Abstract
Neddylation, a post-translational modification that conjugates an ubiquitin-like protein NEDD8 to substrate proteins, is an important biochemical process that regulates protein function. The best-characterized substrates of neddylation are the cullin subunits of Cullin-RING ligases (CRLs), which, as the largest family of E3 ubiquitin ligases, control many important biological processes, including tumorigenesis, through promoting ubiquitylation and subsequent degradation of a variety of key regulatory proteins. Recently, increasing pieces of experimental evidence strongly indicate that the process of protein neddylation modification is elevated in multiple human cancers, providing sound rationale for its targeting as an attractive anticancer therapeutic strategy. Indeed, neddylation inactivation by MLN4924 (also known as pevonedistat), a small molecule inhibitor of E1 NEDD8-activating enzyme currently in phase I/II clinical trials, exerts significant anticancer effects by inducing cell cycle arrest, apoptosis, senescence and autophagy in a cell-type and context dependent manner. Here, we summarize the latest progresses in the field with a major focus on preclinical studies in validation of neddylation modification as a promising anticancer target.
Keywords: Neddylation, Cancer target, Cullin RING Ligase, MLN4924, cell cycle arrest, apoptosis, senescence, autophagy
Introduction
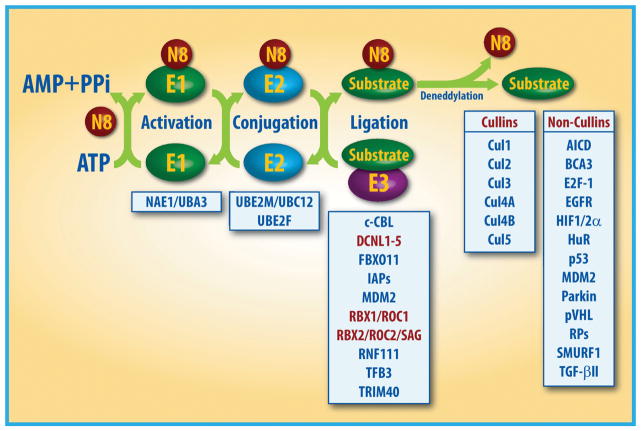
Protein neddylation is a process of conjugating an ubiquitin-like molecule NEDD8 (neuronal precursor cell-expressed developmentally down-regulated protein 8) to targeted proteins via a three-step enzymatic reaction, catalyzed by NEDD8-activating enzyme E1 (NAE), NEDD8-conjugating enzyme E2s (UBC12/UBE2M or UBE2F) and substrate-specific NEDD8-E3 ligases [1–5] (Fig. 1). In this process, the mature NEDD8 is first adenylated and activated in an ATP-dependent manner by the NEDD8-activating enzyme (NAE), a heterodimer consisting of NAE1 (also known as APPBP1) and UBA3 (also known as NAEβ) [6]. The activated NEDD8 is then transferred to one of two NEDD8-conjugating enzymes, UBE2M (also known as UBC12) or UBE2F through a trans-thiolation reaction [7, 8]. The final step in the process involves the transfer of NEDD8 from E2 to its substrate protein on a lysine residue via covalent attachment, catalyzed by an E3 ligase (Fig. 1).
Figure 1. The process of protein modification by neddylation.
Neddylation is a process that conjugates NEDD8, an ubiquitin-like molecule, to targeted protein substrates via enzymatic cascades involving NEDD8-activating enzyme E1, NEDD8-conjuagating enzyme E2 and substrate-specific NEDD8-E3 ligases. Shown are reported neddylation E3s and substrates. The substrates were divided into cullins and non-cullins. N8: NEDD8.
Unlike E3 ubiquitin ligases which have 4 different classes with more than 600 members [9], there are only about 10 different NEDD8 E3 ligases and the most of them contains the Really Interesting New Gene (RING) domain structure. Specifically, these NEDD8 E3 ligases include RING-box proteins 1 (RBX1) and RBX2 [also known as regulators of cullins 1 (ROC1) and ROC2/SAG, respectively] [10–12], murine double minute 2 (MDM2) [13], casitas B-lineage lymphoma (c-CBL) [14, 15], SCFFBXO11 [16], ring fingers protein 111 (RNF111) [17], inhibitor of apoptosis (IAPs) [18], TFB3 [19] and TRIM40 [20]. In yeast and Caenorhabditis elegans, defective in cullin neddylation 1 (DCN1) has been identified as a NEDD8 E3 ligase, which does not contain a RING domain for its catalytic activity, rather it directly interacts with the UBE2M on a surface that overlaps with the E1-binding site [21, 22]. In human cells, DCN1-LIKE proteins (DCNLs) with five family members, DCNL1–DCNL5, have distinct amino-terminal domains, but share a conserved C-terminal potentiating neddylation (PONY) domain, which is necessary and sufficient for cullin neddylation, as shown in yeast DCN1 [21]. Among all NEDD8 E3 ligases, the best characterized ones are RBX family members: RBX1 pairs with neddylation E2 UBE2M to catalyze the neddylation of CUL-1, -2, -3, -4A, and -4B, which is facilitated by DCNL1, whereas RBX2 pairs with E2 UBE2F for CUL-5 neddylation [11, 23]. Interestingly, all currently reported NEDD8 E3 ligases can also function as ubiquitin E3 ligases; and the cullin members, upon activation by neddylation, acting as the largest family of ubiquitin E3s [5]. It will be very interesting to understand under what physiological or stressed conditions these dual E3s will promote either ubiquitylation, neddylation or both, and on what protein substrates and on which lysine residues.
Unexpectedly, a recent study reported that glycyl-tRNA synthetase, an essential enzyme in protein synthesis, plays a critical role in neddylation by binding to the NAE1/APPBP1 subunit of E1 to capture and protect activated UBE2M E2 before it reaches a downstream target [24]. Thus, neddylation enzymatic cascade can be subjected to additional regulation.
Substrates of neddylation
To date, the best-characterized physiological substrates, subjected to neddylation modification, are cullin family members (Cul-1, 2, 3, 4A, 4B, and 5, other two cullins, cullin-7 and -9 are less studied) that each are a scaffold subunit of Cullin-RING ligases (CRLs) [25] (Fig. 1). CRLs, upon activation by neddylation, act as the largest family of multiunit E3 ubiquitin ligases, and are responsible for ubiquitylation and degradation of about 20% of all cellular proteins for targeted degradation via ubiquitin-proteasome system (UPS) [25–27]. Some of CRL substrates are short-lived key regulatory molecules such as signal transducers, cell cycle regulators, transcription factors, tumor suppressors, oncoproteins, etc [25–28]. Through targeted degradation, CRLs regulate many biological processes [5, 25, 26]. The assembling of CRLs requires cullin proteins acting as a molecular scaffold that binds to an adaptor protein (such as SKP1 in CRL1) and a substrate receptor protein (such as F-box protein in CRL1) at the N-terminus, and a RING protein, RBX1 or RBX2 at the C-terminus [25, 29–31]. Activation of CRLs requires the attachment of NEDD8 to a C-terminal lysine residue of cullins [32, 33], which causes structural change in the CRLs complex, adapting an open conformation to facilitate the access of the substrates for ubiquitylation [34–36].
In addition to cullins, several other non-cullin proteins have been identified as the substrates of neddylation (Fig. 1). For example, p53 tumor suppressor can be neddylated by Mdm2, but unlike Mdm2-induced p53 ubiquitylation for targeted degradation [37], p53 neddylation inhibits its transcriptional activity without affecting its degradation [13, 38]. Thus, Mdm2 promotes both ubiquitylation and neddylation of p53, leading to its inactivation via different mechanisms. Interestingly, a recent study showed that Src phosphorylation of Mdm2 on Y281 and Y302 converts Mdm2 from a ubiquitylating to a neddylating E3 ligase [39]. On the other hand, Mdm2 E3 also promotes both ubiquitylation and neddylation of itself [13]. Mdm2 auto-ubiquitylation destabilizes itself; whereas Mdm2 auto-neddylation increases its protein stability [40]. Furthermore, Mdm2 promotes neddylation of few ribosomal proteins, such as L11 and S14, to regulate their stabilization and subcellular location [41–43]. Other reported non-cullin substrates of neddylation include tumor suppressor pVHL [44], oncoproteins Hu antigen R (HuR) [45, 46], receptor proteins such as EGFR [14] and TGF-β type II receptor [15], transcriptional regulators such as HIF1α/HIF2α [47], breast cancer-associated protein 3 (BCA3) [48], APP intracellular domain (AICD) [49], and E2F-1 [50], HECT-domain ubiquitin E3 ligase SMURF1 and RBR ubiquitin E3 ligase Parkin [1, 51] (Fig. 1). The proteomics analysis at the whole genome levels has identified numerous proteins responsive to MLN4924 treatment [52, 53] and some of them could be new non-cullin substrates of neddylation, awaiting experimental validation. It is worthy-noting that while neddylation of cullin family members is well defined mechanistically and structurally [1], neither of non-cullin substrates has been fully characterized as physiological substrate of neddylation, since neddylated form of a non-cullin substrate is hardly detectable under physiological condition due to either its low abundance or non-existence. Moreover, the mechanism by which neddylation modification of non-cullin substrates is poorly defined. Since ubiquitin and NEDD8 could compete for the same lysine residue on a substrate, experimental observations of smeared slow-migrating bands upon NEDD8 overexpression likely reflect a mixture of ubiquitylation and neddylation on a given substrate.
Neddylation modification as an attractive anticancer target
Inhibition of protein neddylation has been recently characterized as an attractive anticancer strategy. A high throughput screening identified N6-benzyl adenosine as an inhibitor of NAE, resulting in the discovery of MLN4924 (also known as Pevonedistat) with additional medicinal chemistry efforts [27]. MLN4924, developed by Millennium Pharmaceuticals, is a potent and highly selective small molecular inhibitor of NAE, inactivating the first step of the neddylation cascade [27]. When bound to the active site of NAE, MLN4924 forms a steady-state covalent adduct with NEDD8, which resembles the adenylate-NEDD8 adduct at the active site of NAE to block further enzymatic process, thus terminating the cascade at this proximal step [1, 54]. Functionally, MLN4924 effectively blocks cullin neddylation to inhibit the activation of CRLs, leading to accumulation of various CRLs substrates and subsequently induction of multiple cell death pathways in cancer cells [27, 54–57].
Preclinical studies showed that MLN4924 has potent antitumor activity with well-tolerated toxicity against a range of solid tumors and hematological malignancies [58–60]. Based on these findings, a series of Phase I and II clinical trials were conducted to assess the tolerable safety, pharmacokinetics (PK), pharmacodynamics (PD) and antitumor activity of MLN4924 in patients suffering from MDS, AML, lymphoma, melanoma and solid tumors (summarized in Table 1) [61–66]. The first four earlier phase I trials, which are now completed, demonstrated the validity of NAE inhibition as a therapeutic target in the clinical setting for the objective effects with prolonged stable disease, partial responses (PRs) and completed responses (CRs) [66] (Table 1).
Table 1.
