Significance
Uncultivated bacteria are hypothesized to represent a large resource of new bioactive natural products and biosynthetic enzymes. Previous work identified uncultivated “Entotheonella” symbionts of the sponge chemotype Theonella swinhoei Y as producers of a broad range of bioactive metabolites unknown from cultured bacteria. Here we present whole-genome data of an Entotheonella variant from a distinct chemotype of T. swinhoei. “Candidatus Entotheonella serta,” obtained from sponges from two oceans, possesses a biosynthetic complement even larger than, and nearly orthogonal to, those of the two previously described Entotheonella symbionts. This includes genes assigned to the misakinolides and theonellamides and suggests numerous additional natural products. The data validate Entotheonella as a rich and varied producer taxon with considerable biotechnological potential.
Keywords: uncultivated bacteria, sponges, single-cell genomics
Abstract
Marine sponges are prolific sources of unique bioactive natural products. The sponge Theonella swinhoei is represented by several distinct variants with largely nonoverlapping chemistry. For the Japanese chemotype Y harboring diverse complex polyketides and peptides, we previously provided genomic and functional evidence that a single symbiont, the filamentous, multicellular organism “Candidatus Entotheonella factor,” produces almost all of these compounds. To obtain further insights into the chemistry of “Entotheonella,” we investigated another phylotype, “Candidatus Entotheonella serta,” present in the T. swinhoei WA sponge chemotype, a source of theonellamide- and misakinolide-type compounds. Unexpectedly, considering the lower chemical diversity, sequencing of individual bacterial filaments revealed an even larger number of biosynthetic gene regions than for Ca. E. factor, with virtually no overlap. These included genes for misakinolide and theonellamide biosynthesis, the latter assigned by comparative genomic and metabolic analysis of a T. swinhoei chemotype from Israel, and by biochemical studies. The data suggest that both compound families, which were among the earliest model substances to study bacterial producers in sponges, originate from the same bacterium in T. swinhoei WA. They also add evidence that metabolic richness and variability could be a more general feature of Entotheonella symbionts.
The sponge Theonella swinhoei ranges throughout the Indo-West Pacific (www.marinespecies.org/porifera) and is a remarkably rich source of biologically active and chemically distinctive natural products (1–3). In the nearly 40 y since T. swinhoei gained the interest of chemists (4), over 150 substances have been reported. These occur in sponge variants forming three major phenotypes with distinct coloration and largely nonoverlapping sets of natural product families (5). Within each phenotype, further chemical variation occurs, resulting in significant overall metabolic complexity that has made T. swinhoei an attractive target for drug discovery and biosynthetic studies.
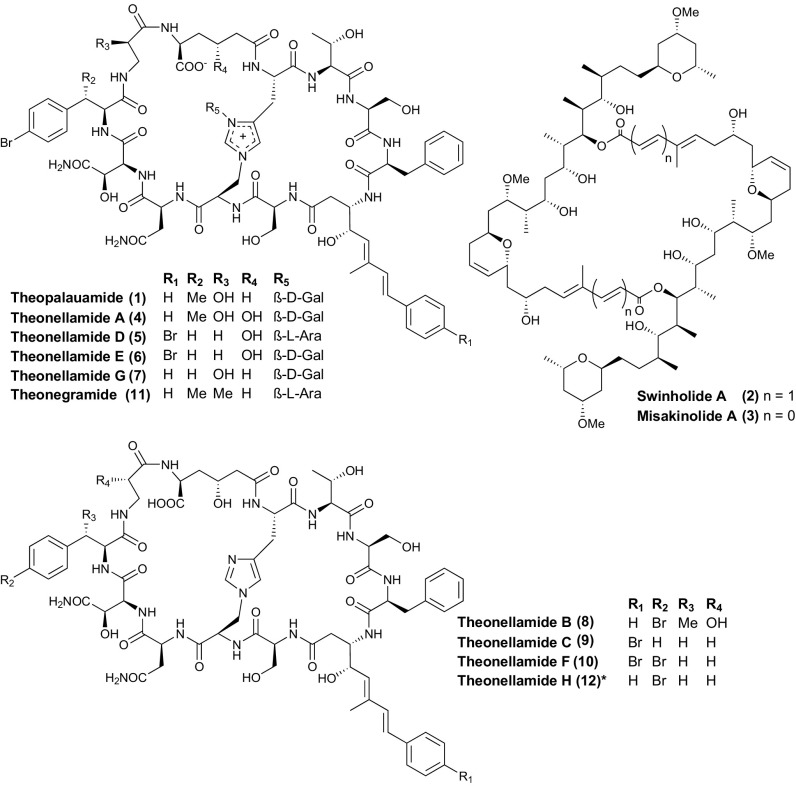
T. swinhoei and many other sponges harbor complex microbiota with similar diversity to that of humans (6, 7), suggesting the possibility that symbionts contribute to the chemistry of sponges (2, 8, 9). Showcase compounds that became early models to understand the origin of sponge metabolites are the antifungal peptide theopalauamide (1) and the actin-inhibiting complex polyketide swinholide A (2) (Fig. 1) (10, 11). In cell preparations of a Palauan T. swinhoei phenotype with white interior (12, 13), 2 was mainly detected in a mixed unicellular bacterial fraction, while amounts of 1 were highest in a fraction enriched with filamentous bacteria, named “Candidatus Entotheonella palauensis.” These chemical data suggested a bacterial origin and laid an important foundation for studies in the field to identify functional links between sponge-derived metabolites and their producers.
Fig. 1.
Major metabolites of T. swinhoei white chemotype sponges. The “*” indicates a metabolite proposed in this study.
