Abstract
Development of optimal vaccines for influenza is challenging, in part as a result of the high antigenic variability in field strains associated with genetic shift from reassortment and genetic drift from point mutations. Discovery of conserved antigenic sites on the hemagglutinin (HA) protein for neutralizing antibodies suggested the possibility that influenza vaccines could be developed that induce focused antibody responses to the conserved neutralizing determinants, especially the HA stem region. Recent studies have focused on the antigenicity and immunogenicity of such domains, using monoclonal antibodies and candidate-engineered HA stem-based vaccines. Much progress has been made, but we still do not fully understand the biology of the immune response to this unique antigenic region.
Great Debates
What are the most interesting topics likely to come up over dinner or drinks with your colleagues? Or, more importantly, what are the topics that don't come up because they are a little too controversial? In Immune Memory and Vaccines: Great Debates, Editors Rafi Ahmed and Shane Crotty have put together a collection of articles on such questions, written by thought leaders in these fields, with the freedom to talk about the issues as they see fit. This short, innovative format aims to bring a fresh perspective by encouraging authors to be opinionated, focus on what is most interesting and current, and avoid restating introductory material covered in many other reviews.
The Editors posed 13 interesting questions critical for our understanding of vaccines and immune memory to a broad group of experts in the field. In each case, several different perspectives are provided. Note that while each author knew that there were additional scientists addressing the same question, they did not know who these authors were, which ensured the independence of the opinions and perspectives expressed in each article. Our hope is that readers enjoy these articles and that they trigger many more conversations on these important topics.
Antibodies are the principal mediator of protection against infection following exposure in subjects with prior vaccination or infection. Protective antibodies are mostly directed to the surface proteins hemagglutinin (HA) and neuraminidase (NA). There is a large body of literature on the molecular basis for recognition of the HA protein with less extensive research on antibodies to NA. Influenza circulates in three serotypes (types) A, B, C, with only viruses of A and B infecting humans. Within the A type, there are at least 18 subtypes of HA, based on antigenic profiling studies, with H1, H2, H3, H5, H7, and H9 viruses having caused significant numbers of infections in humans. Historically, antibody-mediated immunity focused on HA was known to exhibit a predominantly heterosubtype-specific pattern, although some studies in animals or humans with polyclonal immune serum samples or in animal models with virus challenge noted a small degree of heterosubtypic immunity. More recently, various technologies for making human monoclonal antibodies (mAbs) have matured, and investigators began making human mAbs to diverse influenza viruses, including to H5 influenza, which has threatened to cause a pandemic. In the 2009–2011 period, while studying H5 responses, investigators isolated H5-neutralizing antibodies that also recognized seasonal H1 viruses, and found through atomic resolution structures of antigen–antibody complexes that these antibodies bound to a conserved region in the HA stem, the HA2 subunit that contains the fusion peptide (Sui et al. 2009; Ekiert et al. 2011). Further efforts to screen for broadly neutralizing influenza antibodies rapidly identified mAbs that even crossed the major group 1 and 2 collections of type A viruses (Corti et al. 2011). This exciting work has led to an entire field of stem-based antibody and vaccine development efforts aimed at achieving “universal” coverage of influenza. Universal is a loosely held term sometimes intended to mean pertinent to all seasonal (type A H1, type A H3, type B) strains, or both seasonal and likely prepandemic (H2, H5, H7, H9) strains, or even all influenza strains. Regardless of the clinical intent of the meaning of the term “universal,” the discovery of very broad human mAbs to influenza HA has been an exciting development (Fig. 1).
