Significance
Success or failure of CD8+ T cell responses dictates whether an infected patient can control HIV durably. HIV-specific CD8+ T cells from HIV controllers and chronic progressors were shown to differ in their antiviral function. Here, we investigated whether HIV-specific T cell receptors (TCRs) can explain functional differences in CD8+ T cells from patients with differential immunologic control. Through in vitro, in vivo, and in silico studies, we found that HIV-specific TCRs were functionally equivalent. These results suggest that the factors that modulate the CD8+ T cell response to HIV are independent of their TCRs.
Keywords: HIV controllers, killer T cells, T cell receptors
Abstract
HIV controllers (HCs) are individuals who can naturally control HIV infection, partially due to potent HIV-specific CD8+ T cell responses. Here, we examined the hypothesis that superior function of CD8+ T cells from HCs is encoded by their T cell receptors (TCRs). We compared the functional properties of immunodominant HIV-specific TCRs obtained from HLA-B*2705 HCs and chronic progressors (CPs) following expression in primary T cells. T cells transduced with TCRs from HCs and CPs showed equivalent induction of epitope-specific cytotoxicity, cytokine secretion, and antigen-binding properties. Transduced T cells comparably, albeit modestly, also suppressed HIV infection in vitro and in humanized mice. We also performed extensive molecular dynamics simulations that provided a structural basis for similarities in cytotoxicity and epitope cross-reactivity. These results demonstrate that the differential abilities of HIV-specific CD8+ T cells from HCs and CPs are not genetically encoded in the TCRs alone and must depend on additional factors.
HIV infection persists despite the induction of humoral and cell-mediated host immune responses. In most patients, known as chronic progressors (CPs), HIV infection leads to progressive depletion of CD4+ T cells and resultant severe immune deficiency if not treated with combination antiretroviral therapy (cART). HIV-specific immunity in these patients reduces viremia transiently but fails to suppress it durably (1, 2). In contrast, HIV controllers (HCs) durably maintain viral loads of less than 2,000 RNA copies per milliliter of plasma without cART (1, 2). HIV-specific CD8+ T lymphocyte (CTL) responses play a dominant role in controlling viremia in HCs (3–6). During acute infection, CTL responses exert antiviral pressure, the magnitude of which correlates with the viral set point (7). These responses likely lead to enhanced immunologic control in HCs, because HIV-specific CTLs from HCs are functionally superior to those from CPs (4, 6, 8, 9). CTLs from HCs mediate more efficient HIV inhibition in vitro (10–12), polyfunctional secretion of cytokines such as IL-2 and IFN-γ (1, 10, 11), greater antigen-induced proliferation (10, 13), less T cell activation (4), and evasion of regulatory T cell suppression (14). CTLs from HCs may also be phenotypically more inclined to protect from HIV infection for long durations (9, 15) and sustain effector functions without T cell exhaustion (16). Several HLA-I alleles, including B*2705 and B*5701, are enriched in HCs (17, 18), underscoring the role of CTLs in suppressing HIV infection. However, these alleles are neither necessary nor sufficient to confer protection against HIV disease progression (6).
Direct comparison of HIV-specific CTLs isolated from HCs and CPs may elucidate the factors underlying protection. In HLA-B*2705+ individuals, the HIV-specific CTL responses are largely focused on the KK10 (KRWIILGLNK) epitope in the Gag protein (19) and are often dominated by one or two clonotypes, each with a unique T cell receptor (TCR) (8, 12). B27-KK10–specific CTL clonotypes from HCs and CPs were previously shown to diverge in their in vitro functional capabilities (8). The CTL repertoire in HCs was comprised of immunodominant effective clones and subdominant ineffective clones, whereas in CPs both immunodominant and subdominant clones were ineffective at controlling HIV in vitro (8). Therefore, we hypothesized that the TCR, the sole genetic difference between CTL clones within a patient and the sole determinant of antigen specificity, might determine whether a particular CTL clone effectively controls HIV. The avidity and cross-reactivity of a T cell are also functions of its TCR, and these characteristics influence the effectiveness of antiviral immunity (20). In particular, high-avidity B27-KK10–restricted CTLs were shown to be associated with viral control (21). While high functional avidity or antigen sensitivity can correlate with superior T cell function, it can also correlate with heightened senescence or turnover (22). Existence of high-avidity public TCR clonotypes in HCs suggests a TCR-dependent mechanism of viral control (23). Molecular dynamics (MD) simulations of both effective and ineffective clonotypes derived from a single patient suggest that inherent sequential, structural, and dynamical differences between TCRs provide a basis for differences in CTL activity (24). Moreover, superior immunodominant HIV-specific TCRs from CD4+ T cells are also associated with viral control (25). Thus, there is evidence to suggest that immunologic control of HIV may be a function of the TCRs that mediate HIV-specific immunity (26). A recent report based on data from a single HC indicated that clonotype-specific differences between CTLs are not dependent on their TCRs alone (27). However, those results do not address the role of TCRs in HIV control because they did not compare TCRs from HCs and CPs. To elucidate the role of TCRs in HIV control, a direct comparison of TCRs from individuals with different virologic and immunologic status is required. Therefore, in this study, we addressed this question directly by comparing the functional characteristics of B27-KK10–specific TCRs from three types of patients: HCs who maintained low levels of viremia stably, CPs who progressed to high viral loads, and failed controllers (FCs) who suppressed viremia initially but progressed to high viral loads. We isolated immunodominant TCRs from two patients from each group and tested their function in vitro and in vivo. Furthermore, we applied MD simulations to investigate the structural and dynamical features of bound pHLA–TCR complexes. Through these studies, we addressed whether differential function of HIV-specific CTLs in patients with diverse immunologic outcomes is genetically encoded in their TCRs.
Results
Cloning and Expression of Immunodominant B27-KK10–Specific TCRs.
