Abstract
The brain has long been known to display the most complex pattern of alternative splicing, thereby producing diverse protein isoforms compared to other tissues. Recent evidence indicates that many alternative exons are neuron-specific, evolutionarily conserved, and found in regulators of transcription including DNA-binding protein and histone modifying enzymes. This raises a possibility that neurons adopt unique mechanisms of transcription. Given that transcriptional machineries are frequently mutated in neurodevelopmental disorders with cognitive dysfunction, it is important to understand how neuron-specific alternative splicing contributes to proper transcriptional regulation in the brain. In this review, we summarize current knowledge regarding how neuron-specific splicing events alter the function of transcriptional regulators and shape unique gene expression patterns in the brain and the implications of neuronal splicing to the pathophysiology of neurodevelopmental disorders.
Keywords: Alternative splicing, Microexons, Chromatin, Transcription Factors, Neuronal Isoforms, Neurodevelopmental Disorders
1. Introduction
One of the long-standing questions in genetics is how cells achieve cell-type-specific gene expression. The major roles of DNA-binding transcription factors (TFs) in this process have been well established. Master TFs are often expressed in limited cell types and bind to their cognate DNA sequence at promoters and enhancers, thereby activating or repressing gene expression in a cell-type-specific manner (Deplancke, Alpern et al. 2016). In multicellular organisms, DNA is organized into nucleosomes with the four core histones (Luger, Mader et al. 1997), which are generally refractory to actions of RNA polymerase II (Li and Reinberg 2011). To relax the inherently closed chromatin structure, TFs recruit a variety of machineries, including histone-modification enzymes and chromatin-remodeling complexes.
Unlike master TFs, chromatin modifiers and remodelers tend to be ubiquitously expressed. More recent work has begun to reveal exceptions to this rule. Germ-cell-specific assembly of the preinitiation complexes (Goodrich and Tjian 2010), neuron-specific micro-RNA circuitries (Yoo, Sun et al. 2011), and neuron-specific ATP-dependent chromatin-remodeling complexes (Staahl and Crabtree 2013) provided earlier insights into how non-TF agents can contribute to cell-type-specific gene expression. While these mechanisms all rely on the cell-type-restricted presence of transcriptional regulators, recent evidence indicates that alternative splicing of ubiquitously-expressed factors can contribute to cell-type specific transcription, in particular, within neurons.
In this article, we discuss current views on how alternative splicing contributes to complexity of the brain, its link to neurodevelopmental disorders, and how neuron-specific splicing events can influence the roles of transcriptional machineries. A growing amount of literature has begun to support the idea that compromised function of the neuronal isoforms of transcriptional regulators may underlie multiple neurodevelopmental disorders.
2. Alternative Splicing in the Brain
Alternative splicing generates multiple proteins from a single pre-mRNA by including and/or excluding alternative exons, thereby diversifying cellular proteomes. In complex organisms, such as humans, alternative splicing events are estimated to occur in 92–94% of genes (Wang, Sandberg et al. 2008). Throughout vertebrate evolution, alternative splicing programs are notably most complex in the nervous system (Yeo, Holste et al. 2004, Chen and Manley 2009, Barbosa-Morais, Irimia et al. 2012, Merkin, Russell et al. 2012), suggesting that alternative splicing contributes to the complexity of brain anatomy, development, and function. Not only does the brain have a higher number of alternative splicing events relative to other tissues (Xu, Modrek et al. 2002, Yeo, Holste et al. 2004, Pan, Shai et al. 2008), but conservation of the brain-specific alternative splicing program is especially prominent through vertebrate evolution suggesting functionality of spliced products (Barbosa-Morais, Irimia et al. 2012, Merkin, Russell et al. 2012). Recent work has highlighted the neocortex, the center for higher-order cognitive processes, as a hotspot of alternative splicing events that influence cortical development, layering, and cell fate (McKee, Minet et al. 2005, Belgard, Marques et al. 2011, Zhang, Chen et al. 2014, Zhang, Chen et al. 2016). As will be discussed in detail below, dysregulation of this alternative splicing program leads to neurological disease (Licatalosi and Darnell 2006).
3. Mechanisms and Biological Roles of Neuron-Specific Alternative Splicing Factors
Alternative splicing is coordinated by cis-acting RNA elements and trans-acting RNA binding proteins that regulate intron excision. The spliceosome is the major molecular machinery, which controls intron excision and determines which pre-mRNA sequences are to be included or excluded from the mature mRNA. The core spliceosome is a large RNA-protein complex and involves the five subunits defined by the five RNA components, U1, U2, U4, U5, and U6, and the associated small ribonucleoproteins (RNPs). A large number of auxiliary proteins help the spliceosome recognize splice sites (Li, Lee et al. 2007, Chen and Manley 2009, Wahl, Will et al. 2009). While most spliceosome components are constitutively expressed, tissue-specific RNA-binding proteins direct spliceosome machinery to specific splice sites to generate tissue-specific splicing patterns. Neuron-specific alternative splicing is one such example controlled by the coordinate actions of many brain-specific RNA-binding proteins. Several recent review articles have comprehensively discussed the mechanisms of actions and roles in brain development of these splicing regulators (Raj and Blencowe 2015, Lara-Pezzi, Desco et al. 2016, Vuong, Black et al. 2016). Below, we provide a brief summary of the biological roles of key factors that are crucial in generating unique splicing patterns within neurons and also highlight the recent discovery of microexons. We highlight five key splicing factors, nSR100, NOVA, RBFOX family members, PTB, and Hu/ELAV family members, which have been well characterized. It should be noted that other factors including SAM68 family members, TDP-43, and MBNL, also contribute to neuron-specific alternative splicing as reviewed by others (Yap and Makeyev 2013, Raj and Blencowe 2015, Iijima, Hidaka et al. 2016).
3.1 Brain-Specific Spliceosome Recruiting Factor, nSR100
Neural-specific SR-related protein of 100 kDa, nSR100, was identified as a vertebrate and tissue-specific Serine/Arginine-repeat region containing splicing factor that activates inclusion of a large number of brain-specific exons (Calarco, Superina et al. 2009, Raj, Irimia et al. 2014). nSR100 recognizes pyrimidine-rich motifs flanking the 3′ splice site and binds specifically with U2-RNP components to assist in early-acting spliceosome assembly (Raj, Irimia et al. 2014).
Expression of nSR100 increases upon neuronal maturation (Irimia, Weatheritt et al. 2014). In mammalian cell culture and zebrafish models, nSR100 is required for neurogenesis and neuronal differentiation (Calarco, Superina et al. 2009, Raj, Irimia et al. 2014). An nSR100 haploinsufficient mouse model has impaired neurite outgrowth, altered neuronal excitability and synaptic transmission, and behavioral abnormalities that resemble autism spectrum disorder (Quesnel-Vallieres, Irimia et al. 2015, Quesnel-Vallières, Dargaei et al. 2016).
3.2 Position-Dependent Splicing Regulators
3.2.1 NOVA
Neurooncologic ventral antigen (NOVA) was the first described splicing factor that is responsible for neuron-specific exon content (Buckanovich, Yang et al. 1996, Yang, Yin et al. 1998, Jensen, Dredge et al. 2000). NOVA was initially identified as an antigen produced in tumor tissues that leads to an autoimmune neurological disorder, paraneoplastic opsoclonus myoclonus ataxia (POMA) (Luque, Furneaux et al. 1991, Buckanovich, Posner et al. 1993). An initial survey of NOVA-target RNAs identified 34 transcripts regulated by NOVA in mice, but recent high-throughput methods suggest the regulatory network of NOVA may include as many as 700 gene transcripts (Ule, Jensen et al. 2003, Zhang, Frias et al. 2010).
Compared to nSR100, NOVA plays more diverse roles in mRNA regulation. NOVA appears to control both alternative splicing (Ule, Ule et al. 2005) and selection of polyadenylation sites to generate brain-specific 3′-UTR of mRNAs through binding of YCAY clusters, which influences both U2 and U1 recruitment (Licatalosi, Mele et al. 2008). Interestingly, binding of NOVA near 5′ splice sites promotes exon inclusion through U2 recruitment; however, binding of NOVA near 3′ splice sites promotes exon skipping through inhibition of U1 binding (Ule, Stefani et al. 2006, Licatalosi, Mele et al. 2008). The distinct actions at 5′ and 3′ splice sites are referred to as position-dependent control of splicing.
NOVA is expressed specifically in neurons (Buckanovich, Yang et al. 1996, Yang, Yin et al. 1998, Jensen, Dredge et al. 2000), and NOVA targets transcripts encoding synaptic proteins that are important for synaptic plasticity (Ule, Jensen et al. 2003, Ule, Ule et al. 2005). In human and mouse, the NOVA1 and NOVA2 genes encode highly homologous proteins, and mouse reverse genetics has provided insights into their interplay. Nova1-null mice exhibit progressive motor dysfunction, brain stem and spinal cord neuronal apoptosis, and death 1–2 weeks after birth (Jensen, Dredge et al. 2000). Nova2-null mice display a specific deficit in long-term potentiation of slow inhibitory postsynaptic current in hippocampal CA1 neurons (Huang, Shi et al. 2005). Nova1/Nova2-double null mice are born, but are completely paralyzed and die shortly after birth (Ruggiu, Herbst et al. 2009). These mouse models and human genetics studies establish pivotal roles of NOVA in plasticity and development of both central and peripheral nervous systems.
3.2.2 RBFOX
The RNA-binding protein FOX paralogs (RBFOX1, 2, and 3) are another major set of splicing factors that increase in expression during neuronal development and promote neuronal exon inclusion. RBFOX specifically recognizes UGCAUG motifs, which are found at both 5′- and 3′-regions of introns. Similar to NOVA, RBFOX exerts position-dependent splicing control (Auweter, Fasan et al. 2006, Zhang, Zhang et al. 2008). RBFOX binding in 3′ splice site regions inhibits exon inclusion, whereas binding in 5′ splice site regions enhances exon inclusion. Such context-specific function suggests the combinatorial involvement of other splicing regulators to select for the inclusion of neuron-specific exons (Zhang, Zhang et al. 2008).
