Significance
Alzheimer’s disease (AD) is an age-related neurodegenerative disease. Genome-wide association studies predominately focusing on Caucasian populations have identified risk loci and genes associated with AD; the majority of these variants reside in noncoding regions with unclear functions. Here, we report a whole-genome sequencing study for AD in the Chinese population. Other than the APOE locus, we identified common variants in GCH1 and KCNJ15 that show suggestive associations with AD. For these two risk variants, an association with AD or advanced onset of disease can be observed in non-Asian AD cohorts. An association study of risk variants with expression data revealed their modulatory effects on immune signatures, linking the potential roles of these genes with immune-related pathways during AD pathogenesis.
Keywords: Alzheimer’s disease, whole-genome sequencing, GWAS, risk variant, immune
Abstract
Alzheimer’s disease (AD) is a leading cause of mortality among the elderly. We performed a whole-genome sequencing study of AD in the Chinese population. In addition to the variants identified in or around the APOE locus (sentinel variant rs73052335, P = 1.44 × 10−14), two common variants, GCH1 (rs72713460, P = 4.36 × 10−5) and KCNJ15 (rs928771, P = 3.60 × 10−6), were identified and further verified for their possible risk effects for AD in three small non-Asian AD cohorts. Genotype–phenotype analysis showed that KCNJ15 variant rs928771 affects the onset age of AD, with earlier disease onset in minor allele carriers. In addition, altered expression level of the KCNJ15 transcript can be observed in the blood of AD subjects. Moreover, the risk variants of GCH1 and KCNJ15 are associated with changes in their transcript levels in specific tissues, as well as changes of plasma biomarkers levels in AD subjects. Importantly, network analysis of hippocampus and blood transcriptome datasets suggests that the risk variants in the APOE, GCH1, and KCNJ15 loci might exert their functions through their regulatory effects on immune-related pathways. Taking these data together, we identified common variants of GCH1 and KCNJ15 in the Chinese population that contribute to AD risk. These variants may exert their functional effects through the immune system.
Alzheimer’s disease (AD) is an age-related neurodegenerative disease and a leading cause of mortality in the elderly. Its prevalence is increasing rapidly with the aging population, affecting more than 36 million people worldwide. A recent meta-analysis revealed that the number of AD patients in China increased from 1.9 million in 1990 to 5.7 million in 2010 (1). The pathophysiological mechanisms of AD are complex, with genetic factors playing critical roles. Previous genetics studies, including genome-wide association studies (GWAS), candidate gene sequencing, and whole-exome sequencing have identified several disease genes and risk alleles in AD (2). Among the identified genetic risk factors for AD, a substantial proportion of the genes are associated with immune pathways (3–8).
Most existing genetic data on AD are from Caucasian populations, whereas information for the other ethnic populations is limited. Susceptibility to certain genetic risk factors varies among populations (9). Importantly, even for APOE-ε4, the most consistent risk factor for late-onset AD, the risk levels [i.e., odds ratios (ORs)] vary among ethnic groups (10). Furthermore, recent small-scale studies of Chinese populations report that not all of the AD susceptibility SNPs identified in Caucasian populations can be replicated in Chinese AD patients (11). Indeed, variations of the prevalence of disease-associated genes in different populations have also been observed in other neurodegenerative diseases. For example, the Parkinson’s disease susceptibility gene, MAPT, a major contributor to the disease in Caucasian populations, is only weakly associated with the disease in the Asian population (12, 13). Similarly, whereas the multiple nonsynonymous variants of TREM2 are strongly associated with AD in Caucasian populations (3, 4), these associations were not replicated in East Asian populations (14–16). On the other hand, an independent variant in TREM2 (p. H157Y) has been identified as a susceptibility missense mutation for AD in a Chinese cohort (17).
Most genetic risk variants associated with diseases identified from GWAS are located in the noncoding regions, with relatively low disease penetrance. The biological functions of these noncoding variants in diseases such as AD remain largely unknown (18). However, the recent development of genotype–expression analysis can correlate the genotype of AD risk variants with gene transcript level in specific tissues (7); biomarker data, including protein levels (19) or imaging data (20, 21) may provide insights into the roles of these variants in specific biological pathways and predict potential disease risk factors (22). Understanding the effect of these variants in specific cellular contexts enables the study of the functional consequences of these disease-risk genes.
Therefore, it is important to systematically investigate the genetic risk factors for AD in populations of different ethnicities. Furthermore, the successful implementation of genotype–expression analysis for newly identified risk loci may enable us to further investigate the underlying biological mechanism. Our study identified that, in addition to variants in or near the APOE locus, two loci—GCH1 and KCNJ15—are associated with AD. Furthermore, the genotype–expression analyses reveal that these AD risk loci are associated with the regulation of immune-related gene networks in the hippocampus and blood, as well as the changes in plasma biomarkers. These findings implicate a role of immune-related pathways in the disease.
Materials and Methods
Subject Recruitment for the In-House Chinese AD Whole-Genome Sequencing Cohort.
For this study, we included a cohort of Chinese subjects who visited the Department of Neurology or Memory Clinic, Huashan Hospital, Fudan University, Shanghai, China from 2007 to 2016. There were a total of 972 subjects including 489 with AD, 260 with mild cognitive impairment (MCI), and 223 age- and gender-matched normal controls (NCs). AD patients were diagnosed on the basis of the recommendations of the National Institute on Aging and the Alzheimer’s Association workgroup (23) and had an onset age ≥50 y. Patients with MCI were diagnosed according to the Peterson criteria (24). We excluded individuals with any significant neurologic disease or psychiatric disorder. In addition, 250 NCs without subjective memory complaints were recruited from the community in Shanghai. We subjected all subjects to medical history assessment, neuropsychological assessment, and imaging assessment including computed tomography (CT) or magnetic resonance imaging (MRI). Some subjects also underwent positron emission tomography using Pittsburgh compound B. This study was approved by the Ethics Committee of Huashan Hospital, the Hong Kong University of Science and Technology (HKUST), and the HKUST Shenzhen Research Institute. All subjects provided written informed consent for both study enrollment and sample collection.
CONVERGE Chinese Whole-Genome Sequencing Cohort.
We included the CONVERGE (China Oxford and Virginia Commonwealth University Experimental Research on Genetic Epidemiology) whole-genome sequencing (WGS) dataset (n = 10,640) to serve as a multicenter control to generalize the results (25). We applied an age filter of ≥55 y for the elderly population, yielding 1,745 subjects (n = 1,745) for the downstream analysis.
WGS and Variant Calling Method.
Low-coverage WGS (5× coverage) was performed by Novogene. The genomic DNA libraries were sequenced on an Illumina Hiseq × Ten platform, with 150-bp paired-end reads generated. The researchers were blinded to phenotypic labels during the WGS process. For variant detection, the Gotcloud pipeline (26) was adopted to detect variants from our low-pass WGS data, comprising 1,348 samples, including 126 resequenced samples. An average of 15-Gb Illumina sequencing data per subject were generated, and data were subsequently subjected to FastQC (27) for quality checking and Trimmomatic (28) for the trimming and filtering of low-quality reads. Clean data were mapped to the GRCh37 reference genome containing the decoy fragments using BWA-mem. After de-duplication and clipping of the overlapped paired-end reads, BAM files were subjected to samtools-hybrid, a specialized version of samtools, to generate glf files, which store the marginal likelihoods for genotypes. glfFlex was adopted for the population-based SNP calling, with a total of 24,742,555 single-nucleotide variants obtained after variant calling. We applied hard-filtering methods implemented in the Gotcloud pipeline as VcfCooker to filter low-confidant variant calls on the basis of multiple metrics, such as distance, with known insertion/deletion sites, allele balance, and mapping quality. We subjected variants with high-confidence calls in the range of minor allele frequency (MAF) ≥ 5% (n = 5,523,365; 22.3% of raw detected sites, 5,369,369 of which were in autosomal chromosomes) to Beagle (29, 30) for phasing and using the genotype likelihood information in chromosome-separated VCF files (See SI Appendix, SI Materials and Methods for details).
To assess the accuracy of variant detection, we resequenced 126 of 1,222 samples (10.3% of all samples) using the same WGS protocol, together with 96 samples (7.9% of total samples) genotyped using the Axiom Genome-Wide CHB 1 and CHB 2 Array Plate Set (Affymetrix). See SI Appendix, SI Materials and Methods for additional details.
Association Test and Data Visualization for GWAS.
We performed association tests between cases and controls using PLINK software with the following parameters: –keep-allele-order, –assoc, –ci 0.95, –hwe 0.00001, and –maf 0.10 for the stage 1, stage 2, and stage 1+2 analyses. A genomic inflation factor was generated on the basis of the χ2-values obtained from PLINK results using R programming (31). In addition, to correct for population stratification, we performed conditional logistical regression combined with a genetic similarity score matching (GSM) model (32) or logistic regression combined with phenotype-associated principal components generated from EIGENSOFT smartpca (33). For GSM correction, pruned SNP sets with an MAF > 10% were subjected to the software score_match (Linux) (-s alleleibs -k 2 -m 10000 -model additive -w 10 -U 5 -Ut 5), with matched results further subjected to R for conditional regression test using clogit function. To visualize the data, Manhattan plots and quantile–quantile plots were generated using the R qqman package. Regional plots for individual loci were generated using LocusZoom (34). To generate regional plots, association test results were obtained from PLINK, with pairwise linkage disequilibrium (LD) information generated from VCFtools using the –hap-r2 option.
Cohorts and Data for Replication Study.
To verify our findings, we obtained the following data: (i) The International Genomics of Alzheimer’s Project (IGAP) summary statistics from stage 1 data (35); (ii) genotype, transcriptome, and biomarker data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu/) (36); (iii) genotype and phenotype data from the National Institute on Aging Alzheimer’s Disease Centers Cohort (ADC) (phs000372.v2.p1) (37); and (iv) genotype and phenotype data from the Late Onset Alzheimer’s Disease (LOAD) Family Study (phs000168.v2.p2) (38). See SI Appendix, SI Materials and Methods for additional details.
