Abstract
Introduction
Metabolomics is a well-established tool in systems biology, especially in the top–down approach. Metabolomics experiments often results in discovery studies that provide intriguing biological hypotheses but rarely offer mechanistic explanation of such findings. In this light, the interpretation of metabolomics data can be boosted by deploying systems biology approaches.
Objectives
This review aims to provide an overview of systems biology approaches that are relevant to metabolomics and to discuss some successful applications of these methods.
Methods
We review the most recent applications of systems biology tools in the field of metabolomics, such as network inference and analysis, metabolic modelling and pathways analysis.
Results
We offer an ample overview of systems biology tools that can be applied to address metabolomics problems. The characteristics and application results of these tools are discussed also in a comparative manner.
Conclusions
Systems biology-enhanced analysis of metabolomics data can provide insights into the molecular mechanisms originating the observed metabolic profiles and enhance the scientific impact of metabolomics studies.
Keywords: Pathway, Network analysis, Correlation network, Association network, Enrichment analysis
Introduction
The pioneering experimental work of Mamer and Horning (Horning and Horning 1971; Mamer and Crawhall 1971) and the first application by Pauling (1971) laid the bases for metabolomic profiling of samples. These approaches constituted the precursors of today’s metabolomics techniques. It was with the work of Oliver (1998) and Trethewey (1999) that metabolomics established itself as a standalone discipline and then became a core component of systems biology (SB), providing an integrated view of biochemistry in complex organisms (Nicholson and Lindon 2008). The rapid evolution and spreading of metabolomics leveraged the technical developments of Nuclear Magnetic Resonance (NMR) and Mass Spectroscopy (MS), which made metabolomics experiments widely accessible.
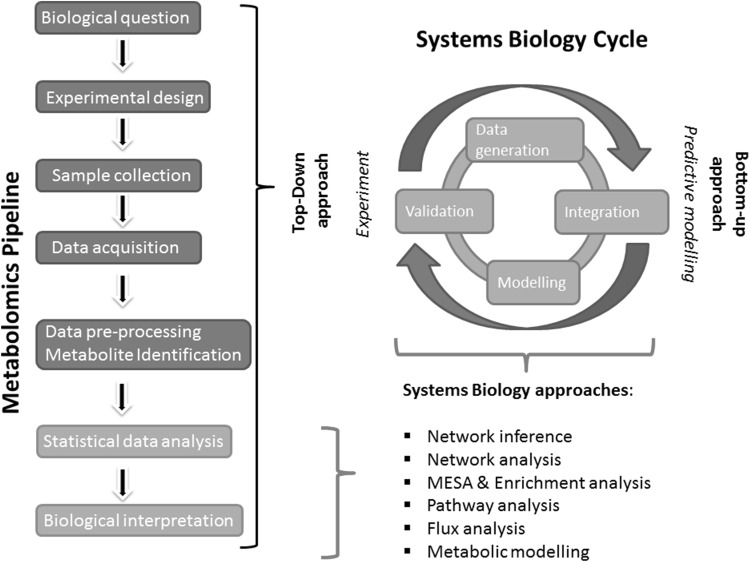
In the top-down approach of SB (see Fig. 1), hypotheses about the regulatory mechanisms are drawn upon the analysis of patterns observed in metabolite profiles. Such hypotheses can be tested in new experiments in an iterative cycle (Bruggeman and Westerhoff 2007). In fact, metabolomics takes a special position among the omics disciplines in the SB top–down approach: the metabolome is the endpoint of biological processes, carrying imprints of genetic, epigenetic and environmental factors, and thus it can provide the link between genotype and phenotype (Fiehn 2002; Griffin 2006; Krumsiek et al. 2016). A crucial demonstration of this concept was the observation that metabolomics measurements can reveal phenotypes for proteins active in metabolic regulation, even if their deletion does not change metabolic fluxes, such as growth rate (Raamsdonk et al. 2001).
Fig. 1.
Relationship between the systems biology cycle and the metabolomics pipeline
Contextually with experimental advancements, researchers soon realized that the potential of metabolomics data could be exploited by deploying multivariate and pattern-recognition methods. The use of components methods, such as principal component analysis and factor analysis was established early (Meuzelaar and Kistemaker 1973; Windig et al. 1980). Then, metabolomics became rapidly intertwined in an almost symbiotic fashion with chemometrics (Trygg et al. 2007; van der Greef and Smilde 2005; Wishart 2007). This alliance has resulted in the development of a vast array of different tools for extracting (bio)chemically relevant information from measured (bio)chemical data, representing and displaying such information, and getting it into databases (Wold 1995; Wold and Sjöström 1998; Spicer et al. 2017).
Chemometrics proved to be pivotal in studies that showcased the potential of metabolomics (Assfalg et al. 2008; Holmes et al. 2008; Nicholson et al. 2011). However, nowadays data analysis based on chemometrics alone may be considered the major bottleneck for further advancement of metabolomics itself. Chemometrics approaches have an intrinsic exploratory nature, and thus their application to metabolomics analyses typically generates novel biological hypotheses that need validation. Moving from research generating hypotheses towards research generating mechanistic insight about biological problems would constitute a major advance for the omics fields (Yates 2016). One way to achieve this is to deploy systems biology approaches, such as network analysis and metabolic modelling, to investigate metabolomics data. This may open new avenues to obtain biological knowledge from transcriptomics, proteomics and metabolomics studies and will allow researchers to leverage all omics to contextualize their results.
In line with the concepts outlined above, in this review we did not cover the approaches to data analysis that are typical of chemometrics and statistical analysis, such as supervised and regression methods (e.g., Partial Least Square Discriminant Analysis, principal component regression) or unsupervised tools (e.g., Principal Component Analysis, cluster analysis). Instead, we focused on systems biology approaches like network inference and metabolic modelling.
Metabolite identification and mapping
An important aspect underlying most if not all the methods for the analysis of metabolomics data that we will address in the next sections is to properly identify the metabolites in the MS or NMR spectra and map them within the metabolic context of the organism. Often the peaks detected in experiments are assigned based on reference spectra contained in large chemical database. However, the analytical methods used in metabolomics do not allow coverage of the whole range of small molecules produced by an organism, introducing possible biases in the interpretation of whole-organism metabolism. Although this is a very broad theme, in this section we will try to summarize the features of some tools for metabolite mapping that can be exploited in the context of systems biology approaches.
Metabolome Searcher (Dhanasekaran et al. 2015) is a web-based application (http://procyc.westcent.usu.edu/cgi-bin/MetaboSearcher.cgi) to directly search genome-constructed metabolic databases. Its aim is to enhance the identification of MS data by using compound databases derived empirically. Incorporating information on genome-encoded metabolism facilitates the identification of MS peaks that may not be present in standard chemical databases. Only the compounds that the organism of interest is able to produce, based on its genome, are investigated for potential matches. The output metabolites are mapped also to known metabolic pathways.
The MassTRIX web server (Suhre and Schmitt-Kopplin 2008) (http://masstrix3.helmholtz-muenchen.de/masstrix3/) addresses the annotation of putative metabolites by providing a hypothesis-driven approach to interpret MS data. MassTRIX processes the submitted list of raw mass peaks by comparing the input experimental masses against all chemical compounds of the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al. 2015, 2017), additionally including 13C, 15N and other isotopes and optionally adding selected lipids. Then it presents the identified chemical compounds in their genomic context as differentially coloured objects on KEGG pathway maps. By adding transcriptomics data or information on differences in the gene complement (e.g. samples from different bacterial strains), the user can interpret the metabolic state of the organism in the context of its actual or potential enzymatic capacities.
A similar approach was also employed in MetaMapp (Barupal et al. 2012). MetaMapp is a tool to integrate biochemical pathways (using the KEGG reactant pair database) and chemical relationships (using the Tanimoto chemical similarity score and the mass spectral similarity score of the National Institute of Standards and Technology, NIST) to map the metabolites detected in MS and/or NMR experiments in a network graph. Such graphs can be displayed in Cytoscape (Shannon et al. 2003). MetaMapp is independent of the experimental technology utilized to identify metabolomics profiles, thus providing a way to integrate and visualize data from different metabolomics platforms.
MetExplore (Cottret et al. 2010) is a computational pipeline designed to map chemical libraries on genome-scale metabolic networks. This tool can be used to obtain statistics on the experimental coverage of organism-specific metabolic networks. The main purpose of MetExplore is to provide an interactive visualization of metabolic networks (or sub-networks) to mine metabolomics (and other “omics”) data. After the mapping is performed, MetExplore permits to visualize metabolites in the context of the whole network, a specific pathway, a selection of pathways or a selection of reactions.
Another recent tool integrating automated analysis of mass spectrometry data and visualization of biological context by linking each metabolite to one or more biological pathways (see also next section) is the Polyomics integrated Metabolomics Pipeline (PiMP) (Gloaguen et al. 2017). This tool annotates metabolites identified in mass spectrometry experiments, providing direct access to the experimental features supporting each annotation, and then allows users to jump directly to the pathway(s) relevant for each metabolite. However, this is a visualization tool and does not perform pathway analysis as described in Sect. 6.
Pre-existing biochemical knowledge about metabolic pathways may provide useful information for the assignment of unknown compounds in large metabolomics datasets. Gipson et al. (2008) exploited this idea by developing a computational protocol to improve UPLS-MS metabolite assignment through the matching of peak correlation pairs (from acquired MS data) with a database of biochemically relevant interaction pairs (pathway data from the KEEG database). A stochastic local search optimization algorithm was implemented to select the putative peak assignment that maximizes both the correlations and the strength of correlations in each cluster of MS peaks, in agreement with the most likely metabolic pathway from the database.
Integrated approaches that combine transcriptome, proteome and metabolome profiling have gained popularity and have proven to provide novel insights in the understanding of the biological systems (Cho et al. 2008; Jiang et al. 2015; Kolbe et al. 2006). A first approach to the interpretation of complex omics experiments is the joined visualization of the data on templates that collect previous knowledge. In this frame, the Paintomics web server (http://www.paintomics.org) (García-Alcalde et al. 2011) provides a simple but effective resource for integrated visualization in studies where transcriptomics and metabolomics data are generated on the same set of samples. The inputs to the server are gene expression and metabolite quantifications, which are then displayed on KEGG maps.
The web-based ProMeTra system (Neuweger et al. 2009) (https://omictools.com/prometra-tool) allows users to combine datasets from heterogeneous multiple-omics sources. This tool visualizes and combines datasets from transcriptomics, proteomics, and metabolomics on user defined metabolic pathway maps. ProMeTra supports pathway maps designed and annotated by the users.
There are only a few tools explicitly devoted to the analysis of metabolomics data. Metscape (Gao et al. 2010) (metscape.ncibi.org) is a plug-in for Cytoscape (Shannon et al. 2003), developed to visualize and interpret metabolomics data in the context of human metabolic networks. Metscape allows users to trace the connections between metabolites and genes, visualize compound networks and display compound structures as well as information for reactions, enzymes, genes, and pathways. Experimental data can be visualized and explored as networks and as a function of time or experimental conditions. A subsequent redesign of Metscape (Metscape 2) (Karnovsky et al. 2012) allows users to enter experimental data and display them in the context of relevant metabolic networks to identify enriched pathways from expression profiling data.
Table 1 presents a list of the tools for mapping metabolites into biochemical pathways mentioned in this section.
Table 1.
