Abstract
Magnetotactic bacteria (MTB) migrate in complex porous sediments where fluid flow is ubiquitous. Here, we demonstrate that magnetotaxis enables MTB to migrate effectively through porous micromodels. Directed MTB can circumvent curved obstacles by traveling along the boundaries and pass flat obstacles by repeatedly switching between forward and backward runs. Magnetotaxis enables directed motion of MTB through heterogeneous porous media, overcoming tortuous flow fields with local velocities as high as 250 μm s−1. Our findings bring new insights into the migration behaviour of MTB in their natural habitats and their potential in vivo applications as microbiorobots.
INTRODUCTION
Magnetotactic bacteria (MTB) live in complex and heterogeneous habitats, such as aquatic sediments,1–3 where fluid flow is ubiquitous.4–6 MTB play a crucial role in biogeochemical processes,7 including the Earth's iron and sulfur cycles.8,9 MTB possess magnetite (Fe3O4) or greigite (Fe3S4) nanocrystals arranged in chains, called magnetosomes, which enable the cells to orient and swim along the geomagnetic field lines to reach optimal oxygen concentrations in the oxic-anoxic transition zone,10 a behaviour termed magnetotaxis.11 The upward movement and downward movement of MTB in their habitats are crucial for their metabolism and survival.3 While migrating through porous environments (e.g., sediments), the motility of MTB is affected by the direction and magnitude of the magnetic field they experience and their interaction with solid surfaces and currents, which ultimately influence their swimming behaviour and navigation strategies.12–18 Recently, MTB have shown great promise as potential drug delivery vehicles, which requires their navigation through physiological environments.19,20 Yazdi et al.21 recently demonstrated that magnetotaxis enables MTB to exhibit directed motion in flows relevant to their natural habitats and biomedical applications. However, the influence of the interplay between surface confinement, flow, and magnetotaxis on the motility of MTB in porous media has remained elusive.
Here, we use microfluidics to study the swimming behaviour of MTB in porous environments, mimicking their natural habitats. We demonstrate that, under an applied magnetic field, MTB circumvent curved obstacles by traveling along the boundaries. When encountering a flat surface, however, MTB switch between forward and backward runs in order to pass the obstacle. We further demonstrate that magnetotaxis enables directed motion of MTB through heterogeneous porous micromodels, overcoming tortuous flow fields with local velocities as high as 250 μm s−1.
MATERIALS AND METHODS
Microfluidic devices were made of polydimethylsiloxane (PDMS) using soft lithography techniques.22,23 Magnetospirillum magneticum strain AMB-1 was grown in an ATCC 1653 revised magnetic spirillum growth medium (MSGM 1653 ATCC® Manassas, USA) at 37 °C as described elsewhere.21 After three days of culture, bacteria were sampled at an optical density in the range of 0.2–0.3 for use in experiments. For control experiments, motile Escherichia coli strain RP437 (Yale stocks CGSC# 12122) was grown and used as describe elsewhere.21 In flow experiments, a syringe pump (Pump 11 Elite, Harvard Apparatus, MA) was used to administrate the flow. An inverted microscope (Olympus IX83, Germany) equipped with a CMOS camera (Zyla-4.2-CL10, ANDOR, Ireland) was used to capture bright-field image sequences of bacteria at 25 frames s−1. Z-stack acquisition and cellSens software (Olympus, Germany) were used to quantify the bacteria distribution across the height of the microchannel. Images were processed using ImageJ24 to track the bacteria manually and extract quantitative information on their swimming characteristics. The progressive velocity was calculated as the displacement vector between the first and last tracking points divided by the duration of the track segment.25 COMSOL Multiphysics (Burlington, MA) was used to simulate the flow field and the magnetic field in the porous micromodel. For statistical evaluation, the P value was determined using a two-sample t-test with unequal variances, and P < 0.05 was considered significant.
