Significance
Polyubiquitination is critically important for many cellular functions from protein degradation to signal transduction. TNF receptor-associated factor 6 (TRAF6) is a RING-type ubiquitin ligase that promotes polyubiquitination required for the signaling of many innate immune receptors. How TRAF6 uses its RING dimer to facilitate polyubiquitination remains elusive. In this paper, we mapped TRAF6 residues on both subunits of the TRAF6 dimer that are required for this process via allosteric activation. We found that in addition to the RING domain itself, residues outside the RING domain play indispensable roles. Moreover, our studies reveal common features for the activation of polyubiquitination by the RING family and related domains, which shed light on the functional mechanisms of both dimeric and monomeric ubiquitin ligases.
Keywords: TRAF6, ubiquitination, RING dimer, innate immunity, ubiquitin ligase
Abstract
Tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) plays a vital role in immune signal transduction pathways by acting as a ubiquitin ligase (E3) for Lys63-linked polyubiquitin chain synthesis. However, the detailed mechanism by which the TRAF6 RING dimer promotes ubiquitin transfer was unknown. Through structural modeling and biochemical analysis, we here show that the TRAF6 RING dimer employs a concerted allosteric mechanism using both subunits of the TRAF6 dimer to promote ubiquitin (Ub) transfer. In particular, we reveal the importance of the C-terminal extension of the TRAF6 RING domain that mediates trans-interactions with the donor-Ub. By analyzing structures and models of E3s in complex with Ub-loaded ubiquitin-conjugating enzymes (E2s), we further highlight the roles of N-terminal and C-terminal extensions beyond the bona fide RING domains in promoting Ub transfer through engagement with a donor-Ub in cis and in trans, respectively.
As a vital posttranslational modification, ubiquitination is achieved by three sequential enzymatic reactions: (i) ATP-dependent attachment of ubiquitin via a thioester bond to a ubiquitin-activating enzyme (E1); (ii) transfer of ubiquitin from E1 to the catalytic cysteine of a ubiquitin-conjugating enzyme (E2), producing ubiquitin-charged E2 (E2∼Ub); and (iii) transfer of ubiquitin from E2 to a substrate catalyzed by a ubiquitin ligase (E3) (1). So far, three major types of E3s have been characterized (2, 3). HECT ubiquitin ligases have catalytic cysteine residues and catalyze substrate ubiquitination via an E3∼Ub intermediate. In contrast, ubiquitin ligases containing RING or U-box domains do not have a catalytic cysteine. These two domains share a similar structural fold, except that RING domains contain two structurally important zinc ions, while U-box domains do not. Although an initial hypothesis was that RING/U-box E3s simply bridge E2s and substrates, it was subsequently shown that E3 modules lacking substrate-binding functions greatly stimulate the rate of ubiquitin release from E2s, suggesting promotion of E2 catalysis by RING/U-box domains (4). A third type of E3s contains the RING-between-RING (RBR) finger, which functions as RING/HECT hybrids. An RBR is composed of a classical RING (RING1), an In-Between-RING (IBR), and a RING2, in which RING1 interacts with an E2∼Ub conjugate and transfers Ub onto a cysteine on the RING2 for its conjugation to a substrate (5).
When structural and biochemical studies were carried out to address how RING-type E3s promote Ub transfer, it became apparent that RING domains bind E2 at a region far away from the E2 active sites (6). An earlier study identified clusters of coevolving and functionally linked residues within an E2 that span from its E3-binding site to the active site, suggesting that RING-type E3s activate E2s through allosteric communication (7). In support of this, E2 mutants were identified which are defective in being stimulated by E3s despite their abilities to bind to these E3s and to release ubiquitin at a basal rate (7). Additionally, a conserved hydrogen bond between RING/U-box domains and E2s appears to contribute to the catalytic activity of RING/U-box–type E3s in general (8). For many E3/E2 pairs, the conserved hydrogen bond is between an arginine in E3s and a main chain carbonyl oxygen in E2s.
Other lines of evidence, which are not mutually exclusive with E3-induced allosteric communication to E2, point to ubiquitin positioning as a mechanism of E2 activation. For dimeric RING/U-box E3s, which require homodimerization or heterodimerization for their ubiquitin ligase activity (1), ternary complex structures of E3/E2∼Ub complexes uncovered that the donor-Ub interacts with both subunits of a RING E3 dimer (in cis and in trans) (9–12). The interactions lock the conformation of the Ub tail, thereby activating the thioester bond for nucleophilic attack. For monomeric RING/U-box E3s with available structures of their E3/E2∼Ub complexes, it appears that the RING domains and regions outside the RING together provide sufficient contacts for donor-Ub positioning (13). Upon such E3/donor-Ub interactions, the E2s also specifically interact with the donor-Ub at and around its I44 hydrophobic patch (8, 14).
Previous biochemical and cellular studies showed that TRAF6 is a RING-type E3 that catalyzes Lys63-linked polyubiquitination using the Ubc13/Uev1A complex as the E2 (6). TRAF6 plays pleiotropic roles in the signal transduction of diverse receptor systems, including Toll-like receptors, members of the TNF receptor superfamily and the IL-1 receptor family, RIG-I–like receptors, and B-cell receptors (15, 16). It is composed of an N-terminal RING domain followed by at least four zinc finger domains (ZF1-4), a central coiled–coil domain, and a C-terminal TRAF-C domain (6) (Fig. 1A). Structural studies dissected that the N-terminal RING and ZF1 domains constitute the minimal unit for catalyzing Lys63 polyubiquitin chain synthesis in vitro, while the C-terminal TRAF-C domain interacts with receptors and adaptor proteins (6, 17). Upon receptor activation, TRAF6 is recruited to upstream signaling complexes and further activates downstream inhibitor of nuclear factor κB (IκB) kinase (IKK) or mitogen-activated protein (MAP) kinases, ultimately leading to the activation of nuclear factor κB (NF-κB) and activator protein 1 (AP-1) transcription factors for immune and inflammatory responses (18, 19). The crystal structures of both TRAF6 RING-ZF1 and of RING-ZF1-3 are homodimeric, and dimerization is required for polyubiquitination (6).
