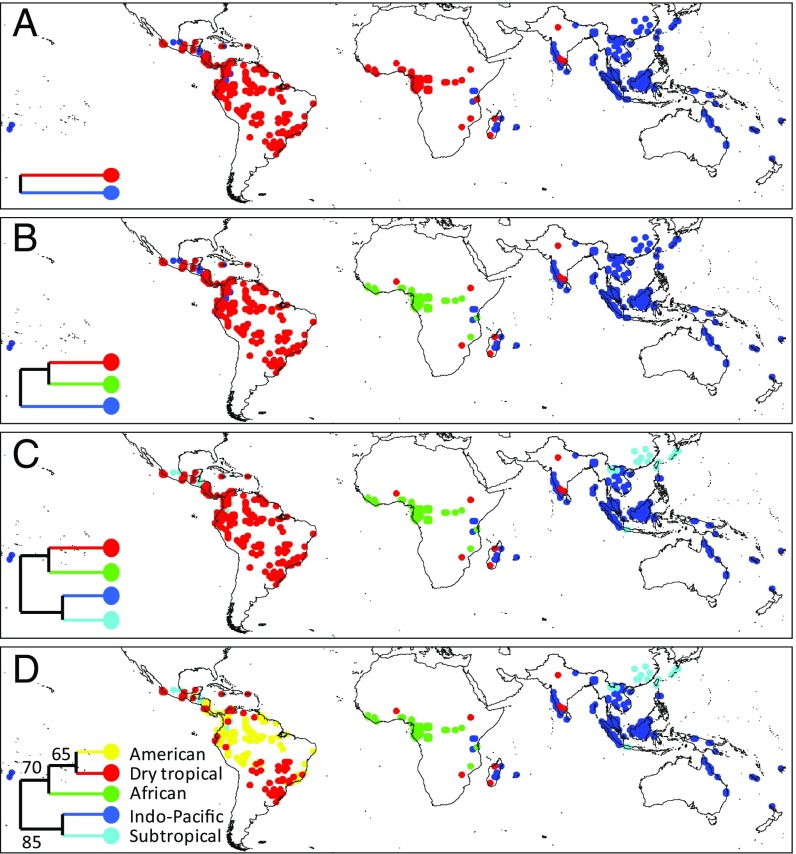
We compiled a standardized dataset of old-growth tropical forest inventories of angiosperm trees (trunk diameter ≥10 cm) for 406 1° latitude/longitude grid cells (hereafter referred to as “locations”) originally dominated by natural forests across the (sub)tropics (Table S1). These locations represented all major tropical forest regions and had broad environmental amplitude, including low to high elevations and dry to wet forests (Fig. 1 and Fig. S1). To determine the phylogenetic distance between locations, we constructed a dated phylogenetic tree that was resolved to genus level and contained all taxa used for our classification analyses (Dataset S1). Location pairwise phylogenetic distance matrices were constructed using 20 randomly drawn tree taxa per location. We used 20 taxa, as this maximized the number of locations that could be included in the classification analyses while still providing a reliable classification result. In total, we generated 20 phylogenetic distance matrices, each with a different set of 20 randomly drawn taxa per location, which served as input for 20 cluster analyses (Fig. S2). The final classification of each location depended on the frequency with which it was classified in a particular cluster across all 20 cluster analyses (Fig. S3). Relationships between the clusters were represented by a majority rule consensus tree (Fig. 1).
Fig. 1.
Classification maps of the world’s tropical forests, showing two (A), three (B), four (C), and five (D) clusters. Cluster result represents a majority rule consensus tree, with the percentage of times that each grouping was observed in the 20 separate cluster analyses shown in D. Only locations that could be classified with certainty (P < 0.05) are shown (n = 392).
Results and Discussion
Mean pairwise phylogenetic distance analysis, which emphasizes ancient lineages in phylogenetic community comparisons, detected almost no spatial patterns in community phylogenetic similarity across the tropics, indicating that all tropical forest locations consist of more or less the same set of ancient plant lineages. This is in accordance with recent findings that the whole present-day tropics are dominated by similar high levels of Late Cretaceous aged phylogenetic lineages (11). Only when we used mean nearest taxon distance, which emphasizes recent lineages in phylogenetic community comparisons, did we detect clear spatial patterns across the tropics. Therefore, current-day biogeographic patterns in the tropics seem to mainly reflect Cenozoic speciation events when Gondwanan breakup was already well on its way.
