Abstract
The nervous system comprises many different cell types including neurons, glia, macrophages, and immune cells, each of which is defined by specific patterns of gene expression, morphology, function, and anatomical location. Establishment of these complex and highly regulated cell fates requires spatial and temporal coordination of gene transcription. Open chromatin (euchromatin) allows transcription factors to interact with gene promoters and activate lineage specific genes, whereas closed chromatin (heterochromatin) remains inaccessible to transcriptional activation. Changes in the genome-wide distribution of euchromatin accompanies transcriptional plasticity that allows the diversity of mature cell fates to be generated during development. In the past 20 years, many new genes and gene families have been identified to participate in regulation of chromatin accessibility. These genes include chromatin remodelers that interact with Trithorax group (TrxG) and Polycomb group (PcG) proteins to activate or repress transcription, respectively. Here we review the role of TrxG proteins in neurodevelopment and disease.
Keywords: Trithorax, Chromatin, Neurodevelopment, Disease
1. Introduction
Embryonic development proceeds from a single multipotent cell to a multicellular complex organism with distinct organs, tissues, and cell types that retain their identities over developmental space and time. While some mature cells and tissues exhibit high levels of proliferative and regenerative potential (i.e, skin and gut epithelial cells), others (i.e, neurons) are quiescent and unable to self-renew upon injury. The mechanisms by which specific cell types maintain their fate or “memory” that instructs profiles of gene expression despite active DNA replication and mitosis remain a mystery; however, much work has been done to identify the genes, molecules, and chromatin-associated factors involved in this process. Chromatin structure has a regulatory role on the transcriptional profile on processes that underlie cellular proliferation and maintenance of cell fate. Identifying the molecular pathways that direct chromatin structure and gene expression is a central goal in developmental biology, and has important relevance for understanding basic mechanisms of developmental disorders.
This review explores mechanisms of human developmental disorders caused by pathogenic variants in human homologs of trithorax group (TrxG) genes encoding histone methyltransferases, demethylases, and chromatin remodelers (Table 1). TrxG is a family of proteins that form large multi-protein complexes exhibiting histone methyltransferase and/or chromatin remodeling functions (Schuettengruber et al., 2011). Drosophila trithorax (trx) was first identified as a spontaneous pathogenic variant in flies with abnormalities of head, thoracic, and abdominal structures, consistent with transformations of body segment identity (Ingham, 1983). In the fly, trx encodes for a histone methyltransferase and acts to suppress the functions of Polycomb group (PcG) genes. TrxG and PcG genes are highly conserved across evolution, and act antagonistically at genetic targets such as the Hox gene cluster to regulate gene expression (Steffen and Ringrose, 2014). In general, PcG genes encode proteins that function as transcriptional repressors, whereas TrxG genes encode proteins that act as transcriptional activators (Fig. 1). This mutual antagonism has led to a model whereby PcG and TrxG proteins switch between stably repressed or activated patterns of gene expression during development.
Table 1. Human genetic diseases associated with Trithorax group related genes.
Human disease associations, and Autism susceptibility according to SFARI gene classification for Trithorax group related genes. Scoring for SFARI gene is as follows: syndromic (S), high confidence (1), and strong candidate (2).
| Trithorax Group Class | Gene Name | Human Disease Association | SFARI Gene Score | SFARI Syndromic |
|---|---|---|---|---|
| Histone Methyltransferases | KMT2F (SET1A) | Association with Schizophrenia and neurodevelopmental disorders | ||
| KMT2A (MLL1) | Wiedemann-Steiner syndrome and Leukemia Myeloid | 2 | S | |
| KMT2D (MLL2) | Kabuki syndrome 1 | |||
| KMT2C (MLL3) | Kleefstra syndrome | 2 | ||
| KMT2B (MLL4) | Dystonia | |||
| Histone Demethylase | KDM6A (UTX) | Kabuki syndrome 2 | ||
| ATP-Dependent Chromatin Remodelers - SWI/SNF | SMARCA1 (SNF2L1) | Schizophrenia, Microcephaly with intellectual disability, Rett-like phenotypes | ||
| SMARCA2 (BRM) | Nicolaides-Baraitser syndrome and Schizophrenia | S | S | |
| SMARCA4 (BRG1) | Coffin-Siris syndrome 4 and Rhabdoid Tumor Predisposition syndrome 2 | |||
| SMARCB1 (SNF5) | Coffin-Siris syndrome 3, somatic Rhabdoid tumors, Rhabdoid Predisposition syndrome 1, and susceptibility to Schwannomatosis-1 | |||
| SMARCE1 (BAF57) | Coffin-Siris syndrome 5, susceptibility to familial Meningioma | |||
| ARID1A (BAF250A) | Coffin-Siris syndrome 2 | |||
| ARID1B (BAF250B) | Coffin-Siris syndrome 1 | 1 | S | |
| ATP-Dependent Chromatin Remodelers - INO80 | YY1AP1 (YAP) | Grange syndrome | ||
| SRCAP (SWR1) | Floating-Harbor syndrome | 2 | ||
| ATP-Dependent Chromatin Remodelers - CHD | CHD1 | Pilarowski-Bjornsson syndrome | ||
| CHD2 | Childhood-onset Epileptic Encephalopathy | 2 | S | |
| CHD4 | Sifram-Hitz-Weiss syndrome | |||
| CHD7 | CHARGE syndrome | S | S | |
| CHD8 | Autism Spectrum Disorder | 1 | S |
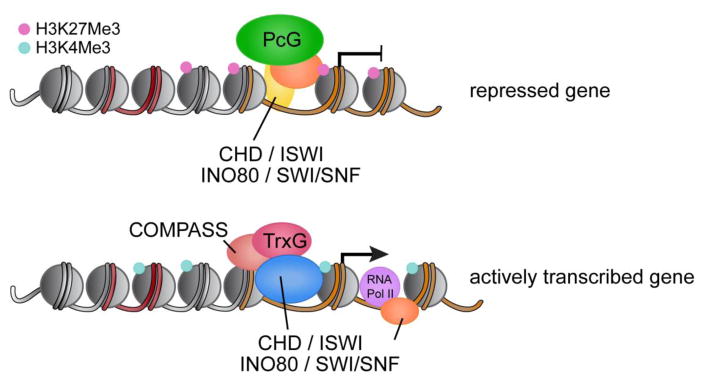
Figure 1. Schematic of Polycomb and Trithorax Related Proteins at Promoters of Repressed and Active Genes.
Repressed genes are bound by Polycomb group proteins (PcG) whereas Trithorax-related proteins (TrxG) localize to actively transcribed genes. COMPASS (complex of proteins associated with Set1) opposes PcG activity to activate transcription. ATP-dependent chromatin remodelers (CHD, ISWI, INO80, and SWI/SNF) regulate DNA accessibility, which influences gene repression and activation during embryonic development.
TrxG proteins generally function as large multi-protein complexes, where they localize to transcription start sites, enhancers, and gene bodies, with variable roles that are influenced largely by their interacting partners and target sites in the genome. Based on their molecular functions, TrxG proteins are categorized into three general classes. The first class of TrxG proteins comprises SET-domain histone methyltransferases. This class includes the COMPASS (complex of proteins associated with Set1) members SET1A, SET1B, and mixed lineage leukemia-1-4 (MLL1, MLL2, MLL3 and MLL4), among others (Piunti and Shilatifard, 2016). The second class of TrxG proteins contains ATP-dependent chromatin remodelers that “read” the histone modifications established by SET domain-containing enzymes. This class includes switch/sucrose non-fermenting (SWI/SNF) proteins, imitation switch (ISWI), inositol auxotroph 80 (INO80), and chromodomain-helicase-DNA binding (CHD) proteins. Chromatin remodelers harness the energy of ATP to slide nucleosomes along DNA, evict nucleosomes from DNA, or exchange histone dimers, thereby altering the chromatin architecture and making it more or less accessible to transcription factors and other regulatory proteins or RNA. The third class of TrxG proteins bind specific DNA sequences called TrxG response elements (TREs), which often coincide with PcG response elements (PREs) that switch status between activation and silencing by mechanisms that involve noncoding RNA transcription (Herzog et al., 2014). This general classification of TrxG proteins is still evolving, as new information is obtained about the functions of this large group of proteins and associated factors.
2. Histone modifications and developmental disorders
2.1 Epigenetic mechanisms
Abundant post-translational modifications of histone tails (phosphorylation, methylation, acetylation, ubiquitination, sumoylation), which regulate accessibility of genetic information, are a distinguishing feature of eukaryotic organisms. Epigenetic regulation of gene expression requires involvement of many different histone modifying enzymes, including “writers” that attach modifications to histone tails, and “erasers” that remove modifications, whereas “readers” recognize modifications distributed in a cell-specific manner across the genome. A function of histone modifications is to coordinate chromatin remodelers and transcriptional machinery for transcriptional regulation. Histone modifications function together with histone variants, chromatin-remodeling activities, DNA methylation, and histone chaperones to contribute to the faithful establishment and maintenance of the chromatin environment.
