Abstract
Secondary metabolite discovery requires an unbiased, comprehensive workflow to detect unknown unknowns for which little to no molecular knowledge exists. Untargeted mass spectrometry-based metabolomics is a powerful platform, particularly when coupled with ion mobility for high-throughput gas-phase separations to increase peak capacity and obtain gas-phase structural information. Ion mobility data are described by the amount of time an ion spends in the drift cell, which is directly related to an ion’s collision cross section (CCS). The CCS parameter describes the size, shape, and charge of a molecule and can be used to characterize unknown metabolomic species. Here, we describe current and emerging applications of ion mobility-mass spectrometry for prioritization, discovery and structure elucidation, and spatial/temporal characterization.
Keywords: Ion mobility, Mass spectrometry, Ion mobility-mass spectrometry, Secondary metabolite, Natural product
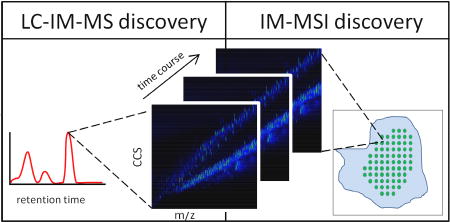
Graphical abstract
Introduction
Advances in microbial genome sequencing have revealed cryptic secondary metabolite gene clusters that are thought to be unexploited sources of new bioactive compounds for discovery [1,2]. Expression of secondary metabolites can be an adaptive response, thus screening an organism under a variety of environmental stimuli to induce silent biosynthetic gene clusters could expand the catalog of natural products (NP) [3]. The analysis of complex chemically-diverse samples and identification of NP candidates is challenging. Untargeted metabolomics, which aims to comprehensively measure all analytes within a biological sample, has the potential to address this chemical diversity. A mass spectrometry (MS)-based metabolomic platform is a powerful approach with high sensitivity and specificity [4]. In a typical experiment, the MS scans over a defined mass range to detect ions related to metabolites of interest. The number of compounds detected depends on the resolving power of the separation platform(s). Pre-ionization separations are routinely interfaced with MS; liquid chromatography (LC) is widely selected [5,6] since a majority of NP are non-volatile. Coupling LC with MS increases peak capacity, although there is a concomitant decrease in throughput. Ion mobility (IM) is a high-throughput separation technique that allows for rapid (µs to ms) separation of molecules [7] and has shown utility for studying a variety of natural products sources [8–10].
Ion mobility is a form of gas-phase electrophoresis that is readily coupled to mass spectrometry. In temporally dispersive IM methods, ions traverse an electric field colliding with a carrier buffer gas. The number of collisions encountered depends upon an ion’s physical properties (charge state, size, and shape), which are described by a collision cross section (CCS). Ions with a low mass and compact structure arrive at the mass analyzer with short drift times, while larger and bulkier molecules elute from the drift cell later. Ultimately, the ion mobility dimension increases the signal-to-noise ratio (S/N) in MS measurements. This report describes emerging applications for IM-MS analyses including feature prioritization strategies for downstream natural product discovery and structural elucidation, as well as ion mobility-MS imaging (MSI) for secondary metabolite spatial characterization. For a much more detailed survey of ion mobility-mass spectrometry, the reader is referred to several comprehensive reviews describing IM-MS theory, instrumentation, and analyses [7,11–13].
IM-MS for prioritization
IM is an appealing technology for NP prioritization efforts owing to its ability to separate molecules based on their gas-phase structural conformation. Unlike primary metabolites which are largely composed of conserved functional groups, secondary metabolite structures have diverse physicochemical properties (e.g., variable molecular weight, atom type, cyclization, degree of oxidation, etc.) [14]. This diversity can affect structural conformation and result in discrete regions of conformational space along trendlines being occupied by various molecular classes in mobility-mass correlations [15,16], as shown in (Figure 1a). Deviations of species from predicted trendlines can be exploited for secondary metabolite prioritization. Recent work has shown that a peptide natural product from cave actinomycetes [17] and halogenated natural products (i.e., particularly attractive lead compounds [18]) derived from crude cyanobacteria extracts [9] deviate from predicted or experimentally-derived mobility trendlines.