We sorted B27-KK10-dextramer (hereafter called dextramer)-labeled CD8+ T cells from peripheral blood of two B*2705+ patients from each of the following groups—HCs, FCs, and CPs—and isolated TCRs from them (Table S1). A unique immunodominant TCR was identified from each patient based on its frequency in the total B27-KK10–specific CTL repertoire. These TCRs showed diverse usage of Vα and Vβ families and CDR3 regions as demonstrated in Table S2. We coexpressed these TCRs with a reporter gene (LNGFR) in activated primary human T cells and measured transduction and surface expression by flow cytometry (Fig. S1 A–C). Despite similar levels of transduction, a range of surface expression as measured by B27-KK10-dextramer staining was observed, presumably due to their differential competitive strengths relative to endogenous TCRs expressed by T cells. To account for this heterogeneity, we diluted TCR-transduced cells with mock-transduced cells to normalize the total number of dextramer+ cells in the effector cell population for each TCR. As a control, we used F5, a TCR that recognizes the HLA-A2–MART1 epitope (28).
HIV-Specific TCRs from HCs, FCs, or CPs Respond to Cognate Antigens Equally Well.
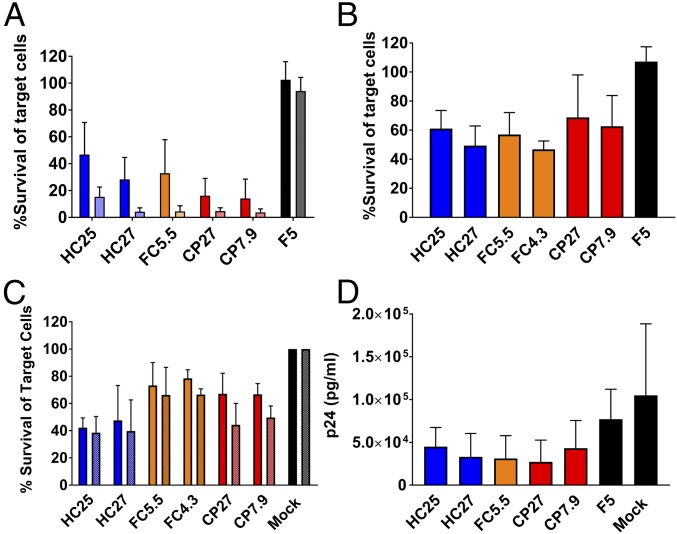
We measured the ability of TCRs to induce cytotoxicity upon recognition of antigen-presenting target cells. To that end, we coincubated TCR-transduced effector T cells with B27-KK10–expressing target cells and counted surviving target cells at 24 h by flow cytometry (Fig. S2A). We used two different types of target cells for these assays: (i) GXR-B27+ cells pulsed with KK10 peptide to provide overexpressed HLA and a short pulse of antigen stimulation (Fig. 1A) and (ii) autologous HLA-B27+ primary T cells pulsed with KK10 peptide for physiological levels of HLA expression and a short pulse of antigen stimulation (Fig. 1B). All B27-KK10–specific TCRs induced cytotoxicity in an antigen-specific manner, as evidenced by lack of cytotoxicity induced either by effector cells transduced with F5 TCR (Fig. 1 A and B) or by target cells pulsed with KY9, a B27-restricted epitope from HIV (Fig. S2B). We confirmed that the transduced cells were loaded equally well with Perforin, irrespective of the transduced TCR, and thus the measured cytotoxicity output was a result of degranulation caused by TCR–peptide–HLA interactions (Fig. S1 E and F). Surprisingly, we observed that the cytotoxic abilities of HC–, FC–, and CP–TCRs were equivalent in both contexts of antigen presentation. To assess whether interference from or mispairing with the endogenous TCR affects cytotoxicity, we constructed variants of these TCRs with murine constant regions. Despite higher surface expression and presumably diminished mispairing in the modified TCRs, the functional equivalence between HC– and CP–TCRs remained intact (Fig. S3). We used the unmodified TCRs for subsequent testing to avoid influence of murine constant regions on their antigen-binding properties.
Fig. 1.
B27-KK10–specific TCRs from HCs, FCs, and CPs induce equivalent antigen-specific responses. (A) GXR-B27+ cells pulsed with KK10 peptide used as target cells. An effector:target (E:T) ratio of 2:1 is indicated by dark bars and E:T ratio 4:1 indicated by light bars. Bars represent mean ± SD from n = 6. (B) Autologous B27+ cells pulsed with KK10 peptide used as targets. Bars represent mean ± SD of n = 3. (C) HIV-infected GXR-B27+ cells used as targets. Cytotoxicity at 24 h postcoincubation is shown. E:T ratio of 8:1 indicated by dark bars, and E:T ratio of 4:1 indicated by light bars. Bars represent mean ± SD from n = 3. (D) Reduction in HIV infection in autologous cocultures by T cells transduced with HC–, FC–, and CP–TCRs at 6 d in culture. Bars represent mean ± SD from n = 3.
