Significance
Mitochondrial genome replication and segregation are essential processes in most eukaryotic cells. While replication has been studied in some detail, much less is known about the molecular machinery required to distribute the replicated genomes. Using superresolution microscopy in combination with molecular biology and biochemistry, we show in which order the segregation machinery is assembled and that it is likely assembled de novo rather than in a semiconservative fashion in the single-celled parasite Trypanosoma brucei. Furthermore, we demonstrate that the mitochondrial genome itself is not required for assembly to occur. It seems that the physical connection of the mitochondrial genome to cytoskeletal elements is a conserved feature in most eukaryotes; however, the molecular components are highly diverse.
Keywords: Trypanosoma brucei, mitochondrial genome segregation machinery, tripartite attachment complex, kDNA, superresolution microscopy
Abstract
In almost all eukaryotes, mitochondria maintain their own genome. Despite the discovery more than 50 y ago, still very little is known about how the genome is correctly segregated during cell division. The protozoan parasite Trypanosoma brucei contains a single mitochondrion with a singular genome, the kinetoplast DNA (kDNA). Electron microscopy studies revealed the tripartite attachment complex (TAC) to physically connect the kDNA to the basal body of the flagellum and to ensure correct segregation of the mitochondrial genome via the basal bodies movement, during the cell cycle. Using superresolution microscopy, we precisely localize each of the currently known TAC components. We demonstrate that the TAC is assembled in a hierarchical order from the base of the flagellum toward the mitochondrial genome and that the assembly is not dependent on the kDNA itself. Based on the biochemical analysis, the TAC consists of several nonoverlapping subcomplexes, suggesting an overall size of the TAC exceeding 2.8 mDa. We furthermore demonstrate that the TAC is required for correct mitochondrial organelle positioning but not for organelle biogenesis or segregation.
Mitochondria are key organelles in almost all eukaryotes. Their ability to generate energy via oxidative phosphorylation depends on a small number of proteins that are encoded on the mitochondrial genome (mt-genome) (1, 2). Consequently, accurate replication and segregation of the mt-genome are essential for cell growth and healthy tissues. While many aspects of the replication have been studied in great detail, the segregation of the organelle’s genome is less well understood.
Trypanosomes are parasitic, single-celled eukaryotes within the supergroup of the Excavates. One of the best studied trypanosomes is Trypanosoma brucei, the causative agent of human African sleeping sickness and nagana in cattle. T. brucei has a complex life cycle, alternating between the mammalian bloodstream and the insect vector, the tsetse fly (3). The bloodstream form (BSF) parasite almost entirely relies on glycolysis for energy generation and lacks oxidative phosphorylation and consequently also cristae formation in the mitochondrion. In the insect, the procyclic form (PCF) of the parasite relies on amino acids for energy generation. Its mitochondrion is structurally and functionally more complex with many cristae and is fully active in oxidative phosphorylation (4).
The single large mitochondrion of T. brucei contains a singular mt-genome that is also known as kinetoplast DNA, or kDNA (5–8). Maintenance of the kDNA is essential for cell survival. However, similar to the petite mutants in yeast, it is possible to generate BSF trypanosomes that are able to survive without kDNA (γL262P cell line) (9, 10). These cells have acquired a mutation in the gamma subunit of the mitochondrial ATP synthase that allows the maintenance of an electrochemical gradient over the mitochondrial inner membrane (IM) in the absence of an otherwise essential kDNA-encoded ATP synthase subunit (9).
In T. brucei, the kDNA consists of 5,000 plasmid-like minicircles, diverse in sequence, each encoding three to five guide RNAs (gRNAs) that are required to edit the cryptic transcripts from the 25 maxicircles, which are the homologous structures of mt-genomes in many other well-studied eukaryotes (11–13). Each minicircle is physically connected to three other minicircles, and the maxicircles are interwoven in this network such that the overall structure of isolated kDNA resembles a knight’s chain mail (14). The kDNA is tightly packed into a disk-like structure of about 450 × 150 nm, localized in the kDNA pocket adjacent to the flagellum basal bodies (15). More than 30 proteins including several different polymerases and helicases are involved in the replication of the kDNA (14, 16). Postreplication, the kDNA is segregated into the developing daughter cells through the movement of the basal bodies (17). In 2003 an electron microscopy study by Ogbadoyi et al. (18) revealed a filamentous structure connecting the basal body and the kDNA that was named the tripartite attachment complex (TAC). The three parts of the TAC are the exclusion zone filaments (EZFs), named for the lack of cytoplasmic ribosomes, that extend from the base of the flagellum to the mitochondrial outer membrane (OM); the differentiated mitochondrial OM and IM, which in this region are resistant to detergent treatment; and the unilateral filaments (ULFs) that connect the mitochondrial IM to the kDNA (18). The ULFs can be subdivided into a region with DNA enriched in basic proteins and a region without DNA with mostly acidic proteins (19).
A number of individual TAC components have been identified and characterized in recent years (Table 1), including TAC102, which is the kDNA most proximal component (see cartoon in Fig. 1). TAC102 is a 102-kDa structural, basic (pI 9.2) protein with a mitochondrial import sequence in the C-terminal region, which is part of the ULFs (20). Depletion of the protein initially leads to missegregation of the kDNA such that the daughter cell with the old basal body retains large parts of the kDNA, while the daughter cell with the new basal body can only bind very small portions of the kDNA (20). Eventually, this leads to few cells with giant kinetoplasts and loss of the kDNA in the majority of the population. The function of TAC102 is restricted to kDNA segregation since the loss of the protein has no impact on kDNA replication, organelle morphology, biogenesis, or segregation (20). Consequently, TAC102 is dispensable in a trypanosome cell line that is able to survive without mt-genome (γL262P; refs. 9 and 20). Further details of the molecular functions of TAC102 remain elusive. The first component of the TAC to be described was p166, a 166-kDa large acidic (pI 5.1) protein, with an N-terminal mitochondrial targeting sequence that localizes to the region between the kDNA and the mitochondrial IM (21). The protein p166 contains a potential transmembrane domain that is not required for localization; however, it remains unclear if it is required for correct function of the protein (21). Similar to TAC102, p166 is stably associated with the TAC in flagellar extracts, isolated with a detergent under high-salt conditions (20, 21). Furthermore, the RNAi phenotypes of both genes are very similar.
Table 1.
TAC proteins and reagents
| Name | Gene ID | Source | Information |
| BBA4 | — | (34) | - Decorates a structure of the basal- and probasal body |
| - Stably associates with isolated flagella | |||
| This study | - Loss of BBA4 after p197 RNAi | ||
| p166 | Tb927.11.3290 | (21) | - First discovered TAC component |
| - Transmembrane domain (residue 1,440–1,462) | |||
| - Localizes between kDNA and basal body (PCF) | |||
| - kDNA missegregation/growth retardation after RNAi (PCF) | |||
| - Stably associates with isolated flagella | |||
| This study | - Phenotype in BSF and not essential in BSF γL262P cells | ||
| Mab22 | — | (28) | - Decorates a protein in the EZFs and mature/probasal body |
| p197 | Tb927.10.15750 | (27) | - Localizes between kDNA and basal body (PCF) |
| - kDNA missegregation/growth arrest after RNAi (PCF) | |||
| This study | - Localizes to the EZFs | ||
| - Phenotype in BSF and not essential in BSF γL262P cells | |||
| TAC40 | Tb927.4.1610 | (22) | - Beta-barrel OM protein |
| - kDNA missegregation/growth arrest after RNAi (BSF, PCF) | |||
| - kDNA missegregation (BSF γL262P) | |||
| - Localizes between kDNA and basal body | |||
| - Stably associates with isolated flagella | |||
| TAC102 | Tb927.7.2390 | (20) | - Localizes to the ULFs |
| - kDNA missegregation/growth arrest after RNAi (BSF, PCF) | |||
| - kDNA missegregation (BSF γL262P) | |||
| - Stably associates with isolated flagella | |||
| TAC65 | Tb927.5.830 | (26) | - Localizes to the EZFs |
| - kDNA missegregation/growth arrest after RNAi (PCF, BSF) | |||
| - kDNA missegregation (BSF γL262P) | |||
| - Stably associates with isolated flagella | |||
| This study | - Localizes to the EZFs | ||
| TAC60 | Tb927.7.1400 | (38) | - OM protein with two transmembrane domains |
| - kDNA missegregation/growth arrest after RNAi (BSF, PCF) | |||
| - kDNA missegregation (BSF γL262P) | |||
| - Localizes between kDNA and basal body | |||
| - In complex with TAC40 | |||
| - Stably associates with isolated flagella |
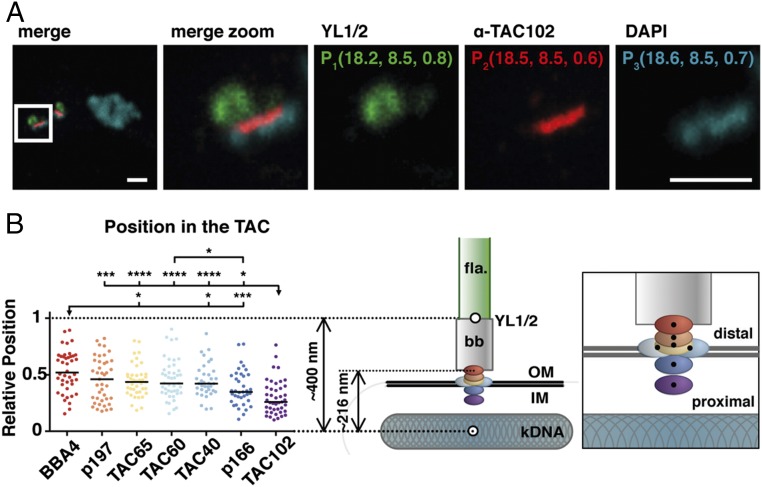
Fig. 1.