RESULTS AND DISCUSSION
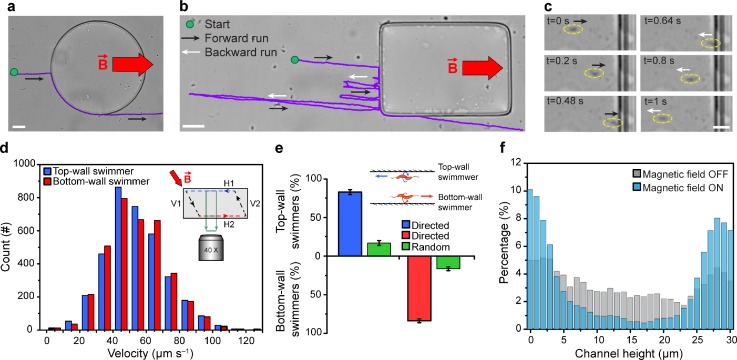
MTB exhibited distinct swimming behaviour when encountering curved and flat obstacles, under an applied magnetic field of 0.3 mT. At curved obstacles (e.g., circles), MTB traveled along the perimeter by maintaining their axial motility direction, pushing against the surface, and continued swimming straight along the field lines after passing the obstacle [Fig. 1(a)]. The ability of MTB to push against a curved boundary is potentially attributed to their interaction angle with the surface and resulting hydrodynamic effects near a curved surface.25,26 In contrast, upon encountering a flat obstacle (e.g., rectangular objects), MTB switched between forward and backward runs repeatedly, until they circumvented the obstacle [Fig. 1(b)]. A representative bacterial reversal event at a flat obstacle is shown in Fig. 1(c). When reaching the obstacle, the bacterium reversed its swimming direction without turning the cell body. This switch in the swimming direction is linked to the tactile response of MTB and their ability to sense magnetic field gradients.12,18,27 Specifically, at a flat obstacle, when the swimming path of MTB is blocked, a tactile response causes MTB to switch their swimming direction.12 However, while swimming towards a lower magnetic field strength (Fig. S1, supplementary material), the magnetosensing mechanism that MTB possess enables them to sense the magnetic field gradient and reverse their swimming direction again to migrate towards regions with higher magnetic field strengths.18
FIG. 1.
Interaction of MTB with (a) curved and (b) flat obstacles, B = 0.3 mT. (c) A bacterial reversal event at a flat obstacle. The bacterium is marked with a yellow dashed ellipse. (d) Histograms of the instantaneous swimming velocity of MTB near the top (blue) and bottom (red) microchannel walls (n = 3573). The inset shows the cross-section of the microchannel, with the dashed line representing the parallelogram path of MTB. (e) Fraction of random and directed MTB near top and bottom microchannel walls (n = 422). Values are reported as mean ± standard deviation. (f) Distributions of MTB across the microchannel height in the absence (gray) and presence (cyan) of the applied magnetic field, B = 5 mT (n = 9844). Scale bars: 30 μm (a) and (b) and 5 μm (c).
By applying a magnetic field of 5 mT oblique to the microchannel horizontal walls [H1 and H2, Fig. 1(d), inset], MTB followed a closed-loop path, with a shape similar to a parallelogram. Bacteria swam with similar average velocities of 53.8 ± 18 μm s−1 and 53.7 ± 17 μm s−1 near the top (H1) and bottom (H2) walls (n = 3573, P = 0.819, not statistically significant), respectively. Notably, these results show that M. magneticum can reach similar velocities using either flagellum at each end of its body for propulsion.28 This looping behaviour was observed in ∼80% of the bacterial population that exhibited directed motion [Fig. 1(e)]. The lack of the directed response for the remaining fraction of randomly swimming bacteria can be due to the phenotypic variation of bacteria, the absence or abnormal arrangement of magnetite nanocrystals,29–31 and defects in the cell's morphology (e.g., malfunctioning flagella).1,32 The looping behaviour of directed MTB was also observed to influence the concentration of bacteria near the horizontal walls. Specifically, for B = 0 mT (magnetic field OFF), only a slightly higher number of bacteria were observed near horizontal walls compared to the number of bacteria in the bulk [Fig. 1(f)] due to the hydrodynamic interaction between the bacteria and the surface.33,34 However, for B = 5 mT (magnetic field ON), the concentration of MTB near the top and bottom walls was considerably higher (∼10-fold) than the bacterial population in the bulk [Fig. 1(f)]. Over 73% of directed MTB population concentrated within 5 μm of the horizontal walls—a 1.7-fold increase compared to the concentration of MTB that swam randomly near horizontal walls in the absence of an applied magnetic field.