Several lines of evidence have indicated important roles of RBFOX family proteins in neuronal development and function. Expression of RBFOX1 was downregulated in post-mortem brains from autistic individuals, and RBFOX1 downregulation was associated with splicing dysregulation of genes relevant to synaptogenesis (Voineagu, Wang et al. 2011). Another study found specific regulation of a calcium channel alternative exon that alters the electrophysiological properties of this channel activation in neurons (Tang, Zheng et al. 2009). Genome-wide mapping of protein-RNA interaction sites revealed that RBFOX1, 2, and 3 directly control splicing of genes that are up-regulated during brain development and whose dysregulation has been linked to autism (Weyn-Vanhentenryck, Mele et al. 2014).
3.3 Negative Regulator of Exon Inclusion, PTB
While NOVA, nSR100, and RBFOX primarily promote inclusion of alternative exons (Ule, Stefani et al. 2006, Zhang, Zhang et al. 2008, Calarco, Superina et al. 2009), Polypyrimidine Tract-Binding protein 1 (PTB or PTBP1) is a well-known negative regulator of exon inclusion. PTB binds CU-rich regions causing a looping-out of the RNA, which prevents assembly of the spliceosome (Oberstrass, Auweter et al. 2005). Furthermore, PTB suppresses expression of its neuron-specific paralog neural-PTB (nPTB, PTBP2) by excluding an exon within nPTB, whereby the absence of this exon leads to a frameshift and degradation of nPTB mRNA by nonsense-mediated decay (Boutz, Stoilov et al. 2007). This inter-isoform suppression mechanism defines undifferentiated neuro-progenitors.
During neuronal differentiation, a canonical PTB is post-transcriptionally repressed, in part by decreased expression of the transcription factor REST (which will be discussed below) and subsequent increased expression of the neuronal microRNA, miR-124, which targets canonical PTB and in turn reduces its protein level (Yoo, Sun et al. 2011). This miR-124-mediated regulatory switch relieves suppression of nPTB. Expression of nPTB in turn initiates a neuronal program of alternative splicing events which is required for the differentiation of progenitors to mature neurons (Boutz, Stoilov et al. 2007, Makeyev, Zhang et al. 2007). Repression of PTB results in trans-differentiation of a variety of cell types to the neural lineage, illuminating this post-transcriptional regulatory circuit as a master key for neuronal cell fate (Xue, Ouyang et al. 2013, Xue, Qian et al. 2016).
3.4 Dual Roles in Alternative Splicing and Polyadenylation, Hu/ELAV
The Hu/ELAV family of splicing factors was identified in a similar way as NOVA as the autoimmune target of a paraneoplastic neurological syndrome (Szabo, Dalmau et al. 1991). The family consists of four proteins in mammals (HuA, HuB, HuC, and HuD; HuA is known as HuR in humans). HuB/C/D are exclusively expressed in neurons with the exception that HuB is also present in germ cells (Okano and Darnell 1997). ELAV (Embryonic Lethal Abnormal Visual system) is the Drosophila homologue of Hu and analogously controls alternative splicing in nervous system development in the fly. Initially, Hu proteins were thought to bind the 3′-UTR of mRNAs and affect their cytoplasmic stability and thus the extent of translation (Jain, Andrews et al. 1997). In this context, Hu proteins bind to AU-rich elements at the 3′-UTR of mRNAs and thereby stabilize them (Wang and Tanaka Hall 2001). However, later studies revealed that Hu plays a role in alternative splicing of neuronal mRNAs including the calcitonin/CGRP transcript (Zhu, Hasman et al. 2006, Zhou, Hinman et al. 2011). Hu proteins compete with positive splicing factors TIA-1/TIAR leading to the interference of U1/U6 snRNP binding and thereby promote differential exon inclusion or alternative polyadenylation (Zhu, Hinman et al. 2008, Zhou, Hinman et al. 2011). Given that the 3′-UTR plays unique roles in mRNA stability, sub-cellular localization, and translation, Hu proteins represent a unique regulatory mechanism that may potentially coordinate mRNA metabolism and proteome diversity in neurons.
The dual roles of Hu in RNA regulation has been linked to both neuronal differentiation and plasticity. HuD-null mice exhibited hind limb clasping, which is associated motor and sensory neuron defects in the cortex and basal ganglia. The HuD-null brains were characterized with a reduced number of cortical neurons despite normal numbers of neural stem cells, suggesting that HuD is crucial for neuronal differentiation (Akamatsu, Fujihara et al. 2005). Various in vivo studies confirmed the role of Hu proteins in alternative splicing and alternative polyadenylation for genes implicated in neuronal function and disease such as Bdnf and Nf1. The differential 3′-UTR generated by Hu-mediated alternative polyadenylation has been shown to stabilize mRNAs in dendrites for local protein synthesis, implicating its roles in synaptic plasticity (Zhou, Hinman et al. 2011, Allen, Bird et al. 2013, Bronicki and Jasmin 2013).
3.5 Cooperation between Neuron-Specific Splicing Factors
Some of these neuronal splicing factors act cooperatively during neuronal development. nSR100 increases expression of nPTB and works cooperatively with nPTB to overcome PTB-mediated repression during neuronal differentiation and consequently promote neural-specific exon splicing (Calarco, Superina et al. 2009, Raj, Irimia et al. 2014). Antagonistic interplay between PTB and RBFOX1 appears to be crucial for the progenitor-to-neuron transition (Zhang, Chen et al. 2016). Interestingly, several studies have also shown that recruitment of splicing machinery, such as Hu and PTB, during gene transcription can lead to altered local histone modifications that would reinforce the same pattern of exon inclusion in future rounds of transcription (Luco, Pan et al. 2010, Zhou, Hinman et al. 2011). Identifying novel regulators of the neural splicing network and their genetic associations with neurological disorders will provide further insights into the core role of splicing in neurodevelopment and brain function.
4. Microexons Are Enriched in the Brain Alternative Splicing Network
Recent work has illuminated the unique complexity of splicing in the brain. By comparing RNA-Seq data across diverse tissues in mouse and human, Irimia et al recently reported an inverse correlation between alternative exon size and their brain enrichment, i.e. as exons decrease in length, their enrichment in the brain increases (Irimia, Weatheritt et al. 2014). The authors identified more than 200 neuron-specific “microexons”, ranging between 3–27 nucleotides. Strikingly, many microexons display a “switch-like” inclusion or exclusion during neuronal maturation, when neurons are beginning to form synapses. The neuron-specific splicing factor nSR100 appears to be a key regulator of microexon inclusion (Irimia, Weatheritt et al. 2014). Another study also identified a neural-program of microexon splicing and defined microexons as exons fewer than 51 nucleotides (Li, Sanchez-Pulido et al. 2015). In contrast to the regulation by nSR100 identified by Irimia and colleagues, Li and colleagues found that intronic sequences near microexons contained RNA motifs of RBFOX and PTB proteins (Li, Sanchez-Pulido et al. 2015). Microexons often encode domains that mediate protein-protein interactions such that target proteins gain novel binding partners and/or altered binding affinity (Buljan, Chalancon et al. 2012, Irimia, Weatheritt et al. 2014). A variety of proteins harbor microexons, such as cytoskeletal proteins, ion channels, and signaling molecules, suggesting that microexons modulate a broad range of cellular processes in neuronal maturation and synaptogenesis. Intriguingly, a significant number of genes that have been implicated in autism spectrum disorder contain microexons, suggesting that neuron-specific splicing events underlie the pathogenesis of autism (Irimia, Weatheritt et al. 2014).
5. Vulnerability of the Brain to Transcriptional Dysregulation
Genetic association studies of neurodevelopmental disorders, including autism, schizophrenia, and intellectual disability syndromes, have identified numerous mutations in transcriptional regulators (Najmabadi, Hu et al. 2011, Ronan, Wu et al. 2013, De Rubeis, He et al. 2014, Iossifov, O’Roak et al. 2014, McCarthy, Gillis et al. 2014). Neurodevelopmental disorders affect 1%–8% of the population and impose the leading health-care cost in the developed world (Ropers 2010). The mutated transcriptional regulators comprise a large fraction of nuclear proteins such as DNA-binding transcription factors (TFs), histone-modification enzymes (which “write” or “erase” post-translational modifications), and their cognate “reader” proteins (Ronan, Wu et al. 2013, De Rubeis, He et al. 2014, Iossifov, O’Roak et al. 2014). In most cases, how mutations in these transcriptional regulators lead to neurodevelopmental disorders is not well understood.
Transcriptional regulation is fundamental to survival and function of all cell types, so why do mutations in transcriptional regulators influence particularly cognitive phenotypes? It can be argued that the complexity of the central nervous system requires even finer transcriptional control than other tissues; therefore, the impact of hypomorphic mutations is more strongly manifested in the brain than other tissues. An alternative but not mutually-exclusive possibility is that there might be unique characteristics of gene regulation in neurons, which confer vulnerability of the brain to mutations in transcriptional regulators.
6. Neuronal Isoforms of Transcription Regulators
Potential vulnerability of the brain to transcriptional and splicing dysregulations prompted us to examine chromatin regulating genes whose transcripts undergo neuron-specific splicing events. Zhang, et al, published the first brain cell-type specific transcriptomic database, which included alternative splicing events (Zhang, Chen et al. 2014). We compared their list of neuron-specific alternative splicing events to a published list of “EpiFactors” that compiles known and putative chromatin regulators (Medvedeva, Lennartsson et al. 2015). This intersection revealed 115 chromatin regulators that exhibit neuronal alternative splicing events of a variety of types (Table 1). Since some of the specific genes that are discussed below are not included in this list, there may be others that were not captured by this analysis.
Table 1.
