Significance
Mountain ranges and highland plateaus in different parts of the world provide an opportunity to investigate the extent to which native species have followed similar or different routes of adaptation to the challenges of life at high altitude. Here we demonstrate that high-altitude songbirds from the Qinghai-Tibet Plateau independently evolved derived increases in hemoglobin–O2 affinity in comparison with their closest lowland relatives in East Asia. In comparisons that also included more distantly related high-altitude avian taxa, site-directed mutagenesis experiments revealed two cases in which convergent increases in hemoglobin–O2 affinity were caused by identical amino acid substitutions at the same sites. However, most adaptive convergence in protein function was attributable to different amino acid substitutions in different species.
Keywords: hemoglobin, hypoxia, mutation bias, biochemical adaptation, convergence
Abstract
When different species experience similar selection pressures, the probability of evolving similar adaptive solutions may be influenced by legacies of evolutionary history, such as lineage-specific changes in genetic background. Here we test for adaptive convergence in hemoglobin (Hb) function among high-altitude passerine birds that are native to the Qinghai-Tibet Plateau, and we examine whether convergent increases in Hb–O2 affinity have a similar molecular basis in different species. We documented that high-altitude parid and aegithalid species from the Qinghai-Tibet Plateau have evolved derived increases in Hb–O2 affinity in comparison with their closest lowland relatives in East Asia. However, convergent increases in Hb–O2 affinity and convergence in underlying functional mechanisms were seldom attributable to the same amino acid substitutions in different species. Using ancestral protein resurrection and site-directed mutagenesis, we experimentally confirmed two cases in which parallel substitutions contributed to convergent increases in Hb–O2 affinity in codistributed high-altitude species. In one case involving the ground tit (Parus humilis) and gray-crested tit (Lophophanes dichrous), parallel amino acid replacements with affinity-enhancing effects were attributable to nonsynonymous substitutions at a CpG dinucleotide, suggesting a possible role for mutation bias in promoting recurrent changes at the same site. Overall, most altitude-related changes in Hb function were caused by divergent amino acid substitutions, and a select few were caused by parallel substitutions that produced similar phenotypic effects on the divergent genetic backgrounds of different species.
When different species experience similar selection pressures in a shared environment, the probability that they will evolve similar adaptations may be influenced by differences in population size (which determines levels of standing genetic variation and the rate of input of new mutations) and/or differences in the duration of residency in that environment (which determines the time available for new mutations to arise). The probability of evolving similar adaptive solutions may also be influenced by legacies of evolutionary history. Prior genetic changes may preclude or potentiate future changes in a particular trait, in which case the “happenstance of a realized beginning” (1) may play an outsize role in channeling subsequent pathways of evolutionary change. Due to lineage-specific changes in genetic background, different species may hit upon idiosyncratic solutions to the same problem simply because they evolved from different ancestral starting points at the onset of selection.
Mountain ranges and highland plateaus in different parts of the world provide an opportunity to investigate the extent to which native species have followed similar or different routes of high-altitude adaptation (2). The two highest elevation plateaus in the world, the Andean Altiplano in South America and the Qinghai-Tibet Plateau in Asia, have very different physiographic and biogeographic histories and are inhabited by members of phylogenetically distinct faunas. In the Andes, the passerine birds that inhabit the highest elevations include representatives of the globally distributed Passeri clade (oscines) as well as representatives of the exclusively Neotropical Tyranni clade (suboscines). The avifauna of the Qinghai-Tibet Plateau has a very different phylogenetic composition, and the passerine birds that inhabit the highest elevations include a highly disproportionate number of tits in the family Paridae and long-tailed tits in the family Aegithalidae (3–7). Tits are widely distributed throughout the northern hemisphere and tropical Africa, whereas long-tailed tits are mainly restricted to Eurasia; both groups have their center of diversity in East Asia (7, 8).
At high altitude, the challenge of matching reduced O2 availability with an undiminished cellular O2 demand is especially acute for small, active endotherms like passerine birds that cannot rely on metabolic suppression as a general strategy of hypoxia tolerance. To compensate for the reduced partial pressure of O2 (PO2) in inspired air, physiological adjustments involving numerous steps in the O2-transport pathway can help sustain O2 flux to the tissue mitochondria in support of aerobic ATP synthesis (9–11). In combination with changes in the cardiorespiratory system and microcirculation, changes in the oxygenation properties of hemoglobin (Hb) can enhance the O2 capacitance of the blood (the total amount of O2 unloaded for a given arterio-venous difference in O2 tension). Under severe hypoxia, an increased Hb–O2 affinity safeguards arterial O2 saturation, thereby securing tissue oxygenation, albeit at a lower pressure gradient for O2 diffusion from the peripheral capillaries to the cells of respiring tissues (12). Evolutionary increases in Hb–O2 affinity may be caused by amino acid mutations that increase the intrinsic O2 affinity of the Hb tetramer and/or mutations that suppress the sensitivity of Hb to the affinity-reducing effects of allosteric cofactors (nonheme ligands such as Cl− ions and organic phosphates) (12, 13).
In the Andes, birds that are high-altitude natives have generally evolved derived increases in Hb–O2 affinity in comparison with their closest lowland relatives (14–17). However, convergent increases in Hb–O2 affinity in different high-altitude species are seldom attributable to convergent or parallel changes at the amino acid level (17). Here we test whether different high-altitude parid and aegithalid species from the Qinghai-Tibet Plateau have independently evolved increased Hb–O2 affinities in comparison with their closest lowland relatives in East Asia, and we examine whether convergent changes in Hb function have a similar molecular basis in different phylogenetic lineages. In this context we make comparisons among different high-altitude parid and aegithalid species in the Sino-Himalayan region, and, using previously published data (14–17), we also make comparisons with a phylogenetically diverse set of high-altitude Andean birds. If lineage-specific changes in genetic background or happenstances of biogeographic history predisposed the Andean and Sino-Himalayan passerines to follow different evolutionary paths, then representatives of the two distinct avifaunas may exhibit qualitatively distinct adaptations to hypoxia. If such contingencies influence the adaptive evolution of Hb function, such that highland representatives of different lineages hit upon different solutions to the same problem, this may be reflected in clade-specific or region-specific patterns of molecular parallelism.