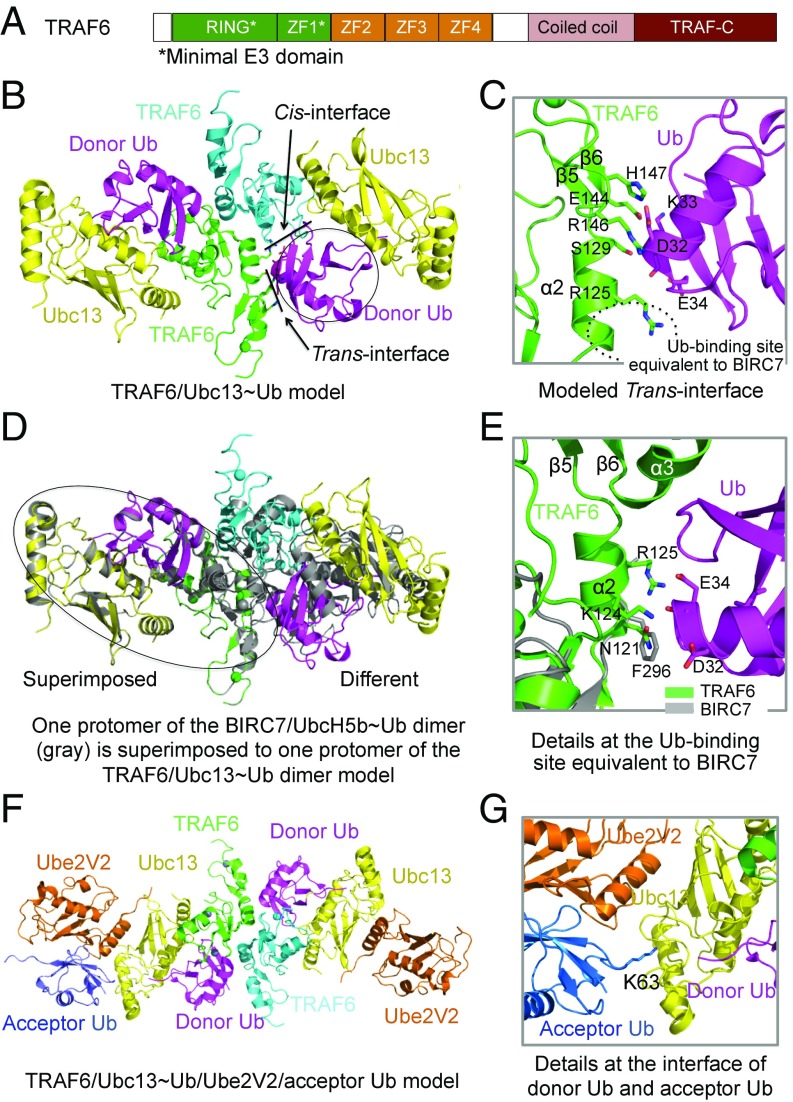
Fig. 1.
Structural model of the TRAF6 RING-ZF1 dimer in complex with the Ubc13∼Ub conjugate. (A) Domain organization of TRAF6. ZF1-ZF4, Zinc Fingers 1–4. (B) Ribbon diagram of the TRAF6/Ubc13-Ub ternary complex, showing the cis- and trans-interactions between RING-ZF1 and a donor-Ub. (C) Detailed interactions at the trans-interface between TRAF6 (green) and Ub (magenta). (D) Superposition of BIRC7/UbcH5b∼Ub onto TRAF6/Ubc13∼Ub. When one protomer of the BIRC7/UbcH5b∼Ub dimer is overlaid with that of the TRAF6/Ubc13∼Ub dimer, the symmetric protomers show different relative positions. (E) Detailed interactions at the Ub-binding site of TRAF6 (green) equivalent to that of BIRC7 (gray). (F) Ribbon diagram of the TRAF6/Ubc13∼Ub/Ube2V2/acceptor Ub complex, showing the structural basis for Lys63 Ub chain synthesis. (G) Detailed view of the donor Ub and acceptor Ub.
How dimeric TRAF6 mediates activation of the Ubc13∼Ub/Uev1A complex remains unexplored. Here, we used previous structures of the TRAF6/Ubc13 complex (6) and of the known E3/E2∼Ub complexes (9–12, 20) to model the TRAF6/Ubc13∼Ub complex and performed structure-based mutagenesis and biochemical assays. Our results showed that the TRAF6/donor-Ub interaction in trans used a surface patch of TRAF6 that is significantly different from the equivalent region used in other dimeric RING-type E3s. Additionally, despite the replacement of the consensus arginine in many E3s with an asparagine in TRAF6, an alanine mutation at this site compromised polyubiquitination, albeit weakly. We further revealed that E3/donor-Ub interactions in trans often use the C-terminal extension beyond the RING, while the E3/donor-Ub interactions in cis are often restricted to the RING itself and its N-terminal extensions. Collectively, these interactions in cis and in trans from multiple contact sites dictate concerted allosteric activation of an E2∼Ub conjugate for Ub transfer, consistent with other previous examples.
Results
Structural Model of the TRAF6/Ubc13∼Ub Ternary Complex.
To gain mechanistic understanding on the role of dimeric TRAF6 E3 in promoting E2 activity, we first superimposed a protomer of the TRAF6 RING-ZF1/Ubc13 complex structure (6) onto a protomer of the BIRC7/UbcH5b∼Ub complex structure (PDB ID: 4AUQ) (10) to generate a monomeric TRAF6 RING-ZF1/Ubc13∼Ub complex model. The monomer was then superimposed onto the dimeric TRAF6 structure, leading to the dimeric model of the TRAF6 RING-ZF1/Ubc13∼Ub ternary complex (Fig. 1B). In the model, a donor-Ub interacts extensively with both the cis-protomer of the TRAF6 dimer with which the Ubc13 directly interacts and the trans-protomer of the TRAF6 dimer (Fig. 1B). For the trans-TRAF6/donor-Ub interaction, we found that surface residues R125, S129, E144, R146, and H147 of TRAF6 are predicted to participate in the interaction with the donor-Ub (Fig. 1C).