Chromatin Regulators that Undergo Neuron-Specific Splicing
| Splicing Event | Genes |
|---|---|
| Exon Skipping |
Actl6b, Adnp, Arid4b, Arntl, Arrb1, Ash2l, Atf2, Banp, Baz2b, Bptf, Brd2, Brd8, Brpf3, Carm1, Chd2, Chd5, Cit, Dpf2, Ep400, Epc2, Exosc1, Eya3, Ezh2, Gatad2a, Gse1, Gtf2i, Gtf3c4, Hdac5, Hp1bp3, Huwe1, Ino80e, Map3k7, Maz, Mecp2, Mllt1, Morf4l2, Mta1, Mtf2, Ncoa1, Ncoa2, Ncor1, Nsd1, Pbrm1, Pcgf6, Phc1, Phf14, Phf20l1, Phf21a, Pkn1, Prkaa1, Prr14, Rps6ka3, Rsf1, Scmh1, Senp1, Setd5, Sirt2, Smarca2, Smarca4, Smarcc2, Smarce1, Spen, Tlk1, Trim33, Ubn1, Uhrf2, Usp16, Vrk1, Whsc1, Yeats2, Ywhaz, Zzz3 |
| Tandem Exon Skipping |
Bptf, Dot1l, Hdac5, Nap1l1, Pbrm1, Phf20, Rps6ka3, Scmh1, Setd3, Smarca2, Smarce1, Vrk1, Zzz3 |
| Mutually Exclusive Exon Usage |
Phf21a, Setd5 |
| Intron Retention |
Aebp2, Asxl1, Brd9, Brms1, Brpf1, Chd4, Chd8, Exosc9, Hcfc1, Hdac7, Hdac10, Hirip3, Lrwd1, Mta2, Nfrkb, Phf1, Prkcd, Prr14, Sf3b1, Sirt7, Tle2, Trim28, Wdr77 |
| Alternative 3′ Splice Site |
Abpp1, Brd9, Chd3, Chd4, Cit, Ehmt1, Eya1, Ezh1, Gtf2i, Mbd6, Mllt10, Ncor1, Ncor2, Nfrkb, Ogt, Prdm4, Ssrp1, Taf1, Tle4, Trim33 |
| Alternative 5′ Splice Site |
Bptf, Chd3, Huwe1, Kat2a, Ncor1, Ncor2, Prmt1, Smarcb1, Trrap |
Table 1: Mouse genes that have neuron specific alternative splicing events as published by Zhang, et al (Zhang, Chen et al. 2014) was compared to the Epifactors list by Medvedeva, et al (Medvedeva, Lennartsson et al. 2015) to produce the above list of epigenetic factors that have neuron-specific alternative splicing events. Some genes are listed twice, as they had multiple types of alternative splicing events. Bolded genes are represented in both Table 1 and 2.
We also intersected the lists of microexons (Irimia, Weatheritt et al. 2014) and EpiFactors (Medvedeva, Lennartsson et al. 2015) and found that 76 transcriptional regulators contain neuron-specific microexons (Table 2). When considering the two tables together, 161 genes are represented (22% of all EpiFactors) as either containing microexons or a neuron-specific splicing event. 30 genes are represented in both tables (bolded in both tables), which likely represent EpiFactors that have neuron-specific and microexon splicing events.
Table 2.
Chromatin Regulators that Contain Microexons
| Class | Subclass | Genes |
|---|---|---|
| Chromatin remodeling/cofactor | ACTL6A, BPTF, CHD1L, CHD9, EP400, HP1BP3, INO80C, MAPKAPK3, MLLT1, MTA1, PHF19, PSIP1, RAD54B, SETD6, SMARCA1, SMARCC2, SMARCD2, SP100, SRCAP, UBR5 | |
| Histone modification | Writer/Cofactor | APBB1, ARRB1, ATXN7, AURKC, BRPF3, CARM1, CIT, DDB2, EP400, EZH2, LAS1L, MBD1, MEAF6, NAT10, OGT, PAXIP1, PHF19, PRDM2, PRKCB, PRMT1, PRMT5, RPS6KA3, SETD6, SUV420H2, TAF1, TEX10, TRRAP, UBR5, WHSC1 |
| Reader | BAZ2B, BRD8, CECR2, L3MBTL3, PBRM1, PHF20L1, PHF21A, ZCWPW1 | |
| Eraser | HDAC1, HDAC10, HDAC3, HDAC6, HDAC7, KDM1A, KDM2B, KDM5B, MORF4L2, NCOR2, SIRT7, SMARCA1, SRCAP, USP16, USP21, USP49, ZMYND8 | |
| Histone chaperone | CHRAC1 | |
| RNA Modification | DND1, EXOSC4, MOV10 | |
| Transcription Factor | E2F6, GTF2I, MBD1 | |
| Polycomb Group | SFMBT1, EZH2 | |
| Protein |
Human microexon list as published by Irimia, et al (Irimia, Weatheritt et al. 2014) was compared to the Epifactors list by Medvedeva, et al (Medvedeva, Lennartsson et al. 2015) to produce the above list of epigenetic factors that contain microexons. Some genes are listed twice, as they fell into multiple categories. Bolded genes are represented in both Table 1 and 2.
Altogether, the large number of genes described in these tables suggests an important role for splicing in the neuron-specific chromatin landscape and transcriptome. Though the impact of microexon inclusion/exclusion on functions of these proteins remains largely undetermined, some studies have begun to reveal marked impact of such splicing events in transcriptional regulation in the brain. Below, we summarize the roles of alternative splicing in DNA-binding TFs and chromatin modification writers and erasers and their implications for neurodevelopmental disorders.
7. DNA-Binding TFs and Neuronal Isoforms
As discussed earlier, the cell-type restricted presence of TFs can be a determinant for cell-type specific gene expression during stem cell differentiation. TFs are able to bind cognate DNA sequences and subsequently dictate target specificity of chromatin regulators which do not generally carry sequence-specific DNA binding domains. This paradigm allows for cell-type specific transcriptional programs. Although the “EpiFactor” database does not cover TFs, this group of transcriptional regulators also appears to be spliced in a brain-specific manner. The examples below represent unique cases, where TFs are expressed ubiquitously yet adopt neuron-specific forms. These examples highlight alternative splicing as a core mechanism of neuron-specific gene regulation and its crucial role in neuronal differentiation, maturation, and function (Figure 1).
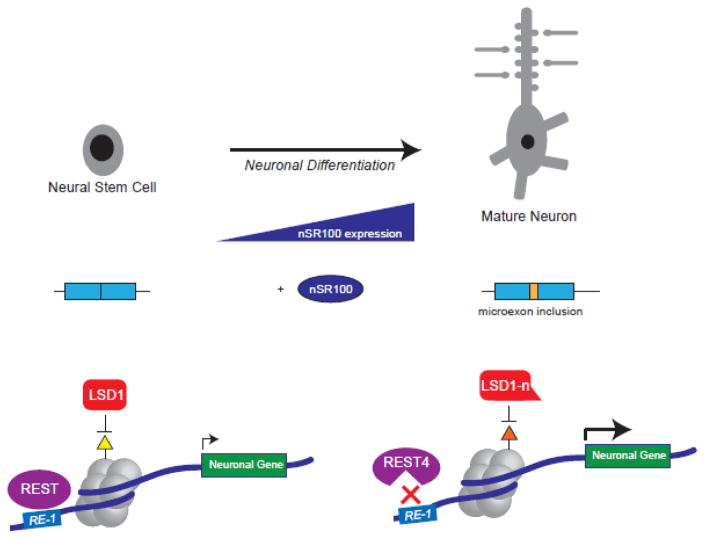
Figure 1.
Microexon inclusion of LSD1/REST, an example of coordinated neuron-specific splicing and the impact on the transcriptional landscape during neuron differentiation and maturation. As neurons differentiate, there is an increase in the expression of neuron-specific splicing factors, such as nSR100. The presence of this splicing factor leads to microexon inclusion in a set of neuronal genes, which includes transcriptional regulators such as REST and LSD1. Whereas in non-neuronal cells, REST and LSD1 act to repress neuronal genes, the neuron-spliced products, REST4 and LSD1-n act in a distinct molecular function in a way that promotes transcription of a set of genes important for neuron development and function.
7.1 REST/NRSF
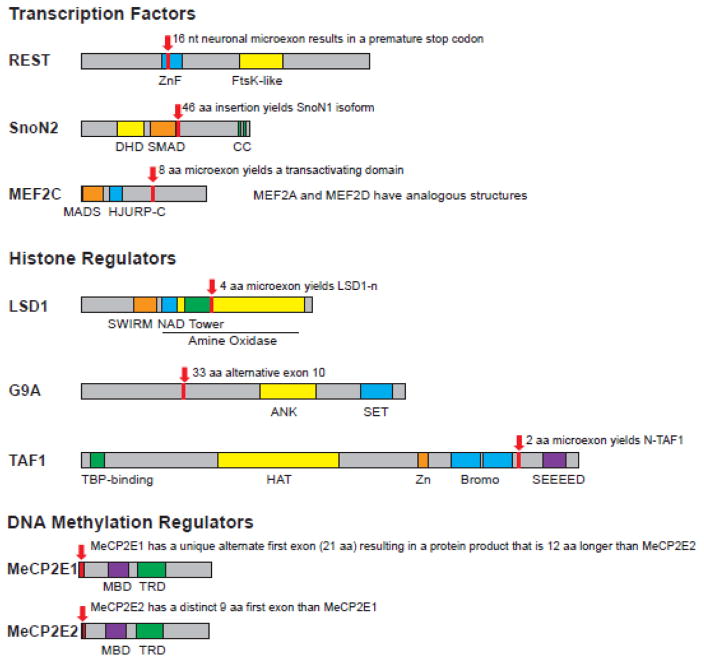
RE-1 Silencing Transcription factor (REST, aka NRSF) was originally identified as a TF which is largely restricted to non-neuronal tissues and represses neuronal genes in non-neuronal cell types (Chong, Tapia-Ramirez et al. 1995). REST binds to RE-1 sequences that lie in the promoter regions of neuronal genes and recruits a variety of co-repressor complexes (Chong, Tapia-Ramirez et al. 1995, Schoenherr and Anderson 1995, Chen, Paquette et al. 1998, Bruce, Donaldson et al. 2004). Within non-neuronal cells, REST directly represses nSR100 expression, and nSR100 overexpression results in REST-target gene de-repression (Raj, O’Hanlon et al. 2011). REST is, in fact, expressed to some extent in neurons but its function is repressed via a neuron-specific alternative splicing event that is controlled by nSR100. nSR100 acts to promote inclusion of a microexon in REST, generating the neuron-specific “REST4” isoform (Palm, Metsis et al. 1999, Raj, O’Hanlon et al. 2011) (Figure 2 shows positions of neuronal alternative splicing events). Interestingly, the 16-nucleotide microexon carries a premature stop codon, and the resultant REST4, which lacks DNA-binding capability, acts as a dominant negative protein which sequesters full-length REST in nonfunctional hetero-oligomers (Shimojo, Paquette et al. 1999). While overall REST expression decreases in neurons, alternative splicing by nSR100 acts as a fail-safe mechanism to ensure complete loss of REST function in neurons. These findings highlight the antagonistic molecular circuitry between the transcriptional repressor, REST, and splicing activator, nSR100, to generate and maintain the identity of neurons and non-neuronal cells (Figure 1).
