Abstract
Significant advances have been made in the application of pharmacogenomic testing for the treatment of patients with psychiatric disorders. Over the past decade, a number of studies have evaluated the utility of pharmacogenomic testing in pediatric patients with psychiatric disorders. The evidence base for pharmacogenomic testing in youth with depressive and anxiety disorders as well as attention/deficit hyperactivity disorder (ADHD) is reviewed in this paper. General pharmacogenomic principles are summarized and functional polymorphisms in P450 enzymes (and associated metabolizer phenotypes), the serotonin transporter promoter polymorphisms, serotonin 2A receptor genes (e.g., HT2AR) and catecholamine pathway genes (e.g., COMT) are reviewed. These commonly tested pharmacogenomic markers are discussed with regard to studies of drug levels, efficacy and side effects.
The translation of pharmacogenomics to individualized/precision medicine in pediatric patients with ADHD, anxiety and depressive disorders has accelerated; however, its application remains challenging given that there are numerous divergent pathways between medication/medication dose and clinical response and side effects. Nonetheless, by leveraging variations in individual genes that may be relevant to medication metabolism or medication target engagement, pharmacogenomic testing may have a role in predicting treatment response, side effects and medication selection in youth with ADHD, depressive and anxiety disorders.
Keywords: Selective Serotonin Reuptake Inhibitor (SSRI), Selective Norepinephrine Reuptake Inhibitor (SNRI), antidepressants, psychopharmacology, stimulants
INTRODUCTION
The promise of personalized medicine and of genetically-informed treatment selection that heralded the beginning of this millennium has yet to be fully-realized. However, great advances have been made with regard to the application of pharmacogenomic testing to the treatment of patients with psychiatric disorders. To date, much of this work has been conducted in adults treated with antidepressants, stimulants, second generation antipsychotics and antiepileptic medications. The last decade has seen an increasing number of studies examining the utility of pharmacogenomic testing as a predictor of treatment response and medication tolerability in pediatric patients.
Pharmacogenetic testing—in its most basic application—leverages variations in individual genes relevant to medication metabolism or targets to predict treatment response and, may guide treatment selection. Ideally, this testing would maximize the likelihood that a specific psychotropic medication produces the best therapeutic benefit while at the same time would minimize adverse effects. Initial use of pharmacogenomic testing focused on single genes (e.g., cytochrome P450 genes) related to medication metabolism and consequently medication exposure. The first FDA-approved pharmacogenetic test, the AmpliChip CYP450 employed a DNA microarray to assess two polymorphisms in CYP2D6 and CYP2C19 (Roche molecular Systems, Inc: AmpliChip CYP450 Test for in vitro diagnostic use). However, more recent strategies utilize combinatorial strategies that rely on (usually proprietary) algorithms; such a test might utilize the genotype for a series of genes (several cytochromes, the serotonin transporter (SLC6A4), and the serotonin 2A receptor (5HT2A), etc.) to generate a profile that aims to guide prescribing for several classes of psychiatric medications.
Given the increase in pharmacogenomic testing and the rapid pace at which data are accumulating in pediatric patients, we sought: (1) to summarize the underlying principles of pharmacogenomic testing in youth with psychiatric disorders; (2) to review current data that support its use and; (3) to discuss potential limitations.
Cytochrome P450 enzymes
The cytochrome P450 system refers to a group of enzymes called monoxygenases that are critical for elimination of many drugs. They are expressed in every tissue although greatest expression is observed in hepatic tissue. Collectively, these enzymes represent Phase I metabolism; they catalyze the transformation of lipophilic drugs into more polar compounds that are then excreted by the kidneys. The genes, and consequently the proteins that they encode, are highly variable as multiple alleles exist for each cytochrome (e.g., CYP2D6—more than 100 alleles have been identified). Specific alleles may code for a fully functional enzyme or an enzyme with decreased or absent activity. In addition, an individual may express multiple copies of an active allele or an allele with enhanced activity, resulting in an increased metabolic rate compared to the wild type (i.e., the prevailing phenotype or “normal” form in the population). A “star” designation is used to refer to specific alleles, with *1 denoting the standard by which the activity of enzymes encoded by other alleles is measured.
An individual’s metabolizer phenotype for a particular cytochrome takes into account the activity of each of the patient’s two alleles (e.g., *1/*2). Given the variation associated with each cytochrome, classifying patients into four main groups based on metabolic phenotype represents the most common approach to assigning phenotypes. Using the current standard terminology from the Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network,1 individuals are classified as extensive (normal), poor, intermediate, and ultrarapid metabolizers (Table 1). This classification system may have significant limitations that are most problematic in the extreme phenotypes (i.e., ultrarapid and poor metabolizers). Thus, some laboratories have reported metabolic phenotypes using a seven-category classification system, that adds phenotypes: enhanced extensive, enhanced intermediate, and reduced intermediate. Examples of allele combinations which could theoretically give rise to these phenotypes are given in Table 2.
TABLE 1.
Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for consideration of genotype and medication dosing or selection for antidepressant medications.
| Medication | Cytochrome | Phenotype | Recommendation | Strength of Recommendation |
|---|---|---|---|---|
| Paroxetine | CYP2D6 | Ultrarapid | Select alternative drug not metabolized by CYP2D6 | Strong |
| Extensive | Initiate therapy at standard starting dose | Strong | ||
| Intermediate | Initiate therapy at standard starting dose | Moderate | ||
| Poor | Consider 50% reduction of recommended starting dose or select alternative drug | Optional | ||
| Fluvoxamine | CYP2D6 | Ultrarapid | No recommendations due to lack of data | |
| Extensive | Initiate therapy at standard starting dose | Strong | ||
| Intermediate | Initiate therapy at standard starting dose | Moderate | ||
| Poor | Consider 25–50% reduction of starting dose or use alternative drug | Optional | ||
| Citalopram, | CYP2C19 | Ultrarapid | Consider using alternative drug | Moderate |
| escitalopram | Extensive | Initiate therapy at standard starting dose | Strong | |
| Intermediate | Initiate therapy at standard starting dose | Strong | ||
| Poor | Consider 50% reduction of standard starting dose or select alternative drug | Moderate | ||
| Sertraline | CYP2C19 | Ultrarapid | Initiate therapy at standard starting dose and consider alternative drug if patient does not respond | Optional |
| Extensive | Initiate therapy at standard starting dose | Strong | ||
| Intermediate | Initiate therapy at standard starting dose | Strong | ||
| Poor | Consider 50% reduction of standard starting dose or select alternative drug | Optional |
TABLE 2.
Metabolizer Phenotypes for P450 Systems Based on “Traditional” and “Seven Category” Categorization. Adapted from Mrazek, 2010.
| Traditional Classification | Ultra-Rapid | Extensive | Intermediate | Poor | |||
|---|---|---|---|---|---|---|---|
| Seven Category Classification | Enhanced extensive | Extensive | Enhanced intermediate | Intermediate | Reduced intermediate | Poor | |
| Example genotypes | >2 alleles with normal activity > 2 alleles with ↑ activity | 1 allele with ↑ activity + 1 allele with normal activity | 2 alleles with normal activity 1 allele with ↑ activity + 1 allele with ↓ activity | 1 allele with ↑ activity + 1 allele with no activity 1 normal allele + 1 allele with ↓ activity | 1 normal allele + 1 allele with no activity 2 alleles with ↓ activity | 1 allele with ↓ activity + 1 allele with no activity | No alleles with any activity |
A patient’s response to a medication is highly dependent on cytochrome P450 enzymatic activity. This phenotypic variance is clinically significant; a patient who is a “poor metabolizer” for a particular P450 enzyme may require lower doses of a medication that is metabolized through that system or he/she will be at increased risk for developing adverse effects. In contrast, ultrarapid metabolizers may not benefit from standard therapeutic doses, they will have sub-therapeutic medication levels and, in some cases with medications with short half-lives, they may experience withdrawal symptoms. This may be particularly relevant for paroxetine in the pediatric population. Phase I metabolism (i.e., oxidation via cytochrome P450, reduction, and hydrolysis reactions that occur in the liver and are responsible for converting medications to more polar, water-soluble, metabolites) is increased in children. This has been hypothesized to underlie the poor tolerability of this medication in pediatric patients with depression and anxiety disorders.2 On the other hand, medications that must be activated through a cytochrome P450 (e.g., codeine and CYP2D6), poor metabolizers receive little pain relief and ultrarapid metabolizers are at increased risk of side effects from supra-therapeutic levels of the active metabolite (morphine). Additionally, allelic frequencies vary widely across ethnic groups (Figure 1), and as such, knowledge of this ethnicity-related variability may inform testing.
FIGURE 1.
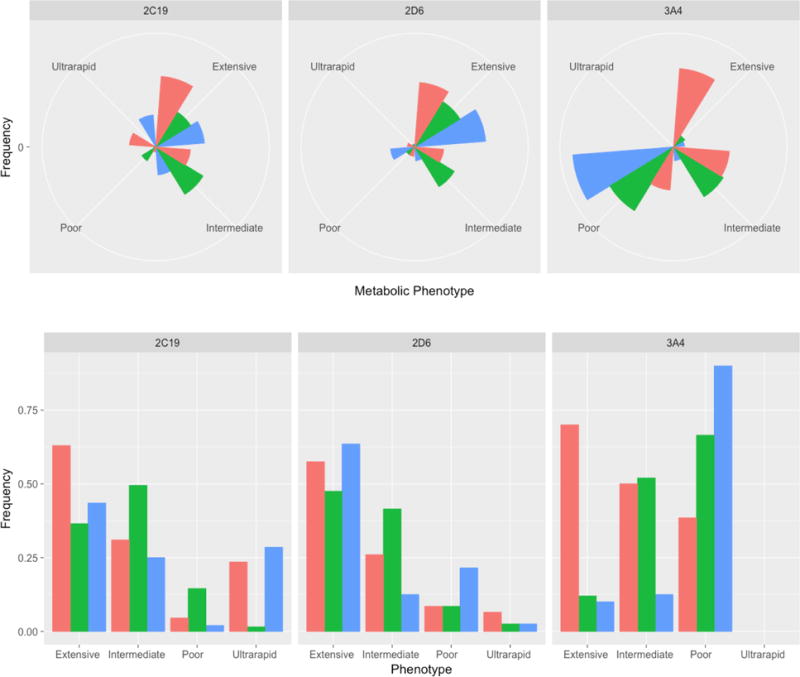
Ethnic variability in CYP450 Phenotypes. Allelic frequencies for phenotypes of 2C19, 2D6 and 3A4 vary across ethnic groups, which may inform testing. These variations are presented for the following ethnicities: (1) African, (2) Asian and (3) Phenotypic frequencies are shown below for Africans (red), Asians (green) and Northern Europeans (blue).
