Abstract
Despite tremendous advances in targeted therapies against lung adenocarcinoma, the majority of patients do not benefit from personalized treatments. A deeper understanding of potential therapeutic targets is crucial to increase the survival of patients. One promising target, ADAR, is amplified in 13% of lung adenocarcinomas and in-vitro studies have demonstrated the potential of its therapeutic inhibition to inhibit tumor growth. ADAR edits millions of adenosines to inosines within the transcriptome, and while previous studies of ADAR in cancer have solely focused on protein-coding edits, > 99% of edits occur in non-protein coding regions. Here, we develop a pipeline to discover the regulatory potential of RNA editing sites across the entire transcriptome and apply it to lung adenocarcinoma tumors from The Cancer Genome Atlas. This method predicts that 1413 genes contain regulatory edits, predominantly in non-coding regions. Genes with the largest numbers of regulatory edits are enriched in both apoptotic and innate immune pathways, providing a link between these known functions of ADAR and its role in cancer. We further show that despite a positive association between ADAR RNA expression and apoptotic and immune pathways, ADAR copy number is negatively associated with apoptosis and several immune cell types' signatures.
Keywords: RNA editing, Lung adenocarcinoma, Cancer, Post-transcriptional regulation, Bioinformatics, ADAR
Highlights
-
•
ADAR potentially regulates the mRNA abundance of thousands of genes.
-
•
Editing of the APOL1 3′ UTR is associated with its upregulation and patient poor overall survival.
-
•
ADAR-regulated genes are enriched for apoptosis and immune pathways.
Lung cancer is the most deadly cancer globally and current targeted treatments only benefit a minority of patients. Inhibiting the ADAR oncogene has shown promising preclinical results; however, little is known about ADAR's functions in cancer. We investigate a key function of ADAR, mRNA regulation via RNA editing, and provide evidence that it is linked to tumor immunity and cell death in human lung adenocarcinoma. Our results provide a motivation to explore combination immunotherapies that include ADAR inhibition.
1. Introduction
Each year, > 500,000 people die from lung adenocarcinoma (LUAD) globally, and only a minority of these tumors harbor alterations targetable by existing personalized therapies (Imielinski et al., 2012, Collisson et al., 2014). Research into targeting new oncogenes is crucial to expanding the proportion of treatable tumors. The genomic locus containing the adenosine deaminase acting on RNA (ADAR) gene is amplified in 13% of LUAD and has been functionally confirmed as an oncogene in lung (Anadón et al., 2015), breast (Fumagalli et al., 2015), stomach (Chan et al., 2016), chronic myeloid leukemia (Jiang et al., 2013) and liver (Chen et al., 2013b) cancers. Knockdown of ADAR in LUAD is associated with reduced in-vitro cell viability, and decreased metastatic potential in xenograft mouse models (Anadón et al., 2015). Therefore, targeted therapies towards and a greater functional understanding of ADAR amplified tumors have the potential to greatly increase the number of treatable cases of LUAD.
Adenosine to inosine RNA editing is mediated predominantly by the ADAR gene and to a lesser extent, the ADARB1 gene. This modification occurs in millions of sites across the transcriptome, mostly within Alu repeats (Bahn et al., 2015). Previous studies of the oncogenic role of ADAR in cancer have focused on the editing of coding sequences of tumor-associated proteins, such as AZIN1 (Chen et al., 2013b) and NEIL1 (Anadón et al., 2015). Although this role has been confirmed in functional studies of hepatocellular carcinoma, < 1% of known RNA editing sites reside in coding sequences (Han et al., 2015). The function of RNA editing in non-protein coding regions is only known for a few edited genes. For example, it has been shown that ADAR-mediated RNA editing changes the accessibility to the HuR RNA binding protein (RBP) within the cathepsin S (CTSS) 3′ UTR, which enhances its mRNA's stability in endothelial cells (Stellos et al., 2016). In addition, Zhang et al. showed that RNA editing of the MDM2 3′ UTR segment can abolish mir-200b mediated repression of the MDM2 mRNA. Recently, RNA editing of non-coding regions by ADAR has been implicated in immune regulation (Mannion et al., 2014, Liddicoat et al., 2015, Pestal et al., 2015, George et al., 2016) and apoptosis (Yang et al., 2017, Sakurai et al., 2017), but it is not clear how these functions are related to ADAR's oncogenic potential.
Here, we investigate the role of mRNA abundance regulation by RNA editing in human LUAD. We create a pipeline that inputs RNA gene abundances and RNA editing frequencies, and outputs potentially regulatory pairs of RNA editing sites and mRNA target genes. This pipeline is applied to The Cancer Genome Atlas (TCGA) LUAD RNA sequencing data to obtain a global picture of the RNA regulatory editome. We identify enrichment of both apoptosis and immunoregulatory genes potentially regulated by ADAR-mediated RNA editing. ADAR is alternatively spliced into two separate isoforms: a short, nuclear, and constitutively expressed p110 isoform; and a long, cytosplasmic, and interferon inducible p150 isoform (Pestal et al., 2015). The p110 isoform is responsible for fine tuning apoptosis, at least in part via modulating accessibility to Staufen 1 binding sites in edited mRNA's (Yang et al., 2017, Sakurai et al., 2017). The p150 isoform edits double stranded dsRNA's and prevents them from activating the MDA5-MAVS interferon response. Neither of these functions of ADAR have been linked to cancer, although it was seen by Fumagalli, et al. that ADAR RNA expression is jointly explained by ADAR genomic copy number and STAT1 expression, a marker of interferon activity (Fumagalli et al., 2015). We further establish that ADAR genomic copy number (CN) is negatively associated with immune and apoptosis pathways, as well as immune cell signatures, establishing potential oncogenic roles for ADAR in LUAD.
2. Materials and Methods
2.1. Data
RNAseq abundances, copy number data, and editing frequencies were downloaded for lung adenocarcinoma (LUAD) matched normal and tumor samples. RNAseq gene level expression (RNAseqV2 RSEM) and copy number data were downloaded from Broad Institute Firehose (https://gdac.broadinstitute.org). Gistic 2.0 copy number data was further processed to gene level as follows. If a gene is contained within a segment as defined in the gistic-processed data, the value of that segment is assigned to the gene. Editing frequency data was downloaded from synapse.org (https://www.synapse.org/#!Synapse:syn2374375/files/) (Han et al., 2015). Editing frequency is the number of edited reads covering a given editing site divided by the total number of reads covering that editing site. Editing sites are annotated in the RADAR (Ramaswami and Li, 2014) database, which collects and curates functionally validated A-I RNA editing sites.
