Abstract
Synovial sarcoma (SS) is genetically characterized by chromosomal translocation, which generates SYT-SSX fusion transcripts. Although SS can occur in any body part, primary gastric SS is substantially rare. Here we describe a detection of the fusion gene sequence of gastric SS in plasma cell-free DNA (cfDNA). A gastric submucosal tumor was detected in the stomach of a 27-year-old woman and diagnosed as SS. Candidate intronic primers were designed to detect the intronic fusion breakpoint and this fusion sequence was confirmed in intron 10 of SYT and intron 5 of SSX2 by genomic polymerase chain reaction (PCR) and direct sequencing. A locked nucleic acid (LNA) probe specific to the fusion sequence was designed for detecting the fusion sequence in plasma and the fusion sequence was detected in preoperative plasma cfDNA, while not detected in postoperative plasma cfDNA. This technique will be useful for monitoring translocation-derived diseases such as SS.
Keywords: Fusion gene, Gastric synovial sarcoma, Plasma, Cell free DNA
Core tip: Synovial sarcoma (SS) is genetically characterized by SYT-SSX fusion transcripts and detection of the fusion gene is necessary for a definitive diagnosis. This study demonstrated the detection of fusion sequence using cfDNA sample. A small gastric SS was detected in the stomach of a 27-year-old woman. Candidate intronic primers were designed and the intronic breakpoint was confirmed by PCR and direct sequencing in frozen tumor sample. A probe specific for fusion sequence was designed and the sequence was detected in preoperative cfDNA and frozen tumor sample. This technique will be useful for monitoring translocation-derived diseases such as SS.
INTRODUCTION
Synovial sarcoma (SS), an aggressive soft tissue tumor, accounts for 7% of human soft tissue sarcomas[1]. Although SS frequently arises in large joint capsules in the extremities of young adults, it can occur in any body part, including the digestive tract. SS morphology resembles the developing synovial tissue; however, its origin remains unknown.
SS is genetically characterized by the reciprocal chromosomal translocation between X and 18 t (X; 18) (p11.2; q11.2) chromosomes, which generates SYT-SSX fusion transcripts[2,3]. SYT (GenBank accession number NC_000018) is normally expressed in human tissues[4], whereas SSX expression is limited to normal testis and thyroid, and some variants are found in human malignancies[4-7]. The SSX family reportedly comprises nine highly homologous synovial sarcoma X genes (SSX1-9)[5]; however, SSX1 or SSX2 (GenBank accession number (SSX1) or NC_000023) account for > 90% of t (X;18) translocations and SSX4 is involved in few cases[7,8]. The breakpoints are frequently found within intron 10 of the SYT gene and intron 5 of the SSX gene[6-8]. The detection of the SYT-SSX fusion gene generally leads to SS diagnosis.
In this time, we could preoperatively diagnose with a primary gastric SS patient by reverse transcription polymerase chain reaction (RT-PCR) of biopsy tissue samples. Although some primary gastric SS cases have been reported, their preoperative diagnosis is rare. Its diagnosis is difficult solely on the basis of clinical or pathological findings[9-11]. Circulating nucleic acids were recently found to reflect the occurrence of cancer or monitoring of many diseases. Regarding SS, the production of the fusion gene SYT-SSX is a highly disease-specific alteration, and detecting the fusion gene in the patient’s circulation will represent a less-invasive diagnostic tool for this disease.
In this study, we report a detection of the fusion gene sequence in plasma circulating cfDNA.
CASE REPORT
Materials and methods
Case of primary gastric SS: A gastric ulcerated submucosal tumor was detected in the lower stomach of a 27-year-old woman using upper gastrointestinal endoscopy. Endoscopic ultrasound-guided biopsy was performed, and a spindle cell sarcoma compatible with SS was found in biopsy samples. The SYT-SSX2 fusion gene was detected in tissue RNA of biopsy samples using RT-PCR. Laparoscopic gastrectomy was performed, and the tumor up to 20 mm diameter was pathologically diagnosed with SS. This patient remained disease free during the 6 mo follow-up.
The patient provided informed written consent for specimen collections and biomarker analyses, and the research was conducted with the approval of the ethics committees of the local institution and according to the Helsinki declaration.
