Significance
We describe an ancient regulatory code that patterns the gene expression required for the efficient C4 pathway. This code is based on two regulatory elements located in exonic sequence that act cooperatively to repress transcription in specific cells. As these regulators are located in gene bodies they determine amino acid sequence as well as gene expression and so are known as duons. They are found in multiple genes and are not restricted to C4 species, but rather found in all land plants and even some algae. The prevalence of these motifs has likely facilitated the repeated evolution of the complex C4 system, and they also provide a mechanism that could be used to engineer photosynthesis.
Keywords: C4 photosynthesis, evolution, duons, gene regulation
Abstract
If the highly efficient C4 photosynthesis pathway could be transferred to crops with the C3 pathway there could be yield gains of up to 50%. It has been proposed that the multiple metabolic and developmental modifications associated with C4 photosynthesis are underpinned by relatively few master regulators that have allowed the evolution of C4 photosynthesis more than 60 times in flowering plants. Here we identify a component of one such regulator that consists of a pair of cis-elements located in coding sequence of multiple genes that are preferentially expressed in bundle sheath cells of C4 leaves. These motifs represent duons as they play a dual role in coding for amino acids as well as controlling the spatial patterning of gene expression associated with the C4 leaf. They act to repress transcription of C4 photosynthesis genes in mesophyll cells. These duons are also present in the C3 model Arabidopsis thaliana, and, in fact, are conserved in all land plants and even some algae that use C3 photosynthesis. C4 photosynthesis therefore appears to have coopted an ancient regulatory code to generate the spatial patterning of gene expression that is a hallmark of C4 photosynthesis. This intragenic transcriptional regulatory sequence could be exploited in the engineering of efficient photosynthesis of crops.
Photosynthesis forms the basis of life on earth. When plants moved onto land they inherited a photosynthetic system developed by bacteria in which ribulose bisphosphate carboxylase oxygenase (RuBisCO) generates phosphoglyceric acid (PGA) (1). As PGA contains three carbon atoms, this is referred to as C3 photosynthesis. A side reaction of RuBisCO fixes O2 rather than CO2 and generates the toxic compound phosphoglycolate. Although plants use photorespiration to remove phosphoglycolate, both carbon and energy are lost in the process (2). Around 30 Mya, some species evolved a system in which CO2 is concentrated around RuBisCO such that oxygenation is minimized, and photosynthetic efficiency increases by around 50% (3, 4). These species now represent the most productive vegetation on the planet (5, 6), and because they initially generate a C4 acid in the photosynthetic process, are known as C4 plants.
The C4 mechanism depends on spatial separation of photosynthetic reactions, most commonly between mesophyll (M) and bundle sheath (BS) cells. How this complex process has evolved in over 66 independent lineages remains a mystery. The finding that expression of multiple NAD-dependent MALIC ENZYME (NAD-ME) genes in the BS of C4 Gynandropsis gynandra is dependent on coding sequence (7) hinted at the importance of exons in this process, which are more highly conserved than intragenic sequences and therefore a potential hotspot underpinning repeated evolution. Although genome-wide studies commonly report transcription factor binding within genes (8, 9), there is little functional validation to support these findings and their general importance remains unclear.
Results
To better understand mechanisms responsible for generating preferential expression in BS cells of C4 plants, analysis was focused on the coding region of GgNAD-ME1. Although a 240-nucleotide fragment under control of the constitutive CaMV35S promoter confers BS accumulation in G. gynandra (Fig. 1 A and B and Figs. S1 and S2) it was unclear whether this was due to transcriptional and posttranscriptional mechanisms. These can be distinguished using an antisense construct that maintains DNA sequence, but when transcribed generates a complementary mRNA. An antisense construct under the constitutive CaMV35S promoter maintained preferential accumulation in the BS (Fig. 1 A and B), indicating that DNA sequence is recognized by trans-acting factors regardless of the orientation of the sequence. The preferential rather than exclusive accumulation of GUS in BS cells reported here is consistent with quantification of transcripts encoding components of the C4 cycle in BS and M cells of C4 leaves (7, 10) and also previous analysis of elements controlling gene expression (11–13). Thus, the coding sequence of GgNAD-ME1 controls transcription, and as it suppresses activity of the constitutive 35SCaMV promoter in M cells, the simplest explanation is that this region interacts with a repressive transcription factor.
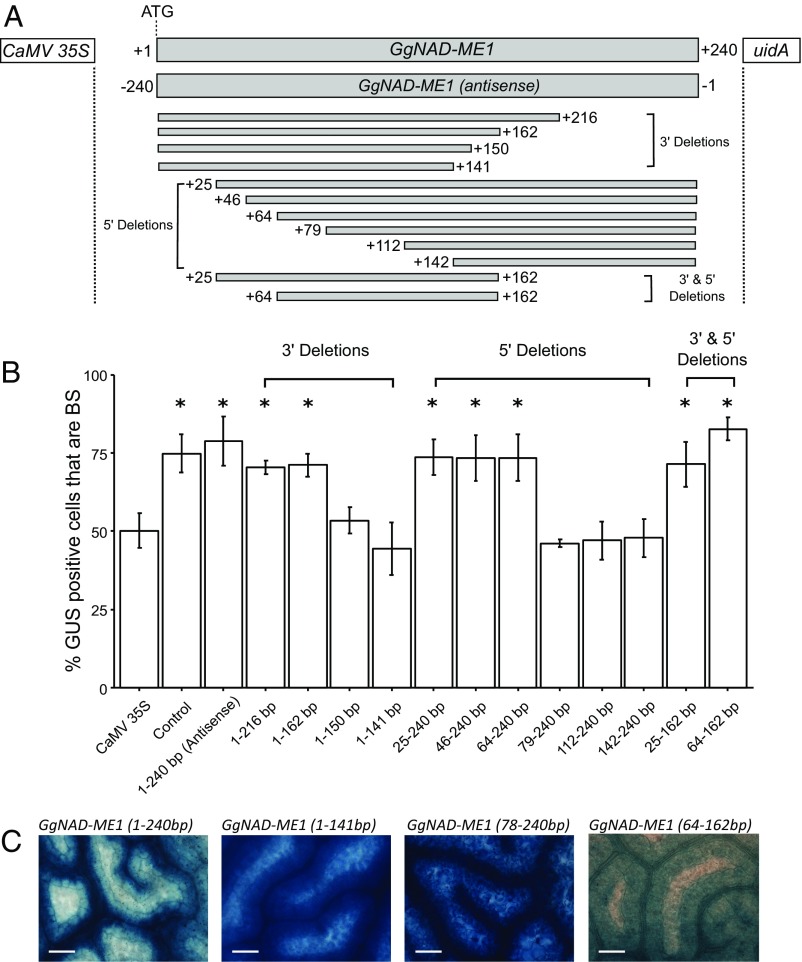
Fig. 1.