By genomic and biosynthetic studies of T. swinhoei Y (14–17), a Japanese variant from Hachijo Island with a yellow interior, we and collaborators recently identified the source of onnamide-/theopederin–type polyketides (18), the ribosomal peptides polytheonamides (3), and the nonribosomal peptides keramamide/orbiculamide (19, 20), cyclotheonamide (21), and nazumamide (22). All of these compounds were linked to a single producer, “Candidatus Entotheonella factor” (16, 23, 24). This remarkable filamentous bacterium contains many additional biosynthetic gene clusters (BGCs) of as-yet unknown function, and is accompanied in the sponge by a second BGC-rich phylotype, “Ca. Entotheonella gemina,” with further BGCs and as-yet uncharacterized chemistry (16). At the Hachijo location, T. swinhoei Y cooccurs with T. swinhoei WA, a chemically distinct white sponge phenotype and the source of the swinholide congener misakinolide A (=bistheonellide A; 3) (25, 26) and the theopalauamide-related theonellamides (4–12) (27, 28). In a recent investigation of the WA variant, we isolated the ∼90-kb misakinolide (mis) BGC from a fosmid library constructed from the holobiont metagenome (16, 29). Analyses based on flanking genes, mass-spectrometric imaging, fluorescence in situ hybridization, and PCR detection of the mis genes in DNA amplified from single-bacterial filaments unexpectedly showed that 3 is likewise produced by Entotheonella (29) rather than unicellular bacteria, as the earlier chemical studies had suggested for 1 (12). Based on its distinct 16S rRNA gene, we proposed the name “Candidatus Entotheonella serta” TSWA1 for the misakinolide producer (29).
Herein, we report results obtained from single-bacterial genome sequencing of Ca. E. serta TSWA1. Although only two compound types are known from the WA sponge chemotype, this symbiont contains a large number of biosynthetic genes, even surpassing that of the talented producer Ca. E. factor TSY1. Using metagenomic data of another white-interior T. swinhoei phenotype from the Red Sea as a scaffold, we also assemble from the fragmented single-bacterial sequence data a large BGC and functionally assign it to the theonellamides, thus linking the peptides and polyketides to the same producer. This study on a distinct T. swinhoei chemotype validates the chemical richness and variability of Entotheonella, corroborates its importance as a source of sponge natural products, and emphasizes the potential of microbial dark matter as metabolic resource.
Results
Single-Cell Genomics of Ca. Entotheonella serta.
Our early efforts to directly sequence Entotheonella serta from Japanese T. swinhoei WA specimens proved unsuccessful, since for unknown reasons metagenomic datasets of even highly enriched filamentous bacterial fractions were virtually devoid of contigs with Entotheonella-type genes (29). In contrast, such contigs were readily obtained by sequencing of single filaments isolated by fluorescence-activated cell sorting (FACS) (29). The basis of the current study was two filaments, sequenced using Illumina technology and named G6 and H6 according to their position in a 96-well FACS plate. De novo assembly of each sequence dataset using the software package SPAdes resulted in 7.7 Mb (5,239 contigs) and 5.1 Mb (4,805 contigs) of genomic information for G6 and H6, respectively (SI Appendix, Table S1). Reciprocal average nucleotide identity (ANI) analysis of the two genomes revealed a 99.96% ANI, indicating that G6 and H6 both belonged to Ca. E. serta TSWA1. Due to the short length of the contigs from the amplified genomes (median length < 2,000 bp), we assembled the G6 and H6 reads into a single draft genome with SPAdes 3.9.0. The assembled and annotated draft genome consisted of 3,146 contigs, totaling 8.97 Mb, similar in size to Ca. E. factor, with an N50 of 6.9 kb (SI Appendix, Table S1). It contained 11,759 predicted genes, including 33 of the 35 bacterial single-copy genes frequently used as phylogenetic markers and 106 of 148 BUSCO single-copy genes, suggesting a relatively high degree of completeness (SI Appendix, Tables S2 and S3).
High Biosynthetic Gene Diversity in Ca. E. serta Confirms Entotheonella as a Talented Producer Taxon.
T. swinhoei WA is the source of misakinolides and theonellamides, of which the misakinolide polyketide synthase (PKS) genes were previously assigned to Ca. E. serta TSWA1 (29). Analysis of the TSWA1 draft genome using the software packages antiSMASH 3.0 (30) and NaPDoS (31), in combination with BLAST-based manual annotation, identified numerous contigs carrying additional natural product genes (Table 1 and SI Appendix, Table S4). These belonged to type I and type III PKS, nonribosomal peptide synthetase (NRPS), ribosomally synthesized and posttranslationally modified peptide (RiPP), terpene, and indolocarbazole alkaloid biosynthesis. Since many contigs were too short to cover more than a few modular PKS and NRPS domains, we performed our initial bioinformatic analysis of such multimodular enzymes at the domain level only. In total, we identified in the dataset 34 KS and 33 ACP domains of PKSs (including 19 KSs and 19 ACPs of the misakinolide PKS), and 46 adenylation (A) and 45 peptidyl carrier protein domains of NRPSs. These numbers were surprising, as they surpass even those of the chemically rich symbiont Ca. E. factor (16), suggesting a much higher natural product diversity in this sponge than previously appreciated (Table 1). Likewise, as a conservative estimate for the number PKS and NRPS clusters, we detected 15 thioesterase domains, exceeding those of Ca. E. factor and Ca. E. gemina combined. It was previously shown that the BGC-rich genomes of Ca. E. factor and Ca. E. gemina contain only two clusters in common, a one-module NRPS and a type III polyketide synthase, both of unknown function (16). The genome of Ca. E. serta contains homologs for both of these clusters, further suggesting that the products are of general importance for Entotheonella. Additionally, a single short Ca. E. serta contig has reasonable homology to a portion of a mixed PKS-NRPS BGC of unknown function in Ca. E. factor. However, none of the other Ca. E. serta BGCs or fragments have significant homology with those of Ca. E. factor or Ca. E. gemina. Ca. E. serta therefore represents a third large, nearly orthogonal Entotheonella biosynthetic repertoire.