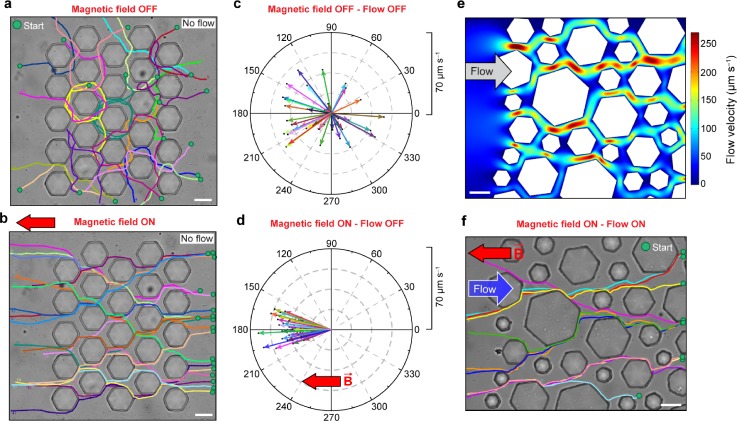
To study the swimming behaviour of MTB through porous media, we used a micromodel that consists of hexagonal posts with a maximal diameter of 45 μm [Figs. 2(a) and 2(b)]. The micromodel was 10 μm in height, and the gap between posts was 10 μm, in order to resemble the typical pore size (∼0.1–200 μm) and porosity (∼0.4) of sediments and soils,35–37 as well as those found in physiological environments, such as tissues and vascular networks.38 In control experiments with motile E. coli, cells swam randomly in all directions through the porous micromodel, both in the absence and presence of an applied magnetic field of 0.3 mT, with similar average progressive velocities of 8 ± 4 μm s−1 and 9 ± 5 μm s−1, respectively (Fig. S2, supplementary material). In the absence of an applied magnetic field, MTB were observed to swim in all directions randomly, and their migration through the porous medium was hindered by their interactions with boundaries, as they repeatedly bounced between posts [Fig. 2(a) and Movie S1, supplementary material]. The average progressive velocity of randomly swimming MTB was 28 ± 14 μm s−1 [Fig. 2(c)]. In contrast, under an applied magnetic field of 0.3 mT, magnetotaxis enabled directed motion of MTB through the micromodel by realigning the bacteria along the field lines after interacting with obstacles [Fig. 2(b) and Movie S2, supplementary material]. Directed MTB navigated through the micromodel comparatively faster than randomly swimming bacteria, with an average progressive velocity of 36 ± 12 μm s−1, and, remarkably, deviated less than 25° from a straight path [Fig. 2(d)]. In a porous micromodel with ∼50% higher porosity (∼0.6), directed MTB navigated faster and straighter than those in a micromodel with a porosity of ∼0.4. Specifically, the average progressive velocity of directed MTB increased by 14% to 41 ± 8 μm s−1, with less than 5° deviation from a straight path (Fig. S3, supplementary material).
FIG. 2.
Swimming trajectories of MTB through a homogenous porous micromodel (a) in the absence and (b) in the presence of the applied magnetic field of B = 0.3 mT, with the corresponding polar graphs of progressive velocities overlaid at the center in (c) and (d), respectively. The radius of the polar graph indicates progressive velocity. (e) COMSOL simulation results of the velocity field in a heterogeneous micromodel. (f) Swimming trajectories of MTB through a heterogeneous micromodel when directed to navigate upstream (B = 0.3 mT). Scale bar: 30 μm.
