Significance
The detection of aneuploidy in clinical samples can be critical for various diagnostic applications in cancer and can also inform cancer genetics. Next-generation sequencing protocols such as whole-genome and exome sequencing are typically used to detect aneuploidy in cancer samples, but amplicon-based protocols achieve high coverage depth at relatively low cost and can be used when only tiny amounts of DNA are available. In this paper, we describe new bioinformatic tools to detect aneuploidy using data generated from amplification with a single primer pair. This approach can be applied to samples containing only a few nanograms of DNA and as little as 1% neoplastic content and has a variety of applications in cancer diagnostics and forensic science.
Keywords: aneuploidy, early cancer detection, liquid biopsy, circulating tumor DNA
Abstract
Aneuploidy is a feature of most cancer cells, and a myriad of approaches have been developed to detect it in clinical samples. We previously described primers that could be used to amplify ∼38,000 unique long interspersed nucleotide elements (LINEs) from throughout the genome. Here we have developed an approach to evaluate the sequencing data obtained from these amplicons. This approach, called Within-Sample AneupLoidy DetectiOn (WALDO), employs supervised machine learning to detect the small changes in multiple chromosome arms that are often present in cancers. We used WALDO to search for chromosome arm gains and losses in 1,677 tumors and in 1,522 liquid biopsies of blood from cancer patients or normal individuals. Aneuploidy was detected in 95% of cancer biopsies and in 22% of liquid biopsies. Using single-nucleotide polymorphisms within the amplified LINEs, WALDO concomitantly assesses allelic imbalances, microsatellite instability, and sample identification. WALDO can be used on samples containing only a few nanograms of DNA and as little as 1% neoplastic content and has a variety of applications in cancer diagnostics and forensic science.
Aneuploidy is defined as an abnormal chromosome number. It was the first genomic abnormality identified in cancers (1, 2), and it has been estimated to be present in >90% of cancers of most histopathologic types (3). Aneuploidy in cancers was first detected by karyotypic studies, later evaluated through microarrays, Sanger sequencing, and most recently, massively parallel sequencing methods (4). Recent sequencing methods include those employing circular binary segmentation, hidden Markov models, expectation maximization, and mean-shift (as reviewed in ref. 5). In addition to their application to cancer genomes, these technologies form the basis for the noninvasive prenatal detection of fetuses with Down’s syndrome and other trisomies (6, 7).
Read-depth–based analytical methods have been widely applied to whole-genome sequencing (WGS) protocols. Under the assumption that reads are uniformly and independently distributed, regions of normal copy number are expected to follow a Poisson or normal distribution (5, 8). Such approaches require correction for biases induced by differing GC nucleotide content across the genome and uneven representation of genomic regions in library preparation (5). Read-depth methods have subsequently been extended to capture-based sequencing protocols, including whole-exome sequencing (WES). Targeted sequencing introduces coverage discontinuities and increased variability due to differences in capture efficiency (5, 9). Their analysis therefore requires modifications of the methods used to evaluate whole genomes such as regression models and principal component analysis (10, 11). Several recent methods for the genome-wide analysis of aneuploidy also leverage randomly distributed off-target reads to improve accuracy (12, 13).
Amplicon-based sequencing protocols can also be used for aneuploidy detection (14). Amplicon-based protocols achieve high coverage depth at relatively low cost, and they are an attractive alternative to WGS, especially when relatively low amounts of template DNA are available. For example, a single primer pair can be used to amplify 38,000 long interspersed nuclear elements (LINEs) by FAST-SeqS (14). The resultant sequencing data can be used to detect trisomies in prenatal diagnostics, even in single cells derived from preimplantation embryos (15). However, aligned reads from amplicon sequencing such as those resulting from FAST-SeqS have properties different from those resulting from WGS and WES. Because these reads are limited to a relatively small number of discrete loci, they are discontinuous. The reads are also not randomly distributed, which makes it difficult to use the statistical models of read-depth coverage designed for WGS and WES. To date, approaches for amplicon-based aneuploidy detection have employed metrics in which the read depth at a locus is compared with an overall mean, and aneuploidy is called when the read depth deviates significantly from the mean (14, 16). In a refinement of this approach, read depth may be converted to a ratio between a test sample and a control at the locus, using either a matched normal or a pool of selected control samples (17).
We describe here Within-Sample AneupLoidy DetectiOn (WALDO), an algorithm for amplicon-based aneuploidy detection. We show that WALDO can be applied to identify chromosome arm gains or losses with improved sensitivity and equivalent specificity compared with previous approaches. Furthermore, we incorporate machine learning to make genome-wide aneuploidy calls, in which samples are classified according to their aneuploidy status. We report WALDO results on thousands of samples, including tissues of 10 different tumor types as well as liquid biopsies of plasma from cancer patients (SI Appendix, Table S1). When two samples are available for comparison, WALDO can be used to assess genetic relatedness or to find somatic mutations within the LINEs. We show that this approach can thereby provide an estimate of somatic mutation load, evaluate carcinogen signatures, and detect microsatellite instability (Fig. 1).
