Abstract
AIM
To study the role of microbial metabolites in the modulation of biochemical and physiological processes in irritable bowel syndrome (IBS).
METHODS
In the current study, using a metabolomic approach, we analyzed the key metabolites differentially excreted in the feces of control mice and mice with IBS, with or without Clostridium butyricum (C. butyricum) treatment. C57BL/6 mice were divided into control, IBS, and IBS + C. butyricum groups. In the IBS and IBS + C. butyricum groups, the mice were subjected to water avoidance stress (WAS) for 1 h/d for ten days. Gas chromatography/mass spectrometry (GC-MS) together with multivariate analysis was employed to compare the fecal samples between groups.
RESULTS
WAS exposure established an appropriate model of IBS in mice, with symptoms of visceral hyperalgesia and diarrhea. The differences in the metabolite profiles between the control group and IBS group significantly changed with the progression of IBS (days 0, 5, 10, and 17). A total of 14 differentially excreted metabolites were identified between the control and IBS groups, and phenylethylamine was a major metabolite induced by stress. In addition, phenylalanine metabolism was found to be the most relevant metabolic pathway. Between the IBS group and IBS + C. butyricum group, 10 differentially excreted metabolites were identified. Among these, pantothenate and coenzyme A (CoA) biosynthesis metabolites, as well as steroid hormone biosynthesis metabolites were identified as significantly relevant metabolic pathways.
CONCLUSION
The metabolic profile of IBS mice is significantly altered compared to control mice. Supplementation with C. butyricum to IBS mice may provide a considerable benefit by modulating host metabolism.
Keywords: Irritable bowel syndrome, Metabolite, Gas chromatography/mass spectrometry, Clostridium butyricum
Core tip: In this study, we analyzed the key metabolites differentially excreted in the feces of control mice and mice with irritable bowel syndrome (IBS). A total of 14 differentially excreted metabolites were identified, and phenylalanine (a major metabolite induced by stress) was found to be the most relevant of these metabolites. Between the IBS group and IBS + C. butyricum group, 10 differentially excreted metabolites were identified, and pantothenate and coenzyme A (CoA) biosynthesis metabolites, as well as steroid hormone biosynthesis metabolites were found to be significantly relevant. Thus, supplementation with C. butyricum to IBS mice had beneficial effects through modulation of host metabolism.
INTRODUCTION
Irritable bowel syndrome (IBS) is a functional disorder of the gastrointestinal (GI) tract. Although the pathology is complicated, it is believed that multiple factors such as genetics, visceral hypersensitivity, gastrointestinal motility, dysregulation of the brain-gut axis, levels of neuropeptides and hormones, as well as inflammatory changes all contribute to IBS development[1]. Recent emerging evidence has demonstrated certain disorders that alter fecal microbiota profiles may cause IBS[2-7]. The adult human intestinal tract contains a high density of microbes (typically 1011-1012 microbes/mL of luminal content)[8], which are predominantly obligate anaerobes[3]. The gut microbiota not only enhances host digestion, nutrient absorption, and energy turnover, but also substantially regulates metabolism, protects against pathogens, and modulates the host immune response[9]. Probiotics are defined as “living nonpathogenic microorganisms that benefit the health of the host by modifying intestinal flora”[10]. A recent study found that both probiotics and symbiotics can benefit host health by improving the body’s nitrogen metabolism[11].
Microbial metabolites positively influence biochemical and physiological processes. Thus, studying the impact of the microbiota on host–microbial interactions is more important than identifying the microbial species present. The metabolome, a complete collection of all metabolites in a biological specimen, is the end-product of the complex interactions between the genome, transcriptome, proteome, and the environment. Metabolomics is defined as “the nonbiased identification and quantification of all metabolites in a biological system”[12]. The metabolome amplifies the metabolic changes caused by a certain biological perturbation[13]. Fecal samples are ideal biospecimen for metabolomics analysis, due to the non-invasive nature of sample collection. Fecal metabolic compositions and variations not only reflect the status of the intestinal microbiota, but also bridge the relationship between symbiotic microbes and host health[14-16]. Thus, the metabonomics signature of IBS fecal samples will be useful to study the IBS pathological process.
The gas chromatography/mass spectrometry (GC-MS) platform presents a unique tool with high sensitivity, high reproducibility, and available spectral libraries. In this study, we investigated the changes of the fecal metabolome in IBS pathology using a murine water-avoidance stress (WAS) model of IBS. The purpose of this study was to compare and verify important metabolites and key pathway alterations between IBS mice and control mice, as well as between IBS mice with and without C. butyricum treatment.
MATERIALS AND METHODS
Probiotic strains
C. butyricum (S20020015) was kindly provided by Shandong Kexing Bioproducts Co. Ltd. (China). Freeze-dried C. butyricum powder contained both viable bacteria [5.6 × 109 colony-forming units (CFU)/g] and spores (4.4 × 109 CFU/g). Before use, the probiotic powder was reconstituted in sterile saline at 37 °C for 15 min. The final concentration of C. butyricum was 1.25 × 109 CFU/mL.
Animals and experimental design
Female C57BL/6 mice (n = 18, aged 5-6 wk) were purchased from the Experimental Animal Center of Zhejiang Province (Zhejiang, China), and housed in the animal maintenance facility at the Zhejiang Chinese Medical University. Mice were randomly divided into three groups (control, IBS, and IBS + C. butyricum), with six mice in each group. Mice in the IBS and IBS + C. butyricum groups were exposed to water-avoidance stress (WAS) for 1 h every day for ten days to induce IBS. All procedures were performed between 8:00 and 10:00 AM to minimize the bias of the circadian rhythm. Briefly, mice were placed on a small platform (3 cm × 6 cm) in a plastic container (56 cm × 50 cm) with warm water (25 °C) 1 cm below the platform. After the ten-day WAS period, mice in the IBS and IBS + C. butyricum groups were given sterile saline (0.4 mL) and C. butyricum (0.4 mL, 5 × 108 CFU), respectively, by gastric gavage daily for a seven-day treatment period. All procedures were approved by the Animal Care Committee of Zhejiang Chinese Medical University, and all methods were performed in accordance with the relevant guidelines and regulations.
Assessment of stool features and visceral sensitivity
Fecal features were evaluated through visual inspection performed by two investigators blinded to the randomization. Presence of semisolid, pasty, or watery stools was considered abnormal[17]. Fresh fecal pellets were collected from all mice in individual metabolic cages within 2 h on days 0, 5, 10, and 17. All fecal samples were then stored at -80 °C until further analysis. The visceral sensitivity of mice was evaluated by measuring the abdominal withdrawal reflex (AWR) in response to colorectal distension (CRD)[18]. Semi-quantitative AWR scores (0-4) were used to grade pain responses at various magnitudes of CRD (20, 40, 60, and 80 mmHg)[17,19,20]. Two investigators blinded to the randomization recorded the AWR scores at each pressure five times to improve accuracy.