Two regions within the coding sequence of GgNAD-ME1 are necessary for preferential gene expression in the bundle sheath (BS). An antisense construct, as well as a deletion series from the 5′ and 3′ ends of GgNAD-ME1 coding sequence were translationally fused to the uidA reporter under the control of the CaMV35S promoter (A). Percentage of cells containing GUS in the BS after microprojectile bombardment of G. gynandra leaves. Bars represent the percentage of stained cells in BS cells, error bars denote the SE. Statistically significant differences with *P values <0.05 and CI = 95% determined by a one-tailed t test (B). GUS in G. gynandra transformants containing uidA fused to 1–240, 1–141, 79–240, and 64–162 base pairs from the translational start site of GgNAD-ME1 (C). (Scale bars, 100 µm.)
Without definition of the motifs within NAD-ME genes specifying expression in the BS, it was not possible to determine whether they control expression of additional genes, or to understand whether the same elements are used in other species. To identify specific nucleotides responsible for BS expression, a deletion series was generated (Fig. 1A). Deletion of 24 or 78 nucleotides from the 3′ end did not affect preferential accumulation in BS cells (Fig. 1 A and B) but removal of 90 nucleotides did (Fig. 1). Similarly, deletion of the first 63 nucleotides from the 5′ end did not abolish preferential accumulation of GUS in BS cells, but removing 78 nucleotides did (Fig. 1). A fragment incorporating bases 64–162 was sufficient for preferential accumulation in the BS after microprojectile bombardment (Fig. 1B) and stable transformation (Fig. 1C and Fig. S2). We conclude that one region composed of nucleotides TTGGGTGAA (64–79 downstream of the translational start codon) and another of GATCCTTG (141–162 nucleotides downstream of the translational start codon) are necessary for preferential accumulation of GgNAD-ME1 in BS cells of C4 G. gynandra. These two regions are separated by 75 nucleotides and will hereafter be referred to as Bundle Sheath Motif 1a (BSM1a) and Bundle Sheath Motif 1b (BSM1b).
To refine efforts to identify the presence of this cis-regulatory logic in other genes, each motif was subjected to site-directed mutagenesis and the spacer separating BSM1a and BSM1b was replaced with exogenous sequence (Fig. S3). This showed that both regions are necessary but that the intervening sequence is not required for preferential accumulation of GUS in BS cells (Fig. S3). The fact that BSM1a and BSM1b alone are sufficient to repress transcription in M cells indicates that they do not function as promoters for long antisense mRNAs. Although the exact sequence separating BSM1a and BSM1b does not impact on their function, the distance separating them does. BSM1a and BSM1b do not generate preferential accumulation in BS cells when fused together directly, or when separated by 999 nucleotides (Fig. S4). However, when the intervening sequence was between 240 and 550 nucleotides, preferential accumulation in BS cells occurred (Fig. S4). We conclude that in the coding region of NAD-ME1, two sequences separated by a spacer are necessary and sufficient to generate strong expression in BS cells.
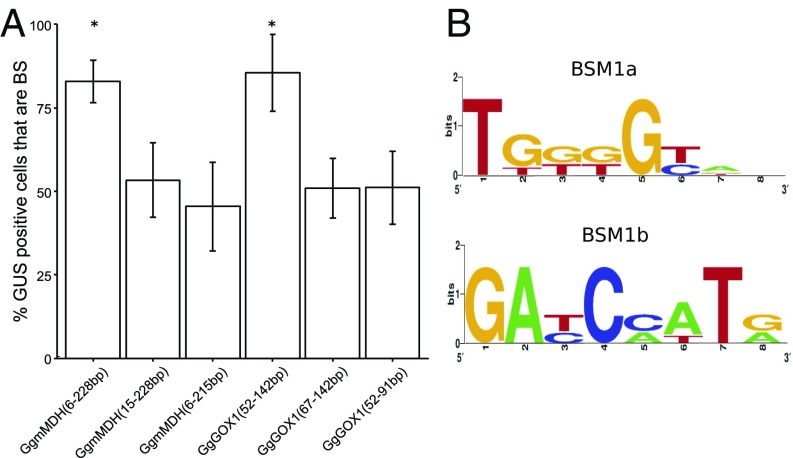
Although thousands of genes are differentially expressed between M and BS cells of C4 plants (11–14), to our knowledge no DNA motifs that determine the patterning of more than one gene in BS cells have been identified. To test whether BSM1a and BSM1b operate more widely to generate preferential expression in BS cells, coding sequence of other genes relevant to C4 photosynthesis was scanned. Sequences similar to BSM1a and BSM1b were identified in two such genes encoding mitochondrial MALATE DEHYDROGENASE (mMDH) and GLYCOLATE OXIDASE 1 (GOX1). Fragments from mMDH and GOX1 containing each motif were sufficient to drive BS accumulation of GUS (Fig. 2A), and when either was deleted preferential accumulation in BS cells was lost (Fig. 2A). The identification of BSM1a and BSM1b in these additional genes allowed consensus sequences to be defined (Fig. 2B). These data suggest that multiple gene families involved in C4 photosynthesis and photorespiration have been recruited into BS expression using a regulatory network based on these two motifs.