Fig. 1.
Overview of WALDO approach. (A) A single primer pair amplifies ∼38,000 LINEs. (B) A test sample is matched to seven euploid samples with genomic DNA of similar size. (C) The genome is divided into 4,361 intervals, each of 500 kb in size. (D) The reads within these 500-kb genomic intervals in the euploid samples are grouped into 4,361 clusters. All of the 500-kb genomic intervals in the clusters have similar read depths. (E) The reads from each of the 500-kb genomic intervals in the test sample are placed into the predefined clusters. (F) Statistical tests, including an SVM-based algorithm, are used to determine whether the total reads from all of the 500-kb genomic intervals on each chromosome arm are distributed as expected if the sample was euploid. The statistical tests are based on the observed distribution of reads within the clusters of the test sample, not by comparison with the reads in euploid samples. (G) Germline sequence variants at sites of known common polymorphisms within the LINEs provide information about arm-level allelic imbalance that can also be used to assess aneuploidy of individual chromosome arms. These same polymorphisms can be used to determine whether any two samples are derived from the same individual. (H) When there is a matched normal sample from the same individual available, WALDO can detect the number and nature of single base substitutions and insertions and deletions within the LINEs.
Results
Statistical Principles Underlying WALDO.
Unlike most previous approaches for assessing copy-number changes, WALDO does not compare normalized read counts from each chromosome arm in a test sample to the fraction of reads in each chromosome arm in other samples. As pointed out by Straver et al. (18), such comparisons are subject to batch effects and other artifacts associated with variables that are difficult to control. Inspired by the approach used by Straver et al. (18) to evaluate whole-genome sequencing data, we attempted to detect aneuploidy by comparing the read counts of LINEs within 4,361 genomic intervals, each containing 500 kb of sequence. The read counts within the 500-kb genomic intervals within a sample are compared only with the read counts of other genomic intervals within the same sample—hence the “within-sample” designation in WALDO.
In euploid samples, the number of LINE reads within each 500-kb genomic interval should track with the number of reads in certain other genomic regions. Genomic intervals that track together do so because the amplicons within them amplify to similar extents. Here, we call such genomic regions that track together “clusters.” We can identify clusters from sequencing data on euploid samples. In a test sample, we determine whether the number of reads in each genomic interval in each predefined cluster is within the expected bound of the other clusters from that same sample. If the reads within a genomic interval are outside the statistically expected bound, and there are many such outsiders on the same chromosome arm, then that chromosome arm is classified as aneuploid. The statistical basis of this test is described in SI Appendix, SI Materials and Methods. In brief, we show that, while the number of reads at each LINE is not randomly distributed across the genome, the distribution of scaled reads within each cluster is approximately normal. A convenient property of normal distributions is that the sum of multiple normal distributions is also a normal distribution. We thus can compute the theoretical mean and variance of the summed reads on each chromosome arm simply by summing the means and variances of all of the clusters represented on that chromosome arm.
WALDO also employs several other innovations that make it applicable to the analysis of PCR-generated amplicons from clinical samples. One of these innovations was controlling amplification bias stemming from the strong dependence of the data on the size of the initial template. Another is the use of a Support Vector Machine (SVM) to enable the detection of aneuploidy in samples containing low neoplastic fractions. The conceptual and statistical bases for WALDO are detailed in SI Appendix, SI Materials and Methods.