Results and Discussion
We conducted a survey of sequence variation in the adult-expressed α- and β-type globin genes in a set of 162 bird specimens representing 13 tit species in the family Paridae (n = 135 specimens) and 4 long-tailed tit species in Aegithalidae (n = 27 specimens). We collected these specimens from localities spanning a broad range of elevations on the Qinghai-Tibet Plateau, the mountains of Southwest China, and in eastern China. Phylogenetic relationships and elevational ranges of focal species are summarized in Fig. S1.
For a subset of six tit species, we experimentally examined functional properties of native Hbs purified from red blood cells. The set of species used in these experiments included extreme high alpine specialists such as the ground tit (Parus humilis, elevational range = 3,100–5,500 m above sea level), which is endemic to the Qinghai-Tibet Plateau (6, 7), as well as predominantly highland species such as the rufous-vented tit (Periparus rubidiventris, 2,400–4,300 m) and gray-crested tit (Lophophanes dichrous, 2,300–4,600 m), and predominantly lowland species such as the oriental tit (Parus minor, sea level–2,000 m) and the marsh tit (Poecile palustris, sea level–2,100 m). We also sampled multiple specimens from high- and low-altitude subspecies of the broadly distributed coal tit [Periparus ater aemodius (2,100–4,600 m) and Periparus ater pekinensis (sea level–1,800 m)]. In addition to analyzing the native Hbs of these seven taxa (six distinct species, including geographically distinct subspecies of Periparus ater), we also functionally tested recombinantly expressed Hbs (rHbs) from an additional pair of high- and low-altitude sister species in the family Aegithalidae: the black-browed bushtit (Aegithalos bonvaloti) and the sooty bushtit (Aegithalos fulginosus), respectively. We performed experiments on rHbs for these two species because we did not have adequate quantities of blood from wild-caught birds to purify native Hbs.
Hb Isoform Composition.
During adulthood, most bird species express two structurally distinct Hb isoforms that incorporate different α-chain subunits but share identical β-chains. The major HbA isoform (αA2βA2) incorporates products of the αA-globin gene, and the minor HbD isoform (αD2βA2) incorporates products of the αD-globin gene (18–20). We used isoelectric focusing (IEF) analysis to characterize Hb isoform composition in the red blood cells of each of the focal species. These analyses revealed that the minor HbD isoform accounted for 20–30% of total Hb (Table S1), consistent with data from the majority of other passerine taxa examined to date (15, 17, 19–21).
Oxygenation Properties of HbA and HbD Isoforms.
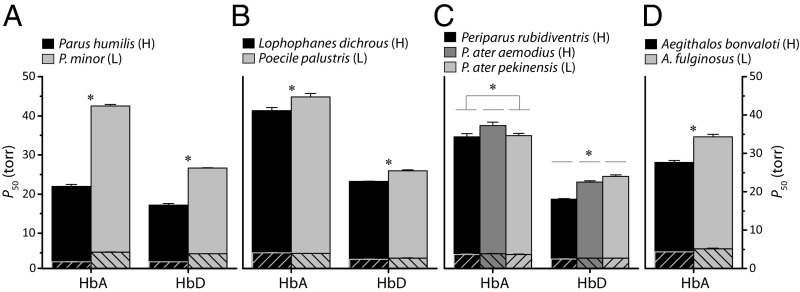
We purified HbA and HbD isoforms from red cell lysates of select specimens with known αA-, αD-, and βA-globin genotypes. We then measured the oxygenation properties of purified HbA and HbD solutions in the presence and absence of two main allosteric cofactors that regulate Hb–O2 affinity: Cl− ions (added as 0.1 M KCl) and inositol hexaphosphate (IHP, a chemical analog of inositol pentaphosphate). Experimental measurements on purified Hb samples from all species revealed that the HbD isoform exhibited a uniformly higher O2 affinity than HbA, both in the absence (“stripped”) and presence of allosteric cofactors (Fig. 1 A–C and Table S2). This is indicated by the lower values of P50 (the PO2 at which heme is 50% saturated) for HbD relative to HbA. This consistent isoform differentiation in Hb–O2 affinity suggests that the up-regulation of HbD could provide a ready means of optimizing blood oxygenation properties in response to changes in O2 availability. However, the absence of altitude-related differences in the HbA/HbD ratio among species (Table S1) indicates that regulatory adjustments in red cell Hb isoform composition do not play an important role in hypoxia adaptation. Both HbA and HbD exhibited cooperative O2 binding, as indicated by Hill coefficients at half-saturation (n50) >2 in the presence of anions (Table S2).
Fig. 1.
O2 affinities of Hbs from high- and low-altitude Asian passerines. (A–C) P50 values (mean ± SE) for purified HbA and HbD isoforms of Parus humilis and Parus minor (A), (B) Lophophanes dichrous and Poecile palustris (B), and Periparus rubidiventris, Periparus ater aemodius, and Periparus ater pekinensis (C). (D) P50 values for purified recombinant HbA isoforms of Aegithalos bonvaloti and Aegithalos fulginosus. Cross-hatched and solid bars show P50 values in the absence (stripped) and presence of anionic cofactors, respectively. Asterisks denote statistically significant differences (P < 0.05) in the presence of KCl + IHP. H, high altitude; L, low altitude.
Evolved Changes in Hb–O2 Affinity and Its Structural Basis.