Relative position of the TAC components within the complex. (A) A representative image of a 2K1N cell stained with DAPI (DNA, cyan) and YL1/2 (basal body, green) is shown. Both channels were acquired with a confocal microscope. TAC102 is stained with a monoclonal antibody (red) and was acquired using STED microscopy. To measure the distance to the kDNA, the center of mass was calculated by using the 3D object counter from ImageJ. The xyz coordinates are shown in single color images. (B) Relative positions of the different components (BBA4, red; p197, orange; TAC65, yellow; TAC60, light blue; TAC40, blue; p166, dark blue; TAC102, purple) are indicated by dots; black line indicates the median (36 ≤ n ≤ 44). The model depicts the relative position within the TAC (right model). The flagellum (fla) is highlighted in green, the basal body (bb) in gray, the kDNA in cyan-gray, and the mitochondrial membrane by two black lines (OM, IM). A zoom-in of the TAC components within the complex is shown next to it. *P ≤ 0.05; ***P ≤ 0.001; ****P ≤ 0.0001. (Scale bar, 1 µm.)
The first mitochondrial OM component of the TAC to be discovered was TAC40, a beta-barrel protein of the porin family with similarities to MDM10 from yeast (22). While the yeast MDM10 is involved in a number of different functions including the endoplasmic reticulum mitochondrial encounter structure (ERMES) complex, nucleoid segregation, and protein import machinery assembly (23–25), the function of TAC40 is restricted to mt-genome segregation (22). Based on localization and biochemical purifications, TAC40 is closely associated with TAC60, which is also embedded in the mitochondrial OM with exclusive function in kDNA segregation. In the region between the OM and the basal body, two proteins have now been described. TAC65 was shown to interact with pATOM36, an OM protein previously described to be involved in the biogenesis of the protein import machinery (26). In the same region, p197 was discovered during proteomic screens to characterize the basal body and bilobe structure of the flagellum (27). Similar to p166, p197 has been suggested to be a TAC component in PCF parasites. For both proteins, it remains unknown if they are also essential in BSF cells and if their function is restricted to mt-genome segregation. Furthermore, Mab22, a monoclonal antibody against an unknown protein, was identified to localize to the EZFs and to the mature and probasal body (28). There are a number of additional proteins that are involved in the TAC. However, these proteins were also shown to be involved in functions other than genome segregation. This includes AEP1, a mitochondrial protein that results from alternative editing of mitochondrial COX3 transcripts and localizes to the TAC in isolated flagella (29), and pATOM36, an OM protein with a dual function in mitochondrial protein import machinery assembly and the kDNA segregation (26). Depletion of the mitochondrial acyl carrier protein, ACP, an enzyme of the fatty acid biosynthesis pathway, leads to missegregated kinetoplasts in BSF but not PCF parasites, where an RNAi knockdown results in a cytochrome-mediated respiration defect (30, 31). The kDNA missegregation in BSF might be due to phospholipid composition changes in the mitochondrial membranes (30). As to why the two life cycle stages are affected differently remains unknown. Furthermore, TBCCD1, a member of the tubulin-binding cofactor C protein family, is involved in bilobe structure formation and correct connection of the TAC to the basal bodies (32). Finally, depletion of the Krebs cycle enzyme α-KDE2 results in an unequal distribution of the replicated kDNA, suggesting a moonlight function for this well-studied protein (33).
Results
p166 and p197 Are Essential TAC Components in BSF Cells.
To verify that p166 and p197 are indeed essential components of the TAC in both major life cycle stages and to test if they are involved in other mitochondrial functions than genome maintenance, we generated cell lines that allowed RNAi depletion in wild-type BSF cells and a BSF cell line that is able to survive without mt-genome but under wild-type conditions retains the kDNA (γL262P; ref. 9).
Knockdown by RNAi of the mitochondrial protein p166 in BSF (NYsm) parasites results in kDNA missegregation whereby daughter cells receive unequal amounts of kDNA, eventually leading to a population of cells without kDNA. The growth of the population is affected 3 d postinduction of RNAi (SI Appendix, Fig. S1A). However, no growth defect is detected in the γL262P cell line while maintaining the kDNA loss phenotype, suggesting the protein is only required in the context of mt-genome segregation (SI Appendix, Fig. S1B). Similarly, RNAi targeting the mRNA of the EZFs protein p197 leads to kDNA missegregation and a growth defect in NYsm cells after 2 d of RNAi induction, while no growth defect is visible in the γL262P BSF cells (SI Appendix, Fig. S2 A and B).
Since the kDNA is physically connected to the basal bodies via the TAC, we investigated any changes in basal body morphology following knockdown by RNAi of p197 by using transmission electron microscopy (TEM). Thin sections of the basal body did not show any obvious differences between wild-type and p197 depleted cells, indicating that the function of p197 is related to TAC biogenesis rather than basal body biogenesis (SI Appendix, Fig. S2C). In this context, we also discovered that the structure decorated by the BBA4 antibody (recognizing an unknown structure in close proximity to the basal body) (34) disappears in the NYsm cell line upon p197 depletion, indicating that the structure is not required for correct growth and/or basal body biogenesis and that BBA4 potentially recognizes a TAC component (SI Appendix, Fig. S2D).
Thus, p166 and p197 are both components of the TAC with essential functions in kDNA maintenance.
Relative Order of the TAC Proteins.
To determine the relative order of the proteins within the TAC, we measured the distance between the individual components using a combination of confocal and superresolution microscopy. The BSF cells were fixed, and proteins of the TAC and the basal body were visualized using different antibodies, while the kDNA and nuclear DNA were detected using the fluorescent stain 4′,6-diamidino-2-phenylindole (DAPI) (Fig. 1A). YL1/2, an antibody targeting tyrosinated alpha-tubulin and the basal body protein TbRP2, localizes to the distal end of the basal body at the transitional fibers (Fig. 1A and refs. 35–37). The individual TAC components were C-terminally tagged (myc, PTP, or HA), except for p197, which was N-terminally tagged. For detection of TAC102, we used a monoclonal anti-TAC102 antibody (20). The distance measurements between the TAC components and the kDNA were normalized to the distance of the kDNA to YL1/2 (see Materials and Methods and Fig. 1B). Based on our measurements, TAC102 is the kDNA most proximal, currently known TAC component with a median relative distance of 0.268 (see cartoon in Fig. 1). It is followed by p166 with a relative distance of 0.352 to the kDNA. Next are TAC40 and TAC60, both of which biochemically localize in the mitochondrial OM (22, 38) and accordingly both show a very similar median relative kDNA distance value of 0.423 (TAC40) and 0.425 (TAC60). The two remaining proteins, TAC65 and p197, are both part of the EZFs (26, 27) and show a relative distance of 0.440 and 0.481, respectively. Thus, p197 is the kDNA most distal, currently characterized TAC protein. Only the unidentified structure recognized by the BBA4 antibody is more distal to the kDNA than p197, with a median value of 0.519.
TAC Assembles in a Hierarchical Order from the Basal Body.