Figure 2.
Domain organization with neuron-specific alternative exons of TFs and chromatin regulators. Length of protein and functional domains are drawn to scale, relative to each other. ZnF: Zinc finger; DHD: Dach Homology Domain; SMAD: SMAD (SMAD refers to homologs of both the C. elegans SMA protein, for small body size, and the Drosophila MAD protein, for mothers against decaptentaplegic) -binding domain; CC: Coiled Coil domain; HJURP-C: Holliday Junction Regulator Protein family C-terminal repeat; NAD: NAD-binding domain; ANK: Ankyrin repeats; Bromo: Bromodomain; HAT: Histone Acetyl-Transferase domain; SEEEED: A conserved sequence with reference to a serine-rich region of AP3B1, a clathrin-adaptor complex; MBD: Methyl-CpG binding domain; TRD: Transcriptional Repression Domain.
7.2 SnoN
SnoN (aka SKI-like proto-oncogene, SKIL) was identified as a TF responsible for axon growth in cerebellar granule neurons, in part by promoting transcription of cytoskeletal regulators (Stegmuller, Konishi et al. 2006, Ikeuchi, Stegmuller et al. 2009). The SnoN gene is alternatively spliced into SnoN1 and SnoN2, where SnoN2 has an alternative splice site in exon 3 leading to a 46 amino acid deletion relative to the SnoN1 isoform (Pelzer, Lyons et al. 1996). This alternative exon is near the SMAD-binding domain, with which SMAD negatively regulates SnoN function (Stroschein, Wang et al. 1999, Wan, Liu et al. 2001) (Fig 2). Whereas SnoN1 promotes axon branching and inhibits migration in the cerebellar cortex, SnoN2 represses branching and promotes migration (Huynh, Ikeuchi et al. 2011). Only the SnoN1 isoform (and not SnoN2) is capable of forming a complex with FOXO1, another TF, which rises in expression during neuronal maturation. Unlike the cell-type specific roles of REST and REST4, both SnoN isoforms operate in neurons, but their functions are restricted to specific layers of the cerebellum — SnoN1 is highly expressed in the inner granular layer, whereas SnoN2 is expressed in the molecular layer of the cerebellum. Furthermore, SnoN1, but not SnoN2, is crucial for repression of the X-linked lissencephaly gene, DCX, in cerebellar granule neurons, providing a potential link between SnoN-mediated neural network assembly and neurodevelopment (Huynh, Ikeuchi et al. 2011).
7.3 MEF2
Mads box transcription enhancer factor 2 (MEF2) is a TF family encoded by four MEF2 genes (MEF2A, MEF2B, MEF2C, and MEF2D) and is involved in nervous system development by promoting neuronal-activity dependent transcription and negatively regulating the number of excitatory synapses (Flavell, Cowan et al. 2006, Flavell, Kim et al. 2008, Lyons, Schwarz et al. 2012, Ataman, Boulting et al. 2016). MEF2 family TFs undergo complex patterns of alternative splicing that involve multiple exon inclusions/exclusions within the single MEF2 gene. Some alternative splicing events modulate binding affinity of MEF2 to other TFs, such as NeuroD, MRF4, and MASH1 (Janson, Chen et al. 2001). Altered expression of specific MEF2 isoforms has been associated with myotonic dystrophy and other neuromuscular disorders (Bachinski, Sirito et al. 2010). Furthermore, three MEF2 family members (MEF2A, MEF2C, and MEF2D) have a conserved 24-nucleotide microexon that is expressed solely in striated-muscle and neuronal cells, particularly in the cerebral cortex (Leifer, Krainc et al. 1993). This microexon is incorporated specifically during myocyte differentiation or neuronal differentiation (Zhu, Ramachandran et al. 2005), and in the case of neuronal differentiation, the splicing factor nSR100 is responsible for its inclusion (Irimia, Weatheritt et al. 2014). This microexon is adjacent to the MEF2 transcriptional activating domains, and the microexon-containing protein product is a much more potent activator of MEF2 target genes as shown through reporter assays (Zhu, Ramachandran et al. 2005) (Fig 2). Mutations within or microdeletions encompassing MEF2C have been found in an intellectual disability syndrome associated with epilepsy, muscular hypotonia, and cerebral malformations (Le Meur, Holder-Espinasse et al. 2010, Zweier, Gregor et al. 2010). A mouse model of Mef2c loss exhibited reduced neuronal differentiation and severe autism-like behavioral deficits (Li, Radford et al. 2008), reinforcing the pivotal roles of MEF2-mediated transcriptional control in brain development.
8. Histone Modifying Enzymes with Neuronal Isoforms
Once TFs occupy target genomic loci, they recruit histone modification enzymes to either relax or compact higher-order chromatin structure, thereby modulating accessibility of RNA polymerase machinery. Genome-wide transcriptome approaches revealed that many histone modifiers are targeted by neuron-specific splicing machineries (Tables 1 and 2). The examples below illustrate how neuron-specific splicing can have profound influences on biochemical functions of histone modifiers and potentially chromatin landscapes within neurons.
8.1 LSD1
Lysine Specific Demethylase 1 (LSD1, aka KDM1A) removes mono- (me1) and di-methylation (me2) specifically from Histone 3 Lysine 4 (H3K4), which are hallmarks of regulatory regions of transcriptionally-engaged genes (Jenuwein and Allis 2001, Heintzman, Stuart et al. 2007). LSD1 was originally identified as a component of the CoREST corepressor complex, which is operated by REST to repress neuronal genes in non-neuronal cells (Shi, Lan et al. 2004, Shi, Matson et al. 2005).
Several groups have reported that LSD1 has a neuronal isoform (LSD1-n), which includes a 4-amino acid alternative microexon in its catalytic amine oxidase domain (Fig. 2). LSD1-n appears to be important for neurite morphogenesis, synaptogenesis, and proper transcriptional response to neuronal depolarization and is transcriptionally upregulated relative to canonical LSD1 as neurons begin to mature (Zibetti, Adamo et al. 2010, Laurent, Ruitu et al. 2015, Rusconi, Paganini et al. 2015, Wang, Telese et al. 2015). However, the biochemical function of the LSD1-n isoform has been debated. The first study reporting this isoform found that LSD1-n acts like the canonical protein to remove H3K4 mono- or di- methylation marks (H3K4me1/2) from a histone peptide with similar efficiency (Zibetti, Adamo et al. 2010). This first study also carried out X-ray crystallography on the histone-interacting segment of LSD1-n and did not find an altered structure compared to the canonical isoform. Another study found that LSD1-n associates with a nuclear factor, Svil, which changes its substrate specificity from H3K4me1/2 to H3K9me1/2, a repressive histone modification (Laurent, Ruitu et al. 2015). A third group reported that LSD1-n demethylates H4K20me1/2, another mark associated with transcriptionally-repressed regions (Wang, Telese et al. 2015). These conflicting data suggest that other unknown regulatory proteins, genomic contexts, or timing in neuron maturation may, in concert, determine the substrate specificity of LSD1-n. The threonine at position 369 of LSD1-n, which is located within the microexon, can be phosphorylated, leading to conformational changes and disassembly of the LSD1/CoREST complex (Toffolo, Rusconi et al. 2014). Thus, the LSD1 microexon might control dynamics of complex assembly instead of, or in addition to, modulating substrate specificity.
Heterozygous missense mutations in LSD1 have been recently implicated in an unnamed neurodevelopmental disorder with clinical features such as developmental delays, craniofacial and palate abnormalities, thinning of the corpus callosum, and hypotonia (Tunovic, Barkovich et al. 2014, Chong, Yu et al. 2016). Interestingly, these features resemble those of Kabuki syndrome, which is primarily associated with haploinsufficiency of MLL2/4 (KMT2D), an H3K4 methyltransferase (Ng, Bigham et al. 2010), which catalyzes the writer reaction reciprocal to the LSD1 eraser reaction. LSD1 mutations fall into the catalytic domain, and they interfere with H3K4 demethylation activity (Pilotto, Speranzini et al. 2016). Although the known LSD1 missense mutations are not located in or around the alternative microexon, it is highly conceivable that these mutations also affect the enzymatic activity of LSD1-n. Given the roles of LSD1-n in neurite morphogenesis, synaptogenesis, and normal excitability, the reported mutations in LSD1-n could alter these parameters in patients, thereby leading to their cognitive dysfunction.
8.2 G9A
While LSD1 was discovered to remove active chromatin marks, G9A (aka EHMT2 and KMT1C) is a histone lysine methyltransferase that deposits H3K9me2, which is enriched in repressive chromatin environments (Tachibana, Matsumura et al. 2008). Despite the enrichment in repressive chromatin, a recent study reported that H3K9me2 can promote transcription within constitutive heterochromatin (Jih, Iglesias et al. 2017). A mouse model of G9a inactivation in forebrain neurons or the hippocampus led to behavioral abnormalities including learning impairment and decreased exploration (Sampath, Marazzi et al. 2007, Schaefer, Sampath et al. 2009, Gupta-Agarwal, Franklin et al. 2012). In the G9a-forebrain null mouse model, non-neuronal genes were de-repressed in neurons leading the authors to hypothesize that this irregular expression underlies the learning and memory impairment and other behavioral abnormalities (Schaefer, Sampath et al. 2009). Although there have been no reported human cases of intellectual disability associated with G9A disruption, haploinsufficiency of the closely related paralog EHMT1 (aka GLP or KMT1D) appears to be responsible for the neurodevelopmental condition, Kleefstra syndrome (Kleefstra, Brunner et al. 2006). G9A and EHMT1 form a stable heteromeric complex in many cell types (Tachibana, Matsumura et al. 2008), including post-mitotic neurons (Benevento, Iacono et al. 2016), raising the possibility that genetic alterations of G9A may be linked to undiagnosed neurodevelopmental conditions.