Using the mean nearest taxon distance, our phylogenetic cluster analyses showed that the world’s tropical forests are divided into two major floristic regions: a combined American-African versus Indo-Pacific region (Fig. 1). This division contradicts previous hypotheses about major global realms, which either recognized neo- versus paleotropical regions or several separate continental regions (4, 12–14). However, Gentry (7) already noted the high generic-level similarity of tropical American and African forests. He attributed this to Cretaceous and Cenozoic plate tectonic history (4, 15). Subsequent studies have shown that despite the severing of direct land connections between the African and South American plates ca. 96 Mya, long-distance dispersal continued throughout the Late Cretaceous and Early Tertiary across the widening Atlantic Ocean (4, 5). The combined effect of shared origin with trans-Atlantic migration may explain the detected connection between South American and African forests.
Within the American-African cluster, the first split separated the African from the American regions (Fig. 1), suggestive of the west Gondwanan breakup associated with the formation of the Atlantic Ocean and, over time, the increasing difficulty for plants to disperse across the Atlantic (1, 15). Interestingly, the African region showed the highest consistency in clustering of all five identified floristic regions. On average, locations belonging to the African region were assigned to this cluster in 91.4% of cases, versus consistency values of 79.5, 63.7, 79.5, and 70.3% for the Indo-Pacific, Subtropical, American, and Dry forest regions, respectively. This clustering consistency indicates high floristic similarity across tropical Africa, which is in accord with the relatively low beta diversity observed for these forests (6). Postulated repeated cycles of contraction and expansion of the tropical African forests from a few small forest refugia in combination with large-scale species shifts during the Pleistocene glaciations may explain the relatively high compositional homogeneity of the forests within the African region (16, 17).
The tropical American forests were further divided into moist and dry forests (Fig. 1 and Fig. S1), indicating that this division is primarily environmental (18). The American floristic region comprises humid forests, including the lowland forests of Central America, the Amazon basin, the Guianas, and the northern half of the Atlantic forest. The Dry forest region encompasses the Caatinga and Cerrado regions as well as other dry forests throughout the Americas but interestingly, and contrary to the nonphylogenetic pan-tropical analysis by Dexter et al. (8), also includes dry forests of Africa, Madagascar, and India. Further research is needed to confirm whether this indicates the existence of a global dry forest region with a shared biogeographic origin, or whether selection for drought and fire resistance has favored the dominance of similar plant lineages in tropical dry forests around the world (8, 18, 19).
The Indo-Pacific floristic region occupies the humid areas of eastern Africa, Madagascar, India, Southeast Asia, Australia, and the Pacific Islands (Fig. 1). With the exception of SE Asia, which is of Laurasian origin, this floristic region combines all areas that once comprised eastern Gondwana (4, 15). Given the diverse geologic history of Asia and the Indo-Pacific (20), it is surprising to find a similar forest type covering most of the region. Nevertheless, there is strong evidence of significant plant migration within this region that likely had a homogenizing effect, notably the biotic exchange between India and Southeast Asia starting from ca. 45 Mya (21), and between Southeast Asia and Australia, New Guinea, and the Pacific Islands that commenced ca. 15 Mya (4). The presence of Indo-Pacific forests in eastern continental Africa may either reflect eastern Gondwanan origin or dispersal within the Indo-Pacific region.
We also identified a group of locations in Asia and America that occupies cooler climates and higher elevations relative to the other identified forest clusters (Fig. 1 and Fig. S1), and which we therefore termed the Subtropical region. This Subtropical floristic region confirms the floristic link between Asia and North America, reflecting a shared boreotropical affinity (22). Within Asia, the Subtropical region is mostly restricted to the subtropics, with the exception of high-elevation forests of Java. In the Americas, by contrast, this floristic region extends from the subtropics deep into the tropics, probably because the cooler montane climate of the Central American highlands and South American Andes has facilitated the southward migration of cold-adapted plant lineages. The absence of continuous, North–South-oriented mountain chains in Asia may have limited the dispersal of such lineages into lower latitudes.
