Abstract
Scap is a polytopic membrane protein that functions as a molecular machine to control the cholesterol content of membranes in mammalian cells. In the 21 years since our laboratory discovered Scap, we have learned how it binds sterol regulatory element-binding proteins (SREBPs) and transports them from the endoplasmic reticulum (ER) to the Golgi for proteolytic processing. Proteolysis releases the SREBP transcription factor domains, which enter the nucleus to promote cholesterol synthesis and uptake. When cholesterol in ER membranes exceeds a threshold, the sterol binds to Scap, triggering several conformational changes that prevent the Scap–SREBP complex from leaving the ER. As a result, SREBPs are no longer processed, cholesterol synthesis and uptake are repressed, and cholesterol homeostasis is restored. This review focuses on the four domains of Scap that undergo concerted conformational changes in response to cholesterol binding. The data provide a molecular mechanism for the control of lipids in cell membranes.
Keywords: cholesterol, Scap, SREBPs, Insig, membrane proteins, conformational changes, ER-to-Golgi transport, COPII vesicles, proteolytic processing, transcriptional regulation
SETTING THE STAGE FOR SCAP
Twenty-one years have elapsed since the discovery of Scap, the key regulatory protein in cholesterol metabolism (1). Scap is the central agent in a feedback system that we have termed the SREBP pathway (Figure 1). SREBP stands for sterol regulatory element-binding protein. SREBPs are membrane-bound transcription factors that activate all of the genes necessary for the synthesis of cholesterol and its receptor-mediated uptake from plasma low-density lipoprotein (LDL) (2, 3). The SREBPs are synthesized as intrinsic proteins of endoplasmic reticulum (ER) membranes, and they must be carried to the Golgi apparatus for processing by two proteases to release the transcriptionally active fragment that enters the nucleus. Scap mediates the ER-to-Golgi transport of SREBPs. Scap also renders the transport process sensitive to inhibition when the level of cholesterol rises above a threshold in ER membranes (4).
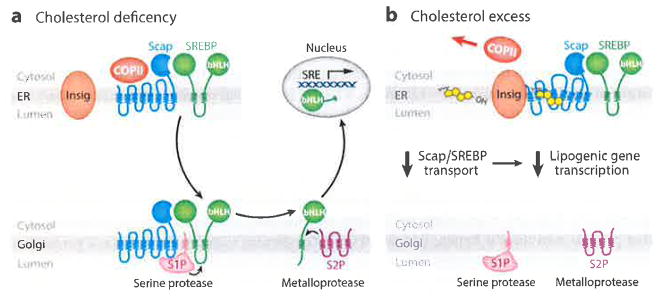
Figure 1.
SREBP pathway under conditions of (a) cholesterol deficiency and (b) cholesterol excess. (a) When mammalian cells are deprived of cholesterol, Scap escorts SREBPs in COPII vesicles from the ER to the Golgi. Two Golgi proteases (S1P and S2P) then sequentially cleave SREBPs, releasing their active NH2-terminal bHLH transcription factor domains, which travel to the nucleus and activate genes involved in cholesterol synthesis and uptake. (b) When cholesterol levels rise, the sterol binds to Scap and triggers Scap’s binding to Insigs, which retain Scap in the ER. Transport of SREBPs to the Golgi and subsequent transcriptional activation of lipogenic genes are halted. Abbreviations: bHLH, basic-helix-loop-helix zipper; COPII, coat protein II; ER, endoplasmic reticulum; S1P, site-1 protease; S2P, site-2 protease; SRE, sterol regulatory element; SREBP, sterol regulatory element-binding protein.
We have learned much about Scap and the SREBP pathway over the past two decades, but major questions remain. The purpose of this retrospective is to chronicle the history of Scap and the elucidation of the functions of its various domains. We focus primarily on work from our laboratory that disclosed the fundamental properties of Scap and then propose a model for the complex sequence of conformational changes that are triggered when cholesterol binds to Scap.
The Beginning: Low-Density Lipoprotein Receptor Pathway and Cholesterol Homeostasis
The story of Scap begins in 1973 when two of us (M.S.B. and J.L.G.) discovered that the synthesis of cholesterol in cultured human fibroblasts is controlled by end product feedback repression (5, 6). This repression is mediated by cholesterol that enters cells through the receptor-mediated endocytosis of cholesterol-carrying plasma LDL (7). Over the next decade, we focused our attention on the LDL receptor (8). With Richard Anderson, we showed that the receptor enters the cell in coated pits and delivers its cholesterol to lysosomes (9). Our studies led to the purification and cloning of the receptor and the documentation of mutations in patients with familial hypercholesterolemia (FH) (reviewed in 10, 11). We also learned that the LDL receptor is subject to end product feedback repression. When cholesterol accumulates in cells, the LDL receptor gene is repressed coordinately with the genes encoding the cholesterol biosynthetic enzymes (12, 13).
In the early 1990s, we turned our attention to the mechanism by which cholesterol represses transcription of the LDL receptor gene and the genes responsible for cholesterol synthesis. Using the LDL receptor gene as a model, postdoctoral fellow Thomas Südhof mapped the cholesterol-responsive regulatory element in the 5′ flanking region (14). We named it the sterol regulatory element (SRE) and showed that this element activates transcription in sterol-depleted cells and loses its activity when excess cholesterol is present.
SREBPs: Membrane-Bound Transcription Factors That Undergo Proteolytic Processing
Using nuclear extracts from sterol-deprived cultured human HeLa cells, two postdoctoral fellows, Xiaodong Wang and Michael Briggs, purified the protein that binds to the SRE (15, 16). Through lack of imagination, we named it sterol regulatory element-binding protein-1 (SREBP-1). Using peptide sequence data, we designed oligonucleotide probes that allowed us to isolate the complementary DNA (cDNA) encoding SREBP-1 (17). DNA sequence data showed that the nuclear protein was a member of the basic-helix-loop-helix-leucine zipper (bHLH-Zip) family of transcription factors. Unlike other members of this family, the bHLH-Zip fragment that we purified from the nucleus (molecular mass ~62 kDa) constituted the NH2-terminal portion of a much larger protein (molecular mass ~125 kDa). This NH2-terminal portion (490 amino acids) was followed by two transmembrane helices, which in turn were followed by a long COOH-terminal portion of 541 amino acids (18, 19). We also found a second closely related transcription factor that we named SREBP-2 (20). It had the same overall membrane-attached structure as SREBP-1.
The SREBPs are synthesized on membrane-bound ribosomes and inserted into the ER membrane with the NH2-terminal bHLH-Zip domain and the COOH-terminal extension facing the cytosol (21). The two transmembrane helices are separated by a short loop of 50 amino acids that projects into the ER lumen. For the bHLH-Zip domain to enter the nucleus, a proteolytic processing step was required (18, 19).
Our subsequent studies relating to cholesterol homeostasis were performed primarily with the SREBP-2 isoform, which is specialized for cholesterol homeostasis. The same control mechanism applies to SREBP-1, but it has additional regulatory features owing to its ability to activate synthesis of fatty acids as well as cholesterol (22). Postdoctoral fellow Juro Sakai found that release of the bHLH-Zip domains of both SREBP-1 and -2 from the membrane requires two sequential proteolytic cleavages (23). The first cleavage occurs within the 50-amino acid luminal loop, separating the SREBP into two halves. The NH2-terminal half remains attached to the membrane by its single transmembrane helix. The second cleavage occurs within this helix, releasing the bHLH-Zip domain so that it can enter the nucleus. We subsequently named the two proteases site-1 protease (S1P) and site-2 protease (S2P) (24, 25). We found that S1P cleaves SREBPs only in cholesterol-depleted cells and that site-2 cleavage requires prior cleavage at site-1. However, we had no idea how cholesterol regulates site-1 cleavage. That is where Scap entered the picture.
DISCOVERY OF SCAP: FIVE-STEP EXPRESSION CLONING STRATEGY
We isolated Scap through the use of somatic cell genetics. In addition to inhibition by cholesterol, the processing of SREBPs is blocked by certain oxygenated derivatives of cholesterol, the most potent of which is 25-hydroxycholesterol. When mammalian cells are grown in the absence of cholesterol, they depend entirely on cholesterol synthesis for survival. Addition of 25-hydroxycholesterol blocks SREBP processing and stops cholesterol synthesis. Inasmuch as 25-hydroxycholesterol cannot replace cholesterol in cell membranes, the cells die. Chang & Limanek (26) mutagenized CHO cells and isolated a clone of cells designated 25-RA that grow in the presence of 25-hydroxycholesterol because the sterol fails to suppress cholesterol synthesis. Through cell fusion experiments, Hasan & Chang (27) showed that the defect in 25-RA cells is dominant, suggesting that the cells produce a mutant protein that blocks the ability of 25-hydroxycholesterol to reduce the activities of cholesterologenic enzymes.