Tissue sample collections and tissue DNA and RNA extractions: Tumor tissues and adjacent normal gastric tissues were obtained from operatively resected specimens and stored at -80 °C before DNA and RNA extractions. Total DNA and RNA were extracted from frozen samples using the AllPrep® DNA/RNA/miRNA universal kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Tissue RNA concentration was determined using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, United States), and the tissue DNA concentration was determined using a Qubit 3.0 fluorometer (Thermo Fisher Scientific).
Plasma sample collections and plasma DNA extractions: Seven-milliliter EDTA blood samples were obtained from the patient at three different times: before operation, 1 mo after the operation, and 6 mo after the operation. Blood samples were also obtained from 10 healthy volunteers. The plasma samples were instantly separated from the cellular fraction using a three-spin protocol (1500 rpm for 30 min, 3000 rpm for 5 min, and 4500 rpm for 5 min) and stored at -80 °C.
Circulating cell-free DNA (cfDNA) was isolated from 2 mL of each plasma sample using the QIAamp Circulating Nucleic Acid kit (Qiagen) according to the manufacturer’s instructions, and the cfDNA concentration was determined using the Qubit 3.0 fluorometer.
Protocols for RT-PCR and genomic PCR of tissue samples: Reverse transcribed reactions were performed with 400 ng tissue RNA in a 20 μL reaction volume using the high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The complementary DNA was subjected to PCR using AmpliTaq DNA polymerase (Thermo Fisher) and our designed primers (Table 1) with the following conditions: 10 min at 95 °C, followed by repetitions of 1 min at 95 °C, 1 min at 60 °C, and 30 s at 72 °C and a final extension step of 30 s at 72 °C.
Table 1.
Overview of primers and probe sequences for all PCR methods
| Primers (5’-3’) | Sequence | Product size (bp) |
| Reverse transcription PCR | ||
| SYT-sense | CCAGCAGAGGCCTTATGGA | 118 |
| SSX2-antisense | GCACAGCTCTTTCCCATCA | |
| Genomic PCR | ||
| SYT1-sense | GTAGTTTGACCGGCTGCAGAA | |
| SYT2-sense | GGTGGTCTGGTTTGTTCACC | |
| SYT3-sense | GCAAATGTTTATTGAGCAACCA | |
| SYT4-sense | GGGAGAAATTAAAAGGGTGGA | |
| SYT5-sense | CACCTGTGAAACCATCAGCA | |
| SYT6-sense | TTTTCTTTATGGATTATGCTTTTGG | |
| SYT7-sense | GCTTACTAGGAGTTTCATTGTAATTG | |
| SYT8-sense | CAGCCTGATAAACTGTATACC | |
| SYT9-sense | GATTTGAATGCGTGATCACAAG | |
| SYT10-sense | GATTGGATTCCAGACATTGTG | |
| SYT11-sense | GCCACCTTGGAATTTGTTAATG | |
| SYT12-sense | CTGATGATTGAAGAAACCGAG | |
| SSX2-1-antisense | ACGGAGAATCAGGGTTCTTTGG | |
| SSX2-2-antisense | TCAGTCTCCACACTGGCAAC | |
| SSX2-3-antisense | TCAAGGCAACATCCGACTCC | |
| SSX2-4-antisense | TGGTTTCCAGGGATAGAATGCT | |
| Real-time PCR | ||
| SYT-specific-sense | CCAGCAACAGTAGTTTACTTTCTATC | 106 |
| SSX2-specific-antisense | AAACATAGGGAGGCGACAAA | |
| SYT-SSX2-FAM probe-sense | ATA+CAA+T+C+CA+G+CAG1 |
+ C, +T, + G = LNA base.
Genomic PCR was performed with 10 ng genomic DNA (gDNA) in a 25 μL reaction volume using 1 unit of AmpliTaq DNA polymerase and 0.2 μmol/L of each primer (Table 1). PCR was run using the GeneAmp PCR System 9700 (Thermo Fisher) at the following conditions: 10 min at 95 °C, followed by repetitions of 1 min at 95 °C, 1 min at 53 °C (40 cycles), and 1 min at 72 °C and a final extension step of 30 s at 72 °C. The accurate length of the PCR products was confirmed using agarose gel electrophoresis. The images were obtained using the BLooK LED transilluminator (BIO-HELIX, Keelung, Taiwan) and a digital camera.