Evaluation of Chromosome Arm Gains and Losses in Primary Tumor Samples.
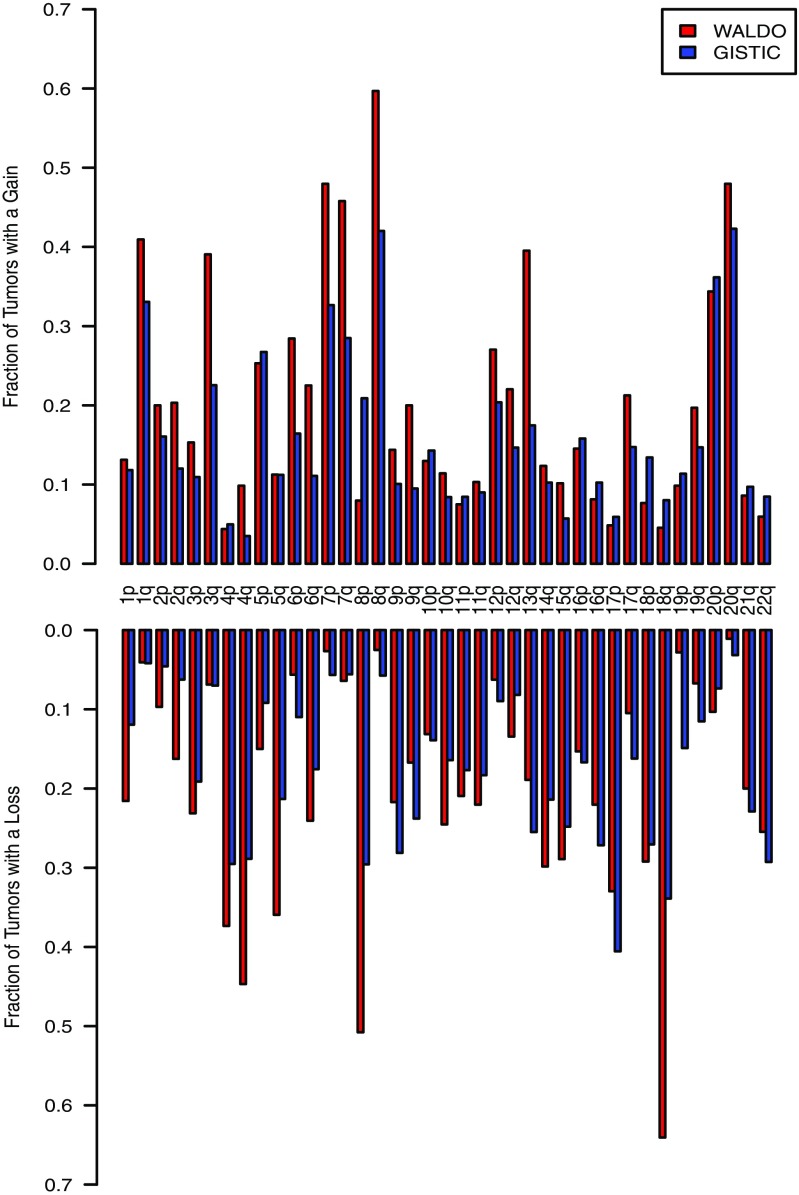
We first used WALDO to study chromosome arm gains and losses in 1,677 primary tumor samples from 10 cancer types. One of the outputs of WALDO is a Z-score for each of the 39 nonacrocentric arms on the autosomal chromosomes. The Z-scores for each of these chromosome arms in each of the primary tumor samples evaluated in this study are provided in Dataset S1 as well as stage, grade, histopathology, and an estimate of tumor purity. These results were compared with those obtained by The Cancer Genome Atlas (TCGA) from independent samples of the same tumor types (19, 20). We identified the fraction of tumor samples having a gain or a loss in each chromosome arm in our data and in TCGA, considering all tumor types together and each tumor type individually. In Fig. 2, Top, the fraction of samples in all cancer types scored as a gain by WALDO or as a gain by TCGA’s algorithm (GISTIC) is shown for each chromosome arm, and the fraction of samples with a loss is shown in Fig. 2, Bottom. The correlations between arm-level gains scored in our study and those in TCGA are shown in SI Appendix, Fig. S1A (R2 = 0.45), and arm-level losses are shown in SI Appendix, Fig. S1B (R2 = 0.39). Considering that the samples were from completely different patients, the specific chromosome arms gained and lost in both datasets were remarkably similar. The chromosome arms with the most gains were 1q, 3q, 7p, 7q, 8q, and 20q, and relatively few losses were observed on these arms. Those with the most losses were 4p, 4q, 8p, and 18q, and relatively few gains were observed on these arms. The arms with fewest gains or losses were 10p, 16p, 19p, and 19q. These results are broadly consistent with other previous reports (reviewed in ref. 21).
Fig. 2.
Individual chromosome arm gains and losses identified in nine cancer types. The average fraction of tumors with a gain or loss in each chromosome arm are depicted in the figure. The same nine tumor types were analyzed in both cohorts, but there was no overlap between the samples assessed by WALDO (red) or GISTIC (blue). WALDO employed the data from LINE sequencing of tumors reported here while GISTIC employed the data from Affymetirx SNP6.0 arrays provided by the TCGA.
We observed similarly high correlations for many of the specific tumor types in those cases in which a sufficient number of cancers were available for comparison (SI Appendix, Fig. S2 and Table S2). The highest correlations were for pancreatic adenocarcinomas and liver cancers (R2 = 0.70 and R2 = 0.64, respectively). One of the most important outcomes of this analysis was the large number of chromosome arms that were aneuploid in the great majority of cancer cases. The median number of chromosome arms that were lost or gained per cancer was 14, with interquartile range of 5–22. This large number was instrumental for the development of the SVM described below.
We also evaluated 32 benign tumors of the colon (colorectal adenomas). We found that 25 of them displayed gains or losses of chromosomes. The median number of chromosome arms that were lost or gained per benign tumor was 4, with an interquartile range of 1–9.75. No benign tumors have yet been studied by TCGA, so comparison was not possible. However, comparison with colorectal cancers showed that the benign tumors had many fewer chromosome arm changes than observed in cancers, which is consistent with the limited data on benign tumors from other studies (22–24). Additionally, the chromosome arms altered in the adenomas overlapped with those in the cancers, and the directionality of the changes (gains vs. losses) was preserved.