G9A has a 33 amino acid alternative exon 10 (E10) (Brown, Campbell et al. 2014). Recently, the G9A E10 isoform was identified to be upregulated upon neuronal differentiation of the mouse neural crest derived cell line, N2A (Fiszbein, Giono et al. 2016). E10 inclusion did not affect methyltransferase enzyme activity in vitro, but rather increased nuclear localization of the E10-containing G9A (Brown, Campbell et al. 2014, Fiszbein, Giono et al. 2016). Thus, Fiszbein, et al hypothesized that the inclusion of E10 promoted nuclear localization by better exposing a nearby nuclear localization signal. Specific siRNA-targeted knockdown of the G9A E10 isoform abolished neurite outgrowth, phenocopying the pan-isoform knockdown of G9A. Importantly, G9A E10 is not uniquely present in differentiated neurons; other non-neuronal cell types, including stimulated lymphocytes and differentiated mammary gland cells, also display increased transcription of the E10 isoform (Martinez, Pan et al. 2012, Fiszbein, Giono et al. 2016). Nonetheless, the switch-like inclusion of E10 in G9A in neurons is reminiscent of the LSD1/LSD1-n dynamics during neuronal differentiation and maturation (Zibetti, Adamo et al. 2010). It is tempting to speculate that functional modulation of a histone methyl writer for an inactive mark (H3K9me) and eraser for an active mark (H3K4me) coordinates to relax or compact chromatin structure for normal development of neurocircuitries.
8.3 TAF1
TAF1 is a histone acetyltransferase component of the TFIID transcriptional initiation complex (Ruppert, Wang et al. 1993, Mizzen, Yang et al. 1996, Jacobson, Ladurner et al. 2000). TFIID directs RNA Polymerase II to transcription start sites. Neuronal TAF1 (N-TAF1) includes a microexon encoding 2 amino acids (A-K) close to one of the two bromodomains (Fig. 2) (Ito, Hendriks et al. 2016). Bromodomains are acetyl-histone reader modules; however, the functional impact of this insertion remains unknown. N-TAF1 is expressed as neurons begin to mature (Jambaldorj, Makino et al. 2012). Knockdown of N-TAF1 in neuroblastoma cells leads to decreased expression of genes related to vesicular transport, synapse function, and dopamine metabolism, implicating is role in synapse development and dopaminergic neurotransmission (Herzfeld, Nolte et al. 2013).
A retrotransposon insertion at the TAF1 locus has been linked to X-linked Dystonia Parkinsonism (XLDP), which is characterized by severe torsion dystonia followed by parkinsonism (Haberhausen, Schmitt et al. 1995). Interestingly, expression of the N-TAF1 isoform is specifically reduced in patient cells suggesting that the retrotransposon could have disrupted a neuron specific cis-element (Makino, Kaji et al. 2007). Another study reported nine families with point mutations in TAF1 that led to X-linked intellectual disability along with facial dysmorphologies, generalized hypotonia, and other variable neurological features (O’Rawe, Wu et al. 2015). O’Rawe et al also identified two families with duplications of TAF1 with phenotypic features overlapping those in the individuals with TAF1 point mutations, but it is unclear how duplication of TAF1 could mechanistically lead to similar consequences. With the exception of XLDP, these genetic lesions alter both canonical and neuron-specific TAF1 isoforms. It would be important to test whether N-TAF1 is specifically responsible for cognitive deficits by knocking out N-TAF1 in mice.
9. Methyl DNA Reader with Neuronal Isoforms
In addition to histone modifications, methyl moieties placed on DNA, including CpG methylation and a variety of non-CpG methylations, also play important roles in transcriptional regulation in higher eukaryotes (Ambrosi, Manzo et al. 2017, Edwards, Yarychkivska et al. 2017). Uniqueness of the brain in methyl DNA regulation was illustrated by the discovery of hydroxymethylation originally in the cerebellum (Kriaucionis and Heintz 2009). Later investigations confirmed a much higher level of this oxidized form of CpG methylation in the brain compared to other tissues (Richa and Sinha 2014). While roles of methyl-DNA regulation in neurodevelopment and plasticity have been intensively studied recently (Bayraktar and Kreutz 2017, Jang, Shin et al. 2017), the impact of neuron-specific splicing on methyl-DNA regulation remains largely unexplored. An exception is MeCP2, a multifunctional protein canonically known for its epigenetic silencing function through binding to methylated CpG sites (Lewis, Meehan et al. 1992).
9.1 MeCP2
X-linked Methyl CpG Binding Protein 2 (MeCP2) is one of the most well-characterized chromatin regulators in the brain (Pohodich and Zoghbi 2015). Heterozygous disruption of MeCP2 is responsible for Rett Syndrome, the progressive neurodevelopmental disorder which primarily affects young females (Amir, Van den Veyver et al. 1999). In contrast, duplication of MeCP2 in males leads to MeCP2 duplication syndrome, characterized by feeding difficulties, poor or absent speech, and muscle stiffness (Ramocki, Tavyev et al. 2010). Thus, precise regulation of MeCP2 dose appears crucial for normal brain function. MeCP2 has two isoforms, E1 and E2, that differ at the N-terminus of the protein (Kriaucionis and Bird 2004, Mnatzakanian, Lohi et al. 2004) (Fig. 2). The E1 isoform is expressed at a much higher level than E2 in postnatal neurons (Dragich, Kim et al. 2007, Zachariah, Olson et al. 2012). DNA CpG methylation levels in the promoter and first intron of MeCP2 correlate with the expression of the two isoforms, suggesting the isoforms regulate themselves through DNA methylation (Olson, Zachariah et al. 2014). The alternate N-terminal exon lies near the Methyl-cytosine-Binding Domain (MBD), and therefore may influence methyl-CpG binding capabilities, but this hypothesis has yet to be tested.
The pan-Mecp2-null mouse is an established model of Rett Syndrome (Chen, Akbarian et al. 2001, Guy, Hendrich et al. 2001). The Mecp2e2-specific knockout mice did not display Rett syndrome-related symptoms (Itoh, Tahimic et al. 2012). On the other hand, Mecp2e1 deletion in mice recapitulated the neurological deficits observed in Rett Syndrome, suggesting that haploinsufficiency of MeCP2E1, but not E2, accounts for Rett Syndrome (Yasui, Gonzales et al. 2014). High levels of MeCP2E2, but not E1, in the mouse brain have been shown to be neurotoxic through functional inhibition and physical interaction with the transcription factor FOXG1, another Rett-associated gene product (Dastidar, Bardai et al. 2012). Thus, in contrast to the strong implication of MeCP2E1 loss in Rett Syndrome, MeCP2E2 might be the causative agent for the MeCP2 duplication syndrome.
10. Conclusions and Perspectives
Neurodevelopmental disorders are not unique in their association with mutations in transcriptional regulators. Many chromatin regulators, including histones (Schwartzentruber, Korshunov et al. 2012), are somatically mutated in a variety of cancers (Allis and Jenuwein 2016). In the context of cancers, recurrent gain-of-function mutations or translocations in transcriptional machinery, in particular, can provide proliferative advantages (Deng, Melnik et al. 2013, Morgan and Shilatifard 2015). However, many germline loss-of-function mutations of transcriptional regulators, which often have functional orthologues with redundant molecular function in the genome, appear to dominate the genetic landscape for neurodevelopmental conditions. It remains unclear why cognitive functions are particularly susceptible to mutations in genes encoding transcriptional regulators.
A number of outstanding questions remain in this field. Although many neuronal isoforms have been identified in transcriptional regulators, the impact of alternative splicing in the context of protein complexes is not understood. As discussed earlier, the transcription factor REST and the histone demethylase LSD1 have both been shown to be present as specific isoforms within neurons (Fig. 1). These two proteins are known to act together in a complex, canonically repressing neuron-specific genes in non-neuronal cells. However, it is unknown how the two neuronal isoforms, REST4 and LSD1-n, act together in complex, if at all. It is possible that different combinations of canonical and neuronal isoforms could influence transcription differently. Some initial studies have used genome-wide interrogation methods, such as ChIP-Seq, to ask whether neuronal isoforms have unique targets within the genome (Laurent, Ruitu et al. 2015, Wang, Telese et al. 2015). However, the data published so far have been contradictory across studies and have not revealed a clear pattern. An important next step is to harness experimental approaches, by which isoforms with subtle sequence differences can be separately analyzed in specific cell-types. These would include genomic-wide approaches on flow-sorted cell types and CRISPR-mediated destruction of neuron-specific exons.
A recurrent observation is the alternative splicing “switch” in maturation processes of post-mitotic neurons rather than earlier developmental processes such as proliferation of neural progenitor cells or cell-type specification. When progenitors are dividing, transcription might primarily satisfy a high demand for generating cellular materials. As cells cease to divide, transcriptional regulations are necessary to allow neurons to respond to extra-cellular cues including synaptic inputs. Given that a transcriptional response to synaptic inputs is required for synaptic plasticity, which is a basis of cognitive function, an important future direction is to determine how neuron-specific splicing events contribute to synaptic plasticity via transcriptional responses. The neuron-specific gene regulatory machineries encompassing splicing and transcription factors might become prime drug targets for cognitive disorders for which we currently have no therapies.
Highlights.