Table 1.
Natural product biosynthetic domains and enzymes of Ca. E. serta TSWA1 compared with the two previously sequenced Entotheonella variants
| Domain type | Ca. E. serta TSWA1 | Ca. E. factor TSY1 | Ca. E. gemina TSY2 |
| NRPS | |||
| Adenylation | 46 | 44 | 11 |
| (Known compounds) | (11) | (26) | (0) |
| Condensation | 43 | 40 | 11 |
| Peptidyl-carrier protein | 45 | 50 | 11 |
| Thioesterase | 11 | 5 | 3 |
| Other NRPS domain | 6 | 8 | 5 |
| PKS | |||
| Ketosynthase | 34 | 25 | 5 |
| (Known compounds) | (22) | (16) | (0) |
| Acyltransferase | 17 | 14 | 6 |
| trans-AT docking | 19 | 12 | 0 |
| Ketoreductase | 21 | 19 | 4 |
| Dehydratase | 11 | 8 | 1 |
| Acyl-carrier protein | 33 | 21 | 3 |
| Thioesterase | 4 | 4 | 2 |
| Type III PKS system | 2 | 1 | 1 |
| Other PKS domain | 12 | 11 | 3 |
| RiPP | |||
| RiPP precursors | 0 | 4 | 5 |
| RiPP maturation enzymes | 1 | 6 | 7 |
| Terpene/Ectoine/Indolocarbazole | 3 | 3 | 1 |
| Total | 308 | 275 | 79 |
Adenylation and ketosynthase domains attributed to known compounds are shown in parentheses. For a more complete breakdown, see SI Appendix, Table S4.
Retrobiosynthetic Prediction and Combined Analysis of Two Sponge Geotypes Permits Identification of Candidate Genes for Theonellamides.
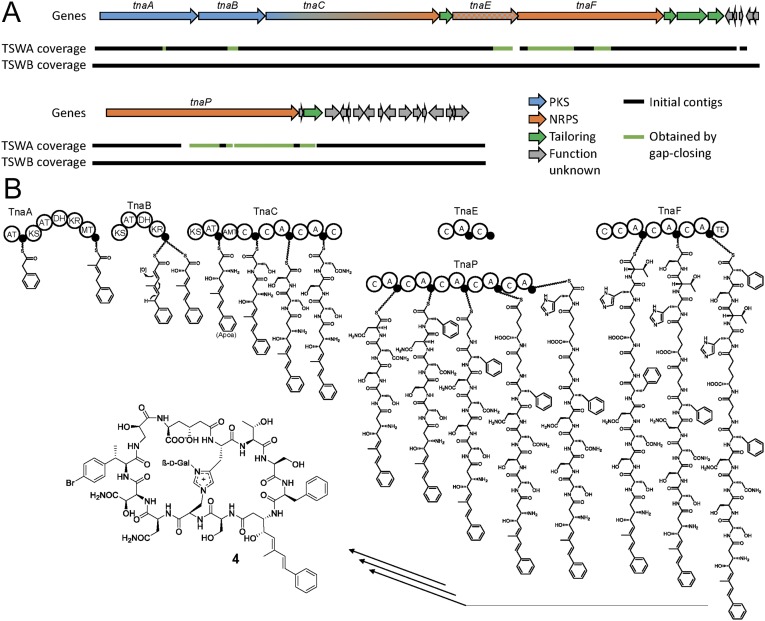
To investigate the function of unassigned natural product genes in Ca. E. serta, we used NRPSpredictor2 (32) to analyze NRPS A domains encoded in our dataset (SI Appendix, Table S5). These domains load amino acid building blocks onto NRPS modules and contain sequence motifs that permit the prediction of their substrates (33, 34). Notably, we identified a 12.7-kb contig from the H6 filament encoding PKS as well as NRPS modules that suggested, based on the presence of an aminotransferase domain and A-domain predictions, a β-amino polyketide moiety connected in series to Ser and Asn residues. This substructure is present in theonellamides and related compounds, suggesting this contig as a strong candidate for further analysis. To identify adjacent regions, we selected contigs with A-domain predictions matching theonellamide residues and designed PCR primers for gap closing using DNA from the mechanically enriched Entotheonella preparation of T. swinhoei WA. The process was guided by retrobiosynthetic prediction under the assumption that theonellamide genes would be collinear with the natural product structure. Although several predicted gaps were closed in this way, most primer combinations failed to generate amplicons, resulting in 40 disconnected contigs. At this stage, we were unable to distinguish whether these negative results were due to the DNA complexity, false biosynthetic assumptions, or sequence artifacts generated during genome amplification (35).