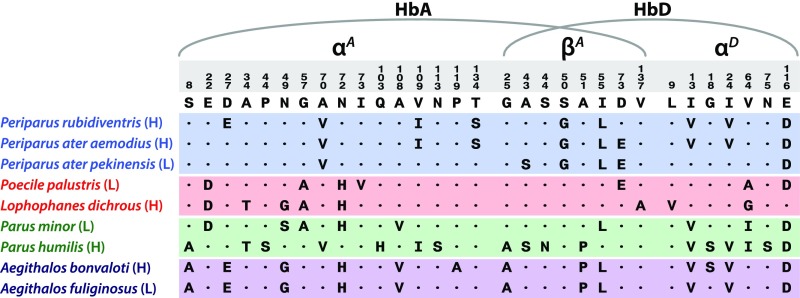
Experimental results for the nine focal taxa revealed that highland species generally have higher Hb–O2 affinities in comparison with their close lowland relatives. Within the genus Parus, the high-altitude Parus humilis and the low-altitude Parus minor provide the basis for an especially informative comparison because the two closely related species have completely nonoverlapping elevational ranges (Fig. S1). The experiments revealed that the HbA and HbD isoforms of Parus humilis exhibited significantly higher O2 affinities than the corresponding isoforms of Parus minor, both in the absence and presence of allosteric cofactors (Fig. 1A and Table S2). In the presence of Cl− and IHP, P50 values for HbA and HbD of Parus humilis were lower than those of Parus minor by factors of 2.0-fold and 1.6-fold, respectively (Table S2). The difference in HbA–O2 affinity is attributable to the independent or combined effects of 17 substitutions: 12 in αA and 5 in βA (Fig. 2). The qualitatively similar species difference in HbD–O2 affinity (Fig. 1A) is associated with eight substitutions: three in αD in addition to the above-mentioned βA substitutions (Fig. 2).
Fig. 2.
Variable residue positions in a multiple alignment of αA-, αD-, and βA-globin sequences from the set of parid and aegithalid species used in the experimental analysis of Hb function. Subunits of the major HbA isoform are encoded by the αA- and βA-globin genes, whereas those of the minor HbD isoform are encoded by the αD- and βA-globin genes. High- and low-altitude natives are denoted by a (H) and (L), respectively.
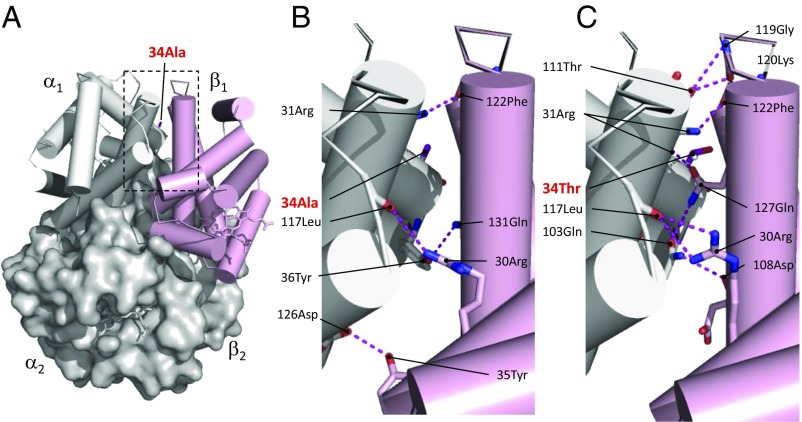
Within the Poecile/Lophophanes clade, the high-altitude Lophophanes dichrous and the low-altitude Poecile palustris also have nonoverlapping elevational ranges (Fig. S1). Our experiments revealed that HbA and HbD of Lophophanes dichrous have significantly higher O2 affinities than the corresponding isoforms of the low-altitude Poecile palustris (Fig. 1B and Table S2). Species differences in HbA–O2 affinity are attributable to the independent or combined effects of five substitutions (three in αA, two in βA) (Fig. 2). A similar difference in HbD–O2 affinity is associated with five substitutions (three in αD in addition to the above-mentioned βA substitutions) (Fig. 2). Among the αA substitutions, it is notable that Lophophanes dichrous shares the same αAA34T substitution as Parus humilis (Fig. 2). This site is located at an intradimer α1β1/α2β2 contact surface. Mutations at such sites are known to affect Hb–O2 affinity and quaternary structural stability (22–26). Homology modeling analysis suggests that the replacement of Ala with Thr at αA34 introduces four new hydrogen bonds between opposing α- and β-chain subunits of the oxygenated R conformation; the same intersubunit α1β1/α2β2 interface is not altered appreciably in the deoxygenated T conformation (Fig. 3 and Table S3). The allosteric equilibrium between the oxygenated (R) and deoxygenated (T) conformations of the Hb tetramer is a primary determinant of Hb–O2 affinity. The differential stabilization of oxygenated Hb therefore increases O2 affinity by shifting the T↔R allosteric equilibrium in favor of the high-affinity R state.
Fig. 3.
The αAA34T substitution in Parus humilis and Lophophanes dichrous increases Hb–O2 affinity by adding new hydrogen bonds that stabilize the oxygenated R conformation of the Hb tetramer relative to the deoxygenated T conformation. (A) Structural model of avian Hb in the liganded R state. (B) A zoomed-in view of the intradimer α1β1 interface of R-state Hb with the ancestral Ala at αA34. Four hydrogen bonds (depicted as dashed lines) are predicted at this interface. (C) Replacing Ala with Thr at α34 produces a twofold increase in the number of hydrogen bonds at the interface (Table S3), thereby increasing the stability of the R state.
Within the genus Periparus, experiments revealed no appreciable differentiation in overall Hb–O2 affinity between the high- and low-altitude subspecies of Periparus ater, as HbA–O2 affinity was slightly higher in the lowland Periparus ater pekinensis, and HbD–O2 affinity was higher in the highland Periparus ater aemodius (Fig. 1C and Table S2). HbA–O2 affinity of the high-altitude Periparus rubidiventris was virtually identical to that of Periparus ater pekinensis and therefore was slightly higher than that of Periparus ater aemodius; HbD–O2 affinity of Periparus rubidiventris was significantly higher than that of both Periparus ater subspecies (Fig. 1C and Table S2).