Metabolite extraction
Fecal samples were collected in 2 mL microfuge tubes, and were extracted with extraction liquid (0.3 mL; methanol: chloroform at 3:1). L-2-chlorophenylalanine (20 μL; 1 mg/mL stock in dH2O) was added as an internal standard. The mixture was vortexed for 30 s and homogenized in a ball mill for 4 min at 45 Hz, and then exposed to ultrasound for 5 min on ice. The supernatant (0.2 mL) was transferred into a fresh 2 mL GC-MS glass vial, and 4 μL from each sample was used to generate a quality control (QC) pooled sample.
Derivatives of metabolites
Fecal samples were dried in a vacuum concentrator without heating. Methoxyamination hydrochloride (70 μL; 20 mg/mL in pyridine) was added and the mixture was incubated at 80 °C for 30 min, followed by addition of the bis(trimethylsilyl)trifluoroacetamide regent (80 μL; 1% trimethylchlorosilane, v/v) to the sample aliquots, which were further incubated for 2 h at 70 °C. A standard mixture of fatty acid methyl esters (10 μL; C8-C16: 1 mg/mL; C18-C24: 0.5 mg/mL in chloroform) was added to the QC sample. Samples were then cooled to room temperature and mixed well prior to GC-MS analysis.
GC-MS analysis
GC-MS analysis was performed using an Agilent 7890 gas chromatograph system coupled with a Pegasus HT time-of-flight mass spectrometer. The system utilized a DB-5MS capillary column coated with 5% diphenyl cross-linked with 95% dimethylpolysiloxane. An aliquot of the analyte (1 μL) was injected in a splitless mode according to the instructions from the kit. The carrier gas was helium, the front inlet purge flow was 3 mL/min, and the gas flow rate through the column was 1 mL/min. The initial temperature for 1 min was 50 °C. The temperature was then raised to 305 °C at a rate of 12 °C/min, and maintained for 7.75 min at 305 °C. The energy was -70 eV in electron impact mode. The mass spectrometry data were acquired in full-scan mode with the mass-to-charge (m/z) range of 50-500 at a rate of 20 spectra per sec after a solvent delay of 6.1 min.
Metabolic profiling analysis
Chroma TOF 4.3× software from LECO Corporation and the LECO-Fiehn Rtx5 database were used for metabolic profiling analysis as described previously[21]. The RI (retention time index) method was used for peak identification, and the RI tolerance was 5000. The metabolic features detected in < 50% of QC samples were removed[22]. Raw GC-MS data were exported to mzData format using MassHunter Workstation Software (Version B.06.00, Agilent Technologies) and subsequently sent to the XCMS package under the R Project. In addition, an internal standard normalization method was used for the analysis of these data. The resulting three-dimensional data involving the peak number, sample name, and normalized peak area were submitted to SIMCA14.1 software package (V14.1, MKS Data Analytics Solutions, Umea, Sweden) for principal component analysis (PCA) and orthogonal projections to latent structures-discriminate analysis (OPLS-DA). PCA provided the distribution of origin data. Subsequently, as an effective approach to sift metabolites, the score plot of OPLS-DA was achieved to detect differences and filter variations between the groups. The quality of the models was assessed by the R2 and Q2 values in OPLS-DA, which represented the variance and the predictability derived from the models. After assessing the data in OPLS-DA, 200 permutations were performed, and the resulting R2 and Q2 values were plotted to further assess the model validity. To refine this analysis, the first principal component of variable importance in the projection (VIP) was obtained. A VIP that exceeded 1 with a P-value less than 0.05 indicated a changed biomarker. In addition, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/) was utilized to link these metabolites to metabolic pathways. MetaboAnalyst, which uses the high-quality KEGG metabolic pathway as the backend knowledge base, as well as integrated enrichment analysis and pathway topology analysis were used to identify the most relevant pathways (http://www.metaboanalyst.ca).
Statistical analysis
Differences between groups were analyzed by Student’s t-test for variables following a normal distribution or Wilcoxon two-sample test for variables without following a normal distribution. A χ2 test was used in the analysis of contingency tables. Data are presented as mean ± SD. A P-value < 0.05 was considered statistically significant. Statistical analyses were conducted using SPSS 22.0 (Chicago, IL, United States) and GraphPad Prism 6.0 software (San Diego, CA, United States).
RESULTS
Impact of C. butyricum treatment on IBS mice
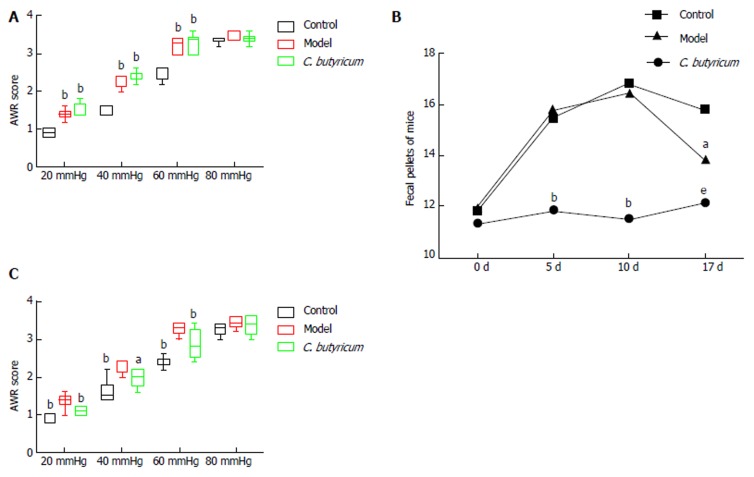
At day 10 following WAS exposure, all 12 IBS mice (IBS and IBS + C. butyricum groups) demonstrated increased mean AWR scores at CRD pressures of 20, 40, 60, and 80 mmHg compared to the six control mice (Figure 1A). A higher proportion of IBS mice showed abnormal stool features and a consistently increased number of fecal pellets compared with control mice (Table 1 and Figure 1B). These results suggested that WAS exposure established an appropriate model of IBS with symptoms of visceral hyperalgesia and diarrhea. Interestingly, when C. butyricum was given for 7 d to WAS-exposed mice, the AWR scores were significantly reduced (Figure 1C), and the stool features and number of pellets were improved (Table 1 and Figure 1B), suggesting that C. butyricum treatment could relieve IBS symptoms in WAS-exposed mice.
Figure 1.