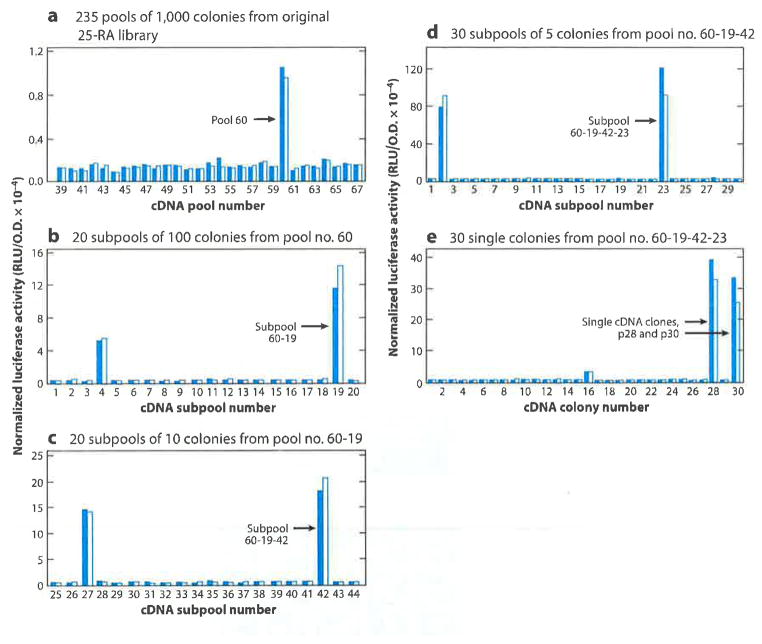
We obtained 25-RA cells from T.Y. Chang and demonstrated that they continue to process SREBPs even when saturating amounts of cholesterol or 25-hydroxycholesterol are present. To our great delight, we were able to convince a brave graduate student, Xianxin Hua, to prepare a cDNA library from the 25-RA cells with the risky aim of identifying its mutant protein by expression cloning in wild-type cells (1). Figure 2 shows Hua’s 5-step screening strategy that led to the discovery of Scap. Hua’s cDNAs from 25-RA cells were inserted into a plasmid vector that allowed transcription driven by a cytomegalovirus promoter. A total of 253 cDNA pools, each containing 1,000 cDNAs, were transfected into wild-type CHO cells together with a reporter plasmid encoding luciferase under control of an artificial promoter containing three copies of the SRE from the LDL receptor gene. Hua incubated the cells with a mixture of cholesterol and 25-hydroxycholesterol, after which the cells were harvested and luciferase activity was measured. In cells that did not receive a cDNA pool, luciferase activity was low, owing to blocking of SREBP processing by the sterols. One of the 253 pools contained a cDNA that allowed high luciferase activity in the presence of sterols. Hua subjected this pool to four sequential fractionations and eventually isolated two identical cDNA clones that conferred sterol resistance.
Figure 2.
The five-step expression cloning strategy that led to discovery of Scap, as originally published in Reference 1. This figure is shown for its historical context. A summary of this experiment is described in the text. Abbreviation: cDNA, complementary DNA.
Hua sequenced the cDNA that conferred sterol resistance and compared it with the sequence of the corresponding cDNA isolated from wild-type cells. He found that the mutant cDNA encoded a protein of 1,276 amino acids that contained a single point mutation that changed amino acid 443 from aspartic acid to asparagine (D443N) (1). When the mutant cDNA was transfected into wild-type CHO cells, it rendered the cells resistant to inhibition of SREBP cleavage by sterols. We hypothesized that wild-type Scap is required for SREBP cleavage and that sterols block its activity. Scap(D443N) retains the ability to stimulate SREBP cleavage, but it has lost sensitivity to inhibition by sterols. When Scap(D443N) is expressed in cells that also express wild-type Scap, wild-type Scap is inhibited but the mutant protein continues to activate SREBP cleavage in the presence of sterols, thus explaining the dominant phenotype. Subsequent studies confirmed this hypothesis, but the mechanism turned out to be a surprise.
SCAP AS A STEROL SENSOR
The sequence of Scap indicated that it is an ~160-kDa polytopic membrane protein with multiple transmembrane helices and a long COOH-terminal extension that includes multiple copies of a WD-repeat sequence known to promote protein-protein interactions (Figure 3). A sequence that includes five transmembrane helices (TMs 2–6) was noted to bear homology to a sequence in the membrane attachment domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMG CoA reductase), the rate-controlling enzyme in cholesterol biosynthesis (1). Previous studies showed that the membrane attachment domain of HMG CoA reductase allows the protein to undergo rapid degradation when cholesterol is present (28). Because Scap is also predicted to be regulated by cholesterol, we named this shared sequence the sterol-sensing domain (SSD). We noted that the D443N mutation that renders Scap insensitive to sterols lies in the SSD (Figure 4c). The function of the SSD of Scap is discussed below in the section titled Domains of Scap.
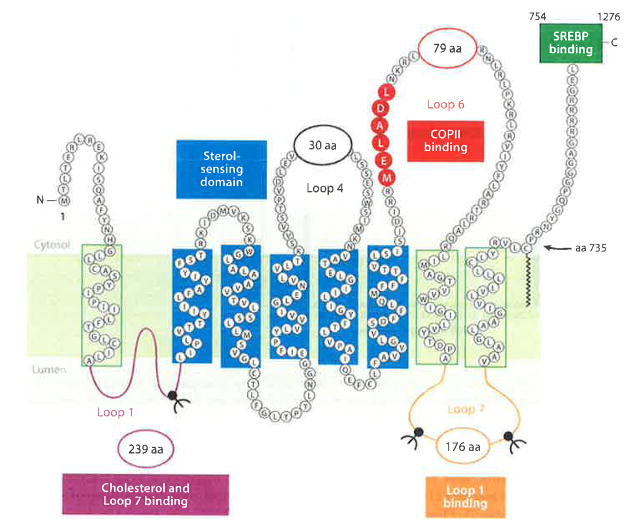
Figure 3.
The domains of Scap. Luminal Loop 1 (purple) binds cholesterol and Loop 7, and it contains one N-linked glycosylation site. The sterol-sensing domain (blue), which is comprised of transmembrane helices 2–6, binds Insigs. Cytoplasmic Loop 6 (red), which contains the hexapeptide MELADL, binds COPII proteins. Loop 7 (orange) binds Loop 1 and contains two N-linked sites. The COOH-terminal 523 amino acids (green box) contain at least five WD-repeat sequences that bind the COOH-terminal domain of SREBPs. Abbreviations: COPII, coat protein II; MELADL sequence, methionine-glutamate-leucine-alanine-aspartate-leucine; SREBPs, sterol regulatory element-binding proteins.
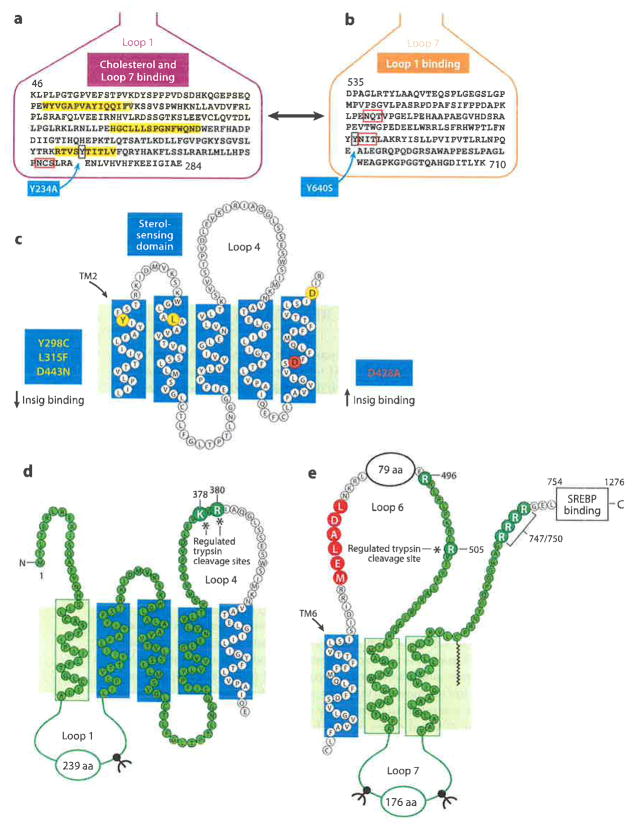
Figure 4.
A more detailed view of the domains of Scap. (a) This sequence of Loop 1 shows three short hydrophobic sequences (yellow), a single N-linked glycosylation site (red box), and tyrosine-234 (blue box). (b) This sequence of Loop 7 shows locations of two N-linked sites (red boxes) and tyrosine-640 (blue box). (c) The sterol-sensing domain (TMs 2–6) contains three residues (Y298, L315, D443) (yellow circles), each of which is required for binding Insigs, and one residue (D428) (red circle), which is required for dissociation from Insigs in the absence of cholesterol. (d) Basic residues in Loop 4 (K378 and R380) that are cleaved by trypsin only when membranes are depleted of cholesterol. This fragment is identified by probing SDS gels with anti-Loop 1 antibody (highlighted in green). (e) Arginine in Loop 6 (R505) that is cleaved by trypsin only in cholesterol-enriched membranes. This fragment is recognized by probing SDS gels with anti-Loop 7 antibody (highlighted in green). Abbreviations: SDS, sodium dodecyl sulfate; SREBP, sterol regulatory element-binding protein; TMs, transmembrane helices.
In addition to Scap and HMG CoA reductase, six other polytopic membrane proteins are now known to contain evolutionarily conserved SSDs (Table 1). One of these proteins, 7-dehydrocholesterol reductase, catalyzes the last step in the cholesterol biosynthetic pathway, converting 7-dehydrocholesterol to cholesterol. Niemann-Pick C1 (NPC1) transports LDL-derived cholesterol out of lysosomes for delivery to the plasma membrane and ER. Niemann-Pick C1-like 1 (NPC1L1) mediates the absorption of cholesterol by the small intestine. Patched and Dispatched are membrane components of a signaling pathway whose major ligand, Hedgehog, contains covalently attached cholesterol at its NH2-terminus. TRC8 is a ubiquitin E3 ligase that participates in the degradation of HMG CoA reductase.
Table 1.
Membrane proteins that contain sterol-sensing domainsa
| Protein | Function | Reference(s) |
|---|---|---|
| Scap | Cholesterol-regulated transporter of SREBPs from the endoplasmic reticulum to the Golgi | 1 |
| HMG CoA reductase | Biosynthesis of cholesterol | 1 |
| 7-DHCR | Biosynthesis of cholesterol | 65 |
| NPC1 | Intracellular cholesterol transport within lysosomes | 33,66 |
| NPC1L1 | Intestinal cholesterol absorption | 67 |
| Patched | Hedgehog signaling pathway | 33,66 |
| Dispatched | Hedgehog signaling pathway | 68 |
| TRC8 | Ubiquitin E3 ligase involved in degradation of HMG CoA reductase and heme oxygenase | 33,69,70 |
For a review of the sterol-sensing domain, see Reference 71.