Similar to each of the other species-level comparisons, experiments on recombinantly expressed HbA isoforms from the high- and low-altitude pair of Aegithalos sister species revealed that the high-altitude Aegithalos bonvaloti had a significantly higher Hb–O2 affinity relative to the low-altitude Aegithalos fulginosus in the presence of Cl− and IHP (Fig. 1D and Table S2). This ∼7 torr difference in Hb–O2 affinity is attributable to a single αAP119A substitution in Aegithalos bonvaloti (Fig. 2). This same substitution is responsible for an evolved increase in Hb–O2 affinity in the bar-headed goose, Anser indicus (27–29), a high-altitude species renowned for its trans-Himalayan migratory flights. The αAP119A substitution eliminates a van der Waals contact between the ancestral Pro αA119 and Met βA55 on opposing subunits of the same αβ dimer. The loss of this intradimer contact destabilizes deoxygenated Hb and shifts the allosteric T↔R equilibrium in favor of the high-affinity R state, thereby increasing overall O2 affinity. Thus, the parallel αAP119A substitutions in Aegithalos bonvaloti and Anser indicus and the parallel αAA34T substitutions in Lophophanes dichrous and Parus humilis are both predicted to increase Hb–O2 affinity by shifting the allosteric equilibrium but via opposite mechanisms: destabilization of the T state in the former case and stabilization of the R state in the latter.
Although the comparison between high- and low-altitude subspecies of Periparus ater revealed no differentiation in overall Hb–O2 affinity, all species-level comparisons between close relatives were consistent in that the highland species always had higher Hb–O2 affinities than the lowland species. In each case, the evolved changes were exclusively attributable to an increase in intrinsic O2 affinity (as revealed by P50 values for stripped Hbs), as there were no appreciable differences in sensitivity to allosteric cofactors (Table S2).
In principle, altitude-related changes in Hb–O2 affinity could be attributable to derived increases in Hb–O2 affinity in high-altitude species, derived reductions in low-altitude species, or a combination of both. These alternatives can be distinguished by deciphering the character polarity of causative amino acid substitutions. For example, if a given high-altitude species has evolved a derived increase in Hb–O2 affinity relative to its low-altitude sister species, then the evolved change in protein function must be attributable to one or more substitutions where the derived amino acid state is fixed in the high-altitude species. As described below, we estimated globin gene trees to identify the phylogenetic intervals in which potentially causative amino acid substitutions occurred in the history of each high-altitude lineage.
Genealogical Discordance of Globin Genes.
To infer the polarity of character state change at substituted sites in HbA and HbD, we estimated DNA-based phylogenies of the αA-, αD-, and βA-globin genes from all 17 species. Estimated phylogenies revealed high levels of genealogical discordance among loci (Fig. S2), likely reflecting a history of incomplete lineage sorting and/or introgressive hybridization.
Molecular Convergence and Parallelism.
It is critically important to account for genealogical discordance when testing for evidence of molecular parallelism and convergence (30, 31). If amino acid substitutions in the αA- and αD-globin genes were mapped onto the branches of the species tree (Fig. S1) rather than the appropriate gene trees (Fig. S2 A and B), it would create the false appearance that multiple substitutions had occurred independently in different lineages.
Estimates of gene tree topologies for the αA-, αD-, and βA-globin genes permit an assessment of the true prevalence of molecular convergence and parallelism in our set of focal taxa. Some degree of homoplasy (due to convergence, parallelism, or mutational reversion) is expected due to chance alone. The key question concerns the number of convergent or parallel substitutions that actually contributed to convergent increases in Hb–O2 affinity in different high-altitude taxa; this applies to substitutions in HbA and HbD of both Parus humilis and Lophophanes dichrous, HbD of Periparus rubidiventris, and HbA of Aegithalos bonvaloti. The sequence data revealed that few derived amino acid states are shared between high-altitude taxa to the exclusion of lowland taxa. Within the set of high-altitude parid and aegithalid taxa that we examined, it appears that there is only one true parallel substitution that is associated with convergent increases in Hb–O2 affinity: A34T in the αA-globin orthologs of Parus humilis and Lophophanes dichrous (Fig. 2).
Functional Test of Adaptive Parallelism.
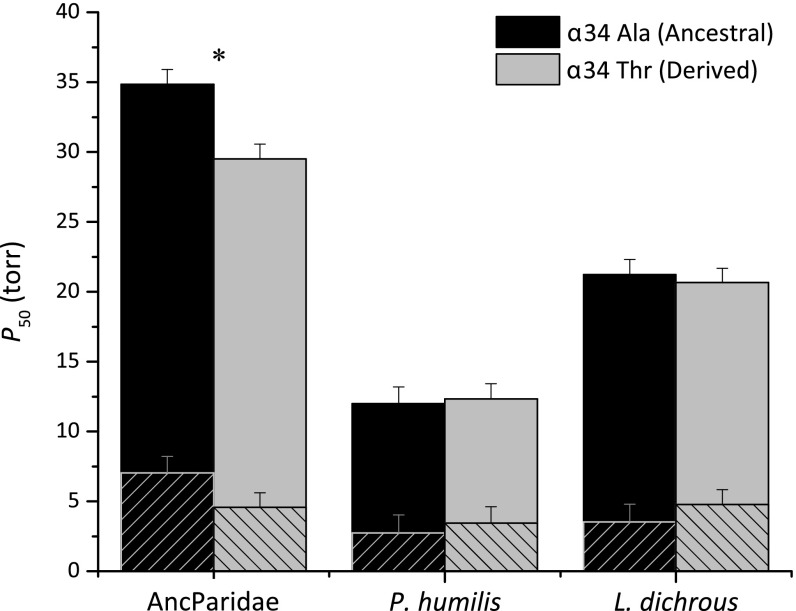
The αA-globin gene tree shown in Fig. S2A indicates that the shared, derived Thr αA34 variants in Parus humilis and Lophophanes dichrous must have had independent mutational origins. To test whether the parallel αAA34T substitutions contributed to derived increases in Hb–O2 affinity in each of these two high-altitude species, we reconstructed the αA and βA sequences of the most recent common ancestor of the family Paridae, “AncParidae,” which is also the most recent common ancestor of Parus and Lophophanes (Fig. S1). This enabled us to test the effect of the αAA34T mutation on an evolutionarily relevant genetic background. Ancestral amino acid states were estimated with a high level of statistical confidence, as site-specific posterior probabilities averaged 0.997 for the α-chain and 1.000 for the β-chain. The reconstructed ancestral Hb was estimated to possess Ala αA34 with a posterior probability of 0.988. Overall, the AncParidae Hb differed from the wild-type Hbs of Parus humilis and Lophophanes dichrous at 12 and 6 amino acid sites, respectively (Fig. S3).