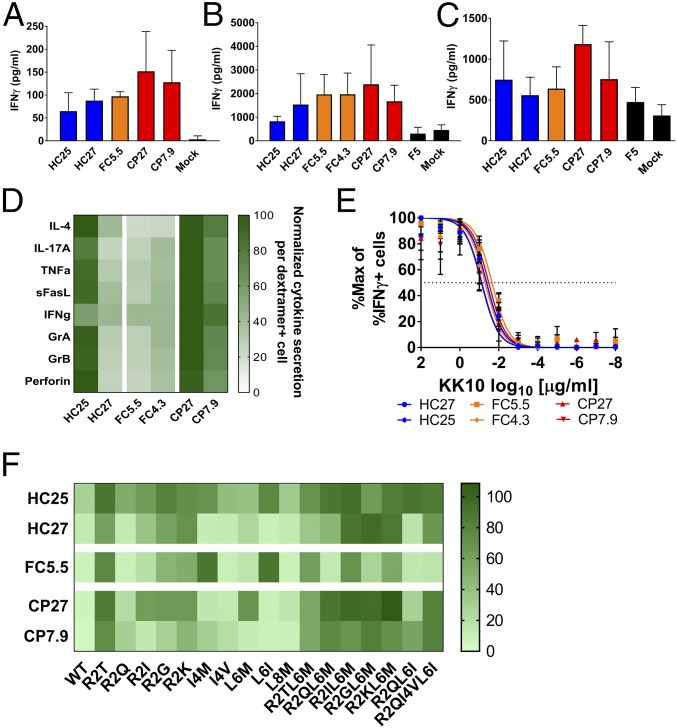
To test whether these TCRs can induce antigen-specific cytotoxicity toward cells infected with HIV (strain NL4-3), we used GXR-B27+ cells, which express GFP upon infection, as targets. HC–, FC–, and CP–TCRs induced equivalent specific killing of infected cells (Fig. 1C) and did not affect uninfected cells (Fig. S2C). Moreover, this effect was dependent on presentation of the wild-type KK10 epitope, as demonstrated by diminished cytotoxicity toward cells infected with NL4-3 encoding a known escape mutant R2T (KTWIILGLNK) (Fig. S4). We also tested if these TCRs induced suppression of HIV in autologous cocultures with HIV-infected HLA-B27+ primary T cells by measuring p24 antigen levels in culture supernatant. HC–, FC–, or CP–TCRs induced similar levels of viral suppression (Fig. 1D). These results indicate that TCRs from all three groups were equally capable of inducing a cytotoxic antiviral response upon recognition of their cognate antigen, in contrast to our previously published observations using bulk CTLs or CTL clones from HCs and CPs (8). However, the ability of transduced T cells to inhibit viral replication was significantly lower than our previous results with the parent CTL clones and in some cases not significantly different from the controls. While this may be due to differences in the assay setup, it may also be due to the transduced cells being less efficient at inhibiting viral replication compared with the original CTL clones. However, the observation that TCRs from patients with distinct immunologic outcomes were similar is consistent with the rest of the results. Another dimension of the CTL response is the ability to produce effector cytokines upon antigen recognition. We first measured IFNγ secretion induced by B27-KK10–specific TCRs upon antigenic stimulation. In agreement with the cytotoxicity results, relative IFNγ secretion among the TCRs was similar when using peptide-pulsed GXR-B27+ cells (Fig. 2A) or peptide-pulsed (Fig. 2B) or HIV-infected (Fig. 2C) autologous HLA-B27+ T cells as target cells. Furthermore, we measured the ability of TCRs to induce secretion of 13 effector cytokines upon stimulation with peptide-pulsed autologous HLA-B27+ T cells as targets. B27-KK10–restricted TCRs stimulated detectable secretion of IL-4, IL-17A, TNFα, sFasL, Granzyme A, Granzyme B, and Perforin (Fig. 2D and Fig. S5). However, all three groups of TCRs induced similar levels of these cytokines, reinforcing the functional equivalence of these TCRs.
Fig. 2.
B27-KK10–specific TCRs from HCs, FCs, and CPs show similar induction of cytokine secretion and antigen-binding properties. (A and B) Cytokine secretion by T cells transduced with B27-KK10–specific TCRs at 24 h after incubation with target cells expressing their cognate antigen. (A) GXR-B27+ cells pulsed with KK10 peptide as target cells. Bars represent mean ± SD from n = 3. (B) Autologous B27+ cells pulsed with KK10 peptide used as target cells. Bars represent mean ± SD from n = 3. (C) Autologous B27+ cells infected with HIV used as target cells. Cytokine secretion was measured at 7 d postcoincubation. Bars represent mean ± SD from n = 6 from two independent experiments. (D) Cytokine secretion by transduced T cells at 24 h after coculture with autologous B27+ target cells pulsed with KK10 peptide. The heatmap indicates relative values of each cytokine quantified as pg/mL secreted per LNGFR+dextramer+ cell. Mean data from five serial dilutions from n = 1 shown. (E) Measurement of functional avidity by staining for intracellular IFNγ-producing cells upon 6 h of stimulation with GXR-B27+ cells pulsed with reducing doses of KK10 peptide (102–10−8 μg/mL). Data points represent mean ± SD of n = 4. The lines represent curves fitted by least squares fit. The dotted line indicates 50% response. (F) Cross-reactivity of TCRs to variants of the KK10 peptide as measured by percentage survival of target cells at 24 h after coincubation. The key indicates percentage survival of target cells relative to mock-transduced T cells. Data represent mean of n = 3. Statistical comparison was done using ANOVA followed by univariate analysis. There was no statistical difference between groups of TCRs.
Antigen-Binding Characteristics of B27-KK10–Specific TCRs.
Functional avidity, antigen sensitivity, and cross-reactivity are properties that often distinguish effective and ineffective T cells. We first compared the functional avidity of TCRs by measuring intracellular IFNγ production in transduced cells upon stimulation with a range of concentrations of KK10 peptide in autologous assays (Fig. 2E). The observed functional avidities of all six TCRs were within 17–60 nM, which is within the expected range for T cell clones (8, 29). Importantly, all three groups of TCRs showed similar functional avidity, highlighting their functional equivalence (Fig. 2E and Fig. S6 A and B). Similar results were also obtained by measuring cytotoxicity upon stimulation with a range of peptide concentrations (Fig. S6C) and by measuring antigen sensitivity by dextramer staining (Fig. S6 D–G). We also compared the cross-reactivity of these TCRs toward 17 documented escape mutants of the KK10 epitope. We measured cytotoxicity and IFNγ secretion induced by TCRs upon recognition of GXR-B27+ cells pulsed with mutant peptides. All of the TCRs showed unique but overlapping profiles of variant recognition. Generally, multiple mutations in the KK10 epitope were not as well tolerated as single mutations. The mutants R2T and R2KL6M were not well-recognized, whereas L6M and R2Q were recognized almost as efficiently as the wild-type peptide (30). Importantly, there was no remarkable global difference between HC– and CP–TCRs (Fig. 2F). These results indicate that the antigen-binding characteristics of the three groups of TCRs were equivalent.