Meta-Analysis and Data Visualization.
We generated association results from three AD cohorts (ADNI, LOAD, and ADC) using logistic regression with phenotype-associated principal components generated from EIGENSOFT smartpca, together with age and gender as covariates to obtain effect sizes (log-ORs) and SEs. The analysis only included “definite AD” cases which were specified in ADC and LOAD cohorts. The results were summarized and processed by METASOFT (39) to estimate the joint risk effects as well as significance levels under a random effects (RE) model (meta-P value or random-effect P value). For transethnic meta-analysis combining both Chinese and non-Asian datasets, Han and Eskin’s Random Effects model (39) was applied. Analysis results were further subjected to ForestPMPlot (40) to generate forest plots for data visualization.
Association Analysis for Candidate Sites in Transcriptome Data and in Plasma and Cerebrospinal Fluid Biomarker Data.
We retrieved genotype and expression data from the Genotype-Tissue Expression (GTEx) project (41, 42) (www.gtexportal.org) in the database of Genotypes and Phenotypes (dbGaP; phs000424.v6.p1). We used the R GenABEL package (43) for data normalization, then mapped the regulatory effects of our candidate variants using R coding and generated network analysis of the top regulatory genes in STRINGdb (44). We examined the association of the candidate variants with the plasma and cerebrospinal fluid biomarker data obtained from the ADNI (SI Appendix, SI Materials and Methods).
Other Statistical Analyses and Data Visualization.
We performed Cox regression with gender and the top five principal components (PCs) to determine the association between AD onset age and candidate variants by using the coxph function from the survival package in R. We performed the Spearman correlation test using the cor.test function in R. We used ggsurvplot from the survMisc package to generate the survival plot. We generated bar charts, scatter plots, and line charts by using GraphPad Prism version 6 (GraphPad software) (45). We used R lm function together with Anova function from car package for ANCOVA analysis. Finally, we obtained gene annotations from database evidence, with annotations for genomic regions obtained from the University of California, Santa Cruz genome browser (46) and annotations for transcript enrichment from the FANTOM CAT data browser (47).
Power Calculation.
According to the study design for the stage 1 analysis (NC: 442; AD: 477), power calculation was performed using Quanto (48). The prevalence of AD was set at 3.3% on the basis of the latest epidemiological report for AD in the Chinese population (5.5 million AD patients of 165.2 million subjects with age ≥60-y old; data were obtained from summary results in 2010 with age ranging between 60 and 99 y) (1). The following parameters were applied: outcome, disease; design, unmatched case–control (1:0.9266); hypothesis, gene only; sample size, 477 cases; significance, 1 × 10−4, two-sided; mode of inheritance, log-additive; population risk, 0.033.
Data Availability.
The summary-level statistics for the reported variants (59 sites) are available at: iplabdatabase.ust.hk/zhou_et_al_2017/GWAS_data.html. A file containing allele frequencies for variants analyzed in this paper (MAF ≥ 10%) is available through application via the above URL.
Results
Identification of AD Susceptibility Loci in the Chinese Population.
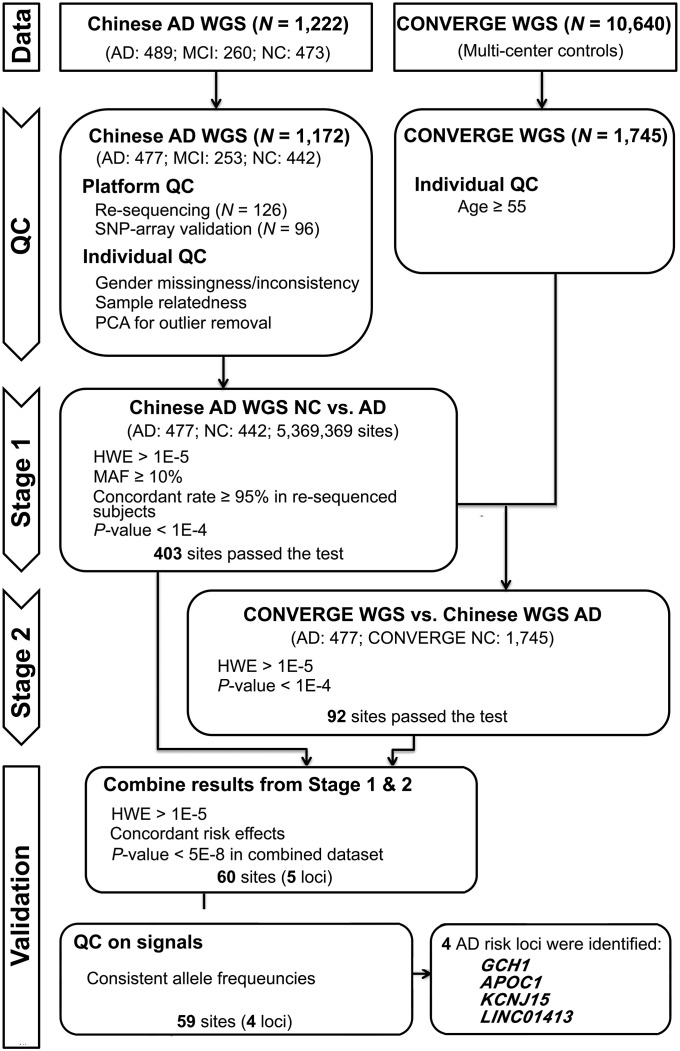
We recruited a Chinese AD cohort comprising 1,222 participants and subjected them to low-pass WGS (5× coverage) (Fig. 1 and SI Appendix, Table S1) (49). Our WGS data yielded 5,369,369 autosomal variants with an MAF ≥ 5%. Genotyping quality was assessed by replicating the WGS with duplicate samples (n = 126), array genotyping (n = 96), and genotyping of APOE-ε4 variants (rs429358 and rs7412) (n = 1,172) using independent aliquots of genomic DNA; overall genotype concordance rates were 0.982, 0.991, and 0.980, respectively (SI Appendix, Tables S2–S4; see also Materials and Methods and SI Appendix, SI Materials and Methods). After 50 subjects were excluded during the quality-control steps (Materials and Methods and SI Appendix, SI Materials and Methods), the study population consisted of 477 AD subjects (AD group, n = 477), 253 with MCI (MCI group, n = 253), and 442 corresponding age- and gender-matched NC subjects (NC group, n = 442). All of the analyzed subjects were from the same center and had a similar demographic background. A detailed analysis of ethnic attributes by comparing the WGS data with 1000 Genomes phase 3 data (www.internationalgenome.org/data) demonstrated that our cohort is part of the East Asian ethnic group (SI Appendix, Figs. S1 and S2).
Fig. 1.
Study design schematic for the discovery of AD susceptibility loci. HWE, Hardy–Weinberg equilibrium; PCA, principal-component analysis; QC, quality control.
The statistical power of our in-house dataset for the identification of AD risk variants with an odds ratio of 1.8 was 0.6073 for an MAF of 0.10, and 0.2078 for an MAF of 0.05 (Materials and Methods). Thus, a variant pool containing 4,082,229 sites with an MAF ≥ 10% after filtering low-quality calls was retained for the analysis of our in-house WGS data (stage 1), with a total of 403 variants showing nominal P < 1 × 10−4. To further increase sample size, enhance statistical power, and verify the findings from stage 1, we conducted the stage 2 analysis separately by using the same AD samples from stage 1 (n = 477) with the CONVERGE samples (n = 1,745, treated as population controls in contrast to the age- and gender-matched cognitive normal controls in stage 1) (Materials and Methods and SI Appendix) in the stage 2 analysis (25). Of 403 sites, we successfully detected 377 concordant sites in the CONVERGE dataset and 92 biallelic variants surviving from the same nominal P value threshold of 1 × 10−4.