CYP2D6
This cytochrome is the most well-characterized P450 enzyme in humans. It is involved in the metabolism of >70 available medications via Phase I oxidation reactions3 and varies significantly across ethnic groups (Figure 1). Multiple SS(N)RIs and tricyclic antidepressants are primarily metabolized by this enzyme (e.g., fluoxetine, paroxetine, fluvoxamine, venlafaxine, amitriptyline, atomoxetine).4 In adults, plasma drug concentrations of several antidepressants are related to genotype.5 Bupropion and duloxetine are substantially, but not exclusively, metabolized by CYP2D6.6 Some medications are metabolized by CYP2D6 and also inhibit the CYP2D6 system (e.g., fluoxetine, paroxetine). Thus, an individual with an extensive metabolizer phenotype can be converted to a poor metabolizer-like phenotype following administration of these drugs, a phenomenon known as phenoconversion. With regard to atomoxetine, an SNRI used in the treatment of ADHD, pediatric patients classified as poor metabolizers based on CYP2D6 genotype were more likely to experience adverse effects from treatment with the medication when compared with extensive metabolizers. However, those patients who were able to tolerate the medication achieved significantly greater improvement in symptom severity.7
CYP2C19
More than 50 medications are metabolized primarily by this enzyme, including citalopram, escitalopram, and many tricyclic antidepressants. Sertraline is metabolized substantially, but not exclusively, by CYP2C19. The incidence of CYP2C19 poor metabolizer phenotypes is much higher in Asians compared to Caucasians3 (Figure 1). Thus, in these patients, 2C19-metabolized medications might be avoided or initiated at lower doses (Hicks et al, 2015). About a third of Caucasians are CYP2C19 ultrarapid metabolizers (Figure 1), and may require higher doses of medications metabolized by this enzyme to achieve equivalent serum concentrations of the medication compared to normal metabolizers.8 The FDA recommends that for CYP2C19 poor metabolizers, the maximum daily dose of citalopram should not exceed 20 mg due to risk of QT prolongation, although because certain patients may require higher doses of citalopram, it is prudent to evaluate cardiac risk and corrected QT interval (QTc) in these patients.
CYP1A2
The only antidepressant metabolized primarily by CYP1A2 is fluvoxamine (Mrazek, 2010), although duloxetine—the only SNRI to be approved for pediatric patients9—is substantially metabolized by this cytochrome. Clearance of duloxetine in pediatric patients is significantly higher in children aged 7-12 years compared to adolescents aged 13-17 years and lowest in patients older than 18 years.10 Median body-weight-normalized clearance is nearly 4 times faster in children and 2 times faster in adolescents. While this is probably a developmentally-related difference in Phase I metabolism rather than genetic variability in CYP1A2 or CYP2D6, this finding highlights the complexity of relying solely on CYP450 metabolizer phenotype in pediatric patients.
Tobacco is a common CYP1A2 inducer, which is relevant in the fact that if an ultrarapid metabolizer starts smoking, he/she may be at risk for loss of medication efficacy if that medication is primarily metabolized by the CYP1A2 cytochrome. Of note, decreased half-lives of some second generation antipsychotics, including olanzapine, have been observed in pediatric patients compared to adults. These findings are hypothesized to be related to reduced first-pass effects, higher bioavailability and possibly to the markedly lower rates of cigarette smoking in children and adolescents with psychotic disorders compared to adults with these conditions.11
CYP3A4
This is one of the most highly expressed P450 enzymes in the liver, and is responsible for metabolizing 50-60% of medications. Expression of this gene in the liver is highly variable,12 and can be influenced by medications, diet and environmental effects. However, very few genetic variants in this gene have been associated with enzyme activity. Antidepressants metabolized by this cytochrome include sertraline, citalopram, escitalopram, fluoxetine, venlafaxine.13 In addition, many second generation antipsychotics and anti-epileptic drugs are also CYP3A4 substrates. Because these medications are infrequently prescribed in the primary care setting, a discussion of the pharmacogenetics of these medications is beyond the scope of this review.
Development and P450 System Activity
CYP450-dependent medication metabolism is not solely influenced by functional polymorphisms, as described above, but it is also influenced by development.14
CYP2C19 activity reaches “adult range” by 6 months of age and then peaks at 1.5-1.8 times adult values by 3-4 years of age and then decreases by the end of puberty to again reach “adult levels.”