Since metagenomic sequencing of the Japanese T. swinhoei WA specimens consistently failed for unknown reasons, we next explored the possibility of analyzing a related sponge phenotype that could help with scaffolding single-filament contigs. Misakinolide-type compounds, such as swinholide A (2), were also reported from a white Israeli T. swinhoei variant (chemotype WB) occurring in the Gulf of Aqaba (11), but theonellamide or congeners were unknown from this sponge. However, PCR tests using the metagenomic DNA and primers specific for Entotheonella 16S rRNA genes generated amplicons with a sequence that was 97.9% identical to that of Ca. E. serta TSWA1, suggesting both sponges might contain symbionts of the same candidate species. We therefore sequenced the metagenome of an enriched filamentous bacterial preparation of T. swinhoei WB, which resulted in successful retrieval of numerous contigs containing Entotheonella-type sequences, including two contigs with 16S rRNA genes. Gratifyingly, we also identified contigs that were almost identical (>99% identity) to the previously cloned misakinolide (mis) gene cluster (29), as well as to candidate contigs for theonellamide biosynthesis from the Japanese single-filament dataset. The extended contigs obtained included two that contained the full biosynthetic architecture predicted to be required for theonellamide (tna) biosynthesis (Fig. 2). When inspecting these contigs, nonbiosynthetic genes or fragments were identified on one edge of the longer contig and two edges of the shorter one, suggesting that the tna genes are located in two distinct, nonclustered genome regions, in the following named regions I and II. The gene architecture of the tna contigs from TSWB allowed the design of a final round of PCR gap closing using total DNA of the Japanese T. swinhoei WA as template. With this, all remaining gaps were closed except for two in region I and three in region II (Fig. 2A).
Fig. 2.
Theonellamide biosynthetic model. (A) The gene clusters from Ca. E. serta, divided into two regions. The bars underneath show the coverage in TSWA and TSWB. The organization of genes and ORFs is the same in both Ca. E. serta strains. (B) The proposed biosynthesis of the linear theonellamide core, followed by cyclization and tailoring.
Architecture of the Putative Theonellamide PKS-NRPS Genes.
The putative tna biosynthetic assembly line comprises 4 PKS and 11 NRPS modules encoded by tnaA-F on region I and tnaP on region II. Bioinformatic domain analysis of PKS modules located on TnaA, TnaB, and the N terminus of TnaC showed close collinearity to the (5E,7E)-3-amino-4-hydroxy-6-methyl-8-(p-bromo)phenyl-5,7-octadienoic acid (Apoa/Aboa) portion of theonellamides (Fig. 2B and SI Appendix, Table S6). Noteworthy is the presence of an acyltransferase (AT) domain in the first module as candidate to load an aromatic starter, which would be atypical but not unprecedented (36). The remaining modules on TnaC and TnaF are NRPS modules with A-domain codes (SI Appendix, Table S5) matching to the first (Ser, Asn) and last three (Thr, Ser, Phe) peptide residues of theonellamides. However, the first NRPS module of TnaC, which positionally corresponds to the first of two consecutive Ser units, lacks an A domain, and a further A domain is missing on the second module of TnaE, an NRPS with unknown function. Similarly absent A domains are known from a few other NRPSs (37, 38). Interestingly, the biosynthetic machinery for five internal residues of the theonellamide peptide chain [Asn, Phe, β-Ala, ε-linked α-aminoadipate (Aad), His] does not seem to be encoded on region I, but a matching gene is present on region II. This gene, tnaP, encodes one large megasynthetase consisting of five NRPS modules with predicted specificities (by NRPSpredictor2) for Asn, Phe/Tyr, β-Ala, Gln, and His, which correspond well to the theonellamide residues. In addition, the next characterized homolog of the fourth A domain is not a canonical NRPS component, but the substrate-activating domain of the α-adipate semialdehyde dehydrogenase Lys2 from fungal lysine biosynthesis (39). This homology matches the unusual α-amino adipate residue of theonellamides.
The structure of theonellamide and congeners suggests several modifications that are not PKS/NRPS-catalyzed. A candidate enzyme for His N-glycosylation is TnaI, which has significant homology to known glycosyltransferases. Hydroxylations on Asn, β-Ala, and/or α-amino adipate might be installed by TnaD and TnaH, which possess similarity to amino acid hydroxylases. TnaQ, resembling radical S-adenosylmethionine methyltransferases, is a candidate for β-methylation of (p-bromo)-Phe. The most intriguing structural feature is the to-date unique C–N bridge between the side chains of His and another amino acid, predicted as Ser by the A-domain specificity code. An enzyme candidate for connecting these units could be TnaH, comprising domains homologous to kinases and tetratricopeptide repeat domains. TnaH might activate Ser by phosphorylation and catalyze cyclodehydration, perhaps with participation of the small unassigned NRPS TnaE. For bromination, no convincing gene was identified.
Identification of Theonellamide-Type Compounds in Ca. E. serta Cell Fractions.