Tools for mapping metabolites into biochemical pathways
| Name | Description | Reference | URL |
|---|---|---|---|
| NA | Refine mass assignments through the intersection of peak correlation pairs with a database of biochemically relevant interaction pairs | Gipson et al. (2008) | NA |
| Metabolome Searcher | Simplify database search in MS databases by limiting the query to genome plausible metabolites | Dhanasekaran et al. (2015) | http://procyc.westcent.usu.edu/cgi-bin/MetaboSearcher.cgi |
| MassTRIX | Presents the MS identified chemical compounds in their genomic context as differentially coloured objects on KEGG pathway maps | Suhre and Schmitt-Kopplin (2008) | http://masstrix3.helmholtz-muenchen.de/masstrix3/ |
| MetaMapp | Map the detected metabolites in a MS experiment in a network graph | Barupal et al. (2012) | NA |
| MetExplore | To provide an interactive visualization of metabolic networks (or sub-networks) to mine metabolomics data | Cottret et al. (2010) | http://metexplore.toulouse.inra.fr/joomla3/index.php |
| Paintomics | Provide a simple but effective resource for integrated visualization in studies where transcriptomics and metabolomics data are generated on the same set of samples | García-Alcalde et al. (2011) | http://www.paintomics.org |
| KaPPa-View | A web-based tool for representing quantitative data for individual transcripts and/or metabolites on plant metabolic pathway maps | Tokimatsu et al. (2005) | http://kpv.kazusa.or.jp/ |
| MapMan | A user-driven tool that displays large data sets onto diagrams of metabolic pathways or other processes | Thimm et al. (2004) | http://mapman.gabipd.org/web/guest |
| ProMeTra | Visualizes and combines datasets from transcriptomics, proteomics, and metabolomics on user defined metabolic pathway maps, with the ability to generate enriched SVG images or animations via a user-friendly web interface | Neuweger et al. (2009) | https://omictools.com/prometra-tool |
| Metscape | Allows users to trace the connections between metabolites and genes, visualize compound networks and display compound structures as well as information for reactions, enzymes, genes, and pathways | Gao et al. (2010) | http://metscape.ncibi.org/ |
Analysis of metabolomics data using network approaches
The most natural extension and complementation of methods based on covariance/correlation for the analysis of multivariate metabolomics data [such as principal component analysis or covariance simultaneous component analysis (Smilde et al. 2015)] is their representation and analysis as networks. Networks constitute a powerful view to understand biological systems where not only the individual components are considered, but also their interconnections and their function as a whole (Ma’ayan 2011; Weckwerth and Fiehn 2002).
A biological network is a graphic representation of objects (called nodes) and their relationships (described by links or edges). It can be conveniently described using a matrix, termed adjacency or connectivity matrix A. The rows and columns of A represent the nodes, i.e. metabolite concentrations or abundances. Here, we refer generically to metabolite concentration. Strictly speaking, this is correct only for targeted metabolomics experiments where the concentrations of metabolites are determined using appropriate standards. In general, MS experiments provide metabolite abundances, which can be considered a proxy for concentrations, whereas NMR provides quantities in arbitrary units that are proportional to concentrations. However, from a numerical point of view this is not relevant for the computational methods presented here, but it might be relevant for the biological interpretation of the data. The non-zero elements of A are real numbers that describe the strength of the relationship between any two nodes. The relationship between two metabolites can be very diverse in nature: for instance, one can postulate the existence of such relationship if their concentration levels are highly correlated, if they participate in the same metabolic pathway, or if they are directly connected through some biochemical reaction. Within this context, it should be noted that metabolomics data can be used to reconstruct metabolic networks at different levels (topology, stoichiometry, directionality and kinetics) using dedicated experiments. In this review, we focus on the application of network approaches to analyze metabolomics data that usually have not been gathered with the aim of reconstructing entire metabolic networks. For the latter purpose, the typical starting point is genome data (see also some of the tools mentioned in the previous section and in Table 1). Nevertheless, some approaches are available to build genome-scale metabolic networks from raw high resolution mass spectroscopy data (Jourdan et al. 2007; Moritz et al. 2017). Methods to reconstruct metabolic networks have been reviewed elsewhere (Frainay and Jourdan 2017; Hendrickx 2013; Hendrickx et al. 2011).
Table 2 presents a list of network-based methods applicable to metabolomics studies. These methods are discussed in the following sections.
Table 2.
List of network inference methods used in metabolomics studies
| Acronym | Name | Reference |
|---|---|---|
| ARACNE | Algorithm for the reconstruction of accurate cellular networks | Margolin et al. (2006) |
| CLR | Context likelihood of relatedness algorithm | Faith et al. (2007) |
| CORR | Correlation | |
| PCLRC | Probabilistic context likelihood of relatedness of correlation algorithm | Saccenti et al. (2014) |
| PIUmet | Prize-collecting Steiner forest algorithm for integrative analysis of untargeted metabolomics | Pirhaji et al. (2016b) |
| WCGNA | Weighted correlation gene network analysis | Zhang and Horvath (2005) |
Association networks
The nodes in a network are associated (connected) based on some similarity measure: in metabolomics the similarity between metabolites, and thus their association, is usually expressed using Pearson or Spearman’s correlation indexes. Consequently, the elements of the corresponding adjacency matrix are in the interval [−1, 1] (Cakır et al. 2009). This kind of networks is sometimes called correlation or relevance networks. Biological information can be derived considering both the magnitude and the sign of correlations: for instance, strong positive correlation () between two metabolites can indicate a condition of rapid equilibrium or enzyme dominance, while strong negative correlation can indicate the presence of a conserved moiety (Camacho et al. 2005). In general, the correlations observed in metabolomics data are the result of the combination of all reactions and regulatory processes in the network (Hendrickx 2013; Stelling et al. 2004; Steuer et al. 2003). Surprisingly, there may be no correlation between metabolites that are close in a metabolic pathway. For instance, in wild type potato tubers, glutamate and glutamine are metabolic neighbors in the glutamine synthase pathway, but appear to be uncorrelated (ρ = 0.0243, Spearman). Instead, valine and methionine are strongly correlated (ρ = 0.951) even if they are not metabolic neighbors (Camacho et al. 2005; Weckwerth et al. 2004). The information encoded in the correlation matrix may be not fully sufficient to reverse engineer the underlying enzymatic system (Steuer et al. 2003). Still, it can be used as a proxy to describe a given physiological state of the system of interest, as the correlation matrix can change with the steady-state concentrations of metabolites (Fukushima et al. 2011). It is then reasonable to assume that differences or communalities in the biological processes are reflected in the characteristics of the inferred correlation networks (Szymanski et al. 2009). This is the rationale for the use of association networks to analyse metabolomics data.
The zero elements of the adjacency matrix can be selected based on the statistical significance of the pairwise metabolite correlations. This was the approach used in (Ursem et al. 2008), one of the first papers to deploy a network approach to the analysis of metabolomics data, where Pearson correlations were calculated among pairs of metabolites measured using gas chromatography–mass spectrometry (GC–MS) in tomato samples. The advantage over principal component analysis (PCA) is that network plots do not focus on the representation of maximum variation in data matrices, which may negatively affects data interpretation. Indeed, the relationships between metabolites whose variation is spread out over several principal axes can be easily overlooked in PCA biplots (Ursem et al. 2008). The work of Ursem et al. (2008) built on previous works, where correlation analysis was used to unravel molecular mechanisms (Kose et al. 2001; Roessner et al. 2001; Steuer et al. 2003; Urbanczyk-Wochniak et al. 2003).
Yang et al. (2012) performed a correlation network analysis on urine metabolomics data from patients suffering of central precocious puberty taking a hybrid approach. First, they identified metabolites discriminating between cases and controls using a Partial Least Squares (PLS) approach and then mapped them on a reconstruction of a global human metabolic network using the KEGG database (Kanehisa et al. 2015, 2017). The discriminating metabolites had significantly higher degree, betweeness and closeness than the global network.
Another commonly used approach is to binarize the adjacency matrix by imposing a threshold τ for the correlation | ρ| between any pair of metabolites and/or a threshold α on the associated P-value. This is usually called hard thresholding, as exemplified below:
| 1 |
The choice of the threshold τ is fundamental since it ultimately drives the topology of the resulting networks. In an analysis of tissue- and/or genotype-dependent metabolomics correlations in Arabidopsis, Fukushima et al. investigated the effect of varying the correlation threshold and found that the number of groups of connected metabolites showed a transition from small to large at τ = 0.5, which they subsequently used (Fukushima et al. 2011). They commented that such a threshold does not guarantee explicit biological significance. However, this value is not far from 0.6, which was indicated as a lower bound for low/weak correlations in metabolomics data (Camacho et al. 2005) and used by other authors (Ghini et al. 2015; Saccenti et al. 2016; Suarez-Diez and Saccenti 2015). Szymanski et al. (2009) applied a threshold α = 0.01 on the P-value of the correlation after Bonferroni correction for multiple testing and used bootstrapping to obtain robust correlation estimation.
The patterns of correlations between metabolites can be compared across different conditions to identify associations that are disrupted or altered by pathophysiological conditions with respect to a healthy or control status, an approach referred to as differential network analysis. Hu et al. (2015) addressed the problem of finding disrupted connections in osteoarthritis by taking a statistical approach that exploited a permutation test to assess the significance of changes in the correlations of two metabolites across different conditions. Similarly, Szymanski et al. (2009) considered metabolite correlation networks from Escherichia coli exposed to different environmental stress conditions and compared network characteristics to pinpoint possible mechanisms underlying stress response.