Direct sequencing analysis: The PCR products obtained were subjected to direct sequencing using the BigDye Terminator version1.1 Cycle Sequencing Kit (Thermo Fisher). Samples were run on ABI 3500 genetic analyzer (Applied Biosystems, Foster, United States).
Detection of SYT-SSX2 fusion gene sequence: To detect the fusion gene sequence in tissue and plasma samples, real-time PCR (rt-PCR) was performed in duplicates with 10 ng tissue DNA or 0.5 ng plasma cfDNA using the StepOne real-time PCR systems (Thermo Fisher). A pair of primers and specific probes using locked nucleic acid (LNA) bases for detecting the SYT-SSX2 fusion gene sequence were designed (Table 1; Integrated DNA Technologies, Coralville, United States). The whole rt-PCR mixture (10 μL reaction volume) comprised DNA, Taqman Fast Advanced Master Mix (Thermo Fisher), RNase-free water, 0.5 μmol/L of each primer, and 0.25 μmol/L of probe solution and was performed with the following conditions: 2 min at 50 °C and 20 s at 95 °C, followed by repetitions of 1 s at 95 °C and 20 s at 60 °C (40 cycles).
Statistical analysis: BLAST software (http://www.ncbi.nlm.nih.gov/BLAST/) was used for sequence data analysis. The detection levels of the fusion gene sequence were compared with those obtained using the ΔΔCt method.
RESULTS
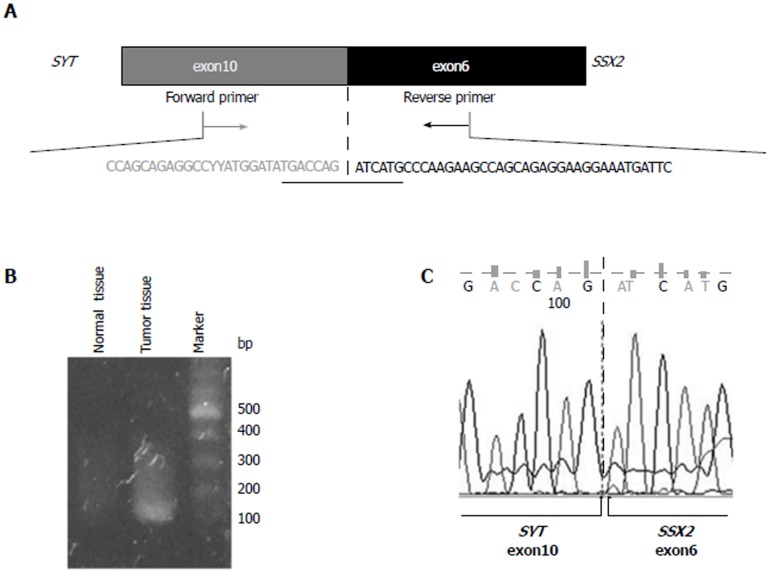
Confirmation of SYT-SSX2 fusion gene occurrence in tumor tissues: To confirm the fusion of the SYT and SSX2 genes, RT-PCR was performed using total RNA extracted from tumor and adjacent normal tissues. The primer set was designed in exon 10 of the SYT gene (SYT-sense) and in exon 6 of the SSX2 gene (SSX2-antisense) (Figure 1A) for an expected PCR product size of 118 base pairs. The sequence data of this primer sets are shown in Table 1. A PCR product of > 100 base pairs was obtained in tumor tissues only (Figure 1B), and direct sequencing of this PCR product confirmed the fusion between exon 10 of the SYT gene and exon 6 of the SSX2 gene (Figure 1A and C).
Figure 1.
Identification of the SYT-SSX2 fusion transcript. A: Structure of the SYT-SSX2 fusion transcript. Exon 10 of the SYT gene and exon 6 of the SSX2 gene are fused together in this transcript. The sequencing of exon is shown. Forward and reverse primers were designed in exon 10 of the SYT gene and exon 6 of the SSX2 gene, respectively; B: The RT-PCR product, including the SYT-SSX2 fusion site, was obtained from tumor tissue samples only. This analysis was performed more than three times, and a representative result of an electrophoresis is shown; C: The result of direct sequencing of the PCR product is shown. Exon 10 of the SYT gene is fused to exon 6 of the SSX2 gene.