Impact of Clostridium butyricum treatment on irritable bowel syndrome mice. A: Comparison of AWR scores between control and IBS groups, as well as between IBS and IBS + C. butyricum groups after WAS exposure (n = 6 per group). aP < 0.05; bP < 0.01. B: Fecal pellets at various time points during the experiment (n = 6 per group). aP < 0.05: IBS vs IBS + C. butyricum; bP < 0.01: control vs IBS, control vs IBS + C. buyricum; eP < 0.001: control vs IBS. C: Comparison of AWR scores between control and IBS groups, as well as between IBS and IBS + C. butyricum groups following the C. butyricum treatment period (n = 6 per group) aP < 0.05; bP < 0.01. AWR: Abdominal withdrawal reflex; C. butyricum: Clostridium butyricum; IBS: Irritable bowel syndrome; WAS: Water avoidance stress.
Table 1.
Assessment of fecal features
| Fecal feature | Control | IBS | IBS + C. butyricum |
| After WAS exposure | 6/0 | 0/6b | 0/6b |
| Normal/Abnormal (n) | |||
| After treatment | 6/0 | 0/6a | 5/1c |
| Normal/Abnormal (n) |
The presence of semisolid, pasty, or watery stools was considered abnormal.
P < 0.01: control vs IBS or IBS + C. butyricum;
P < 0.05, IBS vs IBS + C. butyricum;
P < 0.05, control vs IBS. C. butyricum: Clostridium butyricum; IBS: Irritable bowel syndrome; WAS: Water avoidance stress.
Metabolomic profiles
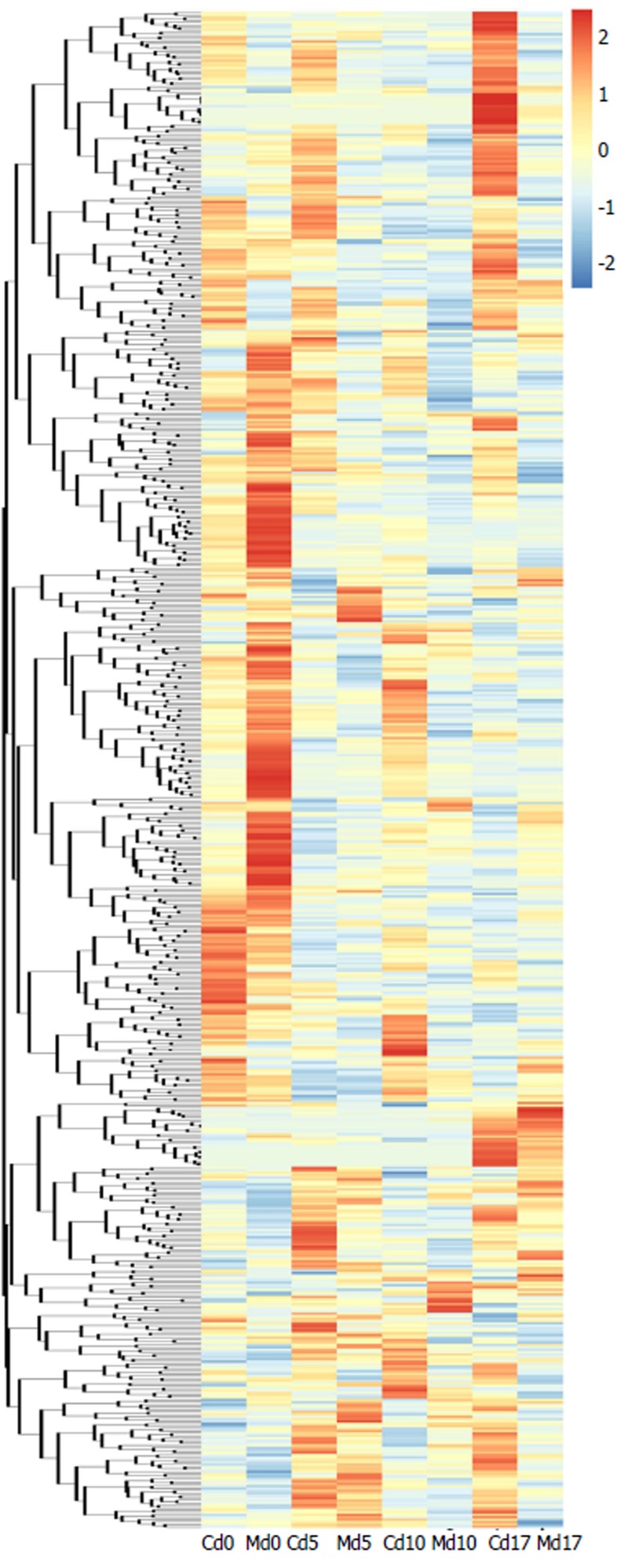
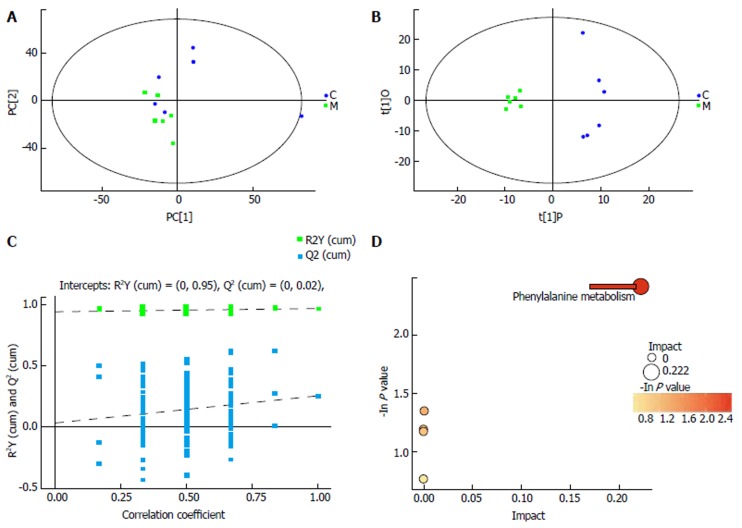
Fecal metabolite profiles in the control and IBS groups were carefully compared. A heat map analysis revealed that metabolomics could be used to distinguish between the control and IBS groups (Figure 2). With the progression of time (days 0, 5, 10, and 17), the differences in the metabolite profiles between these two groups were significant. When comparing the fecal metabolite profiles of the control group with those of the IBS model group on day 17, PCA revealed that metabolites were distinguishable between the groups (Figure 3A). OPLS-DA was used to examine the metabolites that contributed to the group separation (Figure 3B). Permutation testing performed on the OPLS-DA model revealed remarkable positive slopes (Figure 3C), indicating statistical significance in the separation of these two groups. A total of 14 differentially excreted metabolites were distinguishable in the IBS model group compared to the control group, including myristic acid, arachidic acid, octadecanol, N-acetyl-D-galactosamine 1,1-hexadecanol, phenylethylamine, 2-furoic acid, 4,2’,4’-trihydroxychalcone, 24,25-dihydrolanosterol, androsterone 1, chlorogenic acid 1, cysteinylglycine 2, alpha-tocopherol, and 4-hydroxybenzoic acid. Table 2 summarizes the metabolite differences between the IBS model and control samples.