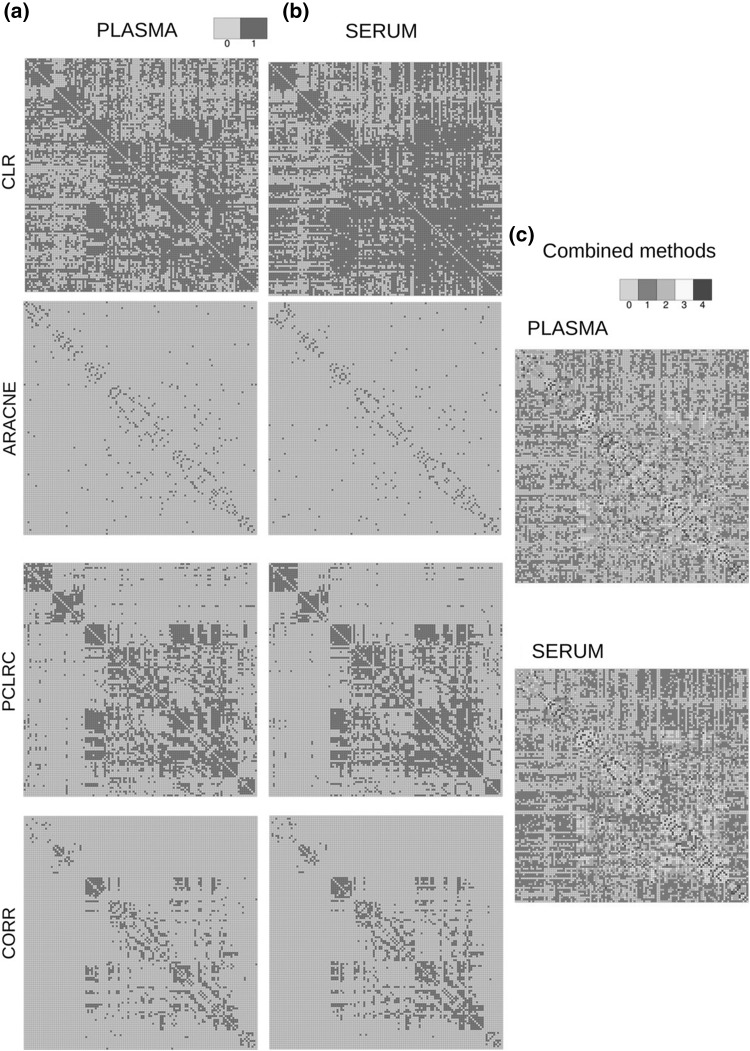
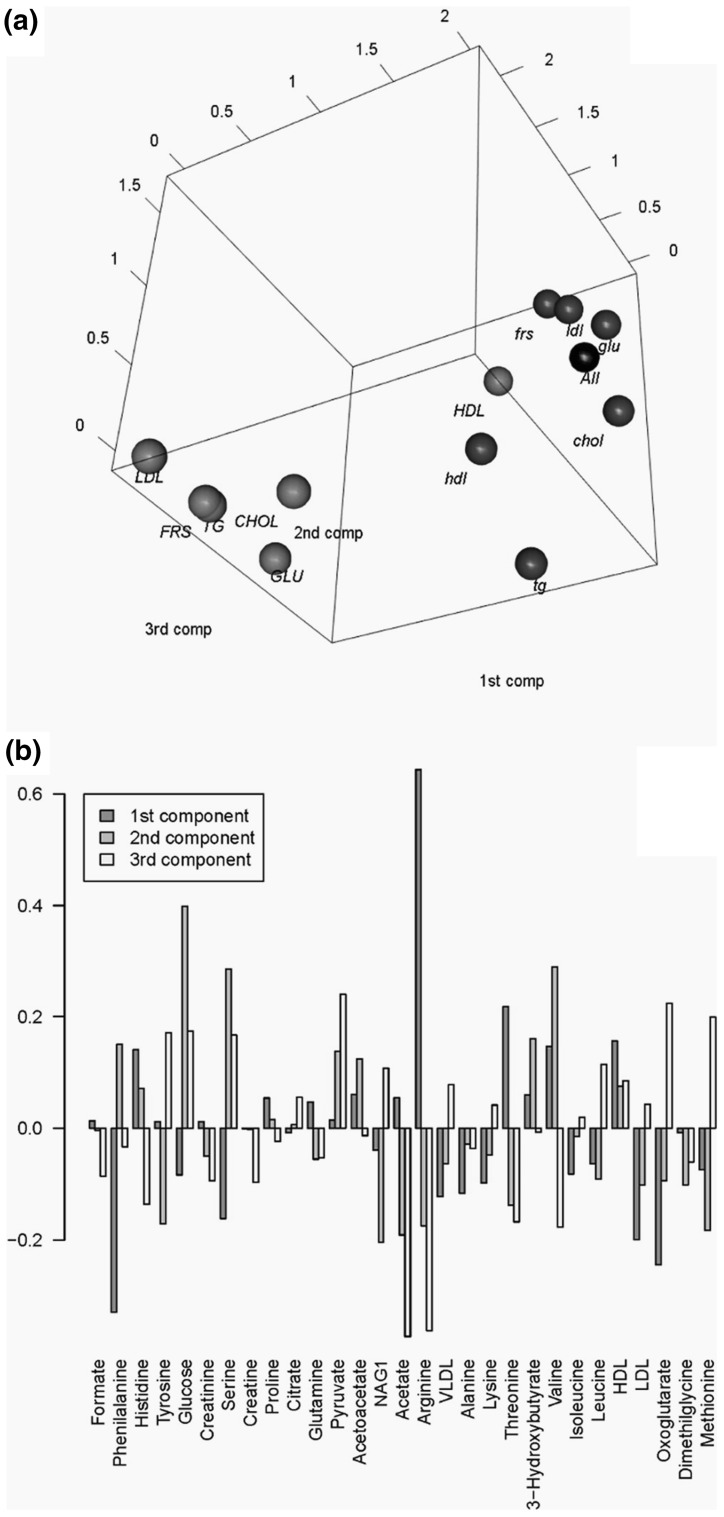
Saccenti et al. (2014) investigated the latent cardiovascular risk of healthy subjects by considering highly connected metabolites, the so called hubs, and reported differential behaviour of Very Low Density Lipoprotein (VLDL) and glucose in high and low risk cardiovascular risk networks. They applied a combined method, by analysing association networks with a multivariate approach to highlight differences among networks pertaining to different risk phenotypes (see Fig. 2). Hubs are nodes that are much more connected than average or typical nodes, and consequently are very likely to play crucial biological roles. The concept of hubs was first introduced within the analysis of yeast protein–protein interaction networks (Jeong et al. 2001).
Fig. 2.
Association network of 133 blood metabolites measured using MS/MS on 2139 subjects. a Plasma metabolites association networks obtained using the four different methods. b Serum metabolites association networks obtained using the four different methods. c Consensus association network for serum and plasma. CLR context likelihood of relatedness, ARACNE algorithm for the reconstruction of accurate cellular networks, PCLRC probabilistic context likelihood of relatedness on correlations, CORR Pearson’s correlation).
Reproduced with permission from Suarez-Diez et al. (2017). Copyright (2017) American Chemical Society
The correlations observed for metabolomics data are usually small () because of the systemic nature of metabolic control. As previously mentioned, two metabolites can be poorly correlated even if they are neighbours in a metabolic pathway because the variance in the enzymes that control them can affect their levels to the same extent and in different directions (Camacho et al. 2005). Metabolites are generated through fast biochemical reactions in an open mass-flow system. Consequently, they can be considered to be in a quasi-steady state when compared to the time scales of the upstream regulatory processes. This results also in indirect, system-wide correlations between distantly connected metabolites (Lee et al. 2008). The latter phenomenon can be taken into account using partial correlations, i.e. considering pairwise correlation between two variables with the effect of a set of controlling random variables removed. Krumsiek et al. (2011) used Gaussian graphical models, a type of undirected network representation where the relationships among metabolites are expressed as partial correlations, to analyse a large human population cohort. They found this approach to generate more sparse and robust networks with modular structure than those based on Pearson’s correlations, and observed that high partial correlation coefficients generally correspond to known metabolic reactions. This is a striking result since associations in a correlation networks do not necessarily correspond to and/or represent metabolic reactions (Marcotte 2001; Steuer et al. 2003). Using the same approach, Krumsiek et al. (2015) investigated sex-related differences in metabolite association networks and found several submodules across different pathways that were strongly gender-regulated.
As a word of caution, it is important to consider that the results of network inference (and data analysis in general) can be affected by data pre-treatment (also known as pre-processing) such as scaling, transformation and normalization. Such pre-treatments are routinely applied to metabolomics data in order to correct for systematic and unwanted variation such as sample-to sample to variability induced by dilution effects (e.g. in the case of urine) or differences in experimental settings (like different sample titration or different number of scans in NMR experiments). The literature on the topic is huge: we refer the reader to Bijlsma et al. (2006), Goodacre et al. (2007), Saccenti (2016), Van Den Berg et al. (2006) and references therein for more information.
Weighted correlation networks
Weighted gene correlation network analysis (WCGNA) is a systems biology method for describing the correlation patterns among genes across microarray samples. WCGNA can be used for finding clusters (modules) of highly correlated genes, for summarizing such clusters using the so-called module eigengene, i.e. a representative gene summarizing the expression profile of the module (Langfelder and Horvath 2007), or an intramodular hub gene, for relating modules to one another and to external sample traits (Langfelder and Horvath 2008). When applied to metabolite profiles rather than to gene expression profiles WCGNA can be considered an extension of correlation network inference. While correlation networks are based on the similarity of metabolites profiles as expressed by the correlation coefficients, WCGNA in based on the dissimilarity profiles obtained from the so-called topological overlap matrix (TOM). Using the TOM makes the networks less sensitive to spurious connections or to connections missing due to random noise (Ravasz et al. 2002; Zhao et al. 2010). However, also the TOM is based on the correlation between any pair of metabolites; indeed, the element wij of TOM is defined as
| 2 |
where
| 3 |
| 4 |
and mi and mj denote metabolite i-th and j-th, ki and kj denote the connectivity of metabolite i-th and j-th. The dissimilarity is defined as 1 − wij, which is a measure of interaction between metabolites weighted by the strength of their correlation. The parameter is chosen to achieve a scale-free topology and its choice is a fundamental step in WCGNA. Clusters of metabolites are obtained by applying a hierarchical clustering algorithm on the dissimilarity matrix in order to assign the metabolites to different modules based on a dynamic branch height cutting algorithm (Langfelder et al. 2007).
DiLeo et al. (2011) applied WCGNA to NMR metabolomics data collected from developmentally staged tomato fruits belonging to several genotypes. With this approach, they could recognize and model systems-level differences in biological networks even where the poorly defined phenotypes precluded the use of PCA or other multivariate approaches.
Lusczek et al. (2013) applied WCGNA to investigate pathophysiologic state associated with traumatic injury and haemorrhagic shock through the analysis of scale-invariant metabolic network which were constructed from NMR urinary metabolic profiles. They could define network modules (i.e. clusters of functionally related metabolites) related, for examples, to tricarboxylic acid (TCA) cycle or to aerobic metabolism. Within those modules they identified hub metabolites related to cellular respiration, highlighting its fundamental role in the pathophysiology of haemorrhagic shock and to late resuscitation time points. They observed that PLS discriminant analysis (PLS-DA) did not capture the significance of several hub metabolites, which emerged only in the network analysis. In the same work (Lusczek et al. 2013), the authors discussed also the limitation of the WCGNA approach. Such limitations rest on the assumptions that the network shows a scale-free topology, that is with few metabolites highly connected and many metabolites with low connectivity; this translates in the connectivity P(k) and the clustering coefficient C(k) to follow a power law. The authors found P(k) to follow a power law but not C(k), indicating the absence of modular structure in the network of urinary metabolites. They suggested that this may be caused by (i) urine being a waste product in which little to no active metabolism occurs and (ii) the limited number of metabolites considered (n = 60) which is less than the content of the full urinary metabolome. A further hypothesis put forward in the same work was that networks constructed from metabolite profiles derived from biological samples that are metabolically active, such as blood or tissue, may exhibit power law (i.e. a few metabolites connected with many metabolites) behaviour in both connectivity and clustering coefficients. However, in contrast to gene regulatory network, expression networks or metabolic networks, the metabolite correlation networks have not been fully characterized in terms of network topology (i.e. the patterns of interconnection among the nodes). Therefore, it is not very clear what are the expected or more likely network properties (e.g. small-world networks, distribution networks). We refer the reader to (Lee et al. 2008; Nikiforova et al. 2005; Weckwerth et al. 2004) and references therein for more on this topic.
Approaches from functional genomics
Since one of the major challenges in systems biology is the reconstruction of gene regulatory networks, many methods have been developed for this scope (Marbach et al. 2012) and some of them have been deployed in metabolomics. Saccenti et al. (Suarez-Diez and Saccenti 2015) compared two methods for the inference of regulatory networks, ARACNE (Algorithm for the Reconstruction of Accurate Cellular Networks) and PCLR (Probabilistic Context Likelihood of Relatedness Algorithm), to reconstruct blood metabolite association networks. Both these methods leverage mutual information. Given two discrete variables A and B (describing, for instance, metabolite concentrations), the mutual information MI(A,B) between A and B is defined as
| 5 |
where p(ai,bj) is the joint probability distribution function of A and B, and p(ai) (respectively p(bj)) indicates the probability that A = ai (respectively B = bj). It should be noted that the mutual information between two variables is not independent from correlations, since, under some conditions, the two variables can be functionally related (Song et al. 2012). The following sections describe the two approaches in some detail.
The algorithm for the reconstruction of Accurate cellular networks (ARACNE)
ARACNE (Algorithm for the Reconstruction of Accurate Cellular Networks) (Margolin et al. 2006) assigns to each pair of metabolites an association weight equal to their mutual information. It then takes into account triplets of edges connecting metabolites i, j and k in the network. The weakest association of each triplet is considered to be indirect (spurious) and pruned, i.e. set to 0, if the difference between the two lowest weights is above a cut-off value ξ. In practice, the following two conditions are evaluated for each triplet i, j, k:
| 6 |
The weighted adjacency matrix is transformed into a binary topological matrix by additionally imposing a threshold on the mutual information. The threshold is usually 0, leading to all non-zero values being transformed to 1. Saccenti et al. (2015) observed that ARACNE produces extremely sparse metabolites association networks; nevertheless, most of the associations deemed relevant by the ARACNE algorithm were also recovered by the other algorithms assessed in the study, indicating that it was able to reconstruct the backbone of the association network.