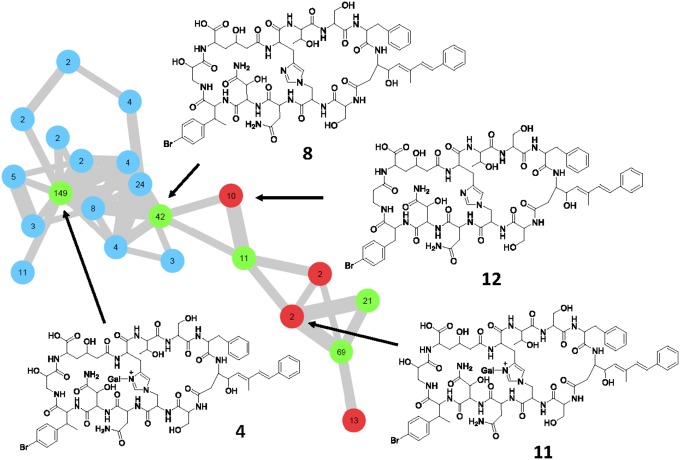
Since theonellamides or related compounds had not been reported from Israeli sponge specimens, we analyzed extracts from such animals by ultrahigh-performance liquid chromatography–high resolution–heated electrospray-data-dependent tandem mass spectrometry (MS2) and molecular networking analysis (40). In the molecular network, 22 different singly charged and 58 doubly charged theonellamide ions were observed (Fig. 3 and SI Appendix, Fig. S1). Due to the chemical composition, structural complexity, and halogenation of the congeners, each theonellamide has multiple highly abundant isotopic signals (SI Appendix, Fig. S2), and as a result each compound is present more than once in the network. In total, these nodes were assigned to 14 different theonellamides present on the basis of MS2 spectra (SI Appendix, Figs. S7–S9 and Table S7). Masses and MS2 fragmentation profiles consistent with three known theonellamides, theonellamide A (4; m/z 882.304, m/z 882.805, and m/z 883.301), theonellamide B (8; m/z 800.779, m/z 801.278, and m/z 801.786), and theonegramide (11; m/z 859.298, m/z 859.791, and m/z 860.299) were observed. Moreover, two compounds related to theonellamide B were observed. These are theonellamide B variants lacking one (m/z 792.271, m/z 793.271, and m/z 793.789) and two hydroxyl groups (m/z 784.276, m/z 784.773, and m/z 785.277). The location of the loss of the hydroxyl groups could not be unambiguously assigned based on MS2 spectra. The ion at m/z 786.272 is consistent with the molecular formula C69H87BrN16O22, which is the same as that of theonellamide C. The MS2 profile, however, shows that the PKS starter molecule is not brominated. Based on the MS2 fragmentation data and the structures of other known theonellamide congeners, the most likely alternative is the p-bromination of phenylalanine, as in the other theonellamides. We term this proposed congener theonellamide H (12; Fig. 1). Additionally, ions at m/z 777.277 and 778.278 represent a theonellamide H analog lacking one hydroxyl group, the location of which could not be determined. We also observed isotopic patterns corresponding to seven additional theonellamide congeners, the structure of which could not be elucidated in this work due to the low amount of theonellamides present, the limited amount of sponge sample available, and poor fragmentation of the metabolites of interest. MS network-based comparison (41) of extracts from the Japanese and Israeli specimens indicates that the most abundant theonellamides largely overlap (Fig. 3), but that both sponges also contain sets of origin-specific theonellamides.
Fig. 3.
Molecular network analysis of theonellamides. Nodes are color-coded according to the sponge sample from which the data were obtained: blue, TSWA (Japan); red, TSWB (Israel); green, metabolites present in both sponges. Numbers within each node indicate the number of MS2 spectra obtained for each metabolite as a proxy for their relative abundance. The edge line width indicates the relatedness between two metabolites (cosine 0.7). This figure represents only singly charged parent ions; for a more complete version see SI Appendix, Fig. S1. The structural assignment of theonellamide B (8) is described in detail in SI Appendix, Figs. S7–S9. The full LC-MS dataset was uploaded to the MASSIVE database (MSV000081318 PW 2017) (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) and individually annotated spectra added to the GNPS library (https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp).
Characterization of the Theonellamide PKS Loading Module.
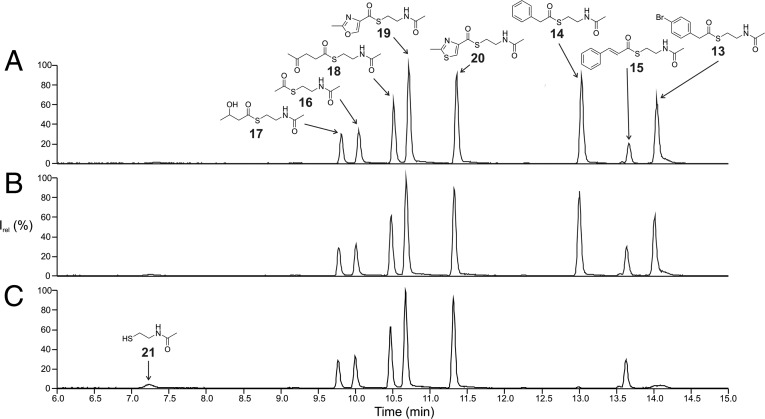
We next wished to obtain functional evidence for the role of the tna genes. The PKS/NRPS architecture suggests that theonellamide biosynthesis is initiated by loading of either phenylacetyl- or 4-bromophenylacetyl-CoA by the N-terminal AT-ACP didomain module of TnaA (Fig. 1). Bioinformatic analyses (SI Appendix, Fig. S3) did not suggest a distinct type of substrate accepted by the AT, aromatic or otherwise, making this module a good candidate to test the function of the tna system. The AT-ACP–encoding region of tnaA was cloned from metagenomic DNA of the filamentous bacterial fraction from T. swinhoei WA and expressed as a His-tagged holo-protein in E. coli BAP1 (41). To examine the specificity of the AT domain, a series of N-acetyl cysteamine (SNAC) thioesters were synthesized (Fig. 4) including the suspected substrates 4-bromophenylacetyl-SNAC (13) and phenylacetyl-SNAC (14), along with six additional test substrates. Competitive depletion assays, in which the AT-ACP didomain was incubated with equimolar concentrations of the substrates, revealed that only the thioesters 13 and 14 were consumed in the reaction (Fig. 4 and SI Appendix, Fig. S4), concomitant with a buildup of free N-acetylcysteamine, but that the similar cinnamoyl-SNAC or other compounds were not, consistent with the biosynthetic model.
Fig. 4.