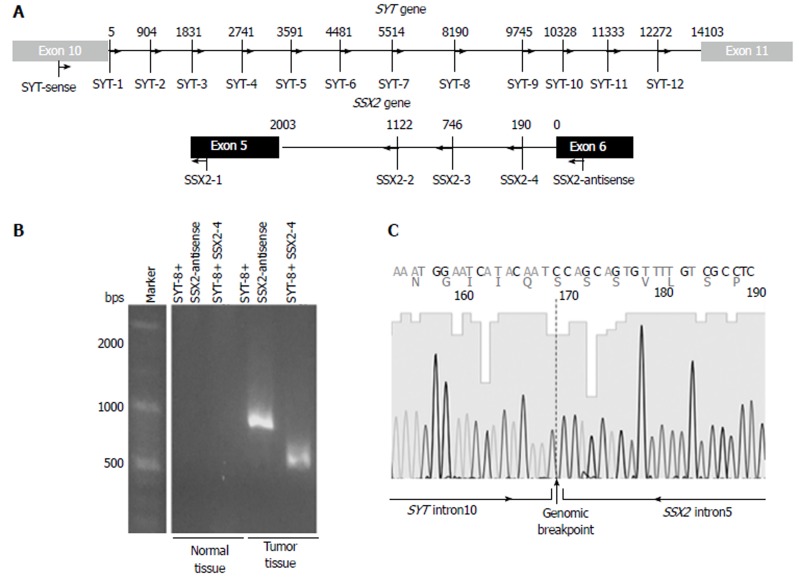
Detection of the genomic breakpoint: To detect a genomic breakpoint in intron 10 of the SYT gene and intron 5 of the SSX2 gene, 12 forward primers in SYT intron 10 and 3 reverse primers in SSX2 intron 5 were designed at intervals of approximately 1000 base pairs (Figure 2A and Table 1). Genomic PCR was performed with each forward and reverse primer using gDNA extracted from tumor and normal tissues. The smallest PCR product, approximately 500 base pairs, was detected with the combination of SYT-8 and SSX2-4 primers in tumor tissues only (Figure 2B). The direct sequencing of the obtained PCR product showed a genomic breakpoint within the SYT and SSX2 introns (Figure 2C).
Figure 2.
Detection of the SYT-SSX2 fusion sequence. A: The intronic primer settings in the SYT and SSX2 genes are shown; B: Genomic PCR products were obtained from tumor tissue samples only. This analysis was performed more than two times, and a representative result of an electrophoresis is shown; C: The intronic breakpoint was confirmed by direct sequencing.
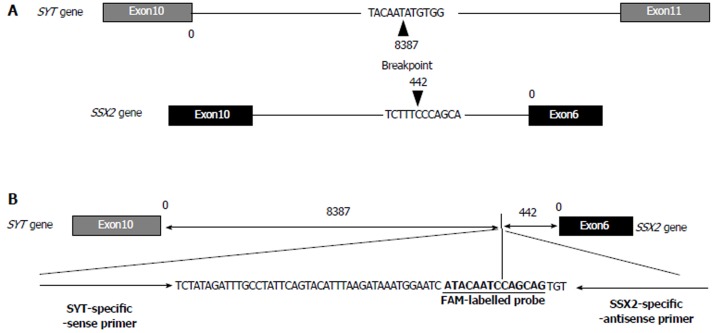
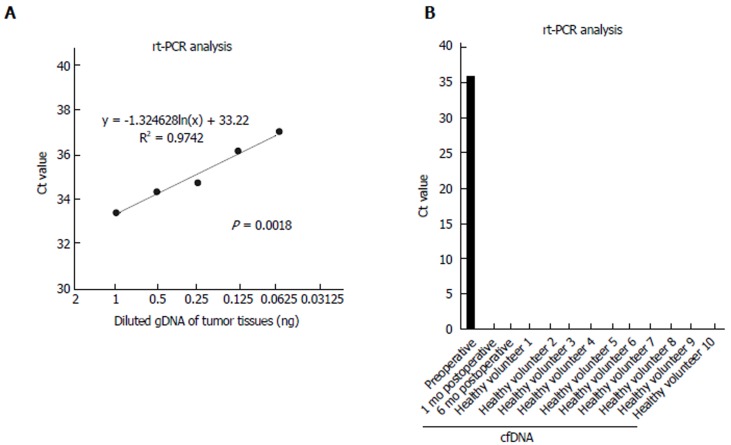
Production of the breakpoint-specific probe: A breakpoint-specific FAM-labeled LNA probe and primer sets for rt-PCR analysis were designed to detect the SYT-SSX2 fusion sequence in DNA samples (Figure 3A and B, Table 1). Using this specific probe and primer sets, the SYT-SSX2 fusion sequence was detected in 10 ng of gDNA extracted from tumor tissues (Ct value; 31.6) (Table 2). Serial dilutions of gDNA extracted from tumor tissue were analyzed by rt-PCR using this specific probe and primer sets to confirm the accurate measurable range. The SYT-SSX2 fusion sequence was detected proportional to gDNA concentrations from 1 to 0.063 ng (range of Ct value, up to 37, R2 = 0.974, P = 0.0018) (Figure 4A).
Figure 3.
Intronic breakpoint and structure of the fusion sequence with specific probe and primer sets. A: The arrowheads indicate the position and sequence of the intronic breakpoint of the SYT and SSX2 genes; B: The structure and sequence of the FAM-labeled probe and primer sets specific to the intronic breakpoint of the SYT-SSX fusion sequence.
Table 2.
Results of rt-PCR analysis by the specific probe and primer sets