2.2. Identification of Regulatory Edits
First, RNA editing sites were matched to the genes containing them. Often, there are multiple RNA editing sites within each gene. Then, for each gene, RNA editing frequencies were associated to RNA abundances with spearman correlation. Significant edits were determined using R's (v1.0.136) built in significance of correlation test and corrected for multiple hypothesis testing (Benjamini Hochberg corrected q-value < 0.1). All correlations performed in this study are spearman correlations.
2.3. RNA Binding Protein and microRNA Motif Analysis and Secondary Structure Visualization
Regulatory edits were grouped by gene and searched for continuously edited regions (CER). A CER was defined as > 5 sequential significant RNA regulatory editing sites, with no two consecutive RNA editing sites being separated by > 100 base pairs. These CERs were then input as a bed file into the RBPmap tool (http://rbpmap.technion.ac.il) (Paz et al., 2014), which searches for RNA binding protein (RBP) motifs within genomic regions. We used the following parameters in our searches: high stringency, hg19 reference, all Human/Mouse motifs, and no conservation filter. MicroRNA motif enrichment was calculated using the targetScan web tool (Grimson et al., 2007), and microRNA's with combined score < − 0.3 were considered for further analysis. Secondary structure prediction was made with the Forna (Kerpedjiev et al., 2015) web app.
2.4. Pathway and Immune Cell Analysis
Pathway scores for tumor samples were calculated using single sample gene set enrichment analysis (SSGSEA) (Barbie et al., 2009). Immune exclusion and tumor purity scores were downloaded from Aran et al. (2015) and converted to immune infiltrate scores (1-SCORE). Immune cell subset analysis was performed using the TIMER web app (https://cistrome.shinyapps.io/timer/) (Li et al., 2016).
2.5. Histological Slide Evaluation of Immune Cell Subsets
Histological slides for TCGA LUAD tumors, were visualized using the digital slide archive (http://cancer.digitalslidearchive.net) (Gutman et al., 2013). We selected a subset of tumors such that there were roughly equal representation of ADAR amplified and normal copy numbers. In total, 97 LUAD tumors, 45 of which had no evidence of ADAR amplification, and 52 of which had high ADAR copy number, were selected for analysis and we conducted the experiment with no knowledge of the ADAR CN status of these tumors. We were able to visually estimate neutrophil, lymphocyte, and macrophage infiltrations, as well as necrosis on a scale of 0–3.
3. Results
3.1. Pipeline for Discovery of Regulatory A-I RNA Editing Sites
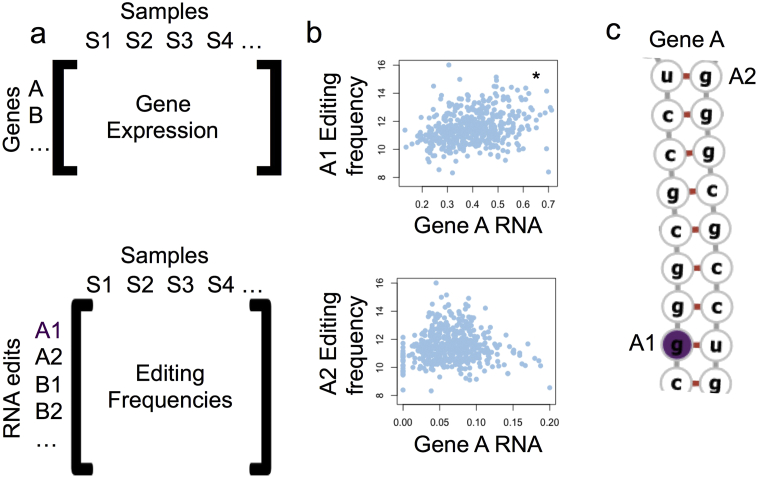
We created a pipeline for discovery of regulatory RNA editing sites as follows. We first matched each RNA editing site with its host RNA abundance (Fig. 1A), then tested all RNA edits for their association with host RNA abundance (Fig. 1B–C). All relevant code can be found at github.com/michaelsharpnack/RNA_edits.
Fig. 1.
Pipeline to discover regulatory RNA editing sites. Editing sites annotated in RADAR database are matched to their host gene (A). The regulatory potential of each editing site is discovered by testing the association between editing frequency and host RNA abundance (B). In this case, one editing site is significantly positively associated with RNA abundance (A1, maroon) and is classified as a potential regulatory editing site (C).
3.2. Landscape of Regulatory Editing Sites in Lung Adenocarcinoma
Previous papers have focused on the potential for protein coding RNA editing sites to modify oncogene and tumor suppressor functions; however, the frequencies of many non-coding RNA editing sites are significantly increased in LUAD. In the TCGA LUAD cohort, 4115 non-protein-coding RNA editing sites were differentially edited (t-test, BH q-value < 0.1) while only 20 protein coding RNA editing sites were differentially edited (Supplementary Fig. 1). We therefore applied our regulatory RNA editing pipeline to the TCGA LUAD RNAseq dataset to discover alternative functions of the ADAR oncogene.
