Summary
Most cells in the liver are polyploid, but the functional role of polyploidy is unknown. Polyploidization occurs through cytokinesis failure and endoreduplication around the time of weaning. To interrogate polyploidy while avoiding irreversible manipulations of essential cell cycle genes, we developed orthogonal mouse models to transiently and potently alter liver ploidy. Premature weaning, as well as knockdown of E2f8 or Anln, allowed us to toggle between diploid and polyploid states. While there was no detectable impact of ploidy alterations on liver function, metabolism, or regeneration, mice with more polyploid hepatocytes suppressed and mice with more diploid hepatocytes accelerated tumorigenesis in mutagen and high-fat induced models. Mechanistically, the diploid state was more susceptible to Cas9-mediated tumor suppressor loss but was similarly susceptible to MYC oncogene activation, indicating that polyploidy differentially protected the liver from distinct genomic aberrations. This suggests that polyploidy evolved in part to prevent malignant outcomes of liver injury.
eTOC Blurb
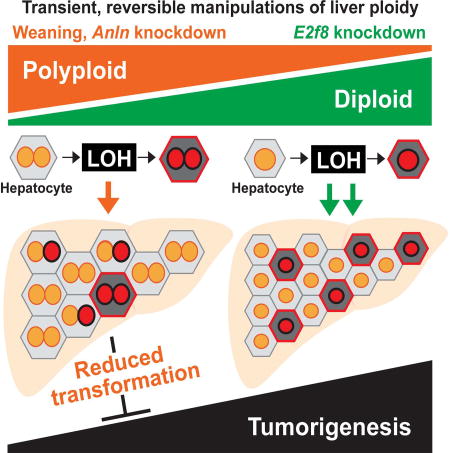
Most liver cells are polyploid, but the functional role of wholesale genome duplications is unknown. To interrogate liver polyploidy function without irreversible manipulations of cell cycle genes, Zhang et al. developed models to transiently alter ploidy, finding that polyploidy reduces tumor development by buffering against tumor suppressor loss of heterozygosity.
Introduction
Polyploid cells and organisms contain more than two homologous sets of chromosomes. Polyploidy is prevalent in plants, fish, and salamanders (Comai, 2005), but rare in mammals except in the cells of the heart, marrow, and liver. Surprisingly, up to 90% of rodent and about 50% of human hepatocytes are polyploid, making the liver one of the largest polyploid organs in mammals (Duncan, 2013; Duncan et al., 2012; Duncan et al., 2010). In rodents, liver ploidy dramatically increases around weaning (P14–P21) and continues to increase with age (Margall-Ducos et al., 2007; Schwartz-Arad et al., 1989). The dominant mechanism for polyploidization is cytokinesis failure that leads to binucleated hepatocytes (Margall-Ducos et al., 2007). To a lesser extent, hepatocyte endoreduplication also contributes to polyploidy via replication of the nuclear genome in the absence of cell division (Gentric and Desdouets, 2014). Thus, polyploid hepatocytes (tetraploids, octoploids, etc.) can be bi- or mononuclear. Postnatal liver polyploidization is developmentally regulated, but ploidy is dynamic and can increase with surgery (Tamura et al., 1992), fatty liver disease, and oxidative stress (Gentric et al., 2015). Given the prevalence and extreme extent to which polyploidy occurs in the liver, it is remarkable that there is little known about the impact of polyploidy on tissue function or cancer risk.
The diverse functions for polyploidy in other contexts can inform our expectations for polyploidy in hepatocytes. In yeast and plants, polyploidy promotes adaptation to environmental stresses, in some cases through the production of genetically diverse aneuploid daughter cells (Chao et al., 2013; Selmecki et al., 2015). Recently, it was shown that a subset of polyploid hepatocytes undergo reductive cell divisions prone to missegregation, leading to the accumulation of aneuploid cells (Duncan et al., 2010). Though the prevalence of aneuploidy in the liver is debated (Knouse et al., 2014), aneuploidy may represent a means by which adaptive genetic diversity or premalignancy could evolve. Whether or not polyploidy in the normal liver is a risk for or protective against cancer is unknown. In support of the premalignancy concept, many cancers show evidence for genome doubling events (Jamal-Hanjani et al., 2017; Zack et al., 2013). In seminal work, Fujiwara et al. showed that tetraploidy in p53 null mouse mammary epithelial cells is a chromosomally unstable state that predisposes to transformation (Fujiwara et al., 2005). Since normal polyploid cells in the liver have intact p53, probably do not proliferate as much, and likely have a much lower risk for genome instability, the relevance to wild-type tetraploid cells in vivo is uncertain.
As an argument against increased cancer risk, the polyploid state in hepatocytes has been associated with terminal differentiation and limited proliferation (Wang et al., 2015). Furthermore, Ganem et al. demonstrated that tetraploid hepatocyte proliferation/transformation is suppressed by the Hippo pathway (Ganem et al., 2014). In other studies, 4c and 8c hepatocytes divide efficiently and contribute as much to proliferation, growth, and regeneration as 2c hepatocytes during liver growth and in transplant assays (Figure S1A)(Duncan et al., 2010). Altogether, it is unclear if there are substantial differences in the proliferative rates of diploid and polyploid hepatocytes. In theory, polyploids could also be protected from cancer due to increased tumor suppressor dosage (Otto, 2007). Although illuminating, these data from diverse organisms and contexts make it difficult to predict how polyploidy might influence the function of normal liver cells and their risk for transformation.
The influence of polyploidy is likely to be context-dependent, mirroring the situation for aneuploidy, which can be tumor suppressive in untransformed cells and oncogenic in cancer cells (Ben-David et al., 2014; Sheltzer et al., 2017; Weaver et al., 2007). Although the relationship between polyploidy and cancer has been interrogated in cell culture (Davoli and de Lange, 2012; Shoshani et al., 2012), it has never been rigorously interrogated in the context of a major organ in vivo. We believe that it is fundamentally important to investigate the role of physiologically programmed polyploidy in one of the largest organs in the body. In this study we specifically focus on the influence of polyploidy on regenerative capacity and transformation risk in the normal liver.
Direct causal connections between polyploidy and tissue function in vivo have not been established because polyploidy is usually not treated as an independent variable. Defects in genes required for cell cycle or cytokinesis (Trp53, Rb, or Cdk1) can dramatically alter ploidy (Diril et al., 2012; Kurinna et al., 2013; Mayhew et al., 2005), but interrogation of mice with these germline mutations cannot distinguish the effects of ploidy from the persistent pleiotropic effects of these mutations. For example, Cdk1 knockout (KO) hepatocytes cannot complete mitosis and are dramatically polyploid (up to 32N or greater). As a consequence, liver-specific Cdk1 KO mice are unable to undergo malignant transformation, but whether this is specifically due to polyploidy or permanent Cdk1 deficiency is unclear (Diril et al., 2012). As a corollary, E2f7/E2f8 double KO livers are almost entirely comprised of diploid hepatocytes, but these mice have no physiological or regenerative phenotypes (Chen et al., 2012; Pandit et al., 2012). Recently, E2f8 KO livers were shown to accelerate tumorigenesis, but it is unknown if this is due to transcriptional effects of E2f8 deficiency or to the diploid state of E2f8 KO hepatocytes (Kent et al., 2016). Similarly, Zhang et al. showed that Yap activation promotes polyploidy and cancer, but it is unclear if the ploidy itself directly impacts tumor biology independent of Yap (Shihao Zhang et al., 2017). Due to the lack of appropriate tools and the inability to assess the chronic impact of ploidy change, the role polyploidy plays in diseases involving long-term cell division cycles remains unclear. Here, we developed multiple methods and in vivo reagents to transiently, reversibly alter ploidy and found that the polyploid state suppressed liver tumorigenesis by buffering against tumor suppressor loss.
Results
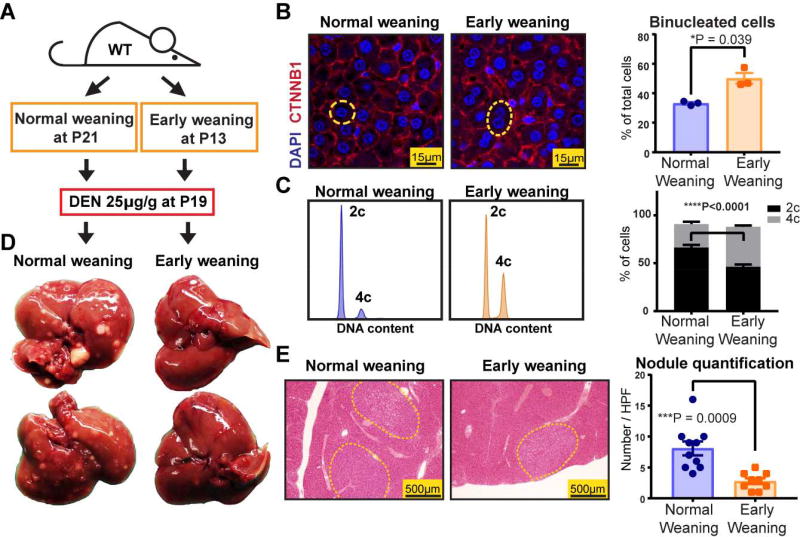
Premature weaning promoted polyploidy and protected against liver tumor development
To answer these questions, we reasoned that the transient, reversible control of ploidy state would represent a fundamental advance to enable the elucidation of ploidy functions. Since the predominant mechanism of widespread hepatocyte polyploidization is cytokinesis failure, a phenomenon temporally associated with weaning in rats (Celton-Morizur et al., 2009), we asked if differential weaning times could transiently influence ploidy in mice. By weaning wild-type (WT) mice at P13 (premature weaning) or P21 (normal weaning) (Figure 1A), we found that prematurely weaned mice had significantly more binucleated hepatocytes (Figure 1B) and increased cellular ploidy (Figure 1C, Figure S1B) at 19 days of age, but not at 2.5 months (Figure S1C). To induce liver tumors when ploidy states were divergent, we gave a single intraperitoneal (IP) dose (25µg/g) of the agent diethylnitrosamine (DEN) to both cohorts at P19. Notably, the expression of Cytochrome P450 and other differentiation genes were not significantly different between premature and normal weaning mice (Figure S1D, E). Five months later, pre-weaned mice with greater ploidy at P19 exhibited significantly reduced gross and microscopic tumor burden (Figure 1D, E), suggesting that polyploidy could exert tumor suppressive effects. Since weaning is confounded by factors other than ploidy, we also used additional methods to control ploidy state.