Among various post-translational modifications, histone H3 Lysine 4 (H3K4) methylation (H3K4me) is evolutionarily conserved and closely associated with transcriptionally active chromatin (Bannister and Kouzarides, 2011; Shilatifard, 2006). Data supports H3K4 methylation in pivotal early steps of the signaling cascade leading to transcriptional activation (Campos and Reinberg, 2009; Ruthenburg et al., 2007). Also, H3K4me recruits basic transcriptional machinery (Tang et al., 2013; Vermeulen et al., 2007), including histone acetyltransferases and ATP-dependent chromatin remodeling proteins of the CHD family such as CHD7 and CHD8 (Ruthenburg et al., 2007; Schnetz et al., 2009; Schnetz et al., 2010; Taverna et al., 2007; Wysocka et al., 2006). Genome-wide analyses show that H3K4me, which is highly enriched at gene promoters and enhancers, positively correlates with transcription rates, occupancy of RNA Pol II, and histone acetylation at 5′ regions of active genes (Barski et al., 2007; Heintzman et al., 2007). H3K4me is also enriched at chromatin regions ‘poised’ for differentiation and lineage specification in embryonic stem cells (ESCs), juxtaposed with the antagonistic H3K27me3 mark (Azuara et al., 2006; Bernstein et al., 2006; Pan et al., 2007). Thus, specific histone marks or combinations of histone marks serve as tags for unique functional regions of the genome.
2.2 Disorders of epigenetic factors
Disruptions of histone modifications and chromatin accessibility comprise an important class of human developmental disorders (Fahrner and Bjornsson, 2014; Lopez and Wood, 2015). Human genetic disorders caused by pathogenic variants in epigenetic modulators include CHARGE, Kabuki, Coffin-Siris, Kleefstra, Wiedemann-Steiner, and Nicolaides-Baraitser syndromes (Table 1)(Jones et al., 2012; Mendelsohn et al., 2014; Strom et al., 2014). Neurodevelopmental disorders have also been associated with pathogenic variants in TrxG genes, COMPASS members, and other ATP-dependent chromatin remodelers. Perhaps not surprisingly, altered dosage and function of TrxG-related proteins leads to a variety of cancers such as leukemia, rhabdoid tumors, and meningioma (Table 1) (Schuettengruber et al., 2017).
3. ATP-dependent chromatin remodelers
3.1 ISWI
Goodwin and Picketts, in this issue, provide a comprehensive review of ISWI and its role in neurodevelopmental disorders, including genes that encode ISWI components BAZ1B and CECR2 in Williams-Beuren and Cat Eye syndrome, respectively (Banting et al., 2005; Bozhenok et al., 2002; Footz et al., 2001; Lu et al., 1998; Mellor, 2006; Peoples et al., 1998). Readers are encouraged to read their paper for more information on this important class of ATP-dependent chromatin remodelers.
3.2 SWI/SNF
SWI/SNF (also known as BRG1/BRM associated factor (BAF)) complexes are comprised of at least 15 different subunits that are enriched at gene promoters, enhancers, and super-enhancers (Sokpor et al., 2017). Dynamic switching among BAF subunits during neuronal development has the potential to generate hundreds of different complexes (Lessard et al., 2007). Five genes encoding subunits of the SWI/SNF family (SMARCA4 (BRG1), SMARCB1 (SNF5), SMARCE1 (BAF57), ARID1A (BAF250A), and ARID1B (BAF250B)) have been implicated in Coffin-Siris syndrome (Santen et al., 2012; Tsurusaki et al., 2012), and pathogenic variants in SMARCA2 (BRM) cause Nicolaides-Baraitser syndrome (Sousa et al., 2014; Van Houdt et al., 2012). Pathogenic variants in SMARCA4 (Coffin-Siris syndrome) and SMARCA2 (Nicolaides-Baraitser syndrome) are predicted to result in functionally inert proteins that retain their abilities to interact with and target specific regions of the genome with other subunits of SWI/SNF. Pathogenic variants in SMARCA1 (SNF2L1) have been reported in individuals with schizophrenia, microcephaly, intellectual disability, and Rett-like phenotypes (Homann et al., 2016; Karaca et al., 2015; Lopes et al., 2016). Mechanistically, SWI/SNF and CHD proteins share the common property of flanking nucleosome-free regions (NFRs) in embryonic stem cells, suggesting that complex interactions between these protein classes are necessary for regulating distinct patterns of chromatin and gene expression (de Dieuleveult et al., 2016).
3.3 INO80
INO80 proteins, unlike other chromatin remodelers, have unique structural features and functions that include regulation of DNA replication and repair. INO80 promotes progression of the DNA replication fork, evicts RNA polymerase II at transcribed genes upon interaction with the replication fork, and releases nucleosomes after oxidative DNA damage (Poli et al., 2017). Recently, INO80 was described as a candidate disease gene for an individual who presented with primary microcephaly and global developmental delay in a cohort of consanguineous families (Alazami et al., 2015). In addition, variants in YY1AP1, a component of the INO80 complex, were identified as a cause of Grange syndrome (Guo et al., 2017). Pathogenic variants in the INO80 homolog SRCAP cause Floating-Harbor syndrome, a neurodevelopmental disorder with expressive language delay, short stature, and abnormal skeletal/craniofacial development (Hood et al., 2012; Hood et al., 2016; Nikkel et al., 2013). Ultimately, evidence from these human genetic studies points to the importance of chromatin remodeling in DNA replication, damage, and transcription as critical during development, and perturbation of these processes leads to overlapping phenotypes that affect neurodevelopment.
3.4 CHD
The CHD family comprises nine chromatin remodeling members characterized by the presence of two chromodomains (chromatin organization modifier), a structural domain of about 40–50 amino acid residues, centrally located DNA helicase domains, and less well-defined carboxyl terminal domains (Shur and Benayahu, 2005; Woodage et al., 1997). Chromodomains are not unique to the CHD family; they are also present in repressive Polycomb protein Pc and heterochromatin associated protein HP1 of D. melanogaster, where they were first described (Paro and Hogness, 1991). CHD proteins, similar to SWI/SNF proteins, regulate access to DNA by using the energy of ATP hydrolysis to alter chromatin structure, slide nucleosomes along DNA, or evict nucleosomes from the DNA strand during transcription or replication (Becker and Hörz, 2002; Eberharter and Becker, 2004; Lusser and Kadonaga, 2003; Manning and Yusufzai, 2017; Narlikar et al., 2002; Smith and Peterson, 2005). Moreover, CHD proteins “read” H3K4 methylation at transcription start sites and enhancers and exhibit pleiotropic functions during development, including regulation of pluripotency, stem cell proliferation, and lineage determination (Dowen et al., 2014; Hnisz et al., 2013; Niederreiter et al., 2015). The CHD family exhibits high evolutionary conservation back to S. cerevisiae and D. melanogaster with one and four members, respectively. In vertebrates, the nine CHD proteins are divided into three distinct subfamilies on the basis of similarities in amino acid sequence and functional protein domains (Liu et al., 2015; Woodage et al., 1997). CHD proteins were also recently shown to target specific nucleosomes near MNase-defined NFRs (de Dieuleveult et al., 2016).
3.4.1 The CHD Family Subclass I
Subclass I of human CHD proteins is comprised of CHD1 and CHD2, both of which are associated with human disease. Notably, subclass I proteins display the ability to interact with histone modifications (methylation of H3K4), through a chromodomain aromatic cage (Flanagan et al., 2007), and the ability to bind DNA through a C-terminal domain structure that resembles a SWI3, ADA2, N-CoR, and TFIIIB (SANT) domain and a SANT-like ISWI domain (SLIDE domain) (Aasland et al., 1996; Delmas et al., 1993; Grune et al., 2003; Ryan et al., 2011; Stokes and Perry, 1995; Woodage et al., 1997). Heterozygous pathogenic variants in CHD1 were recently identified in six individuals with autism, developmental delay, speech apraxia, and craniofacial dysmorphisms (Pilarowski et al., 2017). CHD2 was first implicated in neurodevelopment disease through case reports describing de novo deletions of 15q26 in individuals with epilepsy, developmental delay and craniofacial dysmorphisms (Capelli et al., 2012; Veredice et al., 2009). However, the definitive involvement of this chromatin remodeler in neurodevelopment, independent of other 15q26 genes, was not discovered until cohort studies applied targeted and whole exome sequencing approaches. De novo pathogenic variants and copy number variants (CNVs) in CHD2 were later discovered in epileptic encephalopathy, non-syndromic intellectual disability, and autism spectrum disorder cohorts, suggesting that pathogenic variants in CHD2 cause a spectrum of neurological phenotypes including seizures (Carvill et al., 2013; Chenier et al., 2014; Epi et al., 2013; Lund et al., 2014; O’Roak et al., 2014; Pinto et al., 2014; Rauch et al., 2012; Suls et al., 2013).
The underlying mechanism of CHD2 pathogenic variants has not been precisely defined. Suls et al. utilized morpholino knockdown of chd2 in zebrafish, and reported that larvae with chd2 partial knockdown exhibited epileptiform discharges and abnormal twitching behavior (Suls et al., 2013). Several other phenotypes were also observed in chd2 partial knockdown zebrafish, including microcephaly, growth retardation, curved body appearance, absence of the swim bladder, and pericardial edema. Consistent with multiple abnormalities observed in chd2 mutant zebrafish, Chd2 mutant mice that lack the C-terminal DNA binding domain also exhibit small size, a hunched appearance, and multiple organ abnormalities in the heart, spleen, liver, kidney and lymph nodes (Marfella et al., 2006). Chd2 is expressed in adult mouse brain tissue; however, no brain abnormalities were reported upon necropsy or histopathology examination, nor was epileptic behavior observed (Marfella et al., 2006). It is possible that the lifespan of Chd2 mutant mice (32 to 64 weeks) is too short to observe neurological phenotypes, or neurological abnormities are too subtle to be detected by the macroscopic and histopathology methods used for evaluation. Alternatively, expression of the mutant CHD2 protein (which retains the chromodomains and ATPase domain) may be sufficient to prevent neurological disease. Alternatively, there may be species-specific differences in CHD2 function between humans, zebrafish, and mice.