Abbreviations: 7-DHCR, 7-dehydrocholesterol reductase; HMG CoA reductase, 3-hydro-3-methylglutaryl coenzyme A reductase; NPC1, Niemann-Pick C1; NPC1L1, Niemann-Pick C1-like 1; SREBPs, sterol regulatory element-binding proteins; TRC8, translocation in renal cancer from chromosome 8.
SCAP MOVES FROM THE ENDOPLASMIC RETICULUM TO THE GOLGI
Using protease protection assays and measurements of glycosylation to determine the membrane orientation of Scap, we found that the NH2- and COOH-termini are located in the cytosol and that the protein contains eight transmembrane helices (29) (Figure 3). Two large loops are in the ER lumen (239 and 176 amino acids, respectively), both of which contain N-linked carbohydrate chains. The overall orientation of the Scap membrane domain bears a close resemblance to the orientation of the membrane domain of HMG CoA reductase, which was determined by Olender & Simoni (30). However, HMG CoA reductase does not contain the two long loops that are present in Scap. The presence of N-linked sugars in Scap suggested that it must be synthesized on membrane-bound ribosomes and inserted into the membrane cotranslationally.
Coimmunoprecipitation studies demonstrated that the cytosolic COOH-terminal WD-repeat domain of Scap binds to the cytosolic COOH-terminal extension of SREBPs (31, 32). When SREBP was truncated to remove its COOH-terminal extension, the protein no longer bound to Scap and was no longer processed proteolytically. Addition of cholesterol blocked SREBP processing, but it did not disrupt the Scap–SREBP complex. We concluded that SREBP binding to Scap was essential for the proteolytic processing of SREBPs, but it was not sufficient. Another action of Scap was required. That action turned out to be ER-to-Golgi transport.
The first evidence for ER-to-Golgi movement of Scap came from studies of its N-linked carbohydrate chains that were carried out by Axel Nohturfft, a graduate student who remained as a postdoctoral fellow (33). When CHO cells were grown in the presence of sterols, the N-linked carbohydrates of Scap were mostly in the endoglycosidase H (endo H)–sensitive form, which indicates that the protein had not moved to the Golgi, where N-linked chains are processed by mannosidases to a form that is resistant to digestion by endo H. When cells were switched to sterol depletion medium, the N-linked chains of Scap became endo H resistant, indicating that sterol depletion triggered Scap movement to the Golgi. In the article by Nohturfft et al. (33), we reported the isolation of a second mutant cell line that is resistant to the inhibitory effects of 25-hydroxycholesterol. This second Scap mutation (Y298C), like the original D443N mutation, resides in the SSD (Figure 4c). When cells expressing either of these mutants were grown in the presence of 25-hydroxycholesterol, the N-linked carbohydrates of Scap assumed the endo-H resistant form, indicating that the mutations prevented sterols from blocking ER-to-Golgi transport of Scap. Our colleague, Robert Rawson, subsequently isolated a third sterol-resistant mutation of Scap (L315F). Like the other two mutations, the L315F mutation occurs in the SSD (Figure 4c), providing compelling evidence for the crucial role of this domain in sterol regulation of Scap (34).
To obtain a visual confirmation of sterol-regulated Scap movement from the ER to the Golgi, we transfected cells with a plasmid encoding Scap with green fluorescent protein (GFP) fused at its COOH-terminus (35). As shown by confocal fluorescence microscopy, when the cells were deprived of sterols the protein moved to the Golgi, and this movement was prevented by addition of a mixture of cholesterol and 25-hydroxycholesterol. To identify the mechanism for this movement, we used a system in which ER membranes from mammalian cells are incubated in vitro with cytosol and nucleoside triphosphates (35). When the ER membranes were obtained from sterol-depleted cells, Scap was found in the budding vesicles. When the cells were preincubated with sterols, Scap no longer budded from the ER membranes in vitro.
The demonstration that SREBPs bind to Scap and the demonstration that sterols block the movement of the Scap–SREBP complex to the Golgi raised the hypothesis that the proteases that process SREBPs are located in the Golgi. This hypothesis was strongly supported when postdoctoral fellow Juro Sakai isolated a cDNA encoding S1P, the protease that makes the first cleavage in SREBPs (25). We found that S1P is a membrane-bound serine protease, and its N-linked carbohydrates are in the endo H-resistant form, indicating that the protease resides in the Golgi. To explore the significance of this Golgi localization, postdoctoral fellow Russell DeBose-Boyd used brefeldin A, a compound that is known to cause Golgi proteins to retrotranslocate to the ER. When brefeldin A was added to cells, S1P translocated to the ER and SREBP-2 was cleaved even in the presence of sterols (36). A similar result was obtained when we attached the amino acid sequence motif KDEL to the cytosolic COOH-terminus of S1P. KDEL caused S1P to be retained in the ER, and again SREBP-2 was processed proteolytically even in the presence of sterols (36).
CHOLESTEROL-INDUCED CONFORMATIONAL CHANGE IN SCAP
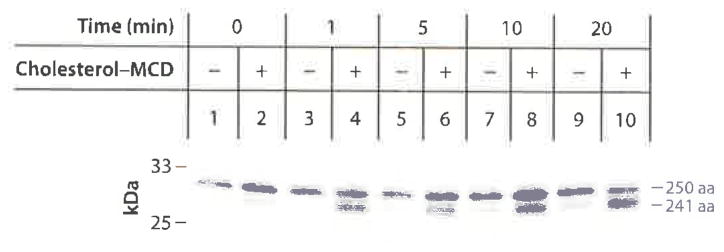
The year 2002 witnessed two fundamental observations that advanced our knowledge of Scap regulation. First, we found that cholesterol causes a conformational change in Scap when added to ER membranes in vitro (37). To detect the change, our postdoctoral fellow Andrew Brown used a protease protection assay. He homogenized cells and isolated sealed ER vesicles that were not permeable to trypsin. He incubated the vesicles with or without cholesterol, digested the cytosolic surface with trypsin, and subjected the proteins to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were then transferred to nitrocellulose and blotted with an antibody to an epitope in Scap Loop 7, which is located in the ER lumen and thus protected from trypsin (Figure 4e). In the absence of cholesterol, the antibody revealed a protected fragment with an apparent molecular mass of 27 kDa (37). When cholesterol was added, the resulting fragment migrated faster—the apparent molecular mass was now 26 kDa (Figure 5). Protein sequencing by Edman degradation revealed that the 26-kDa band (241 amino acids) resulted from trypsin cleavage at arginine-505 and arginine-747, which flank Loop 7 (Figure 4e). When cholesterol was absent, arginine-505 was inaccessible to trypsin. Instead, the larger band resulted from cleavage at arginine-496. Together with the invariant cleavage at arginine-747, trypsin produced a larger peptide of 250 amino acids (Figure 5). When a variety of sterols were added to the membranes, we found that the exposure of arginine-505 occurred only when we added sterols that are capable of suppressing SREBP cleavage in cultured cells. Moreover, epicholesterol, a diastereomer that differs from cholesterol only in the orientation of the hydroxyl group on the A ring of the steroid nucleus, did not change Scap’s conformation or suppress SREBP cleavage.
Figure 5.
Cholesterol-induced conformational change in Loop 6 of Scap as revealed by appearance of a 241-amino acid tryptic fragment after addition of cholesterol to ER membrane vesicles in vitro. CHO cells expressing stably transfected GFP-Scap were deprived of sterols after which sealed membrane vesicles were prepared and incubated with 25 μM cholesterol–MCD for the indicated time at 37°C, followed by cleavage with trypsin. Samples were subjected to 16% SDS-PAGE and immunoblotted with anti-Loop 7 antibody. Abbreviations: ER, endoplasmic reticulum; GFP, green fluorescent protein; MCD, methyl-β-cyclodextrin; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Reprinted from Reference 37.
We also tested the D443N and Y298C mutants of Scap that resist inhibition by sterols. After trypsin digestion, both of these mutant proteins showed only the larger 27-kDa band, even in the presence of cholesterol. The trypsin digestion assay provided the first opportunity to detect an effect of cholesterol on Scap in a cell-free system (37). It provided the first indication that cholesterol acts by changing the conformation of Scap.
INSIGS: PROTEINS THAT TRIGGER THE ENDOPLASMIC RETICULUM RETENTION OF SCAP
The second major discovery of 2002 was the identification of Insigs as proteins that trigger the ER retention of Scap. The search for a retention trigger was stimulated by our observation that overexpression of Scap overcomes the inhibition of SREBP cleavage mediated by cholesterol or 25-hydroxycholesterol (38). Moreover, the endogenous Scap–SREBP complex was no longer retained in the ER when we overexpressed a truncated version of Scap that contained the sequence from the NH2-terminus to the end of transmembrane helix 6 [Scap(TM1–6)] (38). We reasoned that ER retention required that the endogenous Scap–SREBP complex binds to a putative ER protein that recognizes the TM1–6 segment of Scap and somehow triggers ER retention. We postulated that overexpression of wild-type Scap or Scap(TM1–6) saturates the retention protein and liberates the endogenous Scap–SREBP complex to move to the Golgi.
Tong Yang, a postdoctoral fellow, searched for the ER retention protein by expressing an affinity-tagged version of Scap(TM1–6) in human cells (39). As a control, she expressed affinity-tagged Scap(TM1–5), which does not liberate the endogenous Scap–SREBP complex, presumably because it does not bind to the postulated ER retention protein. The cells were grown in the presence of 25-hydroxycholesterol, which promotes ER retention of the Scap–SREBP complex. The membrane proteins were solubilized with a detergent, after which the tagged Scap was purified by tandem affinity chromatography, subjected to SDS-PAGE, and stained with Coomassie. We observed several faintly stained protein bands that were present in the complex isolated from cells transfected with Scap(TM1–6) but not from cells that received Scap(TM1–5). The bands were excised from the gel and sent to Rudi Aebersold (Institute of Systems Biology, Seattle) who had developed a sensitive method to identify proteins by mass spectroscopy. Aebersold identified several proteins that were immunoprecipitated with Scap(TM1–6). For the reason given below, we focused on one of these proteins, Insig-1 (39).
