Significance
Intrinsically disordered proteins (IDPs) have attracted significant attention due to their roles in crucial cellular processes. NMR is the only technique that allows the study of IDPs at atomic-level resolution. However, narrow chemical shift dispersion, rapid exchange with solvent, and high proline content challenge conventional 1H-detected experiments. Here, we report the development of a suite of 3D experiments based on 15N direct detection that harnesses the slow relaxation and the larger chemical shift dispersion of 15N nuclei for complete backbone assignment of IDPs, including proline residues, which are critical to the study of IDPs. Using this approach, we have assigned the regulatory domain of NFATC2 and have identified a likely mechanism by which 14-3-3 proteins regulate NFAT nuclear translocation.
Keywords: 15N detection, IDP, NFAT, NMR resonance assignment, nuclear localization
Abstract
Studies over the past decade have highlighted the functional significance of intrinsically disordered proteins (IDPs). Due to conformational heterogeneity and inherent dynamics, structural studies of IDPs have relied mostly on NMR spectroscopy, despite IDPs having characteristics that make them challenging to study using traditional 1H-detected biomolecular NMR techniques. Here, we develop a suite of 3D 15N-detected experiments that take advantage of the slower transverse relaxation property of 15N nuclei, the associated narrower linewidth, and the greater chemical shift dispersion compared with those of 1H and 13C resonances. The six 3D experiments described here start with aliphatic 1H magnetization to take advantage of its higher initial polarization, and are broadly applicable for backbone assignment of proteins that are disordered, dynamic, or have unfavorable amide proton exchange rates. Using these experiments, backbone resonance assignments were completed for the unstructured regulatory domain (residues 131–294) of the human transcription factor nuclear factor of activated T cells (NFATC2), which includes 28 proline residues located in functionally important serine–proline (SP) repeats. The complete assignment of the NFATC2 regulatory domain enabled us to study phosphorylation of NFAT by kinase PKA and phosphorylation-dependent binding of chaperone protein 14-3-3 to NFAT, providing mechanistic insight on how 14-3-3 regulates NFAT nuclear translocation.
Over the last two decades, proteins or protein segments that lack stable secondary structure have been recognized for their importance. Collectively referred to as intrinsically disordered proteins (IDPs) (1, 2), they orchestrate diverse cellular processes from transcription and translation to self-assembly and signaling (2–8). Consequently, disordered regions occur with greater probability in human proteins associated with cancer (∼79%) and signaling (∼66%). IDPs show remarkable structural flexibility by either transiently forming secondary structure, and/or adopting stable conformations upon binding (9, 10). Some IDPs sample ordered conformations resembling classical secondary structural elements (9), while other retain dynamics in their bound conformation resulting in “fuzzy” complexes (11). The inherently disordered nature of these proteins combined with their structural plasticity renders structural characterization methods such as X-ray crystallography and cryo-EM ineffective.
Solution-state NMR spectroscopy has been the primary structural tool; however, traditional 1H-detected experiments have several limitations for studying IDPs, especially those with large molecular weight and many repeat sequences. Their conformational heterogeneity, prevalence of repetitive sequences, low fraction of hydrophobic amino acids, and high occurrence of charged amino acids homogenize the chemical complexity of IDP residues and lead to extremely narrow chemical shift dispersion (HN; ∼1 ppm). This is further compounded by a high frequency of functionally critical proline residues (12, 13), which cause “sequential connectivity breaks” in the traditional 1HN-detected backbone assignment scheme due to the lack of backbone amide protons in proline. Furthermore, prolines are known to undergo cis/trans isomerization in small unstructured peptides (14, 15) and protein loops (16), which dramatically increases the complexity of the NMR spectra by causing neighboring resonances to exist in multiple chemical environments. In addition, the lack of a stable hydrogen-bonding network and secondary/tertiary structure facilitates the rapid exchange of labile amide protons with bulk water, which broadens the resonances. Exchange broadening of amide protons is exacerbated at physiological pH (7.0–7.5) and ambient temperatures (25–37 °C)—compounding challenges for in-cell NMR experiments. To address these challenges, 13C-detected (17) and higher-dimensional experiments (18–20), as well as experiments that utilize long-range 13C couplings (21–23), have been developed. 15N-detected experiments, with distinct advantages and disadvantages in comparison with 1H and 13C experiments, have also been introduced in recent years for studying IDPs and show comparable sensitivity to their 13C-detected counterparts (24, 25). Furthermore, they are orthogonal to the existing paradigm of 1H and 13C experiments.
Here, we present a suite of six 3D 15N-detected experiments to overcome the challenges of studying IDPs with traditional 1H-detected assignment strategies. These experiments exploit the greater chemical shift dispersion in the 15N dimension relative to that of 1HN (15NH ∼ 30 ppm) and combine it with the chemical shift dispersion in the 13C dimension by encoding the correlated 13C frequency in the indirect dimension. This enables direct detection and assignment of all amino acids, including frequently occurring prolines, as these experiments do not rely on amide protons. Although the low gyromagnetic ratio and the associated lower intrinsic sensitivity are disadvantages of 15N detection, the slower transverse relaxation rate of 15N nuclei results in ultranarrow linewidths and a corresponding increase in signal height and sensitivity. 15N detection also encodes nuclei with the largest dispersion and narrowest intrinsic linewidths in the direct dimension, which allows access to high resolution without sacrificing instrument time. The advantages of 15N detection mean that the 3D experiments developed here can provide essential information necessary for the unambiguous assignment of IDP backbone resonances.
These 15N-detected experiments were used to assign the backbone resonances of human transcription factor nuclear factor of activated T cells (NFATC2) (residues 131–294). In this report, we use NFAT to mean NFATC2, unless otherwise specified. This portion of NFAT is disordered and harbors 28 proline residues (17% of the residues in the region), many of which are located in functionally critical serine–proline (SP)-rich regions. The regulatory region of NFAT (residues 131–294) used in this study harbors phosphorylation sites modified by protein kinase A (PKA). NFAT phosphorylation by PKA is known to create binding sites for 14-3-3 (26); however, the mechanism by which the NFAT–14-3-3 complex regulates transcriptional activity is not understood. Complete backbone assignment of the NFAT (131–294) regulatory domain using 15N-detected experiments enabled us to follow phosphorylation by PKA. Our data demonstrate that, upon PKA phosphorylation, 14-3-3 binds to NFAT and blocks access to the nuclear localization sequence (NLS), which may prevent the nuclear transport protein importin from translocating NFAT to the nucleus, thus preventing transcriptional activation.
Results
Suite of 3D 15N-Detection Experiments.