In Vivo Control of HIV by HC– and CP–TCRs.
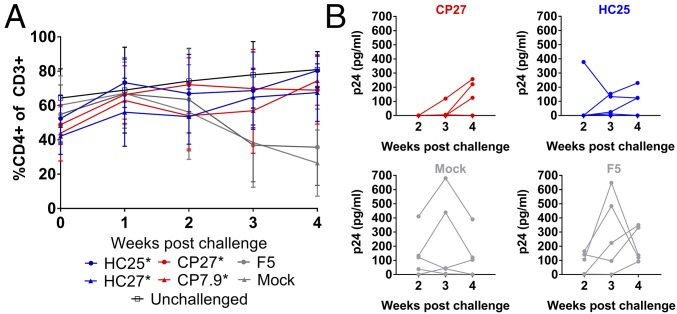
We investigated whether the functional equivalence of HC– and CP–TCRs extends to their ability to control HIV infection in human peripheral blood mononuclear cell (huPBMC) mice as described in Fig. S7A. We generated huPBMC mice engrafted with HLA-B27+ primary T cells transduced with HC– or CP–TCRs, challenged the mice intraperitoneally with NL4-3, and monitored the frequency of CD4+ T cells for 4 wk. As expected, mice engrafted with F5 or mock-transduced T cells showed depletion of CD4+ T cells upon challenge compared with unchallenged mice. Mice receiving T cells transduced with B27-KK10–specific TCRs prevented CD4+ T cell depletion significantly at 3–4 wk postchallenge. In agreement with in vitro results, HC– and CP–TCRs were indistinguishable in their ability to prevent CD4+ T cell depletion in vivo (Fig. 3A and Fig. S7). We performed viral load measurements in a subset of challenged mice. Mice receiving HC25- or CP27-transduced T cells had lower viral loads than controls (Fig. 3B). HC25 and CP27 showed comparable ability to suppress viremia in vivo. Taken together, our results indicate that immunodominant B27-KK10–specific TCRs from HCs and CPs were functionally equivalent in vitro and in vivo.
Fig. 3.
Ability of T cells transduced with HC– and CP–TCRs to suppress HIV infection in vivo. (A) Frequency of CD4+ T cells in mice over 4 wk postchallenge with HIV. Data points show mean ± SD from two independent experiments. The number of mice in each arm was as follows: HC25 = 10, HC27 = 11, CP27 = 17, CP7.9 = 10, F5 = 9, mock = 12, unchallenged = 18. Asterisk indicates a statistically significant difference in %CD4+ cells at week 4 postchallenge compared with “mock” using unpaired t tests. The variances are not significantly different as per F test. (B) Viral loads in plasma of individual mice receiving transduced cells at 2–4 wk postchallenge with HIV. Data points indicate individual mice from one experiment.
Structure and Dynamics of HLA-B27–KK10–TCR Complexes for HC–, FC–, and CP–TCRs.
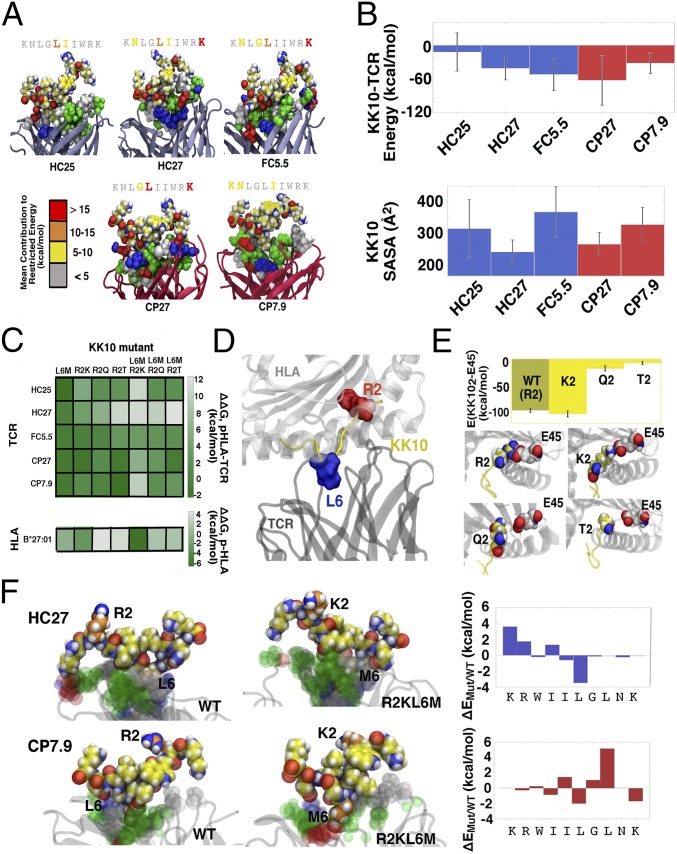
Seeking a physical explanation for the observed functional equivalence among the TCRs under investigation, we next conducted extensive MD simulations of HLA-B27–KK10–TCR structures (pHLA–TCR). These MD simulations describe both the structure and dynamics at the pHLA–TCR interface with atomistic details, sampling the dominant HLA–KK10 and TCR–KK10 interactions that are most likely to correspond with immunological function. Furthermore, free energy perturbation (FEP) calculations are used to assess the effects of peptide and protein mutations in silico. Standard MD trajectories should thus provide insight into the binding properties of wild-type KK10 complexes, while FEP calculations can help deliver mechanistic explanations of mutational escape (22). In general, each pHLA–TCR complex is configured with the KK10 peptide bound inside the HLA-B*27:01 binding groove and with each TCR bound to the still-exposed portion of KK10 (Fig. S8A). As no TCR crystal structures were available for reference, we constructed homology models for each TCR sequence using high-identity crystal templates, aligned these models with B*27:01–KK10 configurations derived from available ternary (B*27:01–KK10–TCR) crystal structures (31), and subjected resulting complexes to optimization with long (1 μs-long) MD trajectories (Table S3). As highlighted in Fig. 4A, we observed that HC27, CP27, and FC5.5 engaged with diverse epitopes consisting of the central, N-, and C-terminal residues of KK10, often with significant (>10 kcal/mol) per-residue interaction energies; CP7.9 interacted primarily with the center and C terminus of KK10, and HC25 promoted only weak interactions with the center of the presented peptide. While epitope characteristics vary within each of the TCRs, no clear distinction emerged among the HC–, FC–, and CP–TCRs. All of the TCRs displayed a broad composition of residue types at the pHLA–TCR interface. HC27, FC5.5, and CP27 presented charged or acidic residues that interact strongly with K1 of KK10, whereas HC25 and CP7.9 presented mostly hydrophobic residues that interact weakly at that position. All five TCRs exhibited a mixture of hydrophobic and polar residues near the middle of KK10, supporting common epitope contributions from the hydrophobic ILGL “core” of KK10 (KRWIILGLNK). The importance of these central hydrophobic residues to TCR binding was established in our previous study of HC FW56, wherein the binding characteristics of TCRs could be distinguished via the KK10 hydrophobic core alone (24). Here, direct comparison among clonotypes is complicated by the fact that TCR sequences are drawn from separate patients, resulting in some additional differences in terminal binding characteristics. Nevertheless, we see that the “ILGL” core also plays a significant and common role in the binding of the five TCRs studied here.