Clinical Trials of MLN4924
| Clinicaltrial. gov identifier | Phase | Tumor type | Time initiated | With conbination | Antitumor activity | Ref |
|---|---|---|---|---|---|---|
| NCT00677170 | I | Solid tumor | May 2008 | Alone | 9 SDs | [61] |
| NCT00722488 | I | HL, HM, MM, lymphoma | July 2008 | Alone | 1 PR | [62] |
| NCT00911066 | I | AML, ALL, MS | May 2009 | Alone With azacitidine |
4 CRs | [63] |
| NCT01011530 | I | Metastatic melanoma | November 2009 | Alone | 9 SDs 1 PR |
[64] |
| NCT01862328 | Ib | Solid tumor | May 2013 | With docetaxel With gemcitabine With carboplatin +paclitaxel |
NR | |
| NCT01814826 | Ib | AML | March 2013 | With azacitidine | 6 CRs 4 PRs |
[65] |
| NCT02610777 | II | AML | November 2015 | with azacitidine versus single-agent azacitidine | NR |
AML: Acute myeloid leukemia; ALL: acute lymphoblastic leukemia; CR: Complete response; HL: Hodgkin lymphoma; HM: hematologic malignancies; MM: multiple myeloma; MS: myelodysplastic syndrome; NR: not reported; PR: partial response; SD: stable disease.
Furthermore, additional preclinical studies have demonstrated that combinations of MLN4924 with chemoradiotherapy increased antitumor activity in AML and solid tumor cell lines and xenograft models [67]. Since 2013, three MLN4924 combinational phase Ib/II trials have been launched. The first phase Ib trial is completed with combination of MLN4924 and azacitidine in patients with AML (NCT01814826). Compared with azacitidine or MLN4924 alone, the combination therapy achieved better therapeutic effects with 6 (33%) CRs and 4 (22%) PRs [65] (Table 1). Given this promising clinical effects, a new randomized phase II study, with MLN4924-azacitidine combination versus azacitidine alone is currently recruiting patients with AML (NCT02610777). Another phase Ib trial is ongoing in combination of MLN4924 with docetaxel, gemcitabine, and carboplatin–paclitaxel in patients with solid tumors (NCT01862328).
Elevated status of neddylation modification in human cancer
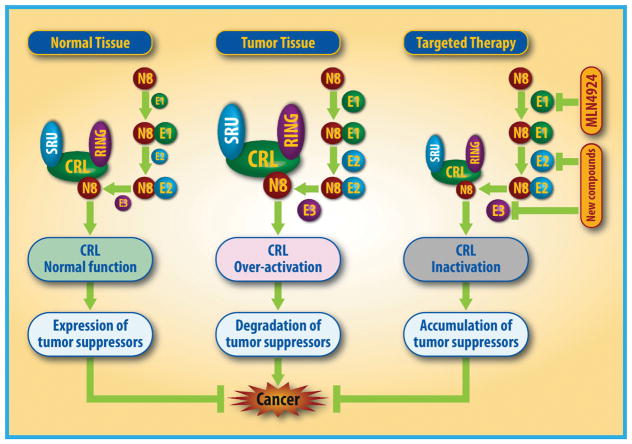
To validate whether neddylation modification is an attractive cancer target, it is important to demonstrate its activation status in various human cancers. Indeed, accumulated experimental data have clearly showed that the levels of neddylation enzymes (e.g. NEDD8 E1, NAE1/UBA3 and NEDD8 E2, UBE2M/UBE2F) are higher in human cancers, including lung cancer, intrahepatic cholangiocarcinoma, liver cancer, colorectal cancer, glioblastoma, nasopharyngeal carcinoma and esophageal squamous cell carcinoma, when compared to adjacent normal tissues [51, 68–73]. Similarly, NEDD8 E3 ligases, such as DCN1 and RBX1/2, are also overexpressed in many human cancers [74, 75]. Moreover, overexpression of these neddylation modifying enzymes is associated with disease progression, conferring a worse overall patient survival [68–70, 72, 73]. Thus, elevated status of neddylation modification may represent an oncogenic event during carcinogenesis, leading to ubiquitylation and degradation of many tumor suppressor substrates of CRLs (e.g. p21 and p27) [68, 76] (Fig. 2). In contrast, the neddylation modification and subsequent CRL activation is tightly regulated and controlled to maintain the homeostasis in normal tissues [5, 68] (Fig. 2). Collectively, elevated status of neddylation modification, as evidenced by overexpression of catalyzing enzymes not only serves as a promising therapeutic cancer target, but also as a useful biomarker for the recruitment of proper cancer patients to receive the treatment with neddylation inhibitors.
Figure 2. Neddylation activates CRLs to regulate tumorigenesis.
In normal cells, neddylation and deneddylation are kept in a balanced status to ensure hemostasis. During tumorigenesis, CRLs are activated to selectively promote ubiquitylation and degradation of tumor suppressors, thus accelerating tumor growth. Blockage of neddylation by MLN4924 or other E2s/E3s inhibitors would inactivate CRLs to cause the accumulation of tumor suppressor substrates to inhibit tumor growth.
Cellular responses to neddylation inhibition
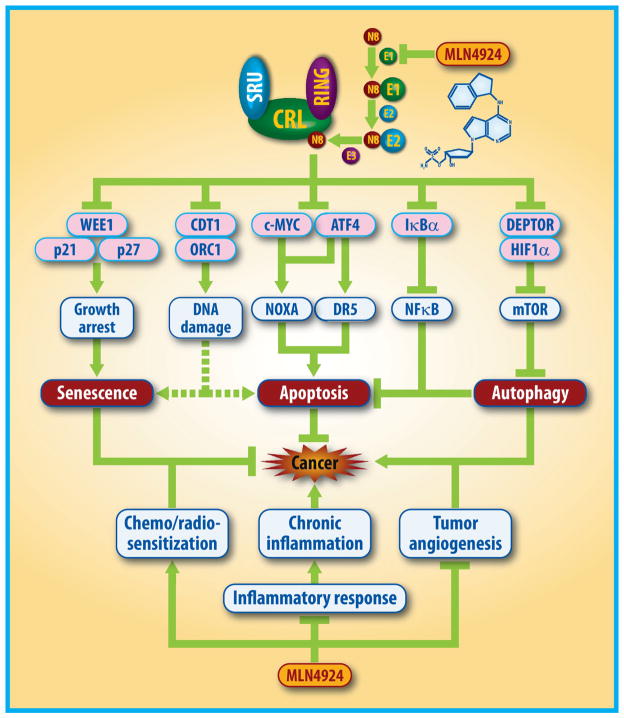
Neddylation inhibition by MLN4924 exerts significant anticancer effects mainly by the inactivation of CRLs and the accumulation of a large number of critical CRLs substrates, thus triggering multiple cellular responses, leading to induction of cell cycle arrest, apoptosis, senescence and autophagy in a cell-type dependent manner.
1. Induction of cell cycle defects
Induction of cell cycle arrest in cancer cells upon drug treatment is an initial response, frequently proceeding apoptosis or senescence [77, 78]. Previous studies showed that MLN4924 induces cell cycle arrest by causing the accumulation of several critical CRLs substrates [27, 55, 56, 58, 79] (Fig. 3). An early study showed that MLN4924-caused accumulation of CRLs substrate CDT1 to trigger DNA re-replication stress and subsequent DNA damage (DD), leading to S phase cell cycle arrest and cell death in some cancer cells [27, 79]. Subsequently, Milhollen et al. reported that MLN4924 induces arrest in different phase of cell cycle in a cell line dependent manner. For example, the S phase arrest was observed in GCB lymphoma cells, whereas in ABC lymphoma cells the arrest occurs in G1 phase, which is associated with dramatic accumulation of the CRLs substrate pIκB and the reduction of nuclear factor-κB (NF-κB) transcriptional activity [58]. Similarly, Czuczman et al. also reported that MLN4924 induces G1 phase arrest in mantle cell lymphoma (MCL) cells [80]. The later studies from our and other’s groups demonstrated that MLN4924 mainly trigger cell cycle arrest at the G2 (or G2/M) phase by inducing the accumulation of cell cycle inhibitors WEE1, p21 and p27, three well-known CRLs substrates in many types of cancer cells [55, 56, 68, 69, 71, 72, 81–89]. Consistently, genetic inhibition of CRLs by RBX1/ROC1 knockdown induces G2 phase arrest in cancer cells as well [74, 90–92]. Recent studies reported that neddylation inhibition by manipulating UBE2M E2 or RNF111 E3 inactivates non-homologous end-joining (NHEJ) and DNA damage responses, leading to changes in cell cycle distribution [17, 93, 94]. Collectively, these findings indicate that neddylation inhibition by MLN4924 or siRNA knockdown approaches could induce cell cycle arrest at different phases in a cell type-dependent manner via various mechanisms.
Figure 3. MLN4924 triggers multiple anticancer mechanisms.
1) MLN4924, acting alone, inactivates CRLs by inhibiting cullin neddylation to cause the accumulation of multiple tumor-suppressive substrates, leading to blockage of tumor angiogenesis and inflammatory responses, induction of senescence or apoptosis in cancer cells, and protective autophagy which serves as an overall survival signal to cancer cells. 2) MLN4924, in combination with chemoradiation, sensitizes resistant cancer cells to conventional therapies.
2. Induction of apoptosis
MLN4924 was firstly reported as an anticancer agent by effectively inducing cell apoptosis in some cancer cell lines [27]. Subsequent studies confirmed this finding with mechanisms all involving the accumulation of CRLs substrates [27, 58, 60, 73, 95] (Fig. 3). Initial reports showed that MLN4924-triggered apoptosis is associated with the inactivation of CRL1-SKP2 and CRL4-CDT2, resulting in the stabilization of the chromatin licensing and DNA replication factor 1 (CDT1), which triggers DNA re-replication to induce apoptosis in HCT-116 cells [27, 79]. Subsequent studies showed that MLN4924-induced apoptosis involved a time-dependent stabilization of the CRLs substrate pIκB to inactivate NF-κB in ABC lymphoma and AML cells [58, 60, 96]. Khalife et al. further reported that MLN4924 decreases the interaction of NF-κB and the miR-155 promoter and downregulates miR-155 in AML cells. It results in the accumulation of the miR-155 targets SHIP1, an inhibitor of the PI3K/Akt pathway, and PU.1, a transcription factor important for myeloid differentiation, leading to monocytic differentiation and apoptosis [97]. Recently, the accumulating studies revealed that MLN4924 causes accumulation of pro-apoptotic proteins (e.g. NOXA, BIK and BIM) and downregulation of anti-apoptotic proteins (e.g. Bcl-xL, Mcl-1 and c-FLIP) to trigger apoptosis [68, 73, 80, 84, 95, 96, 98–101]. Specifically, the MLN4924-induced accumulation of NOXA is attributable to inactivation of a) SAG-CRL5, which promotes NOXA ubiquitylation via K11 linkage for targeted degradation [102, 103], and b) RBX1-CRL1 to cause accumulation of c-Myc and transcription factor 4 (ATF4), which transcriptionally activate NOXA expression [73, 95, 104].
Most recently, we reported a new mechanism of action by which MLN4924 induces apoptosis effectively via the extrinsic (death receptor-mediated) apoptosis pathway [73], which represents an important cytotoxic pathways activated by anticancer agents [105]. Specifically, neddylation inhibition stabilizes activating transcription factor 4 (ATF4), a well-known CRLs substrate, which in turn transactivates CHOP, a transcription factor that induces the expression of Death Receptor5 (DR5), leading to caspase-8 activation to trigger extrinsic apoptosis pathway [73]. Consistently, knockdown of ATF4, CHOP or DR5 remarkably attenuates the activation of Caspase-8 and the MLN4924-induced apoptosis [73]. Thus, MLN4924-induced activation of the ATF4-CHOP-DR5 axis contributes its apoptosis-inducing activity.