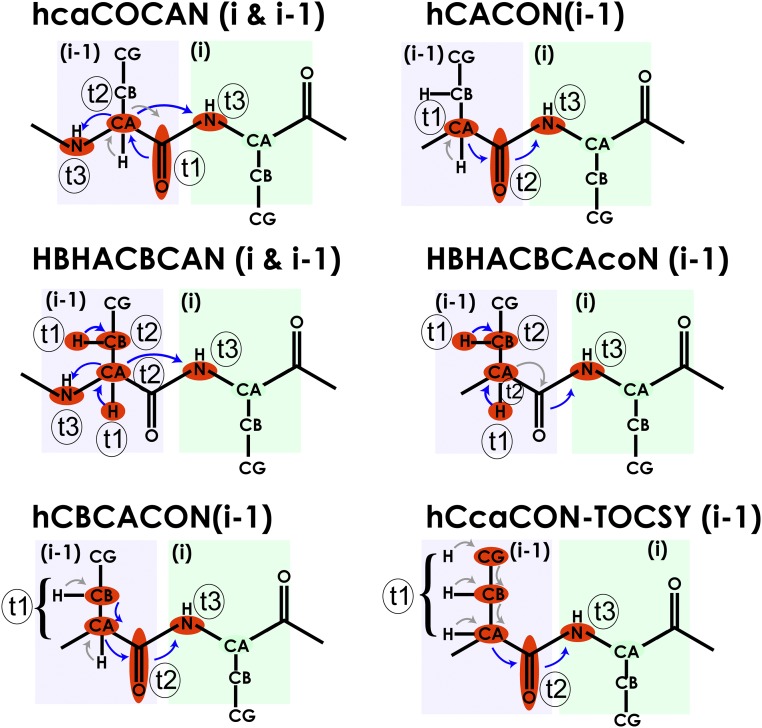
Six separate 3D experiments are reported here that encode the highly resolved 15N chemical shift in the direct dimension. All experiments start from either 1Hα or 1H-aliphatic spins to maximize sensitivity by starting with a sensitive hydrogen nucleus and to minimize the recovery delay after acquisition (24). The experiments are designed to be performed in a D2O solvent to take advantage of the slower relaxation of 15ND compared with that of proton-decoupled 15NH at conventional magnetic fields (25, 27) (SI Appendix, Fig. S1). All of the experiments are designed in an out-and-stay fashion where frequency encoding is done during magnetization transfer using constant-time evolution. The general magnetization flow in these experiments is shown in Fig. 1. Here, in the pulse sequence names, the lowercase letters denote magnetization transfer without frequency encoding and the uppercase letters represent those transfers with frequency encoding. The hcaCOCAN experiment provides the intraresidue and sequential connectivities of 13C′i−1/13Cαi−1 pairs with 15Ni−1 and 15Ni (Fig. 1 and SI Appendix, Fig. S2). The hCACON can be used to disambiguate the 15Ni−1 and 15Ni resonances by isolating the 13Cαi−1 and 15Ni correlations (Fig. 1 and SI Appendix, Fig. S3). The hcaCOCAN and hCACON experiments provide information analogous to the 1H-detected HNCA and HNcoCA, respectively. Assignment ambiguities and degeneracies can be further resolved by incorporating 13Cβ chemical shifts. We designed experiments with three different variations to exploit the dispersion in the 13Cβ chemical shift. First, we designed the hCBCACON experiment (Fig. 1 and SI Appendix, Fig. S4), which provides connectivity between the 13Cαi−1/13Cβi−1 pair and their corresponding 15Ni resonance, dispersed by the 13C′i−1 frequency in the third dimension. This experiment shares two common dimensions (15N and 13C′) with the hcaCOCAN and hCACON experiments and can be used to break degeneracy in the 13Cα-encoded experiments by providing amino acid-specific 13Cβ chemical shift. In addition, we developed the HBHACBCAN experiment (Fig. 1 and SI Appendix, Fig. S5), which provides both sequential and intraresidue connectivities between 1Hαi−113Cαi−1/1Hβi−113Cβi−1 pairs and the corresponding 15Ni−1 and 15Ni. To unambiguously assign the sequential resonances, we use the HBHACBCAcoN experiment (Fig. 1 and SI Appendix, Fig. S6), which provides only the sequential connectivities between 1Hαi−113Cαi−1/1Hβi−113Cβi−1 pairs and 15Ni resonances. These experiments encode the chemical shifts of aliphatic protons in a semiconstant time (SCT) fashion (28) without significant loss of sensitivity. The sixth experiment, the hCcaCON–total correlation spectroscopy (TOCSY), provides amino acid-specific information by correlating side-chain carbon (13Caliphi−1) chemical shifts to the 15Ni resonances, with dispersion by the 13C′i−1 frequency in the third dimension (Fig. 1 and SI Appendix, Fig. S7).
Fig. 1.
Magnetization flow in the panel of 15N-detected experiments. Atoms that are frequency encoded in each experiment are colored in red. t1 and t2 denote the atoms that are frequency encoded in the two indirect dimensions, and t3 is 15N nuclei that is encoded in the direct dimension. Blue arrows indicate frequency labeling during magnetization transfer, and gray arrows indicate magnetization transfer without frequency encoding. Relevant atoms of a given residue i and those of the preceding residue i − 1 are highlighted in light green and light purple boxes, respectively.
The hCACON, hCBCACON, and hCcaCON-TOCSY experiments follow similar magnetization pathways. Initial proton magnetization is transferred to the aliphatic carbons by a simple INEPT step (29) followed by the encoding of the 13Cα/13C-aliphatic carbon frequencies in a constant-time (CT) fashion (30, 31). The CT period eliminates 1JCC homonuclear as well as 1JCH and 1JCN heteronuclear couplings. The 13Cα/13Cβ-aliphatic evolution period is also concatenated with coherence transfer to 13C′. The 13C′ coherence is encoded in a CT manner while being transferred to the 15Ni spins using an INEPT sequence. The antiphase C′zNx coherence is refocused and detected as 15N in-phase coherence. An adiabatic WURST pulse (32) with MLEV-16 supercycle (33) is used to achieve broadband decoupling of 13C′ and 13Cα spins, and 2H is decoupled using a WALTZ-16 composite pulse decoupling scheme (34). The sequential hcaCOCAN experiment uses a slightly modified strategy. The initial 1Hα-to-13Cα INEPT sequence is followed by a second INEPT transfer to 13C′ spins. These two transfer steps are used primarily to gain sensitivity, and there is no frequency encoding during these transfer steps. The antiphase CαzC′x coherence evolved in the transfer step is not refocused until used in the 13C′ to 13Cα back-transfer step. The 13C′ frequency is then encoded in a real-time fashion, followed by a 13C′-to-13Cα INEPT back-transfer sequence. The 13Cα chemical shift is encoded as the second indirect dimension with a CT block while the 13Cα coherence is transferred to 15Ni−1/15Ni. Finally, the antiphase CαzNx coherence is refocused and detected as 15N in-phase coherence with simultaneous 13C and 2H decoupling.
In the HBHACBCAN and HBHACBCAcoN experiments, 1Hαi−1/1Hβi−1 chemical shifts are encoded in a SCT fashion while the coherence is INEPT transferred from the aliphatic protons to the aliphatic carbons to minimize sensitivity loss due to relaxation. The 13Cα/13Cβ-aliphatic carbon frequencies are then encoded in a CT fashion. In the HBHACBCAN experiment, the coherences are transferred to both 15Ni−1/15Ni providing sequential and intraresidue connectivity, whereas the HBHACBCAcoN experiment transfers the magnetization to C′, which results in connectivity only to the 15Ni resonance. 15N is detected after refocusing the antiphase CαzNx or C′zNx coherences to in-phase with simultaneous 13C and 2H decoupling as described above. Combining the HBHACBCAN and the HBHACBCAcoN enables the unambiguous assignment of 13Cα/13Cβ with their backbone 15N resonances. Representative 2D planes from each of these 3D experiments are shown in SI Appendix along with a table describing acquisition parameters (SI Appendix, Figs. S8–S14 and Table S1).
Complete Backbone Assignment of NFAT Regulatory Domain.
To demonstrate the utility of our 15N-detection experiments, we assigned the disordered N-terminal regulatory domain of the human transcription factor NFAT (NFAT131–294). NFAT is ∼1,000 aa in length, and only the DNA binding domain (residues 392–678 in human NFAT) is structured (SI Appendix, Fig. S15A). The N-terminal regulatory domain of NFAT (NFAT131–294), which harbors NLS (residues 251–253), is predicted to be intrinsically disordered (SI Appendix, Fig. S15B) and displays narrow 1HN dispersion (SI Appendix, Fig. S16). This regulatory region of NFAT harbors 18 phosphorylation sites and is targeted by five different kinases (35). In its phosphorylated form, NFAT resides inactive in the cytoplasm. Immunoreceptor and receptor tyrosine kinase (RTK) activation results in the subsequent activation of calcineurin, which dephosphorylates NFAT and induces its translocation to the nucleus, where it initiates transcription of key immune response factors (e.g., IL2, TNFα, INF-γ) (36–38). NFAT131–294 contains 28 prolines (∼17%), as part of the SP-rich regions, which present a major challenge when assigning residues by conventional 1H-detected methods (SI Appendix, Fig. S15C), thus slowing determination of the mechanism of NFAT cytoplasmic sequestration.