| Types of DNA | Sample | Ct value 1 | Ct value 2 | Mean |
| Genomic | Normal tissue | Undetermined | Undetermined | Undetermined |
| Tumor tissue | 31.94651 | 31.30283 | 31.62467 | |
| β-actin | Tumor tissue | 28.97511 | 30.95062 | 29.96287 |
Undetermined: No Ct detection till 40 cycles.
Figure 4.
Detection of the fusion gene sequence in cell-free DNA using specific probe and primer sets. A: The range of reproducibility of rt-PCR with the specific probe and primer sets was confirmed. Diluted serial tumor gDNA of 1-0.063 ng was used. The dotted line indicates an approximate curve (R² = 0.9742, P = 0.0018); B: cfDNA samples of a patient and 10 healthy volunteers were analyzed using rt-PCR. rt-PCR was performed in duplicate, and mean Ct values are indicated. Standard deviation was calculated from duplicate samples. cfDNA: Cell-free DNA.
Detection of the SYT-SSX2 fusion sequence in cfDNA: cfDNA extracted from the plasma was verified by rt-PCR using the specific probe and primer sets. The fusion sequence was detected in the plasma sample collected preoperatively from the patient but not in the plasma sample collected one or six months postoperatively from the patient or in the plasma samples of 10 healthy volunteers (Figure 4B). The Ct value was 35.9 in cfDNA collected preoperatively, whereas it was undetermined in other cfDNA (Not detected till 40 cycles) (Figure 4B).
DISCUSSION
Primary gastric SS is a rare disease, and its diagnosis by clinical or pathological findings is difficult. Detection of the SYT-SSX fusion gene using a molecular biological approach is necessary for a definitive diagnosis. In most cases, this fusion gene or fusion sequence was identified from total RNA or DNA of tumor tissues by PCR method. Many alterations, such as microsatellite alteration[12], DNA methylation[13], and some mutations[14], in cancer cells have been recently detected in circulating cfDNA. Regarding translocation-derived diseases such as SS, it may also be possible to detect the fusion sequence in cfDNA. Indeed, Hayashi et al. reported that the EWS-ETS fusion gene, specific to Ewing sarcoma, was detected in circulating cfDNA extracted from plasma of a patient with Ewing sarcoma using long-range PCR and digital droplet PCR (ddPCR)[15].