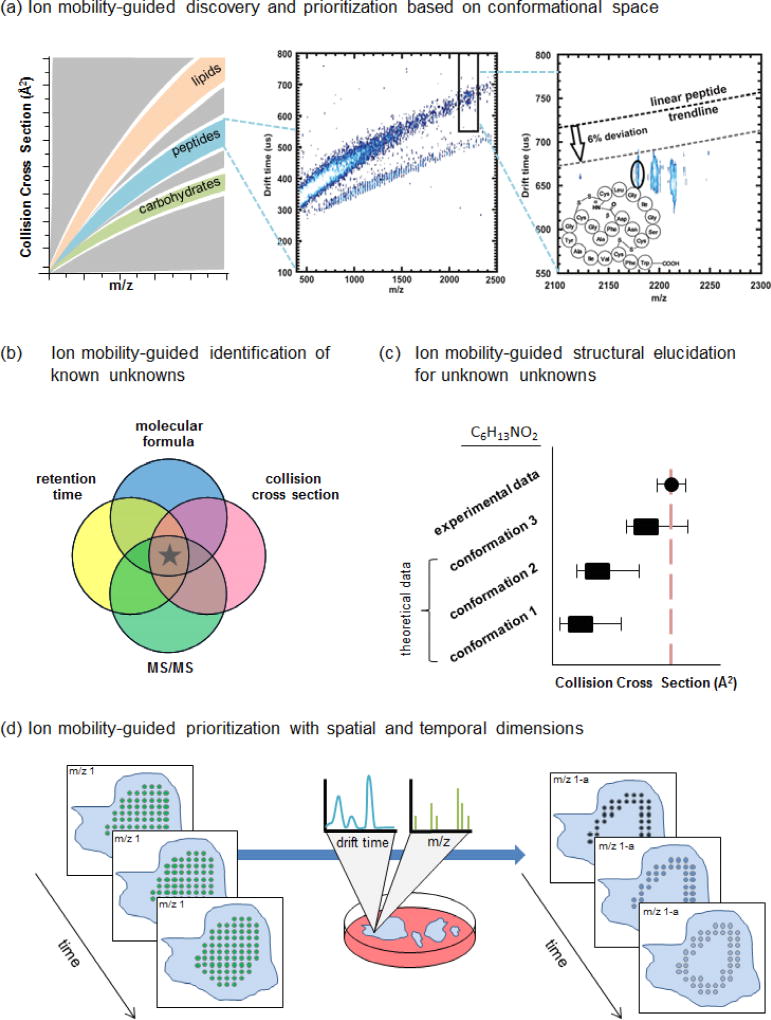
Figure 1.
Applications of metabolomic workflows utilizing ion mobility-mass spectrometry for discovery of secondary metabolite natural products. (a) (Left) IM-MS mobility-mass correlation plots reveal trends of various molecular classes within conformational space. (Middle) Deviations of species from predicted trendlines can be exploited for secondary metabolite prioritization. (Right) This region annotates a peak with dense gas-phase packing efficiency that falls nearly 12% below the linear peptide trendline for this m/z. This compound was identified as siamycin II, a tricyclic peptide, and confirmed by isolation and NMR analyses. Reproduced with permission from [17]. Copyright 2012 American Chemical Society. (b) The Venn diagram represents candidate identifications based on individual data dimensions. Mass spectrometry yields accurate mass and isotope pattern information that can be used to determine molecular formula, liquid chromatography yields retention time, and ion mobility yields drift time/CCS. The goal of including multiple descriptors is to filter candidate space and remove false positives, ultimately leading to a true positive identification (represented here by a star). (c) Computational methods can be used to simulate 3-D structural conformations and calculate theoretical CCS values for comparison with experimental data. Conformations that are consistent with the experimental CCS range are prioritized for structural elucidation. In this mock example, three conformations were predicted for an unknown species with a molecular formula of C6H13NO2. Their theoretical CCS values were compared with the experimental CCS value, revealing conformation 3 as a plausible structure. (d) MSI experiments generate m/z spectra for each spatial coordinate. The integration of an ion mobility dimension enables isobaric species to be separated resulting in more accurate ion images. In this example, m/z 1 appears with uniform signal (green dots) across a sample. Mobility data revealed an isobar, m/z 1-a, with signal (blue dots) that appears as a border around the sample. Further temporal data would enable tracking of this spatial data as a function of time.
Prioritization efforts can also leverage IM’s ability of IM to enhance analytical figures of merit, particularly when combined with LC [19]••[20], although a variety of other front-end separation techniques have been coupled with IM-MS platforms [21]. Basit and coworkers recently showed the ability for an untargeted LC-IM-MS-based lipidomics workflow to detect changes in mouse brain tissue. Specifically, the ion mobility separation dimension uncovered a subclass of dysregulated phospholipids that were not initially revealed by conventional prioritization strategies (i.e., principal component analyses)[22]. Other LC-IM-MS studies have shown the ability to prioritize features via self-organizing maps (SOM) analyses of response trends to either multiplexed environmental stimuli [23] or co-culture with challenger organisms [24] •• designed to elicit potential induction of cryptic or silent gene clusters.