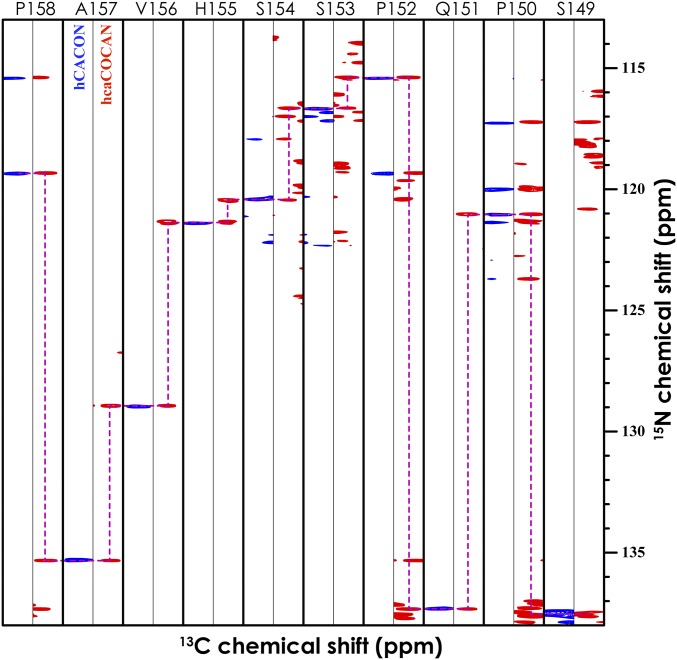
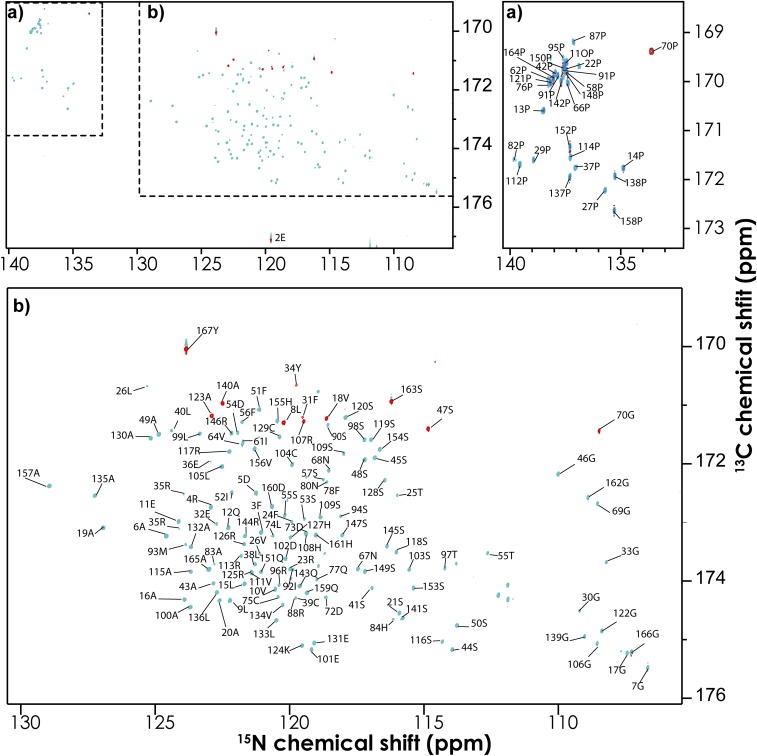
The suite of six 15N-detected experiments were applied to NFAT131–294. Nonuniform sampling (NUS) was used to efficiently acquire high-resolution spectra, with 12% of the Nyquist grid sampled in the two indirect dimensions (39). A striking feature of all of these experiments is the presence of proline 15N resonances at around 138 ppm. Well-separated from all other amino acid types, these “proline islands” can clearly be seen in the overlay of the hCACON and hcaCOCAN spectra (SI Appendix, Fig. S8). Moreover, the hCACON and hcaCOCAN experiments display linewidths that are extremely narrow in the 15N dimension, allowing convenient assignment from 2D planes (CAN) alone. These proline islands are created from their unique 15N frequency coupled with their upfield 13Cα frequency and can facilitate assignment through stretches of amino acids interspaced with prolines (Fig. 2). In this case, we were able to obtain complete 15N, 13Cα, and 13C′ resonance assignments from the 3D hCACON and hcaCOCAN experiments alone. In situations when chemical shift ambiguity and degeneracy stymie the assignment process, the information from the above two experiments can be combined with 13Cβ or 13C-aliphatic chemical shifts obtained from the hCBCACON, HBHACBCAN, HBHACBCAcoN, and hCcaCON-TOCSY experiments. The established assignments of NFAT are best represented in the CON plane of the hCACON experiment (Fig. 3), where there is one resonance for every amino acid in the primary sequence, making it similar to a 1H-15N–heteronuclear single-quantum coherence (HSQC). It should be noted, however, that the CON planes also include proline resonances, which would be absent in a traditional 1H-15N–HSQC (Fig. 3).
Fig. 2.
An example of sequential connectivity through proline residues using 15N-detection experiments for NFAT131–294. Strip plot showing resonances from hCACON (blue) and hcaCOCAN (red) spectra. The experimental time was 3.5 d per experiment on a ∼1 mM sample. NUS was used to collect 12% of the Nyquist grid in the two indirect dimensions (see SI Appendix, Table S1 for detailed acquisition parameters).
Fig. 3.
A 2D-CON plane of the 3D hCOCAN experiment showing the assignments for the regulatory region of NFAT131–294. Residues preceding glycines have negative intensity (red) due to the constant-time nature of the 13Cα encoding. Insets A and B are expanded to display the resonance assignments.
Transfer of Assignments to 1H-Detected Experiments.
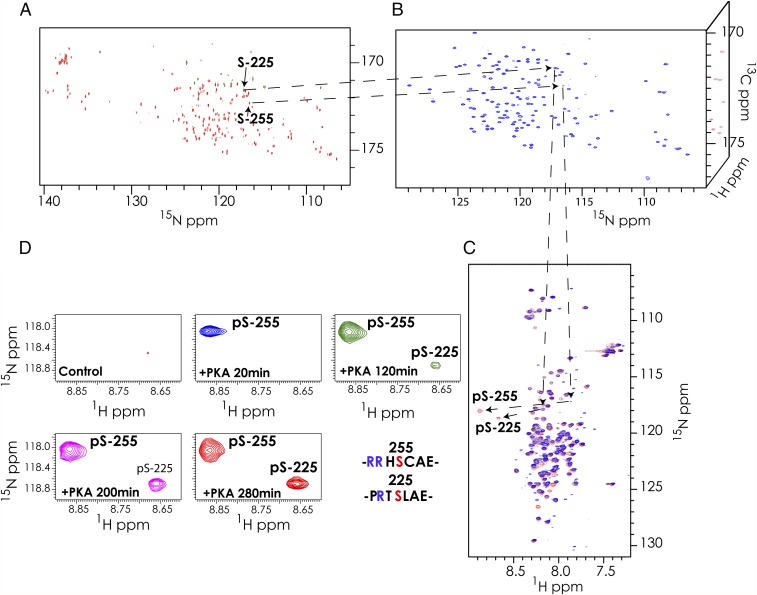
The resonance assignments obtained using 15N detection can be subsequently transferred to 1H-detected experiments such as a 1H-15N–HSQC. This is accomplished by transferring the 15N and 13C′ assignments from a 15N-detected hCACON experiment to the CON plane of a 1H-detected HNCO experiment and then subsequently correlating to the amide proton resonances. Users must account for the isotope shift of experiments conducted in D2O vs. H2O. This shift is typically 0.08 and 0.75 ppm in the 13C′ and 15N dimensions, respectively, and is not sensitive to amino acid type (40, 41). A comparison of the CON planes from 1H- and 15N-detected experiments for NFAT131–294 is shown in Fig. 4. The majority of peaks overlap after correction of the deuterium isotope shift, but a small fraction of the peaks shifted and were assigned by visual inspection. The difference in peak position between the 15N-detected and 1H-detected experiments could be due to partial hydrogen bonding and requires further investigation.
Fig. 4.
Transfer of assignments from 15N-detected to 1H-detected experiments. (A) A 2D-CON plane from the 15N-detected hCACON experiment, and (B) a 2D-CON plane from the 1H-detected 3D-HNCO experiment of the regulatory region of NFAT131–294. Assignments of serine-225 and serine-255 resonances obtained in the 15N-detected experiments were transferred to the CON plane of the 1H-detected HNCO by matching the nitrogen and carbonyl frequencies common to both experiments, thus allowing assignment of the amide 1H resonances. Once the 1H-15N resonances are assigned, conventional 1H-15N–HSQC experiments can be used to follow interactions and modifications such as phosphorylation. (C) NFAT is phosphorylated in vitro by the kinase PKA. An overlay of a 1H-detected 15N-HSQC spectrum of the control sample (blue) and the phosphorylated sample (red) is shown. The two phosphorylated residues are highlighted. (D) Time course of the phosphorylation events on NFAT by the kinase PKA. The region of the 1H-15N–HSQC spectrum harboring the phosphoserine resonances is highlighted. Five panels corresponding to different time periods following initiation of phosphorylation in addition to the control spectrum are shown. Each spectrum was recorded for 20 min, and the data in each spectrum represent the average of events in each 20-min interval. The amino acid sequences of the seven-residue region centered on phosphoserine residues 225 and 255 are shown.