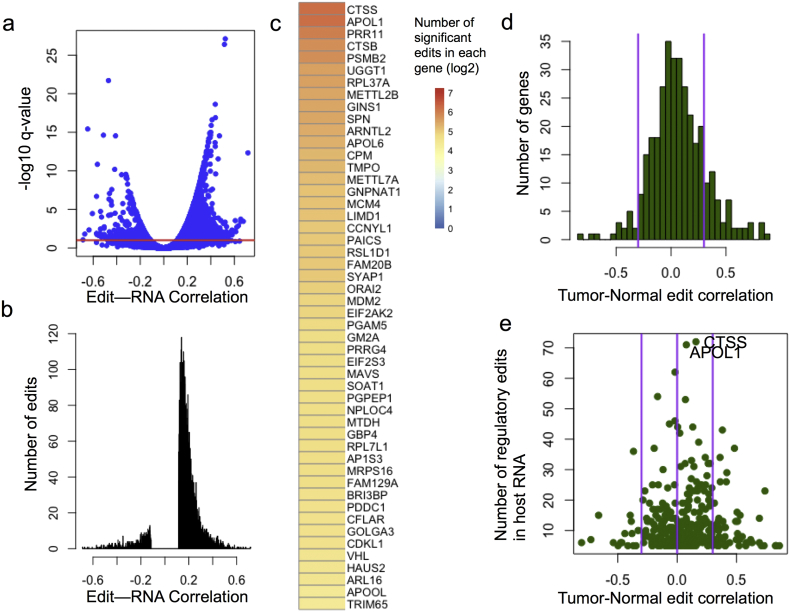
RNAseq data for 488 LUAD tumor and 57 matched normal samples were run through our regulatory RNA editing pipeline. This data includes 54,362 frequently edited sites, and 52,276 RNA editing site-RNA abundance combinations tested for their regulatory potential. 5468 (10%) of the edit-gene combinations were predicted to have regulatory potential across 1413 genes (Fig. 2A). The majority of the significant RNA editing sites had a positive association with RNA abundance (4976 or 91%, Fig. 2B). It is possible that the enrichment of positive RNA editing-mRNA abundance associations is due to a bias in detection; given that we would not be able to measure frequently edited transcripts that are degraded. Gene expression and length are both potential sources of bias in regulatory RNA editing site detection. The expression of each gene with detectable RNA editing sites is, although significantly so, very weakly associated with the ability to detect regulatory RNA editing sites (ρ = 0.059, p = 5 × 10− 4). Gene length and the number of Alu elements within each gene are also not strongly associated with the number of detected regulatory RNA editing sites (ρ = − 0.066, p = 1 × 10− 4 for total gene length; and ρ = 0.028, p = 0.10 for the number of Alu elements present in the gene body).
Fig. 2.
Landscape of regulatory RNA editing sites in LUAD. (A) Plot showing the distribution of RNA editing site-mRNA abundance correlation and the significance of the association. The red line denotes q-value < 0.1 significance threshold. Histogram of regulatory RNA editing sites' association to RNA abundance (B). Genes are ranked by the number of predicted regulatory RNA editing sites within their RNA molecule, and the top 50 are shown in (C). Edits are grouped by their host gene and RNA editing site-RNA spearman correlations are compared between tumor and normal (D). Genes with high Tumor-normal regulatory RNA editing site correlation have regulatory RNA editing sites with similar regulatory potential in both tumor and normal. The tumor-normal edit correlation is plotted against the number of regulatory edits in each host RNA molecule (E).
293 of the genes tested (8.5%) had > 5 predicted regulatory RNA editing sites, and the genes with the top 50 RNA editing sites are shown in Fig. 2C. Regulatory relationships between ADAR and these genes have only been investigated for a small number of these genes, including CTSS (Stellos et al., 2016) and MDM2 (Zhang et al., 2016). Remarkably our method was able to discover these established regulatory relationships. CTSS is the best-studied example of gene regulation by ADAR, and CTSS is the top hit according to our algorithm. For a full list of genes with predicted regulatory RNA editing sites, please see Supplementary Table 1. In addition, we investigated for possible enrichment of putative regulatory relationships that are evolutionarily conserved. 1664/11319 (14.7%) and 4475/40957 (10.9%) of RNA editing sites were significant in rhesus or chimp and non-conserved, respectively, indicating an enrichment of regulatory RNA editing sites in evolutionarily conserved RNA editing sites (fisher's exact test, p < 2.2e− 16).
We next sought to discover which regulatory relationships are present in both tumor and normal, and which are only present in tumor. Since there are so many fewer normal samples, instead of comparing statistical significance of RNA editing sites between tumor and normal, we compared the spearman correlations. For each gene, we calculated the spearman correlation between the tumor and normal regulatory correlations of all the RNA editing sites within that gene. A high tumor-normal correlation indicates that RNA editing mediated regulation of a gene is not cancer specific. Of the 344 genes with at least 5 regulatory RNA editing sites in tumors, 54/344 (16%) of these genes' edits had a positive correlation between tumor and normal regulatory RNA editing sites of ρ > 0.3, while 16 had a negative correlation of ρ < − 0.3. 274/344 (80%) of the regulatory edits are cancer-specific, especially in the genes with the highest numbers of RNA editing sites (Fig. 2E).
The ADAR gene regulates the majority of RNA editing sites, while the ADARB1 gene regulates a relatively small portion (Supplementary Fig. 2). Because ADAR and ADARB1 are upregulated and downregulated in LUAD (Supplementary Fig. 3), respectively, the majority of RNA editing sites is over-edited in tumors (Supplementary Fig. 1B). There are 756 differentially edited regulatory RNA editing sites across 242 differentially expressed genes. Given that the majority of regulatory RNA editing sites are positively associated with target RNA expression, one would expect to see that over-edited genes also tend to be overexpressed. In fact, 558/756 (74%) of the regulatory RNA editing sites are over-edited while their host genes are overexpressed, and 93% of these RNA editing sites are positively associated with RNA abundances in tumors. Of the remaining RNA editing sites, 137/756 (18%) are over-edited with host genes that are underexpressed. Unsurprisingly, 41% of these editing sites are negatively associated with RNA abundances in tumors (Supplementary Fig. 4). Together, these results show that ADAR overexpression in tumors potentially controls cancer-specific differential expression of hundreds of genes in LUAD.
An alternative hypothesis to RNA-editing mediated mRNA abundance regulation is that mRNA abundances are merely associated with RNA editing frequencies. In particular, ADAR is upregulated by interferon signaling, which could also regulate genes that are edited by ADAR. To isolate the effects of ADAR on its target genes, we compared the results from ADAR knockdown experiments on human B-Cells from Wang et al. (2013) to our results in human LUAD. Interferon pathways were not significantly enriched among differentially expressed genes after ADAR knockdown in their study. Wang et al. found 105 genes with evidence of RNA editing had both alterations in RNA editing frequency and mRNA abundance after ADAR knockdown via siRNA. 84/105 (80%) of these genes also displayed evidence of editing in the TCGA LUAD dataset, and 54/84 (64%) of the genes found to be regulated by RNA editing were also putative regulated genes found by our method, representing a significant enrichment. We performed a permutation test to assess the significance of this result by selecting 105 random subsets of 1413 genes from the 3412 genes with evidence of editing. In none of these iterations did a random gene set have a > 64% overlap with the set from Wang et al., a probability of < 1 × 105 that the overlap in our results and the results of Wang et al. occurred randomly. For example, APOL1, CFLAR, DAP3, EIF2AK2, and MAVS were all putative regulated genes found by both our method and Wang et al. For a full comparison, see Supplementary Table 2.