This mapped surface patch of TRAF6 differs from the observed trans-donor-Ub interacting surface on BIRC7 centered at F296 (10) and on RNF4 centered at Y193 (9) (Fig. 1 C–E), both of which have been identified as required for the E3 activity (9, 10). The interaction is also somewhat more extensive than those in BIRC7 and RNF4. This difference is largely because of the distinct dimerization geometry of TRAF6 in comparison with BIRC7 and RNF4. If one protomer of the TRAF6/Ubc13∼Ub complex is superimposed to a protomer of the BIRC7/UbcH5b∼Ub complex, the symmetric protomers of the complexes do not align (Fig. 1D). When TRAF6 is directly superimposed onto BIRC7, the equivalent residue of F296 of BIRC7 is N121 of TRAF6, with K124 and R125 nearby (Fig. 1E), which is significantly shifted from the predicted donor-Ub interacting site in trans (Fig. 1C). In contrast, the donor-Ub interaction in cis should be similar for TRAF6 in comparison with other E3s.
Lys63-specific ubiquitination by TRAF6 requires both Ubc13 and the catalytically dead E2 variant Ube2V2, a human ortholog of yeast Uev1A (21). To reveal the molecular basis for TRAF6-catalyzed synthesis of Lys63 Ub chains, we also generated a structural model of the dimeric TRAF6-Ubc13∼Ub-Ube2V2 complex with a bound acceptor Ub based on the structure of the RNF4-Ubc13∼Ub-Ube2V2 complex (PDB ID: 5AIT) (Fig. 1F) (20). Like in the crystal structure of RNF4-Ubc13∼Ub-Ube2V2, the acceptor Ub in the model binds to Ube2V2, which positions its Lys63 poised for a nucleophilic attack on the thioester bond between donor ubiquitin and Ubc13 to catalyze Lys63 Ub chain synthesis (Fig. 1G).
Mutational Analysis on the Trans-TRAF6/Donor-Ub Interaction Site.
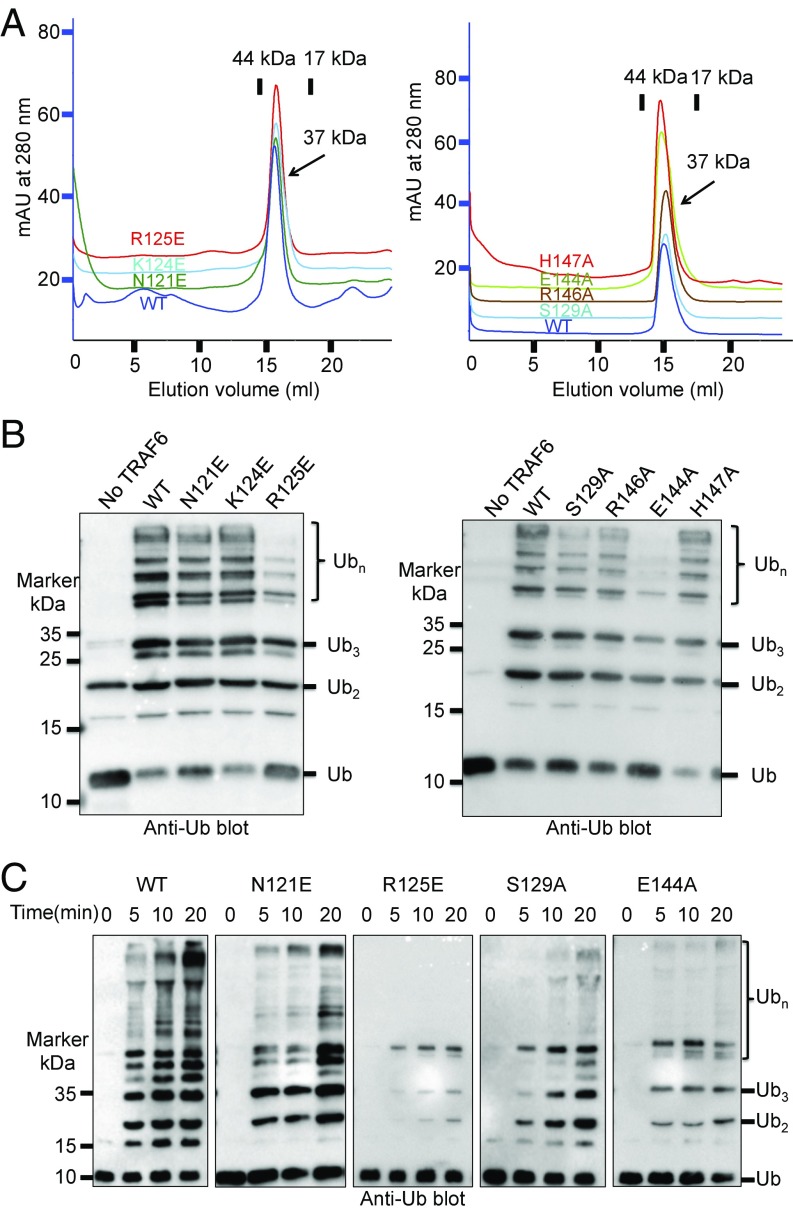
To test the functional role of the predicted TRAF6 residues in donor-Ub interaction in trans, we generated the R125E, S129A, E144A, R146A, and H147A mutants (Fig. 1C) and performed in vitro polyubiquitination assay. As a control, we also mutated TRAF6 residues N121 and K124 (Fig. 1E), which are analogous to the donor-Ub–binding site on BIRC7 and RNF4, to generate the N121E and K124E mutants. While gel filtration chromatography showed that all of the mutant proteins remained as dimers in solution (Fig. 2A), an in vitro polyubiquitination assay as a function of time revealed that the R125E and E144A mutants were severely compromised in promoting ubiquitin chain formation in the presence of the Ubc13/Uev1A complex (Fig. 2 B and C). Despite the small change, the S129A mutation also significantly affected the TRAF6 catalytic activity (Fig. 2 B and C). While R146A caused minimal but visible reduction in polyubiquitination, H147A and the control mutations N121E and K124E behaved similarly as the WT TRAF6 (Fig. 2 B and C and Fig. S1). Among these predicted donor-Ub interacting residues, R125, E144, and S129 are at the center of the interface, while H147 is at the most peripheral part of the interface (Fig. 1C), consistent with the hot-spot hypothesis in protein–protein interactions (22). Therefore, these data supported the predicted donor-Ub interaction site of TRAF6 and confirmed that this site is significantly different from the equivalent site on BIRC7 and RNF4, likely due to differences in the E3 dimer orientation (see below).