2D-CON Correlation Provides a Better Spectral “Fingerprint” for IDPs.
The 2D-CON correlation provides a more representative fingerprint for IDPs compared with the 1H-15N–HSQC. The signals in the 2D-CON are better dispersed than those in the 1H-15N–HSQC for IDPs (18, 42). The CON correlations are less sensitive to exchange broadening even at higher pH conditions, and the proline residues are visible. In addition, there are no problems originating from imperfect water suppression, something occasionally encountered in 1H-detected 1H-15N–HSQC experiments. The CON plane can be recorded either using 15N-detected or 13C-detected experiments [hcaCON for 15N-detected and hcacoNCO[in-phase anti-phase (IPAP)] for 13C-detected experiments, respectively] (43). It should also be noted that having a proline residue right after a phosphorylation site is a rather common feature in IDPs. The phosphorylation of serine–proline/threonine–proline (SP/TP) sequences is often associated with the regulation of functional modules in IDPs, including the control of subcellular localization through activation or inhibition of NLS or nuclear export signal (NES) motifs. The effect of phosphorylation on neighboring proline residues cannot be defined in 1H-detected 1H-15N–HSQC experiments; however, the effect would be evident in a 2D-CON plane. It should be noted that resonances corresponding to the side chains of asparagine and glutamine would also appear in the 15N-detected CON spectrum at around 35 ppm. These can be avoided by using a selective Cα pulse that does not excite these side-chain resonances (Cγ of glutamine and Cβ of asparagine).
Sensitivity Comparison of 15N- and 13C-Detected 2D-CON.
A one-to-one comparison of sensitivity between the 15N- and 13C-detected methods is not straightforward due to the differences in inherent probe design (13C-optimized vs. 15N-optimized TXO probes) and differences in the style of frequency encoding (CT/SCT vs. real time evolution) in the pulse sequence design. Theoretical estimates indicate that the 15N-detected experiment has an advantage in sensitivity (in time-equivalent experiments) compared with its 13C-detected counterpart SI Appendix, Comparison of the Signal Height in15N- vs. 13C-Detected CON Experiments). To directly compare the sensitivities of the 15N- vs. 13C-detected CON experiments, we modified the 15N-detected CON experiment to encode the C′ chemical shift in a semiconstant time fashion, which allowed the acquisition of 13C and 15N experiments with the same resolution in both the direct and indirect dimensions.
To compare the relative sensitivities of the 15N- and 13C-detected CON experiments, we recorded time-equivalent 15N-detected hcaCON and 13C-detected hcacoNCO-IPAP experiments (SI Appendix, Figs. S13 and S14 and Table S2). The spectral width was set to 40 ppm (∼3,422 Hz) in the 15N dimension to include the proline resonances and 10 ppm (∼2,012 Hz) in the 13C′ dimension (SI Appendix, Table S2). These spectral widths are typical for IDPs. The nucleus with the larger spectral width and slower relaxation (15N) requires a larger number of points to be recorded, giving the 15N-detected experiment an immediate advantage over the 13C-detected experiment in which each point in the 15N dimension needs to be recorded as a separate free induction decay (FID). In contrast, in the 15N-detection experiment, high resolution in the 15N dimension can be achieved with a negligible increase in experimental time. We observe that overall the peak heights are larger in the 15N-detected experiment than in the 13C-detected experiment when the noise levels of the spectra are set to be equal. We also observe broader linewidths in the 13C′ dimension in the 13C-detected CON plane, despite the 13C′ evolution times being equal in both experiments. One probable reason for this difference is the use of an IPAP scheme in the 13C-detected experiment. This requires that the in-phase and anti-phase signals be recorded as two different spectra, in an interleaved fashion. The final spectrum is obtained by shifting these two spectra in frequency space by a value equivalent to the scalar coupling between C′-Ca (∼55 Hz) and then adding them together. At this high resolution, even a small discrepancy between the chosen value of the scalar coupling and the actual scalar coupling will lead to broadening. Another advantage of 15N-detected experiment is that it does not require an IPAP scheme, as the 13C-detected experiment does, to remove scalar couplings.
Phosphorylation of NFAT by PKA and Subsequent Binding by 14-3-3.
PKA phosphorylation of NFAT promotes its association with the 14-3-3 chaperone protein, which results in the inhibition of NFAT transcriptional activity (26). The proximity of the 14-3-3 binding site to the NLS of NFAT has led to speculation that 14-3-3 binding could block importin binding to the NLS, thus preventing translocation of NFAT to the nucleus, and inhibiting its activity (26). However, mechanistic details of PKA-dependent NFAT regulation have yet to be elucidated. To understand the importance of phosphorylation by PKA, we performed an in vitro phosphorylation of NFAT by PKA. To follow the effect of phosphorylation on the NFAT regulatory domain, backbone assignments were transferred from the CON plane of a 15N-detected experiment to a conventional 1H-15N–HSQC.
The kinase reaction was performed in an NMR tube by adding unlabeled PKA to 50 μM NFAT. Phosphorylation of NFAT was followed as a function of time by 1H-detected 1H-15N–HSQC experiments. NUS was employed to collect each 1H-15N-HSQC at 20-min intervals. PKA phosphorylation resulted in downfield shifts of the 1H resonances of two serine residues that are assigned to S-225 and S-255. We could readily assign phosphoserine resonances by following the disappearance of the parent serine or nearby residues and the concomitant appearance of a new signal representing the phosphorylated residue (Fig. 4 and SI Appendix, Fig. S17). The phosphoserine assignments were later confirmed by 2D (CAN) planes of the 15N-detected hCACON and hcaCOCAN experiments acquired on the phosphorylated sample (SI Appendix, Fig. S18). It is interesting to note that the rates of phosphorylation of the two serine residues vary significantly. Under the current experimental conditions, S-255 is phosphorylated first and completely, whereas phosphorylation of S-225 is incomplete with a subpopulation of S-225 remaining unphosphorylated (Fig. 4D and SI Appendix, Fig. S17). An overlay of the 1H-15N–HSQC spectra of the control and phosphorylated samples is shown in Fig. 4C, and the phosphoserine resonances are highlighted. Interestingly, some of the unphosphorylated serine residues have weaker intensities in the 1H-detected 1H-15N–HSQC spectrum and the corresponding 1H-detected 3D experiments, as had been reported previously in IDPs (44, 45). However, in the 15N-detected experiments, the intensities of all of the serine residues are comparable with those of the other amino acids.
PKA is a basophilic kinase and has a strong preference for basic residues (R/K) two (i − 2) and three (i − 3) residues before (N-terminal) the phosphorylation site i (46). Although PKA is a serine/threonine kinase, it strongly prefers serine for phosphorylation (47). Therefore, PKA will preferentially target sites with the consensus (R/K)XS or (R/K)XXS for modification. In fact, many of the key substrates of PKA have two basic residues and conform to the consensus (R/K)(R/K)XS (47). A proline at position i + 1 is largely incompatible with PKA modification, which distinguishes the modification targets of PKA from that of proline-directed kinases (48). It has been postulated that GSK3, a kinase that phosphorylates NFAT after transcriptional activation and promotes NFAT nuclear export (49), requires prior phosphorylation by a priming kinase such as PKA (50).
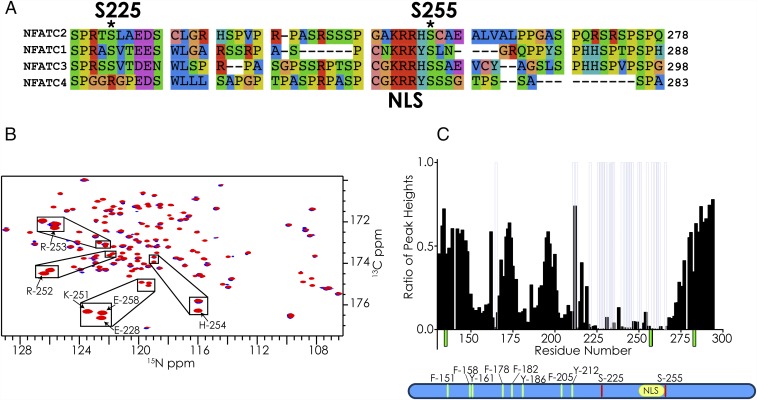
The regulatory region of NFAT (NFAT131–294) used in this study harbors 10 possible phosphorylation sites for a basophilic kinase, namely, T152, S217, S225, S236, S243, S246, S247, S255, S274, and S276. However, only S255 and S225 are actually modified by PKA. S255 is a strong PKA site with two basic residues (Arg) at the i − 2 and i − 3 positions and dominant phosphorylation kinetics in the in vitro PKA phosphorylation assay. Phosphorylation of this site by PKA has been previously observed in NFAT2 and has been shown to control nuclear import (49, 50). S225, which harbors only one basic residue (Arg) at position i − 2, is phosphorylated by PKA after S255 (Fig. 4D). This order of phosphorylation is expected as S255 conforms more closely to the optimal PKA phosphorylation motif. S255 and the arginines/lysines at positions i − 2 and i − 3 are conserved in all four isoforms of NFAT (Fig. 5A). However, S225 and the arginine at position i − 2 are conserved in isoforms 1, 2, and 3, but not in 4. The biological significance of this difference is not known.