The chromatin remodeling role of CHD2 appears to influence the deposition of histone variant H3.3 at genes important for development. H3.3 is generally observed at chromatin associated with active transcription, but also contributes to the chromatin environment at bivalent gene promoters (Goldberg et al., 2010). Interestingly, depletion of H3.3 or Hira, the histone chaperone that deposits H3.3 at genic regions, leads to reduced H3K27me3 at bivalent gene promoters in mouse embryonic stem cells (ESCs), whereas Chd2 depletion results in a greater enrichment of both H3.3 and H3K27me3 (Banaszynski et al., 2013; Semba et al., 2017). These studies highlight the importance of histone chaperones and CHD proteins in balancing repressive and active histone modifications at developmentally critical genes.
3.4.2 The CHD Family Subclass II
The CHD subclass II proteins CHD3, CHD4 and CHD5 are distinguished from the other two subclasses in that they display two plant homeodomain (PHD) zinc finger domains capable of reading lysine 4 methylated H3 (Bienz, 2006; Sanchez and Zhou, 2011). Interestingly, members of the CHD subclass II are components of the nucleosome remodeling and histone deacetylation (NuRD) complex (Tong et al., 1998; Xue et al., 1998; Zhang et al., 1998). The NuRD complex contains at least six subunits histone deacetylase-1 (HDAC1) and -2 (HDAC2), and chromatin remodeling functions from CHD3-5 (Basta and Rauchman, 2015; Bowen et al., 2004; Lai and Wade, 2011). The composition of NuRD during mouse cortical development implicates each of the three-chromatin remodelers (CHD3, CHD4 and CHD5) in a different stage of corticogenesis with generally non-redundant roles (Nitarska et al., 2016). Studies of NuRD in rat postnatal cerebella demonstrate that CHD4 regulates gene repression and drives synaptogenesis of granule neuron parallel fibers and Purkinje cells (Yamada et al., 2014). CHD4 also complexes with Polycomb Repressive Complex 2 (PRC2) catalytic component EZH2 in mouse neural progenitor cells to maintain the sequential order of transcriptional programs, specifically to suppress glial gene expression (Sparmann et al., 2013).
All CHD subclass II members are associated with human disease. Levels of CHD3 (formerly autoantigen Mi-2) and CHD4 were reported to be elevated in sera of patients with arthritis and dermatomyositis (Ge et al., 1995; Seelig et al., 1996). Phenotypes associated with pathogenic CHD4 variants are clinically heterogeneous; however, all individuals are reported to have abnormal neurodevelopment. De novo CHD4 variants, including missense and an in-frame deletion, have been identified in individuals with non-specific neurodevelopmental disorders and in a cohort of individuals with congenital heart defects (Sifrim et al., 2016; Weiss et al., 2016), and some of these individuals have structural brain anomalies such as macrocephaly and ventriculomegaly. The underlying molecular pathologies of the CHD4 pathogenic variants observed in humans are under intense study. Functional studies in HEK293 cells showed that pathogenic CHD4 variants in the C-terminal helicase domain (p.Arg1127Gln and p.Arg1173Leu) do not alter the ability of the mutant CHD4 protein to localize to the nucleus and interact with Histone Deacetylase 1 (HDAC1) (Weiss et al., 2016). Conditional deletion of Chd4 in mice results in mild microcephaly, contrasting the macrocephaly noted in some individuals with CHD4 pathogenic variants (Nitarska et al., 2016). The discordance between human and mouse CHD4 deficiency phenotypes suggests either that this model system does not fully recapitulate human brain development or that the molecular pathology of human CHD4 pathogenic variants deviates from loss-of-function.
CHD5 is highly expressed in the nervous system and is regulated by retinoic acid in neuroblastoma cells (Egan et al., 2013; Higashi et al., 2015). CHD5 is a tumor suppressor in the p19(Arf)/p53 pathway controlling cell proliferation, apoptosis, and senescence (Bagchi et al., 2007).
3.4.3 The CHD Family Subclass III
CHD subfamily III contains CHD6, CHD7, CHD8, and CHD9. Subclass III CHD proteins are unique from the other chromodomain proteins in having a SANT domain and two BRK domains C-terminal to the helicase domain. The SANT domain is conserved among many regulators of transcription and chromatin structure, and is believed to function as a histone tail binding module (Boyer et al., 2004). The BRK domain is found only in CHD subclass III proteins, in the catalytic subunit of the SWI/SNF complex, and in Drosophila brahma and kismet (Daubresse et al., 1999; Dorighi and Tamkun, 2013). BRK domains are proposed to reorganize chromatin structure via formation of a chromodomain aromatic cage similar to CHD subfamily I, suggesting BRK may also participate in binding methylated lysine residues (Daubresse et al., 1999; Doerks et al., 2002).
The Drosophila kismet (kis) gene is highly related to mammalian CHD class III members. The kismet gene was identified in a genetic screen for dominant suppressors of polycomb, a group of genes that act as transcriptional repressors of homeotic genes (Daubresse et al., 1999). Loss of kismet results in homeotic transformations, suggesting that kismet is a member of the TrxG of gene activators. Brahma, the catalytic subunit of the Drosophila SWI/SNF complex, may also function by a similar mechanism (Boyer et al., 2004; Daubresse et al., 1999; Kennison and Tamkun, 1988; Tamkun et al., 1992). Loss of maternal kismet leads to significant defects in larval body segmentation, and expression of the segment polarity gene engrailed is significantly altered in kismet mutant flies, indicating that engrailed and other genes important for proper segmentation require normal kismet function (Daubresse et al., 1999).
Of the four CHD subfamily III members, only CHD7 and CHD8 have been shown to be associated with human genetic disease. In humans, heterozygous CHD7 pathogenic variants cause CHARGE syndrome, a clinically variable, multiple congenital anomaly condition affecting development of the inner ear, eyes, heart, choanae (the region between the oropharynx and nasal passages), genitalia, nervous system, and craniofacial structures including the hard and soft palates, lip, external ear, midface, and olfactory system (Hall, 1979; Vissers et al., 2004). CHARGE is a common cause of deaf-blindness, balance disorders, and congenital heart malformations, with an estimated incidence of 1:8500–1:12,000 in newborns (Harris et al., 1997; Issekutz et al., 2005; Kallen et al., 1999). Heterozygous nonsense, deletion, or missense CHD7 pathogenic variants are estimated to occur up to 90% of patients with CHARGE (Aramaki et al., 2006; Jongmans et al., 2006; Lalani et al., 2006; Sanlaville et al., 2005; Vissers et al., 2004). CHD7 pathogenic variants are distributed throughout the coding sequence and do not correlate with specific aspects of the clinical phenotype. In addition, most human CHD7 pathogenic variants identified thus far are de novo; however, evidence for germline mosaicism has been reported in a family with affected siblings (Jongmans et al., 2006) and in a father of two children with CHARGE syndrome (Pauli et al., 2009).
CHD7 preferentially binds to enhancers and transcription start sites, some of which are marked by methylation of H3K4 (Scacheri et al., 2006). In addition, CHD7 regulates rRNA transcription along with key transcription factors and signaling molecules that control neurogenesis (Basson and van Ravenswaaij-Arts, 2015; Feng et al., 2013; Jones et al., 2015; Layman et al., 2011; Layman et al., 2009; Micucci et al., 2014; Whittaker et al., 2017; Zentner et al., 2010). Ethylnitrosourea (ENU) mutagenesis projects have led to characterization of nine different lines of Chd7 mutant mice, each with an identifiable single base pair Chd7 pathogenic variant (Bosman et al., 2005; Nolan et al., 1995). These Chd7 mutant mice are viable and exhibit hyperactivity, head bobbing, circling behaviors, disrupted lateral semicircular canals, reduced postnatal growth, variable cleft palate, choanal atresia, cardiac septal defects, hemorrhage, prenatal death, genital abnormalities, and keratoconjunctivitis sicca (dry eye) (Bosman et al., 2005; Hawker et al., 2005; Kiernan et al., 2002; Nolan et al., 1995; Pickard et al., 1995). Another mutant mouse (Wheels) has a similar phenotype and maps to the same region of mouse chromosome 4 but is not known to harbor a Chd7 pathogenic variant (Alavizadeh et al., 2001; Bosman et al., 2005; Nolan et al., 1995; Pickard et al., 1995). Our laboratory generated and characterized a lacZ-expressing, Chd7 gene trap null allele (Chd7Gt) that results in homozygous intrauterine lethality by E11.5 and heterozygous phenotypes similar to other Chd7 mutant mice (Hurd et al., 2007). Chd7Gt/+ mice also have the advantage of expressing β-galactosidase (β-gal) from a null Chd7 allele and can be used to track Chd7 mutant cells. Together, these observations demonstrate that CHD7 is involved in epigenetic regulation of gene transcription during development and CHD7 deficiency causes similar phenotypes in mice and humans.
CHD8 was recently identified as a novel candidate gene for Autism Spectrum Disorder (ASD) (Bernier et al., 2014; O’Roak et al., 2012a; O’Roak et al., 2012b; Zahir et al., 2007). Although ASD is genetically heterogeneous, many studies utilizing whole exome sequencing and molecular inversion probe sequencing methods have demonstrated that individuals with ASD show enrichment for both truncating and non-truncating CHD8 variants (Bernier et al., 2014; De Rubeis et al., 2014; Neale et al., 2012; O’Roak et al., 2014; O’Roak et al., 2012a). Interestingly, individuals with CHD8 pathogenic variants often have macrocephaly, facial dysmorphisms and gastrointestinal dysfunction, suggesting important roles for CHD8 related chromatin remodeling in brain and craniofacial development.