Conclusion
We provide a phylogenetic distance-based biogeographic classification of the world’s tropical forests, using the most extensive sampling scheme for the tropics currently in existence. Our results uncover floristic patterns which will help in the development of region-specific models for forest structure, diversity, and dynamics as well as possible responses of tropical forest regions to global environmental change. Our results may necessitate reconsideration of established biogeographic ideas. For example, Madagascar and New Guinea have often been considered two separate major tropical regions, ecologically and biogeographically distinct from tropical America, Africa, and Southeast Asia (23, 24). However, despite their highly endemic species compositions, we show that they are both part of the widespread Indo-Pacific floristic region. Finally, our analysis can serve as a model for classifying regional floras.
Materials and Methods
Tree Inventory Dataset.
Individual angiosperm trees (diameter at breast height ≥10 cm) from old-growth forest inventories throughout the (sub)tropics (between −35°S and 35°N latitudes) were pooled within their respective 1° latitude/longitude grid cells (henceforth called locations). These locations represented all major tropical forest regions and had broad environmental amplitude, including low to high elevations and dry to wet forests (Fig. S1). Monocots and Cactaceae were excluded because these were not consistently surveyed in all datasets. This dataset originally included 439 locations containing 925,009 individual trees belonging to 15,012 taxa. Species names were standardized using The Plant List (www.theplantlist.org), Taxonomic Name Resolution Service (tnrs.iplantcollaborative.org/TNRSapp.html), and Asian Plant Synonym Lookup (phylodiversity.net/fslik/synonym_lookup.htm). On average, 1.4% of individual stems per location remained unidentified. These unidentified individuals were excluded from further analyses.
Community Phylogenetic Tree.
The APG-III classification (25) served as the family-level backbone of our community phylogenetic tree. Recent updates in APG-IV (26) are mostly of nomenclatural nature and did not affect our analyses. This tree was further resolved up to genus level using the species-level phylogeny (32,223 species included) published by Zanne et al. (27), which covered most genera in our dataset (Dataset S1). Genera present in our dataset, but not in Zanne et al. (27), were placed at the base of their respective families. Genera that had disjunct species occurrences in the phylogeny of Zanne et al. (27) were placed at the most basal node connecting the disjunct species. This phylogeny was subsequently dated using the BLADJ function in Phylocom v4.2 (28), using taxon ages given in Magallón et al. (29) for the age file.
Phylogenetic Distance Analysis.
Phylogenetic distance between all pairs of locations was calculated using the options COMDIST and COMDISTNT in Phylocom v4.2 (28). COMDIST uses the mean pairwise phylogenetic distance (MPPD); for each taxon in a location, it finds the average phylogenetic distance to all taxa in the other location, and calculates the mean. COMDISTNT uses the mean nearest taxon distance (MNTD); for each taxon in location 1, it finds the nearest phylogenetic neighbor in location 2, records this, and calculates the mean. Both functions return a symmetrical matrix of locations versus locations with their pairwise phylogenetic distances. Principal coordinate (PCO) analyses (in MultiVariate Statistical Package v3.13; Kovach Computing Services) on resulting location versus location matrices showed that the MPPD matrices had almost no explanatory power (generally the first five PCO axes explained less than 5% of data variance), meaning that detected patterns were mostly random. The MNTD matrices, however, explained considerable amounts of data variance in the first five axes of the PCO. Therefore, we used only MNTD for further analysis.
Correcting for Taxon Richness Bias in MNTD.
Taxon richness differed considerably between locations, varying between 4 and 1,466. MNTD may be sensitive to such differences in taxon richness because the chance of finding a close relative between two locations may increase when their taxon richness increases. Applying MNTD to determine phylogenetic distance between locations with differing taxon numbers could therefore result in taxon-rich locations being grouped together in the cluster analysis simply because they are more taxon-rich. To determine the impact of this effect, we created five “location-by-taxon” matrices, each with a lower number of taxa per location (320, 160, 80, 40, and 20 taxa per location), using the 41 locations containing more than 320 taxa. For each location, taxa were ranked according to abundance, so that the location-by-taxon matrix based on, for example, 320 taxa consisted only of the 320 most abundant taxa per location. Where tied abundances exceeded the predefined number of taxa, we randomly selected the appropriate number of taxa from among those with tied minimum abundance. We then calculated the MNTD matrices for each of these five location-by-taxon matrices and found that with increasing taxon richness of locations, MNTD (as averaged over all locations) decreased with increasing taxon richness per location following a power function [y = 310.4x−0.194 (Fig. S4)], demonstrating that MNTD is indeed sensitive to taxon richness.