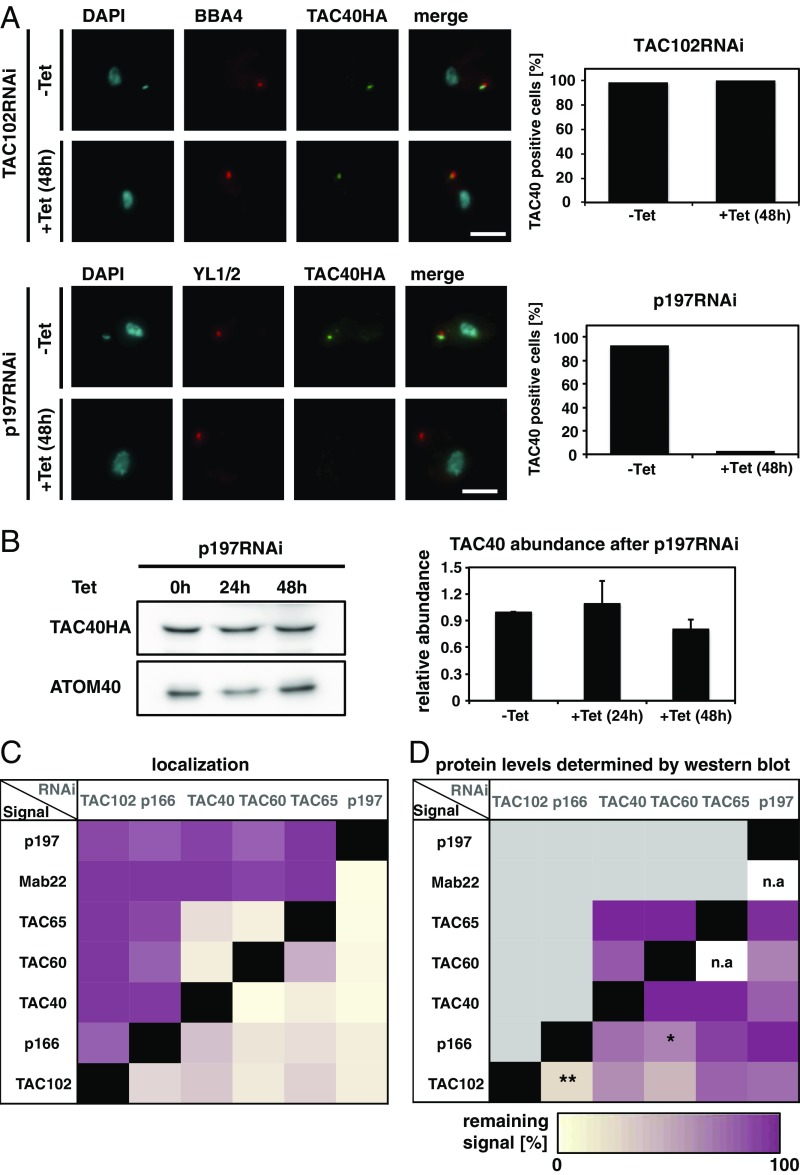
Based on the distance measurements, we were able to test what impact the relative position of each component has on the overall assembly of the TAC structure. For this, we applied RNAi against each of the TAC components and then analyzed after tetracycline (Tet) induction the presence of remaining TAC proteins by epifluorescence microscopy. Depleting the kDNA most proximal TAC component, TAC102, leads to missegregation of kDNA and eventually loss of the mt-genome in the majority of the cells, as described previously (20). However, despite the loss of kDNA, no substantial changes in localization of the OM protein TAC40 was detected (Fig. 2A). Thus, in the absence of TAC102, the localization of the more kDNA distal TAC component TAC40 remains unchanged. On the other hand, if we deplete p197, the kDNA most distal component, we detect a similar kDNA loss phenotype as described for TAC102 RNAi, and we also lose the epifluorescence signal for TAC40 (Fig. 2A). To verify the loss of TAC40 upon p197 RNAi, we also probed for the protein in whole-cell extracts by Western blotting and found the overall levels of TAC40 to be decreased only marginally after 48 h of p197 depletion (Fig. 2B). This indicates that the loss of epifluorescence signal is likely due to mislocalization rather than degradation of HA-tagged TAC40. To test all combinations of RNAi knockdown against a TAC component while analyzing the presence of the remaining TAC signals, we created 23 different cell lines. The results of immunofluorescence analyses are summarized in Fig. 2C and SI Appendix, Figs. S3–S8. Depletion of any currently known TAC component leads to the loss of correct localization of the more kDNA proximal TAC components at the epifluorescence microscopy level. Additionally, the two mitochondrial OM proteins TAC40 and TAC60 also impact each other’s localization such that TAC40 RNAi leads to loss of TAC60 and vice versa. Furthermore, TAC65, which is very close to TAC40 and TAC60 but is not a mitochondrial OM component (26), is also affected by the depletion of TAC40 and TAC60.
Fig. 2.
Dependence of TAC assembly on the individual components. (A) Fluorescence microscopy images of TAC102 RNAi or p197 RNAi uninduced (−Tet) or 48 h-induced (+Tet) cells, stained with DAPI (DNA, cyan) and probed with antibodies BBA4 or YL1/2 (basal body, red) and anti-HA (TAC40HA, green). Next to it, the quantification of TAC40-positive cells [cells without a YL1/2 signal were excluded (n ∼ 100)]. (B) Protein abundance of TAC40 determined by Western blotting from cells uninduced and 24 h and 48 h post-p197 RNAi induction. Bar graph shows protein abundance (n = 3). ATOM40 abundance was used as a loading control. (C) Quantification of each protein as described in A. Purple indicates that in 100% of the cells, the signal can correctly localize, whereas 0% is indicated by yellow. A black box shows that a knockdown of this component results in a loss of the signal for the same protein. (D) Quantification of each protein as described in B. The percentage of the remaining signal after inducing for 48 h in comparison with the uninduced cells was calculated. Yellow indicates loss of signal in Western blotting, while purple indicates no changes. A black box shows that a knockdown of this component results in a loss of the signal for the same protein, and a gray box implies no investigation on Western blot since no changes occurred in localization; n/a, not applicable. The P value was calculated by two-tailed heteroscedastic t test to perform significance measurements. *P ≤ 0.05; **P ≤ 0.01. (Scale bar: 3 µm.)
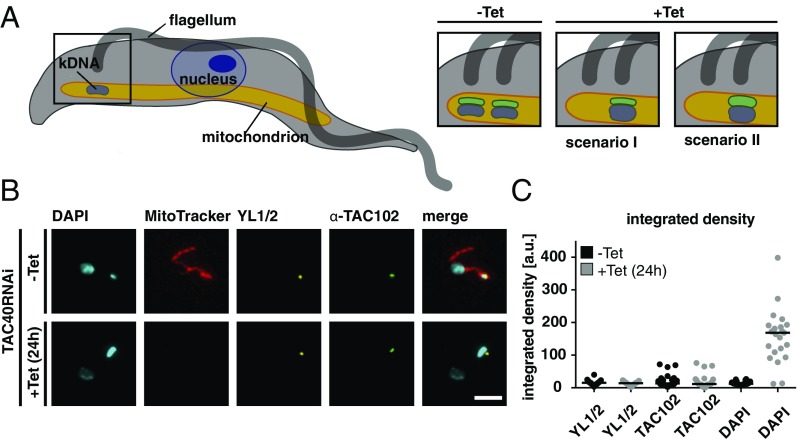
When we evaluated the protein abundance level of the TAC components that showed a loss of immunofluorescence signal (yellow in Fig. 2C) in the same cell lines by Western blotting, we found that, in general, loss of the epifluorescence signal did not correlate with loss of the protein (Fig. 2D and SI Appendix, Figs. S9–S13). However, there are two exceptions. Depletion of p166 leads to a significant loss of the TAC102 protein (0.001 < P ≤ 0.01), and depletion of TAC60 in the mitochondrial OM leads to a significant loss of the p166 protein (0.01 < P ≤ 0.05), which we suspect to reside at the IM (21). One model to explain the data assumes that the TAC is built in a hierarchical order from the basal body toward the kDNA, such that the kDNA proximal components (like, for example, TAC102) require the kDNA distal proteins (like, for example, TAC40) for correct localization (see cartoon in Fig. 1). Alternatively, one could propose that during TAC biogenesis, the new TAC proteins assemble on the old TAC structure. Once positioned correctly, they connect to their proximal and distal partners in the TAC. The movement of the basal bodies would then separate the new from the old TAC structure. If in the alternative model the kDNA distal partner is missing, the kDNA proximal components would remain on the old TAC, and thus, this part would increase in size (Fig. 3A, scenario II). We tested this alternative model by depleting TAC40 and measured if TAC102 would accumulate at the old TAC in cells that show the missegregation phenotype (enlarged kDNAs) (Fig. 3B and ref. 20). Integrated density measurements showed no changes in TAC102 intensity 24 h post-TAC40 depletion, thus supporting the hierarchical model of the TAC assembly (Fig. 3C, scenario I).
Fig. 3.
TAC102 does not accumulate upon TAC40 depletion. (A) Cartoon of a trypanosome; boxed region is enlarged on Right. The mitochondrion is depicted in orange, the flagellum in dark gray, the kDNA in blue, and TAC102 in green. After depletion of TAC40 (OM), two scenarios are shown for cells with an enlarged kDNA. In scenario I, the signal for TAC102 does not increase in size or intensity; in scenario II, the TAC102 signal increases. (B) TAC40 RNAi cells uninduced (−Tet, MitoTracker stained, red) and 24 h-induced (+Tet) were mixed and stained with YL1/2 (basal body, yellow), TAC102 (green), and DAPI (DNA, cyan). (C) The integrated density of the YL1/2, TAC102, and DAPI signal was measured with ImageJ from 1K1N uninduced cells (−Tet) and TAC40-depleted cells (+Tet; only cells with enlarged kDNA were quantified) [n(–Tet). = 30, n(+Tet). = 23]. (Scale bar: 3 µm.)
TAC Consists of Subcomplexes.