CYP3A4 activity is similar to activity in adults by 6-12 months of age and then increases relative to “adult levels” from age 1-4 years and then declines to adult levels by the end of puberty.
CYP1A2 reaches adult levels by 4 months of age, peaks at age 1-2 years, and then declines to adult levels by end of puberty.
CYP2D6 activity reaches developmental “maturity” by 3-5 years of age.15
Pharmacogenomic-Informed Dosing Guidelines
The Clinical Pharmacogenetics Implementation Consortium (CPIC) recently published guidelines regarding dosing recommendations for five commonly prescribed SSRIs (fluvoxamine, paroxetine, citalopram, escitalopram, and sertraline) based on CYP2D6 and CYP2C19 genotypes, given that polymorphisms in these genes can have significant effects on how these drugs are metabolized in different individuals (Table 2) (Hicks et al, 2015).
PHARMCOGENOMIC TESTING & PHARMACODYNAMICS
While pharmacokinetic factors determine, in part, medication absorption, distribution, metabolism, and excretion, pharmacodynamic factors are the most important determinants of medication target engagement (the interaction of the medication with the molecular target, e.g., an SSRI blocking the serotonin transporter). Multiple pharmacodynamic genes have been identified as relevant to the mechanisms of action for antidepressants, antipsychotics, stimulants and anti-adrenergic medications. Variation in these genes is being investigated with regard to psychopharmacologic intervention, in children and adults.
SLC6A4
The SLC6A4 gene is one of the most widely studied serotonergically-relevant genes. Polymorphisms in this receptor are associated with decreased expression of the serotonin transporter and may be associated with increased behavioral responses to threats,16 connectivity within in fear processing circuitry17–20 as well as activity within brain networks that are implicated in mood and anxiety disorders.20 In some studies of adults with major depressive disorder and anxiety disorders predicts antidepressant response.21–23 SLC6A4 encodes the presynaptic serotonin (5-hydroxytrypamine, [5-HT]) transporter. This facilitates the return of synaptic serotonin to the presynaptic terminal; inhibition of this transporter is thought to represent the therapeutic mechanism of the SSRIs. One significant polymorphism involves an insertion/deletion in the promoter of the gene, giving rise to a long (l) and a short (s) variant. The short variant results in decreased expression of the 5-HT transporter,24 eo ipso decreased efficiency of 5-HT reuptake and ostensibly a decrease in the substrate on which SSRIs might exert their therapeutic effects (Figure 2). Greater responses to SSRI treatment in depressed adults with two copies of the (long) allele, although not all studies have observed this association (Kraft et al, 2007) and there is some concern that ethnic differences the regulation of 5-HT transporter expression may underlie these inconsistent results observed in clinical trials. Finally, a meta-analysis of the association between the SLC6A4 polymorphism and SSRI efficacy for depression (15 studies, N=1435) revealed that the s/s variant results in lower remission rates (p<0.0001).19 Other variants in this gene may also be associated with expression of the transporter, adverse reactions and response to SSRIs.
Figure 2.
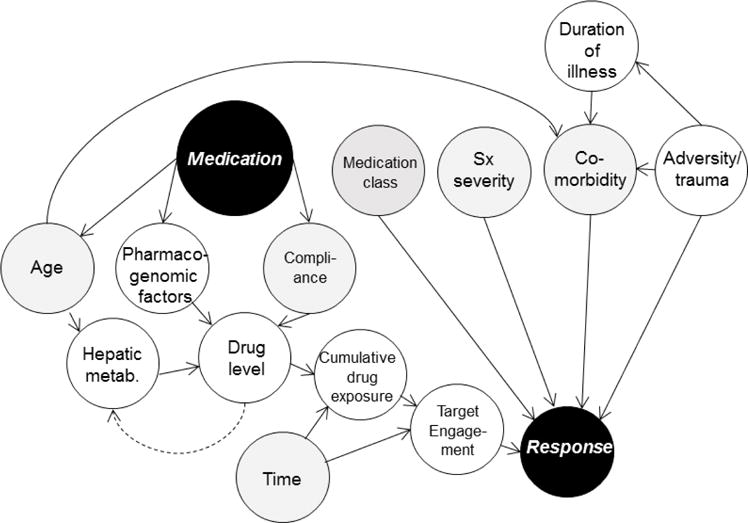
Functional causal model of response in pediatric depressive and anxiety disorders illustrating dependence among variables. The standardized dose of the antidepressant medication (black circle) to response (black circle) relationship is d-separated by the chain involving drug level and by forks including age, pharmacogenomics factors, compliance and hepatic metabolism. Additionally, target engagement refers to myriad factors including polymorphisms within the “drug target” (i.e., serotonin transporter). Gray nodes represent consistently measured variables in clinical trials of youth with mood and anxiety disorders. White nodes represent data that are rarely or inconsistently captured.
To date, no studies have evaluated the predictive utility of SLC6A4 in pediatric patients treated with antidepressants.