3.3. The APOL1 3′ UTR Contains Multiple Cancer-specific Regulatory Editing Sites Controlled by ADAR
The CTSS and Apolipoprotein L1 (APOL1) genes had the largest number of predicted regulatory editing sites in LUAD (Fig. 2C). Since an ADAR-CTSS regulatory relationship has already been established, we investigated the RNA editing patterns within the 3′ UTR of the APOL1 gene. APOL1 functions are not well characterized; however, it circulates with the densest HDL subfraction, HDL3 (Monajemi et al., 2002). APOL1 variants have been implicated in chronic kidney disease (Parsa et al., 2013), and they likely evolved due to their protective effects against T. brucei rhodesiense infections (Vanhamme et al., 2003). APOL1 is upregulated after interferon treatment or TLR3 stimulation via an IRF3-dependent pathway, and high APOL1 expression in HIV-infected patients with an interferon response contributes to chronic kidney disease (Nichols et al., 2015). Despite its role in innate immunity and status as an interferon-regulated gene, APOL1 has not yet been associated to ADAR-mediated regulation.
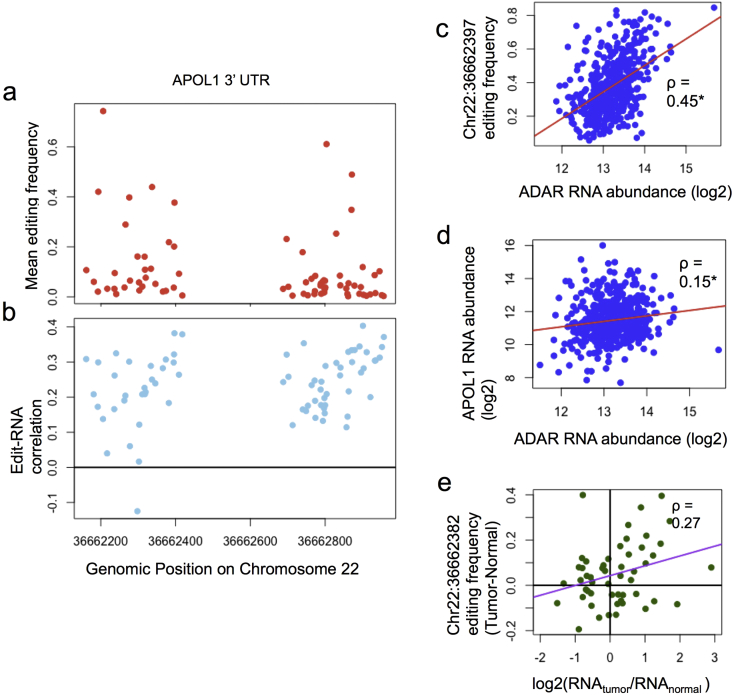
APOL1 editing sites are frequently edited—21/74 (28%) sites have > 10% average editing frequency across all patients in LUAD (Fig. 3A). 71/74 (96%) editing sites in the APOL1 3′ UTR that are consistently edited in LUAD have predicted regulatory roles, and editing of these sites is positively associated with APOL1 RNA abundance (Fig. 3B). To test whether ADAR or ADARB1 controls the editing of APOL1, the editing frequencies of the predicted regulatory sites were correlated with the RNA abundances of ADAR and ADARB1, respectively. The editing frequency of the majority of these sites as well as APOL1 RNA abundance was significantly correlated with ADAR RNA abundance, indicating that ADAR, not ADARB1, controls them (Fig. 3C–D). In addition, Wang et al. (2013) found that after siRNA knockdown of ADAR, both APOL1 editing and gene expression decreased (Supplementary Table 2).
Fig. 3.
The APOL1 3′ UTR contains multiple regulatory editing sites controlled by ADAR. APOL1 3′ UTR contains several RNA editing sites which are edited at a very high frequency (A), and most are significantly positively correlated to APOL1 mRNA abundance (B). APOL1 editing frequencies (C) and RNA abundances (D) are both positively correlated with ADAR RNA abundance. Fig. 3E shows for each patient the difference in chr22:36,662,382 editing frequency and APOL1 mRNA abundance between tumor and normal. APOL1 tends to be over-edited and overexpressed in the same tumors (Rho = 0.27, p = 0.056). *q < 0.1.
We next sought to discover if the regulation of APOL1 could be cancer related. APOL1 is overexpressed in LUAD compared to matched normal tissue samples (q < 0.1), and 26/74 (35%) of its RNA editing sites show increased evidence of editing in cancer. In tumors where APOL1 is over-edited, it also tends to be overexpressed (Fig. 3E). Together, this evidence suggests that ADAR overexpression and associated increased editing in LUAD potentially induces overexpression of APOL1.
Since our method provides nucleotide level regulatory hypotheses, we performed a secondary structure analysis on the edited regions of the APOL1 3′ UTR. We discovered that many of the regulatory RNA editing sites change the predicted secondary structure, and several of these regulatory RNA editing sites potentially effect RBP and microRNA motifs, including numerous predicted PTBP1 binding sites (Supplementary Figs. 5–6). PTBP1 is overexpressed in tumors and correlated with ADAR expression (Supplementary Fig. 7). However, the predictions of RNA secondary structure, and RBP and microRNA motifs are not perfect and require functional validation.