The combination of both ASD and macrocephaly phenotypes has sparked many studies utilizing animal model systems to determine the biological function of CHD8 during neurodevelopment. Knockout of Chd8 is embryonic lethal, and both knockdown and heterozygous germline editing approaches have been utilized to study morphological features attributed to CHD8 depletion (Nishiyama et al., 2004; Nishiyama et al., 2009). Studies utilizing zebrafish with morpholino knockdown of chd8 and generation of chd8 microdeletions by CRISPR-Cas9 were shown to recapitulate macrocephaly, the distinct facial feature of increased distance between the eyes, and a gastrointestinal phenotype observed in humans with CHD8 pathogenic variants (Bernier et al., 2014). Similarly, reports of germline edited heterozygous Chd8 mutant mice have noted increases in brain volume, ASD-related behaviors, increased distance between eyes, and gastrointestinal defects (Katayama et al., 2016; Platt et al., 2017). Taken together, the highly consistent morphological features between individuals with CHD8 pathogenic variants and animal models highlight the importance of CHD8 for building a normal brain.
4. Common Targets and Potential Therapies for Epigenetic Diseases
Collectively, disruptions of histone methyltransferases and chromatin remodelers impact epigenetic regulation of gene transcription, suggesting there may be common genetic targets and potential therapies with broad application for these conditions. One promising example of such a therapy is the use of topoisomerase inhibitors to de-repress the silenced allele in Angelman syndrome (Huang et al., 2012). In CHARGE syndrome, the majority of pathogenic variants result from disruption of a single copy of CHD7 (Janssen et al., 2012). Thus, therapies that (a) alter histone modifying activities, (b) increase CHD7 expression, or (c) counteract changes in downstream gene expression may be relevant for CHARGE and other TrxG protein-related disorders. A recent study showed that CHD7 interacts with Topoisomerase 2b (Feng et al., 2013), suggesting that regulation of Topoisomerase may also be an effective approach for conditions where CHD7 is dysfunctional or absent. For those epigenetic disorders caused by haploinsufficiency, enhancement of expression or functionality of the remaining wild type allele could improve chromatin recruitment and remodeling activities. Pharmacologic agents that target epigenetic regulators may prove to be particularly effective for neurodevelopmental disorders. Additional research is needed to identify novel epigenetic mechanisms underlying brain, craniofacial, neural, and embryonic development. Such studies could provide critical information about chromatin remodeler and histone methyltransferase target genes and regulatory complexes, and will help lay the foundation for further mechanistic studies of histone modifications, nucleosome positioning and basic chromatin biology of cellular proliferation and differentiation.
5. Conclusions
Human homologs of TrxG genes encoding methyltransferases, demethylases, and chromatin remodelers contribute vital regulation of the epigenetic landscape in the cell nucleus. In turn, the corresponding chromatin configurations influence the dynamic states of gene expression, which ultimately dictate proliferative outcomes and cell fates. Pathogenic variants impacting TrxG-related genes occur in a variety of human disorders in which the brain is commonly affected. These genetic disorders display a high degree of phenotypic overlap, suggesting that similar biological pathways or stages of development are impacted. In this review, we have highlighted human studies and animal models which have established the molecular pathologies of disease from altered TrxG, COMPASS, and ATP-dependent chromatin remodelers. Results from these studies have set a foundation for future explorations of the mechanisms controlling the histone code, accessibility of DNA to trans-acting factors, and gene expression.
Highlights.
Epigenetic regulators and chromatin remodelers influence chromatin accessibility.
Human homologs of Trithorax related genes are associated with neurodevelopmental disorders.
Animal models and biochemical studies highlight roles for ATP-dependent chromatin remodeling in brain development.
Acknowledgments
A.M. is supported by the Michigan Predoctoral Training in Genetics (NIH T32GM007544). D.M.M. is supported by NIH R01 DC009410, R01 DC014456, and by the Donita B. Sullivan, MD Research Professorship in Pediatrics and Communicable Disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aasland R, Stewart AF, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- Alavizadeh A, Kiernan AE, Nolan P, Lo C, Steel KP, Bucan M. The Wheels mutation in the mouse causes vascular, hindbrain, and inner ear defects. Dev Biol. 2001;234:244–260. doi: 10.1006/dbio.2001.0241. [DOI] [PubMed] [Google Scholar]
- Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, Hijazi H, Alshammari M, Aldahmesh MA, Salih MA, Faqeih E, Alhashem A, Bashiri FA, Al-Owain M, Kentab AY, Sogaty S, Al Tala S, Temsah MH, Tulbah M, Aljelaify RF, Alshahwan SA, Seidahmed MZ, Alhadid AA, Aldhalaan H, AlQallaf F, Kurdi W, Alfadhel M, Babay Z, Alsogheer M, Kaya N, Al-Hassnan ZN, Abdel-Salam GM, Al-Sannaa N, Al Mutairi F, El Khashab HY, Bohlega S, Jia X, Nguyen HC, Hammami R, Adly N, Mohamed JY, Abdulwahab F, Ibrahim N, Naim EA, Al-Younes B, Meyer BF, Hashem M, Shaheen R, Xiong Y, Abouelhoda M, Aldeeri AA, Monies DM, Alkuraya FS. Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families. Cell Rep. 2015;10:148–161. doi: 10.1016/j.celrep.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Aramaki M, Udaka T, Kosaki R, Makita Y, Okamoto N, Yoshihashi H, Oki H, Nanao K, Moriyama N, Oku S, Hasegawa T, Takahashi T, Fukushima Y, Kawame H, Kosaki K. Phenotypic spectrum of CHARGE syndrome with CHD7 mutations. J Pediatr. 2006;148:410–414. doi: 10.1016/j.jpeds.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, Bredel M, Vogel H, Mills AA. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, Elsasser SJ, Chapgier A, Goldberg AD, Canaani E, Rafii S, Zheng D, Allis CD. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–120. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banting GS, Barak O, Ames TM, Burnham AC, Kardel MD, Cooch NS, Davidson CE, Godbout R, McDermid HE, Shiekhattar R. CECR2, a protein involved in neurulation, forms a novel chromatin remodeling complex with SNF2L. Hum Mol Genet. 2005;14:513–524. doi: 10.1093/hmg/ddi048. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Basson MA, van Ravenswaaij-Arts C. Functional Insights into Chromatin Remodelling from Studies on CHARGE Syndrome. Trends Genet. 2015;31:600–611. doi: 10.1016/j.tig.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basta J, Rauchman M. The nucleosome remodeling and deacetylase complex in development and disease. Transl Res. 2015;165:36–47. doi: 10.1016/j.trsl.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB, Hörz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, Vulto-van Silfhout AT, Schuurs-Hoeijmakers JH, Fichera M, Bosco P, Buono S, Alberti A, Failla P, Peeters H, Steyaert J, Vissers LE, Francescatto L, Mefford HC, Rosenfeld JA, Bakken T, O’Roak BJ, Pawlus M, Moon R, Shendure J, Amaral DG, Lein E, Rankin J, Romano C, de Vries BB, Katsanis N, Eichler EE. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Bosman EA, Penn AC, Ambrose JC, Kettleborough R, Stemple DL, Steel KP. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum Mol Genet. 2005;14:3463–3476. doi: 10.1093/hmg/ddi375. [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Fujita N, Kajita M, Wade PA. Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta. 2004;1677:52–57. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Latek RR, Peterson CL. The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- Bozhenok L, Wade PA, Varga-Weisz P. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 2002;21:2231–2241. doi: 10.1093/emboj/21.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annual review of genetics. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Capelli LP, Krepischi AC, Gurgel-Giannetti J, Mendes MF, Rodrigues T, Varela MC, Koiffmann CP, Rosenberg C. Deletion of the RMGA and CHD2 genes in a child with epilepsy and mental deficiency. Eur J Med Genet. 2012;55:132–134. doi: 10.1016/j.ejmg.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Carvill GL, Heavin SB, Yendle SC, McMahon JM, O’Roak BJ, Cook J, Khan A, Dorschner MO, Weaver M, Calvert S, Malone S, Wallace G, Stanley T, Bye AM, Bleasel A, Howell KB, Kivity S, Mackay MT, Rodriguez-Casero V, Webster R, Korczyn A, Afawi Z, Zelnick N, Lerman-Sagie T, Lev D, Moller RS, Gill D, Andrade DM, Freeman JL, Sadleir LG, Shendure J, Berkovic SF, Scheffer IE, Mefford HC. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet. 2013;45:825–830. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenier S, Yoon G, Argiropoulos B, Lauzon J, Laframboise R, Ahn JW, Ogilvie CM, Lionel AC, Marshall CR, Vaags AK, Hashemi B, Boisvert K, Mathonnet G, Tihy F, So J, Scherer SW, Lemyre E, Stavropoulos DJ. CHD2 haploinsufficiency is associated with developmental delay, intellectual disability, epilepsy and neurobehavioural problems. Journal of neurodevelopmental disorders. 2014;6:9. doi: 10.1186/1866-1955-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubresse G, Deuring R, Moore L, Papoulas O, Zakrajsek I, Waldrip WR, Scott MP, Kennison JA, Tamkun JW. The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development. 1999;126:1175–1187. doi: 10.1242/dev.126.6.1175. [DOI] [PubMed] [Google Scholar]