Fig. 5.
(A) Sequence alignment of the region harboring the NLS and PKA phosphorylation sites for all four isoforms of NFAT. (B) Overlay of the 2D-CON planes (from 1H-detected 3D-HNCO experiments) of PKA-phosphorylated NFAT alone (red) and PKA-phosphorylated NFAT with the addition of unlabeled 14-3-3 in the molar ratio 1:1 (blue). Resonances corresponding to the residues of the NLS are highlighted. (C) The ratios of peak heights of PKA-phosphorylated NFAT with and without the addition of 14-3-3. Transparent gray bars indicate the residues whose resonances are broadened to below five times the noise level of the spectrum. Green bars represent residues that were not assigned in the PKA-phosphorylated sample. The PKA phosphorylation site (red), NLS (yellow), and the aromatic amino acids (green) are shown in the schematic below.
Five out of the eight remaining unmodified sites in NFAT (S217, S236, S247, S274, and S276) have a proline at the i + 1 position after the serine residue, which is disfavored by PKA. These serine sites would be preferred by DYRK or MAP kinases, additional kinases that are known to phosphorylate NFAT. T152 is a suboptimal PKA substrate due to the presence of threonine at the phosphorylation site (i) instead of serine. Residues S243 and S246 have arginine residues at position i − 2; but they are not phosphorylated by PKA. This may be due to the presence of prolines at positions other than the i + 1 position, potentially making these serines unsuitable for PKA modification.
PKA Regulated NFAT–14-3-3 Interaction and NLS Masking.
14-3-3 is a dimeric phosphodependent chaperone that regulates an array of diverse pathways (51, 52). 14-3-3 binding is a common regulatory mechanism to modulate accessibility to functional modules in disordered regions (53), allowing 14-3-3 to act as a regulatory switch that decodes the cell state-dependent phosphorylation pattern generated by active kinases.
There are numerous experimentally characterized examples of 14-3-3 regulating key cellular proteins including cystic fibrosis transmembrane conductance regulator (CFTR) (54), Bcl2-associated agonist of cell death (BAD) (55), and RAF proto-oncogene serine/threonine-protein kinase (RAF1) (56). Often, 14-3-3 acts as a switch to control the localization of a protein by modulating access to subcellular localization signals. Examples of this include 14-3-3 binding to Forkhead box protein O3 (FOXO3) (57), Yes-associated protein (YAP) (58), carbohydrate-response element-binding protein (ChREBP) (59), and protein Mdm4 (60). The classical consensus for 14-3-3 binding is RXXSpXP, which overlaps with the phosphorylation motif consensus of basophilic kinases. However, there is no simple rule that can describe all instances of 14-3-3 engagement (61). Furthermore, 14-3-3 targets can harbor a single or multiple 14-3-3 binding sites (59, 61). As 14-3-3 is a dimer, containing two antiparallel binding pockets (61), multivalent interactions are stronger due to avidity effects (62). Often the 14-3-3 binding sites of a protein are tuned for optimal binding strength corresponding to the desired biological function. This overall binding is mediated by strong and weak sites, based on consensus match, and often associates with a correspondingly strong or weak binding affinity (63).
When the τ isoform of 14-3-3 was added to PKA-phosphorylated NFAT, we observe broadening of resonances from residues 220–300, suggesting that 14-3-3 engages both phosphorylation sites (Fig. 5 and SI Appendix, Figs. S19 and S20). Since we were concentration limited in the amount of phosphorylated NFAT (0.2 mM), where the kinase PKA was the limiting factor, we recorded a 3D 1H-detected HNCO to extract the 2D CON plane. The assignments for 2D CON plane (derived from 1H-detected HNCO) were transferred from the 15N-detected experiments as described above. These results validate the hypothesis that the activity of dimeric 14-3-3 can result from the binding of two sites on one target (64). Furthermore, we see moderate broadening of resonances near aromatic residues in NFAT, suggesting that these residues have additional interaction with 14-3-3 once the phosphorylated consensus sequence is engaged. Of note, the region that broadened upon 14-3-3 binding includes the NLS (251–253) (Fig. 5 B and C), implying that the NLS of NFAT is masked by phosphorylation-dependent binding of 14-3-3. In contrast, when 14-3-3 was added to unphosphorylated NFAT, only minor changes were observed in the resonances corresponding to the NLS, indicating that engagement of NFAT at this site by 14-3-3 is primarily driven by phosphorylation (SI Appendix, Figs. S19A and S20). It should be noted that the NMR results do indicate that 14-3-3 can bind to unphosphorylated NFAT at the concentrations and under the conditions used here. However, the primary binding interface of 14-3-3 to unphosphorylated NFAT is the region corresponding to residues 184–204, which harbor three SP repeats and not the NLS (SI Appendix, Fig. S20). It has been previously shown that 14-3-3 can bind to proteins in a phosphorylation-independent manner (65–67) and that its specificity increases upon phosphorylation of its substrates (26). These results further support the model that NLS masking by 14-3-3 blocks importin binding, thus preventing the translocation of NFAT to the nucleus and the initiation of NFAT-dependent transcription.
Discussion and Conclusions
IDPs display larger chemical shift dispersion in the 13C and 15N dimensions than in the 1H dimension (68, 69). The advantage of obtaining higher resolution outweighs the intrinsic low sensitivity of 13C- and 15N-detected experiments. Here, we present a suite of 3D 15N-detected experiments that leverage the narrow linewidths and wide dispersion of 15N signals in IDPs. These experiments start with aliphatic 1H magnetization and end in 15N detection with the indirect dimension encoded in a constant-time fashion during magnetization transfer. Starting the experiment with 1H magnetization has two distinct advantages: (i) higher sensitivity and (ii) shorter recycling times (compared with an experiment that starts with 13C or 15N magnetization). In the 15N-detected experiments described here, there are no residual couplings in the 13C or 15N dimensions, which provides narrow lines in the 3D spectra. In traditional “out-and-back”–style HNCA or HNCACB experiments, the 13Cα/13Cβ-aliphatic carbon resonances are limited to an approximate digital resolution of 38 Hz because of the 1J13C-13C couplings. Here, the 13Cα/13Cβ coherences are evolved in a CT fashion removing the couplings. The amide protons in IDPs have very narrow dispersion, but in the HBHACBCAN and HBHACBCAcoN experiments we encode both 1Hβ and 1Hα resonances, providing better resolving power. Since the experiments are done using 100% D2O as the solvent, we minimize broadening due to chemical exchange between 15NH and 15ND species. Furthermore, water suppression elements are not required in 15N-detection experiments, and this allows the encoding of all 1Hα resonances. This is in contrast to 1H-detected experiments performed in H2O solvent, where the 1Hα resonances near to and/or overlapping with the water frequency are affected by the various water suppression schemes needed in these experiments.