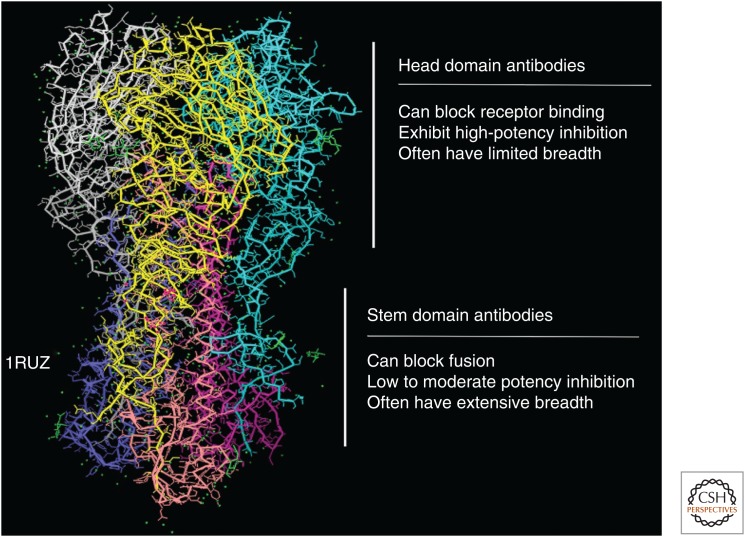
Figure 1.
Structure of the influenza hemagglutinin (HA), with major domains. The structure shown is based on x-ray crystallographic studies of the trimeric soluble 1918 H1N1 influenza HA protein (PDB ID: 1RUZ). The structure contains a globular head domain that is the target for antibodies that bind to the receptor-binding domain and, rarely, the vestigial esterase domain. The stem region is more conserved and binds different classes of antibodies. General characteristics and inhibitory mechanisms of domain-specific human monoclonal antibodies are indicated.
STEM ANTIBODIES
Much has been learned about the genetic and structural basis of influenza stem-reactive antibodies. The initial class of antibodies discovered is quite common in human subjects because one of the human heavy chain variable region gene segments in the germline configuration (VH1-69) encodes a short motif of two hydrophobic amino acids in the HCDR2 loop that are optimal for binding to a hydrophobic pocket in the HA stem. Additional contact residues contribute to the interaction of the overall paratope with stem, but these arise relatively easily with somatic mutations, principally aromatic residues in the heavy-chain framework 3 region. Thus, many humans possess the capacity to make such antibodies. The allele of the VH1-69 does affect the capacity of subjects to make these antibodies, as a biased use of alleles that encode the critical CDR-H2 Phe54 (F-alleles) has been noted in broadly neutralizing antibodies (Avnir et al. 2016). Although the interaction of VH1-69 gene-encoded antibodies perhaps can be considered a canonical interaction, somatic mutations can further optimize the interaction, including broadening the heterosubtypic breadth of recognition (Fu et al. 2016), and multiple antibody clonal lineages can be seen in these responses (Whittle et al. 2014). As additional antibody discovery efforts focused on the stem region, investigators found different classes of antibodies that are encoded by other VH gene segments, and they interrogate the stem region in different manners, with diverse binding poses (Corti et al. 2011).
MECHANISMS OF NEUTRALIZATION
Antibodies to the head domain of HA often block receptor binding. However, stem-directed antibodies do not block receptor binding, and therefore do not exhibit activity in the laboratory assay for blocking sialic acid binding (hemagglutination inhibition). Many stem antibodies do exhibit virus-neutralizing properties in cell culture monolayer assays in vitro. Investigators have pursued detailed studies of the mechanism of neutralization and found several candidate mechanisms. From the very beginning of stem antibody discovery when atomic resolution structures were determined by antigen-antibody complexes, it was apparent that these mAbs bound to the HA subunit containing the fusion peptide. Laboratory studies confirmed that stem antibodies like CR6261 can inhibit HA0 cleavage and pH-dependent conformational changes (Ekiert et al. 2009). In vivo studies have revealed additional functions of stem antibodies that are consistent with Fc-dependent immune-mediated mechanisms. In vivo activity of many stem antibodies requires, or is enhanced by, Fc–FcγR interactions (DiLillo et al. 2014). Functional assays have shown that some protective stem antibodies mediate antibody-dependent cellular cytotoxicity (ADCC) (Jegaskanda et al. 2013, 2014). It is clear that some stem antibodies possess ADCC activity, but not all HA-binding antibodies mediate the activity, and interactions of antibodies to HA head, stem, and NA in polyclonal mixes affect the level of ADCC activity observed (He et al. 2016). Phagocytosis of influenza–antibody immune complexes also can be enhanced by HA stem-specific antibodies in an Fc-dependent manner (Mullarkey et al. 2016).