The context Likelihood of relatedness (CLR) algorithm
The CLR algorithm (Faith et al. 2007) estimates the likelihood of the mutual information MI(i, j) between two metabolites by defining a null model that considers all the possible MI values [MIi] and [MIj] for metabolites i and j. The following equations define the likelihood f
| 7 |
where
| 8 |
and µi and σi are, respectively, the mean and the standard deviation of the distribution of the [MIi] values: a weighted adjacency matrix is built with entries f(zi, zj).
The probabilistic context likelihood of relatedness of correlation algorithm (PCLRC)
Saccenti et al. (2014) developed a novel version of the CLR approach by substituting the mutual information with correlation and using a resampling approach for robust inference of the correlations. In this implementation, two-thirds of the data are used to iteratively estimate pairwise correlations among metabolites retaining only the 30% strongest.
At each iteration a matrix Ait is built in such a way that = 1 if there is an association between metabolites i and j and 0 otherwise; this procedure is repeated K times and the final weighted association network is constructed by averaging the entries of Ait over the K iterations. The weights constitute a probabilistic measurement of edge likeliness on which a threshold can be applied to obtain a binarized association network. This algorithm was used to construct association networks of blood metabolites characteristics of low and high latent cardiovascular risk (Saccenti et al. 2014; Zhao et al. 2010).
The wisdom of crowd approach
Saccenti et al. (2016) proposed a wisdom of crowd approach (Marbach et al. 2012) to define urine metabolite association networks in healthy subjects by considering the consensus obtained from four different approaches (ARACNE, CLR, PCLR and Pearson’s correlations) and deeming relevant only associations inferred by three or more methods. They modelled the subject-specific networks through a statistical mechanics approach (Menichetti et al. 2015), by defining a core network of metabolite–metabolite associations conserved across 31 subjects.
The same approach was used in a study aiming to compare metabolite association networks obtained from serum and plasma samples. The networks were found to be topologically similar but showed local differences as in the case of amino acids (see Fig. 3) (Suarez-Diez et al. 2017). Similarly, Vignoli et al. (2017) studied sex- and age-specific association networks for metabolites in the plasma of healthy subjects. In particular, they investigated the different patterns of interconnectedness and observed sex-related variability in several metabolic pathways (branched-chain amino acids, ketone bodies and propanoate metabolism) as well as reduction in the connectivity of metabolite hubs linked to age in both sex groups.
Fig. 3.
a Weight plot and b loadings plot of the INDSCAL model for the metabolite correlation network obtained using the PCLCR method. Each dot represents a network that corresponds to a given cardiovascular (CVD) risk parameter. Blue dots indicate low latent CVD risk, while red indicate high latent CVD risk. The associated CVD risk parameters are indicated in upper case for high risk and lower case for low risk. A reference network (indicated as “All”, black ball), built using all the subjects in the study, is given as reference.
Reproduced with permission from Saccenti et al. (2014). Copyright (2014) American Chemical Society
Other methods
Pirhaij et al. (2016a) used their algorithm PIUmet (http://fraenkel-nsf.csbi.mit.edu/PIUMet/) to analyse and interpret untargeted liquid chromatography–mass spectrometry (LC–MS) data from lipidomics and phosphoproteomics experiments in a cell-line model of Huntington’s disease. Grounding on database information, the algorithm infers the identity of unassigned metabolites corresponding to features and the molecular mechanisms underlying their dysregulation. This innovative approach helps to reduce the bias towards well-studied metabolites typical of targeted metabolomics. The algorithm takes as input a list of LC–MS peaks that differ between two different conditions and searches for them in a databases containing over 42,000 nodes (either proteins or metabolites) connected by over one million weighted edges representing interactions between proteins as well as enzymatic and transporter reactions. The output is a subnetwork of the database representing metabolic pathways that are dysregulated under the conditions considered.
Kinetic models
The metabolism is a network structure that can be approached as a system of interdependent variables that enable mathematical modelling through kinetic models. These models are defined as systems of ordinary differential equations describing the time course of metabolite concentrations as a function of rate laws that account for enzyme catalysis. The development of these models requires to know both the network structure and the reaction kinetics and parameters (Klipp et al. 2004). On the one hand, there is a large accumulated knowledge regarding the network structure, which is stored in databases like KEGG (Kanehisa et al. 2012), MetaCyc (Caspi et al. 2016) or Biomodels (Chelliah et al. 2015). Although this is a well-studied cellular level, the true structures can be importantly affected by factors like compartmentalization (de Mas et al. 2011; Nicolae et al. 2014) enzyme complexes and metabolic channelling (Castellana et al. 2014; Ovadi 1991). On the other hand, regarding reaction kinetics, there is also an accumulated knowledge, which can be explored in databases such as BRENDA (Scheer et al. 2011; Schomburg et al. 2013) or SABIO-RK (Wittig et al. 2012).
However, the details on enzyme kinetic parameters are available only for a minor part of the latter reactions (Büchel et al. 2013). In addition, the available measurements of the kinetic properties of enzymes historically come from systems reconstituted in vitro using purified enzymes (Savageau 1992). In this setting, the ideal conditions of homogeneity and free diffusion are fulfilled, and consequently the resulting models may neglect some factors affecting the kinetic properties, such as molecular crowding (Schnell and Turner 2004) and limited diffusion (Alekseev et al. 2016). To overcome these limitations, alternative approaches combine sampling methods with the integration of systemic available data and in vivo observations (fluxes, concentrations, perturbation experiments, …) (Andreozzi et al. 2016; Saa and Nielsen 2016; Stanford et al. 2013).
Alternative approaches take advantage of the current availability of data regarding the network structure and of the lineal nature of the system used to describe it, to apply optimization techniques to infer flux distributions (Fouladiha and Marashi 2017). Genome—scale models accounting for thousands of reactions are currently available (Chelliah et al. 2015; King et al. 2016; Swainston et al. 2016).
For those models including only the network structure as well as for complete kinetic models, it is useful to adopt techniques based on stable isotopes to know about the internal distribution of the metabolism. These are addressed in the next section.
Metabolic flux modelling using stable isotope resolved metabolomics data
Although the analysis of metabolite correlative networks may not grasp the complete underlying metabolic mechanisms, it is certainly a valuable tool for the exploration of metabolomics data, as shown by the budding literature on the topic. The use of stable isotopes can provide a greater insight on the mechanisms that underlie the observed metabolomics profiles, permitting a direct analysis of mechanistic changes in metabolism. Each chemical reaction or transport process involved in a metabolic pathway is associated with a rate (flux) of transformation or transport. Mechanistic changes at the level of the metabolism are likely to produce changes in the distribution of fluxes. Intracellular fluxes are not directly measurable, but the use of stable isotope-enriched nutrients, such as 1,2-13C2-glucose or 13C5,15N2-glutamine, in in cell culture media and the application of Stable Isotope Resolved Metabolomics (SIRM) (Fan et al. 2012; Higashi et al. 2014) provides clues about the redistribution of carbon atoms along metabolic pathways. This can be used to estimate information about fluxes, such as their relative or absolute magnitudes (Lee 2006; Zamboni et al. 2005).
The estimation of fluxes based on the measured patterns of stable isotope labeling (especially using 13C) relies upon a combination of different methods, going from the direct interpretation of the labeling patterns to computational model-based approaches (Buescher et al. 2015; Niedenführ et al. 2015). Frequently, direct interpretation of labeling patterns is sufficient to provide information on the relative activities of pathways, on qualitative changes in pathway contributions via alternative metabolic routes, and on nutrient contribution to the production of different metabolites (Buescher et al. 2015). A recent example is the direct interpretation of the contributions of isotopic labeling tracers like 1,2-13C2-glucose to the synthesys of pentoses phosphate (Dong et al. 2017). The entry of this tracer into the oxidative pentose phosphate pathway results in the loss of the 13C tracer in position 1 in 1,2-13C2-glucose, contributing to the synthesis of ribose phosphate molecules that contain only one 13C atom (usually named M+1 pool of ribose-5-phosphate). Instead, the entry into the non-oxidative pentose phosphate pathway results in the synthesis of ribose phosphate molecules that contain two 13C atoms (usually named M+2 pool of ribose-5-phosphate). The subsequent entry of M+1 pentose-phosphate into glycolysis contributes to the synthesis of triose phosphate and lactate molecules with one 13C atom (M+1). An approximate estimation of the relative importance of oxidative versus non-oxidative pentose phosphate pathway fluxes can be inferred from the M+1/M+2 ratio of the RNA-derived ribose. During the last years, the use of this and other isotopic labelling tracers have been applied to unveil the different metabolic pathways activated in cancer cells (see for a review Dong et al. 2017).
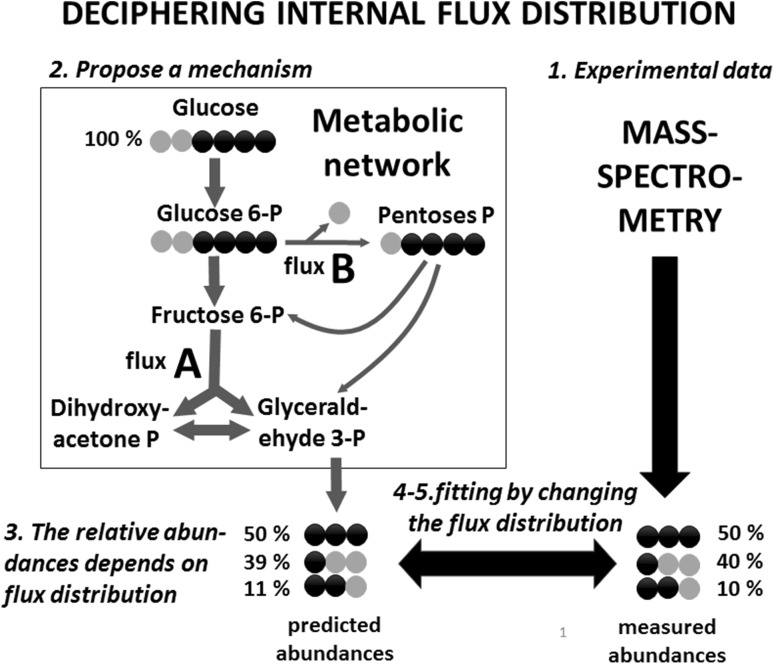
By using computational approaches, all internal metabolic fluxes can be estimated simultaneously by combining the measured labeling patterns resulting from isotope propagation with the measured cellular uptake and secretion rates (Buescher et al. 2015). A reliable model of the relevant network of biochemical reactions is an indispensable input to the computational approach. The reliability of hypotheses regarding flux distributions can be evaluated by comparing measured and predicted isotopologue distributions. (Fig. 4). A variety of different methods are available (Crown and Antoniewicz 2013; Kruger and Ratcliffe 2009; Niedenführ et al. 2015; Sauer 2006; Wiechert and Nöh 2013; Zamboni 2011), together with specific software platforms: FiatFlux (Zamboni et al. 2005); Isodyn (Selivanov et al. 2005); METRAN (Yoo et al. 2008); OpenFlux (Quek et al. 2009); Influx_s (Sokol et al. 2012); 13CFLUX2 (Weitzel et al. 2013); INCA (Young 2014); WUFlux (He et al. 2016). In many cases, a system of balance equations around isotopomers—which depend on specific fluxes—is solved to predict label enrichments. Fluxes are iteratively changed until the difference among measured and predicted label enrichments is reduced.
Fig. 4.