3. Induction of senescence
In addition to apoptosis, neddylation inhibition by MLN4924 also induces senescent cell death in non-apoptotic cells, as evidenced by the enlarged and flattened cellular morphology and positive staining of senescence associated β-Galactosidase [56, 106]. Notably, MLN4924-induced senescence appears to be a universal phenotype in a broad range of cancer types, including carcinoma in the lung and colon, glioblastoma, intrahepatic cholangiocarcinoma, lymphoma, osteosarcoma, multiple myeloma and gastric cancer [68, 69, 71, 81, 82, 106–108]. Moreover, even a minimal dose of MLN4924 is needed to trigger the irreversible senescence, which makes it possible to use low doses of the drug to achieve a therapeutic index [56].
Mechanistic studies revealed that MLN4924-induced senescence occurs in the manner largely independent of pRB/p16 and p53, but mainly dependent on p21, which, as a CUL1SKP2 substrate, is significantly accumulated, and simultaneous p21 knockdown remarkably abrogates MLN4924-induced senescence [56, 109, 110] (Fig. 3). Besides p21, another potential mediator of MLN4924-induced senescence is p27 [56, 60] (Fig. 3), yet another well-known substrate of CUL1SKP2 [111, 112]. Indeed, SKP2 inactivation also profoundly induced cellular senescence in the p21 and p27 dependent manner [113]. Given the fact that MLN4924-induced senescence is largely p53 independent, this activity of MLN4924 provides a promising anticancer prospect regardless of the p53 status, thus with broader applications [56]. Collectively, induction of senescence serves as a mechanism by which MLN4924 suppresses tumor growth.
4. Induction of autophagy
Autophagy plays an important role in response to therapeutic stresses, acting as a pro-death or a pro-survival signals to contribute to either drug effectiveness or drug resistance [114, 115]. MLN4924 was shown to induce autophagy as a cellular response while it triggered apoptosis or senescence in a broad panel of cancer cells [55, 116–119] (Fig. 3). Our mechanistic studies revealed that MLN4924-induced autophagy is attributed mainly to the inhibition of mTOR activity, as evidenced by the reduction of phosphorylation of mTOR itself and two mTORC1 substrates, S6K1 and 4E-BP1 [120]. Specifically, MLN4924-induced mTORC1 inactivation is mediated by accumulation of two proteins (Fig. 3) [116]: i) DEPTOR (DEP domain containing mTOR interacting protein), a CRL1β-TrCP substrate [118, 121, 122] and a natural occurring inhibitor of both mTORC1 and mTORC2 [123]; and ii) HIF1α, a well-known substrate of CRL2VHL [124], which triggers activation of the HIF1-REDD1-TSC1 axis [116]. In addition to mTOR inhibition, MLN4924 could trigger autophagy by enhancing the generation of reactive oxygen species (ROS) [60, 125], which can be partially blocked by N-acetyl cysteine (NAC), a classical ROS scavenger. Taken together, mTORC inactivation and ROS overproduction contribute to autophagy induced by neddylation inhibition [55].
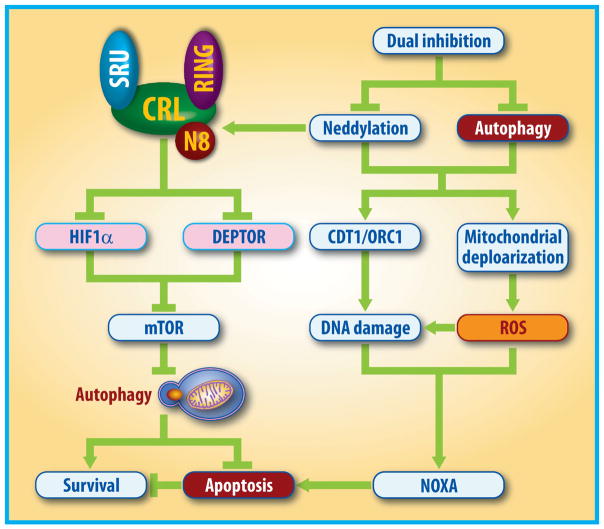
Functionally, autophagy response triggered by neddylation inactivation acts as an pro-survival signal, and likewise autophagy abrogation via genetic and pharmacological means leads to an increased suppression of tumor growth by enhancing apoptosis induction [116, 117, 126]. Specifically, blockage of autophagy with clinically-available autophagy inhibitors (e.g. chloroquine) significantly enhances the efficacy of MLN4924 by triggering NOXA-dependent apoptosis both in vitro and in vivo model of human cancer [55, 116, 119, 127], which is partially attributable to the enhanced DNA damage and the production of excess ROS [127] (Fig. 4). Further mechanistic study revealed that MLN4924 triggers enhanced DNA damage by stabilizing two well-identified CRLs substrates DNA-replication licensing proteins CDT1 and ORC1, whereas enhanced mitochondrial depolarization by MLN4924-chloroquine combination contributes to elevated ROS generation [79, 106, 127–129]. These findings provide the proof-of-concept evidence for combinational anti-cancer therapy with dual inhibition of neddylation and autophagy.
Figure 4. Rational drug combinational design.
Neddylation inactivation by MLN4924 triggers protective autophagy in cancer cells through the blockage of mTOR signals via the accumulation of DEPTOR and the activation of the HIF1α-DDIT4/REDD1-TSC1/2 axis. Thus, combination of MLN4924 with small molecule inhibitors of autophagy (e.g. chloroquine) leads to an increased suppression of tumor growth by enhancing NOXA-dependent apoptosis via triggering DNA damage and ROS generation.
5. Suppression of angiogenesis
Aberrant angiogenesis, an important characteristic of malignant tumors, is governed by tumor microenvironment and served as a promising anticancer target [130–133]. Recent studies showed that the components of neddylation modification, including E1, E2 and E3 enzymes, are expressed and functional in human umbilical vein endothelial cells (HUVECs) [68, 98]. Moreover, MLN4924 treatment significantly decreases the levels of total NEDD8-conjugated proteins and cullin neddylation [98] to suppress the formation of capillary-like tube networks, transwell migration and migrated distance of HUVECs as well as mouse endothelial cells (MS-1) in a dose-dependent manner [98, 134]. The suppressive effect of MLN4924 on angiogenesis was further validated with several classical angiogenic assays, including in vitro rat aortic ring assay, in vivo chick embryo chorioallantoic membrane (CAM) angiogenesis assay and VEGF-induced Matrigel-plug angiogenesis [98]. More importantly, MLN4924 significantly exerts suppressive effects on tumor angiogenesis, tumor growth and metastasis in vivo [98, 134].
Mechanistic studies reveal that short term exposure of MLN4924 triggers the antiangiogenic effect by inducing RhoA accumulation in HUVECs [98, 135, 136]. With prolonged exposure time, MLN4924 induces the accumulation of cell cycle-related CRLs substrates (e.g. p21, p27 and WEE1) and pro-apoptotic proteins (e.g. NOXA), leading to growth arrest and apoptotic cell death [98, 134, 137]. Taken together, these findings revealed that neddylation modification is an important molecular event in regulation of tumor angiogenesis and its blockage may have a therapeutic value, supporting the notion for the development of neddylation inhibitors (e.g. MLN4924) as novel class of antiangiogenic agents [98].
6. Regulation of inflammatory responses
Increased lines of evidence has shown that chronic inflammation, triggered by long-term improper inflammatory responses, plays critical roles in all stages of tumor development from initiation, promotion, and progression to metastasis [138–140]. Thus, targeting the inflammatory responses is likely to be a promising anti-cancer strategy [141, 142]. Several recent studies have implicated a potential role of neddylation modification in regulation of inflammatory responses involving several immune cells, including macrophages, dendritic cells (DC), and T-cells [143–149]. Given that inflammatory responses are mainly regulated by transcription factors, such as NF-κB and HIF-1α, whereas IκB, a negative regulator of NFκB, and HIF-1α are direct substrates of CRLs, it is not unexpected that modulation of neddylation would play a role in cellular inflammatory responses [148, 150, 151].
Indeed, inactivation of neddylation was shown to inhibit macrophage inflammatory responses. Specifically, MLN4924 treatment or siRNA-mediated depletion of Nedd8 or Ubc12 repressed lipopolysaccharides (LPS)-induced cytokines through inducing IκB-α accumulation to block NF-κB translocation and transcriptional activation in human THP-1 and murine macrophages [143, 144]. Manipulation of SAG/RBX2, a neddylation E3, was found to regulate macrophage survival/death and immune response when challenged by pathogen-associated molecular patterns (PAMPs) [152]. Specifically, while SAG knockdown in macrophage caused the accumulation of pro-apoptotic Bax and SARM to induce apoptosis, SAG overexpression triggered upregulation of pro-tumorigenic cytokines (IL-1β, IL-6 and TNFα), and downregulation of anti-tumorigenic cytokine (IL-12p40) and anti-inflammatory cytokine (IL-10) upon exposure to PAMPs [152]. Furthermore, MLN4924 was shown to skew macrophage polarization toward an anti-inflammatory M2 state even in the absence of exogenous M2 polarizing cytokines such as IL-4, highlighting a critical role of neddylation pathway in regulation of the inflammatory macrophages [145].
Similar to macrophages, in DC cells, we showed that by inactivation NF-κB, MLN4924 significantly reduced the release of pro-inflammatory cytokines TNF-α and IL-6 in response to divergent stimuli, and subsequently suppressed DC to stimulate T-cell response [146]. Neddylation inactivation by Ubc12 knockdown or MLN4924 treatment in CD4(+) T cells also impaired T-cell receptor/CD28-induced proliferation and cytokine production by targeting Shc and Erk signaling [147]. We have recently shown that neddylation inactivation by T cell specific Sag/Rbx2 knockout or MLN4924 treatment significantly decreased activation, proliferation, and T-effector cytokine release upon in vitro allogeneic stimulation [153]. Thus, SAG could a novel molecular target that regulates T-cell responses, whereas MLN4924 may be a novel strategy to mitigate T-cell mediated immunopathologies, such as graft-versus host disease [153]. Furthermore, MLN4924 inhibited pro-inflammatory responses while maintaining or increasing the production of anti-inflammatory interleukin (ILs) via blocking NF-κB/MAPK-mediated signaling [154]. Finally, human deneddylase-1 (also known as SENP8) was reported to regulate the inflammatory responses via affecting the translocation of NF-κB and stabilization of HIF-1α [148, 149]. Collectively, these results suggest that inactivation of neddylation via pharmacological (MLN4924) or genetic (manipulation of Sag E3) approaches has anti-inflammatory effect.