WALDO also allows determination of allelic imbalances based on the SNPs with the LINEs that are concomitantly sequenced. This provides a totally independent measure of chromosome arm changes than provided by the number of reads across the 500-kb genomic intervals. Note that measurements of allelic imbalance cannot be used to distinguish gain from loss; these measurements simply represent the ratios between the number of reads of the reference allele vs. those of the variant allele. This ratio will be the same whether the chromosome arm containing the reference allele is gained or the arm containing the variant allele is lost. Nevertheless, one would expect that there would be a strong relationship between the chromosomes exhibiting allelic imbalances and those exhibiting either gains or losses in the same tumor. We found that 63% of chromosome arms with allelic imbalance also had a significant gain or loss at the same chromosome arm. Other uses of the SNPs within the LINEs are described later in this paper.
We next compared the sensitivity and specificity of WALDO to call single chromosome arm gains or losses with the approach previously described in SI Appendix, SI Materials and Methods (14). Both methods were applied to LINE amplicon sequencing data from 677 normal peripheral white blood cell (WBC) samples, with each WBC sample independently amplified and sequenced to an average depth of 9.5 million (M) reads. These experimental data were augmented by 24,570 synthetic samples with single chromosome alterations (SI Appendix, SI Materials and Methods). Sensitivity was computed as the total number of correctly identified altered arms divided by the total number of altered arms in the synthetic samples. Specificity was computed as 1 minus the total number of incorrectly called altered arms divided by the total number of normal arms in the experimental data from the normal WBC samples. For both WALDO and the previous method, we considered three significance thresholds and three neoplastic cell fractions (1, 5, 10%). For all thresholds and neoplastic cell fractions, WALDO had higher specificity and sensitivity (SI Appendix, Tables S3 and S4).
To further evaluate the ability of WALDO to detect single chromosome abnormalities, we also evaluated DNA from patients with trisomy 21. The DNA from individuals with trisomies was physically mixed at a ratio of 2 ng of normal DNA and 0.2 ng of trisomy 21 DNA. The mixtures were created to replicate typical fetal fractions in noninvasive prenatal testing (∼10%). Using polymorphisms in the LINE amplicons, we estimated the trisomy admixture rate of the samples (range: 7.7–10.4%). Using a Z threshold of 2.5, we found that as few as 2 M reads could detect trisomy 21 at fetal fractions typically observed (sensitivity: 95%). We then subsampled 16 normal WBC at various read depths. At 2 M reads using the same threshold, the specificity was 100%. Sensitivities and specificities at other read depths and other admixtures of trisomy 21 samples are summarized in SI Appendix, Fig. S3.
Next, we evaluated whether WALDO can identify smaller amplicons that are present at a much higher copy number than simple aneuploidic gains. We generated 49,141 synthetic samples with amplifications on chromosome 17q. These synthetic samples represented various copy-number changes (1, 5, 10), sizes [5, 10, 20 million base pairs (MB)] and neoplastic cell fractions (1, 5, 10%). We evaluated whether the amplifications could then be detected from the presence of a significant chromosome 17q Z-score and then whether WALDO could be tailored to identify significant subchromosomal windows of interest. Based on our simulations, WALDO is powered to detect 5-MB amplifications in as low as 1% neoplastic content (SI Appendix, Tables S5 and S6).
Aneuploidy Detection in Samples with Low Fractions of Neoplastic Cell DNA.
Many potential applications of aneuploidy detection in cancer require the ability to identify a relatively small fraction of DNA from neoplastic cells within a large pool of DNA derived from normal cells. One notable application is liquid biopsy, i.e., the evaluation of bodily fluids such as urine, saliva, cyst fluid, or sputum for evidence of cancer. Given that aneuploidy is a general feature of cancers of virtually all types (Fig. 2 and ref. 25), detecting aneuploidy could in theory be used for this purpose. Multiple previous studies have shown that aneuploidy can be detected in plasma from advanced cancer patients, in which the proportion of DNA derived from neoplastic cells is often very high (14, 16, 26–28).
To employ WALDO for liquid biopsies, we used a two-stage approach. The first employed a search for individual chromosome arm gains or losses or allelic imbalance, as described above. Simulations with synthetic DNA showed that this approach could detect an individual chromosome arm gain or loss with sensitivities >90% at specificities >99% when the fraction of DNA contributed by the neoplastic cells was >5% of the total DNA. To detect aneuploidy in samples with lower fractions of neoplastic cell DNA, we exploited the fact, noted above in our studies of primary tumors, that the median number of chromosome arm gains or losses per tumor was high (14). We therefore considered a variety of approaches to distinguish samples containing low fractions of neoplastic DNA with multiple chromosome abnormalities from euploid samples. These approaches included counting the number of significant arms (29), combining scores of the most significant arms (27), and summing squared window-based Z-scores (28). Based on synthetic samples, we found that the optimum approach was obtained with an SVM (among many machine-learning algorithms tested). The SVM training was designed to be generally applicable to any cancer type rather than based on patterns of gains and losses typical of specific cancer types. With synthetic samples, the SVM could detect aneuploidy in 78% of samples with a neoplastic cell fraction of 1% at a specificity of 99% as determined by cross-validation. This SVM-based algorithm was therefore incorporated into WALDO for the evaluation of clinical samples with low neoplastic composition (Fig. 3).