- de Dieuleveult M, Yen K, Hmitou I, Depaux A, Boussouar F, Bou Dargham D, Jounier S, Humbertclaude H, Ribierre F, Baulard C, Farrell NP, Park B, Keime C, Carriere L, Berlivet S, Gut M, Gut I, Werner M, Deleuze JF, Olaso R, Aude JC, Chantalat S, Pugh BF, Gerard M. Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature. 2016;530:113–116. doi: 10.1038/nature16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Shih-Chen F, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Curran SR, Dawson G, Duketis E, Fernandez BA, Gallagher L, Geller E, Guter SJ, Hill RS, Ionita-Laza J, Jimenz Gonzalez P, Kilpinen H, Klauck SM, Kolevzon A, Lee I, Lei I, Lei J, Lehtimaki T, Lin CF, Ma’ayan A, Marshall CR, McInnes AL, Neale B, Owen MJ, Ozaki N, Parellada M, Parr JR, Purcell S, Puura K, Rajagopalan D, Rehnstrom K, Reichenberg A, Sabo A, Sachse M, Sanders SJ, Schafer C, Schulte-Ruther M, Skuse D, Stevens C, Szatmari P, Tammimies K, Valladares O, Voran A, Li-San W, Weiss LA, Willsey AJ, Yu TW, Yuen RK, Study DDD, Cook EH, Freitag CM, Gill M, Hultman CM, Lehner T, Palotie A, Schellenberg GD, Sklar P, State MW, Sutcliffe JS, Walsh CA, Scherer SW, Zwick ME, Barett JC, Cutler DJ, Roeder K, Devlin B, Daly MJ, Buxbaum JD Homozygosity Mapping Collaborative for A, Consortium UK. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas V, Stokes DG, Perry RP. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc Natl Acad Sci U S A. 1993;90:2414–2418. doi: 10.1073/pnas.90.6.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T, Copley RR, Schultz J, Ponting CP, Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorighi KM, Tamkun JW. The trithorax group proteins Kismet and ASH1 promote H3K36 dimethylation to counteract Polycomb group repression in Drosophila. Development. 2013;140:4182–4192. doi: 10.1242/dev.095786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, Young RA. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Becker PB. ATP-dependent nucleosome remodelling: factors and functions. J Cell Sci. 2004;117:3707–3711. doi: 10.1242/jcs.01175. [DOI] [PubMed] [Google Scholar]
- Egan CM, Nyman U, Skotte J, Streubel G, Turner S, O’Connell DJ, Rraklli V, Dolan MJ, Chadderton N, Hansen K, Farrar GJ, Helin K, Holmberg J, Bracken AP. CHD5 Is Required for Neurogenesis and Has a Dual Role in Facilitating Gene Expression and Polycomb Gene Repression. Dev Cell. 2013;26:223–236. doi: 10.1016/j.devcel.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Epi KC, Allen AS, Berkovic SF, Cossette P, Delanty N, Dlugos D, Eichler EE, Epstein MP, Glauser T, Goldstein DB, Han Y, Heinzen EL, Hitomi Y, Howell KB, Johnson MR, Kuzniecky R, Lowenstein DH, Lu YF, Madou MR, Marson AG, Mefford HC, Esmaeeli Nieh S, O’Brien TJ, Ottman R, Petrovski S, Poduri A, Ruzzo EK, Scheffer IE, Sherr EH, Yuskaitis CJ, Abou-Khalil B, Alldredge BK, Bautista JF, Berkovic SF, Boro A, Cascino GD, Consalvo D, Crumrine P, Devinsky O, Dlugos D, Epstein MP, Fiol M, Fountain NB, French J, Friedman D, Geller EB, Glauser T, Glynn S, Haut SR, Hayward J, Helmers SL, Joshi S, Kanner A, Kirsch HE, Knowlton RC, Kossoff EH, Kuperman R, Kuzniecky R, Lowenstein DH, McGuire SM, Motika PV, Novotny EJ, Ottman R, Paolicchi JM, Parent JM, Park K, Poduri A, Scheffer IE, Shellhaas RA, Sherr EH, Shih JJ, Singh R, Sirven J, Smith MC, Sullivan J, Lin Thio L, Venkat A, Vining EP, Von Allmen GK, Weisenberg JL, Widdess-Walsh P, Winawer MR Epilepsy Phenome/Genome P. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrner JA, Bjornsson HT. Mendelian disorders of the epigenetic machinery: tipping the balance of chromatin states. Annu Rev Genomics Hum Genet. 2014;15:269–293. doi: 10.1146/annurev-genom-090613-094245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Khan MA, Bellvis P, Zhu Z, Bernhardt O, Herold-Mende C, Liu HK. The Chromatin Remodeler CHD7 Regulates Adult Neurogenesis via Activation of SoxC Transcription Factors. Cell Stem Cell. 2013;13:62–72. doi: 10.1016/j.stem.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Flanagan JF, Blus BJ, Kim D, Clines KL, Rastinejad F, Khorasanizadeh S. Molecular implications of evolutionary differences in CHD double chromodomains. J Mol Biol. 2007;369:334–342. doi: 10.1016/j.jmb.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footz TK, Brinkman-Mills P, Banting GS, Maier SA, Riazi MA, Bridgland L, Hu S, Birren B, Minoshima S, Shimizu N, Pan H, Nguyen T, Fang F, Fu Y, Ray L, Wu H, Shaull S, Phan S, Yao Z, Chen F, Huan A, Hu P, Wang Q, Loh P, Qi S, Roe BA, McDermid HE. Analysis of the cat eye syndrome critical region in humans and the region of conserved synteny in mice: a search for candidate genes at or near the human chromosome 22 pericentromere. Genome Res. 2001;11:1053–1070. doi: 10.1101/gr.154901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Q, Nilasena DS, O’Brien CA, Frank MB, Targoff IN. Molecular analysis of a major antigenic region of the 240-kD protein of Mi-2 autoantigen. J Clin Invest. 1995;96:1730–1737. doi: 10.1172/JCI118218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune T, Brzeski J, Eberharter A, Clapier CR, Corona DF, Becker PB, Muller CW. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell. 2003;12:449–460. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- Guo DC, Duan XY, Regalado ES, Mellor-Crummey L, Kwartler CS, Kim D, Lieberman K, de Vries BB, Pfundt R, Schinzel A, Kotzot D, Shen X, Yang ML, Bamshad MJ, Nickerson DA, Gornik HL, Ganesh SK, Braverman AC, Grange DK, Milewicz DM University of Washington Center for Mendelian G. Loss-of-Function Mutations in YY1AP1 Lead to Grange Syndrome and a Fibromuscular Dysplasia-Like Vascular Disease. Am J Hum Genet. 2017;100:21–30. doi: 10.1016/j.ajhg.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BD. Choanal atresia and associated multiple anomalies. J Pediatr. 1979;95:395–398. doi: 10.1016/s0022-3476(79)80513-2. [DOI] [PubMed] [Google Scholar]
- Harris J, Robert E, Kallen B. Epidemiology of choanal atresia with special reference to the CHARGE association. Pediatrics. 1997;99:363–367. doi: 10.1542/peds.99.3.363. [DOI] [PubMed] [Google Scholar]
- Hawker K, Fuchs H, Angelis MH, Steel KP. Two new mouse mutants with vestibular defects that map to the highly mutable locus on chromosome 4. Int J Audiol. 2005;44:171–177. doi: 10.1080/14992020500057434. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Herzog VA, Lempradl A, Trupke J, Okulski H, Altmutter C, Ruge F, Boidol B, Kubicek S, Schmauss G, Aumayr K, Ruf M, Pospisilik A, Dimond A, Senergin HB, Vargas ML, Simon JA, Ringrose L. A strand-specific switch in noncoding transcription switches the function of a Polycomb/Trithorax response element. Nat Genet. 2014;46:973–981. doi: 10.1038/ng.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi M, Kolla V, Iyer R, Naraparaju K, Zhuang T, Kolla S, Brodeur GM. Retinoic acid-induced CHD5 upregulation and neuronal differentiation of neuroblastoma. Mol Cancer. 2015;14:150. doi: 10.1186/s12943-015-0425-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann OR, Misura K, Lamas E, Sandrock RW, Nelson P, McDonough SI, DeLisi LE. Whole-genome sequencing in multiplex families with psychoses reveals mutations in the SHANK2 and SMARCA1 genes segregating with illness. Molecular psychiatry. 2016;21:1690–1695. doi: 10.1038/mp.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RL, Lines MA, Nikkel SM, Schwartzentruber J, Beaulieu C, Nowaczyk MJ, Allanson J, Kim CA, Wieczorek D, Moilanen JS, Lacombe D, Gillessen-Kaesbach G, Whiteford ML, Quaio CR, Gomy I, Bertola DR, Albrecht B, Platzer K, McGillivray G, Zou R, McLeod DR, Chudley AE, Chodirker BN, Marcadier J, Consortium FC, Majewski J, Bulman DE, White SM, Boycott KM. Mutations in SRCAP, encoding SNF2-related CREBBP activator protein, cause Floating-Harbor syndrome. Am J Hum Genet. 2012;90:308–313. doi: 10.1016/j.ajhg.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RL, Schenkel LC, Nikkel SM, Ainsworth PJ, Pare G, Boycott KM, Bulman DE, Sadikovic B. The defining DNA methylation signature of Floating-Harbor Syndrome. Sci Rep. 2016;6:38803. doi: 10.1038/srep38803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, Taylor-Blake B, Sciaky N, Dutton JW, Jr, Lee HM, Chen X, Jin J, Bridges AS, Zylka MJ, Roth BL, Philpot BD. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481:185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd EA, Capers PL, Blauwkamp MN, Adams ME, Raphael Y, Poucher HK, Martin DM. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm Genome. 2007;18:94–104. doi: 10.1007/s00335-006-0107-6. [DOI] [PubMed] [Google Scholar]