Representative extracted ion chromatograms of the competitive TnaA AT-ACP depletion assay, including the (A) boiled enzyme negative control, (B) no enzyme negative control, and (C) test reaction using all components. Peaks are labeled according to the respective substrates. 13: bromophenylacetyl-SNAC; 14: phenylacetyl-SNAC; 15: cinnamoyl-SNAC; 16: acetyl-SNAC; 17: β-hydroxy-butanyl-SNAC; 18: S-(2-acetamidoethyl) 4-oxopentanethioate; 19: S-(2-acetamidoethyl) 2-methyloxazole-4-carbothioate; 20: S-(2-acetamidoethyl) 2-methylthiazole-4-carbothioate; and 21: free N-acetylcysteamine. Averaged triplicates of this experiment are shown in SI Appendix, Fig. S4. Underlying mass spectra of the corresponding compounds are depicted in SI Appendix, Figs. S5 and S6.
Discussion
This work was motivated by previous studies suggesting Entotheonella bacteria as a rich source of natural products from Theonellid sponges (13, 16). Earlier work by us and others has linked these bacteria to the production of almost all polyketides and modified peptides known from the sponge T. swinhoei Y (16), of misakinolides in T. swinhoei WA (29), and of calyculins and kasumigamides in Discodermia calyx (42, 43). Genomic analysis of the two symbionts Ca. E. factor and Ca. E. gemina in T. swinhoei Y revealed a large diversity of additional biosynthetic gene clusters, of which at least one is functional (16, 24). Entotheonella filaments were also previously described from the discodermolide-containing sponge Discodermia dissoluta (44) and a swinholide- and theopalauamide-containing Palauan chemotype of T. swinhoei (13). Due to the fact that genomic information on Entotheonella was available only from a single sponge, further studies on the metabolic potential and diversity of these symbionts were needed.
In this study on T. swinhoei WA from Japan containing misakinolides and theonellamides, the single-filament dataset of Ca. E. serta revealed an extraordinary richness of natural product genes. They encode NRPS and PKS module numbers that surpass even those of Ca. E. factor, as well as biosynthetic enzymes from diverse additional natural product classes. These results, unexpected considering that only two compound types have been reported from this chemotype, confirm Entotheonella as a metabolically gifted producer taxon, as suggested by the previous study on T. swinhoei Y. The two large misakinolide and theonellamide clusters together account for 46% of the annotated domains, with a wealth of clusters still functionally unassigned. With the exception of an NRPS and a type III PKS cluster of unknown function, so far detected in every Entotheonella phylotype, candidate species that have been sequenced exhibit strikingly little overlap regarding shared biosynthetic genes.
Despite this promise, we found it a considerable challenge to attribute natural products to the highly fragmented BGCs in the single-filament datasets. This was due to repeated failures to obtain data from T. swinhoei WA Entotheonella by complementary, metagenomic sequencing, although filamentous fractions had been prepared in parallel to that of the successfully sequenced Ca. E. factor from cocollected sponges. The reasons for this discrepancy are not known. Nevertheless, assembly of a large PKS-NRPS gene set was ultimately achieved by obtaining a metagenome from the Israeli WB variant that permitted mutual contig scaffolding for gap closure. Through this strategy, we provide evidence that in Japanese T. swinhoei WA sponges a single symbiont, Ca. E. serta TSWA1, produces both misakinolides and theonellamides, close congeners of swinholides and theopalauamides, respectively. For T. swinhoei WB, a model chemotype that was first chemically studied by Kashman and coworkers almost 40 y ago (4), the data also suggest that one or more Entotheonella variants are the source of swinholides, as well as of theonellamides, a compound family previously unknown from this animal. Although our data suggest that the extraction-based assignment of swinholides and theopalauamides to two different bacterial producer types (12) might have to be reevaluated, it cannot be excluded that multiple producers for the same compounds exist in sponges. One argument for this scenario is the identification of cyanobacterial sources of swinholide A and related polyketides (45, 46). However, our experimental results on theonellamide-type antifungal peptides confirm the original proposal of Entotheonella as their producer, one of the early key hypotheses on sources of sponge natural products (12).
Due to their phylogenetic uniqueness, biosynthetic plasticity, high frequency of unusual chemistry and biosynthetic enzymology, and potential for sustainable marine drug development, Entotheonella continue to provide exciting avenues for fundamental and application-oriented research. Future studies will be necessary to better assess the metabolic diversity offered by these bacteria. Entotheonella phylotypes were previously detected in diverse sponges by PCR (16), but their role as symbionts or contaminants remains to be clarified, and it is unknown whether they play a major role in natural product biosynthesis outside the Theonellid sponges. The existence of additional, non-Tectomicrobial producers was previously proposed (47) and recently demonstrated (48) by attributing the production of polybrominated diphenyl ethers in Dysideid sponges to the uncultured cyanobacterium Hormoscilla (Oscillatoria) spongeliae. It is thus likely that further uncultivated and culturable contributors to the remarkable chemistry of sponges exist.
Materials and Methods
Full experimental details are available in SI Appendix, Materials and Methods. Briefly, Entotheonella cells were separated from sponge tissue by differential centrifugation and sequenced as described previously (29). Bioinformatic analysis was performed using antiSMASH (30) and other tools, chemical analysis with Global Natural Products Social Molecular Networking (GNPS) (41), and scaffold gaps were closed by PCR. The DNA sequence encoding the tnaA AT-ACP loading didomain was cloned from metagenomics DNA, expressed from a pCDFDuet-1 vector, and purified with a Ni-NTA affinity resin. Substrate depletion assays were carried out by simultaneous incubation of the protein with 20 µM of eight N-acetylysteamine thioester substrates at 30 °C for 20 min, quenched by addition of 20 μL concentrated formic acid and subsequently analyzed by MS.