3.4. RNA Editing of the APOL1 3′ UTR is Associated With Poor Overall Survival in Lung Adenocarcinoma
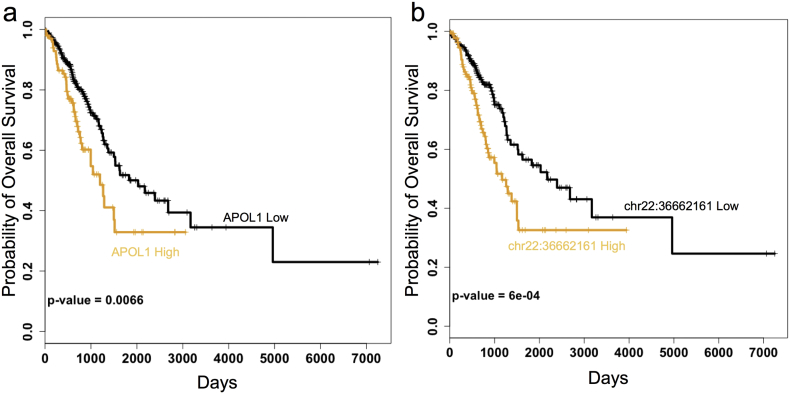
We next investigated the clinical importance of APOL1 in LUAD. We separated the tumors into APOL1 high and APOL1 low based on the mean APOL1 expression across all tumors and performed survival analysis (Fig. 4A). High APOL1 expression is significantly associated with poor prognosis (chi-square, p = 0.0066). In addition, high APOL1 expression is associated with poor survival using the survival meta-analysis tool, precog (precog.stanford.edu) (Gentles et al., 2015), which compiles the results of several gene expression datasets in lung adenocarcinoma (z = 3.37, p < 10− 3). To determine if this effect was due to RNA editing or other modes of APOL1 regulation, we then repeated this survival analysis across all 74 editing sites in the APOL1 3′ UTR. 30/74 (41%) of the RNA editing sites within the 3′ UTR of APOL1 are significantly associated with poor prognosis in LUAD (chi-square, q-value < 0.1). The most significant edit (q = 0.020) was also the closest to the APOL1 coding region, located at chr22:36662161 (Fig. 4B). Editing of this site is predicted to remove a kink in the secondary structure of the APOL1 3′ UTR (Site 20 in Supplementary Fig. 5A–B). An alternative hypothesis is that overall editing rates are prognostic, and not just specific sites; however, Paz-Yaacov et al. (2015) investigated this hypothesis and did not report a significant association between global editing rates and overall survival in LUAD.
Fig. 4.
Survival analysis of APOL1 RNA expression and APOL1 editing sites. Tumors are separated into low vs. high APOL1 expression based on mean expression (A). Tumors are separated into low vs. high chr22:36662161 editing frequency (B). P-values are calculated from Chi-Square tests.
3.5. ADAR Regulated Genes Come From Apoptosis and Innate Immune Related Pathways
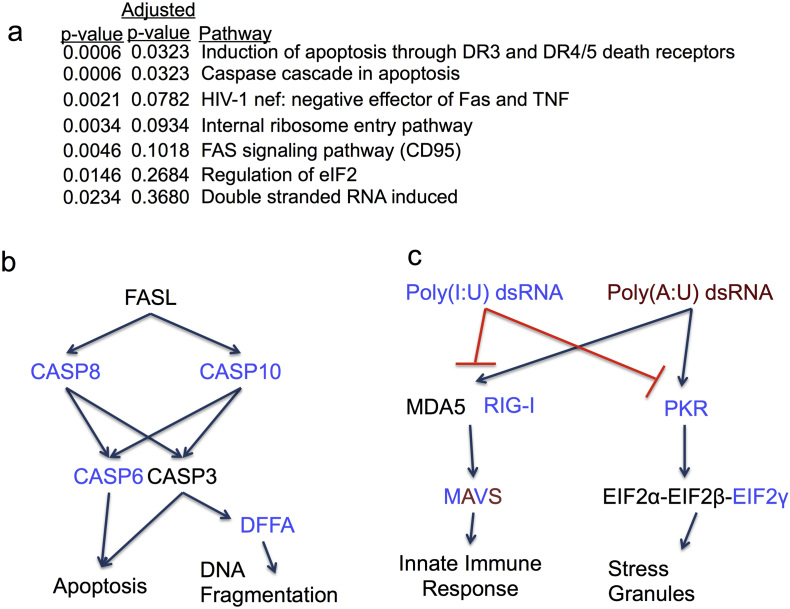
We performed functional enrichment (enrichr.org) (Chen et al., 2013a, Kuleshov et al., 2016) of the genes with five or more regulatory RNA editing sites in LUAD and found enrichment of apoptosis related genes (Fig. 5A). In particular, we found that DFFA, CASP6, CASP8, CASP10, MDM2, and CFLAR all contain multiple predicted regulatory RNA editing sites (shown in parentheses). These genes are inhibitors of the FAS ligand induced apoptotic pathway and except for MDM2 (Zhang et al., 2016), have not yet been linked to ADAR-mediated regulation (Fig. 5B).
Fig. 5.
RNA-editing regulated genes in LUAD are enriched in apoptosis and innate immune-related genes. Biocarta pathway enrichments of genes with 5 or more predicted regulatory RNA editing sites (A). Pathway diagram showing the genes that are enriched in the apoptosis (B) and innate immune-related pathways (C). Blue and red colored proteins indicate positive and negative regulation by ADAR-mediated RNA editing, respectively. The MAVS protein is colored in both blue and red because there are both positive and negative regulatory edits within its 3′ UTR.
In addition, we found enrichment of 2 pathways related to the innate immune functions of ADAR (significant by p-value but not after multiple hypothesis correction). Most importantly, EIF2AK2 (PKR), EIF2S3 (EIF2γ), DDX58 (RIG-I) and MAVS contain numerous regulatory edits (Fig. 2). These proteins are all interferon stimulated, and act in concert to inhibit the replication of dsRNA viruses. PKR phosphorylates EIF2α and leads to inhibition of the EIF2 complex, of which EIF2γ is the core subunit (Carpenter et al., 2014) (Fig. 5C). Inhibition of the EIF2 complex halts translation and leads to stress granule responses (Mccormick and Khaperskyy, 2017). Our results predict that ADAR positively regulates these genes via editing of their 3′ UTR's. In addition, Li Y., et al. noted that ADAR knockdown in interferon stimulated human LUAD A549 cells prevented the expected upregulation of EIF2AK2, suggesting that ADAR is necessary for interferon-mediated EIF2AK2 expression. The authors were unable to explain this finding; however, our method predicts that ADAR upregulates EIF2AK2 via editing of its 3′ UTR (Li et al., 2017).
Interlinked with stress granule formation in the innate immune response to dsRNA, is the MDA5-MAVS complex. MDA5 binds to unedited dsRNA molecules and is the first step of the dsRNA antiviral interferon response (Wu et al., 2013). The MAVS protein binds to MDA5 and forms filaments on the mitochondrial membrane and acts as an adaptor in this process (Hou et al., 2011). MAVS contains both positive and negative regulatory RNA editing sites in LUAD. In fact, RNA editing sites within the MAVS 3′ UTR that have higher mean editing frequency tend to be more negatively associated with MAVS abundance (Rho = − 0.57, p = 6.6 × 10− 11, Supplementary Fig. 8A). This suggests that there is a more complex regulatory process governing MAVS RNA abundance. This pattern is present in other RNA molecules with multiple negative regulatory sites, such as LIMD1 and VHL (Supplementary Fig. 8B—C).