Figure 2.
Heatmap of hierarchical clustering analysis for irritable bowel syndrome group vs control group at various time points. The colors at different positions indicate the relative expression of the corresponding metabolites. C: Control group; M: Model (IBS) group. IBS: Irritable bowel syndrome.
Figure 3.
Separating the irritable bowel syndrome group from the control group by metabolic profiling analysis. A: Score scatter plot of PCA model for IBS group vs control group; B: Score scatter plot of OPLS-DA model for model group vs control group; C: Permutation test of OPLS-DA model for model group vs control group; D: Pathway analysis for IBS group vs control group. C: Control group; M: Model (IBS) group; PCA: Principal component analysis; OPLS-DA: Orthogonal projections to latent structures-discriminant analysis; IBS: Irritable bowel syndrome; C. butyricum: Clostridium butyricum.
Table 2.
Metabolites distinguishing irritable bowel syndrome mice from control mice
| Compound | Formula | Similarity | RT (min) | VIP value | P value | Fold change | Trend compared with controls |
| N-acetyl-D-galactosamine 1 | C8H15NO6 | 886 | 17.1523,0 | 1.167810623 | 0.002673237 | 2.665558179 | Down |
| Myristic acid | C14H28O2 | 878 | 15.2969,0 | 1.454697623 | 0.033949959 | 2.405282471 | Down |
| Arachidic acid | C20H40O2 | 837 | 19.8283,0 | 2.030221054 | 0.033823382 | 0.555519799 | Up |
| 1-Hexadecanol | C16H34O | 761 | 16.2434,0 | 2.137279002 | 0.03239841 | 3.735617796 | Down |
| Octadecanol | C18H38O | 632 | 17.7928,0 | 1.226172539 | 0.039254153 | 3.503198671 | Down |
| Phenylethylamine | C8H11N | 764 | 12.6834,0 | 1.844742352 | 0.040836128 | 0.234956253 | Up |
| 2-Furoic acid | C5H4O3 | 710 | 7.80292,0 | 1.057850377 | 0.044324108 | 1.829643584 | Down |
| 4,2',4'-Trihydroxychalcone | C15H12O4 | 245 | 22.6862,0 | 1.871154718 | 0.049356394 | 0.502503451 | Up |
| 24,25-Dihydrolanosterol | C30H52O | 593 | 25.186,0 | 2.237217404 | 0.041969612 | 0.449343275 | Up |
| Androsterone 1 | C19H30O2 | 476 | 20.6021,0 | 1.51415211 | 0.031210567 | 2.125877191 | Down |
| Chlorogenic acid 1 | C16H18O9 | 402 | 23.8786,0 | 1.950369158 | 0.016252537 | 3.264118801 | Down |
| Cysteinylglycine 2 | C10H18N4O6S2 | 393 | 16.5878,0 | 2.464379346 | 0.003149634 | 11.06227902 | Down |
| Alpha-tocopherol | C29H50O2 | 305 | 24.0995,0 | 1.25237824 | 0.040967864 | 0.406678915 | Up |
| 4-Hydroxybenzoic acid | C7H6O3 | 811 | 13.2781,0 | 1.348316951 | 0.0129929 | 2.36224644 | Down |
IBS: Irritable bowel syndrome; RT: Retention time; VIP: Variable importance in the projection.
To further explore the biological significance associated with IBS morbidity, we used MetaboAnalyst to detect the key relevant pathways influenced by IBS. This pathway analysis revealed that phenylalanine metabolism was the most relevant pathway influenced by IBS with an impact value of 0.222 (Figure 3D).
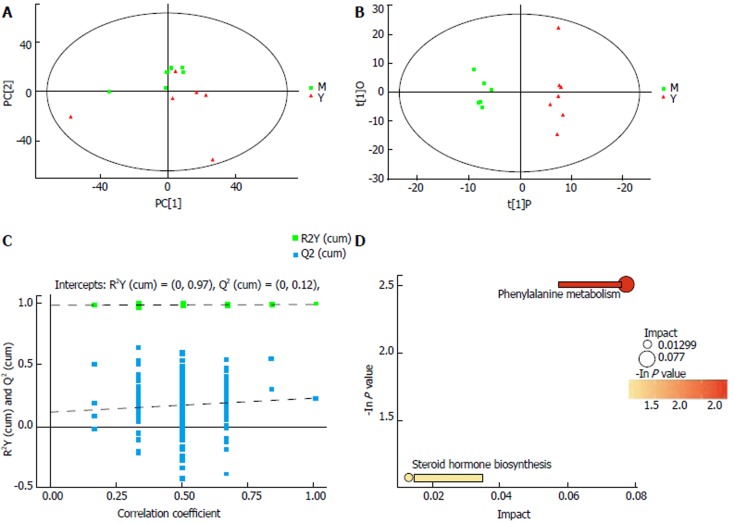
To test whether the metabolomic profile could be used to distinguish the IBS + C. butyricum group from the IBS group, we performed a comparison of metabolomic profiles in these two groups. PCA revealed that the metabolomics were significantly different between these two groups (Figure 4A). OPLS-DA was used to examine the metabolites that contributed to the separation (Figure 4B). Permutation testing performed on the OPLS-DA model revealed significantly positive slopes (Figure 4C), indicating statistical significance for these two groups. The ten differentially excreted metabolites between the IBS and IBS + C. butyricum groups were pantothenic acid, cysteinylglycine 2, d-arabitol, arbutin, 3-hydroxyphenylacetic acid, 4-hydroxybenzaldehyde 2, 1-hexadecanol, octadecanol, 5,6-dihydrouracil 1, and androsterone 1 (Table 3).
Figure 4.
Separating the group treated with Clostridium butyricum from the irritable bowel syndrome only group by metabolic profiling analysis. A: Score scatter plot of PCA model for IBS group vs IBS + C. butyricum group; B: Score scatter plot of OPLS-DA model for IBS group vs IBS + C. butyricum group; C: Permutation test of OPLS-DA model for IBS group vs IBS + C. butyricum group; D: Pathway analysis for IBS group vs IBS + C. butyricum group. M: Model (IBS) group; Y: IBS + C. butyricum group; PCA: Principal component analysis; OPLS-DA: Orthogonal projections to latent structures-discriminant analysis; IBS: Irritable bowel syndrome; C. butyricum: Clostridium butyricum.
Table 3.