Supplementary Material
Acknowledgments
We thank Kentaro Takada and Toshiyuki Wakimoto for a sponge sample, and Silke Reiter and Anna L. Vagstad for advice on experiments. This work was supported by grants from the Swiss National Foundation (205321_165695 and IZLSZ3_149025) and the Helmut Horten Foundation (to J.P.), from the Japan Society for the Promotion of Science Core-to-Core Program, A. Advanced Research Networks (to H.T.), from the Israel Science Foundation (957/15) (to M.I.), and from the Alexander von Humboldt Foundation (to M.C.W.). The bioinformatics support of the Bundesministerium für Bildung und Forschung-funded Bielefeld-Gießen Center for Microbial Bioinformatics (BiGi; Grant 031A533) within the German Network for Bioinformatics Infrastructure (de.NBI) is gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The Ca. E. serta TSWA1 genome and the sequences of the Ca. E. Serta TSWB1 contigs containing the putative theonellamide and misakinolde biosynthetic gene clusters have been deposited at DNA Databank of Japan/European Nucleotide Archive/GenBank (accession nos. PPXO00000000 and MG844357-9, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715496115/-/DCSupplemental.
References
- 1.Winder PL, Pomponi SA, Wright AE. Natural products from the lithistida: A review of the literature since 2000. Mar Drugs. 2011;9:2643–2682. doi: 10.3390/md9122643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bewley CA, Faulkner DJ. Lithistid sponges: Star performers or hosts to the stars. Angew Chem Int Ed. 1998;37:2162–2178. doi: 10.1002/(SICI)1521-3773(19980904)37:16<2162::AID-ANIE2162>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Hamada T, Matsunaga S, Yano G, Fusetani N. Polytheonamides A and B, highly cytotoxic, linear polypeptides with unprecedented structural features, from the marine sponge, Theonella swinhoei. J Am Chem Soc. 2005;127:110–118. doi: 10.1021/ja045749e. [DOI] [PubMed] [Google Scholar]
- 4.Kho E, Imagawa DK, Rohmer M, Kashman Y, Djerassi C. Sterols in marine invertebrates. 22. Isolation and structure elucidation of conicasterol and theonellasterol, two new 4-methylene sterols from the Red Sea sponges Theonella conica and Theonella swinhoei. J Org Chem. 1981;46:1836–1839. [Google Scholar]
- 5.Wegerski CJ, Hammond J, Tenney K, Matainaho T, Crews P. A serendipitous discovery of isomotuporin-containing sponge populations of Theonella swinhoei. J Nat Prod. 2007;70:89–94. doi: 10.1021/np060464w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hentschel U, et al. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol. 2002;68:4431–4440. doi: 10.1128/AEM.68.9.4431-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor MW, Radax R, Steger D, Wagner M. Sponge-associated microorganisms: Evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev. 2007;71:295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piel J. Metabolites from symbiotic bacteria. Nat Prod Rep. 2004;21:519–538. doi: 10.1039/b310175b. [DOI] [PubMed] [Google Scholar]
- 9.Faulkner D, He H, Unson M, Bewley C, Garson M. New metabolites from marine sponges:–Are symbionts important. Gazz Chim Ital. 1993;123:301–307. [Google Scholar]
- 10.Schmidt EW, Bewley CA, Faulkner DJ. Theopalauamide, a bicyclic glycopeptide from filamentous bacterial symbionts of the lithistid sponge Theonella swinhoei from Palau and Mozambique. J Org Chem. 1998;63:1254–1258. [Google Scholar]
- 11.Carmely S, Kashman Y. Structure of swinholide A, a new macrolide from the marine sponge Theonella swinhoei. Tetrahedron Lett. 1985;26:511–514. [Google Scholar]
- 12.Bewley CA, Holland ND, Faulkner DJ. Two classes of metabolites from Theonella swinhoei are localized in distinct populations of bacterial symbionts. Experientia. 1996;52:716–722. doi: 10.1007/BF01925581. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt EW, Obraztsova AY, Davidson SK, Faulkner DJ, Haygood MG. Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel δ-proteobacterium, “Candidatus Entotheonella palauensis. Mar Biol. 2000;136:969–977. [Google Scholar]
- 14.Piel J, et al. Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei. Proc Natl Acad Sci USA. 2004;101:16222–16227. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freeman MF, et al. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science. 2012;338:387–390. doi: 10.1126/science.1226121. [DOI] [PubMed] [Google Scholar]
- 16.Wilson MC, et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 17.Freeman MF, Helf MJ, Bhushan A, Morinaka BI, Piel J. Seven enzymes create extraordinary molecular complexity in an uncultivated bacterium. Nat Chem. 2017;9:387–395. doi: 10.1038/nchem.2666. [DOI] [PubMed] [Google Scholar]
- 18.Sakemi S, Ichiba T, Kohmoto S, Saucy G, Higa T. Isolation and structure elucidation of onnamide A, a new bioactive metabolite of a marine sponge, Theonella sp. J Am Chem Soc. 1988;110:4851–4853. [Google Scholar]
- 19.Fusetani N, Sugawara T, Matsunaga S, Hirota H. Orbiculamide A: A novel cytotoxic cyclic peptide from a marine sponge Theonella sp. J Am Chem Soc. 1991;113:7811–7812. [Google Scholar]
- 20.Ji K, et al. Keramamide A, a novel peptide from the Okinawan marine sponge Theonella sp. J Chem Soc, Perkin Trans 1. 1991:2609–2611. [Google Scholar]