Overview of metabolic flux modelling using stable isotope resolved metabolomics data
Ideally, assuming steady state, the distribution of isotopologues would only depend on the distribution of fluxes and the labeled and non-labeled status of the substrates used in the experiment. However, 13C propagation from tracer precursors to products is a dynamic phenomenon. Initially, all product metabolites are unlabeled (M+0). Progressively, these products are enriched in 13C, with concomitant decrease in M0. Isotopic steady state (Selivanov et al. 2005) is quickly reached for small pools of metabolites but not necessarily for larger pools such as those of fatty acids, glycogen and culture medium metabolites. For these larger pools, M0 values are oversized and may not decrease to the hypothetical value that should be reached at steady state. Accordingly, as an alternative, some software platforms allow for solving the fitting procedure under non isotopic steady state (e.g. Isodyn, INCA among those cited above).
Pathway analysis
Enrichment analysis and overrepresentation analysis: the concept
Enrichment analysis as applied in metabolomics is largely based on the approaches implemented for the analysis of transcriptomes, known as Gene Set Enrichment Analysis (GSEA) (Subramanian et al. 2005). The original idea of GSEA is to focus on «gene sets, that is, groups of genes that share common biological function, chromosomal location, or regulation» instead of performing statistics on individual genes. In practice, the goal of the approach is to detect biological processes, such as metabolic pathways, that differ in the experimental dataset of interest versus control datasets.
Replacing gene transcription level with alterations in metabolite concentrations provides a very straightforward approach to interpret metabolomics experiments in terms of changes in the activity of cellular processes. For the application of the GSEA concept in metabolomics, prior information on the biological relationships between metabolites is needed and can be derived from databases of metabolic pathways and reactions (see Table 3 for a list of databases), such as KEGG (Kanehisa et al. 2015, 2017) or MetaCyc (Caspi et al. 2008), or computed based on the similarity of chemical structures (Moreno et al. 2015).
Table 3.
List of databases of metabolic pathways
| Acronym | Full name | Features | Reference |
|---|---|---|---|
| BiGG | Biochemical genetic and genomic knowledgebase of large scale metabolic reconstructions | A genome-scale metabolic reconstruction of the human metabolism | Schellenberger et al. (2010) |
| BioCyc | BioCyc database collection | A collection of computationally predicted metabolic pathways for nearly 9400 organisms whose genome is available Requires subscription |
Caspi et al. (2016) |
| HumanCyc | Encyclopedia of human genes and metabolism | A partially curated database of metabolic reactions derived from the human genome Requires subscription |
Romero et al. (2004) |
| KEGG | Kyoto encyclopedia of genes and genomes | A collection of manually drawn pathway maps | Kanehisa et al. (2017) |
| MetaCyc | MetaCyc metabolic pathway database | A curated database of experimentally elucidated pathways | Caspi et al. (2016) |
| Reactome | NA | A curated, peer-reviewed knowledgebase of biological pathways, including metabolic pathways. It is mainly focused on human pathways | Fabregat et al. (2016) |
| WikiPathways | NA | A database of biological pathways maintained by and for the scientific community | Kelder et al. (2012) |
A related approach is the so called over-representation analysis (ORA, sometimes called annotation enrichment analysis) where one checks whether a group of differentially expressed genes is enriched for a pathway or ontology term by using overlap statistics such as the cumulative hypergeometric distribution (Doniger et al. 2003; Zhong et al. 2004). In contrast with GSEA, ORA does not involve a quantitative assessment of the change in metabolite concentrations. In practice, the application of a hypergeometric test or Fisher’s exact test, with appropriate corrections for multiple testing (e.g. Bonferroni), allows researchers to evaluate whether specific pathways containing metabolites in an experiment-derived list are overrepresented. If the input list contains metabolites featuring different concentrations in different phenotypes (e.g. healthy versus diseased) then the analysis will identify pathways associated with the phenotype changes.
Metabolite set enrichment analysis (MSEA)
In the application of the GSEA concept to metabolomics, MSEA takes into consideration a quantitative measure associated to each metabolite (e.g. concentration). As the first step of the analysis, metabolites are assigned to specific sets based on one or more reference databases. A group of metabolites are assigned to the same set if they are known to be: (i) involved in the same biological processes (i.e., metabolic pathways, signaling pathways, taken from KEGG) (Kanehisa et al. 2015, 2017); (ii) changed significantly under the same pathological conditions (i.e., various metabolic diseases, taken from the Human Metabolome Database, HMDB) (Wishart et al. 2013) and (iii) present in the same locations such as organs, tissues, or cellular organelles (e.g., also from HMDB).
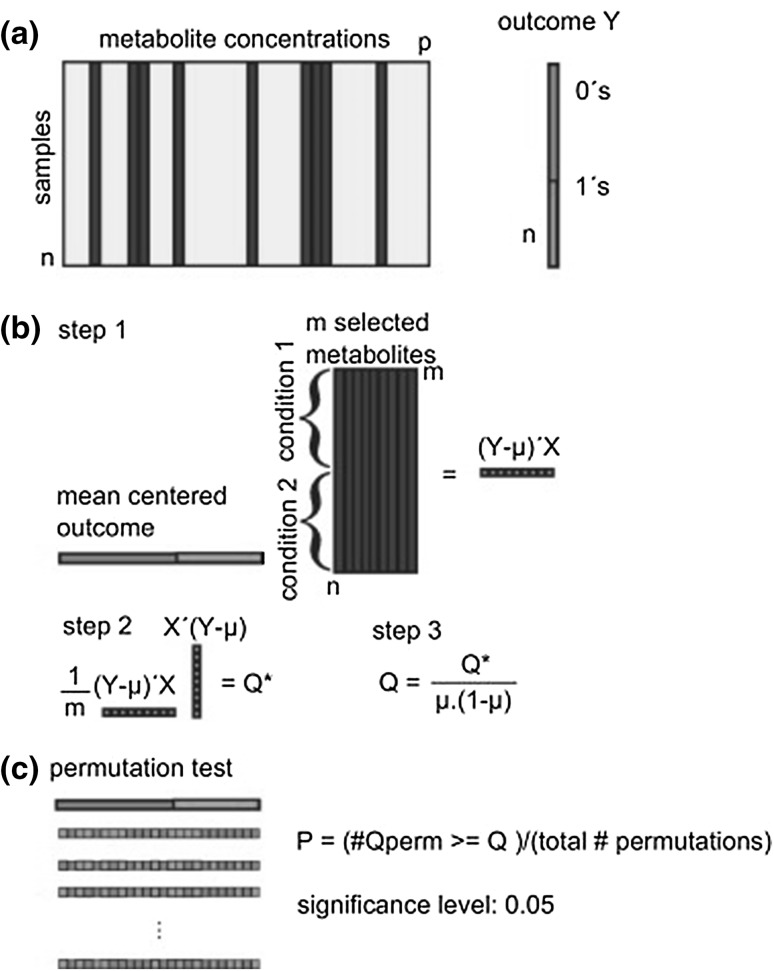
Different strategies exist for performing MSEA depending, among others, on the statistical test applied. In the popular Globaltest method (Goeman et al. 2004) n samples (e.g. individuals) of p metabolites are measured, of which m metabolites belonging to the same pathway are selected. The question whether these metabolites behave differently in the two conditions being compared can be translated into the question whether the metabolite levels are predictive for the outcome (Fig. 5). In other words, the question is “does the knowledge of the metabolite concentrations help to improve the prediction of the phenotype (e.g. group, survival, etc…)?” To answer this question, Globaltest exploits logistic regression, where the regression coefficients indicate whether a certain metabolite affects the difference between the two conditions. The null hypothesis tested is that no metabolite in the pathway has a different concentration in the two conditions. Thus, the regression coefficients are all zero if the group of selected metabolites has no influence on the phenotype. Unfortunately, the number of coefficients is often much larger than the number of samples leaving no room for classical testing procedures. Goeman et al. (2004) dealt with this issue by assuming that all coefficients belong to a common distribution and demonstrated that the covariance of the distribution is zero under the null hypothesis. Thus, the test becomes whether the covariance is zero (null hypothesis) or different from zero (alternative hypothesis). For this purpose, Rao’s score test (Rao 1948), which is very powerful for detecting small deviations from the null hypothesis, can be applied. The quality parameter that is reported is the Q-score statistics, which is based on the differences of metabolite levels between two conditions; a P-value is calculated by using permutations. A correction is needed for multiple hypothesis (pathway) testing (e.g. Bonferroni). The Globaltest detects consistent differences in patterns of metabolite levels between two conditions. It does not test in which direction a pathway is regulated (up or down), nor it determines how many metabolites have changed concentration levels between two conditions. If the tested pathway is activated or inhibited by the tested condition (e.g. healthy versus diseased patients), the differences in metabolite levels will result in a large Q-score and a small P-value. However, the results may change, depending on which metabolites are included, i.e. on the completeness of the database(s) from which prior knowledge has been obtained. If the correlation of the missing metabolite(s) with the outcome is almost equal to the average correlation between the outcome and the metabolites included in the pathway, this has almost no effect on the Q-score. Instead, if a metabolite that has a much higher or lower correlation to the outcome than average is missing then the Q-score will change upon its inclusion. This is an aspect inevitably intrinsic to the MSEA strategy. Databases contain metabolites from only a limited number of pathways, compared to the whole metabolic network of an organism. Consequently, it is possible to test only a relatively small number of pathways and this is an inherent limitation of MSEA.
Fig. 5.
Overview of the Global test. a From the autoscaled data matrix, m metabolites belonging to the same pathway are selected. A binary outcome is defined, coded 0 and 1, for instance healthy versus disease. b A score statistic Q is calculated from the mean centered outcome and the matrix of selected metabolites. c The significance of the relation between the group of metabolites (pathway) and the outcome is determined by performing a permutation test.
Reproduced with permission from Hendrickx et al. (2012); Copyright (2012) Elsevier B. V
Another available method is Global Analisys of Covariace (GlobalANCOVA). GlobalANCOVA exploits linear logistic regression and Analysis of Variance (ANOVA) in the framework of a global assessment for a group of metabolites. GlobalANCOVA aims to evaluate the relationship between the metabolite concentrations and the phenotypic covariates. In particular, the aim of GlobalANCOVA is to prove the relevance of certain covariates in explaining the observed metabolite concentration patterns, called covariates of interest. Therefore, two models are compared: the full model (FM), which contains all covariates and the reduced model (RM), which does not have the covariates of interest. The null hypothesis is that both models explain the data equally well. The relevance of the covariates of interest in explaining the observed pattern is proven if the full model explains the observation better than the reduced model. To do so, a squared error is computed for the fitting of the concentration levels of each metabolite. Subsequently, the residual sum of squares (RSS) over all metabolites in the group is computed. Finally, a multivariate test statistic is built based on the RSS values for the full and reduced models (Hummel et al. 2008; Mansmann and Meister 2005; Smyth 2005). The F-test is applied to test the null hypothesis and a P-value is computed using permutations. A correction for multiple testing is also used. Differently from the Globaltest, GlobalANCOVA evaluates the impact of group membership on the observed metabolite concentration patterns. In other words, GlobalANCOVA practically tests the null hypothesis that the information on the group level does not improve the fitting. The GlobalANCOVA approach allows the inclusion of time-dependent information in a straightforward manner constructed (Hummel et al. 2008).
Hendrickx et al. (2012) first tested the applicability of the Globaltest for metabolomics data and found it effective to highlight the differential behavior of groups of metabolites measured in E. coli and S. cerevisiae under different environmental conditions.
In a recent study on the impact of sequence variability of mitochondrial DNA on metabolism and ageing, MSEA was used to investigate specific pathways in liver and plasma, showing for example significant changes of glutathione metabolism in both organs (Latorre-Pellicer et al. 2016). MSEA is also useful to assess the impact of therapeutic strategies in disease. For example, the inhibition at an early of glutamine metabolism induces extensive changes in the metabolism of other amino acids but also of the oxidation of branched-chain fatty acids in pancreatic ductal adenocarcinoma cells (Biancur et al. 2017).