In vitro functional tests on the purified rHbs confirmed that AncParidae Hb exhibited a significantly lower Hb–O2 affinity than the wild-type Hbs of both Parus humilis and Lophophanes dichrous, indicating that each of the two high-altitude species independently evolved derived increases in Hb–O2 affinity due to the effects of one or more lineage-specific substitutions. Site-directed mutagenesis experiments revealed that αAA34T produced a significant increase in intrinsic Hb–O2 affinity on the AncParidae background, and this affinity difference persisted in the presence of Cl− and IHP (Fig. 4). This result indicates that the parallel αAA34T substitutions in Parus humilis and Lophophanes dichrous contributed to convergent increases in Hb–O2 affinity in each of the two high-altitude species, but the effect of αAA34T alone was not sufficient to completely recapitulate the evolved change in O2 affinity in either species. Thus, additional amino acid substitutions in the α- and/or β-globins of both Parus humilis and Lophophanes dichrous must have contributed to the derived increases in Hb–O2 affinity. Interestingly, although αAA34T produced a significant increase in O2 affinity on the AncParidae background, reversion of this mutation to the ancestral state (αAT34A) did not produce significant reductions in Hb–O2 affinity on the wild-type backgrounds of either Parus humilis or Lophophanes dichrous (Fig. 4). Asymmetry in the effects of forward mutations on ancestral backgrounds and reverse mutations on derived backgrounds, a phenomenon documented in numerous protein-engineering studies, is attributable to epistatic interactions between the focal mutations and residues at other substituted sites that distinguish the two backgrounds (31–36).
Fig. 4.
O2 affinities of recombinantly expressed Hbs representing wild-type genotypes of Parus humilis and Lophophanes dichrous and the reconstructed genotype of their most recent common ancestor AncParidae. Site-directed mutagenesis experiments revealed that αAA34T produces a significant affinity-enhancing effect on the ancestral background (AncParidae), as indicated by the reduction in P50, but does not completely recapitulate the evolved increases in Hb–O2 affinity in Parus humilis or Lophophanes dichrous. Cross-hatched and solid bars show P50 values in the absence (stripped) and presence of anionic cofactors, respectively. Mutational reversions (αAA34T) on the wild-type backgrounds of Parus humilis and Lophophanes dichrous did not produce a symmetrical, affinity-reducing effect. The asterisk denotes a statistically significant difference (P < 0.05) in the presence of KCl + IHP.
The Possible Role of Mutation Bias in Promoting Parallel Substitutions.
Intriguingly, the parallel αAA34T substitutions in both Lophophanes dichrous and Parus humilis are attributable to nonsynonymous substitutions at a CpG dinucleotide. In both species, replacement of the ancestral Ala for Thr at αA34 was caused by CpG→CpA mutations in the first codon position. Depending on the methylation status of the cytosine, transition mutations at CpG sites are expected to occur at a much higher rate than non-CpG point mutations at the same nucleotide position (37–40). This suggests the hypothesis that the αAA34T mutation may be especially likely to contribute to evolved increases in Hb–O2 affinity simply because the underlying CpG→CpA transition mutation will occur at a higher rate than nonsynonymous mutations at non-CpG sites that produce similar affinity-enhancing effects. If adaptation is mutation-limited, an increase in the rate of mutation to a beneficial allele results in a commensurate increase in the allele’s probability of fixation (41–44). The extent to which evolution is mutation-limited in natural populations is not generally known, but a growing body of evidence suggests that mutation bias is an important cause of substitution bias and parallelism in adaptive protein evolution (15, 45, 46).
An examination of αA-globin nucleotide sequences in a phylogenetically diverse set of passerines revealed that the CpG dinucleotide involving the third position of codon 33 and the first position of codon 34 is clearly the ancestral state (and, hence, Ala is the ancestral amino acid at αA34) (Fig. S4). Thus, if the parallel αAA34T substitutions in Lophophanes dichrous and Parus humilis were partly attributable to mutation bias, it is a bias that is not unique to parid species. On the basis of mutational accessibility alone, there is no reason to think that αAA34T substitutions were more likely to contribute to Hb adaptation in parid species than in other passerines.
Broader Patterns of Convergence and Parallelism Involving High-Altitude Birds from the Andes and the Qinghai-Tibet Plateau.
An expansion of the comparative analysis to include other high-altitude birds revealed several convergent and parallel substitutions shared with parid and aegithalid species from the Qinghai-Tibet Plateau. The number of such substitutions is reduced significantly if we restrict our attention to shared, derived replacements that are associated with increases in Hb–O2 affinity in high-altitude lineages. In the Andean birds, convergent and parallel substitutions were not uncommon, but site-directed mutagenesis experiments revealed that a small fraction of such substitutions (i.e., N/G83S, A86S, D94E, and A116S in βA-globin) actually contributed to convergent increases in Hb–O2 affinity in two or more high-altitude species (16, 17). These same sites were all invariant in our sample of parid and aegithalid species.
Conclusions
We documented that high-altitude parid and aegithalid species from the Qinghai-Tibet Plateau have typically evolved increased Hb–O2 affinities in comparison with lowland relatives. This pattern is consistent with data from a large and phylogenetically diverse set of Andean birds (14–17). The data for parid and aegithalid birds further bolster a remarkably strong empirical generalization and demonstrate that repeated increases in Hb–O2 affinity in high-altitude birds represent one of the most striking examples of convergent biochemical adaptation in vertebrates (12).