The Insig-1 mRNA was discovered in 1991 by Taub and coworkers (40), who analyzed mRNAs that rose in abundance when human hepatoma cells were treated with insulin. We had previously encountered Insig-1 as an mRNA that was increased in abundance in livers of transgenic mice that overexpress SREBP-1a (41). Intrigued that a protein produced by an SREBP-target gene would bind to Scap, we thus chose to focus on Insig-1. Using the technique of blue native gel electrophoresis, Yang showed that epitope-tagged Insig-1 binds to Scap but only when Insig-1 is expressed in cells treated with sterols (39). We also found that overexpression of Insig-1 restored sterol inhibition of SREBP processing in cells that overexpress Scap. The most convincing evidence came from experiments with the Y298C mutant of Scap that lacks the ability to be inhibited by sterols. Insig-1 failed to bind to the Y298C mutant protein in sterol-treated cells, indicating that the defective regulation of this mutant protein is attributable to its failure to bind to Insig-1 (39).
In subsequent studies, we identified a gene that produces a protein that is highly related to Insig-1, which we termed Insig-2 (42). Like Insig-1, Insig-2 binds to Scap but only when cells are treated with sterols. Moreover, like the Y298C Scap mutant described above, the two other Scap mutants that resist inhibition by sterols (L315F and D443N) also failed to bind either Insig-1 or Insig-2 in sterol-treated cells (34) (Figure 4c).
Insig-1 is an extremely hydrophobic protein containing 277 amino acids with 6 transmembrane segments separated by short hydrophilic loops ranging from 5 to 16 amino acids (43). It is synthesized and retained in ER membranes. When cells have abundant cholesterol or oxysterols, Insig-1 exists in a complex with Scap (Figure 1). When cholesterol is depleted, Insig-1 dissociates from Scap, thereby allowing Scap to move to the Golgi. After dissociating from Scap, Insig-1 is ubiquitinated and degraded (44). SREBP-2 then enters the nucleus, where it activates the Insig-1 gene. Insig-1 is produced, but it will be rapidly degraded unless cholesterol levels have risen sufficiently so that the Insig-1 can bind to Scap, blocking further SREBP-2 activation and thereby restoring cholesterol balance. Feedback inhibition of cholesterol synthesis therefore requires that sufficient nuclear SREBP-2 must be generated to increase Insig-1 transcription and restore ER cholesterol. SREBP cleavage will not cease until both requirements are met. We termed this “convergent feedback inhibition” (21, 44).
SOLVING THE OXYSTEROL CONUNDRUM
As described above, from the earliest days we knew that cholesterol synthesis could be inhibited by oxysterols such as 25-hydroxycholesterol even more potently than cholesterol (45,46). Christopher Adams, a postdoctoral fellow, showed that both classes of sterols inhibit transport by causing Scap to bind to Insigs and both classes of sterols lose their inhibitory activity when Insigs are reduced by RNA interference (47). A difference between the two classes of sterols was revealed when Adams employed a photoactivatable derivative of cholesterol and showed that it cross-linked to Scap when added to cultured cells. Surprisingly, a similar photoactivatable derivative of 25-hydroxycholesterol failed to cross-link to Scap (47).
The oxysterol conundrum was solved in comparative binding studies of Scap and Insig by Arun Radhakrishnan, then a postdoctoral fellow. He used an insect cell expression system to produce a portion of Scap extending from the NH2-terminus to the end of the eighth transmembrane helix [Scap(TM1–8)]. After purification in the presence of detergents, the protein bound [3H]cholesterol with saturation kinetics but did not bind [3H]25-hydroxycholesterol (48). In competition studies, [3H] cholesterol binding was inhibited by cholesterol derivatives in direct proportion to their ability to produce the conformational change detected by the trypsin protease assay and by their ability to block Scap–SREBP transport. Binding was not inhibited by 25-hydroxycholesterol, further suggesting that oxysterols act differently than cholesterol. Radhakrishnan then expressed and purified one of the Insigs (Insig-2). Remarkably, he showed that Insig-2 bound [3H]25-hydroxycholesterol but not [3H]cholesterol (49). Binding of [3H]25-hydroxycholesterol was inhibited competitively by other oxysterols that inhibit SREBP processing but not by inactive oxysterols.
To identify residues in Insig-2 that are required for regulatory activity, postdoctoral fellow Irina Dobrosotskaya reconstituted the mammalian SREBP transport system by transfection into Drosophila S2 cells (50). When we transfected wild-type human SREBP-2, Scap, and Insig-2 and incubated the cells in the absence of sterols, we observed that SREBP-2 was processed by the Site-1 protease present in the Golgi membranes of the insect cells. (There was no processing by Site-2 protease because the Drosophila enzyme apparently does not recognize human SREBP-2.) Processing by Site-1 protease was blocked when the Drosophila cells were incubated with cholesterol or 25-hydroxycholesterol, and this was dependent on expression of Insig-2. We systematically mutated single residues in Insig-2 and tested the ability of the mutants to inhibit SREBP-2 processing in insect cells treated with 25-hydroxycholesterol or cholesterol (49). We identified five crucial residues. Four of these were in or near the predicted fourth transmembrane helix of Insig-2, and one was in the third transmembrane helix. Two of the mutants showed reduced binding of [3H]25-hydroxycholesterol in vitro. The other three mutants bound [3H]25-hydroxycholesterol normally. All of the mutant Insigs failed to interact with Scap when cells were treated with 25-hydroxycholesterol or cholesterol. We concluded that the third and fourth transmembrane helices of Insig-2 are important for binding to Scap whether binding is induced by cholesterol binding to Scap or by 25-hydroxycholesterol binding to Insig-2, both of which produce a similar conformational change in Scap (51).
MEMBRANE CHOLESTEROL: A DELICATE BALANCE
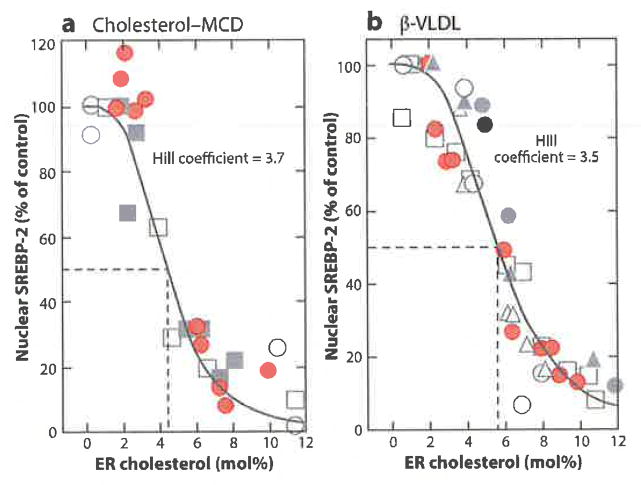
It has been known for some time that the ER contains only a small proportion of the cholesterol in cells (52, 53), and yet Scap, the sterol sensor, is placed in the ER membrane. If ER cholesterol inhibits Scap transport, Scap must be able to respond to low levels of membrane cholesterol. These considerations led us to attempt to determine the concentration of cholesterol in ER membranes that is sufficient to suppress SREBP processing (Figure 6). We devised a method to purify ER membranes from cultured CHO cells (4). We depleted cellular cholesterol by treatment of the cells with hydroxypropyl-β-cyclodextrin, which binds and extracts cholesterol from membranes. We then added back varying amounts of cholesterol either complexed with methyl-β-cyclodextrin, which delivers cholesterol to membranes, or delivered in lipoproteins that enter cells through LDL receptors. As an index of Scap activity, we measured the nuclear content of SREBP-2. We also isolated ER membranes from the same cells and measured their content of cholesterol expressed as molar percentage (mol%) of total ER lipids.
Figure 6.
Threshold response of SREBP-2 processing to changes in cholesterol content of purified ER membranes. ER cholesterol content was varied by treating CHO cells for various times with different amounts of cholesterol (a) complexed to MCD or (b) delivered in β-VLDL. Each symbol represents data from a different experiment. The amount of nuclear SREBP-2 at zero time was normalized to 100%. ER cholesterol concentration is expressed as a molar percentage (mol%) of total ER lipids. Best-fit analysis of the data reveals Hill coefficients of (a) 3.7 ± 0.23 and (b) 3.5 ± 0.13. Abbreviations: β-VLDL, β-migrating very-low-density lipoprotein; ER, endoplasmic reticulum; MCD, methyl-β-cyclodextrin; SREBP, sterol regulatory element-binding protein. Reprinted from Reference 4.
Scap transported SREBP-2 when ER cholesterol was less than 5 mol% of ER lipids. As the cholesterol content of ER membranes rose above a sharp threshold of 5%, Scap transport was inhibited as indicated by an abrupt reduction in nuclear SREBP-2 (Figure 6). The abrupt response of Scap was consistent with a cooperative response with a Hill coefficient of approximately 4. Previous studies suggested that detergent-solubilized Scap behaves as a tetramer as determined by analytical ultracentrifugation (48). This raises the possibility that the threshold response of Scap to cholesterol reflects allosteric interactions within a tetramer as in the classic case of oxygen binding to hemoglobin. The notion that the Scap–SREBP complex is a tetramer is supported by ultrastructural studies of fragments of the yeast equivalents of these proteins isolated from the fission yeast Scbizosacchuromyces pombe. The COOH-terminal domain of the yeast SREBP was shown to form a tetramer when complexed with the COOH-terminal, WD-repeat domain of yeast Scap (54).