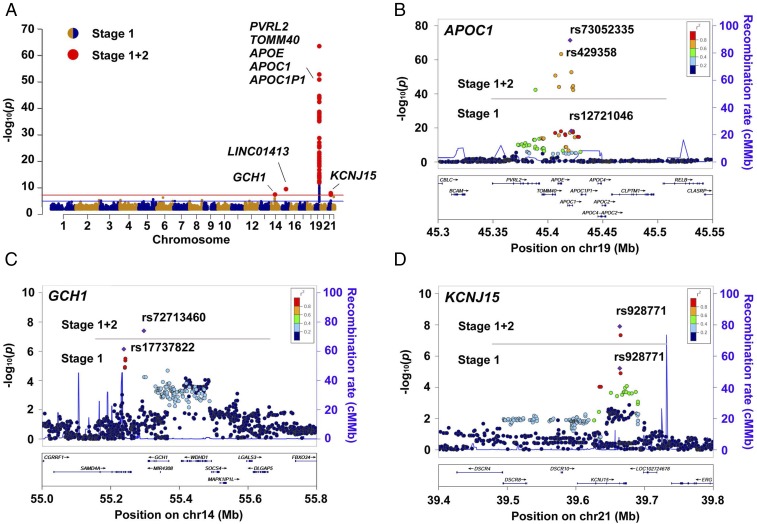
After controlling for the concordance of both allele orders (i.e., ensuring minor alleles are consistent in the two stages) and direction of effect [i.e., log(OR) has the same sign in both stages], we applied a nominal P value threshold of 5 × 10−8 as the final selection criteria in the combined analysis using all samples (477 AD cases and 442 age- and gender-matched cognitive normal controls from stage 1, with 1,745 population controls from CONVERGE) to further enhance power (stages 1+2). Furthermore, we removed one locus with an MAF that deviated strongly in the in-house NC and CONVERGE dataset. Finally, we obtained 59 variants located in the four loci that passed the threshold: APOE, GCH1, LINC01413, and KCNJ15. Other than confirmation of the well-studied APOE-ε4 variant rs429358 (P = 4.1 × 10−64), the four sentinel variants were as follows: GCH1: rs72713460, P = 4.0 × 10−8, OR = 1.74 (95% CI: 1.42–2.12); LINC01413: rs2591054, P = 3.5 × 10−10, OR = 0.61 (95% CI: 0.53–0.71); APOC1: rs73052335, P = 3.5 × 10−72, OR = 4.27 (95% CI: 3.61–5.05); and KCNJ15: rs928771, P = 1.2 × 10−8, OR = 1.59 (95% CI: 1.38–1.93) (Fig. 2A, Table 1, and SI Appendix, Table S5). Besides the APOE-ε4 variant rs429358, the analysis revealed multiple variants including the sentinel variant rs73052335 near the APOE locus (SI Appendix, Table S5). The variant rs73052335 was in LD (R2 = 0.70) with APOE-ε4 rs429358 (Fig. 2B). Regarding the AD susceptibility variants identified in the stage 1 analysis (i.e., APOC1 rs12721046, SAMD4A-GCH1 rs17737822, KCNJ15 rs928771, and LINC01413 rs2591054), the combined analysis, which merged the controls from the stage 1 and 2 datasets, further boosted the signals of these loci (Fig. 2 B–D and SI Appendix, Table S5). Specifically, the sentinel variants for the APOC1 locus shifted from rs12721046 to rs73052335, while the sentinel variants for the GCH1 locus shifted from rs17737822 to rs72713460 (Fig. 2 B and C). Notably, we did not observe any inflation during the stage 1 analysis, as indicated by the quantile–quantile plot and the estimated genomic inflation factor (λGC = 1.011, λ1000 = 1.025) (SI Appendix, Fig. S3). Moreover, we showed that the presence of effect alleles from the newly identified loci (GCH1, LINC01413, and KCNJ15) were not correlated with age, gender, or batch effects in the combined control datasets (SI Appendix, Tables S6 and S7), and was not obviously affected by population stratification after correction using the GSM method (GCH1: rs72713460, adjusted P = 2.5 × 10−7; LINC01413: rs2591054, adjusted P = 3.7 × 10−10; KCNJ15: rs928771, adjusted P = 3.2 × 10−8) (SI Appendix, Table S8) (32). While in the stage 1+2 data, among the top 20 PCs, only the first and third were significantly associated with AD (at nominal level P value threshold of < 0.05). Therefore, we selected phenotype-associated PCs together with age and gender as covariates to adjust for possible population stratification and batch effects (50). Although the overall significance level decreased after covariate adjustment, our candidates still reached the suggestive association threshold (P < 5 × 10−5) (GCH1: rs72713460, adjusted P = 4.36 × 10−5; LINC01413: rs2591054, adjusted P = 3.65 × 10−5; KCNJ15: rs928771, adjusted P = 3.60 × 10−6), implying their associations with AD in Chinese population (Table 1 and SI Appendix, Tables S8 and S9).
Fig. 2.
GWAS results in the Chinese AD WGS study. (A) Manhattan plots showing AD susceptibility loci discovered in the Chinese AD WGS dataset. Horizontal lines represent the suggestive threshold (P = 1E−5, blue) and genome-wide threshold (P = 5E−8, red). The 59 sites that survived the genome-wide threshold in the validation stage are shown as enlarged red dots, with gene symbols marked in the plot. (B–D) Regional association plots of the APOC1 (B), GCH1 (C), and KCNJ15 (D) loci. Horizontal lines separate the association results from the stage 1 and stage 1+2 analyses. Purple diamonds specify the sentinel variant in the corresponding locus. Colors illustrate the LD measured as R2 between the sentinel variant and its neighboring variants. cMMb, centimorgans per megabase.
Table 1.
GWAS results of AD susceptibility loci discovered in the Chinese AD cohort
| CHR:BP (GRCh37) | SNP_ID | Nearest Gene(s) | EA | EAF (AD) | EAF (NC) | Nominal P value | Final association results (adjusted results) | |||
| Stage 1 | Stage 1+2 | Stage 1 | Stage 1+2 | Adjusted P value | Effect size (SE) | |||||
| 14:55297043 | rs72713460 | GCH1 | T | 0.160 | 0.097 | 0.099 | 5.9E−05 | 4.0E−08 | 4.36E−05 | 0.594 (0.145) |
| 15:57612410 | rs2591054 | LINC01413 | C | 0.683 | 0.773 | 0.779 | 1.8E−05 | 3.5E−10 | 3.65E−05 | −0.447 (0.108) |
| 19:45420082 | rs73052335 | APOC1 | C | 0.325 | 0.153 | 0.101 | 7.1E−18 | 3.5E−72 | 1.44E−14 | 0.870 (0.113) |
| 21:39663760 | rs928771 | KCNJ15 | G | 0.238 | 0.154 | 0.161 | 6.0E−06 | 1.2E−08 | 3.60E−06 | 0.555 (0.120) |
Statistical results of four sentinel variants (i.e., variants with the lowest nominal P values among four AD susceptibly loci from the validation stage). Stage 1 (AD: 477, NC: 442); Stage 1+2 (AD: 477, NC: 2,187). BP, base positions in GRCh37 annotation; CHR, chromosome(s); EA, effect alleles; EAF, effect allele frequency.
Replication Study in Non-Asian AD Cohorts.
To justify our finding for the AD risk factors, we first examined the summary metrics from the IGAP stage 1 study (sample size n = 17,008 and 37,154, for AD and NC, respectively) (35). Of the three identified variants, none have been reported to be significantly associated with AD (P = 0.225, 0.793, and 0.349 for GCH1 rs72713460, LINC01413 rs2591054, and KCNJ15 rs928771, respectively). Meanwhile, a concordant sign of β (i.e., effect size) was observed for KCNJ15 and GCH1 variants, implying a possible enrichment of risk alleles of identified risk variants in the non-Asian AD subjects [β = 0.594 (rs72713460-T) and 0.555 (rs928771-G) in the present Chinese study; β = 0.025 (rs72713460-T) and 0.014 (rs928771-G) in the IGAP dataset].
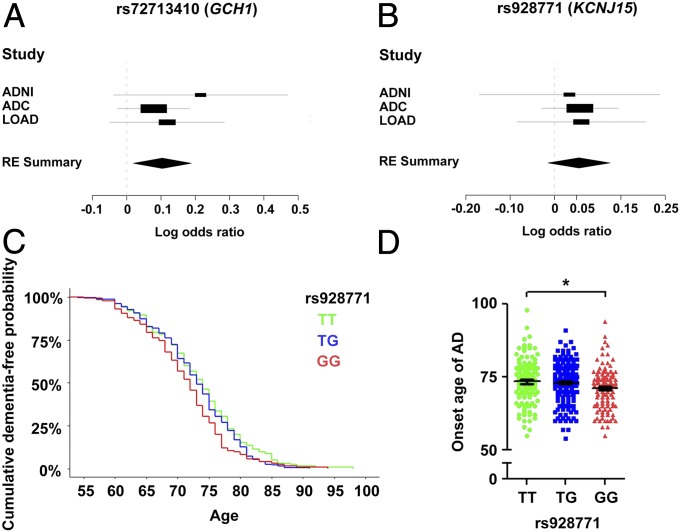
Because the IGAP study included a proportion of AD cases registered as “probable” or “possible” cases (35), we attempted to verify our findings using a subset of cohorts from the IGAP study (i.e., ADNI, ADC, and LOAD) by only retaining the subjects categorized as “definite AD” as cases. Subsequent meta-analysis to summarize the association results of the three sentinel AD risk variants confirmed the AD risk effects of GCH1 rs72713460 in the non-Asian populations (meta-P = 1.55 × 10−2, OR = 1.109) (Fig. 3A and Table 2). Moreover, we observed a concordant trend of the possible risk effect for the G allele of KCNJ15 rs928771 among the three datasets, although this failed to pass the significance threshold in the meta-analysis (meta-P = 1.19 × 10−1) (Fig. 3B and Table 2). Although a concordant risk effect was observed for LINC01413 variant rs2591054 in one of the non-Asian AD cohorts (ADC, P = 8.27 × 10−3, OR = 0.963) (Table 2 and SI Appendix, Fig. S4), an inconsistent risk effect was observed in other cohorts. Therefore, additional genetic evidence is required to validate the association between LINC01413 and AD.
Fig. 3.
Replication study in non-Asian AD cohorts. (A and B) Forest plots representing the meta-analysis results of rs72713460 (GCH1; A) and rs928771 (KCNJ15; B) from non-Asian AD cohorts. Values of effect size (log OR) obtained from independent datasets or metaresults are denoted by rectangles and diamonds, respectively. For the independent dataset, lines indicate the range of 95% confidence intervals, and the sizes of rectangles are proportional to the weights used in the meta-analysis. For the meta-analysis results, the widths of the diamonds cover the range of 95% confidence intervals. ADNI (AD: 515, NC: 339), ADC (AD: 3,946, NC: 1,746), and LOAD (AD: 464, NC: 2,231). GCH1 rs72713460: RE P = 1.55E−02, effect size = 0.1039; KCNJ15 rs928771: RE P = 1.19E−01, effect size = 0.0554. (C and D) Association of KCNJ15 with age at onset of AD. (C) Survival plot of cumulative dementia-free probabilities in AD subjects from the LOAD cohort stratified by rs928771 genotypes (P = 0.0057, Cox proportional hazards model with adjustment for gender and top-5 PCs; HR = 1.1974; n = 126, 204, and 125 for rs928771 genotypes TT, TG, and GG, respectively). (D) Dot plot of individual age at AD onset stratified by KCNJ15 rs928771 genotype. Data are presented as mean ± SEM, *P < 0.05, ANCOVA with Bonferroni correction [n = 126, 204, and 125; average (SD) onset age of dementia = 73.4 (7.6), 73.0 (6.6), and 71.2 (6.8) for rs928771 genotypes TT, TG, and GG, respectively; F = 6.35 for rs928771 genotype TT vs. GG in AD].
Table 2.