Fig. 2.
BSM1a and BSM1b drive the expression of additional genes in C4 photosynthesis and photorespiration. Sequences similar to BSM1a and BSM1b were identified in coding sequences of mMDH and GOX1 genes of G. gynandra. Deleting the motifs resulted in the loss of preferential accumulation of GUS in the bundle sheath (BS) (A). Consensus sequences were defined from motifs operational in NAD-ME1, mMDH, and GOX1 (B). Error bars denote the SE. Statistically significant differences with *P values <0.05 and CI = 95% determined by a one-tailed t test.
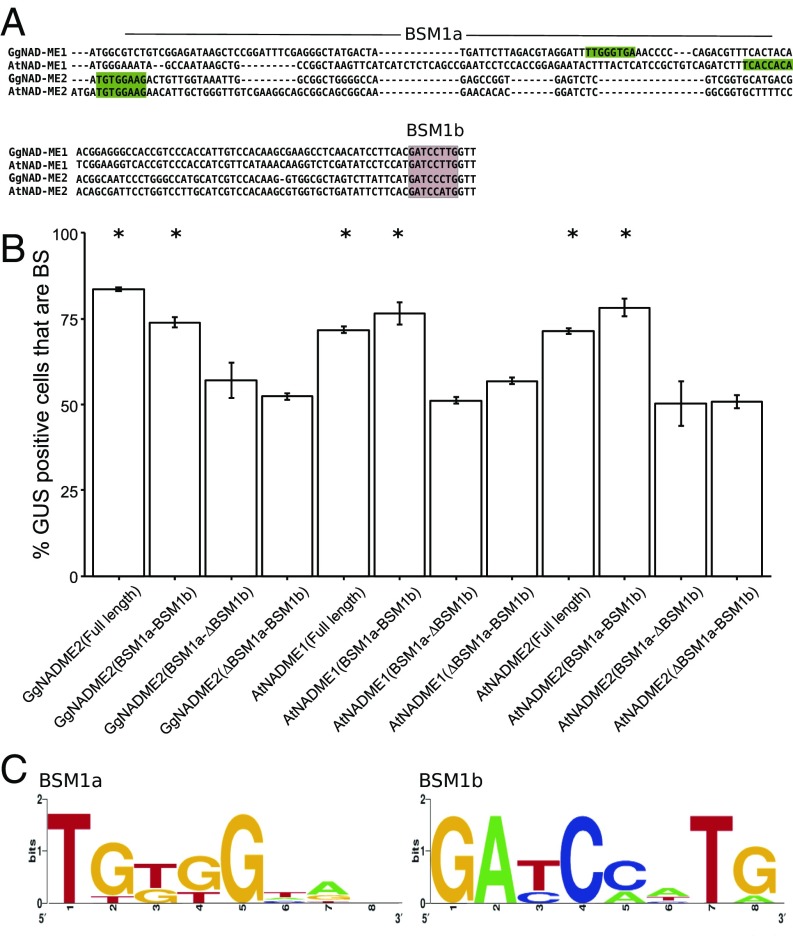
Sequences similar to BSM1a and BSM1b were identified near the predicted translational start sites of GgNAD-ME2, but also in the orthologs AtNAD-ME1 and AtNAD-ME2 from C3 Arabidopsis thaliana (Fig. 3A). In these additional genes, BSM1a is located in the mitochondrial transit peptide and its position varies relative to the translational start site. BSM1b is located in the mature processed protein and its position appears invariant (Fig. 3A). When either motif was removed preferential accumulation in BS cells was lost (Fig. 3B). Thus, sequences defined by BSM1a and BSM1b from eight genes (Fig. 3C and Fig. S6A) are necessary and sufficient to generate BS expression in the C4 leaf. Although synonymous substitutions to the BSM1a and BSM1b sequences from GgNAD-ME1 failed to abolish preferential expression in BS cells (Fig. S5) amino acid sequence encoded by BSM1a and BSM1b vary considerably for two reasons. First, because BSM1a is found on either DNA strand (Fig. S6). Second, in the eight genes studied, amino acids encoded by each motif differ because codons are not in identical frames (Fig. S6). Combined with the fact that both BSM1a and BSM1b are functional when present in antisense orientation, this variation in amino acid sequence supports the notion that these motifs do not act posttranslationally through amino acid sequence, but rather function transcriptionally via transcription factor binding.
Fig. 3.
Functional versions of BSM1a and BSM1b are present in additional NAD-MEs. BSM1a and BSM1b are found in GgNAD-ME2 and in orthologs of GgNAD-ME1 and 2 from the C3 species A. thaliana (A). Translational fusions carrying these fragments confer bundle sheath (BS) preferential expression in G. gynandra leaves. When BSM1a or BSM1b were removed this pattern of GUS was lost (B). Consensus sequences derived from all versions of BSM1a and BSM1b tested experimentally (C). Error bars denote the SE. Statistically significant differences with *P values <0.05 and CI = 95% determined by a one-tailed t test.