In the mammalian system, the cholesterol threshold for inhibition of Scap transport was lowered to 3 mol% when the cells expressed elevated amounts of lnsig-1 (4). This finding is consistent with the notion that inhibition of Scap transport requires binding of Scap to Insig and that the equilibrium of this binding reaction can be shifted in the direction of complex formation when the concentration of Insig is elevated.
DOMAINS OF SCAP
Loop 6 Binds COPII Proteins
In recent years, we have focused on delineating the domains of Scap that mediate its various functions (Figures 3 and 4). We first identified the domain of Scap that is responsible for its incorporation into budding ER vesicles. Peter Espenshade, a postdoctoral fellow, used an in vitro binding assay to show that Scap is incorporated into coat protein II (COPII)-coated vesicles that bud from ER membranes (55). When he isolated ER membranes from sterol-treated cells, vesicles continued to bud, but Scap was no longer incorporated into the vesicles. Using proteins purified from yeast, Espenshade showed that Scap incorporation into vesicles is initiated when the Sec23–Sec24 component of the COPII protein complex binds to Scap.
Following up on Espenshade’s observation, postdoctoral fellow Li-Ping Sun found that Scap budding into COPII-coated vesicles was abolished by addition of cholesterol or 25-hydroxycholesterol to ER membranes in vitro (56). Inhibition was attributed to loss of the binding-of Sec23–Sec24 to Scap. To identify the Sec23–Sec24 binding site on Scap, Sun produced truncated versions of Scap and tested them for binding. Scap fragments that terminated prior to Loop 6 failed to bind Sec23–Sec24. When the fragment was extended to include Loop 6, binding was restored. These results directed attention to cytosolic Loop 6 as the binding site for Sec23–Sec24. Alanine scanning mutagenesis of Loop 6 led to the identification of a sequence of six amino acids that constitute the Sec23–Sec24 binding site. The sequence comprises methionine-glutamate-leucine-alanine-aspartate-leucine, which we abbreviate as MELADL (56) (Figures 4e and 7a). Substitutions in this sequence abolished Sec23–Sec24 binding; therefore, they prevented budding of Scap into COPII-coated vesicles in the absence of sterols.
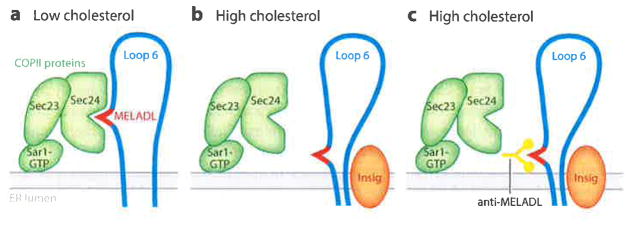
Figure 7.
Proposed mechanism by which sterols relocate the MELADL sequence in Loop 6, thereby preventing binding of COPII proteins and abolishing ER-to-Golgi transport. (a) When ER membrane cholesterol is low, the Sec24 component of the COPII protein complex binds to MELADL in Scap Loop 6, triggering ER-to-Golgi transport. (b) Sterols induce the binding of Insig proteins, which moves the MELADL so that it is no longer accessible to Sec24. (c) Even though Sec24 cannot reach MELADL, an anti-MELADL antibody can still bind. Abbreviations: COPII, coat protein II; ER, endoplasmic reticulum; MELADL sequence, methionine-glutamate-leucine-alanine-aspartate-leucine.
In a further study, we produced a polyclonal rabbit antibody directed against a 16-amino acid peptide that includes the MELADL sequence (51). This anti-MELADL antibody blocked the binding of the Sec23–Sec24 complex to Scap in vitro. When microinjected into CHO cells, a Fab fragment of this antibody blocked the movement of Scap from ER to Golgi. These data provided strong support for the role of the MELADL sequence in Sec23–Sec24 binding. When we added anti-MELADL to Scap-containing membranes in vitro, the anti-MELADL precipitated Scap, indicating that the MELADL sequence was accessible to the antibody. In membranes from cells treated with 25-hydroxycholesterol, anti-MELADL continued to precipitate Scap even though Insig-1 was bound to Scap and the Sec23–Sec24 complex could no longer bind. These data suggested that Insig binding to Scap does not bury the MELADL sequence in a location that is totally inaccessible. Rather, the data suggested that Insig binding causes a conformational change in Scap that relocates the MELADL sequence so that it can no longer be reached by Sec23–Sec24 even though it can still be reached by anti-MELADL (Figure 7b and 7c).
Additional data suggested that the conformational change in Scap alters the distance between MELADL and the ER membrane. The MELADL sequence is at the NH2-terminal end of Loop 6, separated from the membrane by six amino acids (Figure 4e). When the spacing was increased to seven or eight amino acids by addition of one or two alanines or reduced to four or five by deletion of one or two amino acids, Scap could no longer reach the Golgi in sterol-depleted cells. These additions and deletions also prevented the binding of Sec23–Sec24 to Scap in vitro (51). We speculated that the MELADL binding site on Sec23–Sec24 is located at a fixed distance from the ER membrane and that the MELADL sequence must be presented at that location for Sec 23–Sec24 to bind (Figure 7a and 7b).
Sterol-Sensing Domain Binds Insigs
As described above, we identified three point mutations in Scap that prevent the binding of Insigs and render Scap transport resistant to inhibition by cholesterol or oxysterols. All three of these mutations lie in or immediately adjacent to the sequence encompassing transmembrane helices 2–6, in other words, the SSD (Figure 4c). Moreover, we identified a point mutation in transmembrane membrane helix 6 (D428A) that exerts an effect opposite to that of the three sterol-resistant mutants (57). D428A prevents the dissociation of Insigs from Scap in the absence of sterols, blocking Scap transport in sterol-depleted cells. The observation that all of the mutations identified to date that affect Insig binding occur in the SSD provides strong evidence that this domain binds Insigs. This conclusion is consistent with our earlier observation that a truncated version of Scap extending from the NH2-terminus to Loop 6 binds to Insigs. This truncated version of Scap is referred to as Scap(TM1–6). Indeed, Scap(TM1–6) was the bait we used to identify Insig-1 by coimmunoprecipitation. Insigs and the SSD of Scap are almost entirely embedded in the ER membrane with short loops connecting the transmembrane helices. Therefore, the binding is likely to involve interactions between intramembrane segments of the two proteins.
Insig-1 and -2 also bind to the membrane domain of HMG CoA reductase, triggering ubiquitination and degradation of the reductase (58). The only region of sequence identity between Scap and HMG CoA reductase lies in the SSD. The amino acid residue in hamster HMG CoA reductase corresponding to Scap(D443) is valine (59). The other two required residues in Scap(Y298) and Scap(L315) are conserved in HMG CoA reductase. We have yet to determine whether changing Scap(D443) to valine would preserve Insig binding to this protein. Determination as to whether Insigs bind to the other proteins that share SSDs has also not been made (Table 1).
Lumenal Loop 1 Binds Cholesterol
As described above, we had demonstrated that Scap(TM1–8) binds cholesterol with saturation kinetics and a specificity that matches the specificity of the sterol-mediated conformational change in Scap and inhibition of SREBP processing. To localize the cholesterol binding site, Massoud Motamed, a graduate student, prepared a cDNA encoding 245-amino acid luminal Loop 1, one of two long structured loops that project into the ER lumen (Figure 4a). A histidine tag was added to the NH2-terminus of the loop so that it could be purified by nickel chromatography. Previous studies had shown that Loop 1 is glycosylated at a single site near its COOH-terminus (29), which confirms its luminal location. However, when it was expressed as a recombinant protein in insect cells. Loop 1 behaved as an intrinsic membrane protein, requiring detergents for solubilization. Similar membrane attachment was seen when Loop 1 was produced in transfected hamster cells (60). This membrane attachment is likely mediated by three hydrophobic sequences in Loop 1 that are too short to span the membrane but might dip into the membrane, anchoring the protein (Figure 4a).
Motamed purified recombinant Loop 1 solubilized in detergent and showed that it bound [3H] cholesterol with saturation kinetics and an apparent dissociation constant of 67 nM (61), which was similar to the previously measured dissociation constant for [3H]cholesterol binding to Scap(TM1–8) (48). Binding was inhibited competitively by unlabeled cholesterol but not by epicholesterol or 25-hydroxycholesterol. The inhibitory activities of a panel of 21 sterols matched the activities previously measured with Scap(TM1–8) and with the sterol specificity for inhibition of SREBP processing (61). We concluded that cholesterol regulates SREBP processing by binding to luminal Loop 1 of Scap, which must then trigger a conformational change in cytosolic Loop 6 that prevents Sec23–Sec24 binding to the MELADL sequence. Further studies described below were designed to gain insight into this sequence of events.
Loop 1 Binds Loop 7
Recent studies of Scap conformation have provided evidence that the two large luminal loops, Loops 1 and 7, bind to each other in sterol-depleted cells and dissociate from each other when Insig binds to Scap in sterol-replete cells (Figure 8). We hypothesize that the dissociation of the two luminal loops changes the conformation of cytosolic Loop 6, precluding the binding of COPII proteins to the MELADL sequence. The initial evidence for binding of Loop 1 to Loop 7 came from coimmunoprecipitation studies (38). We transfected cells simultaneously with two plasmids—one expressing the NH2-terminal half of Scap extending from the NH2-terminus to the middle of cytosolic Loop 6 [Scap(TM1–6)] and the other expressing the remainder of Scap extending from the middle of Loop 6 to the COOH-terminus [Scap(TM7–end)]. The two halves of Scap bound to each other, as determined by immunoprecipitation (38). Remarkably, they also reconstituted Scap activity, restoring SREBP processing in the Scap-deficient cells and also restoring the ability of Scap-deficient cells to grow in the absence of exogenous cholesterol.