- Ingham PW. Differential expression of bithorax complex genes in the absence of the extra sex combs and trithorax genes. Nature. 1983;306:591–593. doi: 10.1038/306591a0. [DOI] [PubMed] [Google Scholar]
- Issekutz KA, Graham JM, Jr, Prasad C, Smith IM, Blake KD. An epidemiological analysis of CHARGE syndrome: preliminary results from a Canadian study. Am J Med Genet A. 2005;133:309–317. doi: 10.1002/ajmg.a.30560. [DOI] [PubMed] [Google Scholar]
- Janssen N, Bergman JE, Swertz MA, Tranebjaerg L, Lodahl M, Schoots J, Hofstra RM, van Ravenswaaij-Arts CM, Hoefsloot LH. Mutation update on the CHD7 gene involved in CHARGE syndrome. Hum Mutat. 2012;33:1149–1160. doi: 10.1002/humu.22086. [DOI] [PubMed] [Google Scholar]
- Jones KM, Saric N, Russell JP, Andoniadou CL, Scambler PJ, Basson MA. CHD7 maintains neural stem cell quiescence and prevents premature stem cell depletion in the adult hippocampus. Stem Cells. 2015;33:196–210. doi: 10.1002/stem.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WD, Dafou D, McEntagart M, Woollard WJ, Elmslie FV, Holder-Espinasse M, Irving M, Saggar AK, Smithson S, Trembath RC, Deshpande C, Simpson MA. De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am J Hum Genet. 2012;91:358–364. doi: 10.1016/j.ajhg.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongmans MCJ, Admiraal RJ, van der Donk KP, Vissers LELM, Baas AF, Kapusta L, van Hagen JM, Donnai D, de Ravel TJ, Veltman JA, van Kessel AG, De Vries BBA, Brunner HG, Hoefsloot LH, van Ravenswaaij CMA. CHARGE syndrome: the phenotypic spectrum of mutations in the CHD7 gene. Journal of Medical Genetics. 2006;43:306–314. doi: 10.1136/jmg.2005.036061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen K, Robert E, Mastroiacovo P, Castilla EE, Kallen B. CHARGE Association in newborns: a registry-based study. Teratology. 1999;60:334–343. doi: 10.1002/(SICI)1096-9926(199912)60:6<334::AID-TERA5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Karaca E, Harel T, Pehlivan D, Jhangiani SN, Gambin T, Coban Akdemir Z, Gonzaga-Jauregui C, Erdin S, Bayram Y, Campbell IM, Hunter JV, Atik MM, Van Esch H, Yuan B, Wiszniewski W, Isikay S, Yesil G, Yuregir OO, Tug Bozdogan S, Aslan H, Aydin H, Tos T, Aksoy A, De Vivo DC, Jain P, Geckinli BB, Sezer O, Gul D, Durmaz B, Cogulu O, Ozkinay F, Topcu V, Candan S, Cebi AH, Ikbal M, Yilmaz Gulec E, Gezdirici A, Koparir E, Ekici F, Coskun S, Cicek S, Karaer K, Koparir A, Duz MB, Kirat E, Fenercioglu E, Ulucan H, Seven M, Guran T, Elcioglu N, Yildirim MS, Aktas D, Alikasifoglu M, Ture M, Yakut T, Overton JD, Yuksel A, Ozen M, Muzny DM, Adams DR, Boerwinkle E, Chung WK, Gibbs RA, Lupski JR. Genes that Affect Brain Structure and Function Identified by Rare Variant Analyses of Mendelian Neurologic Disease. Neuron. 2015;88:499–513. doi: 10.1016/j.neuron.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y, Nishiyama M, Shoji H, Ohkawa Y, Kawamura A, Sato T, Suyama M, Takumi T, Miyakawa T, Nakayama KI. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016;537:675–679. doi: 10.1038/nature19357. [DOI] [PubMed] [Google Scholar]
- Kennison JA, Tamkun JW. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci U S A. 1988;85:8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE, Erven A, Voegeling S, Peters J, Nolan P, Hunter J, Bacon Y, Steel KP, Brown SD, Guenet JL. ENU mutagenesis reveals a highly mutable locus on mouse Chromosome 4 that affects ear morphogenesis. Mamm Genome. 2002;13:142–148. doi: 10.1007/BF02684018. [DOI] [PubMed] [Google Scholar]
- Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani SR, Safiullah AM, Fernbach SD, Harutyunyan KG, Thaller C, Peterson LE, McPherson JD, Gibbs RA, White LD, Hefner M, Davenport SL, Graham JM, Bacino CA, Glass NL, Towbin JA, Craigen WJ, Neish SR, Lin AE, Belmont JW. Spectrum of CHD7 Mutations in 110 Individuals with CHARGE Syndrome and Genotype-Phenotype Correlation. Am J Hum Genet. 2006;78:303–314. doi: 10.1086/500273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layman WS, Hurd EA, Martin DM. Reproductive dysfunction and decreased GnRH neurogenesis in a mouse model of CHARGE syndrome. Hum Mol Genet. 2011;20:3138–3150. doi: 10.1093/hmg/ddr216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layman WS, McEwen DP, Beyer LA, Lalani SR, Fernbach SD, Oh E, Swaroop A, Hegg CC, Raphael Y, Martens JR, Martin DM. Defects in neural stem cell proliferation and olfaction in Chd7 deficient mice indicate a mechanism for hyposmia in human CHARGE syndrome. Hum Mol Genet. 2009;18:1909–1923. doi: 10.1093/hmg/ddp112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JC, Gerreira CG, Yusufzai T. Human CHD2 Is a Chromatin Assembly ATPase Regulated by Its Chromo- and DNA-binding Domains. The Journal of Biochemistry. 2015;290:25–34. doi: 10.1074/jbc.M114.609156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes F, Barbosa M, Ameur A, Soares G, de Sa J, Dias AI, Oliveira G, Cabral P, Temudo T, Calado E, Cruz IF, Vieira JP, Oliveira R, Esteves S, Sauer S, Jonasson I, Syvanen AC, Gyllensten U, Pinto D, Maciel P. Identification of novel genetic causes of Rett syndrome-like phenotypes. J Med Genet. 2016;53:190–199. doi: 10.1136/jmedgenet-2015-103568. [DOI] [PubMed] [Google Scholar]
- Lopez AJ, Wood MA. Role of nucleosome remodeling in neurodevelopmental and intellectual disability disorders. Frontiers in behavioral neuroscience. 2015;9:100. doi: 10.3389/fnbeh.2015.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Meng X, Morris CA, Keating MT. A novel human gene, WSTF, is deleted in Williams syndrome. Genomics. 1998;54:241–249. doi: 10.1006/geno.1998.5578. [DOI] [PubMed] [Google Scholar]
- Lund C, Brodtkorb E, Oye AM, Rosby O, Selmer KK. CHD2 mutations in Lennox-Gastaut syndrome. Epilepsy Behav. 2014;33:18–21. doi: 10.1016/j.yebeh.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. Bioessays. 2003;25:1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- Manning BJ, Yusufzai T. The ATP-dependent Chromatin Remodeling Enzymes CHD6, CHD7, and CHD8 Exhibit Distinct Nucleosome Binding and Remodeling Activities. J Biol Chem. 2017 doi: 10.1074/jbc.M117.779470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella CG, Ohkawa Y, Coles AH, Garlick DS, Jones SN, Imbalzano AN. Mutation of the SNF2 family member Chd2 affects mouse development and survival. J Cell Physiol. 2006;209:162–171. doi: 10.1002/jcp.20718. [DOI] [PubMed] [Google Scholar]
- Mellor J. Imitation switch complexes. Ernst Schering Res Found Workshop. 2006:61–87. doi: 10.1007/3-540-37633-x_4. [DOI] [PubMed] [Google Scholar]
- Mendelsohn BA, Pronold M, Long R, Smaoui N, Slavotinek AM. Advanced bone age in a girl with Wiedemann-Steiner syndrome and an exonic deletion in KMT2A (MLL) Am J Med Genet A. 2014;164A:2079–2083. doi: 10.1002/ajmg.a.36590. [DOI] [PubMed] [Google Scholar]
