Abstract
There is an urgent need to discover novel antimicrobial therapies. Drug repurposing can reduce the time and cost risk associated with drug development. We report the inhibitory effects of anthelmintic drugs (niclosamide, oxyclozanide, closantel, rafoxanide) against Helicobacter pylori strain 60190