In this study, we developed a highly specific locked nucleic acid (LNA) probe and primer sets for the SYT-SSX2 fusion gene sequence and detected the sequence in cfDNA extracted from the plasma of a primary gastric SS patient. The translocation causes gene rearrangements in tumor tissues only; therefore, this probe and primer sets specific to the fusion sequence are highly unique to identify the intronic breakpoint. Moreover, the normal PCR method instead of highly accurate and sensitive methods such as ddPCR could detect the objective sequence in a small amount of cfDNA.
Nested PCR[8] or long-range PCR[15] techniques were previously reported for identifying the intronic fusion sequences of translocation-related diseases. While the detection of SYT-SSX fusions in tumor tissues has been previously reported, this is the first report regarding the detection of the fusion sequence in circulating cfDNA. Since the discovery of circulating tumor cfDNA in 1994[16,17], investigations on cfDNA molecules have been proven to be useful for diagnosing or monitoring of disease states[11,18-21]. In this study, the fusion sequence could be detected in preoperative circulating cfDNA only, although the primary lesion was only 20 mm in diameter and undetectable by preoperative computed tomography. We expect the detection method of the SYT-SSX fusion sequence in circulating cfDNA to become an effective monitoring tool for SS, because > 90% of SS patients have this reciprocal translocation.
Numerous gene alterations have been recently identified using advanced technologies such as next-generation sequencing, and fusion genes have been reported to contribute to tumorgenesis[22] . For example, recurrent fusions of R-spondin family members RSPO2 and RSPO3, which occur in 10% of colon tumors, have been reported to be involved in the activation of Wnt signaling pathway[23]. NCOA2, a negative growth regulator repressing Wnt signaling pathway, was reported to be disrupted by recurrent fusion with LACTB2 in colorectal cancer[24]. We believe this method can be a less-invasive and personalized monitoring technique with high sensitivity and specificity for translocation-derived diseases, including some cancers.
This technique has some limitations. Sufficient amounts of gDNA from tissue samples need to be obtained to confirm the intronic breakpoint. Frozen tumor samples may be preferred over formalin-fixed tissues to prevent yield decrease because of fragmentation of DNA samples in the latter tissues. Furthermore, although we demonstrated the detection of the fusion sequence in cfDNA of a gastric SS patient, the sensitivity of rt-PCR using cfDNA obtained from 2 mL plasma was not so high (Ct value, 35.9) because of a small amount of cfDNA. A more advanced method to obtain high-quality or large amounts of cfDNA may be more appropriate for this technique in the future regarding a stable application. Finally, while the location of gene fusion is almost same for each disease, the intronic breakpoint may differ in each patient. Thus, a specific probe and primer sets should be designed for each patient per disease. In this regard, if the amounts of collected plasma RNA or sensitivity of the rt-PCR method is improved, a specific probe and primer sets for plasma RNA samples would be more useful and become a valuable diagnostic tool. Large-scale studies will be necessary to confirm the sensitivity and specificity of this method.
In conclusion, we demonstrated the detection of the SYT-SSX fusion sequence in the plasma circulating cfDNA of a rare gastric SS patient. In the future, this method may be useful for translocation-derived diseases such as SS or for other cancers which contain translocations.
ARTICLE HIGHLIGHTS
Case characteristics
A 27-year-old woman presented upper abdominal pain.
Clinical diagnosis
Upper abdominal tenderness associated to the gastric tumor.
Differential diagnosis
Stomach tumor including cancer or gastrointestinal stromal tumor.
Laboratory diagnosis
Blood tests ruled out opportunistic infections and severe anemia.
Imaging diagnosis
Upper gastrointestinal endoscopy and ultrasound endoscopy showed a small submucosal tumor less than 20 mm in diameter in the stomach.
Pathological diagnosis
Histological examination of the biopsies showed a spindle cell sarcoma compatible with SS. SYT-SSX2 fusion gene was detected using biopsy samples by RT-PCR methods.
Treatment
The submucosal tumor was resected by laparosopic and ecdoscopic cooperative surgery.
Related reports
Previous cases of SS whose intronic breakpoints were confirmed using tumor samples have been described.
Term explanation
SYT-SSX fusion gene is SS-specific genetic alternation and detection of SYT-SSX contributes much to the diagnosis of SS.