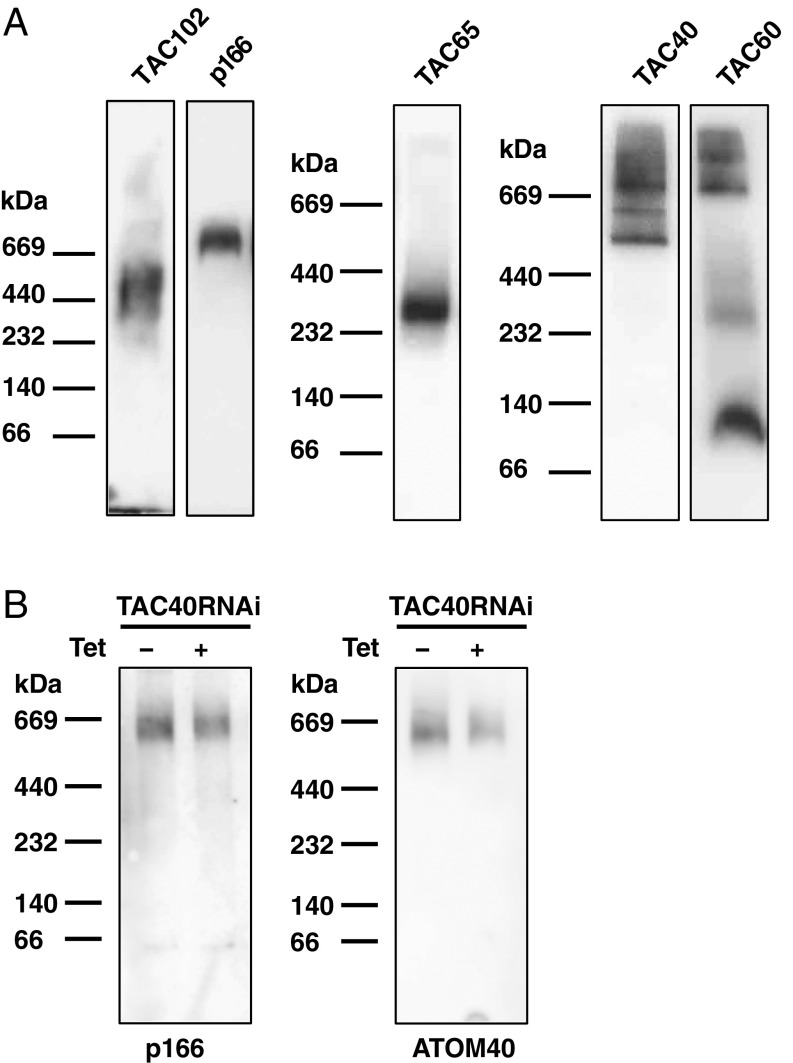
Since the kDNA proximal proteins of the TAC were still present but mislocalized in the RNAi cell lines as described above, we wondered if the individual components might be assembled in subcomplexes that would prevent their proteolysis in the case of incorrect localization. To investigate this, we applied blue native gel electrophoresis in combination with Western blotting to characterize the TAC in BSF trypanosomes. Previously, TAC65 was shown to migrate in a 300-kDa complex in PCF (26). We could confirm the complex size of TAC65 in BSF cells. Under native conditions, TAC102 migrates at around 440 kDa, while p166 is in a distinct complex larger than 670 kDa. The largest currently known complexes seem to be formed by TAC40 (between 500 and 900 kDa) and TAC60, which form several distinct bands, two of which migrate larger than 670 kDa (Fig. 4A). Based on the extraction and native running conditions, there are at least five different subcomplexes that can be identified in BSF cells. There is little overlap in the size of the individual subcomplexes, except for the p166 subcomplex, which partially overlaps with the TAC60 and TAC40 subcomplexes. When we deplete TAC40 and then probe for p166 on blue native gels, we find the complex to be largely unchanged in apparent size and abundance (Fig. 4B). The same could be observed for TAC60 after depleting p197 (SI Appendix, Fig. S14), thus confirming our hypothesis that depletion of a basal body proximal TAC component leads to a mislocalization of the kDNA more proximal partners but the individual subcomplexes remain mostly unaffected.
Fig. 4.
Complex formation of individual TAC components analyzed by blue native PAGE. (A) Western blot from blue native PAGE of mitochondrial protein extract from BSF cells probed with the following antibodies: anti-TAC102, PAP for p166, anti-myc for TAC60 and TAC65, or anti-HA for TAC40. (B) Western blot from blue native PAGE of mitochondrial protein extract from BSF TAC40 RNAi cells (−Tet, uninduced; +Tet, induced). The membrane was either probed with PAP for p166 or anti-ATOM40 as the loading control.
kDNA Is Not Required for TAC Assembly.
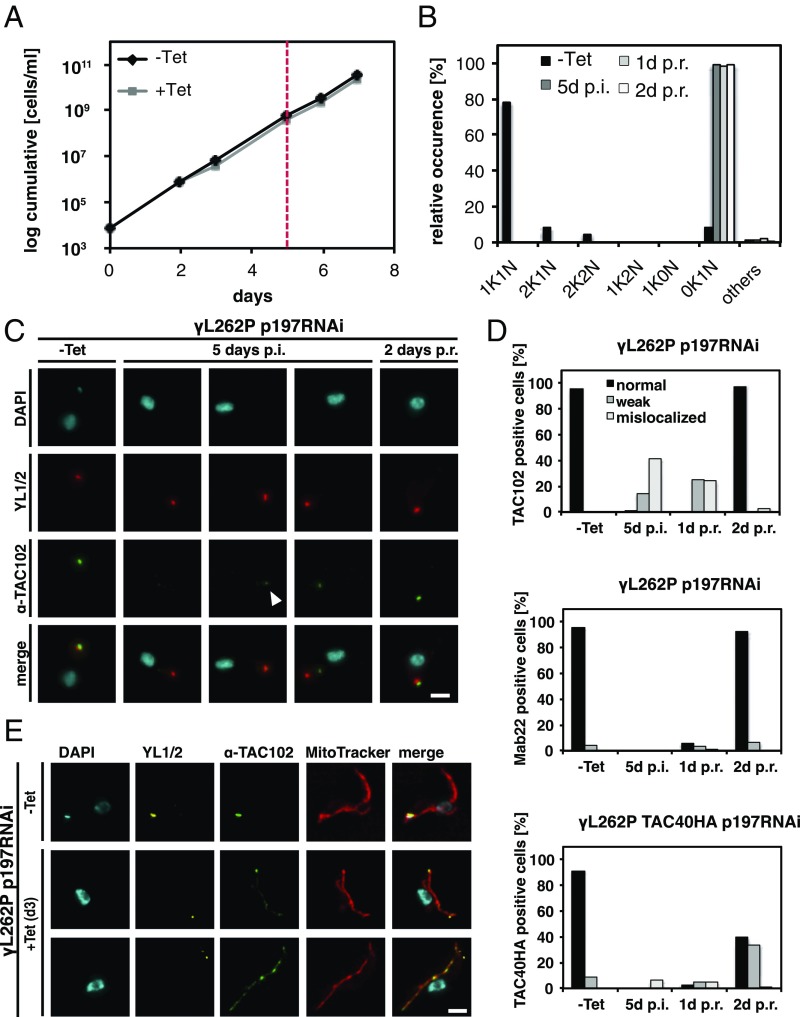
The depletion of individual TAC proteins demonstrated that kDNA more distal components are required for correct localization of the kDNA proximal elements. However, it remained unclear if the TAC itself can reassemble in the absence of kDNA. To investigate this, we used the trypanosome cell line γL262P (9). In this cell line, we depleted the basal body proximal TAC component p197 by RNAi for 5 d until ≥99% of the cells had lost their mt-genome while maintaining wild-type growth rates (Fig. 5 A and B). We then probed for TAC components representing the three regions of the TAC—Mab22 (EZFs), TAC40HA (mitochondrial OM), and TAC102 (ULFs)—by epifluorescence microscopy (Fig. 5C). At day 5 post-RNAi induction, 14% of the cells showed a weak and 42% a mislocalized TAC102 signal; for the remaining 44% of the cells, no signal was detected (Fig. 5D). Costaining with MitoTracker confirmed the mislocalized signal of TAC102 to be mitochondrial (Fig. 5E). At the same time post-p197 depletion, 94% of the cells showed no signal and 6% a mislocalized signal for TAC40; Mab22, the EZFs marker, could not be detected in any cells (Fig. 5D). We then removed Tet from the medium to stop depletion of p197 and investigated the signal for TAC102, TAC40, and Mab22. Two days postrecovery, TAC102 localized to the correct position adjacent to the basal body marker (YL1/2) in 98% of the cells. For Mab22 and TAC40, 99% (92% normal, 7% weak) and 74% (40% normal, 34% weak) of the cells showed a signal at the expected position, respectively. We also depleted p166 in the γL262P cell line (SI Appendix, Fig. S15 A and B). In this case, 44% of the cells had lost the correct signal for TAC102 (13% normal signal, 40% weak signal, 3% mislocalized signal) after 5 d of induction (SI Appendix, Fig. S15 C and D). One day postrecovery, 62% showed a weak signal, and after 2 d, 100% of the cells had recovered the wild-type TAC102 signal. In contrast, upon p166 depletion, no changes for the EZFs marker Mab22 could be detected (SI Appendix, Fig. S15D). Thus, the TAC can assemble in the absence of the mt-genome.
Fig. 5.