7. MLN4924 serves as a chemo/radiosensitizer
In addition to its anticancer effects as a single agent via the mechanism described above, several recent studies showed that MLN4924 could sensitize cancer cells to chemo- or radiation therapy. As for chemotherapy, there are several possible mechanisms triggered by MLN4924 as a potential sensitizer: i) in leukemia cells, MLN4924 sensitizes retinoic acid-induced apoptosis via inducing accumulation of c-Jun and NOXA [155]; ii) In acute myeloid leukemia cells, MLN4924 sensitizes HDAC inhibitor belinostat-induced apoptosis via triggering robust double-stranded breaks, chromatin pulverization and apoptosis [156]. MLN4924 was also shown to increase cellular sensitivity to cytarabine in AML cells, by disrupting nucleotide metabolism [157]; iii) In ovarian cancer cells, MLN4924 sensitizes cisplatin cytotoxicity by enhancing DNA damage and oxidative stress, and by increasing the expression of the pro-apoptotic protein, Bcl-2-interating killer (BIK) [129]. The augment of cisplatin cytotoxicity was also shown in cervical carcinoma cells and urothelial carcinoma cells [158, 159]; iv) In pancreatic cancer cells, MLN4924 sensitizes gemcitabine effect via inducing accumulation of NOXA and ERBIN, a natural occurring inhibitor of RAS-MAPK pathway [160]; v) In multiple myeloma cells, MLN4924 sensitizes bortezomib-induced apoptosis by suppressing AKT and mTOR signaling via upregulation of REDD1 [161]. Intriguingly, combining MLN4924 with the mTOR/PI3K inhibitor GDC-0980 suppresses the growth of NF2-mutant tumor cells in vitro as well as in mouse and patient-derived xenografts [162]; and vi) In several cancer cells, MLN4924 sensitizes DNA interstrand cross-linking (ICLs) agents (e.g. mitomycin C and hydroxyurea) via disrupting DNA damage-induced activation of FANCD2 and CHK1, which are required for the repair of ICLs [163].
Besides chemo-sensitization, MLN4924 also showed radio-sensitization activity. In pancreatic and colorectal cancer cells, MLN4924 effectively sensitizes cells to ionizing radiation both in vitro cell culture and in vivo xenograft models, which can be attributed to the enhancement of radiation-induced DNA damage, aneuploidy, G2/M phase cell cycle arrest and apoptosis, as a result of inducing accumulation of several CRLs substrates, including CDT1, WEE1, NOXA and p27 [164, 165]. In human breast cancer cells, MLN4924 induces G2 arrest, and confers radio-sensitization in a manner dependent of the p21 accumulation [166], whereas combination of MLN4924 with two-deoxy-D-glucose (2DG) enhances the efficacy of radiotherapy [167]. In hormone-resistant prostate cancer cells, MLN4924 acts as a radiosensitizer by inducing accumulation of p21, p27 and WEE1 [168]. Finally, a recent study reported that, in head and neck squamous cells, MLN4924 sensitizes cells to ionizing radiation and enhances IR-induced suppression of cell proliferation in culture and xenografts in mice, which can be attributed mainly to the induction of rereplication via the stabilization of CDT1 [169]. Taken together, MLN4924 can serve either as an anticancer agent alone or being more effective, in combination with chemo or radio-therapy for a variety of chemo/radio-resistant cancers.
Conclusions and perspectives
The findings presented in this review highlight the therapeutic potential in targeting neddylation modification as an effective approach for cancer treatment. Future studies are directed to the following aspects for the advancement of the neddylation field from basic and translational research to clinical application.
First, are there any physiological substrates of neddylation in addition to cullins and how to find them? Currently, cullin family members are the only generally accepted physiological substrates of neddylation and the conversion from neddylated form (slower migrating band) to deneddylated form (faster migrating band) is readily seen in a Western blotting upon MLN4924 treatment. So far, none of reported substrates of neddylation share this feature. In most cases, a slow migrating smear bands were observed as overexpression of exogenous NEDD8 can trigger NEDD8 conjugation through the ubiquitylation machinery, which could represent a mixture of ubiquitin and NEDD8 attachment to a given substrate. A newly developed deconjugation-resistant form of NEDD8, which stabilizes the neddylated form of cullins and other non-cullin substrates [170], may help to resolve this issue.
Second, how does the elevated neddylation modification occur in cancer cells? Does it occur at the transcription levels or post-translation levels? What are the triggers for these changes? If occurred at the transcription levels, but by which transcription factors? This line of study has not been pursued systematically. A recent report showed that NEDD8 E1 UBA3 can be degraded by lysosome-autophagy pathway but not proteasome pathway [171], suggesting a posttranslational regulation. Given that NEDD8 conjugation to a substrate is a dynamic process, yet another regulation could occur at the level of deneddylation system, involving COP9 signalosome, an enzyme with deneddylase activity [172–174], as well as NUB1 and NUB1L, two proteins known to mediate the proteasome-mediated degradation of NEDD8 and NEDD8 conjugates [175–177]. Thus, in-depth elucidation of these mechanisms may provide additional therapeutic targets.
Third, MLN4924 has been shown to suppress tumor cell growth by inducing cell cycle arrest, apoptosis, senescence and autophagy in different cancer types. What determines the cell fate upon MLN4924 exposure --- drug dose, treatment time or intrinsic changes within the cancer cells? Mechanistic understanding of how cancer cells respond to neddylation inhibition with identification of useful biomarkers for cell fate determination will certainly help to maximize the therapeutic efficacy of MLN4924 either acting alone or in combination.
Forth, MLN4924 is a specific inhibitor of NAEβ, a catalytic subunit of NAE E1 [27], which blocks the entire neddylation modification in a cell. It is, therefore, anticipated to suffer from normal cell toxicity as seen in several clinical trials [5, 178] (Table 1). Furthermore, some cancer cells have developed MLN4924 resistance by selecting heterozygous mutations at target NAEβ/UBA3 in preclinical studies [179, 180]. Thus, it is desirable to develop more specific small molecule inhibitors selectively targeting neddylation E2s (UBE2/UBC12 or UBE2F) or E3s (such as RBX1, RBX2, DCN1), if possible. Indeed, two independent groups, including our own discovered small molecule inhibitors targeting UBC12-DCN1 (E2-E3) binding [181, 182], based upon their crystal structure [23, 183]. This specific inhibitor DI-571 we developed in collaboration with Wang lab selectively inhibits CUL3 neddylation to cause accumulation of NRF2 [182], a typical CRL3 substrate [184]. Biological activity of these compounds awaits further investigation.
Finally, the elevated status of the neddylation modification could serve as a potential biomarker for patient recruitment in the clinical trials of neddylation inhibitors. Currently, immunohistochemical (IHC) staining of neddylation enzymes and global NEDD8-conjugated proteins is used to determine the neddylation status [61, 63, 68, 69, 71, 73]. It is desirable to develop more sensitive and convenient method for the measurement of the neddylation status in tumors, ideally without collecting tissue biopsies. Development of sensitive assay using the circulating tumor cells (CTC) as a biomarker could be the future direction.
In summary, the investigation to these pressing issues will not only deepen our understanding of the role of neddylation modification in fundamental cancer biology, but also help to develop biomarkers or more selective targeting inhibitors for clinical application.
Highlights.
Neddylation is one type of post-translational modifications that alters protein function;
Several components that catalyze neddylation modification are over-expressed in multiple human cancers;
Small molecular inhibitor of neddylation modification suppresses cancer cell growth via multiple mechanisms;
Neddylation modification has been validated as a promising anti-cancer target.
Acknowledgments
This work was supported by National Key R&D Program of China (2016YFA0501800), National Natural Science Foundation Grant of China (81172092, 81372196, 81572340, 81572718, 81630076 and 81702244), and “Shuguang Program” supported by Shanghai Education Development Foundation (14SG07).
Footnotes
Competing interests
The authors declare no competing interests.
Authors’ contributions
LSZ and WJZ drafted, LJJ revised, and YS finalized the manuscript. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Enchev RI, Schulman BA, Peter M. Protein neddylation: beyond cullin-RING ligases. Nature reviews Molecular cell biology. 2015;16(1):30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272(45):28557–62. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 3.Rabut G, Peter M. Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9(10):969–76. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xirodimas DP. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans. 2008;36(Pt 5):802–6. doi: 10.1042/BST0360802. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y, Morgan MA, Sun Y. Targeting neddylation pathways to inactivate Cullin-RING ligases for anti-cancer therapy. Antioxid Redox Signal. 2014;21(17):2383–2400. doi: 10.1089/ars.2013.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walden H, Podgorski MS, Huang DT, Miller DW, Howard RJ, Minor DL, Jr, Holton JM, Schulman BA. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Molecular cell. 2003;12(6):1427–37. doi: 10.1016/s1097-2765(03)00452-0. [DOI] [PubMed] [Google Scholar]
- 7.Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274(17):12036–42. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 8.Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8’s E1. Molecular cell. 2005;17(3):341–50. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 9.Zhao Y, Sun Y. Cullin-RING Ligases as attractive anti-cancer targets. Current pharmaceutical design. 2013;19(18):3215–25. doi: 10.2174/13816128113199990300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan H, Wang Y, Aviram M, Swaroop M, Loo JA, Bian J, Tian Y, Mueller T, Bisgaier CL, Sun Y. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol. 1999;19(4):3145–55. doi: 10.1128/mcb.19.4.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, Schulman BA. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33(4):483–95. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13(22):2928–33. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118(1):83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 14.Oved S, Mosesson Y, Zwang Y, Santonico E, Shtiegman K, Marmor MD, Kochupurakkal BS, Katz M, Lavi S, Cesareni G, Yarden Y. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J Biol Chem. 2006;281(31):21640–51. doi: 10.1074/jbc.M513034200. [DOI] [PubMed] [Google Scholar]