To provide orthogonal evidence that BSM1a and BSM1b are indeed the targets for transcription factors binding in vivo, two additional approaches were pursued. First, the consensus motifs for BSM1a and BSM1b were used to search databases that document transcription factor binding specificities (15–17). Six members of the MYOBLASTOMA (MYB) family have been reported to bind sequences that are similar but not identical to the BSM1a motif (P < e−3, Dataset S6) but there were no clear matches for BSM1b. It is therefore possible that a member of the MYB family binds BSM1a, but the cognate factor binding BSM1b remains unclear. Second, DNaseI sequencing of G. gynandra was used to define regions of chromatin that are accessible for transcription factor binding. Genome-wide DNaseI sequencing has previously been used to define the landscape of DNaseI hypersensitive sites (DHSs) as well as digital genomic footprints (DGFs) that mark sequence motifs subject to transcription factor binding (9, 16, 18–20). Consistent with the reporter analysis described above, DNaseI analysis showed that BSM1a and BSM1b from GgNAD-ME1, GgNAD-ME2, GgmMDH, and GgGOX1 were associated with DHSs and so accessible to trans-acting factors (Fig. S7). For GgNAD-ME1, GgmMDH, and GgGOX1, these DHSs were present in germinating seedlings grown in the dark, while for GgNAD-ME2, the DHS appeared within 4 h of transfer from dark to light. The four DHSs remained in adult leaf tissue (Fig. S7). Moreover, BSM1a and BSM1b either overlapped with or were adjacent to DGFs present in these DHSs (Fig. S7). While DNA sequence bound by a transcription factor can be found in the center of a DGF, variation in how individual transcription factors bind and contort DNA and thus make sequence available for DNaseI digestion result in different cleavage profiles (21–24). The similarity of BSM1a to a MYB binding site, the location of both BSM1a and BSM1b in DNA accessible for transcription factor binding, and the presence of DGFs in close proximity to each sequence, support the contention that these motifs function as duons and recruit transcription factors that lead to preferential expression in the BS of C4 G. gynandra.
In contrast, analysis of publicly available DNaseI data for Arabidopsis (9, 21) indicated that for AtNAD-ME1, AtNAD-ME2, or AtGOX1, BSM1a and BSM1b are not located within DHSs (Fig. S8). These data are consistent with a model in which BSM1a and BSM1b are present in the ancestral C3 state, but their relative inaccessibility inhibits binding by cognate transcription factors, and so cell-specific gene expression is not achieved. In contrast, in the C4 leaf, BSM1a and BSM1b are accessible to transcription factor binding and this leads to preferential gene expression in BS compared with M cells.
To determine the extent to which BSM1a and BSM1b could control gene expression more widely, their distribution across the genome was quantified. More than 4,000 genes contained both motifs in G. gynandra, but fewer than half of these genes were separated by 35–550 nucleotides required to repress expression in M cells (Dataset S5). Compared with randomly generated hits of the same size, both BSM1a and BSM1b were overrepresented in coding sequence DHSs (Fig. S9), supporting the notion that they carry out a specialized role in transcription factor binding within gene bodies. Of the genes containing BSM1a and BSM1b separated by 35–550 nucleotides, 250 are found in genes that are preferentially expressed in BS cells of G. gynandra. We therefore estimate that these duons control BS expression of between four and 250 genes in the C4 leaf. It was noticeable that BSM1a and BSM1b were also widespread in C3 A. thaliana (Dataset S5). The simplest explanation for the presence of BSM1a and BSM1b in genes that are not preferentially expressed in BS cells may be that they contain additional cis-elements that override their function. The highly combinatorial nature of transcription factor binding and impact that this has on cell-specific gene expression in the root has previously been documented (25). It is also possible that chromatin environment also influences the function of these motifs and that this can repress their role in restricting expression to BS cells.
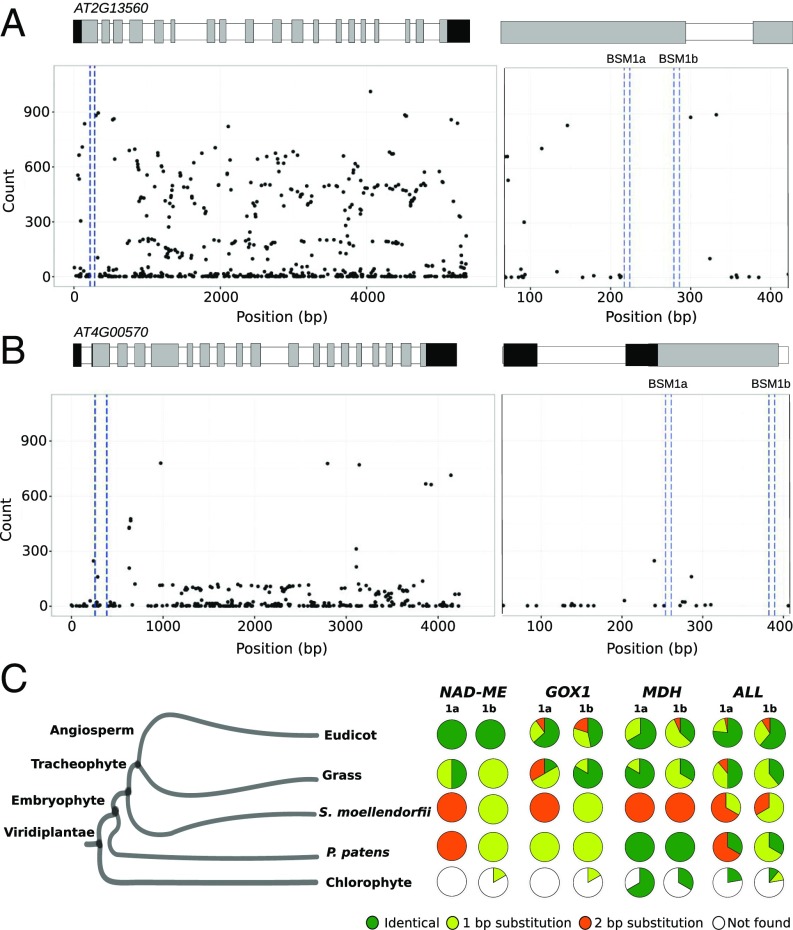
We next investigated the extent to which BSM1a and BSM1b are conserved across 1,135 wild inbred A. thaliana accessions (26). No single nucleotide polymorphisms (SNPs) were detected within either BSM1a or BSM1b (Fig. 4 A and B). Despite the lack of chromatin accessibility observed in the orthologs from A. thaliana (Fig. S7), the BSM1a and BSM1b motifs and sequence around them are highly conserved, suggesting a role for these duons in C3 A. thaliana that is independent of cell preferential expression in the leaf. To determine whether these motifs are also found more widely in NAD-ME genes, homologs were retrieved from all sequenced land plants (Fig. S10). All dicotyledons contained at least one NAD-ME gene carrying sequences that define BSM1a and BSM1b (Fig. 4C). In monocotyledons, BSM1a was completely conserved in rice, Brachypodium, and Panicum. Although BSM1b showed one nucleotide substitution in all monocotyledonous genomes available, its conservation in spikemoss and moss argues for its being ancient (Fig. 4C and Fig. S10). Both BSM1a and BSM1b are highly conserved in GOX1 and MDH genes of land plants, but BSM1b appears more ancient, as it is found in GOX1 genes from all land plants and even chlorophyte algae.