The phylogenetically replicated changes in Hb–O2 affinity provide the opportunity to assess whether such changes consistently involve the same functional mechanisms and, if so, whether such changes are attributable to divergent, convergent, or parallel substitutions at the amino acid level. With regard to functional mechanisms, the fact that interspecific variation in Hb–O2 affinity is exclusively attributable to evolved changes in intrinsic O2 affinity is consistent with comparative data from other high-altitude birds (14–17, 21, 29) and demonstrates that there are multiple possible ways of evolving an increased Hb–O2 affinity without sacrificing allosteric regulatory capacity. In mammals, by contrast, evolutionary changes in Hb–O2 affinity often involve changes in responsiveness to Cl− and/or organic phosphates (47–52).
Despite the pervasive convergence in Hb–O2 affinity (and convergence in the underlying functional mechanisms) among high-altitude avian taxa, the limited amount of convergence and parallelism at the amino acid level demonstrates that changes in Hb–O2 affinity can be produced by numerous possible amino acid substitutions. The analysis of sequence variation in our set of parid and aegithalid species revealed several parallel amino acid substitutions, only one of which contributed to convergent increases in Hb–O2 affinity in different high-altitude tit species: αAA34T in Parus humilis and Lophophanes dichrous. Most parallel substitutions were not uniquely associated with convergent increases in Hb–O2 affinity in highland lineages. In studies of genetic convergence and parallelism, experimental testing of individual substitutions is necessary to distinguish between signal (replicated substitutions that contribute to adaptive convergence in phenotype) and noise (replicated substitutions that have no effect on the selected phenotype).
In broader comparisons involving other avian taxa, we documented one additional example in which parallel substitutions contributed to convergent increases in Hb–O2 affinity in different high-altitude species: the αAP119A substitution in Aegithalos bonvaloti (Aegithalidae: Passeriformes) and the distantly related bar-headed goose, Anser indicus (Anatidae: Anseriformes). Results of site-directed mutagenesis experiments confirm that the parallel αAP119A substitutions have significant affinity-enhancing effects in the Hbs of both species (Fig. 1D and Table S2) (29). The αAP119A mutation also produces a similar affinity-enhancing effect in human Hb (27). All else being equal with respect to effect sizes and mutational pleiotropy, allelic variants that have additive effects across a wide range of genetic backgrounds can be expected to make more consistent contributions to convergent adaptation than those with context-dependent effects. Overall, we did not observe any striking clade-specific patterns of parallelism that could be interpreted as evidence that the high-altitude parid or aegithalid species had hit upon adaptive solutions that were not equally accessible to other high-altitude avian taxa.
Methods
Sample Collection.
We collected a total of 162 bird specimens (representing 17 species) from high-altitude localities on the Qinghai-Tibet Plateau and the mountains of Southwest China and in low-altitude localities throughout eastern China. The collection of all bird specimens was authorized by the Forestry Administrations of Qinghai Province and Xizang Autonormous Region and was in compliance with the National Wildlife Conservation Law of China. All birds were handled in accordance with regulations of the Animal Experimental and Medical Ethics Committee of the Institute of Zoology, Chinese Academy of Sciences. Details are provided in SI Methods.
Hb Isoform Composition and Sequence Variation.
To characterize Hb isoform composition in the red blood cells of wild-caught birds, we separated native Hbs by means of IEF and used densitometric measurements to quantify isoform abundance (20). Details regarding cloning and sequencing protocols are provided in SI Methods. All sequences have been deposited in GenBank (accession nos. MG772099–MG772439).
Protein Purification and in Vitro Analysis of Hb Function.
Using hemolysates of bird specimens with known globin genotypes, we isolated and purified the HbA and HbD isoforms by means of anion-exchange FPLC using a HiTrap Q HP column (GE Healthcare). Details regarding O2-binding experiments are provided in SI Methods.
Vector Construction, Mutagenesis, and Recombinant Expression.
The αA- and βA-globin sequences were synthesized in accordance with Escherichia coli codon preferences. The α-β globin gene cassette was cloned into a custom pGM vector system, and recombinant Hb expression was carried out in the E. coli JM109 (DE3) strain (25, 53). Details regarding mutagenesis and protein purification are provided in SI Methods.
Phylogenetic Analysis.
We used complete exon and intron sequences to estimate phylogenies of each globin gene. Details regarding alignments, substitution models, and ancestral reconstructions are provided in SI Methods.
Structural Modeling.
We modeled structures of avian Hbs using MODELER ver. 9.19 (54). We used Protein Data Bank (PDB) ID 1hho and 3hhb as templates for oxygenated and deoxygenated Hb conformations. We performed additional calculations using PISA (55) and PyMOL ver. 1.8 (Schrödinger).
Supplementary Material
Acknowledgments
We thank N. Zhao, C. Dai, W. Wang, R. Zhang, D. Zhang, C. Zhang, B. Gao, Q. Quan, Y. Wu, and Z. Yin for assistance with field work, E. Petersen for assistance in the laboratory, and P. Alström for help with specimen identification. This study was funded by the Strategic Priority Research Program, Chinese Academy of Sciences Grant XDB13020300 (to F.L.), State Key Program of the National Science Foundation of China Grant 31630069 (to F.L.), Key Laboratory of Zoological Systematics and Evolution Grant O529YX5105 (to F.L.), NIH Grant HL087216 (to J.F.S.), National Science Foundation Grants MCB-1517636 and RII Track-2 FEC-1736249 (to J.F.S.), and Danish Council for Independent Research, Natural Sciences Grant 4181-00094 (to A.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in GenBank (accession nos. MG772099–MG772439).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720487115/-/DCSupplemental.