Replication study of AD susceptibility loci in multiple datasets
| Variants | Genes | Cohorts | P value | Effect size | SE | Meta (REs) | I2 | Q | P value (Q) | Tau2 | ||
| P value | Log OR (±SD) | OR | ||||||||||
| rs72713460* | GCH1 | ADNI | 9.60E−02 | 0.2154 | 0.1294 | 1.55E−02 | 0.1039 (±0.0429) | 1.109 | 0.000 | 0.987 | 0.610 | 0.000 |
| ADC | 1.44E−01 | 0.0788 | 0.0539 | |||||||||
| LOAD | 1.64E−01 | 0.1181 | 0.0849 | |||||||||
| rs928771 | KCNJ15 | ADNI | 7.44E−01 | 0.0338 | 0.1035 | 1.19E−01 | 0.0554 (±0.0356) | 1.057 | 0.000 | 0.051 | 0.975 | 0.000 |
| ADC | 1.93E−01 | 0.0574 | 0.0441 | |||||||||
| LOAD | 4.11E−01 | 0.0607 | 0.0738 | |||||||||
| rs2591054 | LINC01413 | ADNI | 5.59E−01 | 0.0630 | 0.1077 | 5.19E−01 | −0.0375 (±0.0581) | 0.963 | 50.398 | 4.032 | 0.133 | 0.005 |
| ADC† | 8.27E−03 | −0.1167 | 0.0442 | |||||||||
| LOAD | 8.17E−01 | 0.0170 | 0.0733 | |||||||||
Three previously published GWAS AD cohorts with clinically diagnosed AD cases and healthy NCs were included into the meta-analysis: ADNI (AD: 515, NC: 339), ADC (AD: 3,946, NC: 1,746), and LOAD (AD: 464, NC: 2,231). Effect size and SE are shown accordingly. Meta-P values were obtained from the METASOFT program on the basis of the estimation of the RE model. I2, I-square heterogeneity statistic; Q, Cochrane’s Q statistic; Tau2, Tau-square heterogeneity estimator of DerSimonian–Laird.
Variant(s) with P < 5E−2/3 = 1.6E−2 in the meta-analysis results.
Statistical metrics with P < 5E−2/3 = 1.6E−2 in the corresponding cohorts.
Additional transethnic meta-analysis summarizing the results from both the Chinese and non-Asian cohorts showed that our candidates exhibited trends of associations with AD in the meta-analysis (GCH1: rs72713460: meta-P = 2.53 × 10−4; KCNJ15: rs928771, meta-P = 6.41 × 10−4) (Table 2 and SI Appendix, Table S10). The results also showed heterogeneity across the Chinese and non-Asian populations, as indicated by the shifting of values in the heterogeneity measurements (I2 and Cochran’s Q-values) (Table 2 and SI Appendix, Table S10).
Interestingly, although the meta-analysis failed to replicate the risk effects of KCNJ15 rs928771, using the definite AD cases in the LOAD dataset, we showed that KCNJ15 rs928771 exerts an effect on the age of onset in AD. The minor allele G of variant rs928771 was associated with the onset age of AD with a hazard ratio (HR) of 1.197 (P = 0.0057, Cox regression model) (Fig. 3C); that is, AD subjects harboring two copies of the minor allele exhibit an earlier disease onset age compared with subjects with homozygous reference alleles (average onset age of AD: 73.4 and 71.2 y for rs928771 genotypes TT and GG, respectively) (Fig. 3D). This finding further suggests a link between KCNJ15 risk variants or gene with AD pathogenesis.
Functional Implications of Identified Risk Loci from Transcriptome and Biomarker Data.
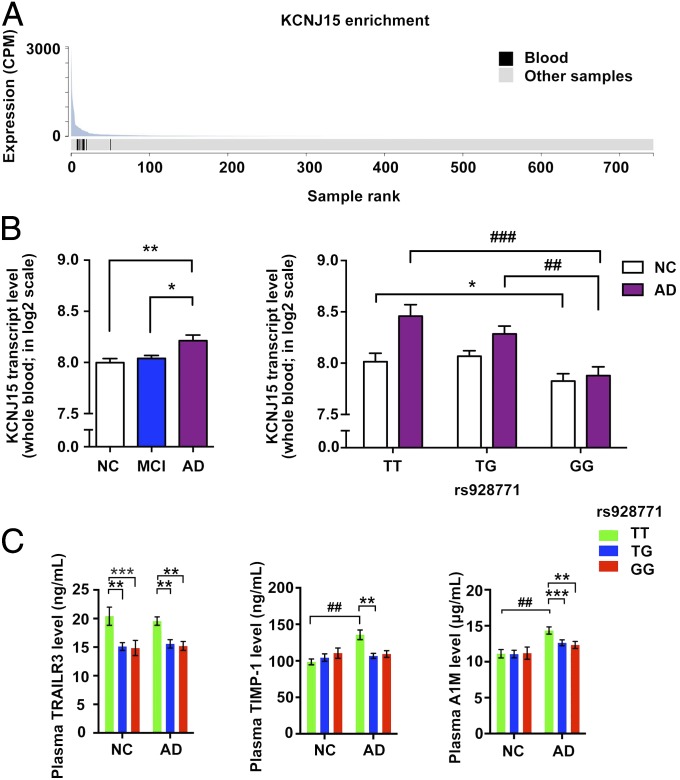
For the KCNJ15 AD risk locus, we found that KCNJ15 transcript level was enriched in blood samples (P = 6.33 × 10−7, fold-enrichment: 10.6) (Fig. 4A), implying that this gene plays roles in the peripheral circulatory system. Genotype and phenotype association analysis of KCNJ15 transcript level in the whole-blood transcriptome dataset revealed that the transcript level was significantly higher in the blood of the AD group than the NC group (Fig. 4B, Left). Nonetheless, the sentinel AD risk variant of KCNJ15, rs928771, was strongly associated with a decrease of KCNJ15 transcript level in both the NC and AD groups (Fig. 4B, Right). Interaction analysis in NC and AD subjects using a linear regression model indicated a hint of a negative interaction effect (effect size = −0.1278, P = 0.1601) between the presence of AD and the rs928771 G allele dosages for KCNJ15 transcript levels (SI Appendix, Fig. S5A). More specifically, a strong association of KCNJ15 transcript level and rs928771 G allele dosage was also observed in the MCI subjects (effect size = −0.2890, P = 3.83 × 10−5) (SI Appendix, Fig. S5B), and a positive correlation between KCNJ15 transcript level and cognitive performance (indicated by Mini-Mental State Examination score) was observed in the AD cases (effect size = 1.6318, P = 0.0294) (SI Appendix, Fig. S5C). These results suggest that intrinsic factors, specifically genomic variations, as well as extrinsic factors, specifically disease context, contribute to the regulation of KCNJ15 gene expression in blood.
Fig. 4.
Functional evidence of KCNJ15 in AD. (A) Among different tissue samples, the KCNJ15 transcript was most abundant in blood as suggested by the lowest P value of enrichment (P = 6.33E−7, fold-enrichment = 10.6; nine blood samples in 735 total samples). The figure was adopted from the FANTOM CAT database. (B) Genotype- and phenotype-dependent regulation of KCNJ15 transcript level in blood transcriptome data. (Left) Increased KCNJ15 transcript level in the whole blood of AD subjects (NC: 244, MCI: 369, AD: 106). Data are presented as mean ± SEM **P < 0.01, *P < 0.05, ANCOVA with Bonferroni correction (F = 10.38 and 5.93, for AD vs. NC, AD vs. MCI, respectively). (Right) The change of KCNJ15 transcript level in whole blood is associated with KCNJ15 rs928771 genotypes in NC and AD subjects; data are presented as mean ± SEM, *P < 0.05 for NC subjects (n = 77, 114, and 53 for rs928771 genotypes TT, TG, and GG, respectively; F = 8.22, for rs928771 TT vs. GG in NC). ##P < 0.01, ###P < 0.001 for AD subjects, ANCOVA with Bonferroni correction (n = 28, 49, and 29 for rs928771 genotypes TT, TG, and GG, respectively; F = 13.55, 10.21 for rs928771 genotypes TT vs. GG, TG vs. GG, respectively). (C) Associations between KCNJ15 rs928771 genotype and biomarker levels in AD cases and NCs. Data are presented as mean ± SEM. Test for genotypes, **P < 0.01, ***P < 0.001; test for phenotypes, ###P < 0.001 (AD: n = 31, 46, and 26; NC: n = 23, 23, and 11 for rs928771 genotypes TT, TG, and GG, respectively), ANCOVA with Bonferroni correction. cpm, counts per million mapped reads.
To further investigate the potential contributions of variant rs928771 in AD subjects for blood-related traits, we examined the association of AD patients’ plasma biomarker levels with rs928771 genotypes. Interestingly, AD subjects showed genotype-dependent reductions of various immune-associated plasma biomarkers, including decreased TNF-related apoptosis-inducing ligand receptor 3 (TRAILR3), metallopeptidase inhibitor 1 (TIMP-1), and α-1-microglobulin (A1M) (Fig. 4C). Further inclusion of control subjects confirmed the effect of the KCNJ15 variant in the modulation of serum TRAILR3 levels, as evidenced by the concordant reduction of TRAILR3 level in AD and NC subjects harboring rs928771 risk alleles. Moreover, for TIMP-1 and A1M, they both exhibited phenotype-dependent elevation of protein levels in AD subjects, implying their possible roles in AD progression, which may be associated with KCNJ15 regulations (Fig. 4C and SI Appendix, Tables S11 and S12).
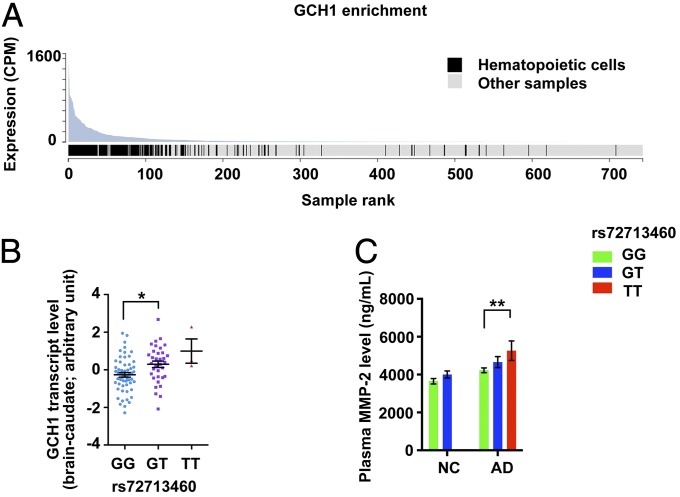
The transcript level of GCH1 was enriched in hematopoietic cells of the myeloid and B lymphoid lineages (P = 6.67 × 10−59, fold-enrichment: 16.9) (Fig. 5A). Moreover, we observed genotype-dependent regulation of GCH1 transcript level in the caudate nucleus region of the brain (Fig. 5B). These findings suggest that multiple systems or cell types in the brain and blood may be associated with GCH1 signaling. Our analysis of the genotype-dependent regulation of plasma biomarkers in AD patients identified the allele-dependent alterations of matrixmetalloproteinase-2 (MMP-2) in AD subjects (Fig. 5C and SI Appendix, Tables S11 and S12).