It is important to note that the same bias that has caused enrichment in positive regulation by RNA editing in a majority of regulatory RNA editing sites may also bias the pathway enrichment analysis we performed. In addition, we performed an analysis to uncouple the upstream effects of interferon pathway expression on potential regulatory RNA editing sites. We split patients into four groups based on interferon pathway expression and RNA editing levels at each RNA editing site, and then compared the gene expression for genes with potential regulatory RNA editing sites between these groups. We tested 6139 regulatory RNA editing sites for potential regulation by RNA editing independent of interferon pathway expression. 2494 regulatory RNA editing sites were measured in a sufficient number of tumors (N > 200) for comparison, and we found evidence of widespread regulation of their host genes by RNA editing independent of interferon pathway expression (full data available in Supplementary Table 3). In addition, there are numerous genes whose expression is both independently associated with RNA editing levels and interferon levels. For example, EIF2AK2 is highly expressed in tumors with high 3′ UTR RNA editing and high interferon pathway expression; moderately expressed in tumors with either high 3′ UTR RNA editing or high interferon pathway expression; and lowly expressed in tumors with low 3′ UTR RNA editing and low interferon pathway expression (Supplementary Fig. 9). This result is consistent with the finding by Li et al. that both ADAR and interferon expression are necessary to induce EIF2AK2 expression (Li et al., 2017).
3.6. ADAR Copy Number is Anti-correlated With Immune and Apoptotic Signatures in Lung Adenocarcinoma
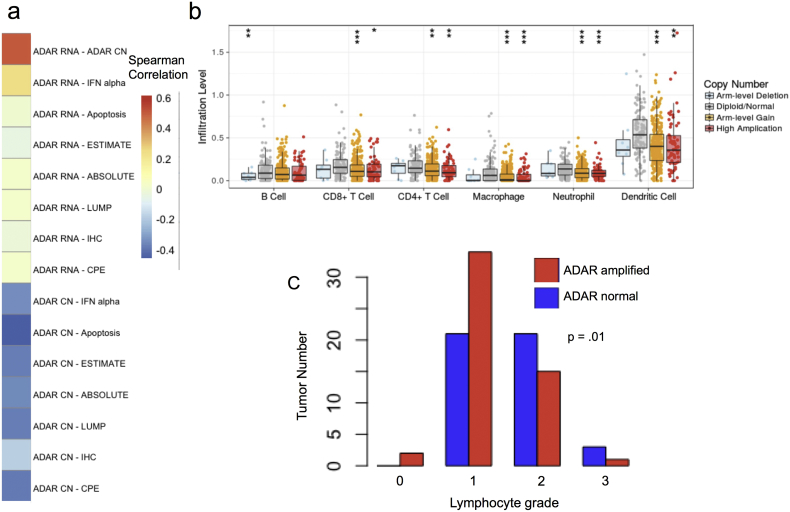
To investigate the role of ADAR in immune and apoptotic pathways we calculated the pathway enrichment in each tumor sample using single sample gene set enrichment analysis (ssGSEA) (Barbie et al., 2009) on the 50 MSigDB hallmark gene sets (Liberzon et al., 2015). ADAR was initially identified as an oncogene due to its frequent amplification in several human cancers (Han et al., 2015). As such, we investigated the association between ADAR CN and pathway activity, and found that for many pathways, the association between ADAR CN and pathway activity is the opposite of the association between ADAR RNA abundance and pathway activity, in spite of a high ADAR CN-RNA correlation (ρ = 0.50). We found that ADAR CN is strongly negatively associated with apoptotic activity in LUAD (Fig. 6A). ADAR RNA abundance was not correlated with the apoptosis pathway in LUAD. Similarly for immune related measures, ADAR RNA is positively correlated with the interferon alpha pathway but ADAR CN is negatively associated with it (Fig. 6A). One possible explanation for this finding is that separate ADAR isoforms have opposing effects on immune and apoptotic pathways. The ADAR p150 isoform contains exon 1 while ADAR p110 does not; however, the exons are too highly correlated to distinguish between their expression. The lowest correlation between exon 1 and exons 2–15 expression is 0.77. Therefore, we cannot distinguish between the expression of ADAR p150 or ADAR p110 in this cohort.
Fig. 6.
ADAR amplification is associated with decreased immune cell concentrations in lung adenocarcinoma. Fig. 6A shows the correlation between ADAR RNA and CN abundances and relevant pathway, tumor purity, and immune infiltrate signatures. The TIMER app was used to estimate immune cell subtype concentrations in LUAD tumors with known ADAR CN status (B). Tumors with ADAR CN gain or amplification had evidence of significantly fewer CD8 + T Cells, CD4 + T Cells, Macrophages, Neutrophils, and Dendritic Cells. Histological slides were scored for their lymphocyte concentrations on a grade 0–3 (C). Significance codes for (B): 0 ≤ *** ≤ 0.001 ≤ ** ≤ 0.01 ≤ * ≤ 0.05.
Given the finding that ADAR CN is negatively associated with signatures of apoptosis and innate immunity, we investigated the association between ADAR CN and immune infiltrates. We first downloaded tumor purity and immune infiltrate data from Aran et al. (2015) and then used the TIMER app (https://cistrome.shinyapps.io/timer/) (LiLi et al., 2016) to test for associations with specific immune cell types. We found that ADAR CN is significantly negatively associated with all markers of immune infiltrate and tumor purity used in Aran et al. (Fig. 6A). In fact, given the strength of the association, we searched the entire genome for genes whose copy numbers negatively correlate with immune infiltrate signatures in LUAD. It was found that genes most negatively associated with immune infiltrates reside on the 1q21 locus (Supplementary Figs. 10–11), and form a coherent amplicon that includes ADAR (Supplementary Fig. 11B). We further discovered that ADAR genomic gains and amplifications have significantly fewer predicted concentrations of CD8 + T cells, CD4 + T cells, macrophages, neutrophils, and dendritic cells (Fig. 6B). It is possible that other genes located at the 1q21 locus mediate this effect, and the causal role of ADAR in immune exclusion requires functional validation.