- 15.Zuo W, Huang F, Chiang YJ, Li M, Du J, Ding Y, Zhang T, Lee HW, Jeong LS, Chen Y, Deng H, Feng XH, Luo S, Gao C, Chen YG. c-Cbl-mediated neddylation antagonizes ubiquitination and degradation of the TGF-beta type II receptor. Mol Cell. 2013;49(3):499–510. doi: 10.1016/j.molcel.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W. FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. The Journal of biological chemistry. 2007;282(3):1797–804. doi: 10.1074/jbc.M609001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma T, Chen Y, Zhang F, Yang CY, Wang S, Yu X. RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Mol Cell. 2013;49(5):897–907. doi: 10.1016/j.molcel.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broemer M, Tenev T, Rigbolt KT, Hempel S, Blagoev B, Silke J, Ditzel M, Meier P. Systematic in vivo RNAi analysis identifies IAPs as NEDD8-E3 ligases. Molecular cell. 2010;40(5):810–22. doi: 10.1016/j.molcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Rabut G, Le Dez G, Verma R, Makhnevych T, Knebel A, Kurz T, Boone C, Deshaies RJ, Peter M. The TFIIH subunit Tfb3 regulates cullin neddylation. Molecular cell. 2011;43(3):488–95. doi: 10.1016/j.molcel.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noguchi K, Okumura F, Takahashi N, Kataoka A, Kamiyama T, Todo S, Hatakeyama S. TRIM40 promotes neddylation of IKKgamma and is downregulated in gastrointestinal cancers. Carcinogenesis. 2011;32(7):995–1004. doi: 10.1093/carcin/bgr068. [DOI] [PubMed] [Google Scholar]
- 21.Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, Peter M, Sicheri F. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Molecular cell. 2008;29(1):23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Kurz T, Ozlu N, Rudolf F, O’Rourke SM, Luke B, Hofmann K, Hyman AA, Bowerman B, Peter M. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435(7046):1257–61. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- 23.Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334(6056):674–8. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mo Z, Zhang Q, Liu Z, Lauer J, Shi Y, Sun L, Griffin PR, Yang XL. Neddylation requires glycyl-tRNA synthetase to protect activated E2. Nat Struct Mol Biol. 2016;23(8):730–7. doi: 10.1038/nsmb.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nature reviews Cancer. 2006;6(5):369–81. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 27.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732–6. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 28.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 29.Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies RJ, Shevchenko A, Deshaies RJ. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13(12):1614–26. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–67. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 31.Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91(2):221–30. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 32.Merlet J, Burger J, Gomes JE, Pintard L. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci. 2009;66(11–12):1924–38. doi: 10.1007/s00018-009-8712-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakata E, Yamaguchi Y, Miyauchi Y, Iwai K, Chiba T, Saeki Y, Matsuda N, Tanaka K, Kato K. Direct interactions between NEDD8 and ubiquitin E2 conjugating enzymes upregulate cullin-based E3 ligase activity. Nat Struct Mol Biol. 2007;14(2):167–8. doi: 10.1038/nsmb1191. [DOI] [PubMed] [Google Scholar]
- 34.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32(1):21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134(6):995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416(6882):703–9. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 37.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Molecular cell. 2006;21(3):307–15. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dohmesen C, Koeppel M, Dobbelstein M. Specific inhibition of Mdm2-mediated neddylation by Tip60. Cell cycle. 2008;7(2):222–31. doi: 10.4161/cc.7.2.5185. [DOI] [PubMed] [Google Scholar]
- 39.Batuello CN, Hauck PM, Gendron JM, Lehman JA, Mayo LD. Src phosphorylation converts Mdm2 from a ubiquitinating to a neddylating E3 ligase. Proc Natl Acad Sci U S A. 2015;112(6):1749–54. doi: 10.1073/pnas.1416656112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watson IR, Li BK, Roche O, Blanch A, Ohh M, Irwin MS. Chemotherapy induces NEDP1-mediated destabilization of MDM2. Oncogene. 2010;29(2):297–304. doi: 10.1038/onc.2009.314. [DOI] [PubMed] [Google Scholar]
- 41.Xirodimas DP, Sundqvist A, Nakamura A, Shen L, Botting C, Hay RT. Ribosomal proteins are targets for the NEDD8 pathway. EMBO reports. 2008;9(3):280–6. doi: 10.1038/embor.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sundqvist A, Liu G, Mirsaliotis A, Xirodimas DP. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009;10(10):1132–9. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Bai D, Ma X, Guan J, Zheng X. hCINAP is a novel regulator of ribosomal protein-HDM2-p53 pathway by controlling NEDDylation of ribosomal protein S14. Oncogene. 2014;33(2):246–54. doi: 10.1038/onc.2012.560. [DOI] [PubMed] [Google Scholar]
- 44.Stickle NH, Chung J, Klco JM, Hill RP, Kaelin WG, Jr, Ohh M. pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Molecular and cellular biology. 2004;24(8):3251–61. doi: 10.1128/MCB.24.8.3251-3261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA. 2010;1(2):214–29. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Embade N, Fernandez-Ramos D, Varela-Rey M, Beraza N, Sini M, Gutierrez de Juan V, Woodhoo A, Martinez-Lopez N, Rodriguez-Iruretagoyena B, Bustamante FJ, de la Hoz AB, Carracedo A, Xirodimas DP, Rodriguez MS, Lu SC, Mato JM, Martinez-Chantar ML. Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology. 2012;55(4):1237–48. doi: 10.1002/hep.24795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryu JH, Li SH, Park HS, Park JW, Lee B, Chun YS. Hypoxia-inducible factor alpha subunit stabilization by NEDD8 conjugation is reactive oxygen species-dependent. The Journal of biological chemistry. 2011;286(9):6963–70. doi: 10.1074/jbc.M110.188706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao F, Cheng J, Shi T, Yeh ET. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFkappaB-dependent transcription. Nature cell biology. 2006;8(10):1171–7. doi: 10.1038/ncb1483. [DOI] [PubMed] [Google Scholar]
- 49.Lee MR, Lee D, Shin SK, Kim YH, Choi CY. Inhibition of APP intracellular domain (AICD) transcriptional activity via covalent conjugation with Nedd8. Biochemical and biophysical research communications. 2008;366(4):976–81. doi: 10.1016/j.bbrc.2007.12.066. [DOI] [PubMed] [Google Scholar]
- 50.Loftus SJ, Liu G, Carr SM, Munro S, La Thangue NB. NEDDylation regulates E2F-1-dependent transcription. EMBO reports. 2012;13(9):811–8. doi: 10.1038/embor.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie P, Zhang M, He S, Lu K, Chen Y, Xing G, Lu Y, Liu P, Li Y, Wang S, Chai N, Wu J, Deng H, Wang HR, Cao Y, Zhao F, Cui Y, Wang J, He F, Zhang L. The covalent modifier Nedd8 is critical for the activation of Smurf1 ubiquitin ligase in tumorigenesis. Nature communications. 2014;5:3733. doi: 10.1038/ncomms4733. [DOI] [PubMed] [Google Scholar]
- 52.Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143(6):951–65. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325–40. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, Ma J, Loke HK, Lingaraj T, Wu D, Hamman KB, Spelman JJ, Cullis CA, Langston SP, Vyskocil S, Sells TB, Mallender WD, Visiers I, Li P, Claiborne CF, Rolfe M, Bolen JB, Dick LR. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37(1):102–11. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 55.Luo Z, Yu G, Lee HW, Li L, Wang L, Yang D, Pan Y, Ding C, Qian J, Wu L, Chu Y, Yi J, Wang X, Sun Y, Jeong LS, Liu J, Jia L. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 2012;72(13):3360–71. doi: 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- 56.Jia L, Li H, Sun Y. Induction of p21-Dependent Senescence by an NAE Inhibitor, MLN4924, as a Mechanism of Growth Suppression. Neoplasia. 2011;13(6):561–569. doi: 10.1593/neo.11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan Y, Xu H, Liu R, Jia L. Induction of cell senescence by targeting to Cullin-RING Ligases (CRLs) for effective cancer therapy. Int J Biochem Mol Biol. 2012;3(3):273–81. [PMC free article] [PubMed] [Google Scholar]
- 58.Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, Manfredi M, Narayanan U, Rolfe M, Staudt LM, Soucy TA, Yu J, Zhang J, Bolen JB, Smith PG. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa}B-dependent lymphoma. Blood. 2010;116(9):1515–23. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 59.Soucy TA, Smith PG, Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin Cancer Res. 2009;15(12):3912–6. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 60.Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O’Dwyer M, Nawrocki ST, Giles FJ, Carew JS. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115(18):3796–800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 61.Sarantopoulos J, Shapiro GI, Cohen RB, Clark JW, Kauh JS, Weiss GJ, Cleary JM, Mahalingam D, Pickard MD, Faessel HM, Berger AJ, Burke K, Mulligan G, Dezube BJ, Harvey RD. Phase I Study of the Investigational NEDD8-Activating Enzyme Inhibitor Pevonedistat (TAK-924/MLN4924) in Patients with Advanced Solid Tumors. Clin Cancer Res. 2016;22(4):847–57. doi: 10.1158/1078-0432.CCR-15-1338. [DOI] [PubMed] [Google Scholar]
- 62.Shah JJ, Jakubowiak AJ, O’Connor OA, Orlowski RZ, Harvey RD, Smith MR, Lebovic D, Diefenbach C, Kelly K, Hua Z, Berger AJ, Mulligan G, Faessel HM, Tirrell S, Dezube BJ, Lonial S. Phase I Study of the Novel Investigational NEDD8-Activating Enzyme Inhibitor Pevonedistat (MLN4924) in Patients with Relapsed/Refractory Multiple Myeloma or Lymphoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22(1):34–43. doi: 10.1158/1078-0432.CCR-15-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swords RT, Erba HP, DeAngelo DJ, Bixby DL, Altman JK, Maris M, Hua Z, Blakemore SJ, Faessel H, Sedarati F, Dezube BJ, Giles FJ, Medeiros BC. Pevonedistat (MLN4924), a First-in-Class NEDD8-activating enzyme inhibitor, in patients with acute myeloid leukaemia and myelodysplastic syndromes: a phase 1 study. British journal of haematology. 2015;169(4):534–43. doi: 10.1111/bjh.13323. [DOI] [PubMed] [Google Scholar]