Determining the Optimal Number of Taxa per Location for Further Analysis.
To avoid taxon richness bias when using MNTD, locations had to be compared based on similar numbers of taxa. Minimum variance clustering, based on the five location-by-taxon matrices described earlier, consistently recovered the same major clusters in the same configuration (African and American locations clustered on one main branch and Asian locations clustered on the other), although the relationships between locations within these main clusters could vary (Fig. S5). Only in the 20-taxon analysis was one American location (location no. 165 from the Brazilian Atlantic Forest) placed in the Asian cluster. The amount of variance captured in the first five axes of a PCO analysis (using the same MNTD matrices) declined by only ∼20%, from 83.3 to 60.7%, between the 320- and 20-taxon analyses, respectively. We decided to use 20 taxa per location in the final analyses (Table S1) because of this limited loss of information in the PCO and similarity of cluster results. In addition, we were able to use most of our locations (406 of the initial 439), including locations on remote islands and extreme habitats that would have been excluded if we had set the minimum number of taxa too high.
Forest Classification Analyses.
For the final analyses, we produced 20 location-by-taxon datasets. In these datasets, each location was represented by 20 randomly drawn taxa (from that location). Random draws were irrespective of taxon abundance, as abundance is a spatially and temporally labile taxon trait that likely reflects contemporary environmental conditions rather than historical biogeographic signal. For each of these 20 location-by-taxon datasets, we calculated the corresponding symmetrical location-by-location matrices with their pairwise phylogenetic distances (MNTD). These matrices were then used as input for cluster analyses.
Locations were grouped in clusters using Ward’s minimum variance method (30), using MultiVariate Statistical Package v3.13. This is a centroid-based clustering technique that identifies cluster centers (centroids) by minimizing the overall squared distances of the objects (in this case locations) to the centroids at each cluster level. This clustering technique identified spatially clearly defined location groupings (Fig. S2). The optimal number of clusters for defining floristic regions across the tropics was determined by calculating the cophenetic correlation coefficient at each cluster level, starting at the first split (K2) in the dendrogram. The cophenetic correlation coefficient calculates the correlation between the distance of the clusters as calculated by the clustering algorithm and the distance based on observed MNTD values between clusters. The higher the cophenetic correlation, the better the cluster result reflects the patterns present in the original distance matrix. We applied this method to each of our 20 datasets, calculated the average cophenetic correlation coefficient for each cluster level, and found a steep increase in cophenetic correlation up to K5, after which it slowly declined (Fig. S6). Therefore, we chose K5 as the optimum level for defining our main floristic regions across the tropics.
For each location, at cluster level K5, we determined the cluster in which it was classified for each of the 20 cluster analyses that we performed. The location was then assigned to the cluster in which it had the highest proportion of observations. A single proportion test (31), which calculates the probability of an observed (sample) proportion (in the range 0 to 1) against a hypothetical proportion, was then used to determine if the observed proportions were significantly higher than expected by random [Paleontological Statistics (PAST) v3.08; https://folk.uio.no/ohammer/past/]. For example, for K5, the expected random proportion of locations per cluster is 0.2. For a sample size of 20, a proportion has to be at least 0.38 to be significantly higher (P < 0.05) than the random expectation. The resulting classification success rates of locations for K5 are shown in Fig. S3 and Table S1. The final classification (K5) of the clusters was based on the majority consensus rule (Fig. 1).