Fig. 2.
Trans-interactions between TRAF6 and donor-Ub are critical for Ub transfer. (A) Superimposed gel filtration profiles of TRAF6 wild-type and mutants, showing the preservation of dimeric states in the mutants. The TRAF6 constructs, containing RING and ZF1-3, have a theoretical dimer molecular mass of 37 kDa. mAU, milli absorption units. (B) Promotion of polyubiquitin chain synthesis by wild-type and mutant TRAF6 in the presence of E1, the E2 complex Ubc13/Uev1A, and ubiquitin. (C) Promotion of polyubiquitin chain synthesis by wild-type and mutant TRAF6 at different time points.
Asymmetric TRAF6 Dimer Is Active in Catalyzing Ub Transfer.
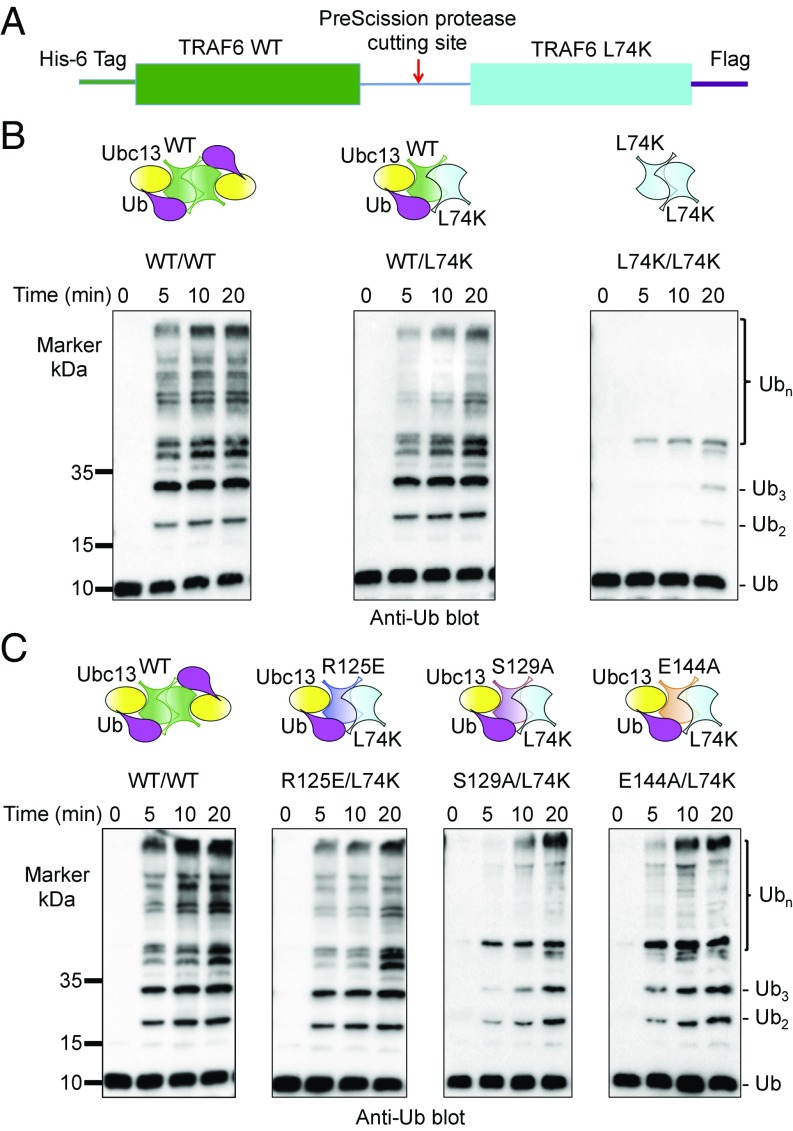
To further validate the role of the trans-interaction between TRAF6 and the donor-Ub in polyubiquitination, we fused in tandem WT TRAF6 and its L74K mutant with an intervening PreScission protease site (Fig. 3A); we previously identified the L74K mutant as defective in Ubc13 binding and in promotion of polyubiquitination (6). Once purified, the fusion protein was treated with the PreScission protease to generate the asymmetric TRAF6 WT/L74K hybrid. While the control L74K homodimer failed to promote polyubiquitination due to deficiency in Ubc13 binding (Fig. 3B), the TRAF6 hybrid catalyzed polyubiquitin chain synthesis slightly less effectively than the WT dimeric TRAF6 (Fig. 3B), consistent with one active site within a hybrid dimer in contrast to two active sites within a WT dimer. These data support that the interaction of donor-Ub with the symmetric TRAF6 L74K in trans promoted the catalytic activity of the E2 complex Ubc13∼Ub/Uev1A.
Fig. 3.
Promotion of Ub chain synthesis by TRAF6 RING hybrid dimer. (A) The construct used for generating TRAF6 wild-type/L74K hybrid. L74K is a previously identified Ubc13-binding deficient mutant of TRAF6 (6). (B) Promotion of polyubiquitin chain synthesis by wild-type, hybrid, and L74K TRAF6 RING dimers in the presence of E1, the E2 complex Ubc13/Uev1A, and ubiquitin. (C) Promotion of polyubiquitin chain synthesis by wild-type, hybrids of R125E/L74K, S129A/L74K, and E144A/L74K TRAF6 RING dimers in the presence of E1, the E2 complex Ubc13/Uev1A, and ubiquitin.
To exclude the possibility that the activity of the hybrid came from reshuffled WT TRAF6 dimer, we generated TRAF6 R125E/L74K, S129A/L74K, and E144A/L74K hybrid dimers and tested their ability in polyubiquitin chain synthesis. These hybrids contain one TRAF6 subunit defective in E2 interaction and the other subunit defective in trans-donor Ub interaction and therefore can only be functional as a heterodimer. As predicted, we found that all of the hybrids were significantly rescued for their polyubiquitination activity (Fig. 3C), in comparison with the homodimers of the respective mutants (Figs. 2C and 3B), further highlighting the importance of the trans-interactions between donor Ub and TRAF6.
A Conserved Hydrogen Bond in cis-TRAF6/Donor-Ub Interaction Contributes to Ub Transfer.