IM-MS for experimental flexibility
Numerous IM-MS instrument designs have been utilized in omics discovery research, and data showcase the benefit of incorporating an IM separation at a variable position within an instrument’s configuration or experimental method. Performing IM prior to MS analysis enables ion signal filtering, for example, using Differential Mobility Spectrometry (DMS) to suppress chemical noise and separate small isobaric molecules [25], and Field Asymmetric waveform Ion Mobility (FAIMS)-MS to detect lower abundance lipid classes by separating a highly abundant protein species [26]•.
Many metabolomics approaches utilize high energy MS fragmentations to generate tandem spectra for identifications. These experiments focus on identifying features of interest by matching MS/MS spectra with reference spectra for candidate annotations. Spectrum matching can be particularly challenging for data independent acquisitions where fragmentation for many ions occurs simultaneously resulting in combined MS/MS spectra. The inclusion of IM prior to fragmentation facilitates deconvolution of MS/MS spectra through correlation of drift time (DT, from the mobility dimension) and retention time (RT, from the LC dimension) with precursor and product ions. This approach has been successfully used for the discovery of novel secondary metabolites and subsequent structure elucidation in mutant Nocardiopsis strains [27]. Mobility-separated spectra are also particularly advantageous for distinguishing isobars in complex samples [19]•[28]•. Recent experiments have shown the utility of an IM dimension for more accurate quantification of individual isobaric species and deconvolution of their combined MS/MS spectra [29].
Performing fragmentation prior to IM analysis also has demonstrated utility. Mobility-separated product ion data was used to guide neutral- and phospholipid identifications [30]•[31], a particularly challenging effort due to the existence of common structural moieties and large number of lipid isomers. Finally, an IM-MS instrument can be configured to enable fragmentation prior to and after mobility separation. This approach aided localization of fatty acyl and double bond positions in phosphatidylcholines [32]. An extension of this configuration incorporates in-source fragmentation to generate MS4 data and has enabled the identification of co-eluting isobaric polycyclic polyprenylated acylphloroglucinols, bioactive components of Garcinia plants, not previously resolved by conventional LC-MS based methods [33]•.
IM–MS for dereplication, identification, and structural elucidation
The process of dereplication (to discriminate novel versus previously elucidated natural products) is crucial in discovery-based MS workflows [34]. Methods to facilitate dereplication efforts via crowd-sourcing analyses have been employed (such as those available through the Global Natural Product Social Molecular Networking (GNPS) website [35]), though many features remain unknown unknowns (i.e., unidentified novel molecules). Analytical advances leading to increased molecular information content should expedite dereplication and elucidation efforts. In omics-based LC-MS/MS experiments, species are characterized with a multi-coordinate set of descriptors (e.g., retention time, accurate mass, isotopic distribution [36,37], MS/MS pattern [38]) to generate candidate identifications. The opportunity to include the IM dimension (i.e., CCS) as a complementary robust measurement to increase specificity and molecular information content is recognized [13,39,40]. Experimental CCSs can be derived directly from drift time measurements [via Drift Tube Ion Mobility Spectrometry (DT IMS)] or by performing a non-linear calibration with an appropriate calibrant under defined conditions [41]•• [via Traveling Wave Ion Mobility Spectrometry (TW IMS)]; inter-lab reproducibility has been established [39].
Though experimental CCS values are valuable physicochemical descriptors, their utility is currently the greatest for known unknown compounds (i.e., known compounds pending identification). Multiple known unknown candidates may meet the accurate mass measurement and isotopic abundance pattern criteria, thus filtering with published CCS values has the potential to reduce the number of candidates to support an identification (Figure 1b). Previously established CCS values are often unavailable. When a compound is labeled as an unknown unknown, CCS alone is not a discriminating identifier. In these cases, experimental IM data can also be used in conjunction with computational methods that simulate 3-D structural conformations and calculate theoretical CCS values [42] [43]• to guide identification of unknown unknowns; predicted conformations consistent with experimental data are prioritized for evaluation (Figure 1c). Recent works incorporating molecular dynamic simulations have allowed for the characterization of a dipeptide-amyloid β complex [44]• and drug metabolite structural isomers [45]•.