- 64.Bhatia S, Pavlick AC, Boasberg P, Thompson JA, Mulligan G, Pickard MD, Faessel H, Dezube BJ, Hamid O. A phase I study of the investigational NEDD8-activating enzyme inhibitor pevonedistat (TAK-924/MLN4924) in patients with metastatic melanoma. Investigational new drugs. 2016;34(4):439–49. doi: 10.1007/s10637-016-0348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ronan MRS, Swords T, Maris Michael B, Erba Harry P, Berdeja Jesus G, Foran James M, Hua Zhaowei, Faessel Hélène M, Dash Ajeeta B, Sedarati Farhad, Dezube Bruce J, Medeiros Bruno C. Pevonedistat (MLN4924), an Investigational, First-in-Class NAE Inhibitor, in Combination with Azacitidine in Elderly Patients with Acute Myeloid Leukemia (AML) Considered Unfit for Conventional Chemotherapy: Updated Results from the Phase 1 C15009 Trial. Blood. 2014;124(21):2313. [Google Scholar]
- 66.Nawrocki ST, Griffin P, Kelly KR, Carew JS. MLN4924: a novel first-in-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert opinion on investigational drugs. 2012;21(10):1563–73. doi: 10.1517/13543784.2012.707192. [DOI] [PubMed] [Google Scholar]
- 67.Abidi N, Xirodimas DP. Regulation of cancer-related pathways by protein NEDDylation and strategies for the use of NEDD8 inhibitors in the clinic. Endocr Relat Cancer. 2015;22(1):T55–70. doi: 10.1530/ERC-14-0315. [DOI] [PubMed] [Google Scholar]
- 68.Li L, Wang M, Yu G, Chen P, Li H, Wei D, Zhu J, Xie L, Jia H, Shi J, Li C, Yao W, Wang Y, Gao Q, Jeong LS, Lee HW, Yu J, Hu F, Mei J, Wang P, Chu Y, Qi H, Yang M, Dong Z, Sun Y, Hoffman RM, Jia L. Overactivated neddylation pathway as a therapeutic target in lung cancer. J Natl Cancer Inst. 2014;106(6):dju083. doi: 10.1093/jnci/dju083. [DOI] [PubMed] [Google Scholar]
- 69.Gao Q, Yu GY, Shi JY, Li LH, Zhang WJ, Wang ZC, Yang LX, Duan M, Zhao H, Wang XY, Zhou J, Qiu SJ, Jeong LS, Jia LJ, Fan J. Neddylation pathway is up-regulated in human intrahepatic cholangiocarcinoma and serves as a potential therapeutic target. Oncotarget. 2014;5(17):7820–32. doi: 10.18632/oncotarget.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barbier-Torres L, Delgado TC, Garcia-Rodriguez JL, Zubiete-Franco I, Fernandez-Ramos D, Buque X, Cano A, Gutierrez-de Juan V, Fernandez-Dominguez I, Lopitz-Otsoa F, Fernandez-Tussy P, Boix L, Bruix J, Villa E, Castro A, Lu SC, Aspichueta P, Xirodimas D, Varela-Rey M, Mato JM, Beraza N, Martinez-Chantar ML. Stabilization of LKB1 and Akt by neddylation regulates energy metabolism in liver cancer. Oncotarget. 2015;6(4):2509–23. doi: 10.18632/oncotarget.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hua W, Li C, Yang Z, Li L, Jiang Y, Yu G, Zhu W, Liu Z, Duan S, Chu Y, Yang M, Zhang Y, Mao Y, Jia L. Suppression of glioblastoma by targeting the overactivated protein neddylation pathway. Neuro Oncol. 2015;17(10):1333–43. doi: 10.1093/neuonc/nov066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie P, Yang JP, Cao Y, Peng LX, Zheng LS, Sun R, Meng DF, Wang MY, Mei Y, Qiang YY, Cao L, Xiang YQ, Luo DH, Yun JP, Huang BJ, Jia LJ, Qian CN. Promoting tumorigenesis in nasopharyngeal carcinoma, NEDD8 serves as a potential theranostic target. Cell death & disease. 2017;8(6):e2834. doi: 10.1038/cddis.2017.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen P, Hu T, Liang Y, Li P, Chen X, Zhang J, Ma Y, Hao Q, Wang J, Zhang P, Zhang Y, Zhao H, Yang S, Yu J, Jeong LS, Qi H, Yang M, Hoffman RM, Dong Z, Jia L. Neddylation Inhibition Activates the Extrinsic Apoptosis Pathway through ATF4-CHOP-DR5 Axis in Human Esophageal Cancer Cells. Clin Cancer Res. 2016;22(16):4145–57. doi: 10.1158/1078-0432.CCR-15-2254. [DOI] [PubMed] [Google Scholar]
- 74.Jia L, Soengas MS, Sun Y. ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2-M arrest, apoptosis, and senescence. Cancer Res. 2009;69(12):4974–82. doi: 10.1158/0008-5472.CAN-08-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sarkaria I, OcP, Talbot SG, Reddy PG, Ngai I, Maghami E, Patel KN, Lee B, Yonekawa Y, Dudas M, Kaufman A, Ryan R, Ghossein R, Rao PH, Stoffel A, Ramanathan Y, Singh B. Squamous cell carcinoma related oncogene/DCUN1D1 is highly conserved and activated by amplification in squamous cell carcinomas. Cancer Res. 2006;66(19):9437–44. doi: 10.1158/0008-5472.CAN-06-2074. [DOI] [PubMed] [Google Scholar]
- 76.Li H, Tan M, Jia L, Wei D, Zhao Y, Chen G, Xu J, Zhao L, Thomas D, Beer DG, Sun Y. Inactivation of SAG/RBX2 E3 ubiquitin ligase suppresses KrasG12D-driven lung tumorigenesis. The Journal of clinical investigation. 2014;124(2):835–46. doi: 10.1172/JCI70297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pucci B, Kasten M, Giordano A. Cell cycle and apoptosis. Neoplasia. 2000;2(4):291–9. doi: 10.1038/sj.neo.7900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gire V, Dulic V. Senescence from G2 arrest, revisited. Cell Cycle. 2015;14(3):297–304. doi: 10.1080/15384101.2014.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Milhollen MA, Narayanan U, Soucy TA, Veiby PO, Smith PG, Amidon B. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res. 2011;71(8):3042–51. doi: 10.1158/0008-5472.CAN-10-2122. [DOI] [PubMed] [Google Scholar]
- 80.Czuczman NM, Barth MJ, Gu J, Neppalli V, Mavis C, Frys SE, Hu Q, Liu S, Klener P, Vockova P, Czuczman MS, Hernandez-Ilizaliturri FJ. Pevonedistat, a NEDD8-activating enzyme inhibitor, is active in mantle cell lymphoma and enhances rituximab activity in vivo. Blood. 2016;127(9):1128–37. doi: 10.1182/blood-2015-04-640920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lan H, Tang Z, Jin H, Sun Y. Neddylation inhibitor MLN4924 suppresses growth and migration of human gastric cancer cells. Sci Rep. 2016;6:24218. doi: 10.1038/srep24218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y, Shi CC, Zhang HP, Li GQ, Li SS. MLN4924 suppresses neddylation and induces cell cycle arrest, senescence, and apoptosis in human osteosarcoma. Oncotarget. 2016;7(29):45263–45274. doi: 10.18632/oncotarget.9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Han K, Wang Q, Cao H, Qiu G, Cao J, Li X, Wang J, Shen B, Zhang J. The NEDD8-activating enzyme inhibitor MLN4924 induces G2 arrest and apoptosis in T-cell acute lymphoblastic leukemia. Oncotarget. 2016;7(17):23812–24. doi: 10.18632/oncotarget.8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tong S, Si Y, Yu H, Zhang L, Xie P, Jiang W. MLN4924 (Pevonedistat), a protein neddylation inhibitor, suppresses proliferation and migration of human clear cell renal cell carcinoma. Scientific reports. 2017;7(1):5599. doi: 10.1038/s41598-017-06098-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Y, Shi CC, Zhang HP, Li GQ, Li SS. MLN4924 suppresses neddylation and induces cell cycle arrest, senescence, and apoptosis in human osteosarcoma. Oncotarget. 2016 doi: 10.18632/oncotarget.9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han K, Wang QY, Cao HL, Qiu GH, Cao JX, Li X, Wang J, Shen BF, Zhang JY. The NEDD8-activating enzyme inhibitor MLN4924 induces G2 arrest and apoptosis in T-cell acute lymphoblastic leukemia. Oncotarget. 2016;7(17):23812–23824. doi: 10.18632/oncotarget.8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Paiva C, Godbersen JC, Berger A, Brown JR, Danilov AV. Targeting neddylation induces DNA damage and checkpoint activation and sensitizes chronic lymphocytic leukemia B cells to alkylating agents. Cell death & disease. 2015;6:e1807. doi: 10.1038/cddis.2015.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuo KL, Ho IL, Shi CS, Wu JT, Lin WC, Tsai YC, Chang HC, Chou CT, Hsu CH, Hsieh JT, Chang SC, Pu YS, Huang KH. MLN4924, a novel protein neddylation inhibitor, suppresses proliferation and migration of human urothelial carcinoma: In vitro and in vivo studies. Cancer Lett. 2015;363(2):127–36. doi: 10.1016/j.canlet.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 89.Mackintosh C, Garcia-Dominguez DJ, Ordonez JL, Ginel-Picardo A, Smith PG, Sacristan MP, de Alava E. WEE1 accumulation and deregulation of S-phase proteins mediate MLN4924 potent inhibitory effect on Ewing sarcoma cells. Oncogene. 2013;32(11):1441–51. doi: 10.1038/onc.2012.153. [DOI] [PubMed] [Google Scholar]
- 90.Jia L, Bickel JS, Wu J, Morgan MA, Li H, Yang J, Yu X, Chan RC, Sun Y. RBX1 (RING box protein 1) E3 ubiquitin ligase is required for genomic integrity by modulating DNA replication licensing proteins. The Journal of biological chemistry. 2011;286(5):3379–86. doi: 10.1074/jbc.M110.188425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang J, Li S, Shang Z, Lin S, Gao P, Zhang Y, Hou S, Mo S, Cao W, Dong Z, Hu T, Chen P. Targeting the overexpressed ROC1 induces G2 cell cycle arrest and apoptosis in esophageal cancer cells. Oncotarget. 2017;8(17):29125–29137. doi: 10.18632/oncotarget.16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang W, Liu Z, Qu P, Zhou Z, Zeng Y, Fan J, Liu Y, Guo Y, Qiu J. Knockdown of regulator of cullins-1 (ROC1) expression induces bladder cancer cell cycle arrest at the G2 phase and senescence. PloS one. 2013;8(5):e62734. doi: 10.1371/journal.pone.0062734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jimeno S, Fernandez-Avila MJ, Cruz-Garcia A, Cepeda-Garcia C, Gomez-Cabello D, Huertas P. Neddylation inhibits CtIP-mediated resection and regulates DNA double strand break repair pathway choice. Nucleic Acids Res. 2015;43(2):987–99. doi: 10.1093/nar/gku1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cukras S, Morffy N, Ohn T, Kee Y. Inactivating UBE2M impacts the DNA damage response and genome integrity involving multiple cullin ligases. PLoS One. 2014;9(7):e101844. doi: 10.1371/journal.pone.0101844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Knorr KL, Schneider PA, Meng XW, Dai H, Smith BD, Hess AD, Karp JE, Kaufmann SH. MLN4924 induces Noxa upregulation in acute myelogenous leukemia and synergizes with Bcl-2 inhibitors. Cell Death Differ. 2015;22(12):2133–42. doi: 10.1038/cdd.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Godbersen JC, Humphries LA, Danilova OV, Kebbekus PE, Brown JR, Eastman A, Danilov AV. The Nedd8-activating enzyme inhibitor MLN4924 thwarts microenvironment-driven NF-kappaB activation and induces apoptosis in chronic lymphocytic leukemia B cells. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20(6):1576–89. doi: 10.1158/1078-0432.CCR-13-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khalife J, Radomska HS, Santhanam R, Huang X, Neviani P, Saultz J, Wang H, Wu YZ, Alachkar H, Anghelina M, Dorrance A, Curfman J, Bloomfield CD, Medeiros BC, Perrotti D, Lee LJ, Lee RJ, Caligiuri MA, Pichiorri F, Croce CM, Garzon R, Guzman ML, Mendler JH, Marcucci G. Pharmacological targeting of miR-155 via the NEDD8-activating enzyme inhibitor MLN4924 (Pevonedistat) in FLT3-ITD acute myeloid leukemia. Leukemia. 2015;29(10):1981–92. doi: 10.1038/leu.2015.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yao WT, Wu JF, Yu GY, Wang R, Wang K, Li LH, Chen P, Jiang YN, Cheng H, Lee HW, Yu J, Qi H, Yu XJ, Wang P, Chu YW, Yang M, Hua ZC, Ying HQ, Hoffman RM, Jeong LS, Jia LJ. Suppression of tumor angiogenesis by targeting the protein neddylation pathway. Cell death & disease. 2014;5:e1059. doi: 10.1038/cddis.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dengler MA, Weilbacher A, Gutekunst M, Staiger AM, Vohringer MC, Horn H, Ott G, Aulitzky WE, van der Kuip H. Discrepant NOXA (PMAIP1) transcript and NOXA protein levels: a potential Achilles’ heel in mantle cell lymphoma. Cell death & disease. 2014;5:e1013. doi: 10.1038/cddis.2013.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leclerc GM, Zheng S, Leclerc GJ, DeSalvo J, Swords RT, Barredo JC. The NEDD8-activating enzyme inhibitor pevonedistat activates the eIF2alpha and mTOR pathways inducing UPR-mediated cell death in acute lymphoblastic leukemia. Leuk Res. 2016;50:1–10. doi: 10.1016/j.leukres.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 101.Wang J, Wang S, Zhang W, Wang X, Liu X, Liu L, Li L, Liang Y, Yu J, Jeong LS, Jia L, Zhao H, Zhang Y. Targeting neddylation pathway with MLN4924 (Pevonedistat) induces NOXA-dependent apoptosis in renal cell carcinoma. Biochemical and biophysical research communications. 2017;490(4):1183–1188. doi: 10.1016/j.bbrc.2017.06.179. [DOI] [PubMed] [Google Scholar]