Fig. 4.
MD simulations of pHLA–TCR structures. (A) Epitope configurations cultivated by various TCRs. Conformations were drawn from high population clusters of MD trajectory data, and energy calculations are restricted to peptide–TCR interactions. For visual clarity, KK10 is represented from the C terminus to N terminus, and interfacial TCR residue types are colored according to the following key: white, hydrophobic; green, polar; red, acidic; blue, basic. For clarity, the presenting HLA molecule (B*27:01) is not rendered but would appear above KK10 in each complex. (B) Global observables (restricted energy and SASA) for epitopic interactions. Error bars represent SDs observed in MD simulations. HC and FC data are presented in blue, while CP data are shown in red. (C) Binding free energy changes of KK10 mutants to HC–, FC–, and CP–TCRs. ΔΔG values were calculated using alchemical FEP cycles. Data specific to TCRs used the KK10-B*27:01 complex as a free state reference, while values related to the HLA invoked isolated KK10 as the free state. (D) Schematic representation of ternary complex featuring residues prone to mutational escape (R2 and L6). (E) Effect of KK10-2 mutations on peptide–HLA interactions. Energy calculations involve only KK102 and E45 of B*27:01. (F) Divergent escape mechanisms for HC27 and CP7.9 specific to KK10-R2KL6M. Mutated KK10 residues (and their WT correspondents) are rendered in orange, and interfacial TCR residues are colored according to the previously mentioned key. ΔE values were computed from energies restricted to KK10–TCR interactions.
We also calculated the interaction energies and solvent-accessible surface areas (SASAs) in the ternary complexes (Fig. 4B). From a physical perspective, low interaction energies (i.e., related to strong binding) and low peptide SASAs (i.e., related to deep burial) should generally correspond to good recognition (32, 33). Overall, the interaction energy and SASA calculations revealed no clear distinction among the TCRs in KK10-restricted interactions at the molecular level. We next modeled TCR cross-reactivity in pHLA–TCR ternary complexes using FEP to quantitatively estimate the binding affinity changes resulting from KK10 mutations. Specifically, we computed binding free energy differences between the wild-type KK10 and five mutants, including both single- and double-point mutations (Fig. 4C). A reasonable statistical correlation (R2 = 0.33) was observed between binding ΔΔG values and raw cytotoxicity values from HC–, FC–, and CP–TCRs (Fig. S8B). Specifically, the well-recognized L6M mutant resulted in mild changes in binding affinities of pHLA–TCR complexes. Notably, Ladell et al. (31) reported a unique role for the TRBV6-5 TRBJ1-1 TCR motif in controlling the L6M escape mutation. This CDR3β motif is not found in any of the five TCRs studied here, suggesting these TCRs independently evolved robustness to L6M-related escape (see SI Methods for further discussion). Among single mutations at KK102, R2Q induced the least dramatic change, on average; R2K and, particularly, R2T yielded higher ΔΔG values. All of the above observations relate well to the functional data (Fig. 2F). The double mutations generally led to even larger binding free energy changes, in agreement with the heightened escape observed in vitro. Mutations at KK102 generally led to diminished HLA interactions as well (Fig. 4 C–E), with the exception of those involving R2K. We did observe two subtly distinct mechanisms for the escape of R2KL6M: In the HC27 complex, mutations pulled the N-terminal residues KK101–4 outward, whereas in the CP7.9 complex, mutations pulled residues 5, 7, and 8 outward. In both cases, KK10–TCR binding was diminished. Overall, these in silico mutagenesis results correlate well with the cross-reactivity data for HC–, FC–, and CP–TCRs. Considered in concert, our structural modeling supports the notion that these TCRs cannot be differentiated with respect to recognizing the KK10 peptide and preventing mutational escape.
Discussion
HCs demonstrate that CD8+ CTLs, under the right conditions, can control HIV infection durably. In contrast, FCs and CPs demonstrate that this ability of CTLs can be limited, leading to failed immunologic control. Here we investigated if clonotype-associated functional differences between B27-KK10–specific CTLs from HCs and CPs are explained by inherent properties of the TCRs derived from them. Surprisingly, TCRs from HCs, FCs, and CPs showed equivalent abilities to induce antigen-specific cytotoxicity, secrete effector cytokines, and suppress HIV infection in vivo and in vitro. Moreover, the antigen-binding characteristics of the TCRs, namely functional avidity and cross-reactivity, were indistinguishable. Therefore, our results suggest that differences in CTL responses in HCs and CPs were not due to differences in functional properties of their TCRs. While our results extend considerably the studies recently reported by Flerin et al. (27), they contrast with previous data showing divergent function in CTL clones from one HC (CTR203) and two CPs (CR338 and FEN33), from which TCRs HC25, CP27, and CP7.9 tested here were derived (8). Unfortunately, the original CTL clones are no longer viable, preventing their direct comparison with transduced T cells in our assays. Our results show that superior CTL function is not explained by superior TCRs and is likely due to some other aspect of the CTL clones. CTL clones could likely reflect the immunological status of the cells from which they were derived and hence could retain their epigenetic characteristics. In the current study, the use of transduced activated T cells from healthy donors in functional assays ensures an epigenetically equivalent background for comparing TCRs.