The brain has complex, conserved splicing patterns, including microexon splicing
Disruption of brain alternative splicing networks underlies neurological disorders
Some transcription factors important for neural fate have neuron-specific isoforms
Neuron-specific isoforms of chromatin regulators play roles in neuron function
Acknowledgments
This work was supported by grants from the University of Michigan Career Training in Reproductive Biology (T32 HD079342 to RP), NIH (F31 NS103377 to RP), University of Michigan Medical School (to SI), NIH (R01 NS089896 to SI), and the Farrehi Research Fund (to SI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akamatsu W, Fujihara H, Mitsuhashi T, Yano M, Shibata S, Hayakawa Y, Okano HJ, Sakakibara S, Takano H, Takano T, Takahashi T, Noda T, Okano H. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc Natl Acad Sci U S A. 2005;102(12):4625–4630. doi: 10.1073/pnas.0407523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M, Bird C, Feng W, Liu G, Li W, Perrone-Bizzozero NI, Feng Y. HuD promotes BDNF expression in brain neurons via selective stabilization of the BDNF long 3′UTR mRNA. PLoS One. 2013;8(1):e55718. doi: 10.1371/journal.pone.0055718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016 doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- Ambrosi C, Manzo M, Baubec T. Dynamics and Context-Dependent Roles of DNA Methylation. J Mol Biol. 2017;429(10):1459–1475. doi: 10.1016/j.jmb.2017.02.008. [DOI] [PubMed] [Google Scholar]
- Amir RE, I, Van den Veyver B, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Ataman B, Boulting GL, Harmin DA, Yang MG, Baker-Salisbury M, Yap EL, Malik AN, Mei K, Rubin AA, Spiegel I, Durresi E, Sharma N, Hu LS, Pletikos M, Griffith EC, Partlow JN, Stevens CR, Adli M, Chahrour M, Sestan N, Walsh CA, Berezovskii VK, Livingstone MS, Greenberg ME. Evolution of Osteocrin as an activity-regulated factor in the primate brain. Nature. 2016;539(7628):242–247. doi: 10.1038/nature20111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. Embo j. 2006;25(1):163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachinski LL, Sirito M, Bohme M, Baggerly KA, Udd B, Krahe R. Altered MEF2 isoforms in myotonic dystrophy and other neuromuscular disorders. Muscle Nerve. 2010;42(6):856–863. doi: 10.1002/mus.21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, Kim T, Misquitta-Ali CM, Wilson MD, Kim PM, Odom DT, Frey BJ, Blencowe BJ. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338(6114):1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Bayraktar G, Kreutz MR. Neuronal DNA Methyltransferases: Epigenetic Mediators between Synaptic Activity and Gene Expression? Neuroscientist. 2017 doi: 10.1177/1073858417707457. 1073858417707457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, Garcia-Moreno F, Molnar Z, Margulies EH, Ponting CP. A transcriptomic atlas of mouse neocortical layers. Neuron. 2011;71(4):605–616. doi: 10.1016/j.neuron.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevento M, Iacono G, Selten M, Ba W, Oudakker A, Frega M, Keller J, Mancini R, Lewerissa E, Kleefstra T, Stunnenberg HG, Zhou H, van Bokhoven H, Nadif Kasri N. Histone Methylation by the Kleefstra Syndrome Protein EHMT1 Mediates Homeostatic Synaptic Scaling. Neuron. 2016;91(2):341–355. doi: 10.1016/j.neuron.2016.06.003. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21(13):1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronicki LM, Jasmin BJ. Emerging complexity of the HuD/ELAVl4 gene; implications for neuronal development, function, and dysfunction. Rna. 2013;19(8):1019–1037. doi: 10.1261/rna.039164.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SE, Campbell RD, Sanderson CM. Novel NG36/G9a gene products encoded within the human and mouse MHC class III regions. Mammalian Genome. 2014;12(12):916–924. doi: 10.1007/s00335-001-3029-3. [DOI] [PubMed] [Google Scholar]
- Bruce AW, I, Donaldson J, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Gottgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A. 2004;101(28):10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Posner JB, Darnell RB. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron. 1993;11(4):657–672. doi: 10.1016/0896-6273(93)90077-5. [DOI] [PubMed] [Google Scholar]
- Buckanovich RJ, Yang YY, Darnell RB. The onconeural antigen Nova-1 is a neuron-specific RNA-binding protein, the activity of which is inhibited by paraneoplastic antibodies. J Neurosci. 1996;16(3):1114–1122. doi: 10.1523/JNEUROSCI.16-03-01114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, Bateman A, Babu MM. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol Cell. 2012;46(6):871–883. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, Superina S, O’Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, Zhen M, Ciruna B, Blencowe BJ. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138(5):898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10(11):741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27(3):327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20(2):136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80(6):949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Chong JX, Yu JH, Lorentzen P, Park KM, Jamal SM, Tabor HK, Rauch A, Saenz MS, Boltshauser E, Patterson KE, Nickerson DA, Bamshad MJ. Gene discovery for Mendelian conditions via social networking: de novo variants in KDM1A cause developmental delay and distinctive facial features. Genet Med. 2016;18(8):788–795. doi: 10.1038/gim.2015.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastidar SG, Bardai FH, Ma C, Price V, Rawat V, Verma P, Narayanan V, D’Mello SR. Isoform-specific toxicity of Mecp2 in postmitotic neurons: suppression of neurotoxicity by FoxG1. J Neurosci. 2012;32(8):2846–2855. doi: 10.1523/JNEUROSCI.5841-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Shih-Chen F, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Curran SR, Dawson G, Duketis E, Fernandez BA, Gallagher L, Geller E, Guter SJ, Hill RS, Ionita-Laza J, Jimenz Gonzalez P, Kilpinen H, Klauck SM, Kolevzon A, Lee I, Lei I, Lei J, Lehtimaki T, Lin CF, Ma’ayan A, Marshall CR, McInnes AL, Neale B, Owen MJ, Ozaki N, Parellada M, Parr JR, Purcell S, Puura K, Rajagopalan D, Rehnstrom K, Reichenberg A, Sabo A, Sachse M, Sanders SJ, Schafer C, Schulte-Ruther M, Skuse D, Stevens C, Szatmari P, Tammimies K, Valladares O, Voran A, Li-San W, Weiss LA, Willsey AJ, Yu TW, Yuen RK, Cook EH, Freitag CM, Gill M, Hultman CM, Lehner T, Palotie A, Schellenberg GD, Sklar P, State MW, Sutcliffe JS, Walsh CA, Scherer SW, Zwick ME, Barett JC, Cutler DJ, Roeder K, Devlin B, Daly MJ, Buxbaum JD D. D. D. Study, A. Homozygosity Mapping Collaborative for, U. K. Consortium. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng B, Melnik S, Cook PR. Transcription factories, chromatin loops, and the dysregulation of gene expression in malignancy. Semin Cancer Biol. 2013;23(2):65–71. doi: 10.1016/j.semcancer.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Deplancke B, Alpern D, Gardeux V. The Genetics of Transcription Factor DNA Binding Variation. Cell. 2016;166(3):538–554. doi: 10.1016/j.cell.2016.07.012. [DOI] [PubMed] [Google Scholar]
- Dragich JM, Kim YH, Arnold AP, Schanen NC. Differential distribution of the MeCP2 splice variants in the postnatal mouse brain. J Comp Neurol. 2007;501(4):526–542. doi: 10.1002/cne.21264. [DOI] [PubMed] [Google Scholar]
- Edwards JR, Yarychkivska O, Boulard M, Bestor TH. DNA methylation and DNA methyltransferases. Epigenetics Chromatin. 2017;10:23. doi: 10.1186/s13072-017-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiszbein A, Giono LE, Quaglino A, Berardino BG, Sigaut L, von Bilderling C, Schor IE, Steinberg JH, Rossi M, Pietrasanta LI, Caramelo JJ, Srebrow A, Kornblihtt AR. Alternative Splicing of G9a Regulates Neuronal Differentiation. Cell Rep. 2016;14(12):2797–2808. doi: 10.1016/j.celrep.2016.02.063. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311(5763):1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60(6):1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat Rev Genet. 2010;11(8):549–558. doi: 10.1038/nrg2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta-Agarwal S, Franklin AV, Deramus T, Wheelock M, Davis RL, McMahon LL, Lubin FD. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J Neurosci. 2012;32(16):5440–5453. doi: 10.1523/JNEUROSCI.0147-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27(3):322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Haberhausen G, Schmitt I, Kohler A, Peters U, Rider S, Chelly J, Terwilliger JD, Monaco AP, Muller U. Assignment of the dystonia-parkinsonism syndrome locus, DYT3, to a small region within a 1.8-Mb YAC contig of Xq13.1. Am J Hum Genet. 1995;57(3):644–650. [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Herzfeld T, Nolte D, Grznarova M, Hofmann A, Schultze JL, Muller U. X-linked dystonia parkinsonism syndrome (XDP, lubag): disease-specific sequence change DSC3 in TAF1/DYT3 affects genes in vesicular transport and dopamine metabolism. Hum Mol Genet. 2013;22(5):941–951. doi: 10.1093/hmg/dds499. [DOI] [PubMed] [Google Scholar]
- Huang CS, Shi SH, Ule J, Ruggiu M, Barker LA, Darnell RB, Jan YN, Jan LY. Common molecular pathways mediate long-term potentiation of synaptic excitation and slow synaptic inhibition. Cell. 2005;123(1):105–118. doi: 10.1016/j.cell.2005.07.033. [DOI] [PubMed] [Google Scholar]
- Huynh MA, Ikeuchi Y, Netherton S, de la Torre-Ubieta L, Kanadia R, Stegmuller J, Cepko C, Bonni S, Bonni A. An isoform-specific SnoN1-FOXO1 repressor complex controls neuronal morphogenesis and positioning in the mammalian brain. Neuron. 2011;69(5):930–944. doi: 10.1016/j.neuron.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima T, Hidaka C, Iijima Y. Spatio-temporal regulations and functions of neuronal alternative RNA splicing in developing and adult brains. Neurosci Res. 2016;109:1–8. doi: 10.1016/j.neures.2016.01.010. [DOI] [PubMed] [Google Scholar]