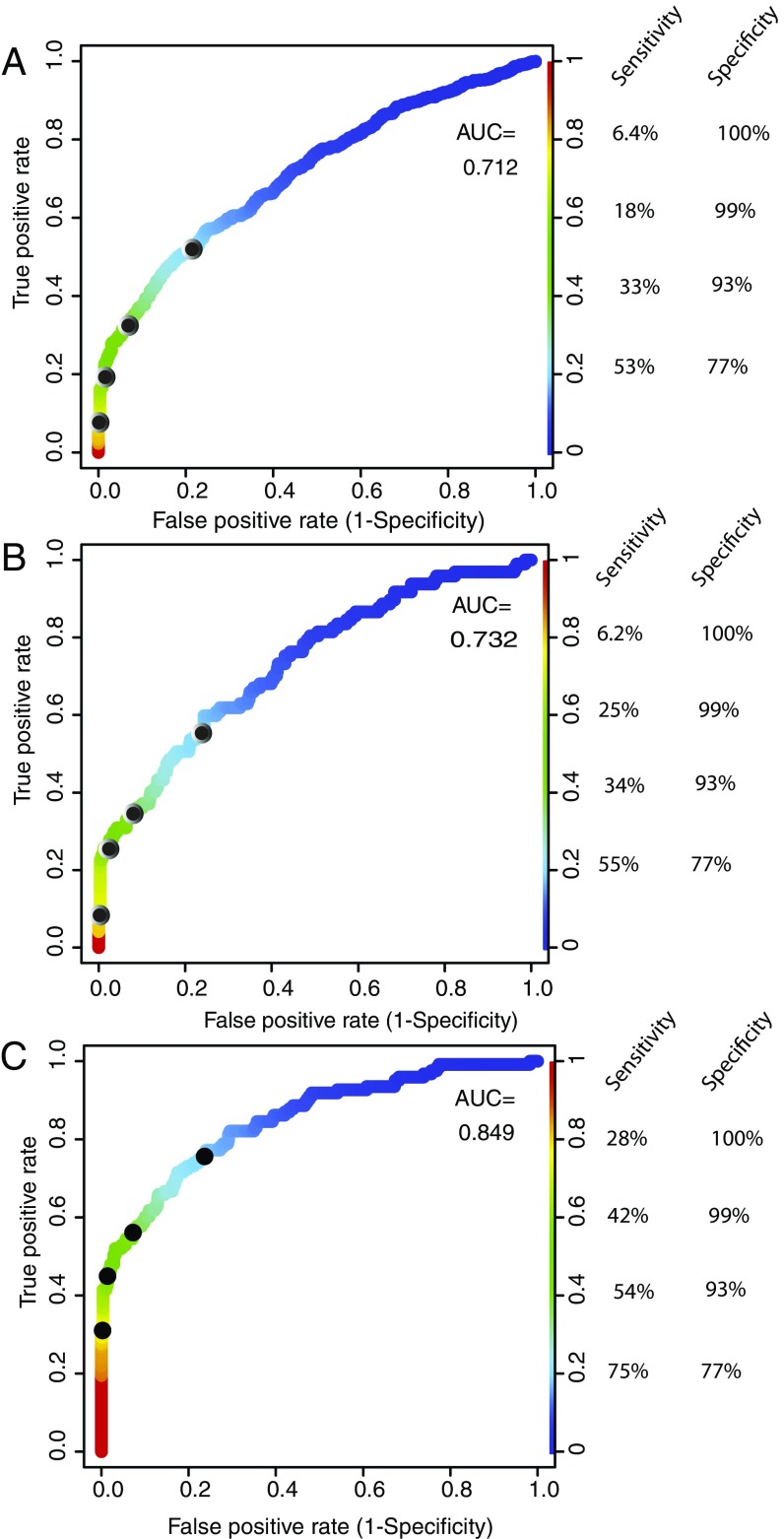
Fig. 3.
Aneuploidy detection in plasma samples from cancer patients. Receiver operating characteristics (ROC) and area under the curve (AUC) are shown for three ranges of neoplastic cell fractions. True positives were defined as those samples from cancer patients scoring positive while false positives were defined as those from normal individuals scoring positive. The neoplastic cell fraction of each plasma samples was estimated from driver gene-sequencing data as described in the text. (A) Samples with neoplastic cell fractions <0.5%. (B) Samples with neoplastic cell fractions ranging from 0.5 to 1%. (C) Samples with neoplastic cell fractions >1%.
WALDO was then used to attempt to evaluate aneuploidy in plasma samples from 961 cancer patients and 566 healthy individuals (Materials and Methods). Early and late-stage cancers of eight different types were evaluated, and a description of grade, stage, and histopathology for each sample is included in Dataset S1. We considered the neoplastic cell fraction of each cancer sample to be the mutant allele fractions determined from deep sequencing data. We divided the samples into those with neoplastic cell fractions >1% (122 samples), between 0.5 and 1% (96 samples), and <0.5% (738 samples). Sensitivity was defined as the proportion of cancer patient samples scored as aneuploid, while specificity was defined as 1 minus the fraction of healthy patient samples scored as aneuploid. Receiver operating curves are shown in Fig. 3 for these three ranges of neoplastic fractions. At stringent specificity (99%), we were able to identify aneuploidy in 42% of samples with neoplastic cell fractions >1% (Fig. 3A). As expected, sensitivity decreased with decreasing neoplastic cell fractions (Fig. 3 B and C). At 99% specificity, WALDO detected aneuploidy in 24% of samples with neoplastic cell fractions of 0.5–1% and in 19% of samples with neoplastic cell fractions of 0–0.5%. The specific cancer type of the patient was not highly correlated with positive aneuploidy calls. However, the number of template molecules that were assessed did correlate with sensitivity (Discussion).