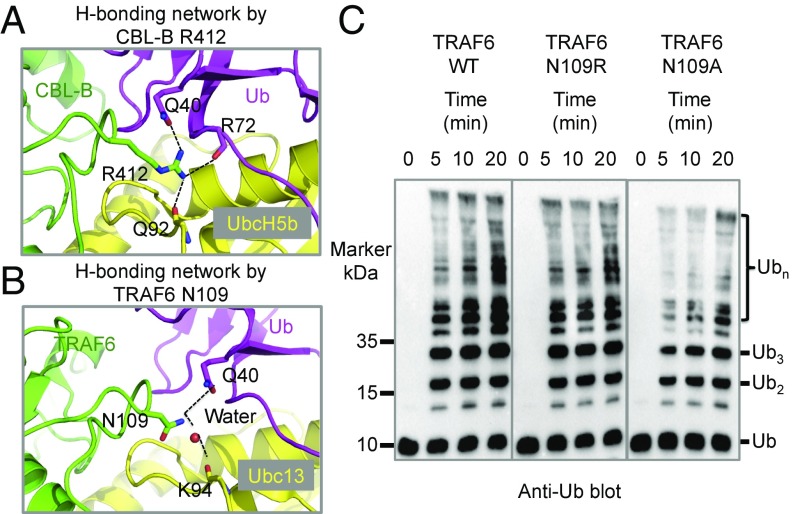
A highly conserved arginine in E3s (such as R412 of CBL-B) that forms a hydrogen bond with a main chain carbonyl oxygen in E2s (such as the UbcH5b Q92) (Fig. 4A) has been implicated as the linchpin for activation of E2∼Ub conjugates (8). However, it was later noted that the arginine also hydrogen bonds with the R72 main chain carbonyl oxygen and the Q40 side chain of ubiquitin (Fig. 4A), raising the question whether the conserved arginine also promotes activation of Ub-charged E2s by direct interaction with the donor-Ub in cis (3).
Fig. 4.
TRAF6 N109, the equivalent residue to a conserved arginine, contributes to Ub transfer. (A) A previously proposed conserved hydrogen-bonding network as shown here for CBL-B R412. CBL-B R412 forms hydrogen bonds with the backbone carbonyl oxygen of UbcH5b Q92, the side chain of Ub Q40 and the backbone carbonyl oxygen of Ub R72. (B) TRAF6 N109, the equivalent residue to the conserved arginine in many other E3s, appears to directly hydrogen bond with the side chain of Ub Q40. It may also form a water-mediated hydrogen bond with the backbone carbonyl oxygen of Ubc13 K94. (C) Promotion of polyubiquitin chain synthesis by TRAF6 wild-type, N109R, and N109A in the presence of E1, the E2 complex Ubc13/Uev1A, and ubiquitin.
In TRAF6, the arginine residue is not conserved, but corresponds to N109. In the TRAF6/Ubc13∼Ub model, as was also noted previously (8), the N109 side chain may form a potential water-mediated hydrogen bond with the K94 main chain carbonyl oxygen of Ubc13 (Fig. 4B). However, because N109 is closest to the Q40 side chain of the donor-Ub in the model (Fig. 4B) and may mediate direct hydrogen bonding in a ternary structure, the interaction of N109 with the donor-Ub may be more dominant than the indirect interaction with Ubc13. We reasoned that if both interactions are equally important for TRAF6 E3 activity, the N109R mutant with N109 mutated back to arginine, which is predicted to directly hydrogen bond with both Ubc13 and donor-Ub, might exhibit an enhanced E2 activation. Surprisingly, the N109R mutant showed similar polyubiquitination activity as the WT TRAF6 (Fig. 4C). In contrast, the N109A mutant exhibited lower catalytic activity than the WT (Fig. 4C), suggesting that the interaction of N109 with the donor-Ub in cis did contribute to TRAF6 E3 activity. As a control, we found that WT TRAF6 and the N109R and N109A mutants all interacted with Ubc13 at similar binding affinities as shown using thermophoresis (Fig. S2).
Coordination Between RING and Other Structural Elements in E2 Activation.
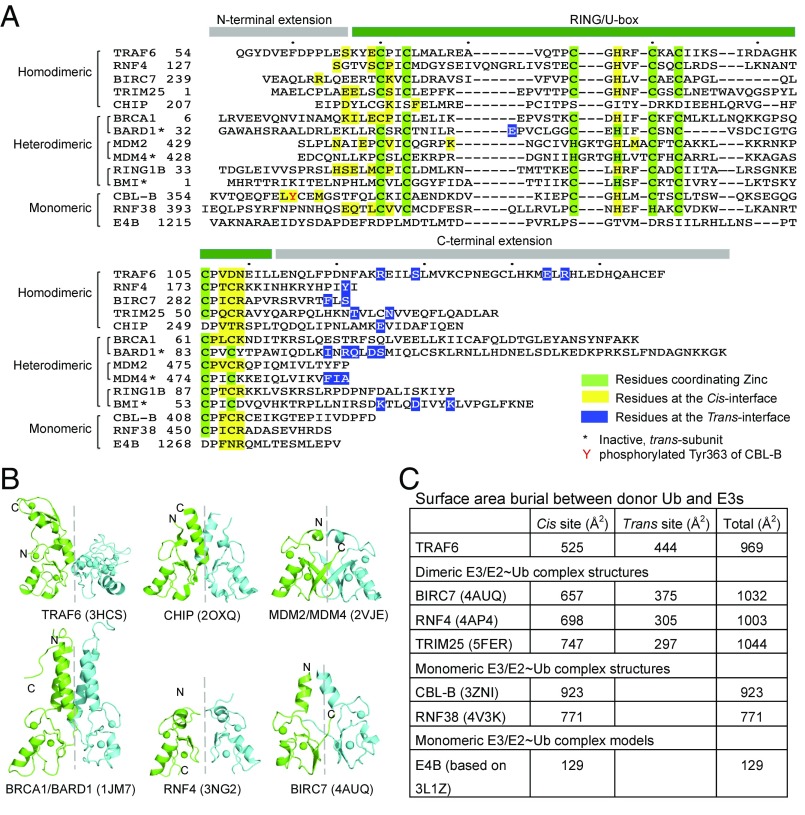
We noticed that the trans-TRAF6 subunit in the dimer uses mainly the C-terminal extension beyond the RING for its interaction with the donor-Ub (Fig. 5A). In contrast, the cis-TRAF6 subunit uses many residues in the bona fide RING element and in the N-terminal extension (Fig. 5A). We therefore asked if these characteristics of the E3/donor-Ub interaction in trans and in cis are conserved. To do this, we first looked at known structures of dimeric E3/E2∼Ub complexes, BIRC7/UbcH5b∼Ub (10), RNF4/UbcH5a∼Ub (9, 23), RNF4/Ubc13∼Ub (20), and TRIM25/UbcH5∼Ub (11, 12). Despite the variable dimeric arrangements as shown by the different orientation of the symmetric subunit when the first subunit is aligned (Fig. 5B), the structural elements involved in the donor-Ub interaction in cis and in trans obey the same characteristics shown for TRAF6 (Fig. 5A). Analysis of the homodimeric U-box protein CHIP using its dimeric E3/E2∼Ub complex model generated from the E3/E2 structure (24) and a UbcH5b∼Ub complex structure (10) revealed the same conclusion (Fig. 5A).