CLINICAL TRIALS
A number of clinical trials are ongoing with human mAbs to the influenza stem. Generally, the antibody infusions have been well tolerated in healthy subjects in phase 1 trials, as expected. Various clinical scenarios are being investigated, including human influenza challenge of healthy volunteers, uncomplicated influenza infection, and hospitalized cases of influenza infection. MAb CR6261 (Crucell) was studied in phase 1 (ClinicalTrials.gov identifier: NCT01406418) and is being studied in the human challenge model (ClinicalTrials.gov identifier: NCT02371668). Crucell also has studied mAb CR8020 for safety (phase 1 trial, ClinicalTrials.gov identifier: NCT01756950, completed) and in a phase 2a challenge study (ClinicalTrials.gov identifier: NCT01938352). The combination of mAbs CR6261 and CR8020 is being tested in hospitalized subjects (ClinicalTrials.gov identifier: NCT01992276). Visterra has studied their mAb VIS410 in phase 1 (ClinicalTrials.gov identifier: NCT02045472). Medimmune has studied the mAb MEDI8852 in phase 1 studies (ClinicalTrials.gov identifier: NCT02350751) and is studying the mAb in a phase 2a trial in uncomplicated influenza (ClinicalTrials.gov identifier: NCT02603952). Genentech is studying a single intravenous dose of their mAb MHAA4549A in combination with oseltamavir (ClinicalTrials.gov identifier: NCT02293863) in adult participants hospitalized with severe influenza A infection. Given this amount of clinical trial activity, it seems likely a significant amount of safety and efficacy data is on the horizon for the concept and implementation of HA stem-specific antibodies. Whether or not any of these antibodies achieves licensure as a therapeutic drug, the data from these trials will inform the design and testing of universal influenza vaccines.
POOR IMMUNOGENICITY OF THE STEM REGION IN NATURAL INFECTION
Most human and murine antibodies isolated historically were directed to the HA head domain, especially neutralizing antibodies for the epitopes that were the easiest to map by escape mutation analysis. High titers in the hemagglutination inhibition test (which measures antibody-mediated blockade of the sialic acid receptor-binding site on the HA head) have correlated relatively well with protection, and is the only correlate of influenza vaccine immunity recognized by regulatory bodies. Even neutralizing antibody tests of human serum samples (which measure neutralizing antibodies directed to head or stem domains) historically have not identified broadly neutralizing patterns, suggesting cross-reactive stem-specific antibodies are a very minor component of the natural human antibody response to influenza. It is curious that mAb discovery campaigns can find broadly neutralizing antibodies specified by circulating B cells or plasmablasts in many or most individuals, but high titers of such heterosubtypic antibodies generally are not observed in human following natural infection or seasonal vaccination. Careful studies of the response after pandemic 2009 H1N1 infection suggested that, in the setting of reexposure to a virus with a mismatched HA head domain but conserved stem domain, immunodominance may shift temporarily to the stem (Wrammert et al. 2011). Even in this setting, however, it appears that the bump in stem-specific responses in a head-mismatch setting may not persist beyond a year or two (Hoa et al. 2016).