Differentially excreted metabolites in response to Clostridium butyricum
| Compound | Formula | Similarity | RT (min) | VIP value | P-value | Fold change | Trend compared with IBS mice |
| Pantothenic acid | C9H17NO5 | 844 | 16.4657,0 | 1.387200939 | 0.016130158 | 0.408462268 | Up |
| Cysteinylglycine 2 | C10H18N4O6S2 | 393 | 16.5878,0 | 1.498082744 | 0.047913423 | 0.166509123 | Up |
| D-Arabitol | C5H12O5 | 807 | 14.118,0 | 2.6949064 | 0.04068909 | 2511115.131 | Down |
| Arbutin | C12H16O7 | 569 | 20.5587,0 | 1.693255625 | 0.019486947 | 0.467550145 | Up |
| 3-Hydroxyphenylacetic acid | C8H8O3 | 545 | 13.0677,0 | 1.462693364 | 0.014333152 | 0.467960253 | Up |
| 4-Hydroxybenzaldehyde 2 | C7H6O2 | 798 | 11.9397,0 | 1.38788733 | 0.01853018 | 0.376921771 | Up |
| 1-Hexadecanol | C16H34O | 761 | 16.2434,0 | 2.206799293 | 0.049776549 | 0.311045618 | Up |
| Octadecanol | C18H38O | 632 | 17.7928,0 | 1.352431526 | 0.033517221 | 0.308621946 | Up |
| 5,6-Dihydrouracil 1 | C4H6N2O2 | 512 | 11.8596,0 | 1.260961668 | 0.022218939 | 0.600255286 | Up |
| Androsterone 1 | C19H30O2 | 476 | 20.6021,0 | 1.62619412 | 0.04838445 | 0.494710622 | Up |
IBS: Irritable bowel syndrome; RT: Retention time; VIP: Variable importance in the projection.
To explain the biological significance associated with C. butyricum administration, we applied MetaboAnalyst to test the important relevant pathways influenced by IBS and IBS + C. butyricum treatment. Pantothenate, coenzyme A (CoA) biosynthesis, and steroid hormone biosynthesis were found to be involved in the most relevant pathways influenced by C. butyricum treatment with impact values of 0.077 and 0.013, respectively (Figure 4D).
DISCUSSION
In this study, we studied the fecal metabolic profile in a mouse model of IBS, and investigated the impact of C. butyricum on metabolic changes using the GC-MS technique. A total of 14 differentially excreted metabolites were identified in the control and IBS groups, and phenylalanine metabolism was the significant metabolic pathway identified in the IBS group. We found that even 7 d following WAS termination, the metabolites in the IBS group were still distinct from those in the control group. The AWR scores, and the number and features of the fecal pellets were also still abnormal in the IBS group compared to the control group, indicating that the pathological processes resulting in IBS-like symptoms did not immediately stop upon stress termination. Of note, phenylethylamine, which is involved in the key metabolism pathway of phenylalanine, was significantly increased in the IBS model group. Interestingly, treatment of IBS mice with C. butyricum decreased the fecal pellet number and AWR scores, and restored the fecal features. A total of ten differentially expressed metabolites were identified in the IBS + C. butyricum and IBS model groups. Among these, pantothenic acid and androsterone 1 were increased in the IBS + C. butyricum group. Both pantothenic acid and androsterone 1 are implicated in the key metabolic pathways of pantothenate and CoA biosynthesis, as well as steroid hormone biosynthesis. Therefore, C. butyricum could change metabolic pathways and reduce visceral sensitivity and diarrhea symptoms in IBS mice.
The digestive tract of the host is colonized with a highly complex array of microorganisms composed mainly of bacteria, with numbers increasing progressively from the proximal to the distal colon[23]. The microbiome includes more than five million genes. Many genes encode biosynthetic enzymes, proteases, and glycosidases to enlarge the host’s biochemical and metabolic capabilities[24]. Such interactions were termed the “microbial-mammalian metabolic axis”, which was defined as “the multi-way exchange and co-metabolism of compounds between the host organism and the gut microbiome resulting in transgenically-regulated secondary metabolites, which have biological activity on both host and microbial compartments”[25,26]. Intestinal bacteria primarily influence the metabolism of amino acids, fatty acids, steroid hormone biosynthesis, and oxidative stress pathways. Any changes in the gut microbiota, such as the diversity, the number, or the stability, could affect metabolic end-products and lead to disease development. These metabolites can harm the host through[27]: (1) toxic and/or carcinogenic metabolites produced directly; (2) disturbances of the homeostatic energy balance, resulting in changes to host physiology; (3) increased proliferation of other potentially harmful bacteria via syntrophy; and (4) direct interaction with intestinal epithelial cells. Gall et al[28] have used proton nuclear magnetic resonance metabolite profiling of fecal extracts to separate IBS patients from healthy controls, and indicated that the gut microbial metabolites were important factors implicated in the pathogenesis of IBS. Dietary probiotics have been shown to improve the health of the host[29]. A recent study found that probiotics altered a diversity of pathways including amino acid metabolism, methylamines, and short-chain fatty acids (SCFA)[30]. Martin et al[31] also found that the application of Lactobacillus paracasei into the intestine of germ free mice changed the concentration of amino acids, anti-oxidants, and creatine in the jejunum and ileum of these mice.