- Ikeuchi Y, Stegmuller J, Netherton S, Huynh MA, Masu M, Frank D, Bonni S, Bonni A. A SnoN-Ccd1 pathway promotes axonal morphogenesis in the mammalian brain. J Neurosci. 2009;29(13):4312–4321. doi: 10.1523/JNEUROSCI.0126-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, Smith JD, Paeper B, Nickerson DA, Dea J, Dong S, Gonzalez LE, Mandell JD, Mane SM, Murtha MT, Sullivan CA, Walker MF, Waqar Z, Wei L, Willsey AJ, Yamrom B, Lee YH, Grabowska E, Dalkic E, Wang Z, Marks S, Andrews P, Leotta A, Kendall J, Hakker I, Rosenbaum J, Ma B, Rodgers L, Troge J, Narzisi G, Yoon S, Schatz MC, Ye K, McCombie WR, Shendure J, Eichler EE, State MW, Wigler M. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, Quesnel-Vallieres M, Tapial J, Raj B, O’Hanlon D, Barrios-Rodiles M, Sternberg MJ, Cordes SP, Roth FP, Wrana JL, Geschwind DH, Blencowe BJ. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159(7):1511–1523. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N, Hendriks WT, Dhakal J, Vaine CA, Liu C, Shin D, Shin K, Wakabayashi-Ito N, Dy M, Multhaupt-Buell T, Sharma N, Breakefield XO, Bragg DC. Decreased N-TAF1 expression in X-linked dystonia-parkinsonism patient-specific neural stem cells. Dis Model Mech. 2016;9(4):451–462. doi: 10.1242/dmm.022590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Tahimic CG, Ide S, Otsuki A, Sasaoka T, Noguchi S, Oshimura M, Goto Y, Kurimasa A. Methyl CpG-binding protein isoform MeCP2_e2 is dispensable for Rett syndrome phenotypes but essential for embryo viability and placenta development. J Biol Chem. 2012;287(17):13859–13867. doi: 10.1074/jbc.M111.309864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288(5470):1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Jain RG, Andrews LG, McGowan KM, Pekala PH, Keene JD. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17(2):954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambaldorj J, Makino S, Munkhbat B, Tamiya G. Sustained expression of a neuron-specific isoform of the Taf1 gene in development stages and aging in mice. Biochem Biophys Res Commun. 2012;425(2):273–277. doi: 10.1016/j.bbrc.2012.07.081. [DOI] [PubMed] [Google Scholar]
- Jang HS, Shin WJ, Lee JE, Do JT. CpG and Non-CpG Methylation in Epigenetic Gene Regulation and Brain Function. Genes (Basel) 2017;8(6) doi: 10.3390/genes8060148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson CG, Chen Y, Li Y, Leifer D. Functional regulatory regions of human transcription factor MEF2C. Brain Res Mol Brain Res. 2001;97(1):70–82. doi: 10.1016/s0169-328x(01)00187-5. [DOI] [PubMed] [Google Scholar]
- Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25(2):359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jih G, Iglesias N, Currie MA, Bhanu NV, Paulo JA, Gygi SP, Garcia BA, Moazed D. Unique roles for histone H3K9me states in RNAi and heritable silencing of transcription. Nature. 2017;547(7664):463–467. doi: 10.1038/nature23267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, Brunner HG, Amiel J, Oudakker AR, Nillesen WM, Magee A, Genevieve D, Cormier-Daire V, van Esch H, Fryns JP, Hamel BC, Sistermans EA, de Vries BB, van Bokhoven H. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet. 2006;79(2):370–377. doi: 10.1086/505693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Bird A. The major form of MeCP2 has a novel N-terminus generated by alternative splicing. Nucleic Acids Res. 2004;32(5):1818–1823. doi: 10.1093/nar/gkh349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Pezzi E, Desco M, Gatto A, Gomez-Gaviro MV. Neurogenesis: Regulation by Alternative Splicing and Related Posttranscriptional Processes. Neuroscientist. 2016 doi: 10.1177/1073858416678604. [DOI] [PubMed] [Google Scholar]
- Laurent B, Ruitu L, Murn J, Hempel K, Ferrao R, Xiang Y, Liu S, Garcia BA, Wu H, Wu F, Steen H, Shi Y. A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol Cell. 2015;57(6):957–970. doi: 10.1016/j.molcel.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Meur N, Holder-Espinasse M, Jaillard S, Goldenberg A, Joriot S, Amati-Bonneau P, Guichet A, Barth M, Charollais A, Journel H, Auvin S, Boucher C, Kerckaert JP, David V, Manouvrier-Hanu S, Saugier-Veber P, Frebourg T, Dubourg C, Andrieux J, Bonneau D. MEF2C haploinsufficiency caused by either microdeletion of the 5q14.3 region or mutation is responsible for severe mental retardation with stereotypic movements, epilepsy and/or cerebral malformations. J Med Genet. 2010;47(1):22–29. doi: 10.1136/jmg.2009.069732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifer D, Krainc D, Yu YT, McDermott J, Breitbart RE, Heng J, Neve RL, Kosofsky B, Nadal-Ginard B, Lipton SA. MEF2C, a MADS/MEF2-family transcription factor expressed in a laminar distribution in cerebral cortex. Proc Natl Acad Sci U S A. 1993;90(4):1546–1550. doi: 10.1073/pnas.90.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev. 2011;21(2):175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, Soussou W, Nie Z, Kang YJ, Nakanishi N, Okamoto S, Roberts AJ, Schwarz JJ, Lipton SA. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci U S A. 2008;105(27):9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007;8(11):819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- Li YI, Sanchez-Pulido L, Haerty W, Ponting CP. RBFOX and PTBP1 proteins regulate the alternative splicing of micro-exons in human brain transcripts. Genome Res. 2015;25(1):1–13. doi: 10.1101/gr.181990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52(1):93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456(7221):464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327(5968):996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luque FA, Furneaux HM, Ferziger R, Rosenblum MK, Wray SH, Schold SC, Jr, Glantz MJ, Jaeckle KA, Biran H, Lesser M, et al. Anti-Ri: an antibody associated with paraneoplastic opsoclonus and breast cancer. Ann Neurol. 1991;29(3):241–251. doi: 10.1002/ana.410290303. [DOI] [PubMed] [Google Scholar]
- Lyons MR, Schwarz CM, West AE. Members of the myocyte enhancer factor 2 transcription factor family differentially regulate Bdnf transcription in response to neuronal depolarization. J Neurosci. 2012;32(37):12780–12785. doi: 10.1523/JNEUROSCI.0534-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27(3):435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Kaji R, Ando S, Tomizawa M, Yasuno K, Goto S, Matsumoto S, Tabuena MD, Maranon E, Dantes M, Lee LV, Ogasawara K, Tooyama I, Akatsu H, Nishimura M, Tamiya G. Reduced neuron-specific expression of the TAF1 gene is associated with X-linked dystonia-parkinsonism. Am J Hum Genet. 2007;80(3):393–406. doi: 10.1086/512129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NM, Pan Q, Cole BS, Yarosh CA, Babcock GA, Heyd F, Zhu W, Ajith S, Blencowe BJ, Lynch KW. Alternative splicing networks regulated by signaling in human T cells. RNA. 2012;18(5):1029–1040. doi: 10.1261/rna.032243.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y, Mistry M, Pavlidis P, Solomon R, Ghiban E, Antoniou E, Kelleher E, O’Brien C, Donohoe G, Gill M, Morris DW, McCombie WR, Corvin A. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 2014;19(6):652–658. doi: 10.1038/mp.2014.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee AE, Minet E, Stern C, Riahi S, Stiles CD, Silver PA. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol. 2005;5:14. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedeva YA, Lennartsson A, Ehsani R, Kulakovskiy IV, Vorontsov IE, Panahandeh P, Khimulya G, Kasukawa T, Drablos F Consortium F. EpiFactors: a comprehensive database of human epigenetic factors and complexes. Database (Oxford) 2015;2015:bav067. doi: 10.1093/database/bav067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338(6114):1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen CA, Yang XJ, Kokubo T, Brownell JE, Bannister AJ, Owen-Hughes T, Workman J, Wang L, Berger SL, Kouzarides T, Nakatani Y, Allis CD. The TAFII250 Subunit of TFIID Has Histone Acetyltransferase Activity. Cell. 1996;87(7):1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- Mnatzakanian GN, Lohi H, Munteanu I, Alfred SE, Yamada T, MacLeod PJ, Jones JR, Scherer SW, Schanen NC, Friez MJ, Vincent JB, Minassian BA. A previously unidentified MECP2 open reading frame defines a new protein isoform relevant to Rett syndrome. Nat Genet. 2004;36(4):339–341. doi: 10.1038/ng1327. [DOI] [PubMed] [Google Scholar]
- Morgan MA, Shilatifard A. Chromatin signatures of cancer. Genes Dev. 2015;29(3):238–249. doi: 10.1101/gad.255182.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, Hosseini M, Behjati F, Haas S, Jamali P, Zecha A, Mohseni M, Puttmann L, Vahid LN, Jensen C, Moheb LA, Bienek M, Larti F, Mueller I, Weissmann R, Darvish H, Wrogemann K, Hadavi V, Lipkowitz B, Esmaeeli-Nieh S, Wieczorek D, Kariminejad R, Firouzabadi SG, Cohen M, Fattahi Z, Rost I, Mojahedi F, Hertzberg C, Dehghan A, Rajab A, Banavandi MJ, Hoffer J, Falah M, Musante L, Kalscheuer V, Ullmann R, Kuss AW, Tzschach A, Kahrizi K, Ropers HH. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478(7367):57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42(9):790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rawe JA, Wu Y, Dorfel MJ, Rope AF, Au PY, Parboosingh JS, Moon S, Kousi M, Kosma K, Smith CS, Tzetis M, Schuette JL, Hufnagel RB, Prada CE, Martinez F, Orellana C, Crain J, Caro-Llopis A, Oltra S, Monfort S, Jimenez-Barron LT, Swensen J, Ellingwood S, Smith R, Fang H, Ospina S, Stegmann S, Den Hollander N, Mittelman D, Highnam G, Robison R, Yang E, Faivre L, Roubertie A, Riviere JB, Monaghan KG, Wang K, Davis EE, Katsanis N, Kalscheuer VM, Wang EH, Metcalfe K, Kleefstra T, Innes AM, Kitsiou-Tzeli S, Rosello M, Keegan CE, Lyon GJ. TAF1 Variants Are Associated with Dysmorphic Features, Intellectual Disability, and Neurological Manifestations. Am J Hum Genet. 2015;97(6):922–932. doi: 10.1016/j.ajhg.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FH. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309(5743):2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J Neurosci. 1997;17(9):3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson CO, Zachariah RM, Ezeonwuka CD, Liyanage VR, Rastegar M. Brain region-specific expression of MeCP2 isoforms correlates with DNA methylation within Mecp2 regulatory elements. PLoS One. 2014;9(3):e90645. doi: 10.1371/journal.pone.0090645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm K, Metsis M, Timmusk T. Neuron-specific splicing of zinc finger transcription factor REST/NRSF/XBR is frequent in neuroblastomas and conserved in human, mouse and rat. Brain Res Mol Brain Res. 1999;72(1):30–39. doi: 10.1016/s0169-328x(99)00196-5. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pelzer T, Lyons GE, Kim S, Moreadith RW. Cloning and characterization of the murine homolog of the sno proto-oncogene reveals a novel splice variant. Dev Dyn. 1996;205(2):114–125. doi: 10.1002/(SICI)1097-0177(199602)205:2<114::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Pilotto S, Speranzini V, Marabelli C, Rusconi F, Toffolo E, Grillo B, Battaglioli E, Mattevi A. LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors. Hum Mol Genet. 2016;25(12):2578–2587. doi: 10.1093/hmg/ddw120. [DOI] [PubMed] [Google Scholar]