Fig. 4.
BSM1a and BSM1b are highly conserved in land plants. Single nucleotide polymorphisms (SNPs) in AtNAD-ME1 (A) and AtNAD-ME2 (B) genes from 1,135 wild inbred A. thaliana accessions. (Left) Positions of BSM1a and BSM1b are highlighted by dashed blue lines; UTRs, exons, and introns are denoted by black, gray, and white bars, respectively, on the x axis. (Right) Expanded area representing exon 1, intron 1, and exon 2 is shown, with BSM1a and BSM1b marked within the blue dashed lines. For both genes, no SNPs were detected in either motif. (C) The presence of each motif was investigated in gene sequences of NAD-ME1, mMDH, and GOX1 retrieved from 44 species in Phytozome (v10.1) (51). Each pie chart shows the percentage of motif instances that were identical (green), or had one base pair (yellow), two base pair (orange) substitutions, or no similarity (white) detected.
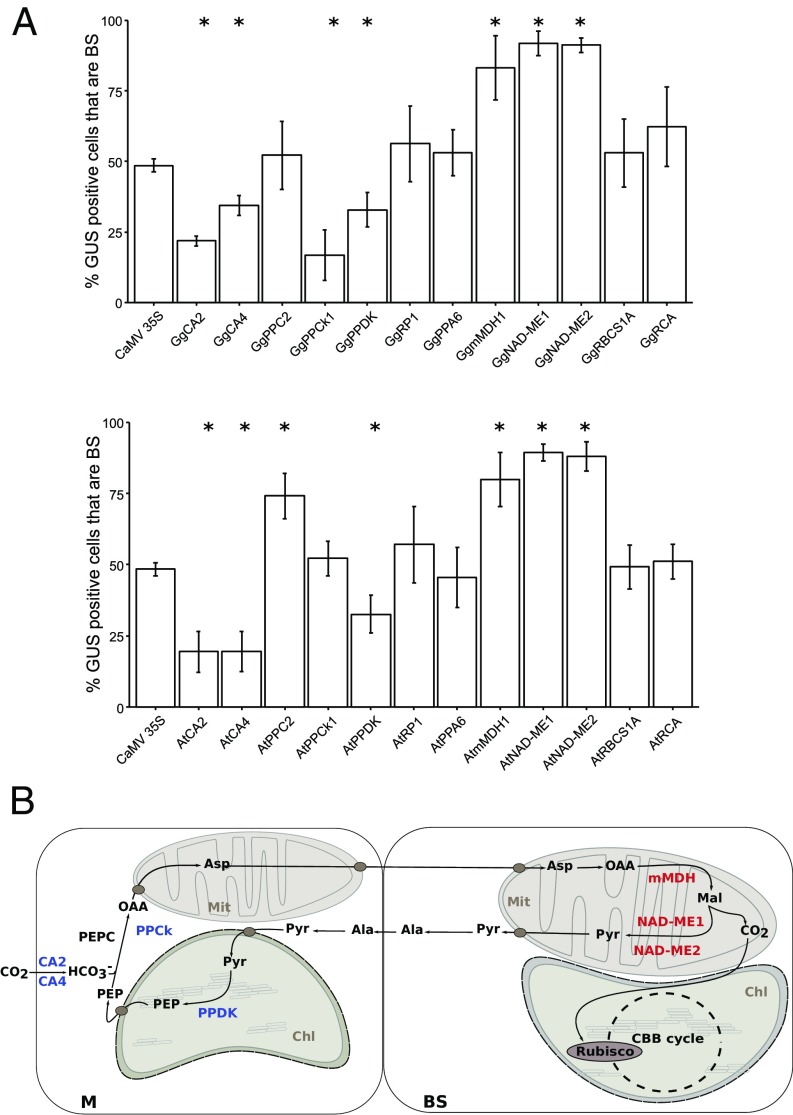
In view of the importance of exonic sequences in regulating BS accumulation of NAD-ME, MDH, and GOX1, the wider importance of exonic regulation in controlling C4 gene expression was investigated. Seven of 12 core C4 cycle genes contained regulatory elements located in transcribed sequence that were sufficient to generate preferential accumulation in M or BS cells (Fig. 5). Except for PPCk1, which encodes the kinase that posttranslationally modifies PEPC in M cells of C4 plants, coding sequences from orthologs of these C4 genes in C3 A. thaliana led to preferential expression in either M or BS cells of the C4 leaf (Fig. 5). Overall, these data indicate that spatial patterning of gene expression in the C4 leaf is largely derived from cis-regulatory elements present in genes found in the ancestral C3 state.
Fig. 5.