TAC biogenesis after recovery from p197 depletion. (A) Growth curve of γL262P 197 RNAi cells. After 5 d of p197 RNAi induction, cells were washed and grown in medium without Tet (red line). (B) Percentage of cells with kDNA and nucleus in p197 RNAi induced and uninduced trypanosomes (n ∼ 100). (C) TAC102 (green), YL1/2 (basal body, red), and DAPI (DNA, cyan) stained cells after 0 and 5 d of Tet induction (postinduction, p.i.) and 2 d postrecovery (p.r.) are shown. White arrowhead points to an example for a weak signal. (D) Quantitative analysis of TAC102 (n ≥ 106), Mab22 (n ≥ 93), or TAC40HA (n ≥ 119) in uninduced (−Tet), 5 d-induced (5 d p.i.), and 1 or 2 d after removing Tet (1 d p.r., 2 d p.r.) in γL262P p197 RNAi cells. As a control for the staining, only cells with a discernable basal body signal were used. A black bar indicates a correct localized signal; a gray bar indicates a weak signal and a light gray a mislocalized signal. (E) p197 RNAi γL262P cells were Tet induced for 3 d and stained with MitoTracker (mitochondrion, red), TAC102 (green), YL1/2 (basal body, yellow), and DAPI (DNA, cyan). (Scale bar: 2 µm.)
Timing of TAC Assembly During the Cell Cycle.
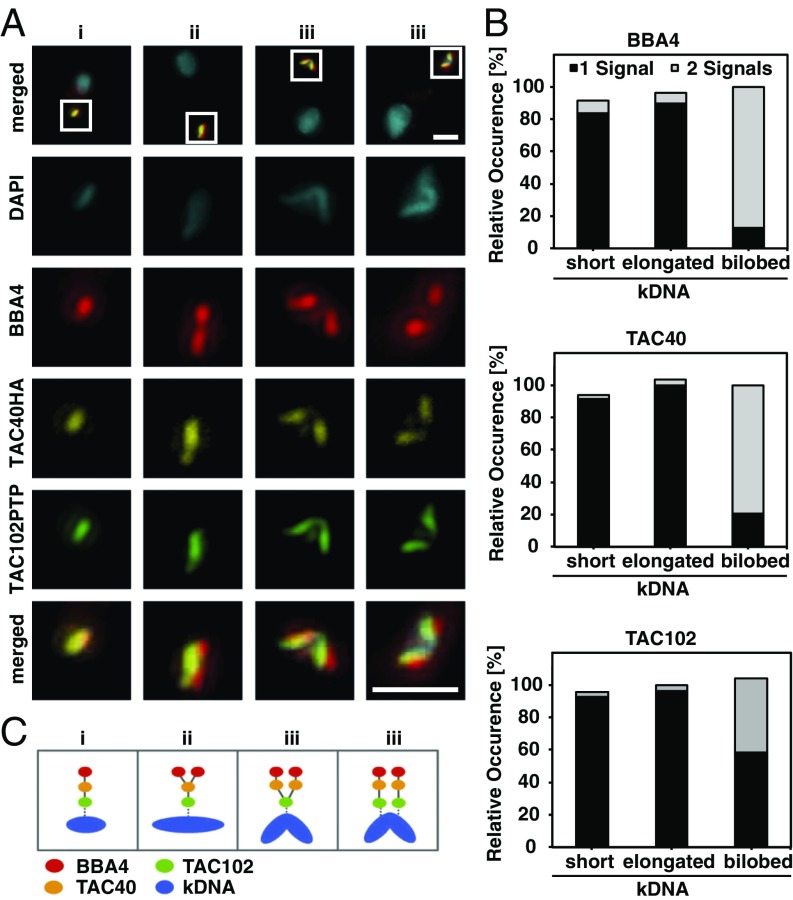
The next question we wanted to address was the timing of TAC assembly during the cell cycle. Previous studies have shown that mt-genome replication is initiated after basal body duplication, but before the replication of the nuclear genome (15, 39, 40). During the replication, the unit size kDNA grows into a bilobed structure that is subsequently segregated into two kinetoplasts. One can easily detect four different stages of kDNA replication: (i) the unit size kDNA, (ii) the enlarged kDNA, (iii) the bilobed kDNA, and the (iv) segregated kDNA connected via the nabelschnur (40). We followed individual markers of the three TAC regions, relative to the mt-genome replication and segregation (Fig. 6A and SI Appendix, Fig. S16). Before kDNA replication, the vast majority of cells show one signal for each of the three markers corresponding to one TAC structure being present. During the replication of the kDNA, first the basal body proximal components like BBA4 and later TAC40 are assembled into a new TAC, clearly separated from the old structure. The last component to be assembled and separated is TAC102, the kDNA most proximal TAC component (Fig. 6 B and C).
Fig. 6.
Dynamics of the TAC proteins during kDNA replication. (A) Tagged BSF cells were stained with DAPI (DNA, cyan), BBA4 (red), anti-HA for TAC40HA (yellow), and anti-ProteinA for TAC102PTP (green) to investigate the replication of the TAC in more detail. The region of the white box is shown enlarged below. (B) Cells were analyzed; cells were divided into stage i (short), ii (enlarged), and iii (bilobed) (40); and the number of signals per staining were counted (n = 125). One signal is depicted by gray and two signals by black bars. (C) A schematic model illustrates the TAC replication. (Scale bar: 1 µm.)
Taken together these results demonstrate that the TAC is assembled throughout the first three stages of kDNA replication and assembly progresses from the basal body toward the kDNA.
Physical Connection of the Basal Body to the Mitochondrial Membranes.
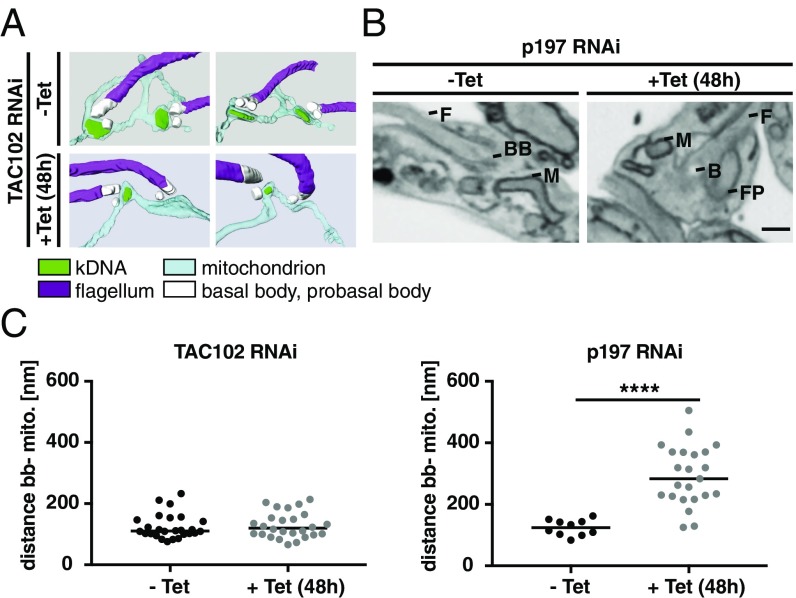
Based on our current hierarchy model, we would predict that severing the TAC in the EZFs would lead to a change in localization of the posterior region of the mitochondrion relative to the basal body, while severing the connection in the ULFs should not affect the basal body–mitochondrial positioning. To test this model, we depleted either p197 or TAC102 and used serial block face–scanning electron microscopy (SBFSEM) to analyze the distance between the basal body and the mitochondrial OM, compared with the wild-type situation (Fig. 7 A and B). After TAC102 depletion, no changes in the distance of the basal body to the mitochondrial membrane could be observed, while the median distance after p197 knockdown significantly increases from 124 nm to 283 nm (Fig. 7C). Thus, indeed the TAC complex also holds the posterior region of the mitochondrion in place.
Fig. 7.
Distance between the mitochondrion and the basal body after TAC102/p197 depletion. (A) 3D reconstructions of TAC102 RNAi-uninduced (−Tet) and 48 h-induced (+Tet) cells are shown. (B) 48 h-induced (−Tet) and uninduced (+Tet) p197 RNAi electron microscopy images are shown. BB, basal body; F, flagella; FP, flagellar pocket; M, mitochondrion. (C) The distance between the basal body and the mitochondrial membrane was measured for TAC102 (n = 26) and p197 after 0 and 48 h of RNAi induction [n(–Tet) = 10; n(+Tet) = 23]. P value using two-tailed heteroscedastic t test. ****P ≤ 0.0001.
Discussion
In this study, we describe the architecture and assembly of the mt-genome segregation machinery, also named the TAC in trypanosomes. The TAC is a large structure about twice the diameter of a nuclear pore complex (>200 nm) and encompasses three regions in the cell: the cytoplasm, the outer and inner mitochondrial membranes, and the mitochondrial matrix (18, 41). It provides the physical connection between the base of the flagellum and the kDNA that allows the segregation of the kDNA in concert with the movement of the basal bodies during the cell cycle. Thus, similar to the centrioles that nucleate the microtubule organizing centers, which in turn are responsible to build the spindles that separate sister chromatids during mitosis, the basal bodies in T. brucei organize the mitochondrial segregation machinery (42, 43).