Figure 1. Premature weaning promoted polyploidy and was protective against HCC development.
(A) WT mice were weaned at P13 and P21 to induce differences in polyploidization. Diethylnitrosamine (DEN) was injected to both cohorts at P19 to induce hepatocellular carcinoma (HCC) formation.
(B) DAPI and CTNNB1 immunofluorescence staining identified nuclei and cell membranes (left panel). Representative mononucleated and binucleated hepatocytes are circled by the yellow dashed lines. The percentages of binucleated hepatocytes are quantified on the right (4 images were analyzed and averaged per mouse, n = 3 mice per group).
(C) Ploidy distribution was analyzed by flow cytometry. Representative histograms show DNA content as stained by PI. Compiled data from the two groups are shown on the right (n = 8 mice in each group).
(D) Gross tumor burden was examined five months after DEN.
(E) Microscopic tumor nodules are circled by yellow dashed lines. The right panel shows nodule quantification (Tumors from one section of an entire lobe was quantified per mouse, n = 10 and n = 9 for normal and early weaning groups).
See also Figure S1.
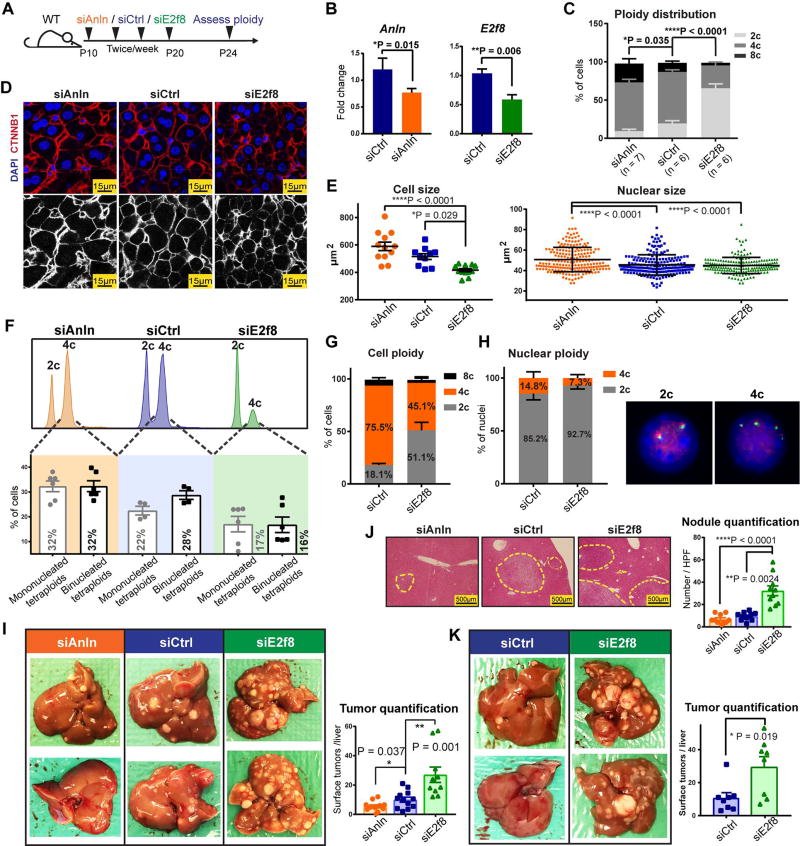
In vivo siRNAs revealed that the polyploid state protected against tumorigenesis
To inducibly toggle polyploidy without introducing permanent genetic lesions, we used siRNA to transiently knockdown genes that affect ploidy. We targeted Anillin (Anln), an actin binding protein required for cytokinesis (Kittler et al., 2007)(Figure S1F, G) and E2f8, a transcription factor required for polyploidization (Pandit et al., 2012). In vivo, Anln and E2f8 siRNA vs. scramble siRNA (siCtrl) delivered in lipid nanoparticles (Zhou et al., 2016) from P10–P20 resulted in significant knockdown of Anln and E2f8 mRNA and protein (Figure 2A, B and Figure S1H). As expected, cellular ploidy was significantly increased after Anln knockdown and decreased after E2f8 knockdown (Figure 2C). Ploidy characterization using confocal imaging revealed that siAnln treated livers had significantly larger cell and nuclear size, while the siE2f8 treated livers showed the opposite (Figure 2D, E). Since cell and nuclear size correlate with DNA content, this suggested significant ploidy changes. To quantitatively distinguish mono- vs. binucleated tetraploids, we integrated flow and confocal imaging data (Figure 2F). This revealed that siAnln hepatocytes were 32% mononuclear diploid, 32% mononuclear tetraploid, and 32% binuclear tetraploid. siE2f8 hepatocytes were 64% mononuclear diploid, 17% mononuclear tetraploid, and 16% binuclear tetraploid. Furthermore, Florescence In Situ Hybridization (FISH) confirmed the existence of mono- and binuclear tetraploids (Figure 2G, H). Overall, this indicated that both mononuclear and binuclear tetraploids were increased in siAnln vs. control livers and in control vs. siE2f8 livers.
Figure 2. In vivo siRNAs that toggled ploidy revealed that the polyploid state protected against tumorigenesis.
(A) Schema for the siRNA experiment: Anln, E2f8, and scramble siRNAs (siCtrl) were encapsulated into lipid nanoparticles and injected into WT C3H strain mice starting at P10. Four total injections (two intraperitoneal and two retro-orbital) were performed twice per week. Between P24–26, hepatocytes were dissociated for ploidy analysis or mice were injected with DEN (75µg/g). Tumor burden was examined six months later.
(B) mRNA knockdown after Anln and E2f8 siRNA treatments. RT-qPCR was performed on the liver 4 days after the last siRNA dose.
(C) Average cellular ploidy distribution within livers, as determined by PI staining and flow cytometry (n = 6 mice in each group were analyzed).
(D) Confocal images of DAPI and CTNNB1 stained siRNA treated livers. The bottom images highlight CTNNB1 staining alone, marking membranes and cellular outlines.
(E) Cross-sectional cell and nucleus size measurements. Each group includes 12 total analyzed image fields from 4 individual mice. For nucleus size quantification, each data point is one nucleus; for cell size quantification, each data point is an average of the cell sizes from one image and 3 images were taken from each mouse.
(F) Representative cellular ploidy distribution of siRNA treated livers (upper panel). The lower panel shows the distribution of mono- and binucleated tetraploid hepatocytes. Calculations were performed as follows: % of mononucleated tetraploid cells = % tetraploid cells by FACS − % binucleated cells by confocal (images from Figure 2D, n = 3 images were counted for each mouse, 2 mice for each group).
(G) Cellular ploidy distribution of siRNA treated livers, as determined by Flow Cytometry (n = 2 mice/group).
(H) On the left is the nuclear ploidy of the above siRNA treated livers, as determined by FISH. 60 nuclei were analyzed for each mouse. On the right are representative images showing a 2c and a 4c nucleus. 2 green and 2 red signals identify a 2c nucleus. 4 green and 4 red signals identify a 4c nucleus.
(I) Gross tumor burden of siRNA treated livers harvested 6 months after DEN injection. Quantification of liver surface tumor numbers (right panel).
(J) H&E liver histology showing tumor nodules circled by dashed lines (left panel). Quantification of microscopic nodules (right panel).
(K) Tumor burden in mice treated with DEN at later time points after siRNA delivery. Mice were treated with four doses of siRNAs. 14 days after the last siRNA injection, one dose of DEN (100µg/g) was given, and 7.5 months later, tumor burden was assessed (n = 8 in each group). The liver surface tumors were quantified in the right panel.
See also Figure S2 and Figure S3.
Altered ploidy did not impact overall health, liver mass, body mass, hepatocyte differentiation, CYP450 expression, or proliferation (Figure S2A–E). Moreover, we challenged these mice to acute regeneration assays such as partial hepatectomy and hepatotoxin treatments. Hepatectomy was performed at 5–6 weeks of age, when ploidy differences were still detected (Figure S2F). Liver/body weight ratios after 2/3rds partial hepatectomy were not significantly different between different ploidy groups (Figure S2G). There were also no differences in necrosis and proliferation after single doses of DEN or carbon tetrachloride (CCl4) (Figure S2H, I). These results suggested that polyploid and diploid hepatocytes were equivalently able to survive, recover, and proliferate after injuries. These findings are consistent with previous studies in mice with ploidy alterations (Chen et al., 2012; Pandit et al., 2012), again demonstrating that ploidy state has minimal influences on post-natal liver growth and regeneration, likely because only 2–4 cell division cycles are required for recovery after these profound, acute injuries.
After establishing that inducible inhibition of Anln and E2f8 alters ploidy without introducing irreversible genetic mutations, we next evaluated the impact of ploidy on tumor development. We injected DEN (75µg/g × 1 dose) into mice four days after the last dose of siCtrl, siAnln, or siE2f8 (siRNA delivery schema in Figure 2A). Six months later, siE2f8 treated livers with more diploid cells had significantly increased gross tumor burden, microscopic tumor nodules, and liver/body weight ratio than siCtrl livers (Figure 2I, J and Figure S3A–C). In contrast, siAnln treated livers with more polyploid cells showed the opposite. To further exclude the possibility of residual siRNA effects, we also introduced DEN at 14 days, rather than 4 days, after the last siE2f8 injection. Polyploidy again demonstrated a potent tumor suppressive effect (Figure 2K and Figure S3D). To generate higher levels of polyploidy, we developed two additional and distinct N-acetylgalactosamine (GalNAc) conjugated siRNAs against Anln. The exposed GalNAc moiety mediates siRNA uptake via Asialogycoprotein receptors (ASGPR), which are specifically expressed by hepatocytes. Subcutaneous injections of these siRNAs (3 × 4mg/kg doses between P8–P20) resulted in high levels of polyploidy when compared to mice treated with Luciferase siRNA (siLuc) (Figure S3E). After DEN (75µg/g at P26), the siAnln mice also exhibited less tumors (Figure S3F). To avoid the possibility of Anln specific effects, we also induced liver polyploidy by knocking down Cdk1 with an in vivo siRNA (Figure S3G). After siRNA injections, mice were dosed with DEN (75µg/g). 6 months after DEN, siCdk1 treated mice exhibited reduced tumor burden (Figure S3H), again supporting the tumor protective role of polyploidy. Altogether, these findings suggested that the degree of liver polyploidy is anti-correlated with the efficiency of carcinogenesis.