Amino acids are major nutrients in the diet and the basic units of protein. The host uses amino acids as metabolic fuel, and gut bacteria are linked to host energy metabolism. Proportions of arginine, proline, and phenylalanine were shown to be high in Streptococcus spp[32]. As an essential aromatic amino acid, phenylalanine is utilized for protein synthesis by cells throughout the body. Peripheral phenylalanine is elevated in phenylketonuria disease, which can lead to mental retardation, seizures, and neurological and psychiatric dysfunctions[33]. Phenylalanine has also been linked to inflammation. Klassen et al[34] have found that the phenylalanine level was increased in patients during the acute phase of dengue fever, suggesting that infection may alter amino acid metabolism. In our study, we found increased phenylethylamine in fecal extracts of IBS mice compared with healthy control mice, indicating that the increased stress in mice led to higher phenylethylamine levels. Phenylethylamine is an endogenous neuroamine that promotes energy and elevates mood[35], and it is a potential mediator of stress[36] . We speculated that the fecal metabolic changes could be associated with increased energy metabolism induced by stress in IBS mice to accommodate the extra energy demand due to muscle hyper-contractility and inflammation. These findings were consistent with a study by Martin et al[37], in which they also indicated that post-infectious IBS was related to changes in energy metabolic intermediates, as well as lipid and amino acid metabolism, to meet the energy demand of intestinal muscular hyper-contractility[37]. In another study, Ponnusamy et al[38] found that amino acids and phenolic compounds were increased in IBS patients. This metabolic signature is similar to that observed in humans experiencing stress and trauma. Therefore, it is likely that there exists an energy metabolism disorder induced by stress in the IBS pathological processes. In our study, we also found that alpha-tocopherol was increased in the feces of IBS mice, likely resulting from its poor absorption due to diarrhea. Alpha-tocopherol was shown to be an important anti-oxidant metabolite with anti-inflammatory activity[39]. Therefore, its poor absorption would further deteriorate the state of IBS. Vitamins are micronutrients that serve as precursors to many important enzymes necessary for vital biochemical reactions in all living cells, and vitamin metabolism has been shown to be important in all types of gut microbiomes[40]. Pantothenic acid is widely distributed in foods, and interestingly, Escherichia coli secretes 15-fold more pantothenic acid than that required for maintaining intracellular CoA biosynthesis[41]. The precursor of CoA is pantothenic acid, and CoA is necessary for 4% of known enzymatic reactions[42]. CoA is involved in cellular functions and mitochondrial energy production associated with fatty acids, cholesterol, heme synthesis, and acetylcholine, and also takes part in mitochondrial aerobic respiration (tricarboxylic acid (TCA) cycle)[42,43]. Increased levels of CoA contribute to cellular repair by diminishing lipid peroxidation and potentiating the synthesis of membrane phospholipids. Even et al[44] found that pantothenic acid had a protective effect by improving the muscular response in a mouse model of muscular dystrophy. Tumors in animals have been found to have low CoA levels[45], which were related to increased glycolytic metabolism but less associated with oxidative phosphorylation and the TCA cycle. In our study, we found that the C. butyricum treatment increased the level of pantothenic acid, suggesting that C. butyricum likely modulates energy metabolism in IBS mice by up-regulating the pantothenate and CoA biosynthesis pathways. Martin et al[46] found that probiotics could improve fecal excretion of glutamine, branched-chain amino acids, glutamate, glycine, and alanine, and therefore suggested that IBS could be treated with probiotics. An additional study has also shown that probiotics could regulate microbial proteolytic activity, and bacterial metabolism of amino acids, SCFAs, and methylamines[47]. In addition, Hong et al[48] revealed a significant increase of acetate, butyrate, and glutamine, together with a decrease of trimethylamine in the feces of lactic acid bacteria (LAB) + dextran sulfate sodium (DSS)-treated mice when compared to DSS-only mice. Thus, it is possible that probiotic treatment is beneficial to energy metabolism, and could help to relieve gut microbial disturbances. The normal intestinal microbiota is involved in sex hormone metabolism of the enterohepatic circulation. Some Clostridium species can convert cortisol to androgen[49]. Our study demonstrated that treatment with C. butyricum increased androsterone 1 in fecal samples, thereby influencing the pathway of steroid hormone biosynthesis. These results suggested that C. butyricum might change the intestinal microbiota and enterohepatic circulation to restore gut homeostasis in IBS mice. IBS is a common disorder in both children and adults. The pathogenesis and pathophysiology of IBS result from multifactorial and multisystemic alterations. The biopsychosocial model describes IBS as a dysfunction of the gut-brain axis that is influenced by genetic susceptibility, as well as physiological, psychological, and environmental variables, in addition to individual coping mechanisms[50]. In the future, we will perform fecal metabolomic studies in IBS patients to explore the gut metabolites in humans.
In summary, this study demonstrated the potential of metabolomic studies to provide new insights into the etiology of IBS. Supplementation with probiotics may provide great future prospects for the treatment of IBS. However, the findings of this study were limited in that the metabolites were only detected by GC-MS. Thus, liquid chromatography (LC)-MS analysis should be considered for future studies.
ARTICLE HIGHLIGHTS
Research background
The prevalence of irritable bowel syndrome (IBS) in Western societies is approximately 10%-20%, and the pathology of IBS is complicated. It is believed that multiple factors such as genetics, visceral hypersensitivity, gastrointestinal motility, dysregulation of the brain-gut axis, levels of neuropeptides and hormones, as well as inflammatory changes all contribute to IBS development.
Research motivation
Microbial metabolites regulate biochemical and physiological processes. Certain disorders that alter fecal microbial profiles may cause IBS. Thus, the key topic we wanted to address in this study is the impact of the microbiota on host–microbial interactions. Fecal metabolic compositions and variations not only reflect the status of the intestinal microbiota, but also bridge the relationship between symbiotic microbes and host health.
Research objectives
The study of fecal metabolomics offers a unique insight to investigate IBS. In the present study, differentially expressed metabolites and key metabolic pathways were found in fecal samples from IBS mice, when compared to the control group. The metabolomic profile in the IBS group was significantly altered following Clostridium butyricum treatment.
Research methods
Fecal samples were analyzed using gas chromatography-mass spectrometry (GC-MS) method. The resulting three-dimensional data involving the peak number, sample name, and normalized peak area were submitted to SIMCA14.1 software package for principal component analysis (PCA) and orthogonal projections to latent structures-discriminate analysis (OPLS-DA). MetaboAnalyst was used to identify the most relevant pathways (http://www.metaboanalyst.ca).
Research results
In this study, we found differentially expressed metabolites between the control and IBS groups. C. butyricum administration modulated metabolic profiles and reduced visceral sensitivity and diarrhea symptoms in IBS mice. This study demonstrated the impact of metabolomic studies on the etiology of IBS. Supplementation with probiotics may provide great prospects for the treatment of IBS. In the future, we will focus fecal metabolomic studies in IBS patients to explore the prevalent pathways and mechanisms in humans.
Research conclusions
Based on the GC-MS analysis, we found that fecal metabolites were changed during the pathological process of IBS. IBS mice demonstrated disorders in fecal microbial profiles, which led to fecal metabolic changes that may affect the development of IBS. This study also demonstrated the potential of metabolomic studies to provide new insights into the etiology of IBS. Probiotics can be used to improve the symptoms of IBS and alter fecal metabolites, and therefore may be used to treat IBS.
Research perspectives
Intestinal microbiota metabolites are very complex. In the future, our research will focus on fecal metabolites in IBS patients to explore the pathophysiological mechanisms in humans. GC-MS combined with liquid chromatography (LC)-MS analysis should be considered for future studies.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Supported by the National Natural Science Foundation of China, No. 81470814 and No. 81400594; and Zhejiang Provincial Natural Science Foundation of China, No. LQ14H160014.
Institutional review board statement: This study was approved by the Ethics Committee of the Zhejiang Chinese Medical University. All procedures in the animal studies were performed in accordance with the ethical standards of the institution or practice.
Institutional animal care and use committee statement: All procedures were approved by the Animal Care Committee of Zhejiang Chinese Medical University, and all methods were performed in accordance with the relevant guidelines and regulations.
Conflict-of-interest statement: The authors declare that they have no conflict of interest.
Data sharing statement: No additional data are available.
Peer-review started: December 13, 2017
First decision: December 27, 2017
Article in press: January 20, 2018
P- Reviewer: Chuah SK, Soares R S- Editor: Gong ZM L- Editor: Wang TQ E- Editor: Ma YJ
Contributor Information
Lei-Min Yu, Department of Gastroenterology, First Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou 310006, Zhejiang Province, China; First Clinical Medical College of Zhejiang Chinese Medical University, Hangzhou 310053, Zhejiang Province, China.