To confirm this finding, we graded LUAD tumors based on their apparent infiltration of different immune cell types on a 0–3 scale in histological slides from TCGA (http://cancer.digitalslidearchive.net) (Gutman et al., 2013). We selected 97 LUAD tumors, 45 of which had no evidence of ADAR amplification, and 52 of which had high ADAR copy number, and conducted the experiment with no knowledge of the ADAR CN status of these tumors. We were able to visually estimate neutrophil, lymphocyte, and macrophage infiltrations, as well as necrosis. While no apparent pattern was observed for neutrophils, necrosis, and macrophages, there was a trend towards fewer lymphocytes in ADAR CN high tumors (t-test, p = 0.01) (Fig. 6C).
The fundamental question is, does RNA abundance regulation by RNA editing account for the differences between ADAR RNA and copy number pathway enrichments? We looked at the independent effects of interferon pathway expression and ADAR copy number on editing levels, by doing a differential editing experiment with one of the variables controlled. The differences in editing between copy number high vs. low and interferon high vs. low are extremely similar (ρ = 0.76). In other words, whether ADAR is upregulated by interferon or copy number amplifications doesn't matter, the same RNA editing sites are changed. This is evidence against RNA abundance regulation by RNA editing as being the driving force behind the intransitive relationship that we see between ADAR RNA, copy number, and interferon expression.
4. Discussion
Despite the prevalence of A-I RNA editing of non-coding regions within mRNAs, very little research has been done to elucidate the functions of these alterations in cancer. To remedy this situation, we perform a comprehensive analysis of RNA abundance regulation by RNA editing. These results are a resource to better understand the regulation of thousands of genes, and a full list of genes with regulatory edits is provided in Supplementary Table 1. Despite confirmation of some of these results in an ADAR knockdown experiment, further functional validation is necessary to confirm the causality of these regulatory relationships. This method can be run on any dataset of matched RNA abundance and RNA editing frequencies, and it provides nucleotide level information to better guide validation experiments. For example, our method could be used as a resource to understand the RNA regulatory landscape of normal tissues from data published by (Tan et al., 2017). A limitation of our method is a lesser ability to detect negative compared to positive regulatory relationships. Detection of negative relationships may require time-lapse functional experiments, where the degradation of heavily edited transcripts can be tracked in real time.
In addition, an integrative analysis of ADAR CN and pathway enrichments showed that despite the high correlation between ADAR CN and RNA, ADAR copy number and RNA tend to have opposite relationships with immune and apoptosis pathways. ADAR is an interferon stimulated gene (Pestal et al., 2015), and positive correlation between ADAR RNA and immune regulated pathways could point towards ADAR being upregulated in a compensatory fashion by interferon stimulation. Sensing of aberrant DNA or RNA, such as by toll like receptors and MDA5 can cause this interferon stimulation. When ADAR is genomically amplified, these patterns could imply that unregulated increases in ADAR expression constitutively repress interferon in the tumor microenvironment and drive tumor immune evasion. It has been shown previously that poly(I:U) dsRNA is sufficient to decrease interferon responses (Vitali and Scadden, 2010). In addition, this finding raises the possibility of a synergism between ADAR inhibition and immunotherapies. Three studies recently discovered that DNA methylation inhibitors induce an interferon response via expression of aberrant dsRNA's, and can even induce sensitivity to immune checkpoint therapies (Chiappinelli et al., 2015, Roulois et al., 2015, Wrangle et al., 2013). In a similar manner, ADAR inhibition could be a new addition to combination immunotherapies.
The following are the supplementary data related to this article.
Predicted regulatory RNA editing sites in lung adenocarcinoma.
Comparison between predicted regulatory RNA editing sites and results from Wang et al.
Results of interferon controlled differential expression experiment.
Supplementary figures for Global Transcriptome Analysis of RNA Abundance Regulation by ADAR in Lung Adenocarcinoma.
Funding Sources
This work was supported by the National Library of Medicine (4 T15 LM011270-05), the National Cancer Institute (ITCR U01 CA188547-03), Leidos (15X040), and the Shenzhen Peacock Plan (KQTD2016053112051497).
Conflicts of Interest
The authors of this manuscript report no conflicts of interest.
Author Contributions
MFS conceived of the study, wrote and implemented the code, and wrote the manuscript. DA assisted with the immune cell infiltrate and pathway enrichment analyses. IK assisted with the RNA binding motif analysis. BC, DPC, PM, and KH all provided mentorship and assisted in the formulation of the manuscript. DDS analyzed the tumor slides for immune cell abundances.
Acknowledgements
We thank Jin Billy Li and Han Leng for providing useful conversations and sharing the RNA editing compilation software.
Contributor Information
Parag Mallick, Email: paragm@stanford.edu.
Kun Huang, Email: kunhuang@iu.edu.