Based on superresolution microscopy, we are able to localize each component within a particular region of the TAC (Fig. 1). Overall the distance measurements are in good agreement with the available biochemical data for TAC40 TAC60, and TAC65. TAC40 and TAC60 are both mitochondrial OM proteins and thus should be positioned between p166, a protein with a canonical mitochondrial targeting sequence, and p197, which is clearly nonmitochondrial (21, 22, 27, 38). TAC65 is not a mitochondrial protein and thus should localize between the OM proteins TAC40/TAC60 and p197 (26). Furthermore, the positioning of TAC102 (pI 9.2) in a region close to the kDNA is consistent with the electron microscopy data that demonstrated a region of basic proteins in the ULFs close to the kDNA (19). Similarly, our superresolution data place p166 (pI 5.1) further away from the kDNA than TAC102 in a region that has been reported to contain acidic proteins close to the mitochondrial IM (19). The only reagent we did not include in our measurements was Mab22, a monoclonal antibody that only recognizes its antigen if the cells are strongly extracted with detergent, which potentially changes the cytoskeletal structure within the cells.
While we are confident in the order of the TAC proteins detected by superresolution microscopy, we are also aware that the epitope tags, depending on the tag position (N or C terminus) and the protein conformation, might introduce a bias in localization relative to the native protein.
To test if TAC biogenesis is organized in a particular hierarchy or if assembly occurs independently in different regions of the complex, we depleted each TAC component and subsequently checked if and how the assembly of the TAC and its components is affected (Fig. 2). The results from these experiments are not consistent with an assembly starting at the kDNA, since depletion of the kDNA most proximal component TAC102 has no impact on the localization of any other TAC component that is more distal to the kDNA. In the random assembly model, we would expect that each component of the TAC could assemble on the old TAC structure irrespective of the presence of neighboring TAC proteins. The connection to the more distal and more proximal TAC components would occur after each protein is at the correct position. In this model, the depletion of any TAC component would lead to a correctly formed and segregated TAC structure distal to the protein that is depleted, while the kDNA proximal part would show an accumulation of the new proteins on the old TAC structure, since they cannot be separated through the basal body movement. We demonstrated that in cells where TAC40 is depleted, the more proximal component TAC102 does not accumulate at the old basal body kDNA connection (Fig. 3). Thus, only the hierarchical model where assembly of the TAC starts at the base of the flagellum and extends through the two-mitochondrial membranes to the kDNA is consistent with our current data. This model is also supported by the imaging data that describe the TAC biogenesis during the replication cycle of the kDNA (Fig. 6). Clearly, the TAC components close to the basal body appear first in two separate structures, while it happens only just before segregation for TAC102, the kDNA most proximal protein. Furthermore, the recovery experiments in the petite yeast-like trypanosome cell line showing TAC assembly in the absence of kDNA also support the hierarchical model since even lack of the kDNA and some parts of the TAC for several generations does not impact TAC assembly (Fig. 5). In our opinion, the most parsimonious model explaining our data is that the TAC is assembled de novo without any template requirement. However, it might be that a small part of the TAC insufficient to correctly segregate the mt-genome remains after RNAi and serves as a template for the assembly.
Interestingly, the complex involved in mt-genome maintenance and segregation in yeast, which has been named two membrane-spanning structure (TMS), can also assemble in the absence of the mt-genome (44). The TMS was identified through colocalization of the mitochondrial matrix protein Mgm101 with a subset of the mitochondrial nucleoids and the OM protein Mmm1 by immunofluorescence microscopy (44), and while it seems clear that both proteins together are required for mitochondrial DNA maintenance, the actual connection to the cytoskeleton remains unknown.
Including previous work and this study, more than 10 proteins have been described to be involved in TAC biogenesis in BSF and PCF trypanosomes. Six of these proteins—p197 and TAC65 in the EZFs, TAC40 and TAC60 in the mitochondrial OM, and p166 and TAC102 in the ULFs—seem to exclusively function in genome maintenance (20–22, 26, 27, 38). The strategy to exclusively employ a number of different proteins for mt-genome segregation seems unique to trypanosomes. Other model systems rely on proteins with multiple functions. In yeast, MDM10/MDM12 and Mgm101, for example, are involved in mt-genome segregation but also in multiple processes like ER mitochondrial connections (MDM10/12), oxidative mtDNA damage repair (Mgm101), and protein import (MDM10) (2, 45, 46). A similar situation can be observed in mammalian cells where the two proteins Mfn1/2 are involved in mitochondrial ER junction formation as well as nucleoid maintenance (47). Why trypanosomes have developed such an elaborate system of specialized proteins remains unknown, but we can speculate that the single-unit nature of the kDNA and its complexity and size were important factors in this development. Homologs of the six TAC components exclusively involved in TAC biogenesis are present in at least 35 of the 41 Kinetoplastea genomes that are currently available on TriTrypDB.org (48). Only a few Leishmania and Trypanosoma cruzi strains seem to miss some homologs, likely due to incomplete genome sequences (SI Appendix, Table S1). Interestingly no homologs of any of the six proteins can be found in the recently completed genome of Perkinsela, an endosymbiotic kinetoplast without flagellum and basal body (48, 49).
Native gel electrophoresis identified high-molecular weight complexes associated with the proteins TAC102, p166, TAC65, as well as TAC40 and TAC60 in BSF cells (Fig. 4). For TAC65, a complex of similar size had previously been shown in the PCF of the parasite, supporting that the TAC is conserved between the two life-cycle stages (26). However, there is very little overlap between the different complexes, indicating that even under the mild detergent conditions several subcomplexes are isolated. The largest subcomplexes seem to be in the mitochondrial OM, where TAC40 and TAC60 have been shown to interact in biochemical immunoprecipitation experiments. The two proteins of the ULFs p166 and TAC102 do not seem to reside in the same subcomplex, which is also supported by the significant distance between the two proteins that place the very basic TAC102 in the region close to the kDNA, while the acidic p166 is potentially associated with the mitochondrial IM. The positioning is further supported by a recent report by Gluenz et al. (19) dividing the ULFs in an acidic region close to the mitochondrial IM and a basic region close to the kDNA. Interestingly, the p166 subcomplex seems to overlap with the TAC60 subcomplex, suggesting that within these subcomplexes the components connecting the inner and outer mitochondrial membrane might be found. The connection of the two subcomplexes is also supported by the finding that depletion of TAC60 leads to a decrease in p166 protein levels, indicating that depletion of the TAC60 subcomplex also impacts the p166 subcomplex.
While we could clearly demonstrate that the EZFs TAC components are required for correct positioning of the kDNA pocket close to the base of the flagellum (Fig. 7), the lack of correct positioning does not seem to influence organelle division, and thus, a separate mechanism for the distribution of the organelle during cell division must exist. Arguably the TAC is now the best described mt-genome segregation machinery; however, there are many aspects that still remain elusive, including the nature of the filamentous structure in the EZFs, the connection between the inner and outer mitochondrial membrane, as well as the connection to the mt-genome itself, just to name a few.
Materials and Methods
Cell Culture.
All experiments were done either with the BSF T. brucei strain New York single marker (NYsm) (50) or the γL262P cell line (9). Cells were grown in HMI-9 medium supplemented with 10% FCS (51) at 37 °C and 5% CO2. Depending on the cell line, 2.5 µg/mL geneticin, 0.5 µg/mL puromycin, 2.5 µg/mL phleomycin, 5 µg/mL blasticidin, or 2.5 µg/mL hygromycin were added. RNAi was induced by the addition of 1 µg/mL Tet.
Transfection with Different Plasmids.
In total, 20 or 40 million cells were transfected with 8–10 µg of the plasmid in 90 mM Na3PO4, 5 mM KCl, 0.15 mM CaCl2, and 50 mM Hepes, pH 7.3, transfection buffer by electroporation using Amaxa Nucleofector II program X-001 (52, 53). The p166 RNAi was targeted against the ORF (3,452–3,952 bp) of the Tb927.11.3290 gene and p197 RNAi against the ORF (2,546–3,083 bp) of Tb927.10.15750. The p166 RNAi construct was created by using the gateway cloning system (54). The p197 RNAi construct was generated using the HindIII/XbaI and XhoI/BamHI restriction sites in the pLEW100 vector with phleomycin resistance (55). Before transfection, both plasmids were linearized by NotI. Cells containing TAC40 RNAi, TAC60 RNAi, or TAC65 RNAi were described previously (22, 26, 38). For tagging p197 and p166, the ORF region 4–1,264 bp or the ORF region 3,204–4,503 bp was PCR-amplified from genomic DNA, respectively. The PCR product for p197 was digested with NotI and ApaI and ligated into these sites of pN-PURO-PTP (56). The resulting plasmid was linearized with BmgBI before transfection. The p166 PCR product was digested with ApaI and EagI and ligated into ApaI and NotI of pC-PTP-PURO (56). This vector is a derivative of pC-PTP-NEO, in which the neomycin resistance gene was replaced by the ORF of the puromycin resistance gene via NdeI and BstBI. The final plasmid was linearized with FspAI and transfected. Cells containing TAC40 HA, TAC65 myc, or TAC60 myc tag were described previously (22, 26, 38). All proteins were C-terminally tagged, except for p197 and the PTP-tagged TAC102. All used primers are summarized in SI Appendix, Table S2.