Fig. 5.
Functional evidence of GCH1 in AD. (A) GCH1 transcript was most abundant in the hematopoietic cells among all samples as suggested by lowest P value of enrichment (P = 6.67E−59, fold-enrichment = 16.9; 163 hematopoietic cell samples in 581 total samples). The figure was adopted from the FANTOM CAT database. (B) Associations between GCH1 transcript levels and GCH1 rs72713460 genotypes in the brain caudate region (n = 57, 34, and 3 for rs72713460 genotypes GG, GT, and TT, respectively). Data are presented as mean ± SEM, *P < 0.05 (F = 6.58); ANCOVA with Bonferroni correction. (C) GCH1 rs72713460 genotype is associated with plasma MMP-2 levels. Levels of MMP-2 in AD cases and NCs [AD: n = 64, 32, and 7; NC: n = 37, 19, and 1 (removed from analysis) for rs72713460 genotypes GG, GT, and TT, respectively]. Data are presented as mean ± SEM **P < 0.01 (F = 8.59); ANCOVA with Bonferroni correction.
Regulatory Roles of AD Susceptibility Loci in Immune-Associated Signatures.
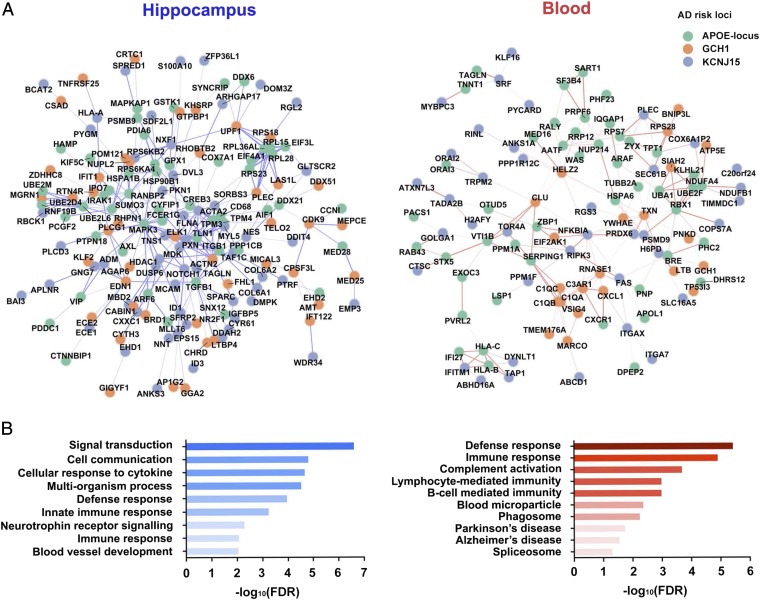
To investigate the possible disease mechanisms of the identified AD risk loci, we subjected the 52 variants at the three AD risk loci (two from KCNJ15, one from GCH1, and the remaining from APOE and the surrounding region) to a global analysis of genotype–expression associations. Because AD is associated with progressive memory loss and immune functions, as evidenced by the identification of immune genes in Caucasian GWAS (3–8), we specifically examined the possible regulatory effects of the identified AD risk loci in the hippocampus and blood. Primary investigations of genotype–expression associations revealed changes in the monocyte markers (APOE-locus for CD68), MHC molecules (KCNJ15 for HLA-A), and epigenetic modifier (GCH1 for HDAC1) in the hippocampus (Fig. 6A, Left). Meanwhile, for GCH1, we observed effects on the regulation of an AD risk gene, clusterin, as well as complement genes (C1QA, C1QB, and C1QC) in blood (Fig. 6A, Right). These findings suggest that the identified AD risk loci regulate immune signatures in the central nervous system and peripheral blood.
Fig. 6.
Enrichment of immune-associated context in the regulatory network of AD risk loci. Association of candidate target genes with AD risk loci by global genotype-expression analysis. (A) Interaction network of the target genes regulated by the AD susceptibility loci in the hippocampus (n = 87, Left) and blood (n = 365, Right). Each circle represents a target gene, with different colors specifying the corresponding AD risk loci. Lines between circles indicate gene–gene interactions. The strength of the pairwise interaction between the target genes is reflected by the color intensity of the lines. (B) GO analysis of the target genes. Representative enriched GO terms for target genes from hippocampal data (Left) and blood data (Right) are shown; x axis indicates the corresponding FDR (in log10 scale), with corresponding ontology categories marked on the y axis.
Furthermore, network analysis revealed enrichment of protein–protein interactions among the genes that showed correlations between their transcript level changes and the identified AD risk variants (P values for protein–protein interactions in the hippocampus and blood: P = 5.64 × 10−4 and 3.14 × 10−4, respectively) (Fig. 6A). Next, we performed gene ontology (GO) analysis for genes in the network. Interestingly, AD risk loci were strongly associated with changes of the immune-associated gene signatures in both the hippocampus and blood, as evidenced by the enrichment of genes involving in cellular response to cytokine stimulus [false-discovery rate (FDR) = 2.0 × 10−5] and innate immune response (FDR = 5.6 × 10−4) in the hippocampal network, as well as defense response (FDR = 3.6 × 10−6), innate immune response (FDR = 1.3 × 10−5), and complement activation (FDR = 2.0 × 10−4) in the blood network (Fig. 6B, Right, and SI Appendix, Table S13). These findings suggest that the identified AD risk loci may exert their effects in AD through the regulation of immune-related pathways.
Replication of Caucasian Risk Loci in the Chinese WGS Dataset.
We examined the contributions of Caucasian AD GWAS risk variants in the Chinese AD cohort using the existing WGS data (SI Appendix, Table S14). To ensure the accuracy of variant detection, among the 21 known risk loci identified in AD meta-analyses (35), we excluded five sites because of their low frequency in the Chinese population (MAF < 5%). Among the remaining 16 sites, based on our current data, only three showed hints of association with AD (BIN1 rs6733839, adjusted P = 4.7 × 10−2; CD2AP rs10948363, adjusted P = 4.5 × 10−2; FERMT2 rs17125944, adjusted P = 3.6 × 10−2) (SI Appendix, Table S14). None of these three variants were located in repetitive regions, indicating good detection quality. Furthermore, concordant risk or protective effects were observed in both the Chinese and Caucasian datasets (Caucasian and Chinese datasets, respectively: BIN1 rs6733839-T, OR = 1.21 and 1.21; CD2AP rs10948363-G, OR = 1.10 and 1.33; FERMT2 rs17125944-C, OR = 0.76 and 0.79) (SI Appendix, Table S14). These findings highlight the role of AD risk variants in multiple ethnic groups and also imply that ethnicity potentially contributes to the genetic basis of AD, as reflected by the observed differences in the population frequencies or disease risk effects for the specific AD risk variants studied herein.
Discussion
In this study, we comprehensively analyzed AD susceptibility loci in WGS data obtained from an AD cohort of Han Chinese ancestry. Our study revealed several common AD genetic risk factors, including APOE, GCH1, and KCNJ15. We revalidated the risk effects of the two identified risk loci, GCH1 (rs72713460) and KCNJ15 (rs928771), either by genotype–phenotype association or onset-age analysis in AD cases. Genotype–expression association analysis enables us to investigate the roles of aforementioned AD risk loci by demonstrating their effects on the regulation of genes in the hippocampus and blood. The associations of the identified AD risk loci and changes in the plasma biomarkers suggest that these loci have functional outcomes in the peripheral immune system of AD patients.
APOE is a well-accepted genetic marker for late-onset AD, and the ε4 allele of the APOE gene is the strongest genetic risk factor for the disease (51, 52). While previous GWAS show the existence of multiple AD-risk variants in and around the APOE region (53–55), adoption of the WGS method enabled a comprehensive examination of this locus, obtaining fine-mapping results of the variants that are associated with AD risk as well as the magnitude of LD between variants (SI Appendix, Tables S5 and S9). A previous fine-mapping study using the Sanger genotyping method conducted in a Japanese AD cohort reported multiple hints and haplotypes in the APOE locus that are associated with AD (56). By utilizing the WGS method, we not only obtained a larger AD-risk variant pool including the sentinel variant rs73052335, but also observed a similar AD-associated genomic structure among Chinese and Japanese populations in this region (indicated by the distributions of both P values and recombination hot spots) (Fig. 2B) (56), demonstrating the advantages of the WGS method in resolving genomic structures in the disease context, and a similar genetic mechanism for AD among Chinese and Japanese for the APOE locus.
In our study, GCH1 and KCNJ15 were identified as genetic risk loci for AD. GCH1 encodes the enzyme GTP cyclohydrolase I, which is a rate-limiting enzyme for the biosynthesis of tetrahydrobiopterin (THB, BH4); the protein is critical for the generation of monoamine neurotransmitters such as serotonin (5-HT) and dopamine, as well as nitric oxide. Mutations of GCH1 are associated with multiple neuronal disorders including dopamine-responsive dystonia (57–59), neuropathic pain (60, 61), and Parkinson’s disease (62–64). GCH1 is also implicated in cardiovascular functions, as suggested by the associations of genetic variants with endophenotypes, including nitric oxide excretion and cardiac autonomic traits (65). The association of GCH1 rs72713460 with the change in the levels of plasma MMP-2 suggests that GCH1 may play a role in the immune system or amyloid-β–associated metabolic pathway in addition to modulating neurotransmitter levels (66, 67).