Intragenic regulatory sequences play a major role controlling C4 photosynthesis genes. Coding sequences encoding core proteins of the C4 pathway from G. gynandra together with orthologs from A. thaliana were translationally fused to uidA and placed under control of the CaMV35S promoter. After introduction into G. gynandra leaves by microprojectile bombardment, mesophyll preferential expression of CA2, CA4, PPDK, and PPCk1, together with bundle sheath (BS) preferential expression of mMDH, NAD-ME1, and NAD-ME2 were observed (A). With the exception of PPCk, these regulatory elements are conserved in orthologues from A. thaliana (B). The contribution of intragenic sequences controlling gene regulation of the C4 pathway is summarized in C, CA2, CA4, PPDK, and PPCk1 (blue) and mMDH, and NAD-ME1 and -2 (red) denote genes where intragenic sequences control cell preferential gene expression. Error bars denote the SE. Statistically significant differences with *P values <0.05 and CI = 95% determined by a one-tailed t test.
Discussion
The data presented here, combined with previous reports (7, 27, 28), portray an overview of the contribution that untranslated regions (UTRs) and coding sequences make to the generation of cell-specific gene expression in leaves of C4 G. gynandra. Seven of the 12 core C4 cycle genes possess regulatory elements in their transcript sequences that are sufficient for preferential accumulation in either M or BS cells of the C4 leaf. These data strongly imply that, in addition to promoters being involved in generating cell specificity in C4 leaves (29, 30), coding sequences and UTRs play a widespread role in the preferential accumulation of C4 transcripts to either M or BS cells. It remains to be seen whether this high degree of regulation from genic sequence is critical for the spatial control of gene expression in other tissues and other species. However, as genome-wide studies of transcription factor recognition sites in organisms as diverse as A. thaliana and human cells (9, 19) have reported that significant binding occurs in genic sequence, we anticipate many more examples of spatial regulation of gene expression being associated with cis-elements outside of promoter sequences.
The accumulation of GgNADME1, GgNADME2, mMDH, and GOX1 transcripts in BS cells is dependent on the cooperative function of two cis-elements that are separated by a spacer sequence, all of which are located in the first exon of these genes. There are a number of options relating to how such regulatory sequence found within genes could operate. For example, they could impact on transcription by either interacting with a transcription factor or by acting as promoters to drive expression of an antisense RNA. In addition, they could act posttranscriptionally via the RNA molecule once the gene is transcribed. As the BSM1a and BSM1b motifs without additional sequence are sufficient to restrict gene expression to BS, this excludes them acting as promoters to antisense RNA molecules. Furthermore, two pieces of evidence indicate that they do not function posttranscriptionally via the transcripts generated from the endogenous gene. First, in different genes BSM1a is found on each strand of DNA so that it would not be present in all of the mRNAs that accumulate preferentially in BS cells. Second, an antisense construct inserted as a transgene into G. gynandra still generates BS expression again indicating that the transcribed RNA is not involved. The simplest hypothesis is that BSM1a and BSM1b therefore act transcriptionally to suppress activity of the constitutive 35SCaMV promoter in M cells. The proposal that BSM1a and BSM1b act as duons does not exclude additional levels of regulation contributing to the cell-specific accumulation of proteins in the C4 leaf. Indeed, it is notable that these cis-elements alone do not completely restrict accumulation of GUS to BS cells, and so posttranscriptional and/or posttranslational regulation may well act to reinforce their function.
BSM1a and BSM1b are conserved both in their sequences and in the number of nucleotides that separates them. In orthologous NAD-ME genes from C3 A. thaliana, which diverged from the Cleomaceae ∼38 Mya (31, 32), although these motifs are present, they are not sufficient to generate cell preferential expression in the C3 leaf (7). This finding indicates that for NAD-ME genes to be preferentially expressed in BS cells of C4 plants, a change in the behavior of one or more trans-factors was a fundamental event. At least in G. gynandra, evolution appears to have repeatedly made use of cis-elements that exist in genes of C3 species that are orthologous to those recruited into C4 photosynthesis (7, 27, 28, 33). The alteration in trans-factors such that they recognize ancestral elements in cis in the M or BS therefore appears to be an important and common mechanism associated with evolution of the highly complex C4 system.
The dual role of exons in protein coding as well as the regulation of gene expression has received significant attention in vertebrates (19, 34–37). Although 11% of transcription factor binding sites are located in exonic sequence in A. thaliana (9), to our knowledge, the identification of BSM1a and BSM1b represents the first functional evidence for cis-elements in plant exons. The fact that these motifs are present in C3 A. thaliana, and, in fact, also found in the genomes of many land plants and some chlorophyte algae, indicates that these duons play ancient and conserved roles in photosynthetic organisms. The role of such regulatory elements within coding sequences has previously been proposed to be associated with constraints on both protein coding function and codon bias. For example, mutation to these cis-elements could be deleterious to both the correct function of the protein, but also to codon usage and so translational efficiency (38–40). If this is the case, BSM1a and BSM1b could be highly conserved across deep phylogeny because of strong selection pressure on these elements that impact on translation, and this conservation is then coopted to also regulate transcription during the evolution of C4 photosynthesis to generate cell-specific gene expression. Establishing the role of BSM1a and BSM1b in C3 plants would provide insight into the extent to which their role has altered during the transition from C3 to C4 photosynthesis.