We further investigated the influence of counter-directional flow on the migration of MTB through the porous micromodel. To account for the heterogeneous flow environments in sediments, a heterogeneous micromodel with hexagonal posts ranging from 30 to 90 μm in diameter was used [Fig. 2(e)]. A laminar flow with an average velocity of 29 μm s−1 was applied in the micromodel. In the absence of an applied magnetic field, MTB were predominantly advected downstream by the flow. In contrast, under an applied magnetic field of 0.3 mT, MTB swam incessantly upstream (against the flow) through the heterogeneous micromodel at an average swimming velocity of 25 ± 7 μm s−1. It is noteworthy that magnetotaxis enabled MTB to swim relatively fast and straight through a rather tortuous flow field with local velocities as high as 250 μm s−1. This effective migration of MTB through the micromodel contrasts with the behaviour of other bacteria previously reported, such as E. coli, Pseudomonas putida, and Bacillus subtilis, where their transport is impeded by the porous media and the flow.39–41 Our results indicate that magnetotaxis dominates over hydrodynamic effects6,42 to guide MTB to swim along the magnetic field lines in porous media.
CONCLUSIONS
In summary, we used microfluidics to observe and quantify the swimming behaviour of MTB in porous environments that mimic their natural habitats. Magnetotaxis enabled MTB to circumvent the curved obstacles in a micromodel, by swimming around the objects' perimeters. At flat obstacles, directed MTB repeatedly switched their motility directions to pass the object. By applying a magnetic field oblique to the horizontal walls of the microchannel, MTB were observed to push against the horizontal surface and follow a closed-loop path with a shape similar to a parallelogram. Magnetotaxis also facilitated the directed migration of MTB through tortuous flow fields in the heterogeneous porous micromodel by realigning the bacteria along the magnetic field lines after interacting with obstacles. These findings, altogether, bring new insights into the migration of MTB in environments similar to aquatic sediments and their potential in vivo applications as microbiorobots.
SUPPLEMENTARY MATERIAL
See supplementary material for videos showing migration of MTB through the micromodel in the absence (Movie S1) and in the presence (Movie S2) of an applied magnetic field of 0.3 mT.
ACKNOWLEDGMENTS
This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grants (No. RGPIN-201-05138) and a Canada Foundation for Innovation Leaders Opportunity Fund Program (No. 31967) provided to CE. The authors would like to thank Professor Peter Davies (Tier I Canada Research Chair in Protein Engineering) from the Department of Biomedical and Molecular Sciences (Queen's University), Juan Gomez-Cruz, and Tyler D. R. Vance for their valuable collaboration, insights, and guidance and for providing the magnetotactic bacteria. The authors also thank the NanoFabrication Kingston (NFK) and Dr. Graham Gibson for the assistance provided on microfabrication. We also gratefully acknowledge the support to R.N. from an NSERC postdoctoral fellowship and a Queen's Postdoctoral Fellowship. C.A.S. was funded by a PGS-D NSERC Scholarship.