Meanwhile, KCNJ15 is a member of the potassium voltage-gated channel family and is located in the Down syndrome chromosome region-1; it has been reported to be associated with type 2 diabetes mellitus (T2DM) in the Japanese population (68, 69). KCNJ15 also plays roles in glucose response, insulin secretion, and blood-related traits (70–72). The effect of sentinel AD risk variant of KCNJ15, rs928771, on the age at onset of AD, suggests its potential effect on AD pathogenesis or progression. Interestingly, KCNJ15 variant rs3746876, the protective variant for T2DM, is in proximity to rs928771 (∼8 kb apart in this study and ref. 69). These two sites are in weak LD (R2 = 0.013, D′ = 1.0; 1000 Genomes data CHB + JPT), and their minor alleles may be located in separate haplotypes. Haplotype analysis of the KCNJ15 gene may help to dissect the contributions of KCNJ15 to AD and T2DM. It would be interesting to examine whether KCNJ15 exerts its effect on AD or T2DM in East Asians through independent or convergent mechanisms.
The genotype–expression association analysis highlights a role of the KCN15 variant rs928771 in the modulation of KCNJ15 transcript level in the blood. Meanwhile, the opposite effect was observed between genotype and phenotype: the KCNJ15 transcript level was elevated in AD but is down-regulated by the risk G allele of rs928771 in all phenotypic groups (Fig. 4). Because the risk allele G is associated with both advanced onset of AD and lower KCNJ15 transcript level, and because the KCNJ15 transcript level is positively associated with individual cognitive performance (Figs. 3 and 4 and SI Appendix, Fig. S5), KCNJ15 expression in the blood may confer a protective effect during AD progression. Specifically, the plasma biomarker analysis in AD subjects revealed the modulatory effects of rs928771 in inflammation (i.e., A1M) (Fig. 4), Aβ metabolism (modulation of ADAM10 activity by TIMP-1) (73), and cell homeostasis (TRAILR3 and TIMP-1) (74, 75). Therefore, it is of interest to study the effect of KCNJ15 variants in inflammatory response, which may contribute to the pathogenesis of AD.
Our transcriptome and biomarker studies suggest that GCH1 and KCNJ15 are highly expressed in the immune system and are involved in immune-related events. This is congruent with previous findings showing an etiological role of the immune system on AD, including the identification of rare TREM2 variants (3, 4), risk genes marked by the common variants from Caucasian GWAS (7, 35), and the latest report for rare coding variants in PLCG2 and ABI3 identified from Caucasian AD patients (8). Modulations of immune signatures or biomarker levels in blood led by identified AD risk variant, as well as enriched expression of GCH1 and KCNJ15 in the blood or blood cells, further supports the roles of immune pathways, specifically in the peripheral circulatory system, in AD. Interestingly, previous studies show that alterations of genetic signatures in the peripheral circulatory system are associated with several brain disorders, including autism (76), Parkinson’s disease (77), and schizophrenia (78). Thus, comprehensive profiling of peripheral biomarkers in AD patients, including transcript, protein, or metabolite levels, may benefit the prediction and monitoring of AD.
In conclusion, we successfully implemented the low-pass WGS method to identify two AD susceptibility loci represented by common disease-associated variants in the Chinese population. Moreover, genotype–expression association analysis suggests that the identification of the genetic variants may not only provide genetic information about AD, but also the information about functional effects of these genetic variants in the pathogenesis of the disease.
Supplementary Material
Acknowledgments
We thank Dr. Fanny C. F. Ip, Dr. Yu Pong Ng, Dr. Maggie M. K. Chu, Dr. Chun T. Kwok, Dr. Zhuoyi Liang, Ye Wang, Ka Chun Lok, Cara Kwong, Yuling Zhang, Saijuan Liu, Shuangshuang Ma, and Yan Ma for their excellent technical assistance; Chi Wai Ng for the construction of data sharing webpage; other members of the N.Y.I. laboratory for many helpful discussions; Dr. Na Cai and Prof. Jonathan Flint (Wellcome Trust Centre for Human Genetics) for their great help with the CONVERGE dataset; and the International Genomics of Alzheimer’s Project (IGAP) for providing summary data for these analyses. The investigators within the IGAP contributed to the design and implementation of the IGAP and/or provided data but did not participate in analysis or writing of this report. The IGAP was made possible by the generous participation of the control subjects, the patients, and their families. This study was supported in part by the National Basic Research Program of China (973 Program; 2013CB530900), the Hong Kong Research Grants Council Theme-based Research Scheme (T13-607/12R), the Areas of Excellence Scheme of the University Grants Committee (AoE/M-604/16), Innovation Technology Commission (ITS/393/15FP and ITCPD/17-9), the National Natural Science Foundation of China (31671047), the Shenzhen Knowledge Innovation Program (JCYJ20151030140325152, JCYJ20151030154629774, JCYJ20160428145818099, JCYJ20170413173717055, and GJHS20140425165746765), International Partnership Program of Chinese Academy of Sciences (172644KYSB20160026), and the Shenzhen Peacock Plan. X.Z. was a recipient of the HKJEBN (Hing Kee Java Edible Bird’s Nest Company Limited) Scholarship for Health and Quality Living. The i-Select chips were funded by the French National Foundation on Alzheimer’s disease and related disorders. The European Alzheimer’s Disease Initiative was supported by the Laboratory of Excellence Program Investment for the Future DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2, and the Lille University Hospital. The Genetic and Environmental Risk in Alzheimer’s Disease (GERAD) was supported by the Medical Research Council (Grant 503480), Alzheimer’s Research UK (Grant 503176), the Wellcome Trust (Grant 082604/2/07/Z), and the German Federal Ministry of Education and Research: Competence Network Dementia Grants 01GI0102, 01GI0711, and 01GI0420. CHARGE was partly supported by the NIH/National Institute on Aging (NIA) Grant R01 AG033193 and NIA AG081220, and AGES Contract N01-AG-12100, National Heart, Lung, and Blood Institute Grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. The Alzheimer’s Disease Genetics Consortium (ADGC) was supported by the NIH/NIA Grants U01 AG032984, U24 AG021886, and U01 AG016976, and Alzheimer’s Association Grant ADGC-10-196728. For the ADNI dataset, data collection and sharing for this project were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (NIH Grant U01-AG024904) and Department of Defense ADNI (DOD Award W81XWH-12-2-0012). The ADNI is funded by the NIA, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following organizations: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company, Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer, Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research provides funds to support ADNI clinical sites in Canada. Private-sector contributions are facilitated by the Foundation for the NIH (https://fnih.org/). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. For the ADGC Genome-Wide Association Study–NIA Alzheimer’s Disease Centers Cohort (ADC dataset), funding support for the Alzheimer’s Disease Genetics Consortium was provided through the NIA Division of Neuroscience (Grant U01-AG032984). For the National Institute on Aging–Late Onset Alzheimer’s Disease Family Study (LOAD dataset), funding support for the “Genetic Consortium for Late Onset Alzheimer’s Disease” was provided through the Division of Neuroscience, NIA. The Genetic Consortium for Late Onset Alzheimer’s Disease includes a genome-wide association study funded as part of the Division of Neuroscience, NIA. Finally, the Genetic Consortium for Late Onset Alzheimer’s Disease provided assistance with phenotype harmonization and genotype cleaning as well as general study coordination. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the NIH, and by the National Cancer Institute, National Human Genome Research Institute, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke.
Footnotes
The authors declare no conflict of interest.
Data deposition: The summary-level statistics for the reported variants (59 sites) are available at: iplabdatabase.ust.hk/zhou_et_al_2017/GWAS_data.html.
See Profile on page 1675.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715554115/-/DCSupplemental.