Fig. 5.
Analysis of diverse RING/U-box ubiquitin ligases. (A) Sequence alignment of representative RING/U-box E3s. Residues of RING/U-box E3s involved in donor ubiquitin engagement were mapped either from real ternary complex structures or from modeled complex structures. Residues at the cis-interface are colored in yellow, while those at the trans-interface are colored in blue. Zinc-coordinating residues are marked in green. (B) Cartoon diagrams of TRAF6 RING dimer (PDB ID: 3HCS), CHIP U-box dimer (PDB ID: 2OXQ), MDM2/MDM4 RING dimer (PDB ID: 2VJE), BRCA1/BARD1 RING dimer (PDB ID: 1JM7), RNF4 RING dimer (PDB ID: 3NG2), and BIRC7 RING dimer (PDB ID: 4AUQ). When the RING/U-box protomers colored in green are superimposed, the symmetric RING protomers colored in cyan do not show similar orientations, indicating differences in RING/U-box dimer architectures. (C) Buried surface areas between donor-Ub and diverse RING/U-box E3 ubiquitin ligases. The buried interfaces were calculated by PDBePISA using crystal structures of BIRC7/UbcH5b∼Ub (PDB ID: 4AUQ), RNF4/UbcH5a∼Ub (PDB ID: 4AP4), TRIM25/UbcH5a∼Ub (PDB ID: 5FER), CBL-B/UbcH5b∼Ub (PDB ID: 3ZNI), and RNF38/UbcH5b∼Ub (PDB ID: 4V3K) and structural models of TRAF6/Ubc13∼Ub and E4B/UbcH5c∼Ub.
The difference between the patterns of donor-Ub interactions in cis and in trans is seen clearer in heterodimeric E3s, for which only one of the subunit can interact with an E2 (Fig. 5A). For example, among the BRCA1/BARD1 (25), Mdm2/Mdm4 (26), and RING1B/BMI (27) complexes, modeling of the corresponding E3/E2∼Ub complex structures revealed that the cis-interactions by BRCA1, Mdm2, and RING1B utilize bona fide RING elements with further extension N-terminally. Of particular note, a few consecutive residues in RING domains at a region corresponding to the last residue involved in zinc-coordination always participate in donor-Ub interaction in cis (Fig. 5A). In contrast, the modeled trans-interactions almost exclusively use C-terminal extensions (Fig. 5A).
The above analysis led us to wonder about the mechanism of E2 activation by monomeric E3s using known E3/E2∼Ub complex structures. An emerging feature also appears to be the participation of additional structural elements beyond the RING, except that these outside RING elements are not from a dimeric partner. CBL-B is a monomeric RING-type E3, and phosphorylation of a strictly conserved tyrosine within a linker helix region at Tyr363 N-terminal to the RING enhances its ligase activity and is required for CBL-mediated ubiquitination of receptor tyrosine kinases (13). In the CBL-B/UbcH5b∼Ub complex structure (3ZNI), phosphorylated Tyr363 (pTyr363) and the pTyr363-induced element interact directly with the donor-Ub, improving the catalytic efficiency of Ub transfer by ∼200-fold (13). For the monomeric RNF38/UbcH5b∼Ub complex, the buried surface area by the E3/donor-Ub interaction is relatively small (∼770 Å2), in comparison with ∼1,000 Å2 for dimeric E3s (Fig. 5C). Here, an additional Ub bound at the backside of the E2 enhances both RING-independent and RING-dependent UbcH5b-catalyzed donor-Ub transfer, but with a more prominent effect in RING-dependent transfer (28). Perhaps the most intriguing is the case for E4B, in which the E3/donor-Ub interaction is predicted to bury only 130 Å2 surface area. Notably, two identified E4B mutations with enhanced allosteric E2 activation map to a region of E4B at or near its predicted donor-Ub interacting surface (29), which supports the role of these residues in donor ubiquitin positioning. Additionally, because a 20-residue N-terminal extension beyond the known E4B structure is required for its efficient E3 activity (8), it is likely that the same outside U-box interaction enhances E2 activity. Hence, interactions outside the canonical RING/U-box domains may be crucial for optimizing Ub transfer for both monomeric and dimeric RING/U-box E3s.
Discussion
The TRAF6/Ubc13∼Ub model and the model-based mutagenesis presented here illustrated that like other dimeric RING E3s such as BIRC7, RNF4, and TRIM25, the symmetric TRAF6 protomer facilitates E2 activation through interaction with the donor-Ub in trans. During our manuscript revision, we noted publication of the crystal structure of zebrafish TRAF6 in complex with Ubc13∼Ub, which revealed an almost identical interaction as predicted by our model (30). Notably, the region of TRAF6 used in this interaction is different from those in BIRC7 and RNF4, suggesting that this trans-E3/donor-Ub interaction can accommodate variable contacts for different dimeric E3s. Notably, our analysis showed that residues C-terminal to the RING, which differ significantly among different E3s, mediate trans-E3/donor-Ub interactions. Furthermore, the exact modes of dimerization of different E3s lead to variability in the trans-E3/donor-Ub contacts, establishing the necessity for the observed plasticity in trans-E3/donor-Ub interactions.