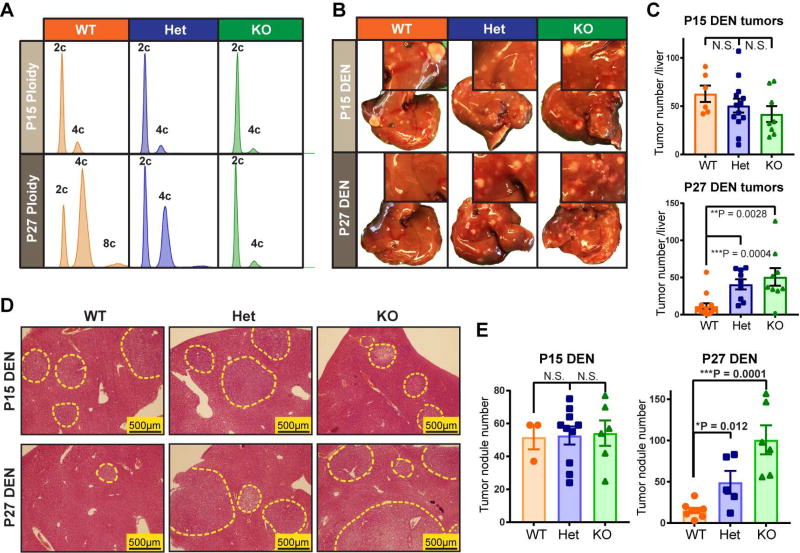
E2f8 knockout and Anln shRNA mice confirmed the protective effects of polyploidy
Given the formal possibility that siRNAs or lipid nanoparticles could have off-target or non-specific effects, we also used Cas9 to generate whole-body E2f8 KO mice (Figure S4A), which have predominantly diploid hepatocytes in adulthood (Figure 3A). Interestingly, we observed that E2f8 WT, Het and KO livers had equivalent levels of ploidy at P15, and only diverged in ploidy state by P27 (Figure 3A and Figure S4B). We hypothesized that if ploidy is a specific and essential factor causing tumor suppression in E2f8 KO livers, then mutagenizing and inducing cancer at P15 would not result in differences in liver tumor development, while inducing at P27 would result in large differences. Indeed, DEN given at P15 (25µg/g) resulted in no cancer differences between the three groups, while DEN at P27 (75µg/g) caused more liver tumors in E2f8 Het and E2f8 KO mice when compared to E2f8 WT mice (Figure 3B–E, Figure S4C). These results further demonstrated that the E2f8 gene itself contributed less to cancer development than the ploidy differences resulting from E2f8 deficiency.
Figure 3. Reduced ploidy accounted for the increased tumorigenesis associated with E2f8 deficiency.
(A) Representative cellular ploidy of E2f8 WT, E2f8 Het, and E2f8 KO livers at P15 and P27, as analyzed by flow cytometry.
(B) DEN was introduced by IP injection in one cohort at P15 (25µg/g) and in another cohort at P27 (75µg/g). Gross tumor burden was evaluated 4.5–5 months later. The inset images show higher magnification images that more clearly exhibit tumor burden.
(C) Liver surface tumor quantification of E2f8 WT, Het, and KO mice given DEN at P15 or P27.
(D) Histology of E2f8 WT, Het, and KO livers show microscopic tumor nodules (circled by dashed yellow outlines).
(E) Quantification of tumor nodules per unit of cross sectional area.
See also Figure S4.
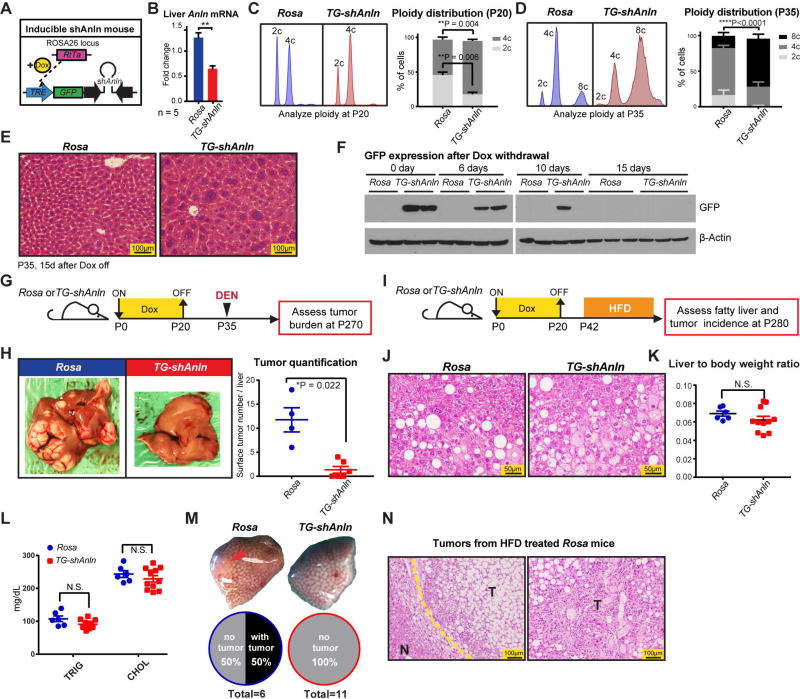
Next, we wanted to generate a more potent and versatile mouse model to increase polyploidy. In addition, our goals were to drive greater levels of polyploidy and to employ a small inhibitory shRNA distinct from the siAnln used above in order to corroborate on target effects on Anln. Thus, we employed a doxycycline (dox)-inducible transgenic mouse expressing an shRNA against Anln (Zhang et al., 2017). Mice were generated from embryonic stem cells containing Rosa-rtTA and a GFP + shAnln cassette under the control of a tetracycline responsive promoter element (TRE) (Figure S4D, E; transgenic design based on Scott Lowe’s group (Premsrirut et al., 2011)). Anln suppression can be induced by Doxycycline (Dox) in a temporally specific fashion (Figure 4A). Rosa-rtTa alone or Rosa-rtTa; TRE-shAnln (hereafter called Rosa and TG-shAnln) transgenic mice exposed to dox water from P0–P20 showed normal growth, development, and liver function (Figure S4F–H). Anln mRNA levels were suppressed by 50% (Figure 4B), which resulted in more polyploid livers at multiple time points after dox withdrawal (Figure 4C–E). These livers were similar to what was seen with Anln siRNA treatment, but had more profound levels of polyploidization. In addition, GFP protein (and likely Anln shRNA) completely disappeared by 15 days after dox withdrawal (Figure 4F), demonstrating the reversibility of Anln suppression.
Figure 4. An inducible transgenic mouse model to temporally control polyploidization shows protection against DEN and high fat induced HCCs.
(A) An inducible double-transgenic mouse model carrying shAnln cassette under the control of a tetracycline responsive promoter element (TRE). These mice carry a Rosa-rtTA knockin construct, allowing induction of Anln suppression with Doxycycline (Dox) water.
(B) Transient Dox induction from P0 to P20 suppressed Anln mRNA levels in the liver.
(C) Representative cellular ploidy distribution of TG-shAnln livers treated with dox from P0–P20 was determined by PI staining and flow cytometry at the age of P20 (left panel). On the right is the average ploidy distribution (n = 3 mice in each group).
(D) Representative cellular ploidy distribution of TG-shAnln livers treated with dox from P0–P20 was determined by PI staining and flow cytometry at the age of P35 (left panel). On the right is the average ploidy distribution (n = 6 mice in each group).
(E) H&E staining showing large, multinucleate hepatocytes in the induced group.
(F) Western blots showing stability of GFP protein expression after dox induction and withdrawal.
(G) Schema for the DEN induced HCC experiment in inducible shRNA mice: Transient dox treatment from P0–P20 in Rosa-rtTa or TG-shAnln mice established ploidy differences. At P35, mice were injected with DEN (100µg/g). Tumor burden was examined 8 months later.
(H) Representative gross tumor burden from the DEN experiment. Quantitation on the right.
(I) Schema for the high fat diet (HFD) induced HCC experiment in inducible TG-shAnln mice: Transient dox treatment mice established ploidy differences. Starting at P42, mice were given ad libitum HFD (60% calories from fat). Tumor burden was examined after 8 months.
(J) H&E of Rosa and TG-shAnln HFD treated livers.
(K) Liver to body weight ratios of HFD treated Rosa and TG-shAnln mice.
(L) Triglyceride and cholesterol levels of Rosa and TG-shAnln mice after 8 months of HFD.
(M) Representative gross tumor burden from the HFD experiment. 50% (3 of 6) of Rosa mice had tumors, while no (0 of 11) TG-shAnln mice carried tumor.
(N) H&E of tumors from Rosa control mice given HFD. Yellow dotted line is the boundary between non-tumor tissue (labeled “N”) and tumor (“T”).
See also Figure S4.
Polyploid livers resulting from transient suppression of Anln were almost completely protected from DEN induced tumorigenesis, confirming the siRNA results (Figure 4G, H). We then wanted to exploit this model to analyze the function of polyploidy in a carcinogenesis model caused by another clinically important mechanism. Steatohepatitis represents an increasingly relevant risk factor for HCC and has been associated with an increase in polyploidy (Gentric et al., 2015). To induce long term fatty liver disease and HCC, we fed mice with high fat diet (HFD) after transiently inducing ploidy changes (Figure 4I). After eight months, Rosa and TG-shAnln mice had similarly high levels of steatosis and liver function abnormalities (Figure 4J–L and Figure S4I), but 50% of control livers while no polyploid livers developed HCC (Figure 4M, N). In summary, multiple murine models increasing and decreasing ploidy corroborated the fact that higher levels of polyploidy suppressed liver tumors in diverse cancer models.