Duons under strong selection pressure may represent a rich resource of cis-elements upon which the C4 pathway has evolved. Although C4 photosynthesis is a complex trait that requires multiple changes to gene expression, the repeated evolution of C4 species across multiple plant lineages suggests that a relatively low number of changes may be required to acquire the C4 syndrome (41–43). A single C4 master switch has been proposed (43) but despite multiple comparative transcriptomic studies (13, 44–46), there is as yet no evidence for it. The only transcription factor associated with photosynthesis in C4 species is Golden-like1 (47), which controls expression of genes associated with chlorophyll biosynthesis and the light harvesting complexes in both C3 and C4 species (48). Given the repeated and highly convergent evolution of the C4 pathway, as well as evidence that separate lineages can arrive at the C4 state via different routes (49), it appears more plausible that C4 photosynthesis made use of a number of gene subnetworks. This is now supported by a number of findings. First, just as core photosynthesis genes encoding the light harvesting complexes and the Calvin–Benson–Bassham cycle are regulated by light, the vast majority of genes that encode proteins of the C4 cycle in C3 A. thaliana are also regulated by light signaling; yet, during the evolution of C4 photosynthesis, there was a significant gain of responsiveness to chloroplast signaling (50). Second, it has been suggested that evolution of the C4 pathway is associated with the recruitment of developmental motifs into leaves that in C3 species operate in roots (46). Lastly, the identification of the cis-element MEM2 (28), which controls preferential expression of multiple genes in C4 M cells, and now BSM1a and BSM1b in four different genes that are strongly expressed in BS cells, indicates that C4 evolution has made use of small-scale recruitment of gene subnetworks in both cell types.
In summary, the data indicate regulatory elements located in transcript sequences are of central importance in patterning gene expression in the C4 leaf. Expression of multiple genes in BS cells is regulated by highly conserved duons that appear to play ancient roles in photosynthetic organisms. Our data also indicate that evolution of the highly complex C4 trait is built upon ancient cis-regulatory architecture that is common to all land plants and some green algae.
Materials and Methods
G. gynandra seeds were germinated in the dark at 30 °C for 24 h. For microprojectile bombardment seedlings were then transferred to Murashige and Skoog (MS) medium with 1% (wt/vol) sucrose and 0.8% (wt/vol) agar (pH 5.8) and grown for a further 13 d in a growth room at 22 °C and 200 μmol⋅m−2⋅s−1 photon flux density (PFD) with a photoperiod of 16 h light. For DNaseI-seq, germinating seeds were placed in the dark to promote hypocotyl extension for 3 d or transferred to a growth cabinet maintained at 25 °C ± 0.5 °C, 60% relative humidity, ambient CO2, and 300 μmol⋅m−2⋅s−1 PFD with a photoperiod of 16 h light. For DNaseI analysis, tissue was flash frozen in liquid nitrogen and stored at −80 °C before processing. After bombardment or stable transformation, plant tissue was GUS stained and then chlorophyll was removed in 70% (vol/vol) ethanol.
Supplementary Material
Acknowledgments
We thank Chris Boursnell for help with data retrieval. I.R.-L. was supported by Consejo Nacional de Ciencia y Tecnologia and Biotechnology and Biological Sciences Research Council (BBSRC) Grant BB/L014130; S.J.B., by a 3to4 grant from the European Union and BB/I002243 from the BBSRC; G.R., by a Gates Cambridge Trust PhD Fellowship; P.S. and S.R.S. by Advanced European Research Council Grant 694733 Revolution (to J.M.H.); and B.P.W., by a BBSRC PhD Studentship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the Sequence Read Archive (accession no. SRP125696).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1720576115/-/DCSupplemental.
References
- 1.Anbar AD, et al. A whiff of oxygen before the great oxidation event? Science. 2007;317:1903–1906. doi: 10.1126/science.1140325. [DOI] [PubMed] [Google Scholar]
- 2.Bauwe H, Hagemann M, Fernie AR. Photorespiration: Players, partners and origin. Trends Plant Sci. 2010;15:330–336. doi: 10.1016/j.tplants.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Hatch MD, Slack CR. Photosynthesis by sugar-cane leaves. A new carboxylation reaction and the pathway of sugar formation. Biochem J. 1966;101:103–111. doi: 10.1042/bj1010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sage RF, Wedin D, Li M. C4 Plant Biology. Elsevier/North–Holland; New York: 1999. The biogeography of C4 photosynthesis: Patterns and controlling factors; pp. 313–373. [Google Scholar]
- 5.Sage RF. Tansley review: The evolution of C4 photosynthesis. New Phytol. 2004;161:30. doi: 10.1111/j.1469-8137.2004.00974.x. [DOI] [PubMed] [Google Scholar]
- 6.Ray DK, Ramankutty N, Mueller ND, West PC, Foley JA. Recent patterns of crop yield growth and stagnation. Nat Commun. 2012;3:1293. doi: 10.1038/ncomms2296. [DOI] [PubMed] [Google Scholar]
- 7.Brown NJ, et al. Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science. 2011;331:1436–1439. doi: 10.1126/science.1201248. [DOI] [PubMed] [Google Scholar]
- 8.Stergachis AB, et al. Exonic transcription factor binding directs codon choice and affects protein evolution. Science. 2013;342:1367–1372. doi: 10.1126/science.1243490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan AM, et al. Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep. 2014;8:2015–2030. doi: 10.1016/j.celrep.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 10.Xu T, Purcell M, Zucchi P, Helentjaris T, Bogorad L. TRM1, a YY1-like suppressor of rbcS-m3 expression in maize mesophyll cells. Proc Natl Acad Sci USA. 2001;98:2295–2300. doi: 10.1073/pnas.041610098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.John CR, Smith-Unna RD, Woodfield H, Covshoff S, Hibberd JM. Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiol. 2014;165:62–75. doi: 10.1104/pp.114.238667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang Y-M, et al. Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol. 2012;160:165–177. doi: 10.1104/pp.112.203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li P, et al. The developmental dynamics of the maize leaf transcriptome. Nat Genet. 2010;42:1060–1067. doi: 10.1038/ng.703. [DOI] [PubMed] [Google Scholar]