Supplementary Material
Acknowledgments
This study benefited greatly from data contributed by Patricia Alvarez-Loayza, Ana Andrade, Peter Ashton, Julian Bayliss, Luis Bernacci, Lilian Blanc, J. Bogaert, Matt Bradford, Mireille Breuer Ndoundou Hockemba, C. De Cannière, Miguel Castillo, Eduardo Catharino, Connie Clark, David Clark, Deborah Clark, Gilles Dauby, Jean-Louis Doucet, Pedro Eisenlohr, Leandro Ferreira, Christine Fletcher, Geraldo Franco, Gabriella M. Fredriksson, Girirai, Nimal Gunatilleke, Terese Hart, Miriam van Heist, Zhila Hemati, M. A. Hernández-Ruedas, David Kenfack, Kanehiro Kitayama, Eileen Larney, Ieda Leao do Amaral, Jean-Remy Makana, Punchi Manage Saranga Amila Ruwan, Antti Marjokorpi, Olga Martha Montiel, Miguel Martínez-Ramos, Henrik Meilby, Jerome Millet, Cao Min, Kazuki Miyamoto, Xiaoxue Mo, Juan Carlos Montero, Badru Mugerwa, Pantaleo Munishi, Helen Murphy, Hidetoshi Nagamasu, David Newbery, Rueben Nilus, Meyner Nusalawo, Susana Ochoa-Gaona, Atila Oliveira, Navendu Page, Andrea Permana, Nigel Pitman, Jean Razafimahaimodison, Rocío Rojas, Hugo Romero, M. Z. Rozainah, Fernanda Santos, Manichanh Satdichanh, Lars Schmidt, Lila Nath Sharma, Kade Sidiyasa, Eduardo da Silva Pinheiro, Peguy Tchouto, Johanna Urtado, Renato Valencia, Luis Valenzuela, Rodolfo Vasquez, Thorsten Wiegand, Guadelupe Williams-Linera, Hansjoerg Woll, Tsuyoshi Yoneda, and Nicole Zweifel. We also acknowledge contributed financial support from the European Union’s Horizon 2020 Research and Innovation Programme under Marie Skłodowska-Curie Grant Agreement 660020, Instituto Bem Ambiental (IBAM), Myr Projetos Sustentáveis, IEF, and CNPq, CAPES FAPEMIG, German Research Foundation (DFG; Grants CRC 552, CU127/3-1, HO 3296/2-2, HO3296/4-1, and RU 816), UNAM-PAPIIT IN218416 and Semarnat-CONACYT 128136, Conselho Nacional de Desenvolvimento Científico e Tecnoloógico (CNPq, Brazil), Fundação Grupo Boticário de Proteção à Natureza/Brazil, PAPIIT-DGAPA-UNAM (Project IN-204215), National Geographic Society, National Foundation for Scientific and Technology Development Vietnam (Grant 106.11-2010.68), Operation Wallacea, and core funding for Crown Research Institutes from the New Zealand Ministry of Business, Innovation and Employment’s Science and Innovation Group. Some data in this publication were provided by the Tropical Ecology Assessment and Monitoring Network, a collaboration between Conservation International, the Missouri Botanical Garden, Smithsonian Institution, and Wildlife Conservation Society, and partially funded by these institutions, The Gordon and Betty Moore Foundation, and other donors.
Footnotes
Conflict of interest statement: V.A.-R., K.B.-G., B.B., F.Q.B., N.F., M.K., W.F.L., S. G. Letcher, C.B.S., D.S., T. Stevart, and S. Wiser have coauthored papers with Jens-Christian Svenning in the past 48 months. A.M.L. and Jens-Christian Svenning are both affiliated with Aarhus University.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714977115/-/DCSupplemental.
References
- 1.Cavender-Bares J, Ackerly DD, Hobbie SE, Townsend PA. Evolutionary legacy effects on ecosystems: Biogeographic origins, plant traits, and implications for management in the era of global change. Annu Rev Ecol Evol Syst. 2016;47:433–462. [Google Scholar]
- 2.Donoghue MJ. A phylogenetic perspective on the distribution of plant diversity. Proc Natl Acad Sci USA. 2008;105:11549–11555. doi: 10.1073/pnas.0801962105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weigelt P, et al. Global patterns and drivers of phylogenetic structure in island floras. Sci Rep. 2015;5:12213. doi: 10.1038/srep12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morley RJ. Interplate dispersal paths for megathermal angiosperms. Perspect Plant Ecol Evol Syst. 2003;6:5–20. [Google Scholar]
- 5.Pennington RT, Dick CW. The role of immigrants in the assembly of the South American rainforest tree flora. Philos Trans R Soc Lond B Biol Sci. 2004;359:1611–1622. doi: 10.1098/rstb.2004.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slik JWF, et al. An estimate of the number of tropical tree species. Proc Natl Acad Sci USA. 2015;112:7472–7477, and correction (2015) 112:E4628–E4629. doi: 10.1073/pnas.1423147112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gentry AH. Changes in plant community diversity and floristic composition on environmental and geographical gradients. Ann Mo Bot Gard. 1988;75:1–34. [Google Scholar]