An array of 13C-detected experiments (17, 18, 23, 42) have been developed especially for IDPs as an alternative to traditional 1H-detected methods. The 15N-detected experiments described here provide correlations and connectivities between the various backbone nuclei similar to those measured by the 13C-detected experiments. Despite the reduced intrinsic sensitivity of 15N compared with 13C due to its lower gyromagnetic ratio [(10/4)1.5 = 3.9], the slower transverse relaxation rate of 15N results in narrower line widths with increased signal height (25). For example, assuming a local rotational correlation time of 8 ns at 500 MHz in D2O, the 15N-detected hCACON signal height is calculated to be slightly better than that of its 13C-detected hCAcoNCO counterpart (i.e., identical 13Cαi-13C′i-15Ni+1 correlations; SI Appendix, Comparison of the Signal Height in15N- vs. 13C-Detected Experiments). This is primarily due to the 3.8 times slower transverse relaxation rate of 15ND (3.7 s−1) compared with that of 13C′N (14.2 s−1). The relative sensitivity gain of 15N detection over 13C-detected experiments should increase with magnet field strength due to the stronger chemical shift anisotropy of 13C′N spins (SI Appendix, Fig. S1). In addition, the increase in rotational correlation time due to transiently forming secondary structures or adoption of stable conformations upon binding would be advantageous to the 15N-detected experiments as the transverse relaxation rate of 15ND is less sensitive to molecular weight. Direct 13C-detection experiments have the additional disadvantage of one-bond 13C-13C scalar couplings that increase spectral crowding and diminish signal intensity. Although these problems can be managed by a judicious choice of detected carbon type (e.g., 13C′ vs. 13Cα), isotope labeling scheme, and experimental design (e.g., IPAP, S3E), these solutions also increase the likelihood of imperfect signal recovery, especially for 13Cα (as reviewed in ref. 27). Here, we report broader resonances for 13C′ in the 13C-detected experiments compared with those in the 15N-detected hcaCON experiment.
Adapting other optimization methods in pulse sequence design will further enhance the sensitivity of these 15N-detection experiments. For example, CT periods can be replaced with 13Cα selective pulses to minimize the relaxation losses during 13Cα encoding. This would improve the sensitivity for most amino acids, except those whose 13Cα chemical shifts are close to 13Cβ resonances, such as glycine, serine, and threonine. The use of the accelerated 1H-longitudinal relaxation of 1Hα spins (1H-flip) would enable the hCACON and hcaCOCAN experiments to be executed with shorter recycling delays (70). The 15N-detected 3D experiments described here can be easily converted to 4D experiments by encoding the 1Hα or 1H-aliphatic chemical shift during the initial INEPT transfer in a SCT fashion. It should be noted that most of the 3D experiments presented here can also be easily converted to high-resolution 2D planes with minimal to no loss in sensitivity, as the frequency encoding in the indirect dimension is incorporated into the magnetization transfer step.
The broadening of the 15N signals that are detected in the direct dimension by the weak two- and three-bond scalar couplings (1–3 Hz) to 1Hα and 1Hβ protons (71) can be removed by 1H decoupling during acquisition. Narrowband 1H decoupling can be employed, targeting the 1Hα and 1Hβ resonances, and thereby reducing the required decoupling power. Although modern cryogenic probes are capable of dissipating heat from simultaneous 13C, 1H, and 2H decoupling, careful attention must be paid to the decoupling power levels and acquisition times. The experiments here were recorded at sample concentrations of 0.5–1 mM on a Bruker TXO cryoprobe where the 13C and 15N coils are positioned closest to the sample. The sensitivity of the 15N-detected experiments will further improve with improvements in coil design in the TXO probes optimized for 15N nuclei. Although the 15N-detected experiments are not as sensitive as the 1H-detected experiments, which can be performed at lower sample concentrations, the advantages in resolution and ability to observe prolines in the 15N-detected experiments offer distinct advantages for characterizing IDPs. The 15N-detected experiments described here can be readily combined with data from 1H- and 13C-detected experiments, especially those that were developed for characterizing IDPs.
Although the 15N-detected experiments described here were applied to the intrinsically disordered region of NFAT131–294, the application is not limited to IDPs. The relaxation rates of 15N in deuterated solvent is comparable to that of the 15N transverse relaxation optimized spectroscopy (TROSY) at low field strengths. Thus, these methods can be applied to structured globular proteins. The recent application of TROSY selection to 15N detection would extend these experiments to larger molecular weights and higher magnetic fields (71, 72). Pulse sequence designs, like the ones presented here, that enable direct 15N detection with TROSY selection (where the TROSY component of 15NH resonance is detected) would facilitate studies of high–molecular-weight proteins, and systems marred by spectral overlap and/or incomplete hydrogen backexchange.
Our newly developed 15N-detected 3D experiments have shed light on the mechanism of the interaction of 14-3-3 with NFAT and its regulation of NFAT through NLS masking. This example illustrates how 15N- and 13C-detected multidimensional experiments open avenues of structural investigation into IDPs. Greater understanding of disordered regions, at atomic resolution, will allow us to dissect the functional and mechanistic details of how IDPs orchestrate cellular functions.
Materials and Methods
Expression and Purification of NFAT Regulatory Domain.
The regulatory domain of human NFATC2 (residues 131–294), NFAT131–294, was expressed as a fusion protein with an N-terminal His-GB1 solubility tag in the Escherichia coli strain BL21 (DE3). Cells were grown at 37 °C to an OD600 of 0.8. Then the temperature was dropped to 20 °C, and expression was induced by 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cells were grown for an additional 15 h at 20 °C before harvesting. For [U-15N13C]-NFAT131–294, the bacterial culture was grown using M9 media containing 6 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 mM MgSO4, and 0.1 mM CaCl2 in H2O supplemented with 2 g/L 13C-glucose and 1 g/L of 15NH4Cl. To obtain [U-15N]-NFAT131–294, 3 g/L of 12C-glucose was used in an otherwise-identical M9 media.
The harvested cells were resuspended in 40 mL of 50 mM Tris⋅HCl (pH 8.0), 350 mM NaCl, 10 mM imidazole, and 5 mM β-mercaptoethanol (β-ME). The suspended cells were then disrupted by sonication, and the insoluble fraction was removed by centrifugation for 40 min at 32,914 × g. The protein was initially purified by gravity-flow affinity chromatography using 5 mL (10 mL of a 50% slurry) of Ni-NTA resin (Qiagen). After washing the resin with 40 mL of 50 mM Tris⋅HCl (pH 8.0), 350 mM NaCl, 40 mM imidazole, and 5 mM β-ME, the protein was eluted in an identical buffer containing 350 mM imidazole. The elution fraction was dialyzed against a buffer containing 30 mM Na2HPO4 (pH 6.7), NaCl (150 mM), and DTT (5 mM), and the His-GB1 solubility tag was removed using TEV protease. The digested NFAT and His-GB1 were separated and further purified using size exclusion chromatography (Superdex 75 10/300 GL; GE Healthcare Life Sciences).
Expression and Purification of 14-3-3.
Human 14-3-3 (τ isoform) was expressed in E. coli as an N-terminally His-tagged protein with a TEV cleavage site using the pNIC28 plasmid (Addgene ID 38931). The τ isoform of the protein 14-3-3 was used in all of the studies described in this manuscript and is referred to as 14-3-3. Cells were grown at 37 °C to an OD600 of 0.8. Then the temperature was dropped to 20 °C, and the expression was induced by 1 mM IPTG. Cells were grown for additional 15 h at 20 °C before harvesting. The harvested cells were resuspended in 40 mL of 50 mM Tris⋅HCl (pH 8.0), 350 mM NaCl, 10 mM imidazole, and 5 mM β-ME. The suspended cells were then disrupted by sonication, and the insoluble fraction was removed by centrifugation for 40 min at 32,914 × g rpm. The protein was initially purified by gravity-flow affinity chromatography using 5 mL (10 mL of a 50% slurry) of Ni-NTA resin (Qiagen). After washing the resin with 40 mL of 50 mM Tris⋅HCl (pH 8.0), 350 mM NaCl, 40 mM imidazole, and 5 mM β-ME, the protein was eluted in an identical buffer containing 350 mM imidazole. The eluted protein was concentrated to a 5-mL volume and further purified using size exclusion chromatography in kinase reaction buffer [50 mM Mes (pH 6.7), 140 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, and 5 mM DTT].
NMR Spectroscopy.
All 3D spectra were collected at 288 K using ∼1 mM protein in 30 mM Na2HPO4 (pH 6.7), 150 mM NaCl, and 5 mM DTT dissolved in 100% D2O. For backbone resonance assignment, 15N-detected NMR spectra (hCACON, hCBCACON, hCcaCON-TOCSY) were acquired on a Bruker Avance I 500 spectrometer equipped with a triple-resonance cryogenic probe (TXO) in which the carbon and nitrogen channels are both on the inner coil and detected with cryogenically cooled preamplifiers. The hcaCOCAN, hCACON (2D planes for 13C-detection comparison), HBHACBCAN, and HBHACBCAcoN experiments, and the 13C-detected hacaCON (2D plane) were recorded on a Bruker Avance III 800 MHz spectrometer with a TXO-style cryogenically cooled probe. The spectral widths for the 13C′, 13Cα, 13Cα/13Cβ, C-aliphatic, and 1Hα/1Hβ were set to 20, 34, 68, 68, and 5 ppm, respectively. The hCcaCON-TOCSY experiment employed the flip-flop spectoscopy (73) mixing sequence with the mixing time set to 18 ms. All 3D spectra were acquired using NUS with 12% sampling of the Nyquist grid using Poisson Gap Sampling (74). The Nyquist grid for the hCACON and hcaCOCAN experiments consisted of 48/64 complex points in the 13Cα/13Cα-13Cβ dimension and 32/64 complex points in the 13C′ dimension. For the hCcaCON-TOCSY experiment, the grid consisted of 64 complex points in the C-aliphatic dimension and 32 complex points in the 13C′ dimension. The NUS spectra were reconstructed and processed with hmsIST (39). Processed spectra were analyzed with CcpNmr-Analysis (version 2.4.1) (75). The average experimental time was 3.5–4 d per 3D experiment (see SI Appendix, Table S1 for detailed acquisition parameters).