- Micucci JA, Layman WS, Hurd EA, Sperry ED, Frank SF, Durham MA, Swiderski DL, Skidmore JM, Scacheri PC, Raphael Y, Martin DM. CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of CHARGE syndrome. Hum Mol Genet. 2014;23:434–448. doi: 10.1093/hmg/ddt435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, Polak P, Yoon S, Maguire J, Crawford EL, Campbell NG, Geller ET, Valladares O, Schafer C, Liu H, Zhao T, Cai G, Lihm J, Dannenfelser R, Jabado O, Peralta Z, Nagaswamy U, Muzny D, Reid JG, Newsham I, Wu Y, Lewis L, Han Y, Voight BF, Lim E, Rossin E, Kirby A, Flannick J, Fromer M, Shakir K, Fennell T, Garimella K, Banks E, Poplin R, Gabriel S, DePristo M, Wimbish JR, Boone BE, Levy SE, Betancur C, Sunyaev S, Boerwinkle E, Buxbaum JD, Cook EH, Jr, Devlin B, Gibbs RA, Roeder K, Schellenberg GD, Sutcliffe JS, Daly MJ. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreiter AR, Varshney A, Parker SCJ, Martin DM. Super Enhancers in Cancers, Complex Disease, and Developmental Disorders. Genes. 2015;6:1183–1200. doi: 10.3390/genes6041183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikkel SM, Dauber A, de Munnik S, Connolly M, Hood RL, Caluseriu O, Hurst J, Kini U, Nowaczyk MJ, Afenjar A, Albrecht B, Allanson JE, Balestri P, Ben-Omran T, Brancati F, Cordeiro I, da Cunha BS, Delaney LA, Destree A, Fitzpatrick D, Forzano F, Ghali N, Gillies G, Harwood K, Hendriks YM, Heron D, Hoischen A, Honey EM, Hoefsloot LH, Ibrahim J, Jacob CM, Kant SG, Kim CA, Kirk EP, Knoers NV, Lacombe D, Lee C, Lo IF, Lucas LS, Mari F, Mericq V, Moilanen JS, Moller ST, Moortgat S, Pilz DT, Pope K, Price S, Renieri A, Sa J, Schoots J, Silveira EL, Simon ME, Slavotinek A, Temple IK, van der Burgt I, de Vries BB, Weisfeld-Adams JD, Whiteford ML, Wierczorek D, Wit JM, Yee CF, Beaulieu CL, Consortium FC, White SM, Bulman DE, Bongers E, Brunner H, Feingold M, Boycott KM. The phenotype of Floating-Harbor syndrome: clinical characterization of 52 individuals with mutations in exon 34 of SRCAP. Orphanet J Rare Dis. 2013;8:63. doi: 10.1186/1750-1172-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Nakayama K, Tsunematsu R, Tsukiyama T, Kikuchi A, Nakayama KI. Early embryonic death in mice lacking the beta-catenin-binding protein Duplin. Mol Cell Biol. 2004;24:8386–8394. doi: 10.1128/MCB.24.19.8386-8394.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Oshikawa K, Tsukada Y, Nakagawa T, Iemura S, Natsume T, Fan Y, Kikuchi A, Skoultchi AI, Nakayama KI. CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nature cell biology. 2009;11:172–182. doi: 10.1038/ncb1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitarska J, Smith JG, Sherlock WT, Hillege MM, Nott A, Barshop WD, Vashisht AA, Wohlschlegel JA, Mitter R, Riccio A. A Functional Switch of NuRD Chromatin Remodeling Complex Subunits Regulates Mouse Cortical Development. Cell Rep. 2016;17:1683–1698. doi: 10.1016/j.celrep.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan PM, Sollars PJ, Bohne BA, Ewens WJ, Pickard GE, Bucan M. Heterozygosity mapping of partially congenic lines: mapping of a semidominant neurological mutation, Wheels (Whl), on mouse chromosome 4. Genetics. 1995;140:245–254. doi: 10.1093/genetics/140.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Stessman HA, Boyle EA, Witherspoon KT, Martin B, Lee C, Vives L, Baker C, Hiatt JB, Nickerson DA, Bernier R, Shendure J, Eichler EE. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nature communications. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, Munson J, Hiatt JB, Turner EH, Levy R, O’Day DR, Krumm N, Coe BP, Martin BK, Borenstein E, Nickerson DA, Mefford HC, Doherty D, Akey JM, Bernier R, Eichler EE, Shendure J. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012a;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, Turner EH, Stanaway IB, Vernot B, Malig M, Baker C, Reilly B, Akey JM, Borenstein E, Rieder MJ, Nickerson DA, Bernier R, Shendure J, Eichler EE. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012b;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Paro R, Hogness DS. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci U S A. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli S, Pieper L, Haberle J, Grzmil P, Burfeind P, Steckel M, Lenz U, Michelmann HW. Proven germline mosaicism in a father of two children with CHARGE syndrome. Clin Genet. 2009;75:473–479. doi: 10.1111/j.1399-0004.2009.01151.x. [DOI] [PubMed] [Google Scholar]
- Peoples RJ, Cisco MJ, Kaplan P, Francke U. Identification of the WBSCR9 gene, encoding a novel transcriptional regulator, in the Williams-Beuren syndrome deletion at 7q11.23. Cytogenet Cell Genet. 1998;82:238–246. doi: 10.1159/000015110. [DOI] [PubMed] [Google Scholar]
- Pickard GE, Sollars PJ, Rinchik EM, Nolan PM, Bucan M. Mutagenesis and behavioral screening for altered circadian activity identifies the mouse mutant, Wheels. Brain Res. 1995;705:255–266. doi: 10.1016/0006-8993(95)01171-4. [DOI] [PubMed] [Google Scholar]
- Pilarowski GO, Vernon HJ, Applegate CD, Boukas L, Cho MT, Gurnett CA, Benke PJ, Beaver E, Heeley JM, Medne L, Krantz ID, Azage M, Niyazov D, Henderson LB, Wentzensen IM, Baskin B, Sacoto MJG, Bowman GD, Bjornsson HT. Missense variants in the chromatin remodeler CHD1 are associated with neurodevelopmental disability. J Med Genet. 2017 doi: 10.1136/jmedgenet-2017-104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, Thiruvahindrapuram B, Xu X, Ziman R, Wang Z, Vorstman JA, Thompson A, Regan R, Pilorge M, Pellecchia G, Pagnamenta AT, Oliveira B, Marshall CR, Magalhaes TR, Lowe JK, Howe JL, Griswold AJ, Gilbert J, Duketis E, Dombroski BA, De Jonge MV, Cuccaro M, Crawford EL, Correia CT, Conroy J, Conceicao IC, Chiocchetti AG, Casey JP, Cai G, Cabrol C, Bolshakova N, Bacchelli E, Anney R, Gallinger S, Cotterchio M, Casey G, Zwaigenbaum L, Wittemeyer K, Wing K, Wallace S, van Engeland H, Tryfon A, Thomson S, Soorya L, Roge B, Roberts W, Poustka F, Mouga S, Minshew N, McInnes LA, McGrew SG, Lord C, Leboyer M, Le Couteur AS, Kolevzon A, Jimenez Gonzalez P, Jacob S, Holt R, Guter S, Green J, Green A, Gillberg C, Fernandez BA, Duque F, Delorme R, Dawson G, Chaste P, Cafe C, Brennan S, Bourgeron T, Bolton PF, Bolte S, Bernier R, Baird G, Bailey AJ, Anagnostou E, Almeida J, Wijsman EM, Vieland VJ, Vicente AM, Schellenberg GD, Pericak-Vance M, Paterson AD, Parr JR, Oliveira G, Nurnberger JI, Monaco AP, Maestrini E, Klauck SM, Hakonarson H, Haines JL, Geschwind DH, Freitag CM, Folstein SE, Ennis S, Coon H, Battaglia A, Szatmari P, Sutcliffe JS, Hallmayer J, Gill M, Cook EH, Buxbaum JD, Devlin B, Gallagher L, Betancur C, Scherer SW. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016;352:aad9780. doi: 10.1126/science.aad9780. [DOI] [PubMed] [Google Scholar]
- Platt RJ, Zhou Y, Slaymaker IM, Shetty AS, Weisbach NR, Kim JA, Sharma J, Desai M, Sood S, Kempton HR, Crabtree GR, Feng G, Zhang F. Chd8 Mutation Leads to Autistic-like Behaviors and Impaired Striatal Circuits. Cell Rep. 2017;19:335–350. doi: 10.1016/j.celrep.2017.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli J, Gasser SM, Papamichos-Chronakis M. The INO80 remodeller in transcription, replication and repair. Philos Trans R Soc Lond B Biol Sci. 2017;372 doi: 10.1098/rstb.2016.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Di Donato N, Dufke A, Cremer K, Hempel M, Horn D, Hoyer J, Joset P, Ropke A, Moog U, Riess A, Thiel CT, Tzschach A, Wiesener A, Wohlleber E, Zweier C, Ekici AB, Zink AM, Rump A, Meisinger C, Grallert H, Sticht H, Schenck A, Engels H, Rappold G, Schrock E, Wieacker P, Riess O, Meitinger T, Reis A, Strom TM. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Ryan DP, Sundaramoorthy R, Martin D, Singh V, Owen-Hughes T. The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. The EMBO journal. 2011;30:2596–2609. doi: 10.1038/emboj.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. The PHD finger: a versatile epigenome reader. Trends Biochem Sci. 2011;36:364–372. doi: 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanlaville D, Etchevers HC, Gonzales M, Martinovic J, Clement-Ziza M, Delezoide AL, Aubry MC, Pelet A, Chemouny S, Cruau C, Audollent S, Esculpavit C, Goudefroye G, Ozilou C, Fredouille C, Joye N, Morichon-Delvallez N, Dumez Y, Weissenbach J, Munnich A, Amiel J, Encha-Razavi F, Lyonnet S, Vekemans M, Attie-Bitach T. Phenotypic spectrum of CHARGE syndrome in fetuses with CHD7 truncating mutations correlates with expression during human development. J Med Genet. 2005 doi: 10.1136/jmg.2005.036160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santen GW, Aten E, Sun Y, Almomani R, Gilissen C, Nielsen M, Kant SG, Snoeck IN, Peeters EA, Hilhorst-Hofstee Y, Wessels MW, den Hollander NS, Ruivenkamp CA, van Ommen GJ, Breuning MH, den Dunnen JT, van Haeringen A, Kriek M. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat Genet. 2012;44:379–380. doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- Scacheri PC, Tie F, Lalani SR, Belmont JW, Colliins FS. The CHD7 protein, mutated in CHARGE styndrome, binds to specific sites on chromatin. 56th Annual Meeting of the Society of Human Genetics; New Orleans. 2006. [Google Scholar]