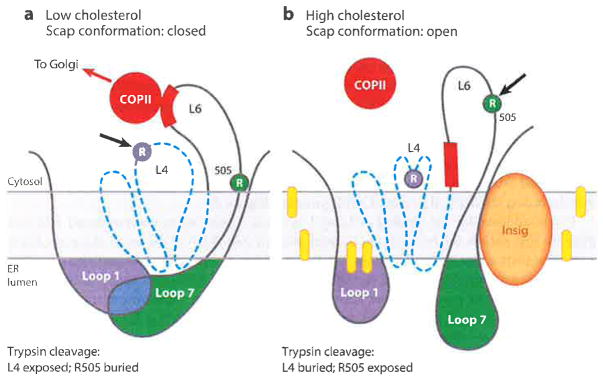
Figure 8.
Model for cholesterol-mediated conformational change in Scap. (a) When in the low-cholesterol state, Loop 1 is bound to Loop 7, creating a closed conformation. The trypsin cleavage site in Loop 4 (L4) is accessible (arrow), whereas the cleavage site at R505 in Loop 6 (L6) is blocked. (b) When in the high-cholesterol state, the trypsin cleavage site in Loop 4 (L4) is not accessible, Insig is bound to the SSD, and Loop 7 is dissociated from Loop 1, creating an open conformation. R505 of Loop 6 (L6) is accessible to trypsin (arrow), and the MELADL (red bar) is not accessible to COPII proteins. Dashed blue line denotes SSD (transmembrane helices 2–6). Yellow ovals denote cholesterol molecules. Abbreviations: COPII, coat protein II; ER, endoplasmic reticulum; MELADL sequence, methionine-glutamate-leucine-alanine-aspartate-leucine; SSD, sterol-sensing domain. Adapted from Reference 63.
To determine the requirements for binding of the two halves of Scap, Yinxin Zhang and Massoud Motamed performed alanine-scanning mutagenesis of luminal Loop 7 (62). They identified tyrosine-640 as a crucial requirement for the binding (Figure 4b). When tyrosine-640 was changed to serine (Y640S mutation), the two halves of Scap no longer bound to each other. When the Y640S mutation was introduced into full-length Scap, the protein lost the ability to transport SREBPs to the Golgi. Control experiments showed that the Y640S mutant protein was folded properly as determined by its normal glycosylation. In contrast to wild-type Scap, which binds Insig only in the presence of sterols, Scap(Y640S) bound Insig-1 even in sterol-depleted cells.
In an earlier alanine-scanning mutagenesis study of luminal Loop 1, we had identified a tyrosine at residue 234 that is essential for Scap exit from the ER (Figure 4a) (61). When Y234 was changed to alanine, Scap behaved just like the version with the Y640S mutation in Loop 7—in other words, it bound Insig even in the absence of cholesterol and could no longer transport SREBP to the Golgi for processing. When the Y234A mutation was introduced into Scap(TAM1–6), it no longer coimmunoprecipitated with Scap(TM7–end) (62). On the basis of these studies, we advanced the hypothesis that luminal Loops 1 and 7 must be bound to each other for the MELADL in Loop 6 to be accessible to COPII proteins. Y234 in Loop 1 and Y640 in Loop 7 are both necessary for binding of Loop 1 to Loop 7. Cholesterol binding to Loop 1 causes Loop 1 to dissociate from Loop 7, and this occurs in concert with Insig binding, altering the conformation of Loop 6 and thereby hiding the MELADL from COPII proteins (Figure 8).
Direct evidence for the binding of Loop 1 to Loop 7 came when we transfected cells with plasmids that encode the two loops as isolated proteins (60). Each of the loops was preceded by a signal sequence that directed it to the ER lumen. When Loop 1 was expressed alone in CHO cells, it remained membrane bound, requiring detergents for solubilization, and it was not secreted from the cell. When we coexpressed Loop 7, it bound to Loop 1, rendering it soluble, and the soluble Loop 1–Loop 7 complex was secreted into the medium. When Loop 1 contained the Y234A mutation or when Loop 7 contained Y640S, the two loops failed to form a complex and Loop 1 remained membrane bound in the cell (60).
To facilitate purification of the Loop 1–Loop 7 complex, we transfected insect cells with a baculovirus encoding the Loop 1–Loop 7 fusion protein with a histidine tag. The two loops were united by a linker containing a site for digestion with tobacco etch virus (TEV) protease. The soluble fusion protein was secreted from insect cells and purified by nickel chromatography. It behaved on gel filtration as a monodisperse species and bound [3H]cholesterol with the same kinetics and specificity as full-length Scap or isolated Loop 1 (60). To determine whether cholesterol caused separation of the loops, we incubated the fusion protein with cholesterol and then digested with TEV protease to separate the loops. Cholesterol did not cause dissociation as judged by the continued coimmunoprecipitation of the two loops after cleavage by TEV protease. We hypothesized that cholesterol-mediated dissociation of the two loops in full-length Scap requires some conformational change in the SSD that separates the two loops. Inasmuch as the isolated loops lack the SSD, cholesterol cannot cause the loops to dissociate.
To detect the postulated cholesterol-induced conformational change in the SSD of Scap, we used a protease protection assay similar to that used to detect the conformation change in Figure 4e, except in the case described below we used an antibody to Loop 1 instead of to Loop 7. Cells were treated with or without cholesterol, and sealed membrane vesicles were prepared and digested with trypsin or proteinase K (63). After SDS-PAGE, we blotted with an antibody to Loop 1 (Figure 9a). When membranes from wild-type CHO cells that had been depleted of cholesterol were digested with trypsin, the Loop 1 antibody revealed two protected fragments with apparent molecular masses of 42 and 40 kDa, referred to as bands L6 and L4, respectively (Figure 9a, lane 1). When membranes from cholesterol-treated cells were digested, the L4 band was dramatically reduced in amount (Figure 9a, lane 2), indicating that a conformational change had rendered a cytosolic cleavage site inaccessible to the proteases. Using systematic mutagenesis and the trypsin assay, we localized the site of regulated cleavage to two closely spaced basic residues (lysine-378 and arginine-380) in Loop 4, the short cytosolic loop that separates TM4 and TM5 (Figure 4d). We concluded that cholesterol binding to Loop 1 causes a conformational change in the SSD, thereby sequestering Loop 4 from protease digestion. The unregulated L6 band arose from cleavage in the next cytosolic loop, Loop 6 (63).
Figure 9.
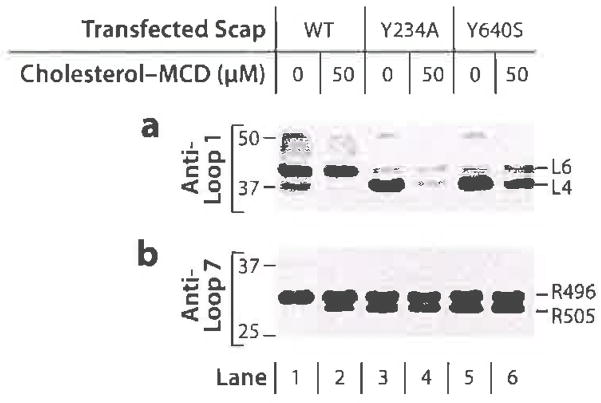
Tryptic digestion of sealed membrane vesicles from cells expressing wild-type (WT) Scap or two mutants (Y234A in Loop 1 and Y640S in Loop 7) that prevent interloop binding. Scap-deficient CHO cells transfected with the indicated Scap plasmid were depleted of sterols and incubated for 4 h without or with 50 μM cholesterol complexed to methyl-β-cyclodextrin (MCD). Sealed membrane vesicles were prepared and treated with trypsin. Samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with either (a) anti-Loop 1 antibody or (b) anti-Loop 7 antibody. The L4 band in panel a denotes trypsin cleavage in Loop 4. The R505 band in panel b denotes cleavage at arginine-505 in Loop 6. Reprinted from Reference 63.
To test for the requirement of Insig proteins for the sterol-induced conformational change in Loop 4, we isolated ER membranes from SRD-15 cells, a line of mutant CHO cells that lacks all Insigs (64). Cholesterol administration to SRD-15 cells induced the disappearance of the L4 band just as in wild-type CHO cells. Whereas 25-hydroxycholesterol caused this disappearance in wild-type CHO cells, it had no effect on Insig-deficient SRD-15 cells (63). We concluded that cholesterol binding to Loop 1 causes a conformational change in the SSD independently of Insig binding. Conversely, oxysterols can induce this conformational change only by binding to Insigs and causing Insigs to bind to Scap (63).
Figure 9 shows the informative results obtained when we compared trypsin digestion of membranes from wild-type CHO cells with Scap-deficient cells expressing the Y234A or Y640S mutations, both of which block the binding of Loop 1 to Loop 7 (63). As described earlier, bands L4 and L6 in Figure 9a indicate cleavage in Loops 4 and 6, respectively, and bands R496 and R505 in Figure 9b indicate different sites of cleavage in Loop 6. Recall that for wild-type Scap, band L4 indicates that the SSD is in the cholesterol-depleted condition, with Loop 1 bound to Loop 7 and with MELADL accessible for COPII binding. Band R505 indicates that the SSD is in the high-cholesterol condition in which MELADL is inaccessible to COPII proteins (Figure 9b, lane 2). For the two mutants in which Loop 1 is separated from Loop 7, trypsin generated band L4 in the absence of cholesterol (Figure 9a, lanes 3 and 5). Cholesterol caused the L4 band to disappear with the two mutants (lanes 4 and 6) just as it did with wild-type Scap (lane 2). This finding indicates that the separation of Loop 1 from Loop 7 does not induce the conformational change in the SSD. Rather, separation of the loops must result from the change in the SSD that is induced by cholesterol binding to Loop 1.