References
- Anadón C. Gene amplification-associated overexpression of the RNA editing enzyme ADAR1 enhances human lung tumorigenesis. Oncogene. 2015:1–7. doi: 10.1038/onc.2016.27. http://www.nature.com/doifinder/10.1038/onc.2015.469 October. Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D., Sirota M., Butte A.J. Systematic pan-cancer analysis of tumour purity. Nat. Commun. 2015;6:8971. doi: 10.1038/ncomms9971. http://www.ncbi.nlm.nih.gov/pubmed/26634437 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn J.H. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways. Nat. Commun. 2015;6:6355. doi: 10.1038/ncomms7355. http://www.ncbi.nlm.nih.gov/pubmed/25751603 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie D.A. Systematic RNA interference reveals that oncogenic KRAS -driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. November. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S. Post-transcriptional regulation of gene expression in innate immunity. Nat. Rev. Immunol. 2014;14(6):361–376. doi: 10.1038/nri3682. http://www.ncbi.nlm.nih.gov/pubmed/24854588 Available at: [DOI] [PubMed] [Google Scholar]
- Chan T.H.M. ADAR-mediated RNA editing predicts progression and prognosis of Gastric Cancer. Gastroenterology. 2016;151(4):637–650. doi: 10.1053/j.gastro.2016.06.043. https://doi.org/10.1053/j.gastro.2016.06.043 e10. Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.Y. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinforma. 2013;14(128) doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013;19(2):209–216. doi: 10.1038/nm.3043. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3783260&tool=pmcentrez&rendertype=abstract Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli K.B. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162(5):974–986. doi: 10.1016/j.cell.2015.07.011. https://doi.org/10.1016/j.cell.2015.07.011 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson E.a. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. http://www.nature.com/doifinder/10.1038/nature13385 Available at: (Accessed July 9, 2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli D. Principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Rep. 2015;13(2):277–289. doi: 10.1016/j.celrep.2015.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015;21(8):1–12. doi: 10.1038/nm.3909. http://www.nature.com/doifinder/10.1038/nm.3909 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- George C.X. Editing of cellular self RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses. J. Biol. Chem. 2016;291(12):6158–6168. doi: 10.1074/jbc.M115.709014. http://www.jbc.org/lookup/doi/10.1074/jbc.M115.709014 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman D. Cancer digital slide archive: an informatics resource to support integrated in silico analysis of TCGA pathology data. J. Am. Med. Inform. Assoc. 2013;20(6):1091–1098. doi: 10.1136/amiajnl-2012-001469. http://www.ncbi.nlm.nih.gov/pubmed/23893318%5Cnhttp://jamia.bmj.com/content/20/6/1091.short Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015;28(4):515–528. doi: 10.1016/j.ccell.2015.08.013. http://linkinghub.elsevier.com/retrieve/pii/S1535610815003049 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146(3):448–461. doi: 10.1016/j.cell.2011.06.041. https://doi.org/10.1016/j.cell.2011.06.041 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–1120. doi: 10.1016/j.cell.2012.08.029. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3557932&tool=pmcentrez&rendertype=abstract Available at: (Accessed July 9, 2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc. Natl. Acad. Sci. U. S. A. 2013;110(3):1041–1046. doi: 10.1073/pnas.1213021110. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3549099&tool=pmcentrez&rendertype=abstract Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerpedjiev P., Hammer S., Hofacker I.L. Forna (force-directed RNA): simple and effective online RNA secondary structure diagrams. Bioinformatics. 2015;31(20):3377–3379. doi: 10.1093/bioinformatics/btv372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(May):90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Bo. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(174):1–16. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Ribonuclease L mediates the cell-lethal phenotype of double-stranded RNA editing enzyme ADAR1 deficiency in a human cell line. eLife. 2017;6(e25687):1–18. doi: 10.7554/eLife.25687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A. The molecular signatures database. Cell Systems. 2015;1(1):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddicoat B.J. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349(6252):1115–1120. doi: 10.1126/science.aac7049. http://www.sciencemag.org/cgi/doi/10.1126/science.aac7049 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion N.M. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9(4):1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccormick C., Khaperskyy D.A. Translation inhibition and stress granules in the antiviral immune response. Nat. Rev. Immunol. 2017;17:647–660. doi: 10.1038/nri.2017.63. https://doi.org/10.1038/nri.2017.63 Available at: [DOI] [PubMed] [Google Scholar]
- Monajemi H. The Apolipoprotein L gene cluster has emerged recently in evolution and is expressed in human vascular tissue. Genomics. 2002;79(4):539–546. doi: 10.1006/geno.2002.6729. [DOI] [PubMed] [Google Scholar]
- Nichols B. Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int. 2015;87(2):332–342. doi: 10.1038/ki.2014.270. https://doi.org/10.1038/ki.2014.270 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa A. APOL1 risk variants, race, and progression of chronic kidney disease. N. Engl. J. Med. 2013;369(23):2183–2196. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz I. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 2014;42:361–367. doi: 10.1093/nar/gku406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Yaacov N. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 2015;13(2):267–276. doi: 10.1016/j.celrep.2015.08.080. http://linkinghub.elsevier.com/retrieve/pii/S2211124715009936 Available at: [DOI] [PubMed] [Google Scholar]
- Pestal K. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity. 2015;43(5):933–944. doi: 10.1016/j.immuni.2015.11.001. https://doi.org/10.1016/j.immuni.2015.11.001 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G., Li J.B. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014;42(D1):109–113. doi: 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulois D. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts article DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162(5):961–973. doi: 10.1016/j.cell.2015.07.056. https://doi.org/10.1016/j.cell.2015.07.056 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai M. ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol. 2017;24(6):534–543. doi: 10.1038/nsmb.3403. https://doi.org/10.1038/nsmb.3403 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellos K. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016;22(10):1140–1150. doi: 10.1038/nm.4172. http://www.ncbi.nlm.nih.gov/pubmed/27595325 Available at: [DOI] [PubMed] [Google Scholar]
- Tan M.H. Dynamic landscape and regulation of RNA editing in mammals. Nature. 2017;550(7675):249–254. doi: 10.1038/nature24041. http://www.nature.com/doifinder/10.1038/nature24041 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhamme L. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422(March):83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- Vitali P., Scadden A.D.J. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat. Struct. Mol. Biol. 2010;17(9):1043–1050. doi: 10.1038/nsmb.1864. https://doi.org/10.1038/nsmb.1864 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I.X. ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep. 2013;5(3):849–860. doi: 10.1016/j.celrep.2013.10.002. https://doi.org/10.1016/j.celrep.2013.10.002 Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrangle J. Alterations of immune response of non-small cell lung cancer with Azacytidine. Oncotarget. 2013;4(11):2067–2079. doi: 10.18632/oncotarget.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152(1–2):276–289. doi: 10.1016/j.cell.2012.11.048. https://doi.org/10.1016/j.cell.2012.11.048 Available at: [DOI] [PubMed] [Google Scholar]
- Yang C. ADAR1-mediated 3′ UTR editing and expression control of antiapoptosis genes fine-tunes cellular apoptosis response. Cell Death Dis. 2017;8:e2833–13. doi: 10.1038/cddis.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Altered RNA editing in 3′ UTR perturbs microRNA-mediated regulation of oncogenes and tumor-suppressors. Sci. Rep. 2016;6:23226. doi: 10.1038/srep23226. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4793219&tool=pmcentrez&rendertype=abstract November 2015. Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Predicted regulatory RNA editing sites in lung adenocarcinoma.
Comparison between predicted regulatory RNA editing sites and results from Wang et al.
Results of interferon controlled differential expression experiment.
Supplementary figures for Global Transcriptome Analysis of RNA Abundance Regulation by ADAR in Lung Adenocarcinoma.