- 102.Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, Xu L, Sun Y. Validation of SAG/RBX2/ROC2 E3 Ubiquitin Ligase as an Anticancer and Radiosensitizing Target. Clin Cancer Res. 2010;16(3):814–24. doi: 10.1158/1078-0432.CCR-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou W, Xu J, Li H, Xu M, Chen ZJ, Wei W, Pan ZQ, Sun Y. Neddylation E2 UBE2F promotes the survival of lung cancer cells by activating CRL5 to degrade NOXA via the K11 linkage. Clin Cancer Res. 2017;23:1104–1116. doi: 10.1158/1078-0432.CCR-16-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pike LR, Phadwal K, Simon AK, Harris AL. ATF4 orchestrates a program of BH3-only protein expression in severe hypoxia. Mol Biol Rep. 2012;39(12):10811–22. doi: 10.1007/s11033-012-1975-3. [DOI] [PubMed] [Google Scholar]
- 105.Fulda S, Debatin KM. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006;25(34):4798–811. doi: 10.1038/sj.onc.1209608. [DOI] [PubMed] [Google Scholar]
- 106.Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 2010;70(24):10310–20. doi: 10.1158/0008-5472.CAN-10-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang Y, Luo Z, Pan Y, Wang W, Zhou X, Jeong LS, Chu Y, Liu J, Jia L. Targeting protein neddylation with an NEDD8-activating enzyme inhibitor MLN4924 induced apoptosis or senescence in human lymphoma cells. Cancer biology & therapy. 2015;16(3):420–9. doi: 10.1080/15384047.2014.1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Huang J, Zhou Y, Thomas GS, Gu Z, Yang Y, Xu H, Tricot G, Zhan F. NEDD8 Inhibition Overcomes CKS1B-Induced Drug Resistance by Upregulation of p21 in Multiple Myeloma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015;21(24):5532–42. doi: 10.1158/1078-0432.CCR-15-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Abbas T, Sivaprasad U, Terai K, Amador V, Pagano M, Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22(18):2496–506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008;22(18):2507–19. doi: 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu ZK, Gervais JL, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc Natl Acad Sci U S A. 1998;95(19):11324–9. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bloom J, Pagano M. Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Semin Cancer Biol. 2003;13(1):41–7. doi: 10.1016/s1044-579x(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 113.Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464(7287):374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Carew JS, Nawrocki ST, Cleveland JL. Modulating autophagy for therapeutic benefit. Autophagy. 2007;3(5):464–7. doi: 10.4161/auto.4311. [DOI] [PubMed] [Google Scholar]
- 115.Buchser WJ, Laskow TC, Pavlik PJ, Lin HM, Lotze MT. Cell-mediated autophagy promotes cancer cell survival. Cancer research. 2012;72(12):2970–9. doi: 10.1158/0008-5472.CAN-11-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhao Y, Xiong X, Jia L, Sun Y. Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell death & disease. 2012;3:e386. doi: 10.1038/cddis.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang D, Li L, Liu H, Wu L, Luo Z, Li H, Zheng S, Gao H, Chu Y, Sun Y, Liu J, Jia L. Induction of autophagy and senescence by knockdown of ROC1 E3 ubiquitin ligase to suppress the growth of liver cancer cells. Cell death and differentiation. 2013;20(2):235–47. doi: 10.1038/cdd.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhao Y, Xiong X, Sun Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF(betaTrCP) E3 ubiquitin ligase and regulates survival and autophagy. Molecular cell. 2011;44(2):304–16. doi: 10.1016/j.molcel.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Luo Z, Pan Y, Jeong LS, Liu J, Jia L. Inactivation of the Cullin (CUL)-RING E3 ligase by the NEDD8-activating enzyme inhibitor MLN4924 triggers protective autophagy in cancer cells. Autophagy. 2012;8(11):1677–9. doi: 10.4161/auto.21484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao Y, Morgan MA, Sun Y. Targeting Neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxidants & redox signaling. 2014;21(17):2383–400. doi: 10.1089/ars.2013.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, Pagano M. mTOR Generates an Auto-Amplification Loop by Triggering the betaTrCP- and CK1alpha-Dependent Degradation of DEPTOR. Mol Cell. 2011;44(2):317–24. doi: 10.1016/j.molcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gao D, Inuzuka H, Tan MK, Fukushima H, Locasale JW, Liu P, Wan L, Zhai B, Chin YR, Shaik S, Lyssiotis CA, Gygi SP, Toker A, Cantley LC, Asara JM, Harper JW, Wei W. mTOR Drives Its Own Activation via SCF(betaTrCP)-Dependent Degradation of the mTOR Inhibitor DEPTOR. Mol Cell. 2011;44(2):290–303. doi: 10.1016/j.molcel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137(5):873–86. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–72. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 125.Scherz-Shouval R, Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends in biochemical sciences. 2011;36(1):30–8. doi: 10.1016/j.tibs.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 126.Yang D, Zhao Y, Liu J, Sun Y, Jia L. Protective autophagy induced by RBX1/ROC1 knockdown or CRL inactivation via modulating the DEPTOR-MTOR axis. Autophagy. 2012;8(12):1856–8. doi: 10.4161/auto.22024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen P, Hu T, Liang Y, Jiang Y, Pan Y, Li C, Zhang P, Wei D, Li P, Jeong LS, Chu Y, Qi H, Yang M, Hoffman RM, Dong Z, Jia L. Synergistic inhibition of autophagy and neddylation pathways as a novel therapeutic approach for targeting liver cancer. Oncotarget. 2015;6(11):9002–17. doi: 10.18632/oncotarget.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Blank JL, Liu XJ, Cosmopoulos K, Bouck DC, Garcia K, Bernard H, Tayber O, Hather G, Liu R, Narayanan U, Milhollen MA, Lightcap ES. Novel DNA damage checkpoints mediating cell death induced by the NEDD8-activating enzyme inhibitor MLN4924. Cancer Res. 2013;73(1):225–34. doi: 10.1158/0008-5472.CAN-12-1729. [DOI] [PubMed] [Google Scholar]
- 129.Nawrocki ST, Kelly KR, Smith PG, Espitia CM, Possemato A, Beausoleil SA, Milhollen M, Blakemore S, Thomas M, Berger A, Carew JS. Disrupting Protein NEDDylation with MLN4924 Is a Novel Strategy to Target Cisplatin Resistance in Ovarian Cancer. Clin Cancer Res. 2013;19(13):3577–90. doi: 10.1158/1078-0432.CCR-12-3212. [DOI] [PubMed] [Google Scholar]
- 130.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29(6 Suppl 16):15–8. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 131.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 132.Weis SM, Cheresh DA. Tumor angiogenesis: molecular pathways and therapeutic targets. Nature medicine. 2011;17(11):1359–70. doi: 10.1038/nm.2537. [DOI] [PubMed] [Google Scholar]
- 133.Kerbel RS. Tumor angiogenesis. The New England journal of medicine. 2008;358(19):2039–49. doi: 10.1056/NEJMra0706596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tan M, Li H, Sun Y. Endothelial deletion of Sag/Rbx2/Roc2 E3 ubiquitin ligase causes embryonic lethality and blocks tumor angiogenesis. Oncogene. 2014;33(44):5211–20. doi: 10.1038/onc.2013.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81(7):1159–70. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 136.Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, Liu L, Ding M, Peng HB, Shao F. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Molecular cell. 2009;35(6):841–55. doi: 10.1016/j.molcel.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 137.Liu X, Jiang Y, Wu J, Zhang W, Liang Y, Jia L, Yu J, Jeong LS, Li L. NEDD8-activating enzyme inhibitor, MLN4924 (Pevonedistat) induces NOXA-dependent apoptosis through up-regulation of ATF-4. Biochemical and biophysical research communications. 2017;488(1):1–5. doi: 10.1016/j.bbrc.2017.04.122. [DOI] [PubMed] [Google Scholar]
- 138.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6(1):24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 139.Liu J, Lin PC, Zhou BP. Inflammation fuels tumor progress and metastasis. Curr Pharm Des. 2015;21(21):3032–40. doi: 10.2174/1381612821666150514105741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest. 2015;125(9):3347–55. doi: 10.1172/JCI80007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Cell. 2005;7(6):513–20. doi: 10.1016/j.ccr.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 142.Brown JM. Tumor microenvironment and the response to anticancer therapy. Cancer Biol Ther. 2002;1(5):453–8. doi: 10.4161/cbt.1.5.157. [DOI] [PubMed] [Google Scholar]
- 143.Li L, Liu B, Dong T, Lee HW, Yu J, Zheng Y, Gao H, Zhang Y, Chu Y, Liu G, Niu W, Zheng S, Jeong LS, Jia L. Neddylation pathway regulates the proliferation and survival of macrophages. Biochem Biophys Res Commun. 2013;432(3):494–8. doi: 10.1016/j.bbrc.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 144.Chang FM, Reyna SM, Granados JC, Wei SJ, Innis-Whitehouse W, Maffi SK, Rodriguez E, Slaga TJ, Short JD. Inhibition of neddylation represses lipopolysaccharide-induced proinflammatory cytokine production in macrophage cells. J Biol Chem. 2012;287(42):35756–67. doi: 10.1074/jbc.M112.397703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Asare Y, Ommer M, Azombo FA, Alampour-Rajabi S, Sternkopf M, Sanati M, Gijbels MJ, Schmitz C, Sinitski D, Tilstam PV, Lue H, Gessner A, Lange D, Schmid JA, Weber C, Dichgans M, Jankowski J, Pardi R, de Winther MP, Noels H, Bernhagen J. Inhibition of atherogenesis by the COP9 signalosome subunit 5 in vivo. Proc Natl Acad Sci U S A. 2017;114(13):E2766–E2775. doi: 10.1073/pnas.1618411114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mathewson N, Toubai T, Kapeles S, Sun Y, Oravecz-Wilson K, Tamaki H, Wang Y, Hou G, Sun Y, Reddy P. Neddylation plays an important role in the regulation of murine and human dendritic cell function. Blood. 2013;122(12):2062–73. doi: 10.1182/blood-2013-02-486373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jin HS, Liao L, Park Y, Liu YC. Neddylation pathway regulates T-cell function by targeting an adaptor protein Shc and a protein kinase Erk signaling. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(2):624–9. doi: 10.1073/pnas.1213819110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ehrentraut SF, Kominsky DJ, Glover LE, Campbell EL, Kelly CJ, Bowers BE, Bayless AJ, Colgan SP. Central role for endothelial human deneddylase-1/SENP8 in fine-tuning the vascular inflammatory response. J Immunol. 2013;190(1):392–400. doi: 10.4049/jimmunol.1202041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ehrentraut SF, Curtis VF, Wang RX, Saeedi BJ, Ehrentraut H, Onyiah JC, Kelly CJ, Campbell EL, Glover LE, Kominsky DJ, Colgan SP. Perturbation of neddylation-dependent NF-kappaB responses in the intestinal epithelium drives apoptosis and inhibits resolution of mucosal inflammation. Mol Biol Cell. 2016 doi: 10.1091/mbc.E16-05-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kanarek N, Ben-Neriah Y. Regulation of NF-kappaB by ubiquitination and degradation of the IkappaBs. Immunol Rev. 2012;246(1):77–94. doi: 10.1111/j.1600-065X.2012.01098.x. [DOI] [PubMed] [Google Scholar]