Experiences and lessons
In patients with SS and other translocation-related diseases, detection of fusion genes specific for those diseases is a powerful diagnostic tool and the detection using liquid sample, such as blood, can be a valuable and less invasive monitoring tool as well.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Informed consent statement: The study participant provided informed written consent prior to their treatments and study enrollment.
Conflict-of-interest statement: All authors declare no conflict of interest related to this study or its publication.
Peer-review started: December 14, 2017
First decision: January 5, 2018
Article in press: January 20, 2018
P- Reviewer: Vorobjova T S- Editor: Gong ZM L- Editor: A E- Editor: Ma YJ
Contributor Information
Shinpei Ogino, Division of Digestive Surgery, Department of Surgery, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan.
Hirotaka Konishi, Division of Digestive Surgery, Department of Surgery, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan. h-koni7@koto.kpu-m.ac.jp.
Daisuke Ichikawa, Division of Digestive Surgery, Department of Surgery, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan.
Junichi Hamada, Division of Digestive Surgery, Department of Surgery, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan.
Katsutoshi Shoda, Division of Digestive Surgery, Department of Surgery, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan.
Tomohiro Arita, Division of Digestive Surgery, Department of Surgery, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan.
Shuhei Komatsu, Division of Digestive Surgery, Department of Surgery, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan.
Atsushi Shiozaki, Division of Digestive Surgery, Department of Surgery, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan.
Kazuma Okamoto, Division of Digestive Surgery, Department of Surgery, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan.
Sanae Yamazaki, Department of Surgical Pathology, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan.
Satoru Yasukawa, Department of Surgical Pathology, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan.
Eiichi Konishi, Department of Surgical Pathology, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan.
Eigo Otsuji, Division of Digestive Surgery, Department of Surgery, Kyoto Prefectural University of Medicine, Kamigyo-ku, Kyoto 602-8566, Japan.
References
- 1.Herzog CE. Overview of sarcomas in the adolescent and young adult population. J Pediatr Hematol Oncol. 2005;27:215–218. doi: 10.1097/01.mph.0000161762.53175.e4. [DOI] [PubMed] [Google Scholar]
- 2.Turc-Carel C, Dal Cin P, Limon J, Rao U, Li FP, Corson JM, Zimmerman R, Parry DM, Cowan JM, Sandberg AA. Involvement of chromosome X in primary cytogenetic change in human neoplasia: nonrandom translocation in synovial sarcoma. Proc Natl Acad Sci USA. 1987;84:1981–1985. doi: 10.1073/pnas.84.7.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Leeuw B, Balemans M, Olde Weghuis D, Geurts van Kessel A. Identification of two alternative fusion genes, SYT-SSX1 and SYT-SSX2, in t(X;18)(p11.2;q11.2)-positive synovial sarcomas. Hum Mol Genet. 1995;4:1097–1099. doi: 10.1093/hmg/4.6.1097. [DOI] [PubMed] [Google Scholar]
- 4.Tamborini E, Agus V, Mezzelani A, Riva C, Sozzi G, Azzarelli A, Pierotti MA, Pilotti S. Identification of a novel spliced variant of the SYT gene expressed in normal tissues and in synovial sarcoma. Br J Cancer. 2001;84:1087–1094. doi: 10.1054/bjoc.2000.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Güre AO, Wei IJ, Old LJ, Chen YT. The SSX gene family: characterization of 9 complete genes. Int J Cancer. 2002;101:448–453. doi: 10.1002/ijc.10634. [DOI] [PubMed] [Google Scholar]