Over representation analysis (ORA)
The most traditional strategy for enrichment analysis in transcriptomics is to take the user’s preselected list of ‘interesting’ genes e.g. genes showing differential expression between two conditions and then iteratively test the enrichment of their annotation terms; Gene Ontology (GO) terms are often used for this purpose. The annotation terms passing the enrichment P-value threshold are then reported in a tabular format, usually ordered by the enrichment probability or P-value. The calculation of the enrichment P-value is related to the number of genes in the list that share the same annotation terms. For example, Gorilla (Eden et al. 2009) enables GO enrichment analysis in ranked lists of genes. Ranking is usually done as a function of expression level or of fold-change in expression. The method identifies, independently for each GO term, the threshold at which the most significant enrichment is obtained. The significance score is corrected for threshold multiple testing. The null assumption is that all configurations of GO term occurrence in the ranked list are equiprobable.
To apply ORA to pathway analysis, the user provides one or more lists of identifiers representing genes/proteins/metabolites significantly associated with the effect of interest. In order to reduce the potential bias when the number of such measured entities is small it is advisable to provide also background lists of all measured genes/proteins/metabolites. Otherwise, all the entities in the predefined pathway database, or in a user-selected sub-ensemble of pathways, are taken into account and used as the background list. Based on the occurrence of its entities within the input lists, the significance of each pathway is assessed by means of a statistical test. ORA analyzes whether, for a given list of metabolites with significantly different concentrations, one particular pathway is overrepresented, i.e. there are more metabolites in the list from that pathway than would be expected by chance. A major difference of ORA with respect to MSEA is that it does not take into account the extent of the fold change of the abundance of metabolites in the list of significant entities: the inclusion of any metabolite in the list typically depends on a fixed arbitrary threshold. In some tools for ORA, however ranked lists are provided, i.e. metabolites are sorted based on the fold-change of their concentration (or their P-values). The analysis focuses on whether common terms tend to occur towards the top or the bottom of the list (Kankainen et al. 2011). An application of ORA to patients with mild cognitive impairment (MCI), a transition phase between normal aging and Alzheimer’s disease (AD), showed that the pentose phosphate pathway was differently regulated in MCI patients who later progressed to AD with respect to patients who remained stable (Oresic et al. 2011).
The common weakness of tools performing ORA is that the linear output of terms can be very large and overwhelming (from hundreds to thousands), and this can make difficult to grasp potential interrelationships of relevant terms. In addition, the quality of the pre-selected metabolite lists has a deep influence on the enrichment analysis, making the output unpredictably sensitive to changing statistical methods or cutoff thresholds. In particular, it is inappropriate to use all the metabolites of the metabolite set library as the reference metabolome, because there is no analytical platform that can measure all these metabolites with the same probability. Thus, the choice of the platform rather than the experimental conditions may cause the observed metabolite enrichment. To tackle this problem, the user may upload a platform-specific reference metabolome. This is an option provided, for example, in the implementation available in MetaboAnalyst (Xia et al. 2015). Finally, since multiple hits on a given pathway are required to achieve statistical significance, ORA is of limited usefulness for small-sized pathways like glutathione biosynthesis pathway, which contains only ten compounds.
Due to their intrinsic differences, MSEA and ORA may not give the same results and potentially lead to unlike biological interpretation of the same experimental data. This has been demonstrated for a small set of microarray data, where different GO terms and therefore different biological processes were identified by Globaltest and GOEAST (a web-tool for the analysis of GO term enrichment) (Hulsegge et al. 2009).
Pathway activity profiling (PAPi)
PAPi allows users to compare the activity of metabolic pathways under different experimental conditions (Aggio et al. 2010). The underlying concept is to associate Activity Scores to each pathway in a set obtained from the KEGG database by averaging the relative abundance of all detected metabolites assigned to that pathway, normalized by a scaling factor that takes into account that not all metabolites are detected. The comparison of Activity Scores under two or more different experimental conditions for the same pathway can pinpoint changes in activity that are statistically significant, as assessed by a two-sample t-test or by ANOVA. PAPi can provide information regarding the impact of environmental conditions and stimuli on metabolite uptake and intracellular metabolic overflow. Metabolic pathway activity is directly related to metabolic flux distribution and thus this kind of analysis can tie directly to fluxomics.
Concluding remarks
The systems biology approach to the interpretation of metabolomics has the potential to unravel the causative mechanisms leading to the observed metabolomics profile. In this way, there is a paradigm shift from the chemometrics framework that makes metabolomics a hypothesis-generating research field to a framework where metabolomics can provide insights into the biological properties of cell and organism functioning. This shift will unlock the potential of metabolomics and related omics disciplines, such as fluxomics and lipidomics, to fully contribute to the advancement of our understanding of health and disease. In this review, we addressed approaches based on association networks and on pathway analysis. These are useful tools to grasp the complexity of metabolomic profiles; however, they are not sufficient to understand fully the intricacies of the metabolism without dedicated experiments.
Many of the methods described here exploit the lessons learned in other, more mature omics, mainly genomics and transcriptomics, e.g. regarding the validation of their theoretical frameworks. As mentioned several times, a major caveat in untargeted metabolomics is the impossibility of measuring all metabolites in the sample, whose consequences are very difficult to predict.
Acknowledgements
We thank José Camacho and Fabien Jourdan for the fruitful comments on the manuscript. MC acknowledges support of CIBERHD (CIBER de enfermedades hepáticas y respiratorias, Madrid) and of the Icrea Academia and 2017-SGR-1033 (AGAUR, Generalitat de Catalunya). This work was supported by the European Commission funded FP7 project INFECT (Contract No. 305280) and by H2020 project PhenoMeNal (Contract No. 654241).
Compliance with ethical standards
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Contributor Information
Antonio Rosato, Email: rosato@cerm.unifi.it.
Edoardo Saccenti, Email: esaccenti@gmail.com.
References
- Aggio RB, Ruggiero K, Villas-Bôas SG. Pathway Activity Profiling (PAPi): From the metabolite profile to the metabolic pathway activity. Bioinformatics. 2010;26:2969–2976. doi: 10.1093/bioinformatics/btq567. [DOI] [PubMed] [Google Scholar]
- Alekseev AE, et al. Restrictions in ATP diffusion within sarcomeres can provoke ATP-depleted zones impairing exercise capacity in chronic obstructive pulmonary disease. Biochimica et Biophysica Acta (BBA)-General Subjects. 2016;1860:2269–2278. doi: 10.1016/j.bbagen.2016.04.018. [DOI] [PubMed] [Google Scholar]
- Andreozzi S, Miskovic L, Hatzimanikatis V. iSCHRUNK–in silico approach to characterization and reduction of uncertainty in the kinetic models of genome-scale metabolic networks. Metabolic Engineering. 2016;33:158–168. doi: 10.1016/j.ymben.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Assfalg M, et al. Evidence of different metabolic phenotypes in humans. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1420–1424. doi: 10.1073/pnas.0705685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barupal DK, et al. MetaMapp: Mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity. BMC Bioinformatics. 2012;13:99. doi: 10.1186/1471-2105-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancur DE, et al. Compensatory metabolic networks in pancreatic cancers upon perturbation of glutamine metabolism. Nature Communication. 2017;8:15965. doi: 10.1038/ncomms15965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma S, et al. Large-scale human metabolomics studies: A strategy for data (pre-) processing and validation. Analytical Chemistry. 2006;78:567–574. doi: 10.1021/ac051495j. [DOI] [PubMed] [Google Scholar]
- Bruggeman FJ, Westerhoff HV. The nature of systems biology. Trends in Microbiology. 2007;15:45–50. doi: 10.1016/j.tim.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Büchel F, Le N, Novère Path2Models: Large-scale generation of computational models from biochemical pathway maps. BMC Systems Biology. 2013;7:116. doi: 10.1186/1752-0509-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher JM, et al. A roadmap for interpreting 13 C metabolite labeling patterns from cells. Current Opinion in Biotechnology. 2015;34:189–201. doi: 10.1016/j.copbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakır T, Hendriks MM, Westerhuis JA, Smilde AK. Metabolic network discovery through reverse engineering of metabolome data. Metabolomics. 2009;5:318–329. doi: 10.1007/s11306-009-0156-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho D, de la Fuente A, Mendes P. The origin of correlations in metabolomics data. Metabolomics. 2005;1:53–63. doi: 10.1007/s11306-005-1107-3. [DOI] [Google Scholar]
- Caspi R, et al. The metaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Research. 2008;36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Research. 2016;44:D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana M, et al. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nature Biotechnology. 2014;32:1011–1018. doi: 10.1038/nbt.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelliah V, et al. BioModels: Ten-year anniversary. Nucleic Acids Research. 2015;43:D542-8. doi: 10.1093/nar/gku1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K, et al. Integrated transcriptomics, proteomics, and metabolomics analyses to survey ozone responses in the leaves of rice seedling. Journal of Proteome Research. 2008;7:2980–2998. doi: 10.1021/pr800128q. [DOI] [PubMed] [Google Scholar]
- Cottret L, et al. MetExplore: A web server to link metabolomic experiments and genome-scale metabolic networks. Nucleic Acids Research. 2010;38:W132–W137. doi: 10.1093/nar/gkq312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown SB, Antoniewicz MR. Parallel labeling experiments and metabolic flux analysis: Past, present and future methodologies. Metabolic Engineering. 2013;16:21–32. doi: 10.1016/j.ymben.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mas IM, et al. Compartmentation of glycogen metabolism revealed from 13C isotopologue distributions. BMC Systems Biology. 2011;5:175. doi: 10.1186/1752-0509-5-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran AR, Pearson JL, Ganesan B, Weimer BC. Metabolome searcher: A high throughput tool for metabolite identification and metabolic pathway mapping directly from mass spectrometry and using genome restriction. BMC Bioinformatics. 2015;16:62. doi: 10.1186/s12859-015-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLeo MV, Strahan GD, den Bakker M, Hoekenga OA. Weighted correlation network analysis (WGCNA) applied to the tomato fruit metabolome. PLoS ONE. 2011;6:e26683. doi: 10.1371/journal.pone.0026683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Keibler MA, Stephanopoulos G. Review of metabolic pathways activated in cancer cells as determined through isotopic labeling and network analysis. Metabolic Engineering. 2017;43:113–124. doi: 10.1016/j.ymben.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: Using gene ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biology. 2003;4:R7–R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Research. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, et al. Large-scale mapping and validation of escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biology. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T W-M, Lorkiewicz PK, Sellers K, Moseley HN, Higashi RM, Lane AN. Stable isotope-resolved metabolomics and applications for drug development. Pharmacology & Therapeutics. 2012;133:366–391. doi: 10.1016/j.pharmthera.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics–the link between genotypes and phenotypes. Plant molecular biology. 2002;48:155–171. doi: 10.1023/A:1013713905833. [DOI] [PubMed] [Google Scholar]
- Fouladiha H, Marashi SA. Biomedical applications of cell- and tissue-specific metabolic network models. Journal of Biomedical Informatics. 2017;68:35–49. doi: 10.1016/j.jbi.2017.02.014. [DOI] [PubMed] [Google Scholar]