In Vitro Phosphorylation and 14-3-3 Binding Experiments.
The 2D NMR experiments were performed on a Varian (DD2 700; Agilent) spectrometer equipped with a cryogenically cooled probe, and the spectrum was recorded at 288 K. The kinase PKA was purchased from Signalchem. The phosphorylation reaction was performed with a sample containing 0.05 mM 15N-labeled NFAT with the addition of 2 μg of PKA in kinase reaction buffer [50 mM Mes (pH 6.7), 140 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, 2 mM ATP, and 5 mM DTT]. First, a control experiment of unphosphorylated NFAT in the same kinase reaction buffer was recorded followed by addition of PKA, and phosphorylation was monitored using 2D 1H-15N–HSQC experiments. In the 1H-detected HNCO experiment recorded for obtaining the CON plane (shown in Fig. 5 and SI Appendix, Fig. S20), the concentration of the PKA-phosphorylated NFAT and the control NFAT sample was held at 0.2 mM and 14-3-3 was added in a 1:1 molar ratio. All spectra were processed using nmrPipe and analyzed with CcpNmr-Analysis (version 2.4.1) (75).
Supplementary Material
Acknowledgments
We thank Kendra E. Leigh for the insightful discussions regarding the work presented here. This work was supported by NIH Grants GM047467 and AI03758 (to G.W.). S.C. acknowledges National Health and Medical Research Council Australia for the C. J. Martin Fellowship. H.A. acknowledges funding from Claudia Adams Barr Program for Innovative Cancer Research. J.J.Z. was supported by NIH Grant K99 GM115814. A.B. thanks Fonds zur Förderung der Wissenschaftlichen Forschung (Project J3872-B21) for support. Maintenance of some of the instruments used for this research was supported by NIH Grant EB002026. Funding was also provided by Japan Science and Technology Agency, Precursory Research for Embryonic Science and Technology (Grant JPMJPR14L5, to K.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717560115/-/DCSupplemental.
References
- 1.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 2.Tompa P. Unstructural biology coming of age. Curr Opin Struct Biol. 2011;21:419–425. doi: 10.1016/j.sbi.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Babu MM, Kriwacki RW, Pappu RV. Structural biology. Versatility from protein disorder. Science. 2012;337:1460–1461. doi: 10.1126/science.1228775. [DOI] [PubMed] [Google Scholar]
- 4.Calabretta S, Richard S. Emerging roles of disordered sequences in RNA-binding proteins. Trends Biochem Sci. 2015;40:662–672. doi: 10.1016/j.tibs.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Flock T, Weatheritt RJ, Latysheva NS, Babu MM. Controlling entropy to tune the functions of intrinsically disordered regions. Curr Opin Struct Biol. 2014;26:62–72. doi: 10.1016/j.sbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 7.Uversky VN, et al. Pathological unfoldomics of uncontrolled chaos: Intrinsically disordered proteins and human diseases. Chem Rev. 2014;114:6844–6879. doi: 10.1021/cr400713r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamiola K, Acar B, Mulder FA. Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J Am Chem Soc. 2010;132:18000–18003. doi: 10.1021/ja105656t. [DOI] [PubMed] [Google Scholar]
- 10.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuxreiter M, Tompa P. Fuzzy complexes: A more stochastic view of protein function. Adv Exp Med Biol. 2012;725:1–14. doi: 10.1007/978-1-4614-0659-4_1. [DOI] [PubMed] [Google Scholar]
- 12.Davis RJ. The mitogen-activated protein kinase signal transduction pathway. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 13.Li H, Rao A, Hogan PG. Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 2011;21:91–103. doi: 10.1016/j.tcb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grathwohl C, Wüthrich K. Nmr studies of the molecular conformations in the linear oligopeptides H-(L-Ala)n-L-Pro-OH. Biopolymers. 1976;15:2043–2057. doi: 10.1002/bip.1976.360151013. [DOI] [PubMed] [Google Scholar]
- 15.Grathwohl C, Wüthrich K. The X-Pro peptide bond as an NMR probe for conformational studies of flexible linear peptides. Biopolymers. 1976;15:2025–2041. doi: 10.1002/bip.1976.360151012. [DOI] [PubMed] [Google Scholar]
- 16.Ivanov D, Stone JR, Maki JL, Collins T, Wagner G. Mammalian SCAN domain dimer is a domain-swapped homolog of the HIV capsid C-terminal domain. Mol Cell. 2005;17:137–143. doi: 10.1016/j.molcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 17.Dziekański P, Grudziąż K, Jarvoll P, Koźmiński W, Zawadzka-Kazimierczuk A. 13C-detected NMR experiments for automatic resonance assignment of IDPs and multiple-fixing SMFT processing. J Biomol NMR. 2015;62:179–190. doi: 10.1007/s10858-015-9932-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bermel W, et al. Speeding up sequence specific assignment of IDPs. J Biomol NMR. 2012;53:293–301. doi: 10.1007/s10858-012-9639-0. [DOI] [PubMed] [Google Scholar]
- 19.Bermel W, et al. High-dimensionality 13C direct-detected NMR experiments for the automatic assignment of intrinsically disordered proteins. J Biomol NMR. 2013;57:353–361. doi: 10.1007/s10858-013-9793-z. [DOI] [PubMed] [Google Scholar]
- 20.Nováček J, et al. 5D 13C-detected experiments for backbone assignment of unstructured proteins with a very low signal dispersion. J Biomol NMR. 2011;50:1–11. doi: 10.1007/s10858-011-9496-2. [DOI] [PubMed] [Google Scholar]
- 21.Bermel W, et al. Protonless NMR experiments for sequence-specific assignment of backbone nuclei in unfolded proteins. J Am Chem Soc. 2006;128:3918–3919. doi: 10.1021/ja0582206. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi K, Gal M, Takahashi H, Shimada I, Wagner G. HNCA-TOCSY-CANH experiments with alternate 13C-12C labeling: A set of 3D experiment with unique supra-sequential information for mainchain resonance assignment. J Biomol NMR. 2011;49:17–26. doi: 10.1007/s10858-010-9456-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshimura Y, Kulminskaya NV, Mulder FA. Easy and unambiguous sequential assignments of intrinsically disordered proteins by correlating the backbone 15N or 13C′ chemical shifts of multiple contiguous residues in highly resolved 3D spectra. J Biomol NMR. 2015;61:109–121. doi: 10.1007/s10858-014-9890-7. [DOI] [PubMed] [Google Scholar]
- 24.Gal M, Edmonds KA, Milbradt AG, Takeuchi K, Wagner G. Speeding up direct 15N detection: hCaN 2D NMR experiment. J Biomol NMR. 2011;51:497–504. doi: 10.1007/s10858-011-9580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeuchi K, Heffron G, Sun ZY, Frueh DP, Wagner G. Nitrogen-detected CAN and CON experiments as alternative experiments for main chain NMR resonance assignments. J Biomol NMR. 2010;47:271–282. doi: 10.1007/s10858-010-9430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chow CW, Davis RJ. Integration of calcium and cyclic AMP signaling pathways by 14-3-3. Mol Cell Biol. 2000;20:702–712. doi: 10.1128/mcb.20.2.702-712.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeuchi K, Gal M, Shimada I, Wagner G. Recent Developments in Biomolecular NMR. Royal Society of Chemistry; London: 2012. Low-[γ] nuclei detection experiments for biomolecular NMR; pp. 25–52. [Google Scholar]
- 28.Grzesiek S, Bax A. Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J Biomol NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 29.Morris GA, Freeman R. Enhancement of nuclear magnetic resonance signals by polarization transfer. J Am Chem Soc. 1979;101:760–762. [Google Scholar]
- 30.Santoro J, King GC. A constant-time 2D overbodenhausen experiment for inverse correlation of isotopically enriched species. J Magn Reson (1969) 1992;97:202–207. [Google Scholar]
- 31.Vuister GW, Bax A. Resolution enhancement and spectral editing of uniformly 13C-enriched proteins by homonuclear broadband 13C decoupling. J Magn Reson (1969) 1992;98:428–435. [Google Scholar]
- 32.Kupce E, Freeman R. Adiabatic pulses for wideband inversion and broadband decoupling. J Magn Reson A. 1995;115:273–276. [Google Scholar]
- 33.Jacobs JWM, Van Os JWM, Veeman WS. Broadband heteronuclear decoupling. J Magn Reson (1969) 1983;51:56–66. [Google Scholar]