Contributor Information
Collaborators: Andrew J. Saykin, Arthur W. Toga, Bret Borowski, Chad Ward, Charles DeCarli, Chet Mathis, Clifford R. Jack, Jr., Danielle Harvey, David Holtzman, David Jones, Devon Gessert, Eli Lilly, Eric M. Reiman, Erin Franklin, Franz Hefti, Greg Sorensen, Gustavo Jimenez, Howard Fillit, Jeff Gunter, Jennifer Salazar, John Hsiao, John Morris, John Q. Trojanowki, Karen Crawford Scott Neu, Kejal Kantarci, Kelley Faber, Kelly Harless, Kewei Chen, Kwangsik Nho, Laurel Beckett, Lean Thal, Leon Thal, Leslie M. Shaw, Lew Kuller, Li Shen, Lindsey Hergesheimer, Lisa Taylor-Reinwald, M. Marcel Mesulam, Magdalena Korecka, Marc Raichle, Maria Carrillo, Marilyn Albert, Matt Senjem, Matthew Bernstein, Michael Donohue, Michael Weiner, Michal Figurski, Neil Buckholtz, Nick Fox, Nigel J. Cairns, Norbert Schuff, Norm Foster, Paul Aisen, Paul Thompson, Peter Davies, Peter J. Snyder, Peter Snyder, Prashanthi Vemuri, Richard Frank, Robert A. Koeppe, Robert C. Green, Ronald Petersen, Sarah Walter, Steven Paul, Steven Potkin, Sungeun Kim, Tatiana M. Foroud, Tom Montine, Virginia Lee, William Jagust, William Potter, Yuliana Cabrera, Zaven Khachaturian, Adam Fleisher, Aimee Pierce, Akiva Mintz, Alan Lerner, Alexander Norbash, Allan I. Levey, Allyson Rosen, Amanda Smith, Anaztasia Ulysse, Andrew E. Budson, Andrew Kertesz, Angela Oliver, Ann Marie Hake, Anna Burke, Antero Sarrael, Anton P. Porsteinsson, Ashley Lamb, Athena Lee, Balebail Ashok Raj, Barton Lane, Beatriz Yanez, Beau Ances, Benita Mudge, Betty Lind, Bojana Stefanovic, Bonnie S. Goldstein, Borna Bonakdarpour, Brandy R. Matthews, Brian R. Ott, Brigid Reynolds, Bruce L. Miller, Bryan M. Spann, Carl Sadowsky, Charles Bernick, Charles D. Smith, Chiadi Onyike, Chris (Chinthaka) Heyn, Chris Hosein, Christi Leach, Christine M. Belden, Christopher H. van Dyck, Christopher M. Clark, Chuang-Kuo Wu, Colleen S. Albers, Connie Brand, Courtney Bodge, Curtis Tatsuoka, Cynthia M. Carlsson, Dana Mathews, Daniel D’Agostino, II, Daniel H.S. Silverman, Daniel Marson, David A. Wolk, David Bachman, David Clark, David Geldmacher, David Hart, David Knopman, David Perry, David Winkfield, Delwyn D. Miller, Diana Kerwin, Dick Drost, Donna M. Simpson, Donna Munic, Douglas W. Scharre, Dr Rob Bartha, Dzintra Celmins, Earl A. Zimmerman, Edmond Teng, Edward Coleman, Edward Zamrini, Effie Mitsis, Elizabeth Finger, Elizabeth Oates, Elizabeth Sosa, Ellen Woo, Emily Rogalski, Evan Fletcher, Francine Parfitt, Gaby Thai, Gad A. Marshall, Gary Conrad, Geoffrey Tremont, George Bartzokis, Ging-Yuek Robin Hsiung, Gloria Chiang, Godfrey D. Pearlson, Greg Jicha, Helen Vanderswag, Hillel Grossman, Horacio Capote, Howard Bergman, Howard Chertkow, Howard Feldman, Howard J. Rosen, Hristina Koleva, Hyungsub Shim, Irina Rachinsky, Jacobo Mintzer, Jaimie Ziolkowski, James Brewer, James J. Lah, Jamika Singleton-Garvin, Janet S. Cellar, Jared R. Brosch, Jared Tinklenberg, Jason H. Karlawish, Javier Villanueva-Meyer, Jeffrey A. Kaye, Jeffrey M. Burns, Jeffrey R. Petrella, Jerome Yesavage, Joanne Allard, Joanne L. Lord, Joel Hetelle, John Brockington, John C. Morris, John Olichney, John Rogers, Joseph Quinn, Joseph S. Kass, Joy L. Taylor, Judith L. Heidebrink, Karen Anderson, Karen Blank, Karen Ekstam Smith, Karen L. Bell, Kathleen Johnson, Kathleen Tingus, Kathryn DeMarco, Kaycee M. Sink, Keith A. Johnson, Kelly M. Makino, Kenneth Spicer, Ki Won Nam, Kim Martin, Kim Poki-Walker, Kris Johnson, Kristin Fargher, Kristine Lipowski, Kyle Womack, Laura A. Flashman, Lawrence S. Honig, Liana Apostolova, Liberty Teodoro, Lisa C. Silbert, Lisa Ravdin, Lon S. Schneider, Lori A. Daiello, M. Saleem Ismail, Marc Seltzer, Marek-Marsel Mesulam, Maria Carroll, Maria Kataki, Maria T. Greig-Custo, Marissa Natelson Love, Mark A. Mintun, Martin R. Farlow, Martin Sadowski, Mary L. Creech, Mary L. Hynes, Mary Quiceno, MaryAnn Oakley, Mauricio Becerra, Megan Witbracht, Melanie Keltz, Melissa Lamar, Mia Yang, Michael Borrie, Michael Lin, Michele Assaly, Michelle Rainka, Mimi Dang, Mohammed O. Sheikh, Mrunalini Gaikwad, Munir Chowdhury, Nadira Trncic, Nancy Johnson, Nancy Kowalksi, Nathaniel Pacini, Neil Kowall, Neill R Graff-Radford, Norman Relkin, Ntekim E. Oyonumo, Nunzio Pomara, Olga James, Olu Ogunlana, Oscar L. Lopez, Owen Carmichael, P. Murali Doraiswamy, Parianne Fatica, Patricia Lynn Johnson, Patricia Samuels, Paul Malloy, Paula Ogrocki, Pauline Maillard, Peter Hardy, Pierre Tariot, Po H. Lu, Pradeep Varma, Rachelle S. Doody, Raina Carter, Raj C. Shah, Randall Griffith, Randy Yeh, Ranjan Duara, Rawan Tarawneh, Raymond Scott Turner, Raymundo Hernando, Reisa A. Sperling, Richard E. Carson, Riham El Khouli, Robert B. Santulli, Ronald Killiany, Rosemarie Rodriguez, Russell H. Swerdlow, Salvador Borges-Neto, Sandra Black, Sandra Weintraub, Sanjay Asthana, Sanjeev Vaishnavi, Sara Dolen, Sara S. Mason, Sarah Kremen, Scott Herring, Sherye A. Sirrel, Smita Kittur, Sonia Pawluczyk, Stacy Schneider, Stephanie Kielb, Stephanie Reeder, Stephen Correia, Stephen Pasternack, Stephen Pasternak, Stephen Salloway, Sterling Johnson, Steven Chao, Steven E. Arnold, Susan K. Schultz, Susan Rountree, T-Y Lee, Terence Z. Wong, Teresa Villena, Thomas O. Obisesan, Valory Pavlik, Vernice Bates, Vesna Sossi, Victoria Shibley, William M. Brooks, William Pavlosky, Yaakov Stern, Adam Simon, Ashok Dongre, Bob Dean, Brad Navia, Dan Spellman, David Lee, David Shera, Eric Siemers, Eve Pickering, Frank Swenson, Fred Immerman, George Nomikos, Holly Soares, Hong Wan, Jeff Seeburger, Jeff Waring, John Trojanowski, Judy Siuciak, Kevin Duffin, Les Shaw, Li-San Wang, Madhav Thambisetty, Marc Walton, Mary Savage, Mats Ferm, Max Kuhn, Neil Buckholtz, Panos Zagouras, Patricia Cole, Ron Hendrickson, Sharon Xie, Sophie Allauzen, Walter Koroshetz, and William Potter
References
- 1.Chan KY, et al. Global Health Epidemiology Reference Group (GHERG) Epidemiology of Alzheimer’s disease and other forms of dementia in China, 1990-2010: A systematic review and analysis. Lancet. 2013;381:2016–2023. doi: 10.1016/S0140-6736(13)60221-4. [DOI] [PubMed] [Google Scholar]
- 2.Guerreiro R, Hardy J. Genetics of Alzheimer’s disease. Neurotherapeutics. 2014;11:732–737. doi: 10.1007/s13311-014-0295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guerreiro R, et al. Alzheimer Genetic Analysis Group TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonsson T, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griciuc A, et al. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 2013;78:631–643. doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 2015;160:1061–1071. doi: 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang KL, et al. International Genomics of Alzheimer’s Project; Alzheimer’s Disease Neuroimaging Initiative A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat Neurosci. 2017;20:1052–1061. doi: 10.1038/nn.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sims R, et al. ARUK Consortium; GERAD/PERADES, CHARGE, ADGC, EADI Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet. 2017;49:1373–1384. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilligan AM, Malone DC, Warholak TL, Armstrong EP. Racial and ethnic disparities in Alzheimer’s disease pharmacotherapy exposure: An analysis across four state Medicaid populations. Am J Geriatr Pharmacother. 2012;10:303–312. doi: 10.1016/j.amjopharm.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Hallman DM, et al. The apolipoprotein E polymorphism: A comparison of allele frequencies and effects in nine populations. Am J Hum Genet. 1991;49:338–349. [PMC free article] [PubMed] [Google Scholar]
- 11.Wang HZ, et al. Validating GWAS-identified risk loci for Alzheimer’s disease in Han Chinese populations. Mol Neurobiol. 2016;53:379–390. doi: 10.1007/s12035-014-9015-z. [DOI] [PubMed] [Google Scholar]
- 12.Guo JF, et al. Polygenic determinants of Parkinson’s disease in a Chinese population. Neurobiol Aging. 2015;36:1765.e1–1765.e6. doi: 10.1016/j.neurobiolaging.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y, et al. SNCA gene, but not MAPT, influences onset age of Parkinson’s disease in Chinese and Australians. BioMed Res Int. 2015;2015:135674. doi: 10.1155/2015/135674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang M, et al. Lack of genetic association between TREM2 and Alzheimer’s disease in East Asian population: A systematic review and meta-analysis. Am J Alzheimers Dis Other Demen. 2015;30:541–546. doi: 10.1177/1533317515577128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma J, et al. Association study of TREM2 polymorphism rs75932628 with late-onset Alzheimer’s disease in Chinese Han population. Neurol Res. 2014;36:894–896. doi: 10.1179/1743132814Y.0000000376. [DOI] [PubMed] [Google Scholar]
- 16.Miyashita A, et al. Lack of genetic association between TREM2 and late-onset Alzheimer’s disease in a Japanese population. J Alzheimers Dis. 2014;41:1031–1038. doi: 10.3233/JAD-140225. [DOI] [PubMed] [Google Scholar]
- 17.Jiang T, et al. A rare coding variant in TREM2 increases risk for Alzheimer’s disease in Han Chinese. Neurobiol Aging. 2016;42:217.e1–3. doi: 10.1016/j.neurobiolaging.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 18.Xiao X, Chang H, Li M. Molecular mechanisms underlying noncoding risk variations in psychiatric genetic studies. Mol Psychiatry. 2017;22:497–511. doi: 10.1038/mp.2016.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruchaga C, et al. GERAD Consortium; Alzheimer’s Disease Neuroimaging Initiative (ADNI); Alzheimer Disease Genetic Consortium (ADGC) GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron. 2013;78:256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramanan VK, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI) GWAS of longitudinal amyloid accumulation on 18F-florbetapir PET in Alzheimer’s disease implicates microglial activation gene IL1RAP. Brain. 2015;138:3076–3088. doi: 10.1093/brain/awv231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen L, et al. Alzheimer’s Disease Neuroimaging Initiative Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. Neuroimage. 2010;53:1051–1063. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 23.McKhann GM, et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med. 2004;256:183–194. doi: 10.1111/j.1365-2796.2004.01388.x. [DOI] [PubMed] [Google Scholar]
- 25.Cai N, et al. Data descriptor: 11,670 whole-genome sequences representative of the Han Chinese population from the CONVERGE project. Sci Data. 2017;4:170011. doi: 10.1038/sdata.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Jun G, Wing MK, Abecasis GR, Kang HM. An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res. 2015;25:918–925. doi: 10.1101/gr.176552.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrews S. 2014 FastQC: A quality control tool for high throughput sequence data. Version 0.11.2. Available at www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed August 12, 2015.