Polyploids were protected from tumor suppressor LOH but not oncogene activation
To probe underlying mechanisms in the DEN model, we first asked if ploidy significantly regulated metabolic properties that would influence tumorigenesis. DEN is first bioactivated by CYP450 family enzymes to become α-hydroxylnitrosamine (Heindryckx et al., 2009). Expression of CYP450 enzymes in general and zonation of Cyp2e1 in particular were unchanged in livers with distinct ploidy (Figure S2C and Figure S5A). Elevated levels of reactive oxygen species (ROS) secondary to hepatotoxins are known to accelerate tumor initiation (Wang et al., 2016), but ROS levels were unchanged between siRNA treated mice and between E2f8 WT/Het/KO mice (Figure S5B–D). It is also known that following the DEN bioactivation, an ethyldiazonium ion is formed, binds DNA, and causes genotoxic damage (Heindryckx et al., 2009). Though DNA damage markers such as p-Brca1, p-p53, and p-γH2A.X increased after DEN, the magnitude of induction was similar between groups (Figure S5E, F). Altogether, livers with altered ploidy did not exhibit differential xenobiotic metabolism, oxidative stress, or DNA damage responses.
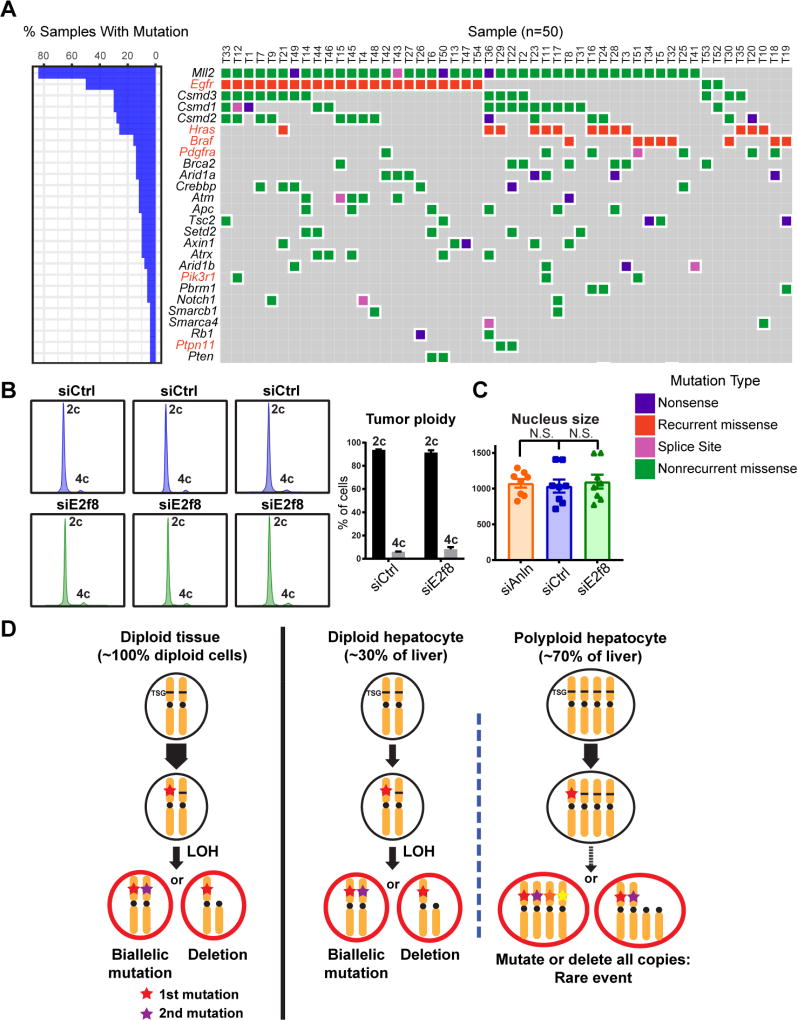
Since the mutagenic activities of DEN were quantitatively similar, it was possible that ploidy differentially influenced tumor suppressor (TSG) loss and proto-oncogene activation. We first performed a theoretical calculation to estimate the risk of TSG loss and oncogene activation in tissues with 10,000 diploid cells vs. 5,000 tetraploid cells (which are ~2× larger). Both tissues contain 20,000 sets of chromosomes (Figure 5A). Assuming 10% of TSGs are mutated in one massive DNA damaging event, then the rate of loss of heterozygosity (LOH) in a diploid cell is 0.12 = 0.01, while for a tetraploid cell it is 0.14 = 0.0001. Thus, 100 cells vs. 0.5 cells on average achieve LOH in diploid vs. tetraploid tissues of the same mass. For simplicity, we can also assume that the oncogene activating mutation rate is also 0.1. The probability this mutation occurs in a diploid cell is 1−0.92 ≈ 0.19, while in a tetraploid cell it is 1−0.94 ≈ 0.34. Although the probability is higher for tetraploid cells, diploid vs. tetraploid tissues have 1900 and 1700 cells at risk once total cell numbers are accounted for. This calculation suggests that polyploid tissues are dramatically protected against TSG LOH but are at similar risk for oncogene activation. This model is oversimplified since tumorigenesis involves more subtle and complex genome alterations (Davoli et al., 2013; Nijhawan et al., 2012), but it is still likely to be informative because TSG LOH is an important part of carcinogenesis.
Figure 5. Polyploids were protected from tumor suppressor LOH but not oncogene activation.
(A) Theoretical calculation of the risk of TSG loss and oncogene activation in tissues with diploid vs. tetraploid cells.
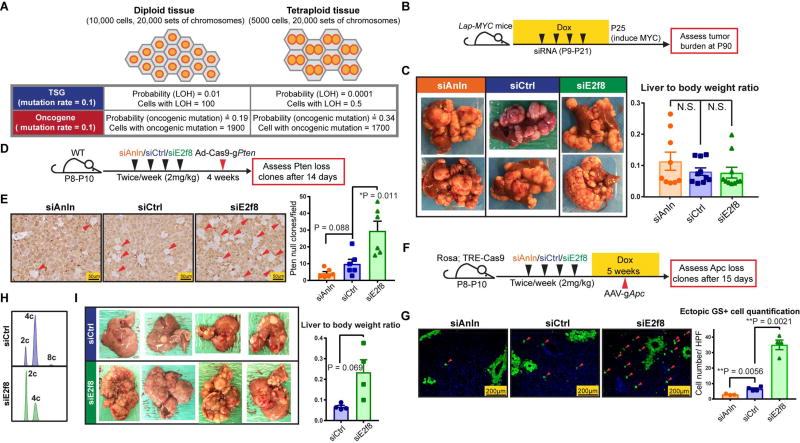
(B) Schema of the LAP-MYC experiment: prior to inducing MYC overexpression by dox withdrawal at P25, LAP-tTA; TRE-MYC mice were given four doses of siRNA to transiently alter ploidy, as described in previously. At P30, ploidy distribution and MYC expression levels were measured; At P90, tumor burden was assessed.
(C) Tumor burden at P90 (left panel). Liver to body weight ratios, another metric of tumor burden, are shown in the right panel.
(D) Schema of the Ad-Cas9-sgPten experiment: WT mice were given four doses of siRNA to alter ploidy; one week later, adenovirus carrying Cas9 and a Pten sgRNA (Ad-Cas9-sgPten) was injected into these mice (109 pfu/mouse); 14 days later, LOH was assessed by PTEN IHC.
(E) PTEN staining (left panel) on siAnln, siCtrl, and siE2f8 treated livers 14 days after virus injection. Red arrowheads point to the cells without PTEN, quantification on the right.
(F) Schema of the AAV-sgApc experiment: Dox inducible Rosa-rtta; TRE-Cas9 mice were given four doses of siRNA to alter ploidy. Five days later, dox (1g/L) was given to induce Cas9 expression. Two weeks after the last dose of siRNA, AAV-sgApc (5 × 1012 pfu/mouse) was retro-orbitally delivered; 15 days later, the Apc LOH was estimated by Glutamine Synthetase (GS) IF.
(G) GS staining (left panel) 15 days after AAV-sgApc injection. Red arrowheads point to ectopic GS positive cells that result from homozygous Apc deletion. Note that the normal, non-ectopic GS staining that surrounds central veins (CV) was not quantified. Ectopic GS positive cells per imaging area were quantified in the right panel.
(H) Representative cellular ploidy distribution of P53fl/fl; Rb1fl/fl; Rbl2fl/fl hepatocytes treated with 4 doses of siRNAs (siCtrl or siE2f8) from P10–P21, as determined by PI staining and flow cytometry at P24.
(I) One week after ploidy was altered with siRNAs, the mice were injected with Ad-Cas9-sgPten (109 pfu/mouse) and AAV-Cre (2.5 × 1010 pfu/mouse). Tumor burden was examined 2.5 months later. Liver to body weight ratios (right).
See also Figure S5 and Figure S6.
To challenge this hypothesis, we first examined the impact of oncogene activation in mice with different levels of polyploidy, we overexpressed MYC using a liver specific driver (LAP-tTA) and a dox-inducible promoter (TRE-MYC) (Figure S6A) (Shachaf et al., 2004). Prior to inducing MYC overexpression, we gave LAP-tTA; TRE-MYC mice four doses of siRNA to transiently alter ploidy (Figure 5B, Figure S6A). At P30, significant differences in ploidy, but not MYC expression levels, were observed (Figure S6B, C). Dox withdrawal at P25 leads to transformation of less than 1% of MYC expressing cells, a dynamic range that allowed us to sensitively quantitate the influence of ploidy on tumor initiation. Nine weeks post-induction, tumor burden, liver to body weight ratios, and tumor ploidy were indistinguishable between siRNA treated ploidy groups (Figure 5C and Figure S6D), showing that ploidy had little impact on MYC oncogene induced tumorigenesis.
Next, we tested whether TSG LOH is more difficult to achieve in hepatocytes with wholesale genome duplications. To quantitate the dynamics of TSG loss, we engineered an adenovirus carrying Cas9 and a guide strand RNA targeting Pten (Ad-Cas9-sgPten) (the Pten sgRNA was validated in reference (Xue et al., 2014)), a commonly inactivated TSG in HCC. 14 days after IV Ad-Cas9-sgPten delivery into mice with different levels of ploidy, Pten LOH was assessed with immunohistochemistry (Figure 5D). The frequency of hepatocytes with complete Pten deletion was inversely proportional to the extent of polyploidy (Figure 5E). To confirm that the degree of ploidy did not change susceptibility to adenoviral infection, we verified that equal numbers of cells expressed GFP after Adenovirus-GFP delivery (Figure S6E). To rule out the possibility of a Pten specific-phenomenon, we also assessed Apc, a TSG in the WNT pathway. We injected an Adeno-associated virus carrying a guide strand against exon 8 of Apc (AAV-sgApc) into dox-inducible Cas9 mice (Rosa-rtTA; TRE-Cas9) subjected to siRNA induced ploidy changes (Figure 5F). In vitro, this particular sgApc effectively mutagenized Apc (Figure S6F) and had previously been used to generate Apc null hepatocytes (Zhang et al., 2014). To quantitate the number of Apc null clones in vivo, we probed for ectopic Glutamine Synthetase (GS), a specific and sensitive marker of aberrant WNT activation in the liver (Colnot et al., 2004). Strikingly, more diploid livers were much more susceptible to Apc LOH than control livers, and more polyploid livers harbored the fewest GS+ cells (Figure 5G). These data support the concept that polyploid livers are protected from TSG loss and are not more sensitive to oncogene activation.