- 34.Shaka AJ, Keeler J, Frenkiel T, Freeman R. An improved sequence for broadband decoupling: WALTZ-16. J Magn Reson (1969) 1983;52:335–338. [Google Scholar]
- 35.Okamura H, et al. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell. 2000;6:539–550. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- 36.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 37.Macian F. NFAT proteins: Key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 38.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: Regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 39.Hyberts SG, Milbradt AG, Wagner AB, Arthanari H, Wagner G. Application of iterative soft thresholding for fast reconstruction of NMR data non-uniformly sampled with multidimensional Poisson gap scheduling. J Biomol NMR. 2012;52:315–327. doi: 10.1007/s10858-012-9611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abildgaard J, Hansen PE, Manalo MN, LiWang A. Deuterium isotope effects on 15N backbone chemical shifts in proteins. J Biomol NMR. 2009;44:119–126. doi: 10.1007/s10858-009-9316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henry GD, Weiner JH, Sykes BD. Backbone dynamics of a model membrane protein: Measurement of individual amide hydrogen-exchange rates in detergent-solubilized M13 coat protein using 13C NMR hydrogen/deuterium isotope shifts. Biochemistry. 1987;26:3626–3634. doi: 10.1021/bi00386a056. [DOI] [PubMed] [Google Scholar]
- 42.Sahu D, Bastidas M, Showalter SA. Generating NMR chemical shift assignments of intrinsically disordered proteins using carbon-detected NMR methods. Anal Biochem. 2014;449:17–25. doi: 10.1016/j.ab.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bermel W, Bertini I, Felli IC, Peruzzini R, Pierattelli R. Exclusively heteronuclear NMR experiments to obtain structural and dynamic information on proteins. ChemPhysChem. 2010;11:689–695. doi: 10.1002/cphc.200900772. [DOI] [PubMed] [Google Scholar]
- 44.Iakoucheva LM, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theillet FX, et al. Cell signaling, post-translational protein modifications and NMR spectroscopy. J Biomol NMR. 2012;54:217–236. doi: 10.1007/s10858-012-9674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujii K, et al. Kinase peptide specificity: Improved determination and relevance to protein phosphorylation. Proc Natl Acad Sci USA. 2004;101:13744–13749. doi: 10.1073/pnas.0401881101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shabb JB. Physiological substrates of cAMP-dependent protein kinase. Chem Rev. 2001;101:2381–2411. doi: 10.1021/cr000236l. [DOI] [PubMed] [Google Scholar]
- 48.Zhu G, et al. Exceptional disfavor for proline at the P + 1 position among AGC and CAMK kinases establishes reciprocal specificity between them and the proline-directed kinases. J Biol Chem. 2005;280:10743–10748. doi: 10.1074/jbc.M413159200. [DOI] [PubMed] [Google Scholar]
- 49.Beals CR, Sheridan CM, Turck CW, Gardner P, Crabtree GR. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 50.Sheridan CM, Heist EK, Beals CR, Crabtree GR, Gardner P. Protein kinase A negatively modulates the nuclear accumulation of NF-ATc1 by priming for subsequent phosphorylation by glycogen synthase kinase-3. J Biol Chem. 2002;277:48664–48676. doi: 10.1074/jbc.M207029200. [DOI] [PubMed] [Google Scholar]
- 51.Aitken A. 14-3-3 proteins: A historic overview. Semin Cancer Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 52.Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Roey K, Dinkel H, Weatheritt RJ, Gibson TJ, Davey NE. The switches.ELM resource: A compendium of conditional regulatory interaction interfaces. Sci Signal. 2013;6:rs7. doi: 10.1126/scisignal.2003345. [DOI] [PubMed] [Google Scholar]
- 54.Stevers LM, et al. Characterization and small-molecule stabilization of the multisite tandem binding between 14-3-3 and the R domain of CFTR. Proc Natl Acad Sci USA. 2016;113:E1152–E1161. doi: 10.1073/pnas.1516631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Datta SR, et al. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- 56.Molzan M, et al. Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling. Mol Cell Biol. 2010;30:4698–4711. doi: 10.1128/MCB.01636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brunet A, et al. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J Cell Biol. 2002;156:817–828. doi: 10.1083/jcb.200112059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 59.Sato S, et al. Metabolite regulation of nuclear localization of carbohydrate-response element-binding protein (ChREBP): Role of AMP as an allosteric inhibitor. J Biol Chem. 2016;291:10515–10527. doi: 10.1074/jbc.M115.708982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pereg Y, et al. Differential roles of ATM- and Chk2-mediated phosphorylations of Hdmx in response to DNA damage. Mol Cell Biol. 2006;26:6819–6831. doi: 10.1128/MCB.00562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dinkel H, et al. ELM 2016–Data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res. 2016;44:D294–D300. doi: 10.1093/nar/gkv1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yaffe MB, et al. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 63.Yaffe MB. How do 14-3-3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 2002;513:53–57. doi: 10.1016/s0014-5793(01)03288-4. [DOI] [PubMed] [Google Scholar]
- 64.Shikano S, Coblitz B, Wu M, Li M. 14-3-3 proteins: Regulation of endoplasmic reticulum localization and surface expression of membrane proteins. Trends Cell Biol. 2006;16:370–375. doi: 10.1016/j.tcb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 65.Bonet R, Vakonakis I, Campbell ID. Characterization of 14-3-3-ζ interactions with integrin tails. J Mol Biol. 2013;425:3060–3072. doi: 10.1016/j.jmb.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fuglsang AT, et al. Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J Biol Chem. 1999;274:36774–36780. doi: 10.1074/jbc.274.51.36774. [DOI] [PubMed] [Google Scholar]
- 67.Waterman MJ, Stavridi ES, Waterman JL, Halazonetis TD. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat Genet. 1998;19:175–178. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]
- 68.Csizmok V, Felli IC, Tompa P, Banci L, Bertini I. Structural and dynamic characterization of intrinsically disordered human securin by NMR spectroscopy. J Am Chem Soc. 2008;130:16873–16879. doi: 10.1021/ja805510b. [DOI] [PubMed] [Google Scholar]
- 69.O’Hare B, Benesi AJ, Showalter SA. Incorporating 1H chemical shift determination into 13C-direct detected spectroscopy of intrinsically disordered proteins in solution. J Magn Reson. 2009;200:354–358. doi: 10.1016/j.jmr.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 70.Bermel W, Bertini I, Felli IC, Pierattelli R. Speeding up 13C direct detection biomolecular NMR spectroscopy. J Am Chem Soc. 2009;131:15339–15345. doi: 10.1021/ja9058525. [DOI] [PubMed] [Google Scholar]
- 71.Takeuchi K, Arthanari H, Imai M, Wagner G, Shimada I. Nitrogen-detected TROSY yields comparable sensitivity to proton-detected TROSY for non-deuterated, large proteins under physiological salt conditions. J Biomol NMR. 2016;64:143–151. doi: 10.1007/s10858-016-0015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takeuchi K, Arthanari H, Shimada I, Wagner G. Nitrogen detected TROSY at high field yields high resolution and sensitivity for protein NMR. J Biomol NMR. 2015;63:323–331. doi: 10.1007/s10858-015-9991-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kadkhodaie M, Rivas O, Tan M, Mohebbi A, Shaka AJ. Broadband homonuclear cross polarization using flip-flop spectroscopy. J Magn Reson (1969) 1991;91:437–443. [Google Scholar]
- 74.Hyberts SG, Takeuchi K, Wagner G. Poisson-gap sampling and forward maximum entropy reconstruction for enhancing the resolution and sensitivity of protein NMR data. J Am Chem Soc. 2010;132:2145–2147. doi: 10.1021/ja908004w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vranken WF, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.