There are a number of caveats that should be considered in evaluating these data. First, the number of patients from which TCRs were derived was small. However, the result that six TCRs from six different patients were functionally similar is compelling, especially given the diversity of the Vα and Vβ families and CDR3 regions among the TCRs. Second, as expected, the expression level of the individual TCRs in primary T cells varied, despite similar levels of transduction. Although this may have influenced functional characteristics, the magnitude of these differences was small and unlikely to mask differences in TCR function. Whether the use of dextramers to capture immunodominant clonotypes may have favored more functional TCRs is also a consideration. Finally, the levels of cytotoxicity and inhibition of replication were much lower than that observed with the parent cells, but unfortunately, the original cells were not available for direct comparison. Despite these caveats, the overall findings strongly indicate functional equivalence of these TCRs.
To address whether structural resemblance plays a role in functional similarity, we performed MD simulations that revealed diverse modes of pHLA–TCR interactions unique to each TCR. Despite diverse residues involved in the interactions, the overall structures predicted similar function, in agreement with our experimental findings. Our previous results reported in Xia et al. (24) showed that structural differences among distinct B27-KK10–specific clones within one patient correlated with functional differences, with results generated in blinded fashion, indicating the ability of the MD simulations to predict functional differences. In the current study, however, the lack of overall structural differences between HC– and CP–TCRs seems to support their functional similarity. The FEP perturbation studies agreed well with in vitro data and were able to predict cross-reactivity of TCRs to the tested mutant epitopes. Taken together, they further support the notion that HC– and CP–TCRs can be equally susceptible to mutational escape of KK10.
Overall, our observations imply that the lack of viral control exhibited by CPs and FCs must be due to loss of their functional capabilities. Although the data shown here are limited to TCRs recognizing the KK10 epitope, it will be interesting to see if they generalize to other epitopes in HIV. In the one HC and two CPs, from whom viral sequence could be obtained, the predominant species of virus bears the wild-type KK10 epitope, precluding mutational escape as the reason for lack of control. Therefore, our data predict that an effective CTL response, while essential, may be insufficient to provide immune control. We postulate that other factors may be acting in concert with CD8+ CTLs to allow HCs to exert greater control over viral replication, prevent exhaustion, and preserve immune function, independent of their TCRs. These factors could involve the innate immunity to the virus (2), fitness of the virus during hyperacute infection (34), NK cell activity associated with protective HLA alleles (35, 36), and cytokine production during acute and chronic phases of infection (37, 38). To address these factors in a TCR-specific manner, experimental systems that allow differentiation of CTLs bearing a particular TCR in an antigen-naïve environment are needed. We are currently investigating if humanized mice engrafted with hematopoietic stem/progenitor cells expressing HC– or CP–TCRs can be used to address these factors. A recent report showed that an additional facet of HIV-specific CD8+ T cell response in HCs was the recognition of epitopes presented by class II HLA alleles, which exhibit antiviral activity. It will be interesting to study the role of TCRs in such activity and whether it contributes to viral control (39). Our results also show potential for developing immunotherapies to treat HIV infection. The results showing that TCR gene transfer conferred HIV-specific cytotoxic activity, ability to recognize epitope variants, and ability to control HIV infection in vivo are encouraging from a therapeutic standpoint. These data highlight the potential of these TCRs for immunotherapy to achieve at least partial control over HIV infection. We postulate that these TCRs can be used in either short-term immunotherapy using transduced autologous primary T cells or as long term-immunotherapy using transduced autologous hematopoietic stem/progenitor cells in HLA-B27+ patients. These results can also be potentially extended to patients with other HLA types. We are currently testing the efficacy of TCR-based immunotherapy approaches to treat HIV infection in humanized mice.
Materials and Methods
In Vitro Functional Assays.
To measure in vitro cytotoxicity, primary human T cells were activated and transduced as described in SI Methods. Surface expression of the TCRs was checked by flow cytometry, and the %dextramer+ was normalized to 2% by mixing dextramer+ cells with mock-transduced cells. In assays using GXR-B27+ cells as targets, they were pulsed at 1 × 106 cells per milliliter with 100 μg/mL peptide for 2 h at 37 °C. Following the incubation, the cells were washed twice with culture medium and used immediately for the assay. In the assays using autologous target cells, T cells were labeled with CFSE (Biolegend) for 15 min at 37 °C, washed twice with cold PBS, and resuspended at 1 × 106 per milliliter in culture medium. The cells were then pulsed with 100 μg/mL peptide for 2 h at 37 °C, washed twice, and used immediately for the assays. To perform the cytotoxicity assays, 5 × 104 target cells and the required number of effector cells were mixed together and plated in 96-well flat-bottom plates and placed in a humidity chamber. The cells were harvested 24 h later and mixed with half volume of PBS + 2% FBS with 30 μg/mL propidium iodide (Life Technologies) and acquired immediately on MACSQuant 10 (Miltenyi). The remaining cells were centrifuged at 500 g for 5 min at 4 °C, and a fraction of the supernatant was collected for measurement of IFNγ. The data were analyzed on Flowjo v10 as described in Fig. S2. To measure functional avidity, GXR-B27+ cells were incubated with 102–10−8 μg/mL peptide for 2 h at 37 °C, followed by coincubation with transduced T cells in the presence of Brefeldin A (Biolegend) for 6 h. Cells were first stained for LNGFR expression, fixed and permeabilized, and then stained for intracellular IFNγ production (see SI Methods for detailed procedures). Each assay was performed using three to four technical replicates, and each assay was repeated independently three to six times. All of the peptides used in coculture experiments were synthesized from Pierce ThermoFisher. To measure antigen sensitivity, TCR-transduced Jurkat cells were stained with B27-KK10-tetramer and anti-TCRαβ antibody, gated on a narrow range of TCR expression to measure dextramer staining intensity (see SI Methods for detailed procedures). TCR-induced secretion of IFNγ was measured using an ELISA kit (Life Technologies/eBiosciences) according to the manufacturer’s protocol. For multiplexed measurement of cytokines, LEGENDPlex human CD8/NK kit (Biolegend) was used according to the manufacturer’s instructions. The samples were acquired on Macsquant 10 and analyzed on the LEGENDPlex analysis software.