Previous studies defined a conserved arginine in RING/U-box E3s as a linchpin to prime Ub transfer through a hydrogen bond to the E2 (8). The equivalent residue in TRAF6 is an asparagine, which appears to form a direct hydrogen bond in cis with the donor-Ub only and likely only indirectly interacts with the E2. Mutation of the asparagine to alanine compromised Ub transfer, suggesting that cis- and transTRAF6/donor-Ub interactions, as well as TRAF6/E2 and E2/Ub interactions, together stabilize donor-Ub in an active conformation for Ub transfer. The complexity in analyzing the linchpin mechanism in facilitating Ub transfer is also reflected in the example of the role of Rbx1 N98 (31), which is at the same position as N109 of TRAF6 and equivalent to the arginine linchpin in other E3s. For Rbx1, the mutational effects of N98A differed dramatically depending on the E2 used: while N98A reduced reactions with UBCH5∼UB, it did not affect reactions with Ubc12∼NEDD8 (31).
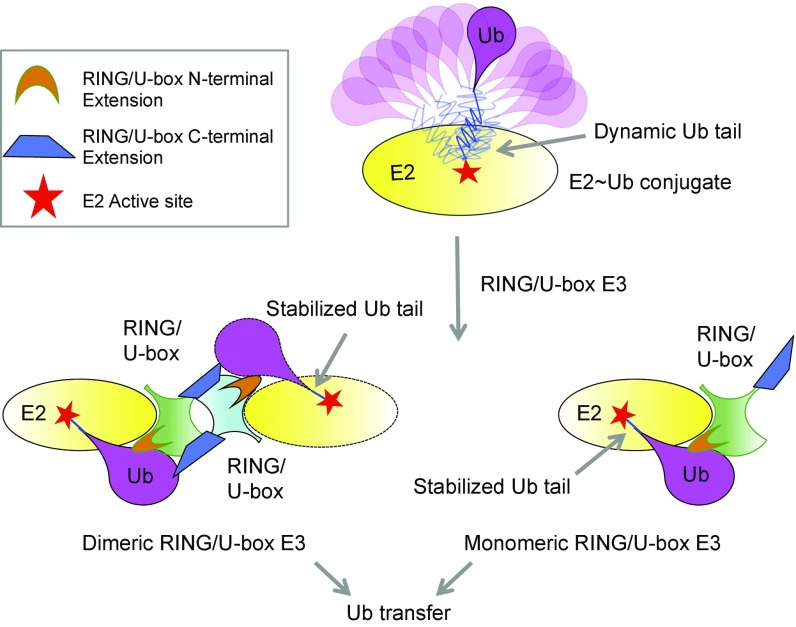
Our studies here point to a unified structural mechanism for dimeric and monomeric RING/U-box E3s to promote Ub transfer using bona fide RING domains and regions beyond the RING (Fig. 6). In the absence of E3s, E2∼Ub complexes are highly dynamic with little interactions between the E2 and the donor-Ub, which is not favorable for Ub transfer (Fig. 6). The binding of an E3 constrains the conformational freedom of the E2∼Ub conjugate and drives it into an active conformation primed for Ub transfer. In the active, closed conformation, the E2∼Ub thioester bond is optimally positioned through extensive E3/donor-Ub interactions for the nucleophilic attack by the incoming lysine. For all RING/U-box E3s, the cis-E3/donor-Ub interactions, both within the RING and at the N-terminal extension, contribute significantly (Fig. 6). For dimeric E3s, the trans-E3/donor-Ub interactions, which mainly come from the C-terminal extension, supplement the cis-interactions to enhance the binding energetics and to position the E2∼Ub thioester (Fig. 6). For monomeric E3s, it appears that the cis-E3/donor-Ub interactions within the RING may be extensively supplemented by the N-terminal extension or by other mechanisms (Fig. 6), reaching the same effect of restraining the donor-Ub into the active, “folded-back” conformation.
Fig. 6.
A schematic summary of Ub transfer catalyzed by RING/U-box E3s. Ubiquitin adopts a variety of conformations relative to E2 in the E2∼Ub conjugate, which does not favor ubiquitin transfer. Upon engagement by RING/U-box E3s, the conformation of the donor ubiquitin is restrained by forming extensive interactions with E2 and RING/U-box E3s, priming the ubiquitin transfer. In the case of dimeric RING/U-box E3s, the two protomers of E3s form cis- and trans-interactions with the donor Ub, mainly via the RING/U-box domain per se and the N-terminal extension for the cis-interaction, and the C-terminal extension for the trans-interaction. Monomeric RING/U-box E3s, which can only form cis-interactions with the donor Ub, may use more extensive interactions at the N-terminal extension or exploit other mechanisms. Therefore, RING/U-box E3s employ a unified mechanism to activate the E2∼Ub conjugate but can adopt variable strategies to engage the donor-Ub. Regions outside the bona fide RING/U-box domain contribute significantly to shift an inactive E2∼Ub conformation to the active conformation.
As previously proposed, our data support that all E3s, the RING/U-box–type or the HECT-type, for Ub or for ubiquitin-like proteins (Ubl) such as Sumo, may share a similar catalytic mechanism for E2 activation. Notably, the crystal structure of the quaternary complex of the E2 Ubc9, E3 Nup358, and Sumo-conjugated substrate RanGAP1, which may mimic the conformation of an E3/E2∼Sumo complex, provided a first hint that the E3 reduces conformational flexibility of Ubc9∼SUMO by extensive interactions with Ubc9 and SUMO and positions the thioester in an optimal orientation to enhance conjugation (32). The crystal structure of the HECT E3 NEDD4L in complex with UbcH5b∼Ub also showed that NEDD4L positions the E2∼Ub thioester linkage for E2-to-HECT Ub transfer (33). Hence, despite the structural diversity, E3s are evolved to promote E2 activation by simultaneously interacting with the E2 and the conjugated donor-Ub to optimally orient the E2∼Ub thioester bond.
Materials and Methods
Structure Modeling.
All of the structure modeling was done using Coot (34). Briefly, TRAF6/Ubc13∼Ub ternary structural model was generated by superimposition of Ubc13 onto UbcH5b of BIRC7/UbcH5b∼Ub dimeric structure (PDB ID: 4AUQ). The TRAF6/Ubc13∼Ub ternary complex was superimposed onto TRAF6 RING dimeric structure to generate the dimeric structure of TRAF6-Ubc13-Ub. The structural model of the TRAF6/Ubc13∼Ub/Ube2V2/acceptor Ub complex was generated based on the RNF4/Ubc13∼Ub/Ube2V2 structure (PDB ID: 5AIT) using a similar method.