To show that polyploidy protects from cancers that arise predominantly from TSG loss, we used a mouse strain with conditional alleles in important tumor suppressor genes: P53fl/fl; Rb1fl/fl; Rbl2fl/fl (Schaffer et al., 2010). First, we changed the ploidy of these mice using control and E2f8 siRNAs (Figure 5H), then mice were injected with Ad-Cas9-sgPten as well as AAV-Cre to delete TSG alleles in a mosaic fashion. 2.5 months after viral injection, siCtrl treated mice exhibited lower tumor burden compared to siE2f8 mice (Figure. 5I). We also used a second HCC model that is caused by TSG loss. We targeted Pten, P53, and Lkb1 in wild-type mice. We used Ad-Cas9-gPten and an AAV containing guide strands against P53 and Lkb1 (AAV-KPL) (Platt et al., 2014). This AAV also contains a guide against Kras as well as a KrasG12D HDR template, so this model is not entirely driven by TSG loss. After ploidy was changed with siRNAs (Figure S6G), mice were injected with the Ad-Cas9-sgPten and AAV-KPL. Two months later, siAnln treated mice exhibited the fewest tumors, followed by siCtrl and siE2f8 treated mice (Figure S6H). These cancer models both support the hypothesis that polyploidy protects from tumorigenesis caused by TSG loss.
Reduced susceptibility to tumor suppressor LOH paired with reduced tumorigenesis in more polyploid livers suggested that DEN-induced tumors were dependent on tumor suppressor loss. Previously, DEN was shown to preferentially select for oncogenic mutations (Hras and Ctnnb1) in studies focused on identifying mutations in these pathways (Buchmann et al., 1989; Yamada et al., 1999). We aimed to more broadly and thoroughly map the mutational landscape of these tumors, so we sequenced 242 of the most commonly mutated genes in human and murine HCCs in 50 individual DEN induced tumors (Figure 6A, Table S2). We did identify a core group of recurrent, mutually exclusive mutations in oncogenes such as Egfr (Phe254Ile), Hras (Gln61Arg), and Braf (Val637Glu), but a majority of the most commonly mutated genes were bona fide TSGs such as Mll2 (Kmt2d), Brca2, Arid1a, Atm, Apc, and Tsc2. Overall, these data suggest that DEN tumor transformation depends on TSG loss in addition to EGFR-RAS-MAPK pathway activation, further supporting the idea that tumor protection in polyploids is in part mediated through retention of WT tumor suppressor alleles.
Figure 6. Tumor suppressor mutations are prevalent in liver tumors from diploid and polyploid mice.
(A) Targeted sequencing on 50 DEN induced liver tumors. 242 of the most commonly mutated genes in human and murine HCC were sequenced. Genes highlighted in red are oncogenes and those in black are TSGs.
(B) Cellular ploidy distribution of DEN induced tumors from siRNA treated mice. Data is averaged on the right (tumor n = 10).
(C) Nucleus sizes of tumors from siRNA treated mice. The nucleus size was quantified using Photoshop based on H&E staining images. 4 mice in each group were quantified, 2 tumors per mouse.
(D) This model depicts how TSGs could be lost in diploid and polyploid cells. Note that the liver contains both diploid and polyploid cells.
See also Table S2.
Given that the mammalian liver is comprised of a mixture of polyploid and diploid cells, these results would predict that HCCs more likely originate from diploid cells. If this was true, it would follow that liver tumors are also more frequently diploid rather than polyploid. We found that even tumors arising from siCtrl mice with polyploid cells were predominantly diploid (Figure 6B, C). In human HCCs examined by flow cytometry, HCCs were more often comprised of diploid as opposed to polyploid cells (Anti et al., 1994; Caselitz et al., 2003; Fujimoto et al., 1991; Nagasue et al., 1993). Altogether, this suggests that human HCCs more likely arise from diploid cells, consistent with the idea that the polyploid state is less compatible with cancer development.
Discussion
The fact that some animals and organs are polyploid, with cells containing whole genome duplications, has perplexed scientists for decades. It is known that 50–90% mammalian hepatocytes are polyploid, and conditions such as chronic hepatitis, steatohepatitis, and oxidative stress have all been associated with increased polyploidy (Gentric et al., 2015; Toyoda et al., 2005), but it is unknown if ploidy changes represent compensation, cause, or bystander effect of such disease states. Analyses of genetically engineered knockout mice with mostly polyploid or diploid hepatocytes have not been able to unequivocally link ploidy with cellular fitness, regeneration, or cancer (Diril et al., 2012; Pandit et al., 2012). In part, this is because the reagents used to assess the response of ploidy states to injuries usually confound the effects of ploidy with the effects of the prominent cancer related genes used to change ploidy.
Here, we devised multiple in vivo methods to alter liver ploidy in a reversible fashion, such that long-term consequences of such changes could be assessed and compared. In agreement with previous studies (Duncan et al., 2010; Pandit et al., 2012), we found that polyploidy had little impact on acute injury or liver regeneration, but chronically, the polyploid state demonstrated tumor suppressive functions in multiple cancer models. In general, the extent of ploidy increase or decrease in different siRNA, GEMM, and inducible transgenic models appeared to correlate with tumor number and burden. Moreover, we believe that this protection is due in large part to buffering against TSG loss rather than limiting proliferation after oncogenic insults. As shown in Figure 6D, cells within predominantly diploid tissues lose their tumor suppressors through classical LOH mechanisms. In contrast, polyploid hepatocytes harbor up to 16 alleles for each TSG, thereby buffering against a second or even third hit. Consistent with this, DEN tumors were most often diploid, suggesting a diploid origin. This is consistent with human data showing that HCCs are predominantly diploid (Anti et al., 1994; Fujimoto et al., 1991; Nagasue et al., 1993). We also showed that MYC induced oncogenesis in the liver was agnostic to ploidy states. It is possible the strong oncogenic effect of MYC overcame tumor protective effects of ploidy, but the effects of other oncogenes may be more influenced by polyploidy.
It is also important to note that we have only altered ploidy in the benign liver and not within established tumors, thus it is possible that increasing ploidy within cancer cells would cause genome destabilizing effects documented by others (Davoli and de Lange, 2012; Fujiwara et al., 2005; Shihao Zhang et al., 2017). This would be in line with aneuploidy studies, which have shown that aneuploidy is either tumor suppressive or oncogenic depending on context (Ben-David et al., 2014; Sheltzer et al., 2017; Silk et al., 2013). It is important to note here that our approaches did not allow us to distinguish euploidy and aneuploidy within polyploid hepatocytes. Polyploidy will probably also demonstrate context dependency, but it is important to underscore that the extensive polyploidy found in the normal liver is an extremely important context to fully understand. Our findings suggest that in the setting of normal hepatocytes, TSG buffering could be an important tumor suppressive function of polyploidy.
We believed that it was essential to use mouse models to interrogate the role of ploidy in cancer because it is the most rigorous way to examine tumor initiation. For example, in vivo and in vitro human HCCs, which are already transformed, cannot be used to study ploidy’s role in the transition state between normal and malignant cells. We also used cancer models that were the most reflective of human disease states. The DEN model generates cancers that are not dependent on singular, artificially strong genetic drivers such as Yap or Akt. Using expression profiling, Snorri Thorgeirsson showed that DEN induced mouse HCCs were more similar to poor survival human HCCs than other mouse cancer models (Lee et al., 2004). Also, Allan Balmain’s group recently argued for the use of chemically induced mouse cancer models because they more accurately reflect the diverse genomics of human cancers (Westcott et al., 2015). Interestingly, Trp53 mutations were not detected in the DEN tumors suggesting that there might be a role for Trp53 in the development of diploid tumors. It would be interesting to test the polyploidy protection hypothesis in a Trp53 null background. Nevertheless, the tumors in our study harbored a wide diversity of oncogenic and tumor suppressor mutations that reflect the biology of human HCCs.
The DEN model may resemble the types of liver cancers caused by acute exposure to mutagens such as aflatoxin. Although HCC in the US is commonly caused by chronic etiologies (HBV, HCV, alcohol, NASH), acute liver injury and mutagenesis by aflatoxin is a common problem in tropical and sub-tropical climates in Africa, Asia and South America (Liu and Wu, 2010). It is conceivable that polyploidy in part evolved to mitigate the cancer risk associated with these types of acutely acting liver mutagens, which frequently induce lethal cancers during the reproductive years. We believe that polyploidy in the context of acute injury events are essential to understand because these may be the types of massive toxic events that mammalian livers have evolved to buffer against.
Despite the importance of acute liver injuries, it is possible that polyploidy might have different implications for chronic injury. The high fat diet model (Figure 4) involves chronic nutritional stress that can contribute to cirrhosis and HCC in patients. Our data suggests that in this chronic context, polyploidy is protective against HCC development. Work on this and other liver cancer models will help to determine if polyploidy in the liver is generally tumor suppressive or is dependent on factors such as the nature of the genetic driver lesion or environmental injury.
In future studies, it would be also interesting to determine if there are specific diploid populations in the liver that have greater cancer risk and if this risk can be modulated with ploidy manipulations. Previously, Wang et al. identified a “stem-like” hepatocyte population around the central vein (Wang et al., 2015). This population of cells repopulated the liver in the absence of injury and were 60% enriched for diploid cells, but it is unknown if the diploid or polyploid cells within this Axin2 compartment have greater self-renewing capacity or tumorigenic potential. It is possible that increasing ploidy in this population might change regenerative capacity or cancer risk. Given that most hepatocytes are capable of extensive proliferation (Duncan et al., 2010; Schaub et al., 2014), it is also possible that other diploid populations, whether or not they are stem cells, could respond to distinct liver injuries and contribute to cancer formation. In this study, we aimed to answer a fundamental question about polyploidy in liver biology, but it is possible that transient or persistent polyploidzation not only serves physiologic functions but could be exploited therapeutically.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Hao Zhu (hao.zhuutsouthwestern.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
H2.35 (ATCC, source: BALB/c female mouse) immortalized hepatocytes were cultured in DMEM (Life Technologies) supplemented with 4% (vol/vol) FBS, 1× Pen/Strep solution (Thermo Scientific HyClone) and 200nM Dexamethasone (Sigma). Cells were cultured at 37°C and 5% CO2 in a humidified incubator.