The molecular basis for the natural immune focusing on the HA head domain are not clear presently. It may be that a large part of this dominance has to do simply with the geometry of the protein, in which the head domain projects into solute and is easily accessible from multiple angles of approach. In contrast, B-cell receptors binding to the stem region on intact HA molecules in viral membrane or cells must negotiate beyond the head domain and interact at a constrained angle. Influenza virion particles contain thousands of copies of the HA trimer, which appear closely spaced in high-resolution images, raising the possibility that closely apposed HA head regions reduce accessibility of B-cell receptors and antibodies to the stem region below. This situation does not appear to be the case, however, as investigators have used cryoelectron tomography to obtain density maps of HA trimers on the surface of the 2009 H1N1 pandemic influenza virus bound by a broadly neutralizing murine stem-specific antibody, C179. The images suggested that the majority of HA stem epitopes were accessible on H1N1 virions, including virions of filamentous or spherical morphology (Harris et al. 2013). The stem epitope also could be considered to have a relatively undesirable structure and biophysical properties for an ideal highly immunogenic target, as it is principally centered on a shallow hydrophobic pocket. As a class, the typical stem-specific mAbs that react with this relatively bland surface structure are low in affinity and potency in neutralization. In contrast, many of the commonly recognized structures in the head domain have a hydrophilic character and interesting structural features such as loops and α-helical elements. The receptor-binding site on the head domain is a recessed epitope too, but there are highly canonical interactions in antibody variable loops (principally aromatic residues or aspartate residues) that commonly occur to facilitate interaction with the base of the receptor-binding domain (Xu et al. 2013). Recently, a new stem-specific mAb was reported that binds to a more complex stem epitope, comprising the highly conserved epitope encompassing a hydrophobic groove in the fusion domain and a large portion of the fusion peptide (Kallewaard et al. 2016). Recognizing the stem in this way appears to achieve better potency and breadth than other stem antibodies; the frequency of such antibodies after natural infection must be low, however, based on their rarity of detection.
A general approach to focus the immune response on the stem domain is to remove the head domain from the HA immunogen completely, thus eliminating any accessibility and head immunodominance limitations (Steel et al. 2010). Such an approach sounds straightforward, but the protein engineering challenges are many. The central issue is expressing a stem antigen that faithfully recapitulates the exact conformation of the naturally occurring stem epitope in the full HA molecule. The presence of the head domain stabilizes the stem conformation, so simply removing the head domain has not served sufficiently to generate a fully conformationally correct antigen. Several approaches have been used to design, express, and present novel stem antigens. Headless protein antigens have been tested as proteins or subviral particles (Graves et al. 1983; Steel et al. 2010; Bommakanti et al. 2012; Lu et al. 2014), and such proteins have been improved by iterative design cycles and linking to nanoparticles (Yassine et al. 2015). Others are using cross-linking approaches to design stabilized headless HA vaccine immunogens. Alternate approaches avoid the complex folding issues of HA fragments by using intact HA molecules containing alterations. For example, one approach is to mask immunogenic regions in the head domain with glycans (Lin et al. 2012, 2014; Eggink et al. 2014). Another interesting new approach is using chimeric HA molecules in which the sequence of the head domain is derived from an “exotic” HA from an avian virus that does not commonly infect humans, and the sequence of the stem is from conserved human seasonal viruses (Hai et al. 2012; Krammer et al. 2013, 2014b). The structure of the stem may differ slightly from that of nonchimeric HA molecules (Tran et al. 2016), but generally the immunogenicity of chimeric molecules appears intact in animal studies.
ENHANCED DISEASE
A limited number of studies have raised the question as to whether antibodies to the HA stem region may be pathogenic in some settings. Typically, prior vaccination or infection with virus antigens contributes to partial protection against infection or disease, but, in rare circumstances, prior vaccination or infection causes enhanced disease during subsequent infection. There are specific instances of enhanced disease and molecular and cellular models of enhancement in the field. For example, several inactivated experimental vaccines for viruses that infect by the airway mucosal route (respiratory syncytial virus or measles virus) were associated in the 1960s with severe enhanced disease in recipients following subsequent natural infection. The pathogenesis of these enhanced infections is incompletely understood, but there is evidence that induction of memory cells associated with unbalanced inflammatory phenotype in the absence of protective antibodies and cellular factors is to blame. Dengue virus infections may be enhanced in severity in the setting of a second infection with a virus of a serotype that differs from that of the first. Likely, preexisting serotype cross-reactive heterologous antibodies that bind live virus during reinfection but do not neutralize that virus facilitate uptake into human cells bearing Fc receptors, with associated increased replication and virus gene expression. Influenza virus differs in many ways from these previous cases, but enhanced disease has been reported in very particular cases of animal model studies with HA stem-based antigens.