Protein Expression and Purification.
Mouse E1 was expressed in Hi5 cells with N-terminal His8 tag and purified by Ni2+-affinity chromatography (Qiagen) followed by gel filtration (Superdex 200; GE healthcare). Ubc13, Uev1A, and Ubiquitin were expressed in Escherichia coli RIPL with a N-terminal His6 tag and purified by Ni2+-affinity chromatography and gel filtration. TRAF6 RING+ZF1-3 (RZ123, residues 50–211) was expressed in E. coli RIPL cells with a C-terminal His6 tag and purified by Ni2+-affinity chromatography and gel filtration.
All of the mutants were generated by site-directed mutagenesis. All of the mutants were expressed in E. coli RIPL and purified as wild-type TRAF6. The N-terminally His-tagged and C-terminally Flag-tagged TRAF6 hybrid protein, containing WT TRAF6 RING+ZF1 (RZ1, residues 50–159) and the E2-binding mutant L74K RZ1 in tandem with an intervening PreScission protease site, was first purified by Ni2+-affinity chromatography and gel filtration followed by PreScission protease digestion overnight at 4 °C. The protein was further purified by tandem procedures of Ni2+-affinity chromatography and Flag-affinity chromatography. The asymmetric dimers of TRAF6-R125E/L74K, TRAF6-S129A/L74K, and TRAF6-E144A/L74K were obtained by coexpression in E. coli of Flag-tagged TRAF6 RING+RZ1 L74K with His-tagged TRAF6 RING+RZ123 R125E, S129A, and E144A, respectively, and purified by tandem procedures of Ni2+-affinity chromatography and Flag-affinity chromatography.
Ubiquitination Assay.
Ubiquitination reactions were performed at 37 °C for 15 min by incubating 100 nM E1, 200 nM Ubc13, 100 nM Uev1A, 20 μM Ub, and 1 μM TRAF6 wild-type or TRAF6 mutants in a reaction buffer of 25 mM Tris⋅HCl at pH 7.5, 0.2 mM DTT, 2.5 mM MgCl2, 2 mM ATP, 5 mM creatine phosphate, 0.6 unit/mL creatine kinase, and 0.6 unit/mL inorganic pyrophosphatase. The reactions were quenched by addition of SDS-loading buffer followed by boiling. The reaction products were applied to SDS/PAGE and transferred to PVDF membranes using a Trans-Blot SD Semi-Dry Transfer Cell (BioRad). The reaction products were probed by an anti-Ub antibody (Santa Cruz Biotechnology).
Supplementary Material
Acknowledgments
We thank Dr. Xuejun Jiang at the Sloan-Kettering Institute for providing the mouse E1 plasmid. This work was supported by the US National Institutes of Health (Award AI050872, to H.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721788115/-/DCSupplemental.
References
- 1.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 2.Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 3.Metzger MB, Pruneda JN, Klevit RE, Weissman AM. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim Biophys Acta. 2014;1843:47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickart CM, Rose IA. Functional heterogeneity of ubiquitin carrier proteins. J Biol Chem. 1985;260:1573–1581. [PubMed] [Google Scholar]
- 5.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin Q, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci USA. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruneda JN, et al. Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell. 2012;47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plechanovová A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koliopoulos MG, Esposito D, Christodoulou E, Taylor IA, Rittinger K. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 2016;35:1204–1218. doi: 10.15252/embj.201593741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez JG, et al. Mechanism of TRIM25 catalytic activation in the antiviral RIG-I pathway. Cell Rep. 2016;16:1315–1325. doi: 10.1016/j.celrep.2016.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nat Struct Mol Biol. 2013;20:982–986. doi: 10.1038/nsmb.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pineda G, Ea CK, Chen ZJ. Ubiquitination and TRAF signaling. Adv Exp Med Biol. 2007;597:80–92. doi: 10.1007/978-0-387-70630-6_7. [DOI] [PubMed] [Google Scholar]
- 16.Wu H. Assembly of post-receptor signaling complexes for the tumor necrosis factor receptor superfamily. Adv Protein Chem. 2004;68:225–279. doi: 10.1016/S0065-3233(04)68007-7. [DOI] [PubMed] [Google Scholar]
- 17.Ye H, et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418:443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 18.Deng L, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 19.Wang C, et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 20.Branigan E, Plechanovová A, Jaffray EG, Naismith JH, Hay RT. Structural basis for the RING-catalyzed synthesis of K63-linked ubiquitin chains. Nat Struct Mol Biol. 2015;22:597–602. doi: 10.1038/nsmb.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 22.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 23.Plechanovová A, et al. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat Struct Mol Biol. 2011;18:1052–1059. doi: 10.1038/nsmb.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M, et al. Chaperoned ubiquitylation–Crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 25.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 26.Lin CC, et al. Tumor necrosis factor-alpha induces MMP-9 expression via p42/p44 MAPK, JNK, and nuclear factor-kappaB in A549 cells. Toxicol Appl Pharmacol. 2008;229:386–398. doi: 10.1016/j.taap.2008.01.032. [DOI] [PubMed] [Google Scholar]
- 27.Bentley ML, et al. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 2011;30:3285–3297. doi: 10.1038/emboj.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buetow L, et al. Activation of a primed RING E3-E2-ubiquitin complex by non-covalent ubiquitin. Mol Cell. 2015;58:297–310. doi: 10.1016/j.molcel.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 29.Starita LM, et al. Activity-enhancing mutations in an E3 ubiquitin ligase identified by high-throughput mutagenesis. Proc Natl Acad Sci USA. 2013;110:E1263–E1272. doi: 10.1073/pnas.1303309110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Middleton AJ, et al. The activity of TRAF RING homo- and heterodimers is regulated by zinc finger 1. Nat Commun. 2017;8:1788. doi: 10.1038/s41467-017-01665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott DC, et al. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell. 2014;157:1671–1684. doi: 10.1016/j.cell.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamadurai HB, et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol Cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.