In Vitro HIV Suppression Assay.
Primary human T cells from an HLA-B27+ donor were activated and transduced as described in SI Methods. To prepare target cells, autologous PBMCs were incubated with 0.5–1 μg p24 of NL4-3 at 1 × 106 cells per milliliter for 4 h at 37 °C in culture medium supplemented with 0.5 μg/mL anti-CD3:8 bispecific antibody [obtained from Johnson Wong and Galit Alter through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), NIH]. The cells were washed and resuspended in culture medium supplemented with 40 U/mL IL-2 and anti-CD3:CD8 antibody for 7 d. At day 7, the cells were harvested, counted, and plated at 5 × 104 cells per well in a flat-bottom 96-well plate. TCR-transduced cells were incubated with target cells at various Effector:Target ratios in a final volume of 200 μL per well. The culture supernatant was collected 7 d later, and the viral load was determined by measuring p24 antigen by ELISA (Zeptometrix) according to the manufacturer’s protocol.
Homology Modeling.
To enable MD simulations of the five TCR clonotypes studied in this work, homology models were constructed for each of the TCR sequences of interest using the MODELER 9.17 software package. In brief, target sequences were aligned with candidate templates using a standard algorithm; template structures were then selected on the basis of maximal sequence identity with constraints on crystal structure resolution, R-free values, and outlier statistics (Table S3). The 3D models were then constructed using single templates, selecting the best of 10 output configurations as determined by discrete optimized protein energy minimization scores. Specific template information and specific methods used for MD simulations and FEP calculations can be found in SI Methods.
Statement of Ethics and Regulations.
PBMCs from HIV+ patients or healthy donors were collected according to protocols approved by Institutional Review Boards of Massachusetts General Hospital and California Institute of Technology. In vitro and in vivo studies were performed as per the regulations of the Caltech Institutional Biosafety Committees. All of the in vivo experiments were performed as per the regulations of the Caltech Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
We thank Caltech Office of Laboratory Animal Research staff for assistance with mouse models, members of the D.B. laboratory for their assistance and suggestions throughout the study, and Michael Leonard and Alexa Parker for their technical assistance. We thank David Gjertson for assistance with statistical analyses. This study was supported by an Innovation Award from Ragon Institute, Caltech Innovation Initiative award, California Institute of Regenerative Medicine Award DISC2-09123, and Leidos Biomedical Research, Inc. Subcontract 11XS287. Support for obtaining primary cells was also provided by NIH-funded Centers for AIDS Research (Grant P30 AI027763, UCLA Center for AIDS Research, and Grant P30 AI060354, Harvard University Center for AIDS Research, which are supported by the following NIH cofunding and participating institutes and centers: NIAID, National Cancer Institute, National Institute of Child Health and Human Development, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, National Institute of Aging, Fogarty International Center, and Office of AIDS Research). HCs were recruited with support from the Mark and Lisa Schwartz Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718659115/-/DCSupplemental.
References
- 1.Zanussi S, et al. CD8+ lymphocyte phenotype and cytokine production in long-term non-progressor and in progressor patients with HIV-1 infection. Clin Exp Immunol. 1996;105:220–224. doi: 10.1046/j.1365-2249.1996.d01-746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker BD. Elite control of HIV infection: Implications for vaccines and treatment. Top HIV Med. 2007;15:134–136. [PubMed] [Google Scholar]
- 3.Migueles SA, et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blankson JN. Effector mechanisms in HIV-1 infected elite controllers: Highly active immune responses? Antiviral Res. 2010;85:295–302. doi: 10.1016/j.antiviral.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ndhlovu ZM, et al. High-dimensional immunomonitoring models of HIV-1-specific CD8 T-cell responses accurately identify subjects achieving spontaneous viral control. Blood. 2013;121:801–811. doi: 10.1182/blood-2012-06-436295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Migueles SA, Connors M. Success and failure of the cellular immune response against HIV-1. Nat Immunol. 2015;16:563–570. doi: 10.1038/ni.3161. [DOI] [PubMed] [Google Scholar]
- 7.Goulder P, et al. Co-evolution of human immunodeficiency virus and cytotoxic T-lymphocyte responses. Immunol Rev. 1997;159:17–29. doi: 10.1111/j.1600-065x.1997.tb01004.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, et al. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat Immunol. 2012;13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sáez-Cirión A, et al. Agence Nationale de Recherches sur le Sida EP36 HIV Controllers Study Group HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc Natl Acad Sci USA. 2007;104:6776–6781. doi: 10.1073/pnas.0611244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendoza D, et al. Comprehensive analysis of unique cases with extraordinary control over HIV replication. Blood. 2012;119:4645–4655. doi: 10.1182/blood-2011-10-381996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendoza D, et al. Cytotoxic capacity of SIV-specific CD8(+) T cells against primary autologous targets correlates with immune control in SIV-infected rhesus macaques. PLoS Pathog. 2013;9:e1003195. doi: 10.1371/journal.ppat.1003195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buckheit RW, 3rd, et al. Host factors dictate control of viral replication in two HIV-1 controller/chronic progressor transmission pairs. Nat Commun. 2012;3:716. doi: 10.1038/ncomms1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Connell KA, et al. Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus. J Virol. 2010;84:7018–7028. doi: 10.1128/JVI.00548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elahi S, et al. Protective HIV-specific CD8+ T cells evade Treg cell suppression. Nat Med. 2011;17:989–995. doi: 10.1038/nm.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrière M, et al. HIV “elite controllers” are characterized by a high frequency of memory CD8+ CD73+ T cells involved in the antigen-specific CD8+ T-cell response. J Infect Dis. 2014;209:1321–1330. doi: 10.1093/infdis/jit643. [DOI] [PubMed] [Google Scholar]