Mice
All mice were handled in accordance with the guidelines of the Institutional Animal Care and Use Committee at UTSW. E2F8 KO mice were made by the Children’s Research Institute Mouse Genome Engineering Core. Cas9 was used to target intron 1 and 3, which led to the deletion of exons 2 and 3. See Key Resource Table for sgRNA sequences. These mice were made and maintained in the C3H/HeJ background. TG-shAnln embryonic stem cells were injected into blastocysts by the UTSW Transgenic Core. The strategy was based on work from Scott Lowe’s group (Premsrirut et al., 2011). Briefly, the shAnln sequence (Mir30 based shAnln sequence: TGCTGTTGACAGTGAGCGCTCCATGCTAAATATAACATTATAGTGAAGCCACA) was cloned into the pCol-TGM vector (Addgene #32715) and electroporated into KH2 embryonic stem cells together with a Flpe vector. Validated KH2 clones were then injected into blastocysts to generate knockin transgenic mice. TRE-Cas9 mice were generated by Dr. Eric Olson’s Laboratory. All other mice used in experiments were WT C3H/HeJ or FVB mice. All experiments were done in an age and sex controlled fashion unless otherwise noted in the figure legends. The experimental mice were randomized using a simple randomization method.
DEN Cancer Model
Diethylnitrosamine (DEN, Sigma) was injected intraperitoneally (IP) at the age of P19 (25µg/g), P24–26 (75µg/g), or P34–36 (100µg/g).
LAP-MYC Liver Cancer Model
In this model, the human c-MYC oncogene was driven by tetracycline response element (TRE). The tetracycline transactivator (tTA) expression was controlled by liver activator protein (LAP). Mice were injected with siRNAs to change their liver ploidy. Doxycycline (Dox; 1g/L in water) was removed at P30 to induce MYC.
High Fat Diet Model
High fat diet (HFD) was purchased from RESEARCH DIETS INC. 60% of calories were from fat. TG-shAnln mice were on Dox from the age of P0 to P20. Then, the mice were kept on HFD from P42 to P280.
P53, Rb1, Rbl2, and Pten Deletion Liver Cancer Model
P53fl/fl; Rb1fl/fl; Rbl2fl/fl mice were injected with siRNAs to change their liver ploidy. Then, mice were injected with Ad-Cas9-sgPten (109 pfu/mouse) and AAV-Cre (2.5 × 1010 pfu/mouse) to delete P53, Rb1, Rbl2 and Pten.
KPL Liver Cancer Model
FVB mice were injected with siRNAs to change their liver ploidy. Then mice were injected with Ad-Cas9-sgPten (109 pfu/mouse) plus AAV-KPL (1012 pfu/mouse) to delete and mutate Trp53, Lkb1, Kras and Pten.
METHOD DETAILS
Cell Transfection
H2.35 (ATCC) cells (2.5×105) were transfected with 25pmol siRNA (Life Technologies) in 6-well plates by using Lipofectamine RNAiMAX and OptiMEM (Life Technologies), as described in the manufacturer’s instructions.
Early Weaning Experiment
Male pups from C3H mothers were randomized into two groups at P13 based on their body weight: an early and a normal weaning group. The early weaned mice were placed into a new cage without their mothers and provided with moist food. The normally weaned mice were kept with their mothers and at P21, placed into a new cage without their mothers. Both groups were given a single dose of DEN at P19 (25ug/g) based on their body weights.
In vivo siRNA Experiments
For in vivo experiments in C3H, LAP-MYC, Rosa-rtta; TRE-Cas9, FVB and P53fl/fl; Rb1fl/fl; Rbl2fl/fl mice, formulated 5A2-SC8 dendrimer lipid nanoparticles (LNPs) were used to package either Negative Control #1, Anln siRNA, Cdk1 siRNA or E2f8 siRNA at 2 mg/kg, two times a week for a total of 4 times (2 intraperitoneal and 2 retroorbital). LNPs containing siRNAs were formulated following previously reported component ratios of 5A2-SC8, DSPC, cholesterol, and lipid PEG2000 (molar ratio of 50:38:10:2) and methods (Zhou et al., 2016) with the aid of a microfluidic rapid mixing instrument (Precision Nanosystems NanoAssemblr) and purified by dialysis in sterile PBS before injection.
GalNAc conjugated Anln siRNAs Experiments
These siRNAs were obtained from Alnylam Pharmaceuticals. 10mg/ml stock for siLuc and siAnln was stored at −20°C. The working concentration of t he siRNA was 4mg/ml, diluted in PBS. We chose two siAnln (#2 and #4) with good knockdown efficiency for the subsequent experiments. C3H mice were subcutaneously injected with siLuc, siAnln#2 or siAnln#4 at 4.0mg/kg starting at P8–10 of age. Injections were given 4 days apart for a total of 3 times.
Partial Hepatectomy
Surgery was performed as previously described (Mitchell and Willenbring, 2008).
Chemical Injury Experiments
CCl4 was diluted 1:10 in corn oil (Sigma), and administered IP at a dose of 0.5 ml/kg of mouse as described previously (Beer et al., 2008). DEN is diluted in saline and administered IP at different doses, depending on mouse age.
Virus Experiments
100µL of Ad-Cas9-sgPten was retro-orbitally injected at a dose of 109 pfu/mouse. 100µL of Adenovirus-GFP (University of Iowa Viral Vector Core) was retro-orbitally injected at a dose of 1.4 × 108 pfu/mouse. 100µL of AAV-sgApc (5 × 1012 pfu/mouse) was retro-orbitally injected. 100µL of Ad-Cas9-sgPten (109 pfu/mouse) plus AAV-Cre (University of Pennsylvania Vector Core, 2.5 × 1010 pfu/mouse) was retro-orbitally injected. 100µL of Ad-Cas9-sgPten (109 pfu/mouse) plus AAV-KPL (1012 pfu/mouse, Boston Children’s Hospital Viral Core) was retro-orbitally injected.
RNA Extraction and RT-qPCR
Total RNA was isolated using Trizol reagent (Invitrogen). For qRT-PCR, cDNA synthesis was performed with 1µg of total RNA using iScript Reverse Transcription Kit (Biorad). Gene expression levels were measured using the ΔΔCt method as described previously (Zhu et al., 2010).
Western Blot Assay
Mouse liver tissues were homogenized and lysed in T-PER Tissue Protein Extraction Reagent (Thermo Scientific Pierce). Western blots were performed in the standard fashion. The following antibodies were used: anti-β-Actin, Anti-Anln, Anti-E2f8, Anti-Phospho-BRCA1, Anti-Phospho-p53 (Ser15), Anti-Phospho-Histone H2A.X (Ser139), Anti-rabbit IgG, HRP-linked Antibody and Anti-mouse IgG, HRP-linked Antibody.
Histology, Immunohistochemistry, and Immunofluorescence
Tissue samples were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin. In some cases, frozen sections were made. Immunohistochemistry was performed as previously described (Zhu et al., 2010). Primary antibodies used: Anti-Ki67, Anti-Cyp2e1, Anti-GS, Anti-Pten, Anti-Ctnnb1, Anti-Phospho-Histone H2A.X. For immunohistochemistry, detection was performed with the Elite ABC Kit and DAB Substrate (Vector Laboratories), followed by Hematoxylin Solution counterstaining (Sigma). For immunofluorescence, second antibodies Alexa Fluor 594 goat anti-mouse IgG1 or Alexa Fluor 488 goat anti-rabbit were used. Confocal images were taken by Zeiss LSM 780 Upright confocal/multiphoton microscope. H&E slides were interpreted by a clinical pathologist with expertise in human liver cancer diagnosis.
Primary Hepatocytes Isolation
Primary hepatocytes were isolated by two-step collagenase perfusion. Liver perfusion medium (Thermo Fisher Scientific, 17701038), liver digest medium (Thermo Fisher Scientific, 17703034) and Hepatocyte wash medium (Thermo Fisher Scientific, 17704024) were used. Cell number and viability were determined by Trypan blue exclusion in a hemocytometer.
ROS Detection
For ROS detection in primary hepatocytes, cells were incubated with 5 µM CellROX Deep Red Reagent at 37°C for 30 min. Then cells were analyze d by Flow Cytometry. For ROS detection on fresh tissue sections (Sun et al., 2017), DHE (Sigma) staining for superoxide was performed. Cryosections (10 µm) were freshly cut and incubated with 10 µM DHE at 37°C for 30 min. Ethidium staining was visualized using laser confocal microscopy (LSM 780, Zeiss) at the UTSW Live Cell Imaging Facility. The red signal corresponded to levels of cellular superoxide anion and the intensities were quantified in 20 cells from five fields for each mouse by ImageJ.
Flow Cytometry
For detection of ploidy populations, primary hepatocytes (2×106/mL) or transfected H2.35 cells (1 × 106 mL) were fixed in 75% ethanol at −20°C, then incubated with 500µL (2×106 mL) of PI/Rnase Staining Buffer (BD Pharmingen) at 25°C for 15 minutes. Cells were analyzed with a BD FACS Aria Fusion machine (BD Biosciences). For Ki67 staining, cells were fixed in 75% ethanol at −20°C, washed twice in staining buffer (PBS with 1% FBS), and incubated with anti-Ki67 antibody (BD#550609, 1:50) at room temperature (RT) for 30 minutes in the dark. Then the cells were incubated with secondary antibody Alexa Fluor 488 goat anti-mouse IgG1 (life technologies) for 20min at RT, and resuspended in PI/Rnase Staining Buffer (BD Pharmingen), and then flow cytometry analyzed.