In plasma samples with higher neoplastic cell content, we could determine which chromosome arms were gained or lost. Among 558 of the plasma samples that had a paired primary tumor, 188 samples had a significant chromosome arm gain or loss and 54% had a concordant gain or loss in the primary tumor. Concordance rose to 63% with respect to all significant chromosome arm gains, losses, and allelic imbalances. In samples with low neoplastic content, none of the individual arms were gained or lost at statistically significant levels but the SVM component of WALDO was presumably able to pick out small deviations in multiple chromosome arms that distinguished them from euploid samples.
Sample Matching.
DNA profiling with short tandem repeats is a well-established forensic technique that is routinely used. Carefully curated SNP panels have also been developed to ensure sample identity, such as between tumor and normal specimens from the same patients (30, 31). The LINEs amplified in FAST-SeqS contain 26,220 common polymorphisms, i.e., variants detected in >1% in 1,000 Genomes (32). These polymorphisms theoretically provide a powerful way to profile DNA samples evaluated for aneuploidy without any additional work or cost. To determine whether such identification was possible in practice, we designed a measure of concordance between any two samples (SI Appendix, SI Materials and Methods). We then used this to measure concordance in replicates of 176 normal WBC samples to one another, using ∼5 replicates per sample, for a total of 676 WBC samples. The input to WALDO was 676 samples, without specifying the sample name, so there were a total of 456,976 (676 × 676) possible matches. WALDO correctly matched all replicate samples with high concordance (>99.9%) without any false matching. Next, we performed this protocol on 970 plasma samples and 1,684 tumor samples. The 558 plasma samples should match the corresponding primary tumor samples, and no other matches should be observed. This produced 7,038,409 [(970 + 1,683) × (970 + 1,683)] possible matches. Nearly all of the 2,653 samples matched as expected, i.e., only to themselves or to the corresponding primary tumors, with concordance >99.8%. However, we found two plasma samples that did not match to the expected primary tumors and 12 plasma samples that matched to other plasma samples that were purportedly derived from different donors. In all these “mismatched cases,” the FastSeqS data indicated high concordances (>99.8%). The mismatches were therefore most likely a result of mislabeling of the samples and illustrated the utility of sample identity checks with WALDO.
Mutation Load, Carcinogenic Signatures, Microsatellite Instability.
When two samples, a normal and a cancer, are available from the same patient, LINE mutations that are in one sample but not the other can conceivably be discerned. For this application, it is imperative to use molecular barcoding to reduce sequencing errors (33), as is achieved through the experimental and bioinformatics components of WALDO (SI Appendix, SI Materials and Methods).
To determine whether somatic mutation detection was feasible, we first evaluated 10 upper tract urothelial carcinomas (UTUCs) and normal tissues from the same patients (SI Appendix, Table S7). These samples had previously been analyzed by exomic sequencing (34). For each tumor sample, we counted the number of somatic mutations and the spectrum of single base substitutions (SBS) (A→T, A→C, etc.). We found that the number of SBS in LINEs was highly correlated with the number of SBS in the exomes of these tumors (R2 = 0.98, P < 2.6 × 10−8). The spectrum of mutations in the LINEs was similarly correlated with the spectrum of mutations in exonic sequences (R2 = 0.95, P < 1.8 × 10−6). Notably, six of these tumors were from patients exposed to aristolochic acid, and the pathognomonic signature (A→T, T→A) of this mutagen was prominent in these six tumors (SI Appendix, Figs. S4–S6). The average estimated tumor content of the UTUC tumors was 73%, and the average mutant allele fraction (MAF) of the somatic mutations was 46%. The high MAFs of all somatic mutations suggested that somatic mutations and mutation signatures would still be detected from tumors with lower neoplastic content. However, we were unable to use this protocol to detect somatic mutations and signatures from plasma samples from relatively early-stage cancer patients, in which there is generally much lower neoplastic content (<1%).
The LINEs assessed by WALDO harbor 17,488 mononucleotide tracts of more than three nucleotides. Because mononucleotide tracts are particularly sensitive to defects in mismatch repair, we determined whether WALDO could be used to assess mismatch repair deficiency. For this purpose, we assessed the number of indels in the 17,488 LINE mononucleotide tracts. We found that the number of indels in six mismatch-repair–deficient colorectal cancers averaged 35 and ranged from 10 to 67. Normal tissues from these patients harbored only zero or one indel, and the difference between the cancers and normal tissues was highly significant (P < 7.2 × 10−4).