The opposite result was obtained when we probed for the cholesterol-induced change in Loop 6 (Figure 9b). In cholesterol-depleted membranes, tryptic digestion of the Y234A and Y640S mutants revealed cleavage at R505, indicating that Loop 6 was in the high-cholesterol conformation (lanes 3 and 5). Cholesterol addition produced no further change (lanes 4 and 6) (63). Moreover, in earlier studies, we found that the Y234A and Y640S mutants failed to transport SREBP-2 even in the absence of Insigs (61, 62). Considered together, these findings indicate that separation of Loop 1 from Loop 7 is sufficient to hide the MELADL sequence in Loop 6, even when the SSD remains in the cholesterol-free conformation and Insig 1 is not bound.
SCAP: MOTIONS OF A MOLECULAR MACHINE
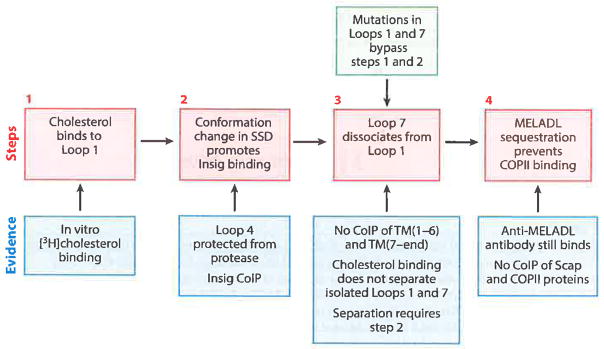
Figure 8 shows Scap in two conformations, namely, the cholesterol free-state (closed) with MELADL accessible to COPII proteins and the cholesterol-bound state (open) with the MELADL no longer accessible. Figure 10 outlines a working model for a proposed sequence of intermediate conformational changes that convert the cholesterol-free state to the cholesterol-bound state. The model is based in part on studies with the various Scap mutants that are summarized in Table 2. When cholesterol is low, Scap is in a closed conformation in which Loop 1 is bound to Loop 7, the SSD is in its cholesterol-free state with Loop 4 exposed to proteases, and the MELADL sequence in Loop 6 is able to bind COPII proteins (Figure 8a). When cholesterol reaches a certain threshold in the ER membranes, the sterol binds to Loop 1, and this leads to a conformational change in the SSD that is marked by the sequestration of Loop 4 from proteases (Figure 8b). The conformational change in the SSD promotes Insig binding, which in turn, causes Loop 7 to dissociate from Loop 1. The dissociation of the luminal loops produces the conformational change in cytosolic Loop 6 that moves the MELADL sequence so that it is no longer accessible to COPII proteins (Figure 8b).
Figure 10.
Scap as a cholesterol-triggered machine: working model of a four-step sequence for cholesterol regulation. Abbreviations: CoIP, coimmimoprecipitation; COPII, coat protein II; MELADL sequence, methionine-glutamate-leucine-alanine-aspartate-leucine; SSD, sterol-sensing domain; TM, transmembrane helix.
Table 2.
Point mutations in domains of Scap that alter SREBP function
| Domain | Mutation | Biochemical defect | ER-to-Golgi transport | Reference(s) |
|---|---|---|---|---|
| Loop 1 | Y234Aa | No binding to Loop 7 | Absent | 61 |
| SSD | Y298Cb L314b D443Nb |
No Insig binding in the presence of sterols | Not suppressed by sterols | 1,33,42 |
| SSD | D428Aa | Insig binding in the presence and absence of sterols | Absent | 57 |
| Loop7 | Y640Sa | No binding to Loop 1 | Absent | 62 |
Identified by alanine-scanning mutagenesis.
Identified by somatic cell genetics in CHO cells.
Abbreviations: ER, endoplasmic reticulum; SSD, sterol-sensing domain.
According to the model presented in Figure 10, the conformational change in Loop 4 of the SSD and the binding of Insig are not sufficient to sequester the MELADL sequence. Rather, these changes must first cause the dissociation of Loop 7 from Loop 1. When Loops 1 and 7 are separated artificially by the Y234A mutation in Loop 1 or the Y640S mutation in Loop 7, the MELADL sequence is not accessible to COPII proteins in cholesterol-depleted membranes even though the SSD is in the cholesterol-free state, as judged by accessibility of Loop 4 to trypsin (see Figure 9).
The models presented here envision the intramolecular binding of Loop 1 to Loop 7 in the same molecule. However, we cannot rule out the possibility that Loop 1 of one Scap molecule binds to Loop 7 of an adjacent Scap molecule. Such intermolecular binding might be favored if mammalian Scap is shown to form tetramers, as has been shown for the detergent-solubilized membrane domain of mammalian Scap (48) and postulated for the equivalent proteins in fission yeast (54).
CHALLENGE FOR THE FUTURE
The challenge for the future is to test the model shown in Figures 8 and 10 through structural analysis using X-ray crystallography and cryo–electron microscopy techniques. It will be necessary to determine the structure of Scap in both the cholesterol-free and cholesterol-bound states. The structures of the Scap–Insig complex and the Scap–COPII complex must also be determined. Our laboratory is making extensive attempts to perform these ultrastructural studies in collaboration with experienced structural biologists. Insight may also be gained by studying the structure of Scap in the fission yeast S. pombe and other fungal organisms. It is our hope that we or others will eventually solve these structures, which will give atomic-level insight into a sophisticated molecular machine that measures the cholesterol content of membranes.
Acknowledgments
We thank David Russell for critical review of the manuscript. The research described in this review was supported by the National Institutes of Health (HL20948 to J.L.G., M.S.B., and A.R.) and The Welch Foundation (I-1793 to A.R.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Hua X, Nohturfft A, Goldstein JL, Brown MS. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage activating protein (SCAP) Cell. 1996;87:415–26. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 2.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–40. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 3.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, et al. Combined analysis of olignucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. PNAS. 2003;100:12027–32. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008;8:512–21. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown MS, Dana SE, Goldstein JL. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in human fibroblasts by lipoproteins. PNAS. 1973;70:2162–66. doi: 10.1073/pnas.70.7.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein JL, Brown MS. Familial hypercholesterolemia: identification of a defect in the regulation of 3-hydroxy-3-methylgultaryl coenzyme A reductase activity associated with overproduction of cholesterol. PNAS. 1973;70:2804–8. doi: 10.1073/pnas.70.10.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown MS, Goldstein JL. Familial hypercholesterolemia: defective binding of lipoproteins to cultured fibroblasts associated with impaired regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity. PNAS. 1974;71:788–92. doi: 10.1073/pnas.71.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein JL, Brown MS. History of discovery; the LDL receptor. Arterioscler Thromb Vasc Biol. 2009;29:431–38. doi: 10.1161/ATVBAHA.108.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson RGW, Brown MS, Goldstein JL. Role of the coated endocytic vesicle in the uptake of receptor-bound low density lipoprotein in human fibroblasts. Cell. 1977;10:351–64. doi: 10.1016/0092-8674(77)90022-8. [DOI] [PubMed] [Google Scholar]
- 10.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein JL, Hobbs HH, Brown MS. Familial hypercholesterolemia. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 1995. pp. 1981–2030. [Google Scholar]
- 12.Brown MS, Goldstein JL. Regulation of the activity of the low density lipoprotein receptor in human fibroblasts. Cell. 1975;6:307–16. doi: 10.1016/0092-8674(75)90182-8. [DOI] [PubMed] [Google Scholar]
- 13.Brown MS, Goldstein JL. Cholesterol feedback: from Schoenheimer’s bottle to Scap’s MELADL. J Lipid Res. 2009;50:S15–27. doi: 10.1194/jlr.R800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Südhof TC, van der Westhuyzen DR, Goldstein JL, Brown MS, Russell DW. Three direct repeats and a TATA-like sequence are required for regulated expression of the human LDL receptor gene. J Biol Chem. 1987;262:10773–79. [PubMed] [Google Scholar]
- 15.Briggs MR, Yokoyama C, Wang X, Brown MS, Goldstein JL. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter: I. Identification of the protein and delineation of its target nucleotide sequence. J Biol Chem. 1993;268:14490–96. [PubMed] [Google Scholar]
- 16.Wang X, Briggs MR, Hua X, Yokoyama C, Goldstein JL, Brown MS. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter: II. Purification and characterization. J Biol Chem. 1993;268:14497–504. [PubMed] [Google Scholar]
- 17.Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, et al. SREBP-1, a basic helix-loop-helix leucine zipper protein that controls transcription of the LDL receptor gene. Cell. 1993;75:187–97. [PubMed] [Google Scholar]
- 18.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 19.Hua X, Sakai J, Ho YK, Goldstein JL, Brown MS. Hairpin orientation of sterol regulatory element binding protein-2 in cell membranes as determined by protease protection. J Biol Chem. 1995;270:29422–27. doi: 10.1074/jbc.270.49.29422. [DOI] [PubMed] [Google Scholar]
- 20.Hua X, Yokoyama C, Wu J, Briggs MR, Brown MS, et al. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. PNAS. 1993;90:11603–7. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 22.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–31. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakai J, Duncan EA, Rawson RB, Hua X, Brown MS, Goldstein JL. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–46. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 24.Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, et al. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 25.Sakai J, Rawson RB, Espenshade PJ, Cheng D, Seegmiller AC, et al. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol Cell. 1998;2:505–14. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- 26.Chang T-Y, Limanek JS. Regulation of cytosolic acetoacetyl coenzyme A thiolase, 3-hydroxy-3-methylglutaryl coenzyme A synthase, 3-hydroxy-3-methylglutaryl coenzyme A reductase, and mevalonate kinase by low density lipoprotein and by 25-hydroxycholesterol in Chinese hamster ovary cells. J Biol Chem. 1980;255:7787–95. [PubMed] [Google Scholar]