- 14.Aubry S, Kelly S, Kümpers BM, Smith-Unna R, Hibberd JM. Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genet. 2014;10:e1004365. doi: 10.1371/journal.pgen.1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franco-Zorrilla JM, et al. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA. 2014;111:2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Malley RC, et al. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016;165:1280–1292. doi: 10.1016/j.cell.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang T, Marand AP, Jiang J. PlantDHS: A database for DNase I hypersensitive sites in plants. Nucleic Acids Res. 2016;44:D1148–D1153. doi: 10.1093/nar/gkv962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song L, Crawford GE. Dnase-seq: A high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010;2010:pdb.prot5384. doi: 10.1101/pdb.prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stergachis AB, et al. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature. 2014;515:365–370. doi: 10.1038/nature13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sung M-H, Guertin MJ, Baek S, Hager GL. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol Cell. 2014;56:275–285. doi: 10.1016/j.molcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piper J, et al. Wellington: A novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res. 2013;41:e201. doi: 10.1093/nar/gkt850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulakovskiy IV, Favorov AV, Makeev VJ. Motif discovery and motif finding from genome-mapped DNase footprint data. Bioinformatics. 2009;25:2318–2325. doi: 10.1093/bioinformatics/btp434. [DOI] [PubMed] [Google Scholar]
- 25.Sparks EE, et al. Establishment of expression in the SHORTROOT-SCARECROW transcriptional cascade through opposing activities of both activators and repressors. Dev Cell. 2016;39:585–596. doi: 10.1016/j.devcel.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.1001 Genomes Consortium 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016;166:481–491. doi: 10.1016/j.cell.2016.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kajala K, et al. Multiple Arabidopsis genes primed for recruitment into C4 photosynthesis. Plant J. 2012;69:47–56. doi: 10.1111/j.1365-313X.2011.04769.x. [DOI] [PubMed] [Google Scholar]
- 28.Williams BP, et al. An untranslated cis-element regulates the accumulation of multiple C4 enzymes in Gynandropsis gynandra mesophyll cells. Plant Cell. 2016;28:454–465. doi: 10.1105/tpc.15.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gowik U, et al. cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell. 2004;16:1077–1090. doi: 10.1105/tpc.019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheen J. C4 gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:187–217. doi: 10.1146/annurev.arplant.50.1.187. [DOI] [PubMed] [Google Scholar]
- 31.Schranz ME, Mitchell-Olds T. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell. 2006;18:1152–1165. doi: 10.1105/tpc.106.041111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Couvreur TLP, et al. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae) Mol Biol Evol. 2010;27:55–71. doi: 10.1093/molbev/msp202. [DOI] [PubMed] [Google Scholar]
- 33.Reyna-Llorens I, Hibberd JM. Recruitment of pre-existing networks during the evolution of C4 photosynthesis. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160386. doi: 10.1098/rstb.2016.0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lang G, Gombert WM, Gould HJ. A transcriptional regulatory element in the coding sequence of the human Bcl-2 gene. Immunology. 2005;114:25–36. doi: 10.1111/j.1365-2567.2004.02073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goren A, et al. Comparative analysis identifies exonic splicing regulatory sequences–The complex definition of enhancers and silencers. Mol Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 36.Tümpel S, Cambronero F, Sims C, Krumlauf R, Wiedemann LM. A regulatory module embedded in the coding region of Hoxa2 controls expression in rhombomere 2. Proc Natl Acad Sci USA. 2008;105:20077–20082. doi: 10.1073/pnas.0806360105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong X, et al. Exonic remnants of whole-genome duplication reveal cis-regulatory function of coding exons. Nucleic Acids Res. 2010;38:1071–1085. doi: 10.1093/nar/gkp1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson M, et al. Codon usage can affect efficiency of translation of genes in Escherichia coli. Nucleic Acids Res. 1984;12:6663–6671. doi: 10.1093/nar/12.17.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci USA. 2010;107:3645–3650. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakahigashi K, et al. Effect of codon adaptation on codon-level and gene-level translation efficiency in vivo. BMC Genomics. 2014;15:1115. doi: 10.1186/1471-2164-15-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinha NR, Kellogg EA. Parallelism and diversity in multiple origins of C4 photosynthesis in grasses. Am J Bot. 1996;83:1458–1470. [Google Scholar]
- 42.Hibberd JM, Sheehy JE, Langdale JA. Using C4 photosynthesis to increase the yield of rice-rationale and feasibility. Curr Opin Plant Biol. 2008;11:228–231. doi: 10.1016/j.pbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Westhoff P, Gowik U. Evolution of C4 photosynthesis–Looking for the master switch. Plant Physiol. 2010;154:598–601. doi: 10.1104/pp.110.161729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brautigam A, et al. An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 2011;155:142–156. doi: 10.1104/pp.110.159442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aubry S, Kelly S, Kümpers BMC, Smith-Unna RD, Hibberd JM. Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genet. 2014;10:e1004365. doi: 10.1371/journal.pgen.1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Külahoglu C, et al. Comparative transcriptome atlases reveal altered gene expression modules between two Cleomaceae C3 and C4 plant species. Plant Cell. 2014;26:3243–3260. doi: 10.1105/tpc.114.123752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rossini L, Cribb L, Martin DJ, Langdale JA. The maize golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell. 2001;13:1231–1244. doi: 10.1105/tpc.13.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waters MT, et al. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams BP, Johnston IG, Covshoff S, Hibberd JM. Phenotypic landscape inference reveals multiple evolutionary paths to C4 photosynthesis. Elife. 2013;2:e00961. doi: 10.7554/eLife.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burgess SJ, et al. Ancestral light and chloroplast regulation form the foundations for C4 gene expression. Nat Plants. 2016;2:16161. doi: 10.1038/nplants.2016.161. [DOI] [PubMed] [Google Scholar]
- 51.Goodstein DM, et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 53.Newell CA, et al. Agrobacterium tumefaciens-mediated transformation of Cleome gynandra L., a C(4) dicotyledon that is closely related to Arabidopsis thaliana. J Exp Bot. 2010;61:1311–1319. doi: 10.1093/jxb/erq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bailey TL, et al. MEME SUITE : Tools for motif discovery and searching. Conflict. 2009;37:202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gendrel AV, Lippman Z, Martienssen R, Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.