Our approach could also distinguish between microsatellite stable tumors and microsatellite unstable as illustrated by the analysis of 10 UTUC tumor/normal samples (SI Appendix, Table S7). None of the 10 normal UTUC samples had statistically significant numbers of somatic indels within their monotract repeats. In the tumors, only one tumor (sample AA 102) had significant numbers of somatic indels within its monotract repeats. We then compared the numbers of somatic indels detected within the monotracts to the numbers of somatic mutations detected in exome sequencing (34). Low numbers of somatic mutations in exome sequencing suggest that a sample is unlikely to have deficient mismatch repair genes or the presence of microsatellite instable (MSI) while large numbers of somatic mutations in exome sequencing suggest deficient mismatch repair genes and the presence of MSI. AA 102 had 2,415 somatic nonsynonymous mutations while the other nine tumors had on average only 284 nonsynonymous mutations (34). These data suggest that this protocol can be extended beyond our initial 10 MSI tumor normal pairs to microsatellite stable (MSS) tumor samples.
Discussion
The computational tools, collectively known as WALDO, that are described in this work enable the facile detection of chromosome arm gains or losses, allelic imbalances, somatic mutation loads, and microsatellite instability in samples derived from tumors. The work flow is exceedingly simple and involves PCR with a single primer pair. The efficiency of PCR copying of DNA is >90%, and because the single primer pair used amplifies 38,000 unique genomic loci, the approach is particularly well-suited for applications wherein only a small amount of DNA is available, such as in certain diagnostic situations in oncology as well as in forensic sciences.
What are the limitations of WALDO? In both synthetic and clinical samples, we show that WALDO can be used to reliably detect aneuploidy when the neoplastic cell fraction in the sample is >1% and can also detect samples with lower cell fractions, albeit less efficiently. Some cancers are more aneuploid than others, and WALDO presumably detects only the most aneuploid samples when the neoplastic cell component is low. Samples containing more aneuploidy will obviously be easier to detect at a given neoplastic cell content. To detect samples with lower neoplastic cell contents, greater sequencing depth will be needed. The average number of templates evaluated in our study was 4.2 M (interquartile range: 1.1–6.6 M; the number of templates can be precisely determined directly from the number of molecular barcodes, as described in ref. 33). Increasing the number of template molecules evaluated could be achieved in two ways. First, in the current study we used DNA from only 250 μL of plasma. The amplification could easily be performed on DNA from 10 mL of plasma without any major changes in the protocol. Second, the primers that we used amplify only ∼38,000 of the more than 500,000 LINEs in the genome. Using additional primers that amplified the remaining LINEs (or other repeats), coupled with the amplification of DNA from more plasma, should enable amplification of >500 million template molecules from plasma. At that point, aneuploidy should be detectable in samples containing <0.1% neoplastic cell fractions. Hopefully, continued improvements in sequencing technology will make such screening assays economically viable.
Materials and Methods
Detailed materials and methods are available in SI Appendix, SI Materials and Methods. A total of 1,678 tumors were evaluated in this study (SI Appendix, Table S1). The number of cancers of each histopathologic subtype are listed in Dataset S1. The tumors were formalin-fixed and paraffin-embedded (FFPE). In all cases, DNA was purified using QIAsymphony (catalog #937255). Peripheral WBCs were purified from the blood of 176 healthy individuals. Plasma was purified from 566 healthy individuals and 982 patients with cancer. DNA was purified from WBCs and plasma using Qiagen kit numbers (catalog #1091063) and (catalog #937255), respectively. The majority of the plasma samples used in this study have been independently evaluated for mutations in 1 of 12 commonly mutated genes. The fraction of mutant alleles in these plasma samples was used as an estimate of their neoplastic cell content (35). All individuals participating in the study provided written informed consent after approval by the institutional review board at The Johns Hopkins Medical Institutions.
Supplementary Material
Acknowledgments
We thank C. Blair and K. Judge for their assistance with sample collection and N. Silliman, M. Popoli, J. Ptak, L. Bobbyn, and J. Schaefer for their expert technical assistance. This work was supported by National Institutes of Health Grant 5F31HG007804 and by The Conrad N. Hilton Foundation.
Footnotes
Conflict of interest statement: The sponsor (B.V.) is a member of the Scientific Advisory Boards of Sysmex and Exelixis GP and is a founder of PapGene and Personal Genome Diagnostics. Sysmex, PapGene, Personal Genome Diagnostics, and other companies have licensed previously described technologies related to the work described in this paper from Johns Hopkins University. These licenses are associated with equity or royalty payments to B.V. K.W.K. and N.P. are members of the Scientific Advisory Board of Sysmex and are founders of PapGene and Personal Genome Diagnostics. I.K. is an employee of PapGene. Sysmex, PapGene, Personal Genome Diagnostics, and other companies have licensed previously described technologies related to the work described in this paper from Johns Hopkins University. These licenses are associated with equity or royalty payments to K.W.K., N.P., and I.K. Additional patent applications on the work described in this paper may be filed by Johns Hopkins University. The terms of all these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies.