Immunofluorescence Microscopy.
Cells were spread on a slide and fixed with 4% PFA in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4) for 4 min. After washing with PBS, the cells were permeabilized with 0.2% TritonX-100 for 5 min. After 30 min of blocking with blocking solution (4% BSA in PBS), slides were incubated for 45 or 60 min with the primary antibody followed by the secondary antibody incubation for 45 or 60 min at room temperature. The antibodies were diluted in blocking solution. For a double staining, either the primary antibodies and then the secondary antibodies were mixed, or they were used one after the other with an additional blocking step between the first secondary and second primary antibody. All used antibodies are summarized in SI Appendix, Table S3. For the rat α- TAC102 antibody, the secondary antibody had to be diluted 1:500. Cells were mounted with ProLong Gold Antifade Mountant with DAPI (Molecular Probes). The acquisition was performed with the epifluorescence DM5500 microscope from Leica or a DFC360 FX monochrome camera (Leica Microsystems) mounted on a DMI6000B microscope (Leica Microsystems). Image analysis was done using LASX software (Leica Microsystems), ImageJ, and Imaris.
MitoTracker Staining.
Cells were stained in medium with 200 nM MitoTracker Red CMXRos (Molecular Probes) as described in the manufacturer’s instructions. For mixing unstained and stained cells, additional washing steps are crucial. A subsequent immunofluorescence staining was performed as described above. All images were taken from the same microscopic slide using the same acquisition settings. Images were deconvolved using the deconvolution software from Leica (LAS AF 2.6.1.7314). In ImageJ, maximum intensity z projections were made. Binary masks were generated manually for each channel separately using the same linear signal intensity threshold values. The integrated density (Area × Mean Gray Intensity) of each particle was evaluated using the “measure particles” functionality provided by ImageJ. In the settings, the original 16-bit signals were redirected to obtain the values.
Dynamics of the TAC.
Immunofluorescence analysis was performed as described above. Before blocking with 4% BSA in PBS, an additional incubation step for 30 min with Image-iT FX Signal Enhancer (Thermo Fisher) with a following PBS/T washing step was implemented. In ImageJ, maximum intensity z projections were made. Binary masks were generated manually for each channel separately using the same linear signal intensity threshold values. Kinetoplasts were recognized automatically as binary particles inside the DAPI channel. The center of mass was used to generate a squared cropping mask, big enough to contain the particles from all channels. The binary image stacks were quantified in a subsequent step. Based on the area size and binary shape descriptors, three classes of kDNA were assigned: short, enlarged, and bilobed shaped. The number of particles was counted for each kDNA. The combination of the number of particles was computed and automatically grouped and summarized for each kDNA class.
Stimulated Emission Depletion (STED) Microscopy and Distance Measurements.
Cover glasses (no. 1.5) were glow discharged for 30 s with the FEMTO SCIENCE CUTE discharger. Cells were spread on the cover glass, and the fixing, permeabilization, and staining were performed as described above. Images were acquired by using the SP8 STED microscope (Leica) as z stacks with a z step size of 120 nm. To minimize differences that might occur during the cell cycle, we only used cells that already had duplicated and segregated the mt-genome (2K1N, 2K2N). To obtain the distance of the TAC components to the kDNA, the center of mass was determined by using the 3D object counter in ImageJ. With this plug-in, it is possible to reckon the xyz coordinates for the center of mass of an object (57). The distance between two objects can be calculated by using the Pythagorean theorem. To achieve the relative position, the measurements of the TAC component to the kDNA distance were normalized to the distance of the kDNA to the basal body.
Cytoskeleton Extraction.
For the antibody Mab22, a cytoskeleton extraction needed to be performed. For this, total cells were washed with PBS and spread on a slide. After removing the liquid, cells were incubated for 1 min with extraction buffer (100 mM Pipes, pH 6.8, and 1 mM MgCl2) containing 0.05% Nonidet P-40. Afterward, the cells were washed with extraction buffer, and the staining was completed as described above.
Western Blot Analysis.
We mixed 5 × 106 cells with 1× Laemmli buffer (0.4% SDS, 12 mM Tris⋅HCl, pH 6.8, 4.8% glycerol, 1% β-mercaptoethanol, and bromophenol blue in PBS) and loaded them per lane on a 6%, 8%, or 10% SDS–polyacrylamide gel. Blocking (5% or 10% milk in PBS/T) after transferring onto a PVDF membrane was performed for 1 h at room temperature. Primary antibodies were incubated for 1 h at room temperature or overnight at 4 °C, besides PAP, which was incubated for 30 min at room temperature. Secondary antibodies were incubated for 1 h at room temperature. All used antibodies are summarized in SI Appendix, Table S3 and were diluted in blocking solution. The acquisition was performed with the ODYSSEY Infrared imaging system (LI-COR), LAS1000 (Fuji Medical Systems), or Amersham Imager 600 (GE Healthcare).
Blue Native Gel Electrophoresis.
For an enriched mitochondria fraction, 0.025% or 0.015% digitonin in SoTE (0.6 M Sorbitol, 20 mM Tris⋅HCl, pH 7.5, and 2 mM EDTA) was used. With this fraction, it proceeded as described previously (58, 59). Instead of 1.5% digitonin, 1% was used and incubated for 15 min on ice. After a centrifugation step, the supernatant was loaded onto a native gradient gel. Afterward, the gel was soaked in SDS buffer (25 mM Tris, 192 mM glycine, and 0.1% SDS) and transferred onto a PVDF membrane by semidry Western blotting. The probing with the antibodies was performed as described above.
TEM of Thin Sections.
After harvesting, the cells were fixed and embedded as described previously (20). Thin sections were imaged with a transmission electron microscope (Tungsten cathode; FEI Morgani). The microscope was equipped with a digital camera (Morada, 12 megapixel; Soft Imaging System) and the AnalySIS iTEM image analysis software.
SBFSEM.
Sample preparation, data processing, and analysis were performed as described previously for TAC102 RNAi-uninduced and 48 h-induced cells (26). Serial images of the block face were recorded at an accelerating voltage of 4 kV, a spot size of 1, and pressure of 0.33 (+Tet) or 0.3 (−Tet) Torr. Pixel size and the dwell time for each micrograph was 5 nm and 3.2 µs, respectively, and slice thickness was 100 nm. For p197 RNAi 48 h-induced and uninduced cells, block staining, dehydration, and embedding were performed as described previously (15). The shortest distance between the basal body and the mitochondrial membrane was measured manually with IMOD. To do so, the image was orientated to get the basal body and the flagellum as well as the kDNA pocket in one plane. This was not possible for the induced samples since the kDNA pocket was not preserved. To measure, the basal body and the flagellum were oriented in one plane and the nearest mitochondrial tube was used for measuring. Because of this, the measurement had to be done throughout several slices.
Supplementary Material
Acknowledgments
We thank Keith Gull and Derrick Robinson for the BBA4/YL1/2 and Mab22 antibodies, respectively. We acknowledge Bernd Schimanski, Adolfo Odriozola, Evelyne Vonwyl, and Nicolas Niklaus for technical assistance. Imaging was supported by the Microscopy Imaging Center of the University of Bern, Switzerland and the Bioimaging Unit at Oxford Brookes University, United Kingdom. The T.O. laboratory was supported by grants from the Novartis Foundation and the Swiss National Science Foundation (SNF) (160264). The A.S. laboratory was supported by SNF Grant 138355 and in part by the SNF-funded National Centre of Competence in Research “RNA & Disease.” The B.Z. laboratory was supported by SNF Grant 163761. The S.V. laboratory was supported by a grant from the Biological & Biotechnology Research Council (BB/L014122/1’).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716582115/-/DCSupplemental.