- Frainay C, Jourdan F. Computational methods to identify metabolic sub-networks based on metabolomic profiles. Briefings in bioinformatics. 2017;18:43–56. doi: 10.1093/bib/bbv115. [DOI] [PubMed] [Google Scholar]
- Fukushima A, Kusano M, Redestig H, Arita M, Saito K. Metabolomic correlation-network modules in Arabidopsis based on a graph-clustering approach. BMC System Biology. 2011 doi: 10.1186/1752-0509-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, et al. Metscape: A Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. Bioinformatics. 2010;26:971–973. doi: 10.1093/bioinformatics/btq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Alcalde F, García-López F, Dopazo J, Conesa A. Paintomics: A web based tool for the joint visualization of transcriptomics and metabolomics data. Bioinformatics. 2011;27:137–139. doi: 10.1093/bioinformatics/btq594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghini V, Saccenti E, Tenori L, Assfalg M, Luchinat C. Allostasis and resilience of the human individual metabolic phenotype. Journal of Proteome Research. 2015;14:2951–2962. doi: 10.1021/acs.jproteome.5b00275. [DOI] [PubMed] [Google Scholar]
- Gipson GT, Tatsuoka KS, Sokhansanj BA, Ball RJ, Connor SC. Assignment of MS-based metabolomic datasets via compound interaction pair mapping. Metabolomics. 2008;4:94–103. doi: 10.1007/s11306-007-0096-9. [DOI] [Google Scholar]
- Gloaguen Y, et al. PiMP my metabolome: An integrated, web-based tool for LC-MS metabolomics data. Bioinformatics. 2017 doi: 10.1093/bioinformatics/btx499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman JJ, Van De Geer SA, De Kort F, Van Houwelingen HC. A global test for groups of genes: Testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- Goodacre R, et al. Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics. 2007;3:231–241. doi: 10.1007/s11306-007-0081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin JL. The Cinderella story of metabolic profiling: Does metabolomics get to go to the functional genomics ball? Philosophical Transactions of the Royal Society of London B. 2006;361:147–161. doi: 10.1098/rstb.2005.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Wu SG, Zhang M, Chen Y, Tang YJ. WUFlux: An open-source platform for 13 C metabolic flux analysis of bacterial metabolism. BMC Bioinformatics. 2016;17:444. doi: 10.1186/s12859-016-1314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx DM. Network inference from time-resolved metabolomics data. Amsterdam: University of Amsterdam; 2013. [Google Scholar]
- Hendrickx DM, Hendriks MM, Eilers PH, Smilde AK, Hoefsloot HC. Reverse engineering of metabolic networks, a critical assessment. Molecular BioSystems. 2011;7:511–520. doi: 10.1039/C0MB00083C. [DOI] [PubMed] [Google Scholar]
- Hendrickx DM, Hoefsloot HCJ, Hendriks MMWB, Canelas AB, Smilde AK. Global test for metabolic pathway differences between conditions. Analytica Chimica Acta. 2012;719:8–15. doi: 10.1016/j.aca.2011.12.051. [DOI] [PubMed] [Google Scholar]
- Higashi RM, Fan TW-M, Lorkiewicz PK, Moseley HN, Lane AN. Stable isotope-labeled tracers for metabolic pathway elucidation by GC-MS and FT-M. In: Raftery D, editor. Mass spectrometry in metabolomics: Methods and protocols. New York: Humana Press; 2014. pp. 147–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horning EC, Horning MG. Metabolic Profiles: Gas-Phase Methods for Analysis of Metabolites. Clinical Chemistry. 1971;17:802–809. [PubMed] [Google Scholar]
- Hu, T., et al. (2015). Metabolomics differential correlation network analysis of osteoarthritis biocomputing 2016. In Pacific symposium on biocomputing 2016 (pp.120–131). World Scientific, Singapore. [PubMed]
- Hulsegge, I., Kommadath, A., & Smits, M. A. (2009). Globaltest and GOEAST: Two different approaches for gene ontology analysis. In BMC proceedings (Vol. 3, p. S10). London: BioMed Central [DOI] [PMC free article] [PubMed]
- Hummel M, Meister R, Mansmann U. GlobalANCOVA: Exploration and assessment of gene group effects. Bioinformatics. 2008;24:78–85. doi: 10.1093/bioinformatics/btm531. [DOI] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabási A-L, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Jiang J, Wolters JE, van Breda SG, Kleinjans JC, de Kok TM. Development of novel tools for the in vitro investigation of drug-induced liver injury. Expert Opinion on Drug Metabolism & Toxicology. 2015;11:1523–1537. doi: 10.1517/17425255.2015.1065814. [DOI] [PubMed] [Google Scholar]
- Jourdan F, Breitling R, Barrett MP, Gilbert D. MetaNetter: Inference and visualization of high-resolution metabolomic networks. Bioinformatics. 2007;24:143–145. doi: 10.1093/bioinformatics/btm536. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Research. 2015;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankainen M, Gopalacharyulu P, Holm L, Orešič M. MPEA—metabolite pathway enrichment analysis. Bioinformatics. 2011;27:1878–1879. doi: 10.1093/bioinformatics/btr278. [DOI] [PubMed] [Google Scholar]
- Karnovsky A, et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28:373–380. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelder T, et al. WikiPathways: Building research communities on biological pathways. Nucleic Acids Research. 2012;40:D1301–D1307. doi: 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King ZA, et al. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016;44:D515-22. doi: 10.1093/nar/gkv1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipp E, Liebermeister W, Wierling C. Inferring dynamic properties of biochemical reaction networks from structural knowledge. Genome Informatics. 2004;15:125–137. [PubMed] [Google Scholar]
- Kolbe A, Oliver SN, Fernie AR, Stitt M, van Dongen JT, Geigenberger P. Combined transcript and metabolite profiling of Arabidopsis leaves reveals fundamental effects of the thiol-disulfide status on plant metabolism. Plant Physiology. 2006;141:412–422. doi: 10.1104/pp.106.081208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose F, Weckwerth W, Linke T, Fiehn O. Visualizing plant metabolomic correlation networks using clique–metabolite matrices. Bioinformatics. 2001;17:1198–1208. doi: 10.1093/bioinformatics/17.12.1198. [DOI] [PubMed] [Google Scholar]
- Kruger NJ, Ratcliffe RG. Insights into plant metabolic networks from steady-state metabolic flux analysis. Biochimie. 2009;91:697–702. doi: 10.1016/j.biochi.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Krumsiek J, et al. Gender-specific pathway differences in the human serum metabolome. Metabolomics. 2015;11:1815–1833. doi: 10.1007/s11306-015-0829-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumsiek J, Bartel J, Theis FJ. Computational approaches for systems metabolomics. Current Opinion in Biotechnology. 2016;39:198–206. doi: 10.1016/j.copbio.2016.04.009. [DOI] [PubMed] [Google Scholar]
- Krumsiek J, Suhre K, Illig T, Adamski J, Theis FJ. Gaussian graphical modeling reconstructs pathway reactions from high-throughput metabolomics data. BMC Systems Biology. 2011;5:21. doi: 10.1186/1752-0509-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Systems Biology. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: The dynamic tree cut library for R. Bioinformatics. 2007 doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- Latorre-Pellicer A, et al. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature. 2016;535:561–565. doi: 10.1038/nature18618. [DOI] [PubMed] [Google Scholar]
- Lee D-S, Park J, Kay K, Christakis N, Oltvai Z, Barabási AL. The implications of human metabolic network topology for disease comorbidity. Proceedings of the National Academy of Sciences. 2008;105:9880–9885. doi: 10.1073/pnas.0802208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Gianchandani EP, Eddy JA, Papin JA. Dynamic analysis of integrated signaling, metabolic, and regulatory networks. PLOS Computational Biology. 2008;4:e1000086. doi: 10.1371/annotation/5594348b-de00-446a-bdd0-ec56e70b3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WNP. Characterizing phenotype with tracer based metabolomics. Metabolomics. 2006;2:31–39. doi: 10.1007/s11306-006-0017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusczek E, Lexcen D, Witowski N, Mulier K, Beilman G. Urinary metabolic network analysis in trauma, hemorrhagic shock, and resuscitation. Metabolomics. 2013;9:223–235. doi: 10.1007/s11306-012-0441-5. [DOI] [Google Scholar]
- Ma’ayan A. Introduction to network analysis in systems biology. Science Signaling. 2011;4:tr5. doi: 10.1126/scisignal.2001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamer O, Crawhall J. The identification of urinary acids by coupled gas chromatography-mass spectrometry. Clinica Chimica Acta. 1971;32:171–184. doi: 10.1016/0009-8981(71)90329-9. [DOI] [PubMed] [Google Scholar]
- Mansmann U, Meister R. Testing differential gene expression in functional groups Goeman’s global test versus an ANCOVA approach. Methods Archive. 2005;44:449–453. [PubMed] [Google Scholar]
- Marbach D, et al. Wisdom of crowds for robust gene network inference. Nature Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte EM. The path not taken. Nature biotechnology. 2001;19:626–628. doi: 10.1038/90222. [DOI] [PubMed] [Google Scholar]
- Margolin AA, et al. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a Mammalian cellular context. BMC Bioinformatics. 2006;7:S7–S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menichetti G, Bianconi G, Castellani G, Giampieri E, Remondini D. Multiscale characterization of ageing and cancer progression by a novel network entropy measure. Molecular BioSystems. 2015;11:1824–1831. doi: 10.1039/C5MB00143A. [DOI] [PubMed] [Google Scholar]
- Meuzelaar HC, Kistemaker PG. Technique for fast and reproducible fingerprinting of bacteria by pyrolysis mass spectrometry. Analytical Chemistry. 1973;45:587–590. doi: 10.1021/ac60325a051. [DOI] [PubMed] [Google Scholar]
- Moreno P, et al. BiNChE: A web tool and library for chemical enrichment analysis based on the ChEBI ontology. BMC Bioinformatics. 2015;16:56. doi: 10.1186/s12859-015-0486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz F, Kaling M, Schnitzler JP, Schmitt-Kopplin P. Characterization of poplar metabotypes via mass difference enrichment analysis. Plant, Cell & Environment. 2017;40:1057–1073. doi: 10.1111/pce.12878. [DOI] [PubMed] [Google Scholar]
- Neuweger H, et al. Visualizing post genomics data-sets on customized pathway maps by ProMeTra–aeration-dependent gene expression and metabolism of Corynebacterium glutamicum as an example. BMC Systems Biology. 2009;3:82. doi: 10.1186/1752-0509-3-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson G, et al. Human metabolic profiles are stably controlled by genetic and environmental variation. Molecular Systems Biology. 2011;7:525. doi: 10.1038/msb.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK, Lindon JC. Systems biology: Metabonomics. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- Nicolae A, Wahrheit J, Bahnemann J, Zeng AP, Heinzle E. Non-stationary 13C metabolic flux analysis of Chinese hamster ovary cells in batch culture using extracellular labeling highlights metabolic reversibility and compartmentation. BMC Systems Biology. 2014;8:50. doi: 10.1186/1752-0509-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenführ S, Wiechert W, Nöh K. How to measure metabolic fluxes: A taxonomic guide for 13 C fluxomics. Current Opinion in Biotechnology. 2015;34:82–90. doi: 10.1016/j.copbio.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Daub CO, Hesse H, Willmitzer L, Hoefgen R. Integrative gene-metabolite network with implemented causality deciphers informational fluxes of sulphur stress response. Journal of Experimental Botany. 2005;56:1887–1896. doi: 10.1093/jxb/eri179. [DOI] [PubMed] [Google Scholar]
- Oliver SG, Winson MK, Kell DB, Baganz F. Systematic functional analysis of the yeast genome. Trends in Biotechnology. 1998;16:373–378. doi: 10.1016/S0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- Oresic M, et al. Metabolome in progression to Alzheimer’s disease. Translational Psychiatry. 2011;1:e57. doi: 10.1038/tp.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovadi J. Physiological significance of metabolic channelling. Journal of Theoretical Biology. 1991;152:1–22. doi: 10.1016/S0022-5193(05)80500-4. [DOI] [PubMed] [Google Scholar]
- Pauling L, Robinson AB, Teranishi R, Cary P. Quantitative analysis of urine vapor and breath by gas-liquid partition chromatography. Proceedings of the National Academy of Sciences. 1971;68:2374–2376. doi: 10.1073/pnas.68.10.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirhaji L, et al. Revealing disease-associated pathways by network integration of untargeted metabolomics. Nature Methods. 2016;13:770–776. doi: 10.1038/nmeth.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quek L-E, Wittmann C, Nielsen LK, Krömer JO. OpenFLUX: Efficient modelling software for 13 C-based metabolic flux analysis. Microbial Cell Factories. 2009;8:25. doi: 10.1186/1475-2859-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raamsdonk LM, et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nature Biotechnology. 2001;19:45–50. doi: 10.1038/83496. [DOI] [PubMed] [Google Scholar]
- Rao, C. R. (1948). Large sample tests of statistical hypotheses concerning several parameters with applications to problems of estimation. In Mathematical Proceedings of the Cambridge Philosophical Society (Vol. 44, pp. 50–57). Cambridge: Cambridge University Press.