References
- 1.Gould SJ. The Structure of Evolutionary Theory. Belknap; Cambridge, MA: 2002. [Google Scholar]
- 2.Beall CM. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc Natl Acad Sci USA. 2007;104:8655–8660. doi: 10.1073/pnas.0701985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCallum DA, Gill FB, Gaunt SLL. Community assembly patterns of parids along an elevational gradient in western China. Wilson Bull. 2001;113:53–64. [Google Scholar]
- 4.Martens J, Tietze DT, Packert M. Phylogeny, biodiversity, and species limits of passerine birds in the Sino-Himalayan region:–A critical review. Ornithol Monogr. 2011;70:64–94. [Google Scholar]
- 5.Packert M, et al. Horizontal and elevational phylogeographic patterns of Himalayan and Southeast Asian forest passerines (Aves: Passeriformes) J Biogeogr. 2012;39:556–573. [Google Scholar]
- 6.Lei FM, Qu YH, Song G. Species diversification and phylogeographical patterns of birds in response to the uplift of the Qinghai-Tibet plateau and quaternary glaciations. Curr Zool. 2014;60:149–161. [Google Scholar]
- 7.Packert M, Martens J, Sun YH, Tietze DT. Evolutionary history of passerine birds (Aves: Passeriformes) from the Qinghai-Tibetan plateau: From a pre-quarternary perspective to an integrative biodiversity assessment. J Ornithol. 2015;156:S355–S365. [Google Scholar]
- 8.Tietze DT, Borthakur U. Historical biogeography of tits (Aves: Paridae, Remizidae) Org Divers Evol. 2012;12:433–444. [Google Scholar]
- 9.Scott GR, Milsom WK. Flying high: A theoretical analysis of the factors limiting exercise performance in birds at altitude. Respir Physiol Neurobiol. 2006;154:284–301. doi: 10.1016/j.resp.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Storz JF, Scott GR, Cheviron ZA. Phenotypic plasticity and genetic adaptation to high-altitude hypoxia in vertebrates. J Exp Biol. 2010;213:4125–4136. doi: 10.1242/jeb.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott GR. Elevated performance: The unique physiology of birds that fly at high altitudes. J Exp Biol. 2011;214:2455–2462. doi: 10.1242/jeb.052548. [DOI] [PubMed] [Google Scholar]
- 12.Storz JF. Hemoglobin-oxygen affinity in high-altitude vertebrates: Is there evidence for an adaptive trend? J Exp Biol. 2016;219:3190–3203. doi: 10.1242/jeb.127134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber RE. High-altitude adaptations in vertebrate hemoglobins. Respir Physiol Neurobiol. 2007;158:132–142. doi: 10.1016/j.resp.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Projecto-Garcia J, et al. Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc Natl Acad Sci USA. 2013;110:20669–20674. doi: 10.1073/pnas.1315456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galen SC, et al. Contribution of a mutational hotspot to adaptive changes in hemoglobin function in high-altitude Andean house wrens. Proc Natl Acad Sci USA. 2015;112:13958–13963. doi: 10.1073/pnas.1507300112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Natarajan C, et al. Convergent evolution of hemoglobin function in high-altitude Andean waterfowl involves limited parallelism at the molecular sequence level. PLoS Genet. 2015;11:e1005681. doi: 10.1371/journal.pgen.1005681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Natarajan C, et al. Predictable convergence in hemoglobin function has unpredictable molecular underpinnings. Science. 2016;354:336–339. doi: 10.1126/science.aaf9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann FG, Storz JF. The alphaD-globin gene originated via duplication of an embryonic α-like globin gene in the ancestor of tetrapod vertebrates. Mol Biol Evol. 2007;24:1982–1990. doi: 10.1093/molbev/msm127. [DOI] [PubMed] [Google Scholar]
- 19.Grispo MT, et al. Gene duplication and the evolution of hemoglobin isoform differentiation in birds. J Biol Chem. 2012;287:37647–37658. doi: 10.1074/jbc.M112.375600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Opazo JC, et al. Gene turnover in the avian globin gene families and evolutionary changes in hemoglobin isoform expression. Mol Biol Evol. 2015;32:871–887. doi: 10.1093/molbev/msu341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheviron ZA, et al. Integrating evolutionary and functional tests of adaptive hypotheses: A case study of altitudinal differentiation in hemoglobin function in an Andean Sparrow, Zonotrichia capensis. Mol Biol Evol. 2014;31:2948–2962. doi: 10.1093/molbev/msu234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yasuda Jp, et al. The α 1 β 1 contact of human hemoglobin plays a key role in stabilizing the bound dioxygen. Eur J Biochem. 2002;269:202–211. doi: 10.1046/j.0014-2956.2002.02635.x. [DOI] [PubMed] [Google Scholar]
- 23.Shikama K, Matsuoka A. Human haemoglobin: A new paradigm for oxygen binding involving two types of alphabeta contacts. Eur J Biochem. 2003;270:4041–4051. doi: 10.1046/j.1432-1033.2003.03791.x. [DOI] [PubMed] [Google Scholar]
- 24.Bellelli A, Brunori M, Miele AE, Panetta G, Vallone B. The allosteric properties of hemoglobin: Insights from natural and site directed mutants. Curr Protein Pept Sci. 2006;7:17–45. doi: 10.2174/138920306775474121. [DOI] [PubMed] [Google Scholar]
- 25.Natarajan C, et al. Epistasis among adaptive mutations in deer mouse hemoglobin. Science. 2013;340:1324–1327. doi: 10.1126/science.1236862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar A, et al. Stability-mediated epistasis restricts accessible mutational pathways in the functional evolution of avian hemoglobin. Mol Biol Evol. 2017;34:1240–1251. doi: 10.1093/molbev/msx085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jessen TH, Weber RE, Fermi G, Tame J, Braunitzer G. Adaptation of bird hemoglobins to high altitudes: Demonstration of molecular mechanism by protein engineering. Proc Natl Acad Sci USA. 1991;88:6519–6522. doi: 10.1073/pnas.88.15.6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber RE, Jessen TH, Malte H, Tame J. Mutant hemoglobins (α 119-Ala and β 55-Ser): Functions related to high-altitude respiration in geese. J Appl Physiol (1985) 1993;75:2646–2655. doi: 10.1152/jappl.1993.75.6.2646. [DOI] [PubMed] [Google Scholar]
- 29.Natarajan C, et al. 2018. Molecular basis of hemoglobin adaptation in the high-flying bar-headed goose. bioRxiv:10.1101/222182.