Fluorescence In Situ Hybridization (FISH) assay
Two FISH probes for mChr12 were provided by Dr. Hongtao Yu’s lab. Bacterial artificial chromosomes were nick translated with the CGH Nick Translation Kit (Abbott Molecular) using Red 580 dUTP and Green 496 dUTP (Enzo Life Sciences). Isolated primary hepatocytes were treated with 0.56% KCL and fixed with cold 3:1 methonal: acetic acid solution. Then the fixed cells were dropped onto slides, and allowed to dry. Then the slides were incubated with probes, covered with coverslips sealed with rubber cement. Slides were incubated at 80°C for 5 min and 37°C overnight. Following incubation, slides were washed in 0.5× SSC+0.1% at 70°C for 5 min, 1×SSC for 3×5 min at room temparature, 4×SSC+0.1% Tween for 5 min at room temperature, and 2×SSC for 5 min, and then mounted with DAPI (Vectorlabs). Samples were analyzed and scored under an Olympus I×83 microscope. For analysis of nuclei ploidy, 2 green and 2 red dots was counted as a 2c nucleus, and 4 green and 4 red dots was counted as a 4c nucleus. 60 nuclei were counted for each sample.
Genomic DNA Isolation and Targeted Sequencing
50 flash-frozen tumors free of visible normal tissue were used for library preparation. The genomic DNA was extracted using QIAGEN AllPrep DNA/RNA Kit (Cat. 80204). DNA integrity was assessed by electrophoresis on 1% agarose gels, and concentration was determined by nanodrop. Genomic DNA was sonicated into 500bp fragments and purified using Genomic DNA Clean & Concentrator Kit (ZYMO RESEARCH). The DNA library was prepared using Ovation Target Enrichment System (NuGEN) following the manufacturer’s instructions. Target genes were selected from human HCC and mouse sequencing studies (Bard-Chapeau et al., 2014; Keng et al., 2009; Shen et al., 2016). The target probes were synthesized by NuGEN. More than 99.5% of the probes had >90% coverage. The sequencing was performed using a 150 bp single-end protocol on Illumina NextSeq 500 platform by the CRI Sequencing Core.
QUANTIFICATION AND STATISTICAL ANALYSIS
Sequence Processing
BCL files from Illumina Nextseq 500 sequencing were converted to FASTQ files by bcl2fastq (Illumina). After trimming by trim_galore package, BWA-MEM (version 0.7.15) was used to align FASTQ files to reference genome GRCm38 with subsequent processing by Samtools (version 1.3) and Nudup.py (Nugen) to ensure proper file formatting and remove duplicates. Alignments were then recalibrated and realigned by GATK (version 3.5). We acquired 42 million uniquely mapped reads on average for the 53 samples we sequenced with an average on target coverage at 128X and more than 86.6% region has more than 50× coverage.
Identification and Annotation of Somatic SNVs
To detect somatic variants in tumor samples, we use the somatic variant detection program Mutect (version 1.1.7). 50 tumor samples were called against a panel of 3 normal liver samples from two WT C3H/HeJ mice to the reference genome GRCm38. Variants passed the Mutect high-confidence somatic mutation filters were selected and further filtered against from known SNVs in C3H/HeJ mice provided by http://ftp-mouse.sanger.ac.uk (mgp.v3). SNVs were further annotated using snpEff (version 4.2) and Variant Effect Predictor. SNVs predicted to have High or Moderate impact were compared to a stringent list of 125 driver genes in human cancer by Vogelstein et al (Vogelstein et al., 2013) to map putative driver genes and mutations. The mutation landscape was graphed by using GenVisR (a package from R Bioconductor) waterfall plot algorithm on putative driver genes for all 50 tumor samples (Figure 6A).
Statistical Analysis
The sample size was determined by producing a confidence interval estimate with a specified margin of error to ensure that a test of hypothesis has a high probability of detecting a meaningful difference in the parameter. The data in most figure panels reflect multiple experiments performed on different days using mice derived from different litters. Variation is indicated using standard error presented as mean ± SEM. For each dataset, we first performed the Shapiro-Wilk normality test and the D’Agostino’s K-squared test. If the dataset passed the normality test, we then conducted the F-test to see whether the variances of the two groups were equal or not. If variances were unequal, we conducted the hypothesis test using t-test with Welch’s correction (assuming unequal variances); if variances were equal, we conducted the hypothesis test using the two-tailed student’s t-test. If the dataset did not pass the normality test, we then conducted the hypothesis test using non-parametric test Mann-Whitney U test. Statistical significance is displayed as p < 0.05 (*) or p < 0.01 (**) unless specified otherwise. The investigators were blinded during the processes of mice treatments and data analysis. Image analysis for the quantification of cell proliferation, Pten deletion, GFP+ cells, liver surface tumor and malignant nodule numbers were performed in a blinded fashion.
DATA AND SOFWARE AVAILABILITY
The DEN tumor DNA targeted sequencing data reported in Figure 6A and Table S2 has been deposited into the NCBI GEO database with the accession number GSE108645.
Supplementary Material
Table S2. Target gene list of the DNA targeted sequencing (designed by NuGEN). Related to Figure 6.
Highlights.
Premature weaning, E2f8 knockdown, and Anln knockdown altered hepatocyte ploidy.
Ploidy alterations did not alter liver function, metabolism, or regeneration.
Livers with more polyploid cells had suppressed tumorigenesis
Polyploid livers were protected from tumor suppressor loss of heterozygosity.
Acknowledgments
We thank H. Yu, M. Buszczak, S. Morrison, and H. Sadek for critical input and advice; E. Choi for assistance with FISH; Jian Xu and the CRI sequencing core for genome sequencing; Kate Luby-Phelps and Abhijit Bugde for confocal imaging. We thank Alnylam’s Bioinformatics, Oligo synthesis and RNAi Lead Discovery team for designing, generating and screening for the Anln GalNAc siRNAs. We thank Eric Olson and John McAnally for advice and assistance with transgenic mouse generation. D.J.S. was supported by CPRIT (R1212), the Welch Foundation (I-1855), and the American Cancer Society (RSG-17-012-01). X.L. is supported by Cancer Prevention and Research Institute of Texas (RP150596). H.Z. was supported by the Pollack Foundation, an American Cancer Society pilot grant (IRG-02-196), a NIH/NCI R01 grant (R01CA190525), a Burroughs Welcome Career Award for Medical Scientists, and a CPRIT New Investigator grant (R1209).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
S.Z. and H.Z. conceived the project, performed the experiments and wrote the manuscript. L.L., S.Z., L.N., assisted with experiments and mouse husbandry. K.Z. and D.S. assisted with the chemistry experiments. Y.Z. helped to make the mouse model. X.L. performed bioinformatics analysis. H.T. and A.S. provided the GalNAc conjugated siRNAs. P.G. performed pathologic interpretation. B.T. assisted with writing and project design.
Declaration of Interest
Ho-Chou Tu and Alfica Sehgal were employees of Alnylam Pharmaceuticals when the work was done.
References
- Anti M, Marra G, Rapaccini GL, Rumi C, Bussa S, Fadda G, Vecchio FM, Valenti A, Percesepe A, Pompili M, et al. DNA ploidy pattern in human chronic liver diseases and hepatic nodular lesions. Flow cytometric analysis on echo-guided needle liver biopsy. Cancer. 1994;73:281–288. doi: 10.1002/1097-0142(19940115)73:2<281::aid-cncr2820730208>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Bard-Chapeau EA, Nguyen AT, Rust AG, Sayadi A, Lee P, Chua BQ, New LS, de Jong J, Ward JM, Chin CK, et al. Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nature genetics. 2014;46:24–32. doi: 10.1038/ng.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer S, Komatsubara K, Bellovin DI, Kurobe M, Sylvester K, Felsher DW. Hepatotoxin-induced changes in the adult murine liver promote MYC-induced tumorigenesis. PloS one. 2008;3:e2493. doi: 10.1371/journal.pone.0002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Arad G, Weissbein U, Mandefro B, Maimon A, Golan-Lev T, Narwani K, Clark AT, Andrews PW, Benvenisty N, Carlos Biancotti J. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nature communications. 2014;5:4825. doi: 10.1038/ncomms5825. [DOI] [PubMed] [Google Scholar]
- Buchmann A, Mahr J, Bauerhofmann R, Schwarz M. Mutations at Codon 61 of the Ha-Ras Proto-Oncogene in Precancerous Liver-Lesions of the B6c3f1 Mouse. Mol Carcinogen. 1989;2:121–125. doi: 10.1002/mc.2940020303. [DOI] [PubMed] [Google Scholar]
- Caselitz M, Masche N, Bleck JS, Gebel M, Atay Z, Stern C, Manns MP, Kubicka S. Increasing sensitivity of morphological diagnosis in hepatocellular carcinoma (HCC) by combination of cytological and fine-needle histological examination after ultrasound guided fine needle biopsy. Zeitschrift fur Gastroenterologie. 2003;41:559–564. doi: 10.1055/s-2003-39637. [DOI] [PubMed] [Google Scholar]
- Celton-Morizur S, Merlen G, Couton D, Margall-Ducos G, Desdouets C. The insulin/Akt pathway controls a specific cell division program that leads to generation of binucleated tetraploid liver cells in rodents. Journal of Clinical Investigation. 2009;119:1880–1887. doi: 10.1172/JCI38677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao DY, Dilkes B, Luo HB, Douglas A, Yakubova E, Lahner B, Salt DE. Polyploids Exhibit Higher Potassium Uptake and Salinity Tolerance in Arabidopsis. Science. 2013;341:658–659. doi: 10.1126/science.1240561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HZ, Ouseph MM, Li J, Pecot T, Chokshi V, Kent L, Bae S, Byrne M, Duran C, Comstock G, et al. Canonical and atypical E2Fs regulate the mammalian endocycle. Nat Cell Biol. 2012;14:1192-+. doi: 10.1038/ncb2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colnot S, Decaens T, Niwa-Kawakita M, Godard C, Hamard G, Kahn A, Giovannini M, Perret C. Liver-targeted disruption of Apc in mice activates beta-catenin signaling and leads to hepatocellular carcinomas. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17216–17221. doi: 10.1073/pnas.0404761101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nature reviews Genetics. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Davoli T, de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21:765–776. doi: 10.1016/j.ccr.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155:948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diril MK, Ratnacaram CK, Padmakumar VC, Du T, Wasser M, Coppola V, Tessarollo L, Kaldis P. Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3826–3831. doi: 10.1073/pnas.1115201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW. Aneuploidy, polyploidy and ploidy reversal in the liver. Seminars in cell & developmental biology. 2013;24:347–356. doi: 10.1016/j.semcdb.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Duncan AW, Hanlon Newell AE, Smith L, Wilson EM, Olson SB, Thayer MJ, Strom SC, Grompe M. Frequent aneuploidy among normal human hepatocytes. Gastroenterology. 2012;142:25–28. doi: 10.1053/j.gastro.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, Grompe M. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto J, Okamoto E, Yamanaka N, Toyosaka A, Mitsunobu M. Flow cytometric DNA analysis of hepatocellular carcinoma. Cancer. 1991;67:939–944. doi: 10.1002/1097-0142(19910215)67:4<939::aid-cncr2820670414>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Cornils H, Chiu SY, O'Rourke KP, Arnaud J, Yimlamai D, Thery M, Camargo FD, Pellman D. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell. 2014;158:833–848. doi: 10.1016/j.cell.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentric G, Desdouets C. Polyploidization in liver tissue. The American journal of pathology. 2014;184:322–331. doi: 10.1016/j.ajpath.2013.06.035. [DOI] [PubMed] [Google Scholar]