- 21.Fusetani N, Matsunaga S, Matsumoto H, Takebayashi Y. Bioactive marine metabolites. 33. Cyclotheonamides, potent thrombin inhibitors, from a marine sponge Theonella sp. J Am Chem Soc. 1990;112:7053–7054. [Google Scholar]
- 22.Fusetani N, Nakao Y, Matsunaga S. Nazumamide A, a thrombin-inhibitory tetrapeptide, from a marine sponge, Theonella sp. Tetrahedron Lett. 1991;32:7073–7074. [Google Scholar]
- 23.Lackner G, Peters EE, Helfrich EJ, Piel J. Insights into the lifestyle of uncultured bacterial natural product factories associated with marine sponges. Proc Natl Acad Sci USA. 2017;114:E347–E356. doi: 10.1073/pnas.1616234114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helf MJ, Jud A, Piel J. Enzyme from an uncultivated sponge bacterium catalyzes S-methylation in a ribosomal peptide. ChemBioChem. 2017;18:444–450. doi: 10.1002/cbic.201600594. [DOI] [PubMed] [Google Scholar]
- 25.Sakai R, Higa T, Kashman Y. Misakinolide A, an antitumor macrolide from the marine sponge Theonella sp. Chem Lett. 1986;15:1499–1502. [Google Scholar]
- 26.Kato Y, et al. Antitumor macrodiolides isolated from a marine sponge Theonella sp.: Structure revision of misakinolide A. Tetrahedron Lett. 1987;28:6225–6228. [Google Scholar]
- 27.Matsunaga S, Fusetani N, Hashimoto K, Walchli M. Theonellamide F. A novel antifungal bicyclic peptide from a marine sponge Theonella sp. J Am Chem Soc. 1989;111:2582–2588. [Google Scholar]
- 28.Matsunaga S, Fusetani N. Theonellamides A-E, cytotoxic bicyclic peptides, from a marine sponge Theonella sp. J Org Chem. 1995;60:1177–1181. [Google Scholar]
- 29.Ueoka R, et al. Metabolic and evolutionary origin of actin-binding polyketides from diverse organisms. Nat Chem Biol. 2015;11:705–712. doi: 10.1038/nchembio.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber T, et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ziemert N, et al. The natural product domain seeker NaPDoS: A phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS One. 2012;7:e34064. doi: 10.1371/journal.pone.0034064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Röttig M, et al. NRPSpredictor2–A web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011;39:W362–W367. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 34.Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 35.Zhang K, et al. Sequencing genomes from single cells by polymerase cloning. Nat Biotechnol. 2006;24:680–686. doi: 10.1038/nbt1214. [DOI] [PubMed] [Google Scholar]
- 36.Wilkinson CJ, Frost EJ, Staunton J, Leadlay PF. Chain initiation on the soraphen-producing modular polyketide synthase from Sorangium cellulosum. Chem Biol. 2001;8:1197–1208. doi: 10.1016/s1074-5521(01)00087-4. [DOI] [PubMed] [Google Scholar]
- 37.Yin X, Zabriskie TM. The enduracidin biosynthetic gene cluster from Streptomyces fungicidicus. Microbiology. 2006;152:2969–2983. doi: 10.1099/mic.0.29043-0. [DOI] [PubMed] [Google Scholar]
- 38.Guenzi E, Galli G, Grgurina I, Gross DC, Grandi G. Characterization of the syringomycin synthetase gene cluster. A link between prokaryotic and eukaryotic peptide synthetases. J Biol Chem. 1998;273:32857–32863. doi: 10.1074/jbc.273.49.32857. [DOI] [PubMed] [Google Scholar]
- 39.Ehmann DE, Gehring AM, Walsh CT. Lysine biosynthesis in Saccharomyces cerevisiae: Mechanism of α-aminoadipate reductase (Lys2) involves posttranslational phosphopantetheinylation by Lys5. Biochemistry. 1999;38:6171–6177. doi: 10.1021/bi9829940. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen DD, et al. MS/MS networking guided analysis of molecule and gene cluster families. Proc Natl Acad Sci USA. 2013;110:E2611–E2620. doi: 10.1073/pnas.1303471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang M, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wakimoto T, et al. Calyculin biogenesis from a pyrophosphate protoxin produced by a sponge symbiont. Nat Chem Biol. 2014;10:648–655. doi: 10.1038/nchembio.1573. [DOI] [PubMed] [Google Scholar]
- 43.Nakashima Y, Egami Y, Kimura M, Wakimoto T, Abe I. Metagenomic analysis of the sponge Discodermia reveals the production of the cyanobacterial natural product kasumigamide by ‘Entotheonella’. PLoS One. 2016;11:e0164468. doi: 10.1371/journal.pone.0164468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brück WM, Sennett SH, Pomponi SA, Willenz P, McCarthy PJ. Identification of the bacterial symbiont Entotheonella sp. in the mesohyl of the marine sponge Discodermia sp. ISME J. 2008;2:335–339. doi: 10.1038/ismej.2007.91. [DOI] [PubMed] [Google Scholar]
- 45.Andrianasolo EH, et al. Isolation of swinholide A and related glycosylated derivatives from two field collections of marine cyanobacteria. Org Lett. 2005;7:1375–1378. doi: 10.1021/ol050188x. [DOI] [PubMed] [Google Scholar]
- 46.Humisto A, et al. The swinholide biosynthetic gene cluster from a terrestrial cyanobacterium Nostoc sp. UHCC 0450. Appl Environ Microbiol. November 17, 2017 doi: 10.1128/AEM.02321-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unson MD, Holland ND, Faulkner DJ. A brominated secondary metabolite synthesized by the cyanobacterial symbiont of a marine sponge and accumulation of the crystalline metabolite in the sponge tissue. Mar Biol. 1994;119:1–11. [Google Scholar]
- 48.Agarwal V, et al. Metagenomic discovery of polybrominated diphenyl ether biosynthesis by marine sponges. Nat Chem Biol. 2017;13:537–543. doi: 10.1038/nchembio.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.