IM-MSI for spatial characterization
Multi-dimensional IM-MS metabolomics data allow for separations of ions based on their structural and physicochemical attributes, including hydrophobicity, mass, and collision cross section along with fragmentation data (Figure 1b). Mass spectrometry imaging (MSI) generates data with yet another level of knowledge, positional context, to interrogate spatial distributions of molecules and has been successfully combined with IM separations for added sensitivity. The ionization source of MSI instruments must be amenable to sampling the surface of a biological substrate (e.g., tissue, agar, etc.) and coincide with experimental goals (sample preparation, image resolution, etc.) [46]. Matrix Assisted Laser Desorption Ionization (MALDI), Desorption Electrospray Ionization (DESI), and Liquid-Microjunction-Surface Sampling Probe (LMJ-SSP) have demonstrated success for IM-MSI applications [46–51] [52]•. After matrix application on the surface, MALDI imaging analyses are typically performed in-vacuo with relatively high spatial resolution (dictated by the size and shape of the laser); ambient ionization techniques, such as DESI and LMJ-SSP, require minimal to no sample preparation allowing samples to be analyzed in native state, but have more moderate spatial resolution [53]. Fernandez and coworkers used an IM separation in an effort to compensate for the limited spatial resolution of DESI [50]. The incorporation of DMS as a front end ion filter enabled increased sensitivity by improving S/N, effectively rendering higher quality images. A recent study used an interchangeable source to compare DESI and MALDI ionization of lipids in brain tissue on a single TWIM-MS instrument [52] •. The DESI IM configuration was particularly advantageous for this work as a trendline comprised of fragile lipid species was revealed, though several species with lower DESI ionization efficiencies were ionized better by MALDI, rendering complementary data.
Inherently, MSI does not enable chromatographic separation prior to imaging. Identification of ions detected in an MSI experiment is therefore challenging, owing to the m/z interferences (isobaric compounds and in-source fragmentation) [54] that confound the spatial data interpretation (Figure 1d). Further, the lack of an LC RT descriptor results in the use of accurate mass and isotopic abundance pattern alone for identification efforts. The ability of IM-MSI to address these limitations has been explored. Applications have shown the ability to reduce background chemical noise and increase sensitive and specificity, for example, to resolve poly-sialylated gangliosides in murine brain sections [52] and enable high-throughput localization and identification of recombinant biocatalysts in live bacterial colonies [55]•. Advances in IM-MSI technology will continue to benefit discovery-based research, particularly for 3-D imaging approaches to capture spatial profiles of molecular interactions [34] and investigations of native environments for biological systems exhibiting steep surface topologies [56].
Concluding remarks
IM-MS analyses can facilitate biological discovery research by providing an additional dimension of sensitive and specific physicochemical data for metabolite prioritization. LC-IM-MS measurements are particularly advantageous for natural product discovery where multiple descriptors (see Fig 1b.) are used to minimize dereplication efforts. Structural elucidation is a more arduous task. While IM will certainly augment identification confidence, pre-ionization separations/chromatography and MS/MS are still necessary for MS-based elucidation.
The full potential for massive data sets generated using IM-MS-based spatial and/or temporal experiments is tantalizing, though hurdles related to the mining the data must be overcome. New software tools need to be developed to extract and process high-dimensionality data in order for researchers to be able to interpret and manage findings. Additionally, data storage for copious large datasets is a legitimate concern. The IM field is currently determining the boundaries with which we can utilize CCS measurements for discovery and structural elucidations. Innovations in IM technology, specifically leading to high mobility resolution analyses [57–60], MSI technology, and automated data analysis workflows incorporating both theoretical and experimental CCS data, should have far reaching implications in the natural product, and thus human health, research communities. Though we focused many examples on natural product-based secondary metabolite research, the utility of IM is applicable for primary and secondary metabolites in any biological system.
Highlights.
IM techniques separate ions in the gas-phase based on size, shape, and charge