- Pohodich AE, Zoghbi HY. Rett syndrome: disruption of epigenetic control of postnatal neurological functions. Hum Mol Genet. 2015;24(R1):R10–16. doi: 10.1093/hmg/ddv217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesnel-Vallières M, Dargaei Z, Irimia M, Gonatopoulos-Pournatzis T, Ip JY, Wu M, Sterne-Weiler T, Nakagawa S, Woodin MA, Blencowe BJ, Cordes SP. Misregulation of an Activity-Dependent Splicing Network as a Common Mechanism Underlying Autism Spectrum Disorders. Molecular Cell. 2016;64(6):1023–1034. doi: 10.1016/j.molcel.2016.11.033. [DOI] [PubMed] [Google Scholar]
- Quesnel-Vallieres M, Irimia M, Cordes SP, Blencowe BJ. Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development. Genes Dev. 2015;29(7):746–759. doi: 10.1101/gad.256115.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, Blencowe BJ. Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron. 2015;87(1):14–27. doi: 10.1016/j.neuron.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Raj B, Irimia M, Braunschweig U, Sterne-Weiler T, O’Hanlon D, Lin ZY, Chen GI, Easton LE, Ule J, Gingras AC, Eyras E, Blencowe BJ. A global regulatory mechanism for activating an exon network required for neurogenesis. Mol Cell. 2014;56(1):90–103. doi: 10.1016/j.molcel.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, O’Hanlon D, Vessey JP, Pan Q, Ray D, Buckley NJ, Miller FD, Blencowe BJ. Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Mol Cell. 2011;43(5):843–850. doi: 10.1016/j.molcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Ramocki MB, Tavyev YJ, Peters SU. The MECP2 duplication syndrome. Am J Med Genet A. 2010;152A(5):1079–1088. doi: 10.1002/ajmg.a.33184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richa R, Sinha RP. Hydroxymethylation of DNA: an epigenetic marker. Excli j. 2014;13:592–610. [PMC free article] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14(5):347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers HH. Genetics of early onset cognitive impairment. Annu Rev Genomics Hum Genet. 2010;11:161–187. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- Ruggiu M, Herbst R, Kim N, Jevsek M, Fak JJ, Mann MA, Fischbach G, Burden SJ, Darnell RB. Rescuing Z+ agrin splicing in Nova null mice restores synapse formation and unmasks a physiologic defect in motor neuron firing. Proc Natl Acad Sci U S A. 2009;106(9):3513–3518. doi: 10.1073/pnas.0813112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert S, Wang EH, Tjian R. Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature. 1993;362(6416):175–179. doi: 10.1038/362175a0. [DOI] [PubMed] [Google Scholar]
- Rusconi F, Paganini L, Braida D, Ponzoni L, Toffolo E, Maroli A, Landsberger N, Bedogni F, Turco E, Pattini L, Altruda F, De Biasi S, Sala M, Battaglioli E. LSD1 Neurospecific Alternative Splicing Controls Neuronal Excitability in Mouse Models of Epilepsy. Cereb Cortex. 2015;25(9):2729–2740. doi: 10.1093/cercor/bhu070. [DOI] [PubMed] [Google Scholar]
- Sampath SC, Marazzi I, Yap KL, Sampath SC, Krutchinsky AN, Mecklenbrauker I, Viale A, Rudensky E, Zhou MM, Chait BT, Tarakhovsky A. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell. 2007;27(4):596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, Surmeier DJ, Tarakhovsky A, Greengard P. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64(5):678–691. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267(5202):1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19(6):857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Shimojo M, Paquette AJ, Anderson DJ, Hersh LB. Protein kinase A regulates cholinergic gene expression in PC12 cells: REST4 silences the silencing activity of neuron-restrictive silencer factor/REST. Mol Cell Biol. 1999;19(10):6788–6795. doi: 10.1128/mcb.19.10.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staahl BT, Crabtree GR. Creating a neural specific chromatin landscape by npBAF and nBAF complexes. Curr Opin Neurobiol. 2013;23(6):903–913. doi: 10.1016/j.conb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmuller J, Konishi Y, Huynh MA, Yuan Z, Dibacco S, Bonni A. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1-APC target SnoN. Neuron. 2006;50(3):389–400. doi: 10.1016/j.neuron.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K. Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science. 1999;286(5440):771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, Posner JB, Furneaux HM. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991;67(2):325–333. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. Embo j. 2008;27(20):2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang ZZ, Zheng S, Nikolic J, Black DL. Developmental control of CaV1.2 L-type calcium channel splicing by Fox proteins. Mol Cell Biol. 2009;29(17):4757–4765. doi: 10.1128/MCB.00608-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffolo E, Rusconi F, Paganini L, Tortorici M, Pilotto S, Heise C, Verpelli C, Tedeschi G, Maffioli E, Sala C, Mattevi A, Battaglioli E. Phosphorylation of neuronal Lysine-Specific Demethylase 1LSD1/KDM1A impairs transcriptional repression by regulating interaction with CoREST and histone deacetylases HDAC1/2. J Neurochem. 2014;128(5):603–616. doi: 10.1111/jnc.12457. [DOI] [PubMed] [Google Scholar]
- Tunovic S, Barkovich J, Sherr EH, Slavotinek AM. De novo ANKRD11 and KDM1A gene mutations in a male with features of KBG syndrome and Kabuki syndrome. Am J Med Genet A. 2014;164A(7):1744–1749. doi: 10.1002/ajmg.a.36450. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302(5648):1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444(7119):580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, Zeeberg BR, Kane D, Weinstein JN, Blume J, Darnell RB. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37(8):844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong CK, Black DL, Zheng S. The neurogenetics of alternative splicing. Nat Rev Neurosci. 2016;17(5):265–281. doi: 10.1038/nrn.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wan Y, Liu X, Kirschner MW. The anaphase-promoting complex mediates TGF-beta signaling by targeting SnoN for destruction. Mol Cell. 2001;8(5):1027–1039. doi: 10.1016/s1097-2765(01)00382-3. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Telese F, Tan Y, Li W, Jin C, He X, Basnet H, Ma Q, Merkurjev D, Zhu X, Liu Z, Zhang J, Ohgi K, Taylor H, White RR, Tazearslan C, Suh Y, Macfarlan TS, Pfaff SL, Rosenfeld MG. LSD1n is an H4K20 demethylase regulating memory formation via transcriptional elongation control. Nat Neurosci. 2015;18(9):1256–1264. doi: 10.1038/nn.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol. 2001;8(2):141–145. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, Krainer AR, Darnell RB, Zhang C. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 2014;6(6):1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Modrek B, Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30(17):3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, Jiang L, Cai Z, Sun H, Zhang K, Zhang Y, Chen J, Fu XD. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152(1–2):82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Qian H, Hu J, Zhou B, Zhou Y, Hu X, Karakhanyan A, Pang Z, Fu XD. Sequential regulatory loops as key gatekeepers for neuronal reprogramming in human cells. Nat Neurosci. 2016;19(6):807–815. doi: 10.1038/nn.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YY, Yin GL, Darnell RB. The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc Natl Acad Sci U S A. 1998;95(22):13254–13259. doi: 10.1073/pnas.95.22.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K, Makeyev EV. Regulation of gene expression in mammalian nervous system through alternative pre-mRNA splicing coupled with RNA quality control mechanisms. Mol Cell Neurosci. 2013;56:420–428. doi: 10.1016/j.mcn.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Yasui DH, Gonzales ML, Aflatooni JO, Crary FK, Hu DJ, Gavino BJ, Golub MS, Vincent JB, Carolyn Schanen N, Olson CO, Rastegar M, Lasalle JM. Mice with an isoform-ablating Mecp2 exon 1 mutation recapitulate the neurologic deficits of Rett syndrome. Hum Mol Genet. 2014;23(9):2447–2458. doi: 10.1093/hmg/ddt640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5(10):R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476(7359):228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariah RM, Olson CO, Ezeonwuka C, Rastegar M. Novel MeCP2 isoform-specific antibody reveals the endogenous MeCP2E1 expression in murine brain, primary neurons and astrocytes. PLoS One. 2012;7(11):e49763. doi: 10.1371/journal.pone.0049763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 2010;329(5990):439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22(18):2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen MH, Wu X, Kodani A, Fan J, Doan R, Ozawa M, Ma J, Yoshida N, Reiter JF, Black DL, Kharchenko PV, Sharp PA, Walsh CA. Cell-Type-Specific Alternative Splicing Governs Cell Fate in the Developing Cerebral Cortex. Cell. 2016;166(5):1147–1162. e1115. doi: 10.1016/j.cell.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34(36):11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HL, Hinman MN, Barron VA, Geng C, Zhou G, Luo G, Siegel RE, Lou H. Hu proteins regulate alternative splicing by inducing localized histone hyperacetylation in an RNA-dependent manner. Proc Natl Acad Sci U S A. 2011;108(36):E627–635. doi: 10.1073/pnas.1103344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Ramachandran B, Gulick T. Alternative pre-mRNA splicing governs expression of a conserved acidic transactivation domain in myocyte enhancer factor 2 factors of striated muscle and brain. J Biol Chem. 2005;280(31):28749–28760. doi: 10.1074/jbc.M502491200. [DOI] [PubMed] [Google Scholar]
- Zhu H, Hasman RA, Barron VA, Luo G, Lou H. A nuclear function of Hu proteins as neuron-specific alternative RNA processing regulators. Mol Biol Cell. 2006;17(12):5105–5114. doi: 10.1091/mbc.E06-02-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Hinman MN, Hasman RA, Mehta P, Lou H. Regulation of neuron-specific alternative splicing of neurofibromatosis type 1 pre-mRNA. Mol Cell Biol. 2008;28(4):1240–1251. doi: 10.1128/MCB.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zibetti C, Adamo A, Binda C, Forneris F, Toffolo E, Verpelli C, Ginelli E, Mattevi A, Sala C, Battaglioli E. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J Neurosci. 2010;30(7):2521–2532. doi: 10.1523/JNEUROSCI.5500-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweier M, Gregor A, Zweier C, Engels H, Sticht H, Wohlleber E, Bijlsma EK, Holder SE, Zenker M, Rossier E, Grasshoff U, Johnson DS, Robertson L, Firth HV, Cornelia K, Ekici AB, Reis A, Rauch A. Mutations in MEF2C from the 5q14.3q15 microdeletion syndrome region are a frequent cause of severe mental retardation and diminish MECP2 and CDKL5 expression. Hum Mutat. 2010;31(6):722–733. doi: 10.1002/humu.21253. [DOI] [PubMed] [Google Scholar]