A study by U.S. Department of Agriculture (USDA) investigators found that pigs immunized with whole inactivated vaccine containing a human-like H1N2 and challenged later with pandemic 2009 H1N1 virus exhibited enhanced respiratory disease, including compared to nonvaccinated animals exposed to pH1N1 (Gauger et al. 2011). This group has used the term vaccine-associated enhanced respiratory disease (VAERD) to categorize this observation. The hypothesis is that pigs vaccinated with other influenza antigen vaccines or with mismatched inactivated viruses can develop worse disease when challenged with H1 viruses. The mechanism for this observation is unknown, but antibodies have been implicated (Heinen et al. 2002; Vincent et al. 2008). U.S. Food and Drug Administration (FDA) investigators have found that the binding of these antibodies to the HA2 subunit, which contains the fusion peptide, facilitated more efficient fusion of pandemic 2009 H1N1 viruses with lung epithelial cells (Khurana et al. 2013). It is challenging to know how generalizable this concern should be. The studies to date find this mechanism only with porcine vaccines, in the porcine influenza model, and the studies are reported by a limited number of investigator groups. It has been observed that inactivated porcine vaccines induce narrower responses than do live virus vaccines or infections. The enhancement studies occurred during studies using adjuvanted antigen. Ferrets studied for similar antigenic exposures using intranasally applied nonadjuvanted vaccines did not show enhanced disease (Krammer et al. 2014a).
Human data on this type of observation is sparse. The unusual prevalence of severe disease in otherwise healthy young adults during major pandemics could be consistent with influence of some immune factors induced by prior heterologous immunity, but this phenomenon is poorly understood. Human subjects who had been immunized with trivalent 2008–2009 inactivated influenza vaccine exhibited an increase of illness in some reports following infection with the pandemic 2009 H1N1 virus (Janjua et al. 2010; Skowronski et al. 2010). Studies of lung tissues of fatal cases in Argentina during the 2009 H1N1 virus pandemic suggested that cross-reactive antibodies may have played a role in severe disease seen in otherwise healthy young adults, as evidenced by C4d deposition in lung, a marker of complement activation mediated by immune complexes (Monsalvo et al. 2011). Those investigators also found similar patterns of C4d deposition in archival lung tissues from fatal cases of influenza in the United States during the 1957 H2N2 influenza pandemic. On the whole, it appears that enhanced disease associated with antibody responses to the HA stem may occur under certain experimental circumstances, but it is unclear whether this phenomenon is likely to pertain to use of stem-based HA vaccine candidates in humans. Given the species-specific nature of the responses observed to date, it is unlikely any influenza animal model could “prove” the safety of stem vaccine candidates for humans. Thus, as with any new vaccine concept, safety will need to be evaluated in humans in a step-wise careful manner.
CONCLUDING REMARKS
The discovery of broadly neutralizing antibodies to the influenza HA stem region is an exciting discovery that raises the conceptual possibility that broadly cross-reactive antibodies might be induced by vaccines designed to be more “universal” than current strategies. Studies are ongoing concerning the safety and efficacy of stem-specific antibodies as treatments and stem-only immunogens as candidate vaccines. It appears that induction of stem-specific antibodies in model systems mediates substantial levels of protection and may contribute to human immunity. Overall, studies to date have shown the relatively poor immunogenicity of the stem region in natural infection or seasonal vaccination of humans, the relatively low potency and durability of stem-specific antibodies in humans, and alternative mechanisms of neutralization used by stem antibodies. One way to interpret these general observations is that stem-based immunity may become an important component in the influenza treatment and prevention arsenal, but might be best used in a complementary fashion in combination with head domain-based antigens or therapies, which as a group exhibit higher potency but more limited breadth.
ACKNOWLEDGMENTS
The author’s work on influenza is supported by National Institute of Allergy and Infectious Diseases (NIAID) Grant U19 AI117905, NIAID Contract HHSN272201400024C, NIAID Grant R43 AI118087, and NIAID Contract HHSN2720100007C (a Center of Excellence for Influenza Research and Surveillance).