- Gentric G, Maillet V, Paradis V, Couton D, L'Hermitte A, Panasyuk G, Fromenty B, Celton-Morizur S, Desdouets C. Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. The Journal of clinical investigation. 2015;125:981–992. doi: 10.1172/JCI73957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindryckx F, Colle I, Van Vlierberghe H. Experimental mouse models for hepatocellular carcinoma research. International journal of experimental pathology. 2009;90:367–386. doi: 10.1111/j.1365-2613.2009.00656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. The New England journal of medicine. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- Keng VW, Villanueva A, Chiang DY, Dupuy AJ, Ryan BJ, Matise I, Silverstein KA, Sarver A, Starr TK, Akagi K, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nature biotechnology. 2009;27:264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent LN, Rakijas JB, Pandit SK, Westendorp B, Chen HZ, Huntington JT, Tang X, Bae S, Srivastava A, Senapati S, et al. E2f8 mediates tumor suppression in postnatal liver development. The Journal of clinical investigation. 2016;126:2955–2969. doi: 10.1172/JCI85506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Pelletier L, Heninger AK, Slabicki M, Theis M, Miroslaw L, Poser I, Lawo S, Grabner H, Kozak K, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007;9:1401–1412. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- Knouse KA, Wu J, Whittaker CA, Amon A. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:13409–13414. doi: 10.1073/pnas.1415287111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurinna S, Stratton SA, Coban Z, Schumacher JM, Grompe M, Duncan AW, Barton MC. p53 Regulates a Mitotic Transcription Program and Determines Ploidy in Normal Mouse Liver. Hepatology. 2013;57:2004–2013. doi: 10.1002/hep.26233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, Thorgeirsson SS. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nature genetics. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wu F. Global burden of aflatoxin-induced hepatocellular carcinoma: a risk assessment. Environmental health perspectives. 2010;118:818–824. doi: 10.1289/ehp.0901388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margall-Ducos G, Celton-Morizur S, Couton D, Bregerie O, Desdouets C. Liver tetraploidization is controlled by a new process of incomplete cytokinesis. Journal of cell science. 2007;120:3633–3639. doi: 10.1242/jcs.016907. [DOI] [PubMed] [Google Scholar]
- Mayhew CN, Bosco EE, Fox SR, Okaya T, Tarapore P, Schwemberger SJ, Babcock GF, Lentsch AB, Fukasawa K, Knudsen ES. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer research. 2005;65:4568–4577. doi: 10.1158/0008-5472.CAN-04-4221. [DOI] [PubMed] [Google Scholar]
- Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nature protocols. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- Nagasue N, Kohno H, Hayashi T, Yamanoi A, Uchida M, Takemoto Y, Makino Y, Ono T, Hayashi J, Nakamura T. Lack of intratumoral heterogeneity in DNA ploidy pattern of hepatocellular carcinoma. Gastroenterology. 1993;105:1449–1454. doi: 10.1016/0016-5085(93)90150-b. [DOI] [PubMed] [Google Scholar]
- Nijhawan D, Zack TI, Ren Y, Strickland MR, Lamothe R, Schumacher SE, Tsherniak A, Besche HC, Rosenbluh J, Shehata S, et al. Cancer vulnerabilities unveiled by genomic loss. Cell. 2012;150:842–854. doi: 10.1016/j.cell.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Pandit SK, Westendorp B, Nantasanti S, van Liere E, Tooten PC, Cornelissen PW, Toussaint MJ, Lamers WH, de Bruin A. E2F8 is essential for polyploidization in mammalian cells. Nature cell biology. 2012;14:1181–1191. doi: 10.1038/ncb2585. [DOI] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, Scuoppo C, Zuber J, Dickins RA, Kogan SC, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer BE, Park KS, Yiu G, Conklin JF, Lin C, Burkhart DL, Karnezis AN, Sweet-Cordero EA, Sage J. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer research. 2010;70:3877–3883. doi: 10.1158/0008-5472.CAN-09-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub JR, Malato Y, Gormond C, Willenbring H. Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell reports. 2014;8:933–939. doi: 10.1016/j.celrep.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz-Arad D, Zajicek G, Bartfeld E. The streaming liver IV: DNA content of the hepatocyte increases with its age. Liver. 1989;9:93–99. doi: 10.1111/j.1600-0676.1989.tb00385.x. [DOI] [PubMed] [Google Scholar]
- Selmecki AM, Maruvka YE, Richmond PA, Guillet M, Shoresh N, Sorenson AL, De S, Kishony R, Michor F, Dowell R, Pellman D. Polyploidy can drive rapid adaptation in yeast. Nature. 2015;519:349-+. doi: 10.1038/nature14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- Sheltzer JM, Ko JH, Replogle JM, Habibe Burgos NC, Chung ES, Meehl CM, Sayles NM, Passerini V, Storchova Z, Amon A. Single-chromosome Gains Commonly Function as Tumor Suppressors. Cancer Cell. 2017;31:240–255. doi: 10.1016/j.ccell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Tsoi H, Liang Q, Chu ES, Liu D, Yu AC, Chan TF, Li X, Sung JJ, Wong VW, Yu J. Oncogenic mutations and dysregulated pathways in obesity-associated hepatocellular carcinoma. Oncogene. 2016;35:6271–6280. doi: 10.1038/onc.2016.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihao Zhang QC, Liu Qingxu, Li Yuxi, Sun Xiufeng, Hong Lixin, Ji Suyuan, Liu Chengyan, Jing Geng WZ, Lu Zhonglei, Yin Zhen-Yu, Zeng Yuanyuan, Lin Kwang-Huei, Wu Qiao, Li Qiyuan, KIN Keiko Nakayama, Deng Xianming, Johnson Randy L, Zhu Liang, Gao Daming, Chen Lanfen, Zhou aD. Hippo Signaling Suppresses Cell Ploidy and Tumorigenesis through Skp2. Cancer Cell. 2017;31:669–684. doi: 10.1016/j.ccell.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoshani O, Massalha H, Shani N, Kagan S, Ravid O, Madar S, Trakhtenbrot L, Leshkowitz D, Rechavi G, Zipori D. Polyploidization of murine mesenchymal cells is associated with suppression of the long noncoding RNA H19 and reduced tumorigenicity. Cancer research. 2012;72:6403–6413. doi: 10.1158/0008-5472.CAN-12-1155. [DOI] [PubMed] [Google Scholar]
- Silk AD, Zasadil LM, Holland AJ, Vitre B, Cleveland DW, Weaver BA. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E4134–4141. doi: 10.1073/pnas.1317042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wang SC, Wei Y, Luo X, Jia Y, Li L, Gopal P, Zhu M, Nassour I, Chuang JC, et al. Arid1a Has Context-Dependent Oncogenic and Tumor Suppressor Functions in Liver Cancer. Cancer cell. 2017;32:574–589. e576. doi: 10.1016/j.ccell.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura J, Tanaka J, Fujita K, Yoshida M, Kasamatsu T, Arii S, Tobe T. Cell kinetics of regenerating liver after 70% hepatectomy in rats-2-color flow cytometric analysis. HPB surgery : a world journal of hepatic, pancreatic and biliary surgery. 1992;5:103–114. doi: 10.1155/1992/51372. discussion 114-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda H, Bregerie O, Vallet A, Nalpas B, Pivert G, Brechot C, Desdouets C. Changes to hepatocyte ploidy and binuclearity profiles during human chronic viral hepatitis. Gut. 2005;54:297–302. doi: 10.1136/gut.2004.043893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature. 2015;524:180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li Z, Ye Y, Xie L, Li W. Oxidative Stress and Liver Cancer: Etiology and Therapeutic Targets. Oxid Med Cell Longev. 2016;2016:7891574. doi: 10.1155/2016/7891574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Westcott PM, Halliwill KD, To MD, Rashid M, Rust AG, Keane TM, Delrosario R, Jen KY, Gurley KE, Kemp CJ, et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature. 2015;517:489–492. doi: 10.1038/nature13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Yoshimi N, Sugie S, Suzui M, Matsunaga K, Kawabata K, Hara A, Mori H. Beta-catenin (Ctnnb1) gene mutations in diethylnitrosamine (DEN)-induced liver tumors in male F344 rats. Japanese journal of cancer research : Gann. 1999;90:824–828. doi: 10.1111/j.1349-7006.1999.tb00822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J, Mermel CH, et al. Pan-cancer patterns of somatic copy number alteration. Nature genetics. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Li L, Kendrick SL, Gerard RD, Zhu H. TALEN-mediated somatic mutagenesis in murine models of cancer. Cancer research. 2014;74:5311–5321. doi: 10.1158/0008-5472.CAN-14-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Nguyen LH, Zhou K, Tu HC, Sehgal A, Nassour I, Li L, Gopal P, Goodman J, Singal AG, et al. Knockdown of Anillin Actin Binding Protein Blocks Cytokinesis in Hepatocytes and Reduces Liver Tumor Development in Mice without Affecting Regeneration. Gastroenterology. 2017 doi: 10.1053/j.gastro.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Nguyen LH, Miller JB, Yan Y, Kos P, Xiong H, Li L, Hao J, Minnig JT, Zhu H, Siegwart DJ. Modular degradable dendrimers enable small RNAs to extend survival in an aggressive liver cancer model. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:520–525. doi: 10.1073/pnas.1520756113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nature genetics. 2010;42:626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Target gene list of the DNA targeted sequencing (designed by NuGEN). Related to Figure 6.