Ke-Jia Zhao, Department of Gastroenterology, First Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou 310006, Zhejiang Province, China; First Clinical Medical College of Zhejiang Chinese Medical University, Hangzhou 310053, Zhejiang Province, China.
Shuang-Shuang Wang, Department of Gastroenterology, First Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou 310006, Zhejiang Province, China; First Clinical Medical College of Zhejiang Chinese Medical University, Hangzhou 310053, Zhejiang Province, China.
Xi Wang, Key Laboratory of Digestive Pathophysiology of Zhejiang Province, First Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou 310053, Zhejiang Province, China.
Bin Lu, Department of Gastroenterology, First Affiliated Hospital of Zhejiang Chinese Medical University, Hangzhou 310006, Zhejiang Province, China. lvbin@medmail.com.cn.
References
- 1.Ohman L, Simrén M. New insights into the pathogenesis and pathophysiology of irritable bowel syndrome. Dig Liver Dis. 2007;39:201–215. doi: 10.1016/j.dld.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 2.Madden JA, Hunter JO. A review of the role of the gut microflora in irritable bowel syndrome and the effects of probiotics. Br J Nutr. 2002;88 Suppl 1:S67–S72. doi: 10.1079/BJN2002631. [DOI] [PubMed] [Google Scholar]
- 3.Malinen E, Rinttilä T, Kajander K, Mättö J, Kassinen A, Krogius L, Saarela M, Korpela R, Palva A. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–382. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 4.Si JM, Yu YC, Fan YJ, Chen SJ. Intestinal microecology and quality of life in irritable bowel syndrome patients. World J Gastroenterol. 2004;10:1802–1805. doi: 10.3748/wjg.v10.i12.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noor SO, Ridgway K, Scovell L, Kemsley EK, Lund EK, Jamieson C, Johnson IT, Narbad A. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol. 2010;10:134. doi: 10.1186/1471-230X-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonfrate L, Tack J, Grattagliano I, Cuomo R, Portincasa P. Microbiota in health and irritable bowel syndrome: current knowledge, perspectives and therapeutic options. Scand J Gastroenterol. 2013;48:995–1009. doi: 10.3109/00365521.2013.799220. [DOI] [PubMed] [Google Scholar]
- 7.Krogius-Kurikka L, Lyra A, Malinen E, Aarnikunnas J, Tuimala J, Paulin L, Mäkivuokko H, Kajander K, Palva A. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9:95. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 10.Sengül N, Aslím B, Uçar G, Yücel N, Işik S, Bozkurt H, Sakaoğullari Z, Atalay F. Effects of exopolysaccharide-producing probiotic strains on experimental colitis in rats. Dis Colon Rectum. 2006;49:250–258. doi: 10.1007/s10350-005-0267-6. [DOI] [PubMed] [Google Scholar]
- 11.Bergen WG, Wu G. Intestinal nitrogen recycling and utilization in health and disease. J Nutr. 2009;139:821–825. doi: 10.3945/jn.109.104497. [DOI] [PubMed] [Google Scholar]
- 12.Ellis DI, Dunn WB, Griffin JL, Allwood JW, Goodacre R. Metabolic fingerprinting as a diagnostic tool. Pharmacogenomics. 2007;8:1243–1266. doi: 10.2217/14622416.8.9.1243. [DOI] [PubMed] [Google Scholar]
- 13.Bertini I, Calabrò A, De Carli V, Luchinat C, Nepi S, Porfirio B, Renzi D, Saccenti E, Tenori L. The metabonomic signature of celiac disease. J Proteome Res. 2009;8:170–177. doi: 10.1021/pr800548z. [DOI] [PubMed] [Google Scholar]
- 14.Vanden Bussche J, Marzorati M, Laukens D, Vanhaecke L. Validated High Resolution Mass Spectrometry-Based Approach for Metabolomic Fingerprinting of the Human Gut Phenotype. Anal Chem. 2015;87:10927–10934. doi: 10.1021/acs.analchem.5b02688. [DOI] [PubMed] [Google Scholar]
- 15.Zeng H, Grapov D, Jackson MI, Fahrmann J, Fiehn O, Combs GF. Integrating Multiple Analytical Datasets to Compare Metabolite Profiles of Mouse Colonic-Cecal Contents and Feces. Metabolites. 2015;5:489–501. doi: 10.3390/metabo5030489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saric J, Wang Y, Li J, Coen M, Utzinger J, Marchesi JR, Keiser J, Veselkov K, Lindon JC, Nicholson JK, et al. Species variation in the fecal metabolome gives insight into differential gastrointestinal function. J Proteome Res. 2008;7:352–360. doi: 10.1021/pr070340k. [DOI] [PubMed] [Google Scholar]
- 17.Chen B, Zhu S, Du L, He H, Kim JJ, Dai N. Reduced interstitial cells of Cajal and increased intraepithelial lymphocytes are associated with development of small intestinal bacterial overgrowth in post-infectious IBS mouse model. Scand J Gastroenterol. 2017;52:1065–1071. doi: 10.1080/00365521.2017.1342141. [DOI] [PubMed] [Google Scholar]
- 18.Jones RC 3rd, Otsuka E, Wagstrom E, Jensen CS, Price MP, Gebhart GF. Short-term sensitization of colon mechanoreceptors is associated with long-term hypersensitivity to colon distention in the mouse. Gastroenterology. 2007;133:184–194. doi: 10.1053/j.gastro.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 19.Yang B, Zhou X, Lan C. Changes of cytokine levels in a mouse model of post-infectious irritable bowel syndrome. BMC Gastroenterol. 2015;15:43. doi: 10.1186/s12876-015-0272-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu YB, Zuo XL, Zhao QJ, Chen FX, Yang J, Dong YY, Wang P, Li YQ. Brain-derived neurotrophic factor contributes to abdominal pain in irritable bowel syndrome. Gut. 2012;61:685–694. doi: 10.1136/gutjnl-2011-300265. [DOI] [PubMed] [Google Scholar]
- 21.Kind T, Wohlgemuth G, Lee DY, Lu Y, Palazoglu M, Shahbaz S, Fiehn O. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81:10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, Nicholls AW, Wilson ID, Kell DB, Goodacre R; Human Serum Metabolome (HUSERMET) Consortium. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 23.Macfarlane GT, Macfarlane S. Bacteria, colonic fermentation, and gastrointestinal health. J AOAC Int. 2012;95:50–60. doi: 10.5740/jaoacint.sge_macfarlane. [DOI] [PubMed] [Google Scholar]