References
- 1. Uebe R. and Schüler D., Nat. Rev. Microbiol. 14, 621 (2016). 10.1038/nrmicro.2016.99 [DOI] [PubMed] [Google Scholar]
- 2. Fassbinder J. W. E., Stanjekt H., and Vali H., Nature 343, 161 (1990). 10.1038/343161a0 [DOI] [PubMed] [Google Scholar]
- 3. Xu C., Zhang W., Pan H., Du H., and Xiao T., J. Soils Sediments 1, 1 (2018) 10.1007/s11368-018-1912-8. [DOI] [Google Scholar]
- 4. Son K., Brumley D. R., and Stocker R., Nat. Rev. Microbiol. 13, 761 (2015). 10.1038/nrmicro3567 [DOI] [PubMed] [Google Scholar]
- 5. Persat A., Nadell C. D., Kim M. K., Ingremeau F., Siryaporn A., Drescher K., Wingreen N. S., Bassler B. L., Gitai Z., and Stone H. A., Cell 161, 988 (2015). 10.1016/j.cell.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rusconi R. and Stocker R., Curr. Opin. Microbiol. 25, 1 (2015). 10.1016/j.mib.2015.03.003 [DOI] [PubMed] [Google Scholar]
- 7. Lin W. and Pan Y., FEMS Microbiol. Lett. 302, 85 (2010). 10.1111/j.1574-6968.2009.01838.x [DOI] [PubMed] [Google Scholar]
- 8. Bazylinski D. A. and Frankel R. B., Nat. Rev. Microbiol. 2, 217 (2004). 10.1038/nrmicro842 [DOI] [PubMed] [Google Scholar]
- 9. Schüler D., Magnetoreception and Magnetosomes in Bacteria ( Springer; Berlin Heidelberg, Berlin, Heidelberg, 2007). [Google Scholar]
- 10. Faivre D. and Schüler D., Chem. Rev. 108, 4875 (2008). 10.1021/cr078258w [DOI] [PubMed] [Google Scholar]
- 11. Blakemore R., Science 190, 377 (1975). 10.1126/science.170679 [DOI] [PubMed] [Google Scholar]
- 12. Spormann A. M. and Wolfe R. S., FEMS Microbiol. Lett. 22, 171 (1984). 10.1111/j.1574-6968.1984.tb00720.x [DOI] [Google Scholar]
- 13. Mao X., Egli R., Petersen N., Hanzlik M., and Zhao X., Geochem., Geophys. Geosyst. 15, 255 (2014). 10.1002/2013GC005034 [DOI] [Google Scholar]
- 14. Da Zhang S., Petersen N., Zhang W. J., Cargou S., Ruan J., Murat D., Santini C. L., Song T., Kato T., Notareschi P., Li Y., Namba K., Gué A. M., and Wu L. F., Environ. Microbiol. Rep. 6, 14 (2014). 10.1111/1758-2229.12102 [DOI] [PubMed] [Google Scholar]
- 15. Mao X., Egli R., Petersen N., Hanzlik M., and Liu X., PLoS One 9, e102810 (2014). 10.1371/journal.pone.0102810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu J., Zhang W., Li X., Li X., Chen X., Li J.-H., Teng Z., Xu C., Santini C.-L., Zhao L., Zhao Y., Zhang H., Zhang W.-J., Xu K., Li C., Pan Y., Xiao T., Pan H., and Wu L.-F., Sci. Rep. 7, 17964 (2017). 10.1038/s41598-017-17445-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Waisbord N., Lefèvre C. T., Bocquet L., Ybert C., and Cottin-Bizonne C., Phys. Rev. Fluids 1, 53203 (2016). 10.1103/PhysRevFluids.1.053203 [DOI] [PubMed] [Google Scholar]
- 18. González L. M. M. M., Ruder W. C. C. C., Mitchell A. P. P. P., Messner W. C. C. C., and LeDuc P. R. R. R., ISME J. 9, 1 (2014) 10.1038/ismej.2014.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Felfoul O., Mohammadi M., Taherkhani S., de Lanauze D., Zhong Xu Y., Loghin D., Essa S., Jancik S., Houle D., Lafleur M., Gaboury L., Tabrizian M., Kaou N., Atkin M., Vuong T., Batist G., Beauchemin N., Radzioch D., and Martel S., Nat. Nanotechnol. 11, 941 (2016). 10.1038/nnano.2016.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stanton M. M., Park B.-W., Vilela D., Bente K., Faivre D., Sitti M., and Sánchez S., ACS Nano 11, 9968 (2017). 10.1021/acsnano.7b04128 [DOI] [PubMed] [Google Scholar]