References
- 1.Ernster L, Schatz G. Mitochondria: A historical review. J Cell Biol. 1981;91:227s–255s. doi: 10.1083/jcb.91.3.227s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 3.Fenn K, Matthews KR. The cell biology of Trypanosoma brucei differentiation. Curr Opin Microbiol. 2007;10:539–546. doi: 10.1016/j.mib.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Priest JW, Hajduk SL. Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J Bioenerg Biomembr. 1994;26:179–191. doi: 10.1007/BF00763067. [DOI] [PubMed] [Google Scholar]
- 5.Meyer H, Porter KR. A study of Trypanosoma cruzi with the electron microscope. Parasitology. 1954;44:16–23. doi: 10.1017/s0031182000018722. [DOI] [PubMed] [Google Scholar]
- 6.Meyer H, De Oliveira Musacchio M, De Andrade Mendonça I. Electron microscopic study of Trypanosoma cruzi in thin sections of infected tissue cultures and of blood-agar forms. Parasitology. 1958;48:1–8. doi: 10.1017/s0031182000021028. [DOI] [PubMed] [Google Scholar]
- 7.Steinert G, Firket H, Steinert M. [Synthesis of desoxyribonucleic acid in the parabasal body of Trypanosoma mega] Exp Cell Res. 1958;15:632–635. doi: 10.1016/0014-4827(58)90117-4. [DOI] [PubMed] [Google Scholar]
- 8.Steinert M. Mitochondria associated with the kinetonucleus of Trypanosoma mega. J Biophys Biochem Cytol. 1960;8:542–546. doi: 10.1083/jcb.8.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean S, Gould MK, Dewar CE, Schnaufer AC. Single point mutations in ATP synthase compensate for mitochondrial genome loss in trypanosomes. Proc Natl Acad Sci USA. 2013;110:14741–14746. doi: 10.1073/pnas.1305404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mounolou JC, Jakob H, Slonimski PP. Mitochondrial DNA from yeast “petite” mutants: Specific changes in buoyant density corresponding to different cytoplasmic mutations. Biochem Biophys Res Commun. 1966;24:218–224. doi: 10.1016/0006-291x(66)90723-6. [DOI] [PubMed] [Google Scholar]
- 11.Hajduk S, Ochsenreiter T. RNA editing in kinetoplastids. RNA Biol. 2010;7:229–236. doi: 10.4161/rna.7.2.11393. [DOI] [PubMed] [Google Scholar]
- 12.Read LK, Lukeš J, Hashimi H. Trypanosome RNA editing: The complexity of getting U in and taking U out. Wiley Interdiscip Rev RNA. 2016;7:33–51. doi: 10.1002/wrna.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Jensen RE, Englund PT. Network news: The replication of kinetoplast DNA. Annu Rev Microbiol. 2012;66:473–491. doi: 10.1146/annurev-micro-092611-150057. [DOI] [PubMed] [Google Scholar]
- 15.Jakob M, et al. Mitochondrial growth during the cell cycle of Trypanosoma brucei bloodstream forms. Sci Rep. 2016;6:36565. doi: 10.1038/srep36565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Povelones ML. Beyond replication: Division and segregation of mitochondrial DNA in kinetoplastids. Mol Biochem Parasitol. 2014;196:53–60. doi: 10.1016/j.molbiopara.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Robinson DR, Gull K. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature. 1991;352:731–733. doi: 10.1038/352731a0. [DOI] [PubMed] [Google Scholar]
- 18.Ogbadoyi EO, Robinson DR, Gull K. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol Biol Cell. 2003;14:1769–1779. doi: 10.1091/mbc.E02-08-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gluenz E, Shaw MK, Gull K. Structural asymmetry and discrete nucleic acid subdomains in the Trypanosoma brucei kinetoplast. Mol Microbiol. 2007;64:1529–1539. doi: 10.1111/j.1365-2958.2007.05749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trikin R, et al. TAC102 is a novel component of the mitochondrial genome segregation machinery in trypanosomes. PLoS Pathog. 2016;12:e1005586, and erratum (2016) 12:e1005750. doi: 10.1371/journal.ppat.1005586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Z, Lindsay ME, Roy Chowdhury A, Robinson DR, Englund PT. p166, a link between the trypanosome mitochondrial DNA and flagellum, mediates genome segregation. EMBO J. 2008;27:143–154. doi: 10.1038/sj.emboj.7601956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnarwiler F, et al. Trypanosomal TAC40 constitutes a novel subclass of mitochondrial β-barrel proteins specialized in mitochondrial genome inheritance. Proc Natl Acad Sci USA. 2014;111:7624–7629. doi: 10.1073/pnas.1404854111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flinner N, et al. Mdm10 is an ancient eukaryotic porin co-occurring with the ERMES complex. Biochim Biophys Acta. 2013;1833:3314–3325. doi: 10.1016/j.bbamcr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Boldogh IR, et al. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol Biol Cell. 2003;14:4618–4627. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Käser S, et al. Outer membrane protein functions as integrator of protein import and DNA inheritance in mitochondria. Proc Natl Acad Sci USA. 2016;113:E4467–E4475. doi: 10.1073/pnas.1605497113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gheiratmand L, Brasseur A, Zhou Q, He CY. Biochemical characterization of the bi-lobe reveals a continuous structural network linking the bi-lobe to other single-copied organelles in Trypanosoma brucei. J Biol Chem. 2013;288:3489–3499. doi: 10.1074/jbc.M112.417428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonhivers M, Landrein N, Decossas M, Robinson DR. A monoclonal antibody marker for the exclusion-zone filaments of Trypanosoma brucei. Parasit Vectors. 2008;1:21. doi: 10.1186/1756-3305-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochsenreiter T, Anderson S, Wood ZA, Hajduk SL. Alternative RNA editing produces a novel protein involved in mitochondrial DNA maintenance in trypanosomes. Mol Cell Biol. 2008;28:5595–5604. doi: 10.1128/MCB.00637-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clayton AM, et al. Depletion of mitochondrial acyl carrier protein in bloodstream-form Trypanosoma brucei causes a kinetoplast segregation defect. Eukaryot Cell. 2011;10:286–292. doi: 10.1128/EC.00290-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guler JL, Kriegova E, Smith TK, Lukeš J, Englund PT. Mitochondrial fatty acid synthesis is required for normal mitochondrial morphology and function in Trypanosoma brucei. Mol Microbiol. 2008;67:1125–1142. doi: 10.1111/j.1365-2958.2008.06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.André J, et al. The tubulin cofactor C family member TBCCD1 orchestrates cytoskeletal filament formation. J Cell Sci. 2013;126:5350–5356. doi: 10.1242/jcs.136515. [DOI] [PubMed] [Google Scholar]
- 33.Sykes SE, Hajduk SL. Dual functions of α-ketoglutarate dehydrogenase E2 in the Krebs cycle and mitochondrial DNA inheritance in Trypanosoma brucei. Eukaryot Cell. 2013;12:78–90. doi: 10.1128/EC.00269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woods A, et al. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- 35.Kilmartin JV, Wright B, Milstein C. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephan A, Vaughan S, Shaw MK, Gull K, McKean PG. An essential quality control mechanism at the eukaryotic basal body prior to intraflagellar transport. Traffic. 2007;8:1323–1330. doi: 10.1111/j.1600-0854.2007.00611.x. [DOI] [PubMed] [Google Scholar]
- 37.Andre J, et al. An alternative model for the role of RP2 protein in flagellum assembly in the African trypanosome. J Biol Chem. 2014;289:464–475. doi: 10.1074/jbc.M113.509521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Käser S, et al. Biogenesis of a mitochondrial DNA inheritance machinery in the mitochondrial outer membrane. PLoS Pathog. 2017;12:e1006808. doi: 10.1371/journal.ppat.1006808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKean PG. Coordination of cell cycle and cytokinesis in Trypanosoma brucei. Curr Opin Microbiol. 2003;6:600–607. doi: 10.1016/j.mib.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Gluenz E, Povelones ML, Englund PT, Gull K. The kinetoplast duplication cycle in Trypanosoma brucei is orchestrated by cytoskeleton-mediated cell morphogenesis. Mol Cell Biol. 2011;31:1012–1021. doi: 10.1128/MCB.01176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 42.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 44.Meeusen S, Nunnari J. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J Cell Biol. 2003;163:503–510. doi: 10.1083/jcb.200304040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westermann B. Mitochondrial inheritance in yeast. Biochim Biophys Acta. 2014;1837:1039–1046. doi: 10.1016/j.bbabio.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 46.Kaufman BA, et al. In organello formaldehyde crosslinking of proteins to mtDNA: Identification of bifunctional proteins. Proc Natl Acad Sci USA. 2000;97:7772–7777. doi: 10.1073/pnas.140063197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 48.Aslett M, et al. TriTrypDB: A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38:D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanifuji G, et al. Genome sequencing reveals metabolic and cellular interdependence in an amoeba-kinetoplastid symbiosis. Sci Rep. 2017;7:11688. doi: 10.1038/s41598-017-11866-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wirtz E, Leal S, Ochatt C, Cross GAM. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 51.Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- 52.Burkard G, Fragoso CM, Roditi I. Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol Biochem Parasitol. 2007;153:220–223. doi: 10.1016/j.molbiopara.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 53.Schumann Burkard G, Jutzi P, Roditi I. Genome-wide RNAi screens in bloodstream form trypanosomes identify drug transporters. Mol Biochem Parasitol. 2011;175:91–94. doi: 10.1016/j.molbiopara.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 54.Kalidas S, Li Q, Phillips MA. A Gateway® compatible vector for gene silencing in bloodstream form Trypanosoma brucei. Mol Biochem Parasitol. 2011;178:51–55. doi: 10.1016/j.molbiopara.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bochud-Allemann N, Schneider A. Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J Biol Chem. 2002;277:32849–32854. doi: 10.1074/jbc.M205776200. [DOI] [PubMed] [Google Scholar]
- 56.Schimanski B, Nguyen TN, Günzl A. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot Cell. 2005;4:1942–1950. doi: 10.1128/EC.4.11.1942-1950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 58.Mani J, et al. Mitochondrial protein import receptors in Kinetoplastids reveal convergent evolution over large phylogenetic distances. Nat Commun. 2015;6:6646. doi: 10.1038/ncomms7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pusnik M, et al. Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr Biol. 2011;21:1738–1743. doi: 10.1016/j.cub.2011.08.060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.