- 28.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. CRAN (2017) R (Short summer). Version 3.4.2. Available at https://www.r-project.org/. Accessed November 2, 2017.
- 32.Guan W, Liang L, Boehnke M, Abecasis GR. Genotype-based matching to correct for population stratification in large-scale case-control genetic association studies. Genet Epidemiol. 2009;33:508–517. doi: 10.1002/gepi.20403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 34.Pruim RJ, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambert JC, et al. European Alzheimer's Disease Initiative (EADI); Genetic and Environmental Risk in Alzheimer's Disease; Alzheimer's Disease Genetic Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiner MW, et al. The Alzheimer’s disease neuroimaging initiative: Progress report and future plans. Alzheimers Dement. 2010;6:202.e7–211.e7. doi: 10.1016/j.jalz.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris JC, et al. The Uniform Data Set (UDS): Clinical and cognitive variables and descriptive data from Alzheimer disease centers. Alzheimer Dis Assoc Disord. 2006;20:210–216. doi: 10.1097/01.wad.0000213865.09806.92. [DOI] [PubMed] [Google Scholar]
- 38.Lee JH, Cheng R, Graff-Radford N, Foroud T, Mayeux R. National Institute on Aging Late-Onset Alzheimer’s Disease Family Study Group Analyses of the National Institute on Aging Late-Onset Alzheimer’s Disease family study: Implication of additional loci. Arch Neurol. 2008;65:1518–1526. doi: 10.1001/archneur.65.11.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet. 2011;88:586–598. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang EY, et al. ForestPMPlot: A flexible tool for visualizing heterogeneity between studies in meta-analysis. G3 (Bethesda) 2016;6:1793–1798. doi: 10.1534/g3.116.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Consortium GT. GTEx Consortium Human genomics. The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carithers LJ, et al. GTEx Consortium A novel approach to high-quality postmortem tissue procurement: The GTEx project. Biopreserv Biobank. 2015;13:311–319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karssen L, Team G. New software and developments in the GenABEL project. Hum Hered. 2013;76:111. [Google Scholar]
- 44.Szklarczyk D, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basham B. Graphpad prism. Biotechnol Softw J. 1997;14:14–17. [Google Scholar]
- 46.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hon CC, et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gauderman W, Morrison J. 2009 QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. Available at biostats.usc.edu/Quanto.html. Accessed April 25, 2017.
- 49.Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: Implications for design of complex trait association studies. Genome Res. 2011;21:940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Epstein MP, et al. Stratification-score matching improves correction for confounding by population stratification in case-control association studies. Genet Epidemiol. 2012;36:195–205. doi: 10.1002/gepi.21611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corder EH, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 52.Genin E, et al. APOE and Alzheimer disease: A major gene with semi-dominant inheritance. Mol Psychiatry. 2011;16:903–907. doi: 10.1038/mp.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helisalmi S, et al. Promoter polymorphism (-491A/T) in the APOE gene of Finnish Alzheimer’s disease patients and control individuals. J Neurol. 1999;246:821–824. doi: 10.1007/s004150050461. [DOI] [PubMed] [Google Scholar]
- 54.Roses AD, et al. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer’s disease. Pharmacogenomics J. 2010;10:375–384. doi: 10.1038/tpj.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carrasquillo MM, Morgan K. Commentary on ‘Functional analysis of APOE locus genetic variation implicates regional enhancers in the regulation of both TOMM40 and APOE’. J Hum Genet. 2012;57:3–4. doi: 10.1038/jhg.2011.135. [DOI] [PubMed] [Google Scholar]
- 56.Takei N, et al. Japanese Genetic Study Consortium for Alzheimer Disease Genetic association study on in and around the APOE in late-onset Alzheimer disease in Japanese. Genomics. 2009;93:441–448. doi: 10.1016/j.ygeno.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 57.Steinberger D, et al. High penetrance and pronounced variation in expressivity of GCH1 mutations in five families with dopa-responsive dystonia. Ann Neurol. 1998;43:634–639. doi: 10.1002/ana.410430512. [DOI] [PubMed] [Google Scholar]
- 58.Steinberger D, et al. German Dystonia Study Group Dopa-responsive dystonia: Mutation analysis of GCH1 and analysis of therapeutic doses of L-dopa. Neurology. 2000;55:1735–1737. doi: 10.1212/wnl.55.11.1735. [DOI] [PubMed] [Google Scholar]
- 59.Shi WT, Cai CY, Li MS, Ling C, Li WD. Han Chinese patients with dopa-responsive dystonia exhibit a low frequency of exonic deletion in the GCH1 gene. Genet Mol Res. 2015;14:11185–11190. doi: 10.4238/2015.September.22.12. [DOI] [PubMed] [Google Scholar]
- 60.Taylor JG, et al. A GCH1 haplotype associated with susceptibility to vasoocclusive pain and impaired vascular function in sickle cell anemia. Blood. 2009;114:575. [Google Scholar]
- 61.Belfer I, et al. A GCH1 haplotype confers sex-specific susceptibility to pain crises and altered endothelial function in adults with sickle cell anemia. Am J Hematol. 2014;89:187–193. doi: 10.1002/ajh.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cobb SA, et al. GCH1 in early-onset Parkinson’s disease. Mov Disord. 2009;24:2070–2075. doi: 10.1002/mds.22729. [DOI] [PubMed] [Google Scholar]
- 63.Webb J, Willette AA. Aging modifies the effect of GCH1 RS11158026 on DAT uptake and Parkinson’s disease clinical severity. Neurobiol Aging. 2017;50:39–46. doi: 10.1016/j.neurobiolaging.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mencacci NE, et al. International Parkinson’s Disease Genomics Consortium and UCL-exomes consortium Parkinson’s disease in GTP cyclohydrolase 1 mutation carriers. Brain. 2014;137:2480–2492. doi: 10.1093/brain/awu179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang L, et al. Discovery of common human genetic variants of GTP cyclohydrolase 1 (GCH1) governing nitric oxide, autonomic activity, and cardiovascular risk. J Clin Invest. 2007;117:2658–2671. doi: 10.1172/JCI31093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yin KJ, et al. Matrix metalloproteinases expressed by astrocytes mediate extracellular amyloid-beta peptide catabolism. J Neurosci. 2006;26:10939–10948. doi: 10.1523/JNEUROSCI.2085-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Detry B, et al. Matrix metalloproteinase-2 governs lymphatic vessel formation as an interstitial collagenase. Blood. 2012;119:5048–5056. doi: 10.1182/blood-2011-12-400267. [DOI] [PubMed] [Google Scholar]
- 68.Okamoto K, et al. Identification of KCNJ15 as a susceptibility gene in Asian patients with type 2 diabetes mellitus. Am J Hum Genet. 2010;86:54–64. doi: 10.1016/j.ajhg.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fukuda H, et al. Replication study for the association of a single-nucleotide polymorphism, rs3746876, within KCNJ15, with susceptibility to type 2 diabetes in a Japanese population. J Hum Genet. 2013;58:490–493. doi: 10.1038/jhg.2013.28. [DOI] [PubMed] [Google Scholar]
- 70.Gosset P, et al. A new inward rectifier potassium channel gene (KCNJ15) localized on chromosome 21 in the Down syndrome chromosome region 1 (DCR1) Genomics. 1997;44:237–241. doi: 10.1006/geno.1997.4865. [DOI] [PubMed] [Google Scholar]
- 71.Okamoto K, et al. Inhibition of glucose-stimulated insulin secretion by KCNJ15, a newly identified susceptibility gene for type 2 diabetes. Diabetes. 2012;61:1734–1741. doi: 10.2337/db11-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JB, Yoo CK, Park HB, Cho IC, Lim HT. Association of the single nucleotide polymorphisms in RUNX1, DYRK1A, and KCNJ15 with blood related traits in pigs. Asian-Australas J Anim Sci. 2016;29:1675–1681. doi: 10.5713/ajas.16.0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Amour A, et al. The in vitro activity of ADAM-10 is inhibited by TIMP-1 and TIMP-3. FEBS Lett. 2000;473:275–279. doi: 10.1016/s0014-5793(00)01528-3. [DOI] [PubMed] [Google Scholar]
- 74.Sheridan JP, et al. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science. 1997;277:818–821. doi: 10.1126/science.277.5327.818. [DOI] [PubMed] [Google Scholar]
- 75.Rho SB, Chung BM, Lee JH. TIMP-1 regulates cell proliferation by interacting with the ninth zinc finger domain of PLZF. J Cell Biochem. 2007;101:57–67. doi: 10.1002/jcb.21127. [DOI] [PubMed] [Google Scholar]
- 76.Ashwood P, Wakefield AJ. Immune activation of peripheral blood and mucosal CD3+ lymphocyte cytokine profiles in children with autism and gastrointestinal symptoms. J Neuroimmunol. 2006;173:126–134. doi: 10.1016/j.jneuroim.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 77.Masliah E, Dumaop W, Galasko D, Desplats P. Distinctive patterns of DNA methylation associated with Parkinson disease: Identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics. 2013;8:1030–1038. doi: 10.4161/epi.25865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bowden NA, et al. Preliminary investigation of gene expression profiles in peripheral blood lymphocytes in schizophrenia. Schizophr Res. 2006;82:175–183. doi: 10.1016/j.schres.2005.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The summary-level statistics for the reported variants (59 sites) are available at: iplabdatabase.ust.hk/zhou_et_al_2017/GWAS_data.html. A file containing allele frequencies for variants analyzed in this paper (MAF ≥ 10%) is available through application via the above URL.