- 21. Rismani Yazdi S., Nosrati R., Stevens C. A., Vogel D., Davies P. L., and Escobedo C., Small 14, 1702982 (2018). 10.1002/smll.201702982 [DOI] [PubMed] [Google Scholar]
- 22. Rismani Yazdi S., Shadmani A., Bürgel S. C., Misun P. M., Hierlemann A., and Frey O., Lab Chip 15, 4138 (2015). 10.1039/C5LC01000D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eamer L., Nosrati R., Vollmer M., Zini A., and Sinton D., Biomicrofluidics 9, 44113 (2015). 10.1063/1.4928129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schneider C. A., Rasband W. S., and Eliceiri K. W., Nat. Methods 9, 671 (2012). 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nosrati R., Driouchi A., Yip C. M., and Sinton D., Nat. Commun. 6, 8703 (2015). 10.1038/ncomms9703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kantsler V., Dunkel J., Polin M., and Goldstein R. E., Proc. Natl. Acad. Sci. U. S. A. 110, 1187 (2013). 10.1073/pnas.1210548110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Greenberg M., Canter K., Mahler I., and Tornheim A., Biophys. J. 88, 1496 (2005). 10.1529/biophysj.104.047068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murat D., Hérisse M., Espinosa L., Bossa A., Alberto F., and Wu L. F., J. Bacteriol. 197, 3275 (2015). 10.1128/JB.00172-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blakemore R. P., Maratea D., and Wolfe R. S., J. Bacteriol. 140, 720 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Faivre D., Menguy N., Pósfai M., and Schüler D., Am. Miner. 93, 463 (2008). 10.2138/am.2008.2678 [DOI] [Google Scholar]
- 31. Klumpp S., Kiani B., Vach P., and Faivre D., Phys. Scr. T165, 14044 (2015). 10.1088/0031-8949/2015/T165/014044 [DOI] [Google Scholar]
- 32. Paterson J. S., J. Pathol. Bacteriol. 48, 25 (1939). 10.1002/path.1700480104 [DOI] [Google Scholar]
- 33. Berke A., Turner L., Berg H., and Lauga E., Phys. Rev. Lett. 101, 38102 (2008). 10.1103/PhysRevLett.101.038102 [DOI] [PubMed] [Google Scholar]
- 34. Li G. and Tang J. X., Phys. Rev. Lett. 103, 78101 (2009). 10.1103/PhysRevLett.103.078101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Minagawa H., Nishikawa Y., Ikeda I., Miyazaki K., Takahara N., Sakamoto Y., Komai T., and Nairta H., J. Geophys. Res. Solid Earth 113, B07210, https://doi.org/10.1029/2007JB005403 (2008). [Google Scholar]
- 36. Yamamoto T. and Turgut A., J. Acoust. Soc. Am. 83, 1744 (1988). 10.1121/1.396507 [DOI] [Google Scholar]
- 37. Nimmo J. R., Encyclopedia of Soils in the Environment ( Elsevier, 2005), pp. 295–303. [Google Scholar]
- 38. Annabi N., Nichol J. W., Zhong X., Ji C., Koshy S., Khademhosseini A., and Dehghani F., Tissue Eng. Part B 16, 371 (2010). 10.1089/ten.teb.2009.0639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ford R. M. and Harvey R. W., Adv. Water Resour. 30, 1608 (2007). 10.1016/j.advwatres.2006.05.019 [DOI] [Google Scholar]
- 40. Barton J. W. and Ford R. M., Biotechnol. Bioeng. 53, 487 (1997). [DOI] [PubMed] [Google Scholar]
- 41. Rusconi R., Guasto J. S., and Stocker R., Nat. Phys. 10, 212 (2014). 10.1038/nphys2883 [DOI] [Google Scholar]
- 42. Marcos, Fu H. C., Powers T. R., and Stocker R., Proc. Natl. Acad. Sci. 109, 4780 (2012). 10.1073/pnas.1120955109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for videos showing migration of MTB through the micromodel in the absence (Movie S1) and in the presence (Movie S2) of an applied magnetic field of 0.3 mT.