- 27.Hasan MT, Chang TY. Somatic cell genetic analysis of two classes of CHO cell mutants expressing opposite phenotypes in sterol-dependent regulation of cholesterol metabolism. Somat Cell Mol Genet. 1994;20:481–91. doi: 10.1007/BF02255839. [DOI] [PubMed] [Google Scholar]
- 28.Gil G, Faust JR, Chin DJ, Goldstein JL, Brown MS. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 1985;41:249–58. doi: 10.1016/0092-8674(85)90078-9. [DOI] [PubMed] [Google Scholar]
- 29.Nohturfft A, Brown MS, Goldstein JL. Topology of SREBP cleavage-activating protein, a polytopic membrane protein with a sterol-sensing domain. J Biol Chem. 1998;273:17243–50. doi: 10.1074/jbc.273.27.17243. [DOI] [PubMed] [Google Scholar]
- 30.Olender EH, Simoni RD. The intracellular targeting and membrane topology of 3-hydroxy-3-methylglutaryl-CoA reductase. J Biol Chem. 1992;267:4223–35. [PubMed] [Google Scholar]
- 31.Sakai J, Nohturfft A, Cheng D, Ho YK, Brown MS, Goldstein JL. Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein (SCAP) J Biol Chem. 1997;272:20213–21. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- 32.Sakai J, Nohturfft A, Goldstein JL, Brown MS. Cleavage of sterol regulatory element binding proteins (SREBPs) at site-1 requires interaction with SREBP cleavage-activating protein. Evidence from in vivo competition studies. J Biol Chem. 1998;273:5785–93. doi: 10.1074/jbc.273.10.5785. [DOI] [PubMed] [Google Scholar]
- 33.Nohturfft A, Brown MS, Goldstein JL. Sterols regulate processing of carbohydrate chains of wild-type SREBP cleavage-activating protein (SCAP), but not sterol-resistant mutants Y298C or D443N. PNAS. 1998;95:12848–53. doi: 10.1073/pnas.95.22.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yabe D, Xia Z-P, Adams CM, Rawson RB. Three mutations in sterol-sensing domain of SCAP block interaction with Insig and render SREBP cleavage insensitive to sterols. PNAS. 2002;99:16672–77. doi: 10.1073/pnas.262669399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nohturfft A, Yabe D, Goldstein JL, Brown MS, Espenshade PJ. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 2000;102:315–23. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- 36.DeBose-Boyd RA, Brown MS, Li W-P, Nohturfft A, Goldstein JL, Espenshade PJ. Transport-dependent proteolysis of SREBP: Relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99:703–12. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- 37.Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell. 2002;10:237–45. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- 38.Yang T, Goldstein JL, Brown MS. Overexpression of membrane domain of SCAP prevents sterols from inhibiting SCAP/SREBP exit from endoplasmic reticulum. J Biol Chem. 2000;275:29881–86. doi: 10.1074/jbc.M005439200. [DOI] [PubMed] [Google Scholar]
- 39.Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, et al. Cruicial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 40.Mohn KL, Laz TM, Hsu J-C, Melby AE, Bravo R, Taub R. The immediate-early growth response in regenerating liver and insulin-stimulated II-35 cells: comparison with serum-stimulated 3T3 cells and identification of 41 novel immediate-early genes. Mol Cell Biol. 1991;11:381–90. doi: 10.1128/mcb.11.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luong A. PhD Thesis. Univ. Tex. Southwest. Med. Cent; Dallas: 2000. Identification of three novel SREBP-activated target genes: acetyl CoA synthetase, 3-β-hydroxysterol dehydrogenase, and CL-6/Insig1. [Google Scholar]
- 42.Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. PNAS. 2002;99:12753–58. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feramisco JD, Goldstein JL, Brown MS. Membrane topology of human Insig-1, a protein regulator of lipid synthesis. J Biol Chem. 2004;279:8487–96. doi: 10.1074/jbc.M312623200. [DOI] [PubMed] [Google Scholar]
- 44.Gong Y, Lee JN, Lee PCW, Goldstein JL, Brown MS, Ye J. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell Metab. 2006;3:15–24. doi: 10.1016/j.cmet.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 45.Kandutsch AA, Chen HW. Inhibition of sterol synthesis in cultured mouse cells by cholesterol derivatives oxygenated in the side chain. J Biol Chem. 1974;249:6057–61. [PubMed] [Google Scholar]
- 46.Brown MS, Goldstein JL. Suppression of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity and inhibition of growth of human fibroblasts by 7-ketocholesterol. J Biol Chem. 1974;249:7306–14. [PubMed] [Google Scholar]
- 47.Adams CM, Reitz J, DeBrabander JK, Feramisco JD, Li L, et al. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J Biol Chem. 2004;279:52772–80. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- 48.Radhakrishnan A, Sun L-P, Kwon HJ, Brown MS, Goldstein JL. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol Cell. 2004;15:259–68. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 49.Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Oxysterols block transport by binding to Insig. PNAS. 2007;104:6511–18. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobrosotskaya I, Goldstein JL, Brown MS, Rawson RB. Reconstitution of sterol-regulated endoplasmic reticulum-to-Golgi transport of SREBP-2 in insect cells by co-expression of mammalian SCAP and Insigs. J Biol Chem. 2003;278:35837–43. doi: 10.1074/jbc.M306476200. [DOI] [PubMed] [Google Scholar]
- 51.Sun L-P, Seemann J, Brown MS, Goldstein JL. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. PNAS. 2007;104:6519–26. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zambrano F, Fleischer S, Fleischer B. Lipid composition of the Golgi apparatus of rat kidney and liver in comparison with other subcellular organelles. Biochim Biophys Acta. 1975;380:357–69. doi: 10.1016/0005-2760(75)90104-6. [DOI] [PubMed] [Google Scholar]
- 53.Lange Y, Steck TL. Quantitation of the pool of cholesterol associated with acyl-CoA: cholesterol acyltransferase in human fibroblasts. J Biol Chem. 1997;272:13103–8. doi: 10.1074/jbc.272.20.13103. [DOI] [PubMed] [Google Scholar]
- 54.Gong X, Qian H, Shao W, Li J, Wu J, et al. Complex structure of the fission yeast SREBP-SCAP binding domains reveals an oligomeric organization. Cell Res. 2016;26:1197–211. doi: 10.1038/cr.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Espenshade PJ, Li W-P, Yabe D. Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. PNAS. 2002;99:11694–99. doi: 10.1073/pnas.182412799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun L-P, Li L, Goldstein JL, Brown MS. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J Biol Chem. 2005;280:26483–90. doi: 10.1074/jbc.M504041200. [DOI] [PubMed] [Google Scholar]
- 57.Feramisco JD, Radhakrishnan A, Ikeda Y, Reitz J, Brown MS, Goldstein JL. Intramembrane aspartic acid in SCAP protein governs cholesterol-induced conformational change. PNAS. 2005;102:3242–47. doi: 10.1073/pnas.0500206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sever N, Yang T, Brown MS, Goldstein JL, DeBose-Boyd RA. Accelerated degradation of HMG CoA reductase mediated by binding of Insig-1 to its sterol-sensing domain. Mol Cell. 2003;11:25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 59.Sever N, Song B-L, Yabe D, Goldstein JL, Brown MS, DeBose-Boyd RA. Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. J Biol Chem. 2003;278:52479–90. doi: 10.1074/jbc.M310053200. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Lee KM, Kinch LN, Clark L, Grishin NV, et al. Direct demonstration that Loop1 of Scap binds to Loop7, a crucial event in cholesterol homeostasis. J Biol Chem. 2016;291:12888–96. doi: 10.1074/jbc.M116.729798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Motamed M, Zhang Y, Wang ML, Seemann J, Kwon HJ, et al. Identification of luminal Loop 1 of Scap protein as the sterol sensor that maintains cholesterol homeostasis. J Biol Chem. 2011;286:18002–12. doi: 10.1074/jbc.M111.238311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Motamed M, Seemann J, Brown MS, Goldstein JL. Point mutation in luminal Loop 7 of Scap blocks interaction with Loop 1 and abolishes movement to Golgi. J Biol Chem. 2013;288:14059–67. doi: 10.1074/jbc.M113.469528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao Y, Zhou Y, Goldstein JL, Brown MS, Radhakrishnan A. Cholesterol-induced conformational changes in the sterol-sensing domain of the Scap protein suggest feedback mechanism to control cholesterol synthesis. J Biol Chem. 2017;292:8729–37. doi: 10.1074/jbc.M117.783894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee PCW, Sever N, DeBose-Boyd RA. Isolation of sterol-resistant Chinese hamster ovary cells with genetic deficiencies in both Insig-1 and Insig-2. J Biol Chem. 2005;280:25242–49. doi: 10.1074/jbc.M502989200. [DOI] [PubMed] [Google Scholar]
- 65.Prabhu AV, Luu W, Li D, Sharpe LJ, Brown AJ. DHCR7: a vital enzyme switch between cholesterol and vitamin D production. Prog Lipid Res. 2016;64:138–51. doi: 10.1016/j.plipres.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 66.Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, et al. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 1997;277:228–31. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- 67.Altmann SW, Davis HR, Jr, Zhu L-J, Yao X, Hoos LM, et al. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 2004;303:1201–4. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- 68.Burke R, Nellen D, Bellotto M, Hafen E, Senti K-A, et al. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 1999;99:803–15. doi: 10.1016/s0092-8674(00)81677-3. [DOI] [PubMed] [Google Scholar]
- 69.Jo Y, Lee PCW, Sguigna PV, DeBose-Boyd RA. Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and Trc8. PNAS. 2011;108:20503–8. doi: 10.1073/pnas.1112831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin PH, Lan WM, Chau LY. TRC8 suppresses tumorigenesis through targeting heme oxygenase-1 for ubiquitination and degradation. Oncogene. 2013;32:2325–34. doi: 10.1038/onc.2012.244. [DOI] [PubMed] [Google Scholar]
- 71.Kuwabara PE, Labouesse M. The sterol-sensing domain: multiple families, a unique role? Trends Genet. 2002;18:193–201. doi: 10.1016/s0168-9525(02)02640-9. [DOI] [PubMed] [Google Scholar]