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabási A-L. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- Roessner U, et al. Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. The Plant Cell. 2001;13:11–29. doi: 10.1105/tpc.13.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, Wagg J, Green ML, Kaiser D, Krummenacker M, Karp PD. Computational prediction of human metabolic pathways from the complete human genome. Genome Biology. 2004;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saa PA, Nielsen LK. Construction of feasible and accurate kinetic models of metabolism: A Bayesian approach. Scientific Reports. 2016;6:29635. doi: 10.1038/srep29635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccenti E. Correlation patterns in experimental data are affected by normalization procedures: Consequences for data analysis and network inference. Journal of Proteome Research. 2016 doi: 10.1021/acs.jproteome.6b00704. [DOI] [PubMed] [Google Scholar]
- Saccenti E, Menichetti G, Ghini V, Remondini D, Tenori L, Luchinat C. Entropy-based network representation of the individual metabolic phenotype. Journal of Proteome Research. 2016;15:3298–3307. doi: 10.1021/acs.jproteome.6b00454. [DOI] [PubMed] [Google Scholar]
- Saccenti E, Suarez-Diez M, Luchinat C, Santucci C, Tenori L. Probabilistic networks of blood metabolites in healthy subjects as indicators of latent cardiovascular risk. Journal of Proteome Research. 2014;14:1101–1111. doi: 10.1021/pr501075r. [DOI] [PubMed] [Google Scholar]
- Sauer U. Metabolic networks in motion: 13C-based flux analysis. Molecular Systems Biology. 2006;2:62. doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savageau MA. Critique of the enzymologist’s test tube. In: Bittar EE, editor. Fundamentals of medical cell biology. Greenwich, CT: JAI Press; 1992. [Google Scholar]
- Scheer M, et al. BRENDA, the enzyme information system in 2011. Nucleic Acids Research. 2011;39:D670–D676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberger J, Park JO, Conrad TM, Palsson B. BiGG: A biochemical genetic and genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinformatics. 2010;11:213. doi: 10.1186/1471-2105-11-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell S, Turner TE. Reaction kinetics in intracellular environments with macromolecular crowding: Simulations and rate laws. Progress in Biophysics and Molecular Biology, 2004;85:235–260. doi: 10.1016/j.pbiomolbio.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Schomburg I, et al. BRENDA in 2013: Integrated reactions, kinetic data, enzyme function data, improved disease classification: New options and contents in BRENDA. Nucleic Acids Research. 2013;41:D764–D772. doi: 10.1093/nar/gks1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selivanov VA, et al. Rapid simulation and analysis of isotopomer distributions using constraints based on enzyme mechanisms: An example from HT29 cancer cells. Bioinformatics. 2005;21:3558–3564. doi: 10.1093/bioinformatics/bti573. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilde AK, Timmerman ME, Saccenti E, Jansen JJ, Hoefsloot HCJ. Covariances simultaneous component analysis: A new method within a framework for modeling covariances. Journal of Chemometrics. 2015;29:277–288. doi: 10.1002/cem.2707. [DOI] [Google Scholar]
- Smyth G. Limma: Linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and computational biology solutions using R and bioconductor. New York: Springer; 2005. [Google Scholar]
- Sokol S, Millard P, Portais J-C. Influx_s: Increasing numerical stability and precision for metabolic flux analysis in isotope labelling experiments. Bioinformatics. 2012;28:687–693. doi: 10.1093/bioinformatics/btr716. [DOI] [PubMed] [Google Scholar]
- Song L, Langfelder P, Horvath S. Comparison of co-expression measures: Mutual information, correlation, and model based indices. BMC Bioinformatics. 2012;13:1–21. doi: 10.1186/1471-2105-13-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer R, Salek RM, Moreno P, Cañueto D, Steinbeck C. Navigating freely-available software tools for metabolomics analysis. Metabolomics. 2017;13:106. doi: 10.1007/s11306-017-1242-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford NJ, Lubitz T, Smallbone K, Klipp E, Mendes P, Liebermeister W. Systematic construction of kinetic models from genome-scale metabolic networks. PLoS ONE. 2013;8:e79195. doi: 10.1371/journal.pone.0079195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelling J, Sauer U, Szallasi Z, Doyle FJ, Doyle J. Robustness of cellular functions. Cell. 2004;118:675–685. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Steuer R, Kurths J, Fiehn O, Weckwerth W. Observing and interpreting correlations in metabolomic networks. Bioinformatics. 2003;19:1019–1026. doi: 10.1093/bioinformatics/btg120. [DOI] [PubMed] [Google Scholar]
- Suarez-Diez M, et al. Plasma and serum metabolite association networks: Comparability within and between studies using NMR and MS profiling. Journal of Proteome Research. 2017 doi: 10.1021/acs.jproteome.7b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Diez M, Saccenti E. Effects of sample size and dimensionality on the performance of four algorithms for inference of association networks in metabonomics. Journal of Proteome Research. 2015 doi: 10.1021/acs.jproteome.5b00344. [DOI] [PubMed] [Google Scholar]
- Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Schmitt-Kopplin P. MassTRIX: Mass translator into pathways. Nucleic Acids Research. 2008;36:W481–W484. doi: 10.1093/nar/gkn194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainston N, et al. Recon 2.2: From reconstruction to model of human metabolism. Metabolomics. 2016;12:109. doi: 10.1007/s11306-016-1051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski J, et al. Stability of metabolic correlations under changing environmental conditions in Escherichia coli—A systems approach. PLoS ONE. 2009;4:e7441. doi: 10.1371/journal.pone.0007441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, et al. Mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal. 2004;37:914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Tokimatsu T, et al. KaPPA-View. A web-based analysis tool for integration of transcript and metabolite data on plant metabolic pathway maps. Plant Physiology. 2005;138:1289–1300. doi: 10.1104/pp.105.060525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trethewey RN, Krotzky AJ, Willmitzert L. Metabolic profiling: A Rosetta Stone for genomics? Current Opinion in Plant Biology. 1999;2:83–85. doi: 10.1016/S1369-5266(99)80017-X. [DOI] [PubMed] [Google Scholar]
- Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. Journal of Proteome Research. 2007;6:469–479. doi: 10.1021/pr060594q. [DOI] [PubMed] [Google Scholar]
- Urbanczyk-Wochniak E, et al. Parallel analysis of transcript and metabolic profiles: A new approach in systems biology. EMBO Reports. 2003;4:989–993. doi: 10.1038/sj.embor.embor944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursem R, Tikunov Y, Bovy A, van Berloo R, van Eeuwijk F. A correlation network approach to metabolic data analysis for tomato fruits. Euphytica. 2008;161:181. doi: 10.1007/s10681-008-9672-y. [DOI] [Google Scholar]
- Van Den Berg RA, Hoefsloot HCJ, Westerhuis JA, Smilde AK, Van Der Werf MJ. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Greef J, Smilde AK. Symbiosis of chemometrics and metabolomics: Past, present, and future. Journal of Chemometrics. 2005;19:376–386. doi: 10.1002/cem.941. [DOI] [Google Scholar]
- Vignoli A, Tenori L, Luchinat C, Saccenti E. Age and sex effects on plasma metabolite association networks in healthy subjects. Journal of Proteome Research. 2017;17:97–107. doi: 10.1021/acs.jproteome.7b00404. [DOI] [PubMed] [Google Scholar]
- Weckwerth W, Fiehn O. Can we discover novel pathways using metabolomic analysis? Current Opinion in Biotechnology. 2002;13:156–160. doi: 10.1016/S0958-1669(02)00299-9. [DOI] [PubMed] [Google Scholar]
- Weckwerth W, Loureiro ME, Wenzel K, Fiehn O. Differential metabolic networks unravel the effects of silent plant phenotypes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7809–7814. doi: 10.1073/pnas.0303415101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel M, Nöh K, Dalman T, Niedenführ S, Stute B, Wiechert W. 13CFLUX2—High-performance software suite for 13C-metabolic flux analysis. Bioinformatics. 2013;29:143–145. doi: 10.1093/bioinformatics/bts646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiechert W, Nöh K. Isotopically non-stationary metabolic flux analysis: Complex yet highly informative. Current Opinion in Biotechnology. 2013;24:979–986. doi: 10.1016/j.copbio.2013.03.024. [DOI] [PubMed] [Google Scholar]
- Windig W, Kistemaker PG, Haverkamp J, Meuzelaar HL. Factor analysis of the influence of changes in experimental conditions in pyrolysis—mass spectrometry. Journal of Analytical and Applied Pyrolysis. 1980;2:7–18. doi: 10.1016/0165-2370(80)80041-6. [DOI] [Google Scholar]
- Wishart DS. Current progress in computational metabolomics. Briefings in Bioinformatics. 2007;8:279–293. doi: 10.1093/bib/bbm030. [DOI] [PubMed] [Google Scholar]
- Wishart DS, et al. HMDB 3.0—The human metabolome database in 2013. Nucleic Acids Research. 2013;41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig U, et al. SABIO-RK–database for biochemical reaction kinetics. Nucleic Acids Research. 2012;40:D790–D796. doi: 10.1093/nar/gkr1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S. Chemometrics; what do we mean with it, and what do we want from it? Chemometrics and Intelligent Laboratory Systems. 1995;30:109–115. doi: 10.1016/0169-7439(95)00042-9. [DOI] [Google Scholar]
- Wold S, Sjöström M. Chemometrics, present and future success. Chemometrics and Intelligent Laboratory Systems. 1998;44:3–14. doi: 10.1016/S0169-7439(98)00075-6. [DOI] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Research. 2015 doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, et al. Potential metabolic mechanism of girls’ central precocious puberty: A network analysis on urine metabonomics data. BMC Systems Biology. 2012;6:S19. doi: 10.1186/1752-0509-6-S3-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR. Change. Journal of Proteome Research. 2016;15:2355–2355. doi: 10.1021/acs.jproteome.6b00640. [DOI] [PubMed] [Google Scholar]
- Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. Journal of Biological Chemistry. 2008;283:20621–20627. doi: 10.1074/jbc.M706494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD. INCA: A computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics. 2014;30:1333–1335. doi: 10.1093/bioinformatics/btu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni N. 13 C metabolic flux analysis in complex systems. Current Opinion in Biotechnology. 2011;22:103–108. doi: 10.1016/j.copbio.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Zamboni N, Fischer E, Sauer U. FiatFlux—A software for metabolic flux analysis from 13 C-glucose experiments. BMC Bioinformatics. 2005;6:209. doi: 10.1186/1471-2105-6-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene Co-expression network analysis. Statistical Applications in Genetics and Molecular Biology. 2005 doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- Zhao W, Langfelder P, Fuller T, Dong J, Li A, Hovarth S. Weighted gene coexpression network analysis: State of the art. Journal of Biopharmaceutical Statistics. 2010;20:281–300. doi: 10.1080/10543400903572753. [DOI] [PubMed] [Google Scholar]
- Zhong S, Storch K-F, Lipan O, Kao M-C J, Weitz CJ, Wong WH. GoSurfer: A graphical interactive tool for comparative analysis of large gene sets in Gene Ontology space. Applied Bioinformatics. 2004;3:261–264. doi: 10.2165/00822942-200403040-00009. [DOI] [PubMed] [Google Scholar]