- 8.Dexter KG, et al. Floristics and biogeography of vegetation in seasonally dry tropical regions. Int For Rev. 2015;17:10–22. [Google Scholar]
- 9.Holt BG, et al. An update of Wallace’s zoogeographic regions of the world. Science. 2013;339:74–78. doi: 10.1126/science.1228282. [DOI] [PubMed] [Google Scholar]
- 10.Webb CO, et al. Phylogenies and community ecology. Annu Rev Ecol Syst. 2002;33:475–505. [Google Scholar]
- 11.Procheş Ş, Ramdhani S, Perera SJ, Ali JR, Gairola S. Global hotspots in the present-day distribution of ancient animal and plant lineages. Sci Rep. 2015;5:15457. doi: 10.1038/srep15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takhtajan AL. The Floristic Regions of the World. Univ of California Press; Berkeley: 1986. [Google Scholar]
- 13.Chang HT. An outline on the regionalisation of the global flora. Acta Sci Nat Univ Sunyatseni. 1994;33:73–80. [Google Scholar]
- 14.Cox CB. The biogeographic regions reconsidered. J Biogeogr. 2001;28:511–523. [Google Scholar]
- 15.McLoughlin S. The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism. Aust J Bot. 2001;49:271–300. [Google Scholar]
- 16.McClean CJ, et al. African plant diversity and climate change. Ann Mo Bot Gard. 2005;92:139–152. [Google Scholar]
- 17.Anhuf D, et al. Paleo-environmental change in Amazonian and African rainforest during the LGM. Palaeogeogr Palaeoclimatol Palaeoecol. 2006;239:510–527. [Google Scholar]
- 18.Banda-R K, et al. DRYFLOR Plant diversity patterns in neotropical dry forests and their conservation implications. Science. 2016;353:1383–1387. doi: 10.1126/science.aaf5080. [DOI] [PubMed] [Google Scholar]
- 19.Ratnam J, Tomlinson KW, Rasquinha DN, Sankaran M. Savannahs of Asia: Antiquity, biogeography, and an uncertain future. Philos Trans R Soc Lond B Biol Sci. 2016;371:20150305. doi: 10.1098/rstb.2015.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall R. Cenozoic reconstructions of SE Asia and the SW Pacific: Changing patterns of land and sea. In: Metcalfe I, Smith JMB, Morwood M, Davidson ID, editors. Faunal and Floral Migrations and Evolution in SE Asia-Australasia. Swets & Zeitlinger; Lisse, The Netherlands: 2001. pp. 35–56. [Google Scholar]
- 21.Klaus S, Morley RJ, Plath M, Zhang YP, Li JT. Biotic interchange between the Indian subcontinent and mainland Asia through time. Nat Commun. 2016;7:12132. doi: 10.1038/ncomms12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian H, Jin Y, Ricklefs RE. Phylogenetic diversity anomaly in angiosperms between eastern Asia and eastern North America. Proc Natl Acad Sci USA. 2017;114:11452–11457. doi: 10.1073/pnas.1703985114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Primack RB, Corlett RT. Tropical Rainforests: An Ecological and Biogeographical Comparison. Wiley-Blackwell; Hoboken, NJ: 2005. [Google Scholar]
- 24.Corlett RT, Primack RB. Tropical rainforests and the need for cross-continental comparisons. Trends Ecol Evol. 2006;21:104–110. doi: 10.1016/j.tree.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 25.APG-III An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 2009;161:105–121. [Google Scholar]
- 26.APG-IV An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 2016;181:1–20. [Google Scholar]
- 27.Zanne AE, et al. Three keys to the radiation of angiosperms into freezing environments. Nature. 2014;506:89–92. doi: 10.1038/nature12872. [DOI] [PubMed] [Google Scholar]
- 28.Webb CO, Ackerly DD, Kembel SW. Phylocom: Software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics. 2008;24:2098–2100. doi: 10.1093/bioinformatics/btn358. [DOI] [PubMed] [Google Scholar]
- 29.Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 2015;207:437–453. doi: 10.1111/nph.13264. [DOI] [PubMed] [Google Scholar]
- 30.Ward JH., Jr Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58:236–244. [Google Scholar]
- 31.Clopper C, Pearson ES. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. [Google Scholar]
- 32.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.