Deviations from mobility-mass trend lines can be prioritized for identification
The inclusion of IM enables flexible MS experimental designs
Collision cross section values can be used as physicochemical descriptors
IM enhances imaging MS with post-ionization separation of isobaric species
Acknowledgments
We thank Jody C. May, James N. Dodds, Simona G. Codreanu, Randi L. Gant-Branum and our collaborators for stimulating discussions that have influenced our thoughts in these areas. This work was supported by the National Institutes of Health (NIH Grant R01GM092218), the Vanderbilt University Center for Innovative Technology, Vanderbilt Institute of Chemical Biology; the Vanderbilt Institute for Integrative Biosystems Research and Education; and Vanderbilt University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 2.Krug D, Müller R. Secondary metabolomics: the impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat Prod Rep. 2014;31:768–783. doi: 10.1039/c3np70127a. [DOI] [PubMed] [Google Scholar]
- 3.Covington BC, McLean JA, Bachmann BO. Comparative mass spectrometry-based metabolomics strategies for the investigation of microbial secondary metabolites. Nat Prod Rep. 2017;34:6–24. doi: 10.1039/c6np00048g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, McLean JA. Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J Am Soc Mass Spectrom. 2016;27:1897–1905. doi: 10.1007/s13361-016-1469-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eldridge GR, Vervoort HC, Lee CM, Cremin PA, Williams CT, Hart SM, Goering MG, O'Neil-Johnson M, Zeng L. High-throughput method for the production and analysis of large natural product libraries for drug discovery. Anal Chem. 2002;74:3963–3971. doi: 10.1021/ac025534s. [DOI] [PubMed] [Google Scholar]
- 6.Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4:206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 7.May JC, McLean JA. Ion mobility-mass spectrometry: time-dispersive instrumentation. Anal Chem. 2015;87:1422–1436. doi: 10.1021/ac504720m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W, Zhang X, Knochenmuss R, Siems WF, Hill HH. Multidimensional Separation of Natural Products Using Liquid Chromatography Coupled to Hadamard Transform Ion Mobility Mass Spectrometry. J Am Soc Mass Spectrom. 2016;27:810–821. doi: 10.1007/s13361-016-1346-8. [DOI] [PubMed] [Google Scholar]
- 9.Esquenazi E, Daly M, Bahrainwala T, Gerwick WH, Dorrestein PC. Ion mobility mass spectrometry enables the efficient detection and identification of halogenated natural products from cyanobacteria with minimal sample preparation. Bioorg Med Chem. 2011;19:6639–6644. doi: 10.1016/j.bmc.2011.06.081. [DOI] [PubMed] [Google Scholar]
- 10.Harvey DJ, Scarff CA, Edgeworth M, Crispin M, Scanlan CN, Sobott F, Allman S, Baruah K, Pritchard L, Scrivens JH. Travelling wave ion mobility and negative ion fragmentation for the structural determination of N-linked glycans. Electrophoresis. 2013;34:2368–2378. doi: 10.1002/elps.201200669. [DOI] [PubMed] [Google Scholar]
- 11.Lanucara F, Holman SW, Gray CJ, Eyers CE. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nature Chemistry. 2014;6:281–294. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- 12.Wyttenbach T, Pierson NA, Clemmer DE, Bowers MT. Ion mobility analysis of molecular dynamics. Annu Rev Phys Chem. 2014;65:175–196. doi: 10.1146/annurev-physchem-040513-103644. [DOI] [PubMed] [Google Scholar]
- 13.May JC, Morris CB, McLean JA. Ion Mobility Collision Cross Section Compendium. Anal Chem. 2017;89:1032–1044. doi: 10.1021/acs.analchem.6b04905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stow SM, Lareau NM, Hines KM, McNees CR, Goodwin CR, Bachmann BO, McLean JA. Natural Products Analysis: Instrumentation, Methods, and Applications. Spizek VHaJ: John Wiley & Sons, Inc; 2014. Structural Separations for Natural Product Characterization by Ion Mobility Mass Spectrometry: Fundamental Theory to Emerging Applications; pp. 397–432. [Google Scholar]
- 15.Fenn LS, McLean JA. Biomolecular structural separations by ion mobility-mass spectrometry. Anal Bioanal Chem. 2008;391:905–909. doi: 10.1007/s00216-008-1951-x. [DOI] [PubMed] [Google Scholar]
- 16.Fenn LS, Kliman M, Mahsut A, Zhao SR, McLean JA. Characterizing ion mobility-mass spectrometry conformation space for the analysis of complex biological samples. Anal Bioanal Chem. 2009;394:235–244. doi: 10.1007/s00216-009-2666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodwin CR, Fenn LS, Derewacz DK, Bachmann BO, McLean JA. Structural mass spectrometry: rapid methods for separation and analysis of peptide natural products. J Nat Prod. 2012;75:48–53. doi: 10.1021/np200457r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandes MZ, Cavalcanti SM, Moreira DR, de Azevedo Junior WF, Leite AC. Halogen atoms in the modern medicinal chemistry: hints for the drug design. Curr Drug Targets. 2010;11:303–314. doi: 10.2174/138945010790711996. [DOI] [PubMed] [Google Scholar]