Data deposition: Summaries of the sequencing data are provided in Dataset S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717846115/-/DCSupplemental.
References
- 1.Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci. 2008;121:1–84. doi: 10.1242/jcs.025742. [DOI] [PubMed] [Google Scholar]
- 2.Nowell PC. The clonal equestioon of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 3.Knouse KA, Davoli T, Elledge SJ, Amon A. Aneuploidy in cancer: Seq-ing answers to old questions. Annu Rev Cancer Biol. 2017;1:335–354. [Google Scholar]
- 4.Wang T-L, et al. Digital karyotyping. Proc Natl Acad Sci USA. 2002;99:16156–16161. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao M, Wang Q, Wang Q, Jia P, Zhao Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: Features and perspectives. BMC Bioinformatics. 2013;14(Suppl 11):S1. doi: 10.1186/1471-2105-14-S11-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianchi DW, et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA. 2015;314:162–169. doi: 10.1001/jama.2015.7120. [DOI] [PubMed] [Google Scholar]
- 7.Zhao C, et al. Detection of fetal subchromosomal abnormalities by sequencing circulating cell-free DNA from maternal plasma. Clin Chem. 2015;61:608–616. doi: 10.1373/clinchem.2014.233312. [DOI] [PubMed] [Google Scholar]
- 8.Pirooznia M, Goes FS, Zandi PP. Whole-genome CNV analysis: Advances in computational approaches. Front Genet. 2015;6:138. doi: 10.3389/fgene.2015.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lonigro RJ, et al. Detection of somatic copy number alterations in cancer using targeted exome capture sequencing. Neoplasia. 2011;13:1019–1025. doi: 10.1593/neo.111252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rigaill GJ, et al. A regression model for estimating DNA copy number applied to capture sequencing data. Bioinformatics. 2012;28:2357–2365. doi: 10.1093/bioinformatics/bts448. [DOI] [PubMed] [Google Scholar]
- 11.Fromer M, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Copy number detection and visualization for targeted sequencing using off-target reads. bioRxiv. 2014 doi: 10.1101/010876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuilman T, et al. CopywriteR: DNA copy number detection from off-target sequence data. Genome Biol. 2015;16:49. doi: 10.1186/s13059-015-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinde I, Papadopoulos N, Kinzler KW, Vogelstein B. FAST-SeqS: A simple and efficient method for the detection of aneuploidy by massively parallel sequencing. PLoS One. 2012;7:e41162. doi: 10.1371/journal.pone.0041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardner DK, Simón C. Handbook of in Vitro Fertilization. CRC Press; Boca Raton, FL: 2017. [Google Scholar]
- 16.Belic J, et al. Rapid identification of plasma DNA samples with increased ctDNA levels by a modified FAST-SeqS approach. Clin Chem. 2015;61:838–849. doi: 10.1373/clinchem.2014.234286. [DOI] [PubMed] [Google Scholar]
- 17.Grasso C, et al. Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J Mol Diagn. 2015;17:53–63. doi: 10.1016/j.jmoldx.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Straver R, et al. WISECONDOR: Detection of fetal aberrations from shallow sequencing maternal plasma based on a within-sample comparison scheme. Nucleic Acids Res. 2014;42:e31. doi: 10.1093/nar/gkt992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim T-M, et al. Functional genomic analysis of chromosomal aberrations in a compendium of 8000 cancer genomes. Genome Res. 2013;23:217–227. doi: 10.1101/gr.140301.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eizuka M, et al. Molecular alterations in colorectal adenomas and intramucosal adenocarcinomas defined by high-density single-nucleotide polymorphism arrays. J Gastroenterol. 2017;52:1158–1168. doi: 10.1007/s00535-017-1317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Vinci A, et al. Correlation between 1p deletions and aneusomy in human colorectal adenomas. Int J Cancer. 1998;75:45–50. doi: 10.1002/(sici)1097-0215(19980105)75:1<45::aid-ijc8>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 24.Drost J, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 25.Heim S, Mitelman F. Cancer Cytogenetics: Chromosomal and Molecular Genetic Aberrations of Tumor Cells. John Wiley & Sons; Hoboken, NJ: 2015. [Google Scholar]
- 26.Chan KC, et al. Cancer genome scanning in plasma: Detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59:211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 27.Leary RJ, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4:162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heitzer E, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5:30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen PA, et al. Abnormal plasma DNA profiles in early ovarian cancer using a non-invasive prenatal testing platform: Implications for cancer screening. BMC Med. 2016;14:126. doi: 10.1186/s12916-016-0667-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kidd KK, et al. Developing a SNP panel for forensic identification of individuals. Forensic Sci Int. 2006;164:20–32. doi: 10.1016/j.forsciint.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 31.Pengelly RJ, et al. A SNP profiling panel for sample tracking in whole-exome sequencing studies. Genome Med. 2013;5:89. doi: 10.1186/gm492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abecasis GR, et al. 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoang ML, et al. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci Transl Med. 2013;5:197ra102. doi: 10.1126/scitranslmed.3006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen JD, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. January 18, 2018 doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.