- 6.Crew AJ, Clark J, Fisher C, Gill S, Grimer R, Chand A, Shipley J, Gusterson BA, Cooper CS. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J. 1995;14:2333–2340. doi: 10.1002/j.1460-2075.1995.tb07228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otsuka S, Nishijo K, Nakayama T, Aoyama T, Ishibe T, Shibata KR, Shima Y, Nakamura T, Otsuka T, Toguchida J. A variant of the SYT-SSX2 fusion gene in a case of synovial sarcoma. Cancer Genet Cytogenet. 2006;167:82–88. doi: 10.1016/j.cancergencyto.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Törnkvist M, Brodin B, Bartolazzi A, Larsson O. A novel type of SYT/SSX fusion: methodological and biological implications. Mod Pathol. 2002;15:679–685. doi: 10.1038/modpathol.3880587. [DOI] [PubMed] [Google Scholar]
- 9.Torres Rivas HE, Fernández S, Fresno MF. Primary gastric synovial sarcoma. Pathology. 2014;46:253–256. doi: 10.1097/PAT.0000000000000078. [DOI] [PubMed] [Google Scholar]
- 10.Michot N, Robert PE, De Muret A, Marques F, de Calan L, Benchellal Z. Gastric synovial sarcoma: case report and systematic review of literature. J Gastrointest Cancer. 2014;45 Suppl 1:129–131. doi: 10.1007/s12029-014-9591-1. [DOI] [PubMed] [Google Scholar]
- 11.Chen XQ, Stroun M, Magnenat JL, Nicod LP, Kurt AM, Lyautey J, Lederrey C, Anker P. Microsatellite alterations in plasma DNA of small cell lung cancer patients. Nat Med. 1996;2:1033–1035. doi: 10.1038/nm0996-1033. [DOI] [PubMed] [Google Scholar]
- 12.Wong IH, Lo YM, Zhang J, Liew CT, Ng MH, Wong N, Lai PB, Lau WY, Hjelm NM, Johnson PJ. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 1999;59:71–73. [PubMed] [Google Scholar]
- 13.Le Calvez-Kelm F, Foll M, Wozniak MB, Delhomme TM, Durand G, Chopard P, Pertesi M, Fabianova E, Adamcakova Z, Holcatova I, et al. KRAS mutations in blood circulating cell-free DNA: a pancreatic cancer case-control. Oncotarget. 2016;7:78827–78840. doi: 10.18632/oncotarget.12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi M, Chu D, Meyer CF, Llosa NJ, McCarty G, Morris CD, Levin AS, Wolinsky JP, Albert CM, Steppan DA, et al. Highly personalized detection of minimal Ewing sarcoma disease burden from plasma tumor DNA. Cancer. 2016;122:3015–3023. doi: 10.1002/cncr.30144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev. 1994;3:67–71. [PubMed] [Google Scholar]
- 16.Vasioukhin V, Anker P, Maurice P, Lyautey J, Lederrey C, Stroun M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br J Haematol. 1994;86:774–779. doi: 10.1111/j.1365-2141.1994.tb04828.x. [DOI] [PubMed] [Google Scholar]
- 17.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 18.McDermott U, Downing JR, Stratton MR. Genomics and the continuum of cancer care. N Engl J Med. 2011;364:340–350. doi: 10.1056/NEJMra0907178. [DOI] [PubMed] [Google Scholar]
- 19.Siravegna G, Bardelli A. Genotyping cell-free tumor DNA in the blood to detect residual disease and drug resistance. Genome Biol. 2014;15:449. doi: 10.1186/s13059-014-0449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shoda K, Ichikawa D, Fujita Y, Masuda K, Hiramoto H, Hamada J, Arita T, Konishi H, Komatsu S, Shiozaki A, et al. Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer. Gastric Cancer. 2017;20:126–135. doi: 10.1007/s10120-016-0599-z. [DOI] [PubMed] [Google Scholar]
- 21.Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman V, Jaiswal BS, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, Wu WK, Liang Q, Zhang N, He J, Li X, Zhang X, Xu L, Chan MT, Ng SS, et al. Disruption of NCOA2 by recurrent fusion with LACTB2 in colorectal cancer. Oncogene. 2016;35:187–195. doi: 10.1038/onc.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urakami K, Shimoda Y, Ohshima K, Nagashima T, Serizawa M, Tanabe T, Saito J, Usui T, Watanabe Y, Naruoka A, et al. Next generation sequencing approach for detecting 491 fusion genes from human cancer. Biomed Res. 2016;37:51–62. doi: 10.2220/biomedres.37.51. [DOI] [PubMed] [Google Scholar]
- 24.Yu J, Wu WK, Liang Q, Zhang N, He J, Li X, Zhang X, Xu L, Chan MT, Ng SS, et al. Disruption of NCOA2 by recurrent fusion with LACTB2 in colorectal cancer. Oncogene. 2016;35:187–195. doi: 10.1038/onc.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]