- 19.Paglia G, Astarita G. Metabolomics and lipidomics using traveling-wave ion mobility mass spectrometry. Nat Protoc. 2017;12:797–813. doi: 10.1038/nprot.2017.013. •• Comprehensive protocol describing procedures for acquiring and processing LC-TWIMS-MS data for metabolomics and lipidomic applications. [DOI] [PubMed] [Google Scholar]
- 20.Rainville PD, Wilson ID, Nicholson JK, Issacs G, Mullin L, Langridge JI, Plumb RS. Ion mobility spectrometry combined with ultra performance liquid chromatography/mass spectrometry for metabolic phenotyping of urine: Effects of column length, gradient duration and ion mobility spectrometry on metabolite detection. Anal Chim Acta. 2017;982:1–8. doi: 10.1016/j.aca.2017.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng X, Wojcik R, Zhang X, Ibrahim YM, Burnum-Johnson KE, Orton DJ, Monroe ME, Moore RJ, Smith RD, Baker ES. Coupling Front-End Separations, Ion Mobility Spectrometry, and Mass Spectrometry For Enhanced Multidimensional Biological and Environmental Analyses. Annu Rev Anal Chem. 2017;10:71–92. doi: 10.1146/annurev-anchem-061516-045212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basit A, Pontis S, Piomelli D, Armirotti A. Ion mobility mass spectrometry enhances low-abundance species detection in untargeted lipidomics. Metabolomics. 2016;12:50. doi: 10.1007/s11306-016-0971-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodwin CR, Covington BC, Derewacz DK, McNees CR, Wikswo JP, McLean JA, Bachmann BO. Structuring Microbial Metabolic Responses to Multiplexed Stimuli via Self-Organizing Metabolomics Maps. Chem Biol. 2015;22:661–670. doi: 10.1016/j.chembiol.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derewacz DK, Covington BC, McLean JA, Bachmann BO. Mapping Microbial Response Metabolomes for Induced Natural Product Discovery. ACS Chem Biol. 2015;10:1998–2006. doi: 10.1021/acschembio.5b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galhena AS, Harris GA, Kwasnik M, Fernandez FM. Enhanced direct ambient analysis by differential mobility-filtered desorption electrospray ionization-mass spectrometry. Anal Chem. 2010;82:9159–9163. doi: 10.1021/ac102340h. [DOI] [PubMed] [Google Scholar]
- 26.Griffiths RL, Dexter A, Creese AJ, Cooper HJ. Liquid extraction surface analysis field asymmetric waveform ion mobility spectrometry mass spectrometry for the analysis of dried blood spots. Analyst. 2015;140:6879–6885. doi: 10.1039/c5an00933b. • LESA coupled with FAIMS-MS for gas-phase separation and detection of lipid and protein in a single dried blood spot extraction. This work also shows potential to multiplex clinical screening assays. [DOI] [PubMed] [Google Scholar]
- 27.Derewacz DK, Goodwin CR, McNees CR, McLean JA, Bachmann BO. Antimicrobial drug resistance affects broad changes in metabolomic phenotype in addition to secondary metabolism. Proc Natl Acad Sci U S A. 2013;110:2336–2341. doi: 10.1073/pnas.1218524110. •• Nocardiopsis strain challenge by individual co-cultures induces natural product generation. Self organizing map (SOM) analytics are used to interrogate metabolomic data, revealing a novel macrolactam polyketide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyle JE, Zhang X, Weitz KK, Monroe ME, Ibrahim YM, Moore RJ, Cha J, Sun X, Lovelace ES, Wagoner J, et al. Uncovering biologically significant lipid isomers with liquid chromatography, ion mobility spectrometry and mass spectrometry. Analyst. 2016;141:1649–1659. doi: 10.1039/c5an02062j. • LC-DTIMS-MS characterization of pregnant mouse uterine tissue extract samples. The method distinguished lipid isomers and isobars not resolved by LC and also provided structural knowledge to guide more confident identifications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Romm M, Zheng X, Zink EM, Kim Y-M, Burnum-Johnson KE, Orton DJ, Apffel A, Ibrahim YM, Monroe ME, et al. SPE-IMS-MS: An automated platform for sub-sixty second surveillance of endogenous metabolites and xenobiotics in biofluids. Clin Mass Spec. 2016;2:1–10. doi: 10.1016/j.clinms.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hankin JA, Barkley RM, Zemski-Berry K, Deng Y, Murphy RC. Mass Spectrometric Collisional Activation and Product Ion Mobility of Human Serum Neutral Lipid Extracts. Anal Chem. 2016;88:6274–6282. doi: 10.1021/acs.analchem.6b00292. • Concerted tandem and traveling wave ion mobility mass spectrometry (CTS) approach is used to obtain mobility measurements of product ions leading to characterization and identification of lipid species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berry KA, Barkley RM, Berry JJ, Hankin JA, Hoyes E, Brown JM, Murphy RC. Tandem Mass Spectrometry in Combination with Product Ion Mobility for the Identification of Phospholipids. Anal Chem. 2017;89:916–921. doi: 10.1021/acs.analchem.6b04047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castro-Perez J, Roddy TP, Nibbering NM, Shah V, McLaren DG, Previs S, Attygalle AB, Herath K, Chen Z, Wang SP, et al. Localization of fatty acyl and double bond positions in phosphatidylcholines using a dual stage CID fragmentation coupled with ion mobility mass spectrometry. J Am Soc Mass Spectrom. 