HTR2A
Data suggest that the HTR2A gene influences the clinical response to antidepressant treatment25 in adults and may modulate the likelihood of adverse drug reactions with certain SSRIs. Additionally, administration of paroxetine downregulates 5-HT2A receptor expression.26 To date, multiple functional polymorphisms of the HTR2A gene have been identified: a silent point mutation 102T>C that is completely linked to a -1438G>A promoter polymorphism, and a polymorphism in the coding region causing a His452Tyr amino-acid substitution. Greater treatment responses have been observed in antidepressant-treated patients with ≥1 C-alleles of the 102T>C polymorphism relative to patients who were T/T homozygous.27
There are currently no available data regarding treatment response alleles in pediatric patients but they have been evaluated for association with side effects during citalopram treatment in pediatric patients.28
COMT
Catechol-o-methyltransferase is the primary enzyme that inactivates circulating catecholamines (i.e., NE/DA), which are pathophysiologically implicated in ADHD and represent targets for stimulant pharmacotherapy in youth with ADHD. One specific polymorphism in this gene involves an amino acid substitution from valine to methionine that results in a 40% reduction in enzymatic activity29 and thus higher synaptic levels of catecholamines in the brain. Several studies have found an association between this COMT substitution and decreased response rates to methylphenidate30,31 although not all studies have observed this relationship.32,33 There was a strong trend observed in the study by Froehlich and colleagues33 towards COMT substitutions predicting treatment response. Similarly, studies have found an association between the val/val genotype and a more favorable response to amphetamines.34
Pharmacogenomic Testing & Side Effects and Tolerability
The ability to predict adverse effects with the use of pharmacogenetic testing may increase the success of psychopharmacologic interventions in youth. Moreover, adverse effects such as activation, gastrointestinal symptoms, and dysomnia in youth create stress and uncertainty for families and patients and may negatively impact the relationship between the clinician and the patient/family. The ability to predict side effects would enhance clinical outcomes. Bohm and Cascorbi35 classify adverse drug reactions as Type A and B. Type A reactions are mediated by the concentration of the medication, and therefore associated with pharmacogenetic polymorphisms. Type B reactions are drug hypersensitivities mediated by the immune system and not concentration-related.
At present, there are few pharmacogenomics studies that report associations between specific polymorphisms or phenotypes and specific side effects. Side effects due to increased plasma concentrations probably occur in youth who are poor metabolizers. The best evidence for this has been observed in youth treated with atomoxetine7. Poor metabolizers of CYP2D6 exhibited greater treatment-response, but they also experienced more side effects, including increases in heart rate and blood pressure and decreased velocity of weight gain.7 A similar association of poor metabolizers having greater treatment response and higher incidence of side effects was demonstrated in a study of psychiatrically-hospitalized pediatric patients treated with psychotropic medications.36 A study of autistic children treated with risperidone monotherapy revealed polymorphisms in the 5-HT2A receptor as well as the 5-HT2C receptor. A BDNF-related polymorphism produced treatment-related increases in prolactin while polymorphisms in the 5-HT2C receptor as well as CYP2D6 phenotype predicted risperidone-related increases in BMI and/or waist circumference (Correia et al. 2010). Two large studies have evaluated the association between specific polymorphisms in the dopamine transporter and COMT and methylphenidate-related side effects. In youth, aged 5-16 years of age, COMT-related polymorphisms were associated with more somatic symptoms during treatment with methylphenidate. In the other study, McGuff and colleagues37 found that individuals who were val/val with regard to COMT had significantly more irritability associated with treatment. There were also strong trends for a relationship between DRD4PROM polymorphisms and the COMT val/val with somatic symptoms in methylphenidate-treated patients. In the same study, McGuff and colleagues37 noted a strong association between the SCL6A4 and vegetative symptoms with individuals who were long/long have a decreased incidence of vegetative symptoms.37
Limitations of Pharmacogenetic Testing in Children & Adolescents
While the study of pharmacogenetics appears promising to predict drug levels for some medications as well as side effects for some individuals, there are several limitations regarding its implementation into standard clinical practice. At this time, prospective studies are needed that will examine the benefit of medication selection and/or dose adjustments based on a patient’s individual genotype prior to starting treatment. It is possible to use genetic information to calculate theoretical dose adjustments based on a patient’s individual phenotype, although it is unclear if this provides a true therapeutic benefit for children and adolescents.
While SSRIs are first line treatment for pediatric anxiety and depressive disorders, this class of medications has a wide therapeutic window and it is unclear if higher plasma drug concentrations are associated with improved efficacy. Antipsychotic-related weight gain may be more common in pediatric patients than adults, and also needs further study.38 Most studies to date have been performed exclusively in adults, with results extrapolated to pediatric populations. Most studies have also been performed on one gene at a time, whereas it may take a combination of pharmacokinetic and pharmacodynamic genes to predict treatment response more accurately.