- 151.Curtis VF, Ehrentraut SF, Campbell EL, Glover LE, Bayless A, Kelly CJ, Kominsky DJ, Colgan SP. Stabilization of HIF through inhibition of Cullin-2 neddylation is protective in mucosal inflammatory responses. FASEB J. 2015;29(1):208–15. doi: 10.1096/fj.14-259663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chang SC, Ding JL. Ubiquitination by SAG regulates macrophage survival/death and immune response during infection. Cell Death Differ. 2014;21(9):1388–98. doi: 10.1038/cdd.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mathewson ND, Fujiwara H, Wu SR, Toubai T, Oravecz-Wilson K, Sun Y, Rossi C, Zajac C, Sun Y, Reddy P. SAG/Rbx2-Dependent Neddylation Regulates T-Cell Responses. The American journal of pathology. 2016;186(10):2679–91. doi: 10.1016/j.ajpath.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Deng Q, Zhang J, Gao Y, She X, Wang Y, Wang Y, Ge X. MLN4924 protects against bleomycin-induced pulmonary fibrosis by inhibiting the early inflammatory process. Am J Transl Res. 2017;9(4):1810–1821. [PMC free article] [PubMed] [Google Scholar]
- 155.Tan M, Li Y, Yang R, Xi N, Sun Y. Inactivation of SAG E3 Ubiquitin Ligase Blocks Embryonic Stem Cell Differentiation and Sensitizes Leukemia Cells to Retinoid Acid. PLoS One. 2011;6(11):e27726. doi: 10.1371/journal.pone.0027726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhou L, Chen S, Zhang Y, Kmieciak M, Leng Y, Li L, Lin H, Rizzo KA, Dumur CI, Ferreira-Gonzalez A, Rahmani M, Povirk L, Chalasani S, Berger AJ, Dai Y, Grant S. The NAE inhibitor pevonedistat interacts with the HDAC inhibitor belinostat to target AML cells by disrupting the DDR. Blood. 2016;127(18):2219–30. doi: 10.1182/blood-2015-06-653717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nawrocki ST, Kelly KR, Smith PG, Keaton M, Carraway H, Sekeres MA, Maciejewski JP, Carew JS. The NEDD8-activating enzyme inhibitor MLN4924 disrupts nucleotide metabolism and augments the efficacy of cytarabine. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015;21(2):439–47. doi: 10.1158/1078-0432.CCR-14-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lin WC, Kuo KL, Shi CS, Wu JT, Hsieh JT, Chang HC, Liao SM, Chou CT, Chiang CK, Chiu WS, Chiu TY, Pu YS, Ho IL, Wang ZH, Chang SC, Liu SH, Jeng YM, Huang KH. MLN4924, a Novel NEDD8-activating enzyme inhibitor, exhibits antitumor activity and enhances cisplatin-induced cytotoxicity in human cervical carcinoma: in vitro and in vivo study. American journal of cancer research. 2015;5(11):3350–62. [PMC free article] [PubMed] [Google Scholar]
- 159.Ho IL, Kuo KL, Liu SH, Chang HC, Hsieh JT, Wu JT, Chiang CK, Lin WC, Tsai YC, Chou CT, Hsu CH, Pu YS, Shi CS, Huang KH. MLN4924 Synergistically Enhances Cisplatin-induced Cytotoxicity via JNK and Bcl-xL Pathways in Human Urothelial Carcinoma. Scientific reports. 2015;5:16948. doi: 10.1038/srep16948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li H, Zhou W, Li L, Wu J, Liu X, Zhao L, Jia L, Sun Y. Inhibition of Neddylation Modification Sensitizes Pancreatic Cancer Cells to Gemcitabine. Neoplasia. 2017;19(6):509–518. doi: 10.1016/j.neo.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gu Y, Kaufman JL, Bernal L, Torre C, Matulis SM, Harvey RD, Chen J, Sun SY, Boise LH, Lonial S. MLN4924, an NAE inhibitor, suppresses AKT and mTOR signaling via upregulation of REDD1 in human myeloma cells. Blood. 2014;123(21):3269–76. doi: 10.1182/blood-2013-08-521914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cooper J, Xu Q, Zhou L, Pavlovic M, Ojeda V, Moulick K, de Stanchina E, Poirier JT, Zauderer M, Rudin CM, Karajannis MA, Hanemann CO, Giancotti FG. Combined Inhibition of NEDD8-Activating Enzyme and mTOR Suppresses NF2 Loss-Driven Tumorigenesis. Molecular cancer therapeutics. 2017;16(8):1693–1704. doi: 10.1158/1535-7163.MCT-16-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kee Y, Huang M, Chang S, Moreau LA, Park E, Smith PG, D’Andrea AD. Inhibition of the Nedd8 system sensitizes cells to DNA interstrand cross-linking agents. Mol Cancer Res. 2012;10(3):369–77. doi: 10.1158/1541-7786.MCR-11-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wei D, Li H, Yu J, Sebolt JT, Zhao L, Lawrence TS, Smith PG, Morgan MA, Sun Y. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer Res. 2012;72(1):282–93. doi: 10.1158/0008-5472.CAN-11-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wan J, Zhu J, Li G, Zhang Z. Radiosensitization of Human Colorectal Cancer Cells by MLN4924: An Inhibitor of NEDD8-Activating Enzyme. Technology in cancer research & treatment. 2016;15(4):527–34. doi: 10.1177/1533034615588197. [DOI] [PubMed] [Google Scholar]
- 166.Yang D, Tan M, Wang G, Sun Y. The p21-dependent radiosensitization of human breast cancer cells by MLN4924, an investigational inhibitor of NEDD8 activating enzyme. PloS one. 2012;7(3):e34079. doi: 10.1371/journal.pone.0034079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Oladghaffari M, Shabestani Monfared A, Farajollahi A, Baradaran B, Mohammadi M, Shanehbandi D, Asghari Jafar Abadi M, Pirayesh Islamian J. MLN4924 and 2DG combined treatment enhances the efficiency of radiotherapy in breast cancer cells. Int J Radiat Biol. 2017:1–10. doi: 10.1080/09553002.2017.1294272. [DOI] [PubMed] [Google Scholar]
- 168.Wang X, Zhang W, Yan Z, Liang Y, Li L, Yu X, Feng Y, Fu S, Zhang Y, Zhao H, Yu J, Jeong LS, Guo X, Jia L. Radiosensitization by the investigational NEDD8-activating enzyme inhibitor MLN4924 (pevonedistat) in hormone-resistant prostate cancer cells. Oncotarget. 2016;7(25):38380–38391. doi: 10.18632/oncotarget.9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vanderdys V, Allak A, Guessous F, Benamar M, Read PW, Jameson MJ, Abbas T. The Neddylation Inhibitor Pevonedistat (MLN4924) Suppresses and Radiosensitizes Head and Neck Squamous Carcinoma Cells and Tumors. Molecular cancer therapeutics. 2017 doi: 10.1158/1535-7163.MCT-17-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Coleman KE, Bekes M, Chapman JR, Crist SB, Jones MJ, Ueberheide BM, Huang TT. SENP8 limits aberrant neddylation of NEDD8 pathway components to promote cullin-RING ubiquitin ligase function. Elife. 2017;6 doi: 10.7554/eLife.24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cao H, Xie J, Guo L, Han K, Pei Y, Li X, Qiu G, Jin J, Hua L, Jing Z, Wu H, Liu J, Wang J, Shen B, Tang S, Zhang J, Zhang J, Chen H, Wang Q. All-trans retinoic acid induces autophagic degradation of ubiquitin-like modifier activating enzyme 3 in acute promyelocytic leukemia cells. Leuk Lymphoma. 2017:1–9. doi: 10.1080/10428194.2017.1365850. [DOI] [PubMed] [Google Scholar]
- 172.Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298(5593):608–11. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 173.Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292(5520):1382–5. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- 174.Lee MH, Zhao R, Phan L, Yeung SC. Roles of COP9 signalosome in cancer. Cell Cycle. 2011;10(18):3057–66. doi: 10.4161/cc.10.18.17320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu S, Yang H, Zhao J, Zhang YH, Song AX, Hu HY. NEDD8 ultimate buster-1 long (NUB1L) protein promotes transfer of NEDD8 to proteasome for degradation through the P97UFD1/NPL4 complex. J Biol Chem. 2013;288(43):31339–49. doi: 10.1074/jbc.M113.484816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tanaka T, Kawashima H, Yeh ET, Kamitani T. Regulation of the NEDD8 conjugation system by a splicing variant, NUB1L. J Biol Chem. 2003;278(35):32905–13. doi: 10.1074/jbc.M212057200. [DOI] [PubMed] [Google Scholar]
- 177.Kamitani T, Kito K, Fukuda-Kamitani T, Yeh ET. Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J Biol Chem. 2001;276(49):46655–60. doi: 10.1074/jbc.M108636200. [DOI] [PubMed] [Google Scholar]
- 178.Nawrocki ST, Griffin P, Kelly KR, Carew JS. MLN4924: a novel first-in-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert Opin Investig Drugs. 2012;21(10):1563–73. doi: 10.1517/13543784.2012.707192. [DOI] [PubMed] [Google Scholar]
- 179.Toth JI, Yang L, Dahl R, Petroski MD. A gatekeeper residue for NEDD8-activating enzyme inhibition by MLN4924. Cell reports. 2012;1(4):309–16. doi: 10.1016/j.celrep.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Milhollen MA, Thomas MP, Narayanan U, Traore T, Riceberg J, Amidon BS, Bence NF, Bolen JB, Brownell J, Dick LR, Loke HK, McDonald AA, Ma J, Manfredi MG, Sells TB, Sintchak MD, Yang X, Xu Q, Koenig EM, Gavin JM, Smith PG. Treatment-emergent mutations in NAEbeta confer resistance to the NEDD8-activating enzyme inhibitor MLN4924. Cancer cell. 2012;21(3):388–401. doi: 10.1016/j.ccr.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 181.Scott DC, Hammill JT, Min J, Rhee DY, Connelly M, Sviderskiy VO, Bhasin D, Chen Y, Ong SS, Chai SC, Goktug AN, Huang G, Monda JK, Low J, Kim HS, Paulo JA, Cannon JR, Shelat AA, Chen T, Kelsall IR, Alpi AF, Pagala V, Wang X, Peng J, Singh B, Harper JW, Schulman BA, Guy RK. Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase. Nat Chem Biol. 2017;13(8):850–857. doi: 10.1038/nchembio.2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhou H, Lu J, Liu L, Bernard D, Yang C-Y, Fernandez-Salas E, Chinnaswamy K, Layton S, Stuckey J, Yu Q, Zhou W, Pan Z-Q, Sun Y, Wang S. A Potent Small-Molecule Inhibitor of the DCN1-UBC12 Interaction that Selectively Blocks Cullin 3 Neddylation. Nat Communs. 2017 doi: 10.1038/s41467-017-01243-7. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Scott DC, Sviderskiy VO, Monda JK, Lydeard JR, Cho SE, Harper JW, Schulman BA. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell. 2014;157(7):1671–84. doi: 10.1016/j.cell.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24(19):8477–86. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]