- 16.Shasha D, et al. Elite controller CD8+ T cells exhibit comparable viral inhibition capacity, but better sustained effector properties compared to chronic progressors. J Leukoc Biol. 2016;100:1425–1433. doi: 10.1189/jlb.4A0915-422R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magierowska M, et al. Combined genotypes of CCR5, CCR2, SDF1, and HLA genes can predict the long-term nonprogressor status in human immunodeficiency virus-1-infected individuals. Blood. 1999;93:936–941. [PubMed] [Google Scholar]
- 18.Kiepiela P, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 19.Payne RP, et al. Efficacious early antiviral activity of HIV Gag- and Pol-specific HLA-B 2705-restricted CD8+ T cells. J Virol. 2010;84:10543–10557. doi: 10.1128/JVI.00793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varela-Rohena A, et al. Control of HIV-1 immune escape by CD8 T cells expressing enhanced T-cell receptor. Nat Med. 2008;14:1390–1395. doi: 10.1038/nm.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almeida JR, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Appay V, Douek DC, Price DA. CD8+ T cell efficacy in vaccination and disease. Nat Med. 2008;14:623–628. doi: 10.1038/nm.f.1774. [DOI] [PubMed] [Google Scholar]
- 23.Iglesias MC, et al. Escape from highly effective public CD8+ T-cell clonotypes by HIV. Blood. 2011;118:2138–2149. doi: 10.1182/blood-2011-01-328781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia Z, et al. The complex and specific pMHC interactions with diverse HIV-1 TCR clonotypes reveal a structural basis for alterations in CTL function. Sci Rep. 2014;4:4087. doi: 10.1038/srep04087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benati D, et al. Public T cell receptors confer high-avidity CD4 responses to HIV controllers. J Clin Invest. 2016;126:2093–2108. doi: 10.1172/JCI83792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lissina A, et al. The link between CD8+ T-cell antigen-sensitivity and HIV-suppressive capacity depends on HLA restriction, target epitope and viral isolate. AIDS. 2014;28:477–486. doi: 10.1097/QAD.0000000000000175. [DOI] [PubMed] [Google Scholar]
- 27.Flerin NC, et al. T-cell receptor (TCR) clonotype-specific differences in inhibitory activity of HIV-1 cytotoxic T-cell clones is not mediated by TCR alone. J Virol. 2017;91:e02412–e02416. doi: 10.1128/JVI.02412-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson LA, et al. Gene transfer of tumor-reactive TCR confers both high avidity and tumor reactivity to nonreactive peripheral blood mononuclear cells and tumor-infiltrating lymphocytes. J Immunol. 2006;177:6548–6559. doi: 10.4049/jimmunol.177.9.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger CT, et al. High-functional-avidity cytotoxic T lymphocyte responses to HLA-B-restricted Gag-derived epitopes associated with relative HIV control. J Virol. 2011;85:9334–9345. doi: 10.1128/JVI.00460-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goulder PJ, Walker BD. HIV and HLA class I: An evolving relationship. Immunity. 2012;37:426–440. doi: 10.1016/j.immuni.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ladell K, et al. A molecular basis for the control of preimmune escape variants by HIV-specific CD8+ T cells. Immunity. 2013;38:425–436. doi: 10.1016/j.immuni.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 32.Cole DK, et al. Human TCR-binding affinity is governed by MHC class restriction. J Immunol. 2007;178:5727–5734. doi: 10.4049/jimmunol.178.9.5727. [DOI] [PubMed] [Google Scholar]
- 33.Holmberg K, Mariathasan S, Ohteki T, Ohashi PS, Gascoigne NR. TCR binding kinetics measured with MHC class I tetramers reveal a positive selecting peptide with relatively high affinity for TCR. J Immunol. 2003;171:2427–2434. doi: 10.4049/jimmunol.171.5.2427. [DOI] [PubMed] [Google Scholar]
- 34.Miura T, et al. Impaired replication capacity of acute/early viruses in persons who become HIV controllers. J Virol. 2010;84:7581–7591. doi: 10.1128/JVI.00286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamya P, et al. Canadian Cohort of HIV Infected Slow Progressors Receptor-ligand requirements for increased NK cell polyfunctional potential in slow progressors infected with HIV-1 coexpressing KIR3DL1*h/*y and HLA-B*57. J Virol. 2011;85:5949–5960. doi: 10.1128/JVI.02652-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuri-Cervantes L, de Oca GS, Avila-Ríos S, Hernández-Juan R, Reyes-Terán G. Activation of NK cells is associated with HIV-1 disease progression. J Leukoc Biol. 2014;96:7–16. doi: 10.1189/jlb.0913514. [DOI] [PubMed] [Google Scholar]
- 37.Freeman BE, Hammarlund E, Raué HP, Slifka MK. Regulation of innate CD8+ T-cell activation mediated by cytokines. Proc Natl Acad Sci USA. 2012;109:9971–9976. doi: 10.1073/pnas.1203543109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhen A, et al. Targeting type I interferon-mediated activation restores immune function in chronic HIV infection. J Clin Invest. 2017;127:260–268. doi: 10.1172/JCI89488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ranasinghe S, et al. Antiviral CD8+ T cells restricted by human leukocyte antigen class II exist during natural HIV infection and exhibit clonal expansion. Immunity. 2016;45:917–930. doi: 10.1016/j.immuni.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.