DISCUSSION
Psychiatric illnesses are common in children and adolescents and, despite available psychopharmacologic and psychotherapeutic interventions, often result in significant morbidity including decreases in quality of life.39,40 It is common for pediatric patients with psychiatric disorders to experience trials of multiple psychotropic medications prior to identifying a medication that is well tolerated and reduces symptoms. Due to the high frequency of toxicity-related side effects (observed in 20-70% of patients), pediatricians, child and adolescent psychiatrists and other clinicians often initiate pharmacotherapy at low doses and slowly titrate the dose until the patient either experiences an intolerable side effect or achieves symptomatic remission.41 The practice of slow titration of medication may minimize the emergence of side effects, it may increase the risk of under-treatment and may lead to a medication change due to the lack of treatment response (especially in ultrarapid metabolizers). Pediatricians as well as child and adolescent psychiatrists have multiple medications from which to choose for treating anxiety and depressive disorders as well as ADHD;42–45 however, many pediatricians report that they have limited data to guide their choice of medication for an individual patient.41
Pharmacogenetically-guided dosing is common in pediatric oncology and gastroenterology (e.g., TPMT-based dosing of thiopurines) and preemptive pharmacogenetic testing is being implemented in several institutions (some of which treat pediatric patients) that include clinical decision support for psychotropic medications based on CPIC guidelines (CYP2D6 and CYP2C19).46 Evidence is accumulating for the inclusion of pharmacodynamic genes in these testing panels as well. While there are definite limitations to pharmacogenetic testing in pediatric patients, testing has the potential to decrease morbidity, decrease treatment-emergent side effects, improve treatment response, decrease inpatient admissions and readmissions due to lack of efficacy or side effects, and cost of care for the patient and his or her family. Ultimately, more research is needed to understand the contributions of pharmacogenomics testing to the clinical treatment of children and adolescents.
Acknowledgments
Dr. Strawn has received research support from the National Institutes of Health, Edgemont, Forest Research Laboratories/Allergan, Lundbeck, Neuronetics, and Shire. He has received material support from Assurex/Genesight—a company which performs pharmacogenomics testing. He has not received any direct payment from Assurex/Genesight, nor has he engaged in any promotional speaking or consultation for this or any other company. Finally, Dr. Strawn receives royalties, from Springer Publishing, for the publication of two textbooks as well as UpToDate.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The other authors report no biomedical conflicts of interest.
Contributor Information
Anna M. Wehry, University of Cincinnati, College of Medicine Cincinnati, Ohio 45267.
Laura Ramsey, Departments of Pediatrics, Division of Clinical Pharmacology, Cincinnati Children’s Hospital Medical Center.
Shane E. Dulemba, Departments of Pediatrics, Division of Child & Adolescent, Cincinnati Children’s Hospital.
Sarah A. Mossman, University of Cincinnati, College of Medicine Cincinnati, Ohio 45267.
Jeffrey R. Strawn, University of Cincinnati, College of Medicine Cincinnati, Ohio 45267.
References
- 1.Kalman LV, Agúndez JAG, Appell ML, et al. Pharmacogenetic Allele Nomenclature: International Workgroup Recommendations for Test Result Reporting. Clin Pharmacol Ther. 2016;99(2):172–185. doi: 10.1002/cpt.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner KD, Berard R, Stein MB, et al. A multicenter, randomized, double-blind, placebo-controlled trial of paroxetine in children and adolescents with social anxiety disorder. Arch Gen Psychiatry. 2004;61(11):1153–1162. doi: 10.1001/archpsyc.61.11.1153. [DOI] [PubMed] [Google Scholar]
- 3.Poolsup, Po LW, Knight Pharmacogenetics and psychopharmacotherapy. J Clin Pharm Ther. 2000;25(3):197–220. doi: 10.1046/j.1365-2710.2000.00281.x. [DOI] [PubMed] [Google Scholar]
- 4.Spina E, Scordo MG, D’Arrigo C. Metabolic drug interactions with new psychotropic agents. Fundam Clin Pharmacol. 2003;17(5):517–538. doi: 10.1046/j.1472-8206.2003.00193.x. [DOI] [PubMed] [Google Scholar]
- 5.Charlier C, Broly F, Lhermitte M. Polymorphisms in the CYP 2D6 Gene . Ther Drug Monit. 2003;25(6):738–742. doi: 10.1097/00007691-200312000-00014. [DOI] [PubMed] [Google Scholar]
- 6.Skinner MH, Kuan HY, Pan A, et al. Duloxetine is both an inhibitor and a substrate of cytochrome P4502D6 in healthy volunteers. Clin Pharmacol Ther. 2003;73(3):170–177. doi: 10.1067/mcp.2003.28. [DOI] [PubMed] [Google Scholar]