2011;22:1552–1567. doi: 10.1007/s13361-011-0172-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, Zheng D, Li HH, Wang H, Tan HS, Xu HX. Diagnostic filtering to screen polycyclic polyprenylated acylphloroglucinols from Garcinia oblongifolia by ultrahigh performance liquid chromatography coupled with ion mobility quadrupole time-of-flight mass spectrometry. Anal Chim Acta. 2016;912:85–96. doi: 10.1016/j.aca.2016.01.039. • Multistage ion mobility-mass spectrometry technique (insource collision-induced dissociation combined with time aligned parallel fragmentation) enabled both MS4 and CCS measurements for identifying and resolving co-eluting isobaric PPAPs. [DOI] [PubMed] [Google Scholar]
- 34.Bouslimani A, Sanchez LM, Garg N, Dorrestein PC. Mass spectrometry of natural products: current, emerging and future technologies. Nat Prod Rep. 2014;31:718–729. doi: 10.1039/c4np00044g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kind T, Fiehn O. Metabolomic database annotations via query of elemental compositions: mass accuracy is insufficient even at less than 1 ppm. BMC Bioinformatics. 2006;7:234. doi: 10.1186/1471-2105-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kind T, Fiehn O. Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics. 2007;8:105. doi: 10.1186/1471-2105-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohimani H, Pevzner PA. Dereplication, sequencing and identification of peptidic natural products: from genome mining to peptidogenomics to spectral networks. Nat Prod Rep. 2016;33:73–86. doi: 10.1039/c5np00050e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paglia G, Williams JP, Menikarachchi L, Thompson JW, Tyldesley-Worster R, Halldorsson S, Rolfsson O, Moseley A, Grant D, Langridge J, et al. Ion mobility derived collision cross sections to support metabolomics applications. Anal Chem. 2014;86:3985–3993. doi: 10.1021/ac500405x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paglia G, Angel P, Williams JP, Richardson K, Olivos HJ, Thompson JW, Menikarachchi L, Lai S, Walsh C, Moseley A, et al. Ion mobility-derived collision cross section as an additional measure for lipid fingerprinting and identification. Anal Chem. 2015;87:1137–1144. doi: 10.1021/ac503715v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hines KM, May JC, McLean JA, Xu L. Evaluation of Collision Cross Section Calibrants for Structural Analysis of Lipids by Traveling Wave Ion Mobility-Mass Spectrometry. Anal Chem. 2016;88:7329–7336. doi: 10.1021/acs.analchem.6b01728. •• Evidence that structurally matched calibrants are necessary for accurate collision cross section measurements (of lipids) to be made using traveling wave ion-mobility mass spectrometry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stow SM, Goodwin CR, Kliman M, Bachmann BO, McLean JA, Lybrand TP. Distance geometry protocol to generate conformations of natural products to structurally interpret ion mobility-mass spectrometry collision cross sections. J Phys Chem B. 2014;118:13812–13820. doi: 10.1021/jp509398e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lapthorn C, Pullen FS, Chowdhry BZ, Wright P, Perkins GL, Heredia Y. How useful is molecular modelling in combination with ion mobility mass spectrometry for 'small molecule' ion mobility collision cross-sections? Analyst. 2015;140:6814–6823. doi: 10.1039/c5an00411j. • Quantative assessment comparing experimental and theoretical collision cross section values for a group of twenty pharmaceutical compounds ranging from 122–609 Da. [DOI] [PubMed] [Google Scholar]
- 44.Soper-Hopper MT, Eschweiler JD, Ruotolo BT. Ion Mobility-Mass Spectrometry Reveals a Dipeptide That Acts as a Molecular Chaperone for Amyloid β. ACS Chem Biol. 2017;12:1113–1120. doi: 10.1021/acschembio.7b00045. •Ion mobility-mass spectrometry and molecular dynamics simulations are used to structurally characterize the interactions between key residues of wild-type or variant forms of amyloid β and the dipeptide, FL, from leucine enkephalin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reading E, Munoz-Muriedas J, Roberts AD, Dear GJ, Robinson CV, Beaumont C. Elucidation of Drug Metabolite Structural Isomers Using Molecular Modeling Coupled with Ion Mobility Mass Spectrometry. Anal Chem. 2016;88:2273–2280. doi: 10.1021/acs.analchem.5b04068. • Ion mobility-derived collision cross section data combined with molecular modeling to guide structural isomer identification. The authors also address the requirement that molecules must exhibit dissimilar CCS values for the ability to differentiate structures. [DOI] [PubMed] [Google Scholar]