- 30.Hahn MW, Nakhleh L. Irrational exuberance for resolved species trees. Evolution. 2016;70:7–17. doi: 10.1111/evo.12832. [DOI] [PubMed] [Google Scholar]
- 31.Storz JF. Causes of molecular convergence and parallelism in protein evolution. Nat Rev Genet. 2016;17:239–250. doi: 10.1038/nrg.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bridgham JT, Ortlund EA, Thornton JW. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461:515–519. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollock DD, Thiltgen G, Goldstein RA. Amino acid coevolution induces an evolutionary Stokes shift. Proc Natl Acad Sci USA. 2012;109:E1352–E1359. doi: 10.1073/pnas.1120084109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah P, McCandlish DM, Plotkin JB. Contingency and entrenchment in protein evolution under purifying selection. Proc Natl Acad Sci USA. 2015;112:E3226–E3235. doi: 10.1073/pnas.1412933112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaltenbach M, Jackson CJ, Campbell EC, Hollfelder F, Tokuriki N. Reverse evolution leads to genotypic incompatibility despite functional and active site convergence. eLife. 2015;4:e06492. doi: 10.7554/eLife.06492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCandlish DM, Shah P, Plotkin JB. Epistasis and the dynamics of reversion in molecular evolution. Genetics. 2016;203:1335–1351. doi: 10.1534/genetics.116.188961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat. 2003;21:12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- 39.Webster MT, Axelsson E, Ellegren H. Strong regional biases in nucleotide substitution in the chicken genome. Mol Biol Evol. 2006;23:1203–1216. doi: 10.1093/molbev/msk008. [DOI] [PubMed] [Google Scholar]
- 40.Ellegren H. The evolutionary genomics of birds. Annu Rev Ecol Evol Syst. 2013;44:239–259. [Google Scholar]
- 41.Yampolsky LY, Stoltzfus A. Bias in the introduction of variation as an orienting factor in evolution. Evol Dev. 2001;3:73–83. doi: 10.1046/j.1525-142x.2001.003002073.x. [DOI] [PubMed] [Google Scholar]
- 42.Stoltzfus A. Mutation-biased adaptation in a protein NK model. Mol Biol Evol. 2006;23:1852–1862. doi: 10.1093/molbev/msl064. [DOI] [PubMed] [Google Scholar]
- 43.Stoltzfus A, Yampolsky LY. Climbing mount probable: Mutation as a cause of nonrandomness in evolution. J Hered. 2009;100:637–647. doi: 10.1093/jhered/esp048. [DOI] [PubMed] [Google Scholar]
- 44.McCandlish DM, Stoltzfus A. Modeling evolution using the probability of fixation: History and implications. Q Rev Biol. 2014;89:225–252. doi: 10.1086/677571. [DOI] [PubMed] [Google Scholar]
- 45.Stoltzfus A, McCandlish DM. Mutational biases influence parallel adaptation. Mol Biol Evol. 2017;34:2163–2172. doi: 10.1093/molbev/msx180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sackman AM, et al. Mutation-driven parallel evolution during viral adaptation. Mol Biol Evol. 2017;34:3243–3253. doi: 10.1093/molbev/msx257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storz JF, et al. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc Natl Acad Sci USA. 2009;106:14450–14455. doi: 10.1073/pnas.0905224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Storz JF, Runck AM, Moriyama H, Weber RE, Fago A. Genetic differences in hemoglobin function between highland and lowland deer mice. J Exp Biol. 2010;213:2565–2574. doi: 10.1242/jeb.042598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Runck AM, Weber RE, Fago A, Storz JF. Evolutionary and functional properties of a two-locus β-globin polymorphism in Indian house mice. Genetics. 2010;184:1121–1131. doi: 10.1534/genetics.109.113506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Revsbech IG, et al. Hemoglobin function and allosteric regulation in semi-fossorial rodents (family Sciuridae) with different altitudinal ranges. J Exp Biol. 2013;216:4264–4271. doi: 10.1242/jeb.091397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janecka JE, et al. Genetically based low oxygen affinities of felid hemoglobins: Lack of biochemical adaptation to high-altitude hypoxia in the snow leopard. J Exp Biol. 2015;218:2402–2409. doi: 10.1242/jeb.125369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Natarajan C, et al. Intraspecific polymorphism, interspecific divergence, and the origins of function-altering mutations in deer mouse hemoglobin. Mol Biol Evol. 2015;32:978–997. doi: 10.1093/molbev/msu403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Natarajan C, et al. Expression and purification of recombinant hemoglobin in Escherichia coli. PLoS One. 2011;6:e20176. doi: 10.1371/journal.pone.0020176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics. 2016;54:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 56.Qu Y, et al. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat Commun. 2013;4:2071. doi: 10.1038/ncomms3071. [DOI] [PubMed] [Google Scholar]
- 57.Weber RE. Use of ionic and zwitterionic (Tris/BisTris and HEPES) buffers in studies on hemoglobin function. J Appl Physiol (1985) 1992;72:1611–1615. doi: 10.1152/jappl.1992.72.4.1611. [DOI] [PubMed] [Google Scholar]
- 58.Weber RE, Fago A, Malte H, Storz JF, Gorr TA. Lack of conventional oxygen-linked proton and anion binding sites does not impair allosteric regulation of oxygen binding in dwarf caiman hemoglobin. Am J Physiol Regul Integr Comp Physiol. 2013;305:R300–R312. doi: 10.1152/ajpregu.00014.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Storz JF, et al. Oxygenation properties and isoform diversity of snake hemoglobins. Am J Physiol Regul Integr Comp Physiol. 2015;309:R1178–R1191. doi: 10.1152/ajpregu.00327.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tufts DM, et al. Epistasis constrains mutational pathways of hemoglobin adaptation in high-altitude pikas. Mol Biol Evol. 2015;32:287–298. doi: 10.1093/molbev/msu311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 65.Johansson US, et al. A complete multilocus species phylogeny of the tits and chickadees (Aves: Paridae) Mol Phylogenet Evol. 2013;69:852–860. doi: 10.1016/j.ympev.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 66.Li X, et al. Taxonomic status and phylogenetic relationship of tits based on mitogenomes and nuclear segments. Mol Phylogenet Evol. 2016;104:14–20. doi: 10.1016/j.ympev.2016.07.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.