- 7.Michelson D, Read HA, Ruff DD, Witcher J, Zhang S, McCracken J. CYP2D6 and clinical response to atomoxetine in children and adolescents with ADHD. J Am Acad Child Adolesc Psychiatry. 2007;46(2):242–251. doi: 10.1097/01.chi.0000246056.83791.b6. [DOI] [PubMed] [Google Scholar]
- 8.Chang M, Tybring G, Dahl ML, Lindh JD. Impact of Cytochrome P450 2C19 Polymorphisms on Citalopram/Escitalopram Exposure: A Systematic Review and Meta-Analysis. Clin Pharmacokinet. 2014;53(9):801–811. doi: 10.1007/s40262-014-0162-1. [DOI] [PubMed] [Google Scholar]
- 9.Strawn JR, Prakash A, Zhang Q, et al. A Randomized, Placebo-Controlled Study of Duloxetine for the Treatment of Children and Adolescents With Generalized Anxiety Disorder. J Am Acad Child Adolesc Psychiatry. 2015;54(4):283–293. doi: 10.1016/j.jaac.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 10.M JS, E GJ, K CJ, et al. An open-label study of tolerability, safety, and pharmacokinetics of duloxetine in children (7–11 years) and adolescents (12–17 years) with MDD. J Child Adolesc Psychopharmacol. 2009;19(6):782–783. [Google Scholar]
- 11.Strawn JR, Delbello MP. Olanzapine for the treatment of bipolar disorder in children and adolescents. Expert Opin Pharmacother. 2008;9(3):1–8. doi: 10.1517/14656566.9.3.467. [DOI] [PubMed] [Google Scholar]
- 12.Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2012;64(SUPPL):256–269. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 13.Moore TR, Hill AM, Panguluri SK. Pharmacogenomics in psychiatry: implications for practice. Recent Pat Biotechnol. 2014;8(2):152–159. doi: 10.2174/1872208309666140904113615. [DOI] [PubMed] [Google Scholar]
- 14.Kearns GL, Abdel-Rahman SM, Alander SW, Blowey DL, Leeder JS, Kauffman RE. Developmental pharmacology–drug disposition, action, and therapy in infants and children. N Engl J Med. 2003;349(12):1157–1167. doi: 10.1056/NEJMra035092. [DOI] [PubMed] [Google Scholar]
- 15.Leeder JS, Kearns GL. Pharmacogenetics in pediatrics: Implications for practice. Pediatr Clin North Am. 1997;44(1):55–77. doi: 10.1016/S0031-3955(05)70463-6.. [DOI] [PubMed] [Google Scholar]
- 16.Kwang T, Wells TT, McGeary JE, Swann WB, Beevers CG. Association of the serotonin transporter promoter region polymorphism with biased attention for negative word stimuli. Depress Anxiety. 2010;27(8):746–751. doi: 10.1002/da.20708.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinz A, Braus DF, Smolka MN, et al. Amygdala-prefrontal coupling depends on a genetic variation of the serotonin transporter. Nat Neurosci. 2005;8(1):20–21. doi: 10.1038/nn1366. [DOI] [PubMed] [Google Scholar]
- 18.Kobiella A, Reimold M, Ulshöfer DE, et al. How the serotonin transporter 5-HTTLPR polymorphism influences amygdala function: the roles of in vivo serotonin transporter expression and amygdala structure. Transl Psychiatry. 2011;1(8):e37. doi: 10.1038/tp.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serretti A, Kato M, De Ronchi D, Kinoshita T. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with selective serotonin reuptake inhibitor efficacy in depressed patients. Mol Psychiatry. 2007;12(3):247–257. doi: 10.1038/sj.mp.4001926. [DOI] [PubMed] [Google Scholar]
- 20.Hariri AR, Mattay VS, Tessitore A, et al. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297(5580):400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- 21.Stein MB, Seedat S, Gelernter J. Serotonin transporter gene promoter polymorphism predicts SSRI response in generalized social anxiety disorder. Psychopharmacology (Berl) 2006;187:68–72. doi: 10.1007/s00213-006-0349-8.. [DOI] [PubMed] [Google Scholar]
- 22.Smeraldi E, Zanardi R, Benedetti F, Di Bella D, Perez J, Catalano M. Polymorphism within the promoter of the serotonin transporter gene and antidepressant efficacy of fluvoxamine. Mol Psychiatry. 1998;3:508–511. doi: 10.1038/sj.mp.4000425. [DOI] [PubMed] [Google Scholar]
- 23.Serretti A, Cusin C, Rausch JL, Bondy B, Smeraldi E. Pooling pharmacogenetic studies on the serotonin transporter: A mega-analysis. Psychiatry Res. 2006;145(1):61–65. doi: 10.1016/j.psychres.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 24.Heils A, Teufel A, Petri S, et al. Allelic variation of human serotonin transporter gene expression. J Neurochem. 1996;66(6):2621–2624. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- 25.Lucae S, Ising M, Horstmann S, et al. HTR2A gene variation is involved in antidepressant treatment response. Eur Neuropsychopharmacol. 2010;20:65–68. doi: 10.1016/j.euroneuro.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Meyer JH, Kapur S, Eisfeld B, et al. The effect of paroxetine on 5-HT2A receptors in depression: An [18F]setoperone PET imaging study. Am J Psychiatry. 2001;158(1):78–85. doi: 10.1176/appi.ajp.158.1.78. [DOI] [PubMed] [Google Scholar]
- 27.Kirchheiner J, Kirchheiner J, Nickchen K, et al. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry. 2004;9:442–473. doi: 10.1038/sj.mp.4001494. [DOI] [PubMed] [Google Scholar]