- 46.Petras D, Jarmusch AK, Dorrestein PC. From single cells to our planet-recent advances in using mass spectrometry for spatially resolved metabolomics. Curr Opin Chem Biol. 2017;36:24–31. doi: 10.1016/j.cbpa.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 47.McLean JA, Ridenour WB, Caprioli RM. Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J Mass Spectrom. 2007;42:1099–1105. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- 48.Trim PJ, Henson CM, Avery JL, McEwen A, Snel MF, Claude E, Marshall PS, West A, Princivalle AP, Clench MR. Matrix-assisted laser desorption/ionization-ion mobility separation-mass spectrometry imaging of vinblastine in whole body tissue sections. Anal Chem. 2008;80:8628–8634. doi: 10.1021/ac8015467. [DOI] [PubMed] [Google Scholar]
- 49.Xu L, Kliman M, Forsythe JG, Korade Z, Hmelo AB, Porter NA, McLean JA. Profiling and Imaging Ion Mobility-Mass Spectrometry Analysis of Cholesterol and 7-Dehydrocholesterol in Cells Via Sputtered Silver MALDI. J Am Soc Mass Spectrom. 2015;26:924–933. doi: 10.1007/s13361-015-1131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bennett RV, Gamage CM, Galhena AS, Fernandez FM. Contrast-enhanced differential mobility-desorption electrospray ionization-mass spectrometry imaging of biological tissues. Anal Chem. 2014;86:3756–3763. doi: 10.1021/ac5007816. [DOI] [PubMed] [Google Scholar]
- 51.Feider CL, Elizondo N, Eberlin LS. Ambient Ionization and FAIMS Mass Spectrometry for Enhanced Imaging of Multiply Charged Molecular Ions in Biological Tissues. Anal Chem. 2016;88:11533–11541. doi: 10.1021/acs.analchem.6b02798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Škrášková K, Claude E, Jones EA, Towers M, Ellis SR, Heeren RM. Enhanced capabilities for imaging gangliosides in murine brain with matrix-assisted laser desorption/ionization and desorption electrospray ionization mass spectrometry coupled to ion mobility separation. Methods. 2016;104:69–78. doi: 10.1016/j.ymeth.2016.02.014. • Description of the utility for both MALDI and DESI sources for imaging mass spectrometry with ion mobility separations, comparing lipidomics data acquired with each ionization source and showing complementarity of findings. [DOI] [PubMed] [Google Scholar]
- 53.Bodzon-Kulakowska A, Suder P. Imaging mass spectrometry: Instrumentation, applications, and combination with other visualization techniques. Mass Spectrom Rev. 2016;35:147–169. doi: 10.1002/mas.21468. [DOI] [PubMed] [Google Scholar]
- 54.Xu YF, Lu W, Rabinowitz JD. Avoiding misannotation of in-source fragmentation products as cellular metabolites in liquid chromatography-mass spectrometry-based metabolomics. Anal Chem. 2015;87:2273–2281. doi: 10.1021/ac504118y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan C, Parmeggiani F, Jones EA, Claude E, Hussain SA, Turner NJ, Flitsch SL, Barran PE. Real-Time Screening of Biocatalysts in Live Bacterial Colonies. J Am Chem Soc. 2017;139:1408–1411. doi: 10.1021/jacs.6b12165. • Innovative example of an ion-mobility imaging mass spectrometry application for screening biotransformation reactions in agar in real-time. [DOI] [PubMed] [Google Scholar]
- 56.Sica VP, Raja HA, El-Elimat T, Kertesz V, Van Berkel GJ, Pearce CJ, Oberlies NH. Dereplicating and Spatial Mapping of Secondary Metabolites from Fungal Cultures in Situ. J Nat Prod. 2015;78:1926–1936. doi: 10.1021/acs.jnatprod.5b00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merenbloom SI, Glaskin RS, Henson ZB, Clemmer DE. High-resolution ion cyclotron mobility spectrometry. Anal Chem. 2009;81:1482–1487. doi: 10.1021/ac801880a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deng L, Ibrahim YM, Hamid AM, Garimella SV, Webb IK, Zheng X, Prost SA, Sandoval JA, Norheim RV, Anderson GA, et al. Ultra-High Resolution Ion Mobility Separations Utilizing Traveling Waves in a 13 m Serpentine Path Length Structures for Lossless Ion Manipulations Module. Anal Chem. 2016;88:8957–8964. doi: 10.1021/acs.analchem.6b01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ujma J, Giles K, Morris M, Barran PE. New High Resolution Ion Mobility Mass Spectrometer Capable of Measurements of Collision Cross Sections from 150 to 520 K. Anal Chem. 2016;88:9469–9478. doi: 10.1021/acs.analchem.6b01812. [DOI] [PubMed] [Google Scholar]
- 60.Hernandez DR, Debord JD, Ridgeway ME, Kaplan DA, Park MA, Fernandez-Lima F. Ion dynamics in a trapped ion mobility spectrometer. Analyst. 2014;139:1913–1921. doi: 10.1039/c3an02174b. [DOI] [PMC free article] [PubMed] [Google Scholar]