Footnotes
Editors: Shane Crotty and Rafi Ahmed
Additional Perspectives on Immune Memory and Vaccines: Great Debates available at www.cshperspectives.org
REFERENCES
- Avnir Y, Watson CT, Glanville J, Peterson EC, Tallarico AS, Bennett AS, Qin K, Fu Y, Huang CY, Beigel JH, et al. 2016. IGHV1-69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity. Sci Rep 6: 20842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommakanti G, Lu X, Citron MP, Najar TA, Heidecker GJ, ter Meulen J, Varadarajan R, Liang X. 2012. Design of Escherichia coli-expressed stalk domain immunogens of H1N1 hemagglutinin that protect mice from lethal challenge. J Virol 86: 13434–13444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333: 850–856. [DOI] [PubMed] [Google Scholar]
- DiLillo DJ, Tan GS, Palese P, Ravetch JV. 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med 20: 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggink D, Goff PH, Palese P. 2014. Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J Virol 88: 699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324: 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, et al. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333: 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Zhang Z, Sheehan J, Avnir Y, Ridenour C, Sachnik T, Sun J, Hossain MJ, Chen LM, Zhu Q, et al. 2016. A broadly neutralizing anti-influenza antibody reveals ongoing capacity of haemagglutinin-specific memory B cells to evolve. Nat Commun 7: 12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauger PC, Vincent AL, Loving CL, Lager KM, Janke BH, Kehrli ME Jr, Roth JA. 2011. Enhanced pneumonia and disease in pigs vaccinated with an inactivated human-like (delta-cluster) H1N2 vaccine and challenged with pandemic 2009 H1N1 influenza virus. Vaccine 29: 2712–2719. [DOI] [PubMed] [Google Scholar]
- Graves PN, Schulman JL, Young JF, Palese P. 1983. Preparation of influenza virus subviral particles lacking the HA1 subunit of hemagglutinin: Unmasking of cross-reactive HA2 determinants. Virology 126: 106–116. [DOI] [PubMed] [Google Scholar]
- Hai R, Krammer F, Tan GS, Pica N, Eggink D, Maamary J, Margine I, Albrecht RA, Palese P. 2012. Influenza viruses expressing chimeric hemagglutinins: Globular head and stalk domains derived from different subtypes. J Virol 86: 5774–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AK, Meyerson JR, Matsuoka Y, Kuybeda O, Moran A, Bliss D, Das SR, Yewdell JW, Sapiro G, Subbarao K, et al. 2013. Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc Natl Acad Sci 110: 4592–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Tan GS, Mullarkey CE, Lee AJ, Lam MM, Krammer F, Henry C, Wilson PC, Ashkar AA, Palese P, et al. 2016. Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc Natl Acad Sci 10.1073/pnas.1609316113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen PP, Rijsewijk FA, de Boer-Luijtze EA, Bianchi AT. 2002. Vaccination of pigs with a DNA construct expressing an influenza virus M2-nucleoprotein fusion protein exacerbates disease after challenge with influenza A virus. J Gen Virol 83: 1851–1859. [DOI] [PubMed] [Google Scholar]
- Hoa LNM, Mai le Q, Bryant JE, Thai PQ, Hang Nle K, Yen NT, Duong TN, Thoang DD, Horby P, Werheim HF, et al. 2016. Association between hemagglutinin stem-reactive antibodies and influenza A/H1N1 virus infection during the 2009 pandemic. J Virol 90: 6549–6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janjua NZ, Skowronski DM, Hottes TS, Osei W, Adams E, Petric M, Sabaiduc S, Chan T, Mak A, Lem M, et al. 2010. Seasonal influenza vaccine and increased risk of pandemic A/H1N1-related illness: First detection of the association in British Columbia, Canada. Clin Infect Dis 51: 1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegaskanda S, Job ER, Kramski M, Laurie K, Isitman G, de Rose R, Winnall WR, Stratov I, Brooks AG, Reading PC, et al. 2013. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J Immunol 190: 1837–1848. [DOI] [PubMed] [Google Scholar]
- Jegaskanda S, Reading PC, Kent SJ. 2014. Influenza-specific antibody-dependent cellular cytotoxicity: Toward a universal influenza vaccine. J Immunol 193: 469–475. [DOI] [PubMed] [Google Scholar]
- Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, Wachter-Rosati L, Palmer-Hill FJ, Yuan AQ, Walker PA, et al. 2016. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell 166: 596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Loving CL, Manischewitz J, King LR, Gauger PC, Henningson J, Vincent AL, Golding H. 2013. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci Transl Med 5: 200ra114. [DOI] [PubMed] [Google Scholar]
- Krammer F, Pica N, Hai R, Margine I, Palese P. 2013. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol 87: 6542–6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Hai R, Yondola M, Tan GS, Leyva-Grado VH, Ryder AB, Miller MS, Rose JK, Palese P, Garcia-Sastre A, et al. 2014a. Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J Virol 88: 3432–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Margine I, Hai R, Flood A, Hirsh A, Tsvetnitsky V, Chen D, Palese P. 2014b. H3 stalk-based chimeric hemagglutinin influenza virus constructs protect mice from H7N9 challenge. J Virol 88: 2340–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lin YF, Chong P, Wu SC. 2012. Broader neutralizing antibodies against H5N1 viruses using prime-boost immunization of hyperglycosylated hemagglutinin DNA and virus-like particles. PLoS ONE 7: e39075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Liu WC, Jan JT, Wu SC. 2014. Glycan masking of hemagglutinin for adenovirus vector and recombinant protein immunizations elicits broadly neutralizing antibodies against H5N1 avian influenza viruses. PLoS ONE 9: e92822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Welsh JP, Swartz JR. 2014. Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc Natl Acad Sci 111: 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalvo AC, Batalle JP, Lopez MF, Krause JC, Klemenc J, Hernandez JZ, Maskin B, Bugna J, Rubinstein C, Aguilar L, et al. 2011. Severe pandemic 2009 H1N1 influenza disease due to pathogenic immune complexes. Nat Med 17: 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullarkey CE, Bailey MJ, Golubeva DA, Tan GS, Nachbagauer R, He W, Novakowski KE, Bowdish DM, Miller MS, Palese P. 2016. Broadly neutralizing hemagglutinin stalk-specific antibodies induce potent phagocytosis of immune complexes by neutrophils in an Fc-dependent manner. MBio 7: e01624–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski DM, De Serres G, Crowcroft NS, Janjua NZ, Boulianne N, Hottes TS, Rosella LC, Dickinson JA, Gilca R, Sethi P, et al. 2010. Association between the 2008–09 seasonal influenza vaccine and pandemic H1N1 illness during Spring-Summer 2009: Four observational studies from Canada. PLoS Med 7: e1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, Garcia-Sastre A, Palese P. 2010. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio 1: e00018–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, et al. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16: 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EE, Podolsky KA, Bartesaghi A, Kuybeda O, Grandinetti G, Wohlbold TJ, Tan GS, Nachbagauer R, Palese P, Krammer F, et al. 2016. Cryo-electron microscopy structures of chimeric hemagglutinin displayed on a universal influenza vaccine candidate. MBio 7: e00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent AL, Lager KM, Janke BH, Gramer MR, Richt JA. 2008. Failure of protection and enhanced pneumonia with a US H1N2 swine influenza virus in pigs vaccinated with an inactivated classical swine H1N1 vaccine. Vet Microbiol 126: 310–323. [DOI] [PubMed] [Google Scholar]
- Whittle JR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, et al. 2014. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J Virol 88: 4047–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, et al. 2011. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med 208: 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Krause JC, McBride R, Paulson JC, Crowe JE Jr, Wilson IA. 2013. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol 20: 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, et al. 2015. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 21: 1065–1070. [DOI] [PubMed] [Google Scholar]