- 28.Amitai M, Kronenberg S, Carmel M, et al. Pharmacogenetics of citalopram-related side effects in children with depression and/or anxiety disorders. J Neural Transm. 2016;123(11):1347–1354. doi: 10.1007/s00702-016-1585-7. [DOI] [PubMed] [Google Scholar]
- 29.Lachman HM, Papolos DF, Saito T, Yu YM, Szumlanski CL, Weinshilboum RM. Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics. 1996;6(3):243–250. doi: 10.1097/00008571-199606000-00007. [DOI] [PubMed] [Google Scholar]
- 30.Cheon K-A, Jun J-Y, Cho D-Y. Association of the catechol-O-methyltransferase polymorphism with methylphenidate response in a classroom setting in children with attention-deficit hyperactivity disorder. Int Clin Psychopharmacol. 2008;23(5):291–298. doi: 10.1097/YIC.0b013e328306a977. [DOI] [PubMed] [Google Scholar]
- 31.Kereszturi E, Tarnok Z, Bognar E, et al. Catechol-O-methyltransferase Val158Met polymorphism is associated with methylphenidate response in ADHD children. Am J Med Genet Part B Neuropsychiatr Genet. 2008;147(8):1431–1435. doi: 10.1002/ajmg.b.30704. [DOI] [PubMed] [Google Scholar]
- 32.Mills S, Langley K, Van den Bree M, et al. No evidence of association between Catechol-O-Methyltransferase (COMT) Val158Met genotype and performance on neuropsychological tasks in children with ADHD: a case-control study. BMC Psychiatry. 2004;4:15. doi: 10.1186/1471-244X-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Froehlich TE, Epstein JN, Nick TG, et al. Pharmacogenetic predictors of methylphenidate dose-response in attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2011;50(11):1129–1139. doi: 10.1016/j.jaac.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattay VS, Goldberg TE, Fera F, et al. individual variation in the brain response to amphetamine. Proc Natl Acad Sci. 2003;100(10):6186–6191. doi: 10.1073/pnas.0931309100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Böhm R, Cascorbi I. Pharmacogenetics and predictive testing of drug hypersensitivity reactions. Front Pharmacol. 2016 Oct;7 doi: 10.3389/fphar.2016.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prows CA, Nick TG, Saldaña SN, et al. Drug-metabolizing enzyme genotypes and aggressive behavior treatment response in hospitalized pediatric psychiatric patients. J Child Adolesc Psychopharmacol. 2009;19(4):385–394. doi: 10.1089/cap.2008.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGough JJ, McCracken JT, Loo SK, et al. A candidate gene analysis of methylphenidate response in attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2009;48(12):1155–1164. doi: 10.1097/CHI.0b013e3181bc72e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maayan L, Correll CU. Weight Gain and Metabolic Risks Associated with Antipsychotic Medications in Children and Adolescents. J Child Adolesc Psychopharmacol. 2011;21(6):517–535. doi: 10.1089/cap.2011.0015.. [DOI] [PubMed] [Google Scholar]
- 39.Bastiaansen D, Koot HM, Ferdinand RF, Verhulst FC. Quality of life in children with psychiatric disorders: self-, parent, and clinician report. J Am Acad Child Adolesc Psychiatry. 2004;43(2):221–230. doi: 10.1097/00004583-200402000-00019. [DOI] [PubMed] [Google Scholar]
- 40.Ginsburg GS, Riddle MA, Davies M. Somatic symptoms in children and adolescents with anxiety disorders. J Am Acad Child Adolesc Psychiatry. 2006;45(10):1179–1187. doi: 10.1097/01.chi.0000231974.43966.6e. [DOI] [PubMed] [Google Scholar]
- 41.Tulisiak AK, Klein JA, Harris E, et al. Antidepressant Prescribing by Pediatricians: A Mixed-Methods Analysis. Curr Probl Pediatr Adolesc Health Care. 2017;47(1):15–24. doi: 10.1016/j.cppeds.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faraone SV, Biederman J, Roe C. Comparative efficacy of Adderall and methylphenidate in attention-deficit/hyperactivity disorder: a meta-analysis. J Clin Psychopharmacol. 2002;22(5):468–473. doi: 10.1097/00004714-200210000-00005. [DOI] [PubMed] [Google Scholar]
- 43.Punja S, Shamseer L, Hartling L, et al. Amphetamines for attention deficit hyperactivity disorder (ADHD) in children and adolescents. Cochrane Database Syst Rev. 2016;2016(2) doi: 10.1002/14651858.CD009996.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strawn JR, Welge JA, Wehry AM, Keeshin B, Rynn MA. Efficacy and tolerability of antidepressants in pediatric anxiety disorders: a systematic review and meta-analysis. Depress Anxiety. 2015;32(3):149–157. doi: 10.1002/da.22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wehry AM, Beesdo-Baum K, Hennelly MM, Connolly SD, Strawn JR. Assessment and treatment of anxiety disorders in children and adolescents. Curr Psychiatry Rep. 2015;17(7):591. doi: 10.1007/s11920-015-0591-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunnenberger HM, Crews KR, Hoffman JM, et al. Preemptive clinical pharmacogenetics implementation: currentprograms in five United States medical centers. Annu Rev Pharmacol Toxicol. 2015;55:89–106. doi: 10.1146/annurev-pharmtox-010814-124835. [DOI] [PMC free article] [PubMed] [Google Scholar]