- Schnetz MP, Bartels CF, Shastri K, Balasubramanian D, Zentner GE, Balaji R, Zhang X, Song L, Wang Z, Laframboise T, Crawford GE, Scacheri PC. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz MP, Handoko L, Akhtar-Zaidi B, Bartels CF, Pereira CF, Fisher AG, Adams DJ, Flicek P, Crawford GE, Laframboise T, Tesar P, Wei CL, Scacheri PC. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010;6:e1001023. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- Seelig HP, Renz M, Targoff IN, Ge Q, Frank MB. Two forms of the major antigenic protein of the dermatomyositis-specific Mi-2 autoantigen. Arthritis Rheum. 1996;39:1769–1771. doi: 10.1002/art.1780391029. [DOI] [PubMed] [Google Scholar]
- Semba Y, Harada A, Maehara K, Oki S, Meno C, Ueda J, Yamagata K, Suzuki A, Onimaru M, Nogami J, Okada S, Akashi K, Ohkawa Y. Chd2 regulates chromatin for proper gene expression toward differentiation in mouse embryonic stem cells. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Shur I, Benayahu D. Characterization and Functional Analysis of CReMM, a Novel Chromodomain Helicase DNA-binding Protein. J Mol Biol. 2005 doi: 10.1016/j.jmb.2005.06.049. [DOI] [PubMed] [Google Scholar]
- Sifrim A, Hitz MP, Wilsdon A, Breckpot J, Turki SH, Thienpont B, McRae J, Fitzgerald TW, Singh T, Swaminathan GJ, Prigmore E, Rajan D, Abdul-Khaliq H, Banka S, Bauer UM, Bentham J, Berger F, Bhattacharya S, Bu’Lock F, Canham N, Colgiu IG, Cosgrove C, Cox H, Daehnert I, Daly A, Danesh J, Fryer A, Gewillig M, Hobson E, Hoff K, Homfray T, Study I, Kahlert AK, Ketley A, Kramer HH, Lachlan K, Lampe AK, Louw JJ, Manickara AK, Manase D, McCarthy KP, Metcalfe K, Moore C, Newbury-Ecob R, Omer SO, Ouwehand WH, Park SM, Parker MJ, Pickardt T, Pollard MO, Robert L, Roberts DJ, Sambrook J, Setchfield K, Stiller B, Thornborough C, Toka O, Watkins H, Williams D, Wright M, Mital S, Daubeney PE, Keavney B, Goodship J, Consortium UK, Abu-Sulaiman RM, Klaassen S, Wright CF, Firth HV, Barrett JC, Devriendt K, FitzPatrick DR, Brook JD, Hurles ME Deciphering Developmental Disorders S. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet. 2016;48:1060–1065. doi: 10.1038/ng.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Current topics in developmental biology. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- Sokpor G, Xie Y, Rosenbusch J, Tuoc T. Chromatin Remodeling BAF (SWI/SNF) Complexes in Neural Development and Disorders. Front Mol Neurosci. 2017;10:243. doi: 10.3389/fnmol.2017.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa SB, Hennekam RC Nicolaides-Baraitser Syndrome International C. Phenotype and genotype in Nicolaides-Baraitser syndrome. Am J Med Genet C Semin Med Genet. 2014;166C:302–314. doi: 10.1002/ajmg.c.31409. [DOI] [PubMed] [Google Scholar]
- Sparmann A, Xie Y, Verhoeven E, Vermeulen M, Lancini C, Gargiulo G, Hulsman D, Mann M, Knoblich JA, van Lohuizen M. The chromodomain helicase Chd4 is required for Polycomb-mediated inhibition of astroglial differentiation. The EMBO journal. 2013;32:1598–1612. doi: 10.1038/emboj.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol. 2014;15:340–356. doi: 10.1038/nrm3789. [DOI] [PubMed] [Google Scholar]
- Stokes DG, Perry RP. DNA-binding and chromatin localization properties of CHD1. Mol Cell Biol. 1995;15:2745–2753. doi: 10.1128/mcb.15.5.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom SP, Lozano R, Lee H, Dorrani N, Mann J, O’Lague PF, Mans N, Deignan JL, Vilain E, Nelson SF, Grody WW, Quintero-Rivera F. De Novo variants in the KMT2A (MLL) gene causing atypical Wiedemann-Steiner syndrome in two unrelated individuals identified by clinical exome sequencing. BMC Med Genet. 2014;15:49. doi: 10.1186/1471-2350-15-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suls A, Jaehn JA, Kecskes A, Weber Y, Weckhuysen S, Craiu DC, Siekierska A, Djemie T, Afrikanova T, Gormley P, von Spiczak S, Kluger G, Iliescu CM, Talvik T, Talvik I, Meral C, Caglayan HS, Giraldez BG, Serratosa J, Lemke JR, Hoffman-Zacharska D, Szczepanik E, Barisic N, Komarek V, Hjalgrim H, Moller RS, Linnankivi T, Dimova P, Striano P, Zara F, Marini C, Guerrini R, Depienne C, Baulac S, Kuhlenbaumer G, Crawford AD, Lehesjoki AE, de Witte PA, Palotie A, Lerche H, Esguerra CV, De Jonghe P, Helbig I, Euro ERESC. De novo loss-of-function mutations in CHD2 cause a fever-sensitive myoclonic epileptic encephalopathy sharing features with Dravet syndrome. Am J Hum Genet. 2013;93:967–975. doi: 10.1016/j.ajhg.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Tang Z, Chen WY, Shimada M, Nguyen UT, Kim J, Sun XJ, Sengoku T, McGinty RK, Fernandez JP, Muir TW, Roeder RG. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell. 2013;154:297–310. doi: 10.1016/j.cell.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Tsurusaki Y, Okamoto N, Ohashi H, Kosho T, Imai Y, Hibi-Ko Y, Kaname T, Naritomi K, Kawame H, Wakui K, Fukushima Y, Homma T, Kato M, Hiraki Y, Yamagata T, Yano S, Mizuno S, Sakazume S, Ishii T, Nagai T, Shiina M, Ogata K, Ohta T, Niikawa N, Miyatake S, Okada I, Mizuguchi T, Doi H, Saitsu H, Miyake N, Matsumoto N. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44:376–378. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- Van Houdt JK, Nowakowska BA, Sousa SB, van Schaik BD, Seuntjens E, Avonce N, Sifrim A, Abdul-Rahman OA, van den Boogaard MJ, Bottani A, Castori M, Cormier-Daire V, Deardorff MA, Filges I, Fryer A, Fryns JP, Gana S, Garavelli L, Gillessen-Kaesbach G, Hall BD, Horn D, Huylebroeck D, Klapecki J, Krajewska-Walasek M, Kuechler A, Lines MA, Maas S, Macdermot KD, McKee S, Magee A, de Man SA, Moreau Y, Morice-Picard F, Obersztyn E, Pilch J, Rosser E, Shannon N, Stolte-Dijkstra I, Van Dijck P, Vilain C, Vogels A, Wakeling E, Wieczorek D, Wilson L, Zuffardi O, van Kampen AH, Devriendt K, Hennekam R, Vermeesch JR. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat Genet. 2012;44:445–449. S441. doi: 10.1038/ng.1105. [DOI] [PubMed] [Google Scholar]
- Veredice C, Bianco F, Contaldo I, Orteschi D, Stefanini MC, Battaglia D, Lettori D, Guzzetta F, Zollino M. Early onset myoclonic epilepsy and 15q26 microdeletion: observation of the first case. Epilepsia. 2009;50:1810–1815. doi: 10.1111/j.1528-1167.2009.02078.x. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Vissers LELM, van Ravenswaaij CMA, Admiraal R, Hurst JA, de Vries BBA, Janssen IM, van der Vliet WA, Huys EHLPG, de Jong PJ, Hamel BCJ, Schoenmakers EFPM, Brunner HG, Veltman JA, van Kessel AG. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nature Genetics. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- Weiss K, Terhal PA, Cohen L, Bruccoleri M, Irving M, Martinez AF, Rosenfeld JA, Machol K, Yang Y, Liu P, Walkiewicz M, Beuten J, Gomez-Ospina N, Haude K, Fong CT, Enns GM, Bernstein JA, Fan J, Gotway G, Ghorbani M, Study DDD, van Gassen K, Monroe GR, van Haaften G, Basel-Vanagaite L, Yang XJ, Campeau PM, Muenke M. De Novo Mutations in CHD4, an ATP-Dependent Chromatin Remodeler Gene, Cause an Intellectual Disability Syndrome with Distinctive Dysmorphisms. Am J Hum Genet. 2016;99:934–941. doi: 10.1016/j.ajhg.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker DE, Riegman KL, Kasah S, Mohan C, Yu T, Sala BP, Hebaishi H, Caruso A, Marques AC, Michetti C, Smachetti ME, Shah A, Sabbioni M, Kulhanci O, Tee WW, Reinberg D, Scattoni ML, Volk H, McGonnell I, Wardle FC, Fernandes C, Basson MA. The chromatin remodeling factor CHD7 controls cerebellar development by regulating reelin expression. J Clin Invest. 2017;127:874–887. doi: 10.1172/JCI83408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodage T, Basrai MA, Baxevanis AD, Hieter P, Collins FS. Characterization of the CHD family of proteins. Proc Natl Acad Sci U S A. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Molecular cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Yamada T, Yang Y, Hemberg M, Yoshida T, Cho HY, Murphy JP, Fioravante D, Regehr WG, Gygi SP, Georgopoulos K, Bonni A. Promoter decommissioning by the NuRD chromatin remodeling complex triggers synaptic connectivity in the mammalian brain. Neuron. 2014;83:122–134. doi: 10.1016/j.neuron.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahir F, Firth HV, Baross A, Delaney AD, Eydoux P, Gibson WT, Langlois S, Martin H, Willatt L, Marra MA, Friedman JM. Novel deletions of 14q11.2 associated with developmental delay, cognitive impairment and similar minor anomalies in three children. Journal of medical genetics. 2007;44:556–561. doi: 10.1136/jmg.2007.050823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Hurd EA, Schnetz MP, Handoko L, Wang C, Wang Z, Wei C, Tesar PJ, Hatzoglou M, Martin DM, Scacheri PC. CHD7 functions in the nucleolus as a positive regulator of ribosomal RNA biogenesis. Hum Mol Genet. 2010;19:3491–3501. doi: 10.1093/hmg/ddq265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]