- 24.Sommer F, Bäckhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 25.Martin FP, Dumas ME, Wang Y, Legido-Quigley C, Yap IKS, Tang H, Zirah S, Murphy GM, Cloarec O, Lindon JC, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicholson JK, Holmes E, Lindon JC, Wilson ID. The challenges of modeling mammalian biocomplexity. Nat Biotechnol. 2004;22:1268–1274. doi: 10.1038/nbt1015. [DOI] [PubMed] [Google Scholar]
- 27.Scanlan PD, Shanahan F, Clune Y, Collins JK, O’Sullivan GC, O’Riordan M, Holmes E, Wang Y, Marchesi JR. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ Microbiol. 2008;10:789–798. doi: 10.1111/j.1462-2920.2007.01503.x. [DOI] [PubMed] [Google Scholar]
- 28.Le Gall G, Noor SO, Ridgway K, Scovell L, Jamieson C, Johnson IT, Colquhoun IJ, Kemsley EK, Narbad A. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res. 2011;10:4208–4218. doi: 10.1021/pr2003598. [DOI] [PubMed] [Google Scholar]
- 29.Madsen K, Cornish A, Soper P, McKaigney C, Jijon H, Yachimec C, Doyle J, Jewell L, De Simone C. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121:580–591. doi: 10.1053/gast.2001.27224. [DOI] [PubMed] [Google Scholar]
- 30.Martin FP, Collino S, Rezzi S, Kochhar S. Metabolomic applications to decipher gut microbial metabolic influence in health and disease. Front Physiol. 2012;3:113. doi: 10.3389/fphys.2012.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin FP, Wang Y, Sprenger N, Holmes E, Lindon JC, Kochhar S, Nicholson JK. Effects of probiotic Lactobacillus paracasei treatment on the host gut tissue metabolic profiles probed via magic-angle-spinning NMR spectroscopy. J Proteome Res. 2007;6:1471–1481. doi: 10.1021/pr060596a. [DOI] [PubMed] [Google Scholar]
- 32.Dai ZL, Wu G, Zhu WY. Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front Biosci (Landmark Ed) 2011;16:1768–1786. doi: 10.2741/3820. [DOI] [PubMed] [Google Scholar]
- 33.Berry MD. The potential of trace amines and their receptors for treating neurological and psychiatric diseases. Rev Recent Clin Trials. 2007;2:3–19. doi: 10.2174/157488707779318107. [DOI] [PubMed] [Google Scholar]
- 34.Klassen P, Fürst P, Schulz C, Mazariegos M, Solomons NW. Plasma free amino acid concentrations in healthy Guatemalan adults and in patients with classic dengue. Am J Clin Nutr. 2001;73:647–652. doi: 10.1093/ajcn/73.3.647. [DOI] [PubMed] [Google Scholar]
- 35.Sabelli HC, Javaid JI. Phenylethylamine modulation of affect: therapeutic and diagnostic implications. J Neuropsychiatry Clin Neurosci. 1995;7:6–14. doi: 10.1176/jnp.7.1.6. [DOI] [PubMed] [Google Scholar]
- 36.Snoddy AM, Heckathorn D, Tessel RE. Cold-restraint stress and urinary endogenous beta-phenylethylamine excretion in rats. Pharmacol Biochem Behav. 1985;22:497–500. doi: 10.1016/0091-3057(85)90054-1. [DOI] [PubMed] [Google Scholar]
- 37.Martin FP, Verdu EF, Wang Y, Dumas ME, Yap IK, Cloarec O, Bergonzelli GE, Corthesy-Theulaz I, Kochhar S, Holmes E, et al. Transgenomic metabolic interactions in a mouse disease model: interactions of Trichinella spiralis infection with dietary Lactobacillus paracasei supplementation. J Proteome Res. 2006;5:2185–2193. doi: 10.1021/pr060157b. [DOI] [PubMed] [Google Scholar]
- 38.Ponnusamy K, Choi JN, Kim J, Lee SY, Lee CH. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J Med Microbiol. 2011;60:817–827. doi: 10.1099/jmm.0.028126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiter E, Jiang Q, Christen S. Anti-inflammatory properties of alpha- and gamma-tocopherol. Mol Aspects Med. 2007;28:668–691. doi: 10.1016/j.mam.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24:160–168. doi: 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 41.Jackowski S, Rock CO. Regulation of coenzyme A biosynthesis. J Bacteriol. 1981;148:926–932. doi: 10.1128/jb.148.3.926-932.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hutschenreuther A, Birkenmeier G, Bigl M, Krohn K, Birkemeyer C. Glycerophosphoglycerol, Beta-alanine, and pantothenic Acid as metabolic companions of glycolytic activity and cell migration in breast cancer cell lines. Metabolites. 2013;3:1084–1101. doi: 10.3390/metabo3041084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Depeint F, Bruce WR, Shangari N, Mehta R, O’Brien PJ. Mitochondrial function and toxicity: role of the B vitamin family on mitochondrial energy metabolism. Chem Biol Interact. 2006;163:94–112. doi: 10.1016/j.cbi.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 44.Even PC, Decrouy A, Chinet A. Defective regulation of energy metabolism in mdx-mouse skeletal muscles. Biochem J. 1994;304(Pt 2):649–654. doi: 10.1042/bj3040649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leonardi R, Zhang YM, Rock CO, Jackowski S. Coenzyme A: back in action. Prog Lipid Res. 2005;44:125–153. doi: 10.1016/j.plipres.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 46.Martin FP, Sprenger N, Yap IK, Wang Y, Bibiloni R, Rochat F, Rezzi S, Cherbut C, Kochhar S, Lindon JC, et al. Panorganismal gut microbiome-host metabolic crosstalk. J Proteome Res. 2009;8:2090–2105. doi: 10.1021/pr801068x. [DOI] [PubMed] [Google Scholar]
- 47.Martin FP, Wang Y, Sprenger N, Yap IK, Lundstedt T, Lek P, Rezzi S, Ramadan Z, van Bladeren P, Fay LB, et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4:157. doi: 10.1038/msb4100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong YS, Ahn YT, Park JC, Lee JH, Lee H, Huh CS, Kim DH, Ryu DH, Hwang GS. 1H NMR-based metabonomic assessment of probiotic effects in a colitis mouse model. Arch Pharm Res. 2010;33:1091–1101. doi: 10.1007/s12272-010-0716-1. [DOI] [PubMed] [Google Scholar]
- 49.Bokkenheuser VD, Morris GN, Ritchie AE, Holdeman LV, Winter J. Biosynthesis of androgen from cortisol by a species of Clostridium recovered from human fecal flora. J Infect Dis. 1984;149:489–494. doi: 10.1093/infdis/149.4.489. [DOI] [PubMed] [Google Scholar]
- 50.Chogle A, Mintjens S, Saps M. Pediatric IBS: an overview on pathophysiology, diagnosis and treatment. Pediatr Ann. 2014;43:e76–e82. doi: 10.3928/00904481-20140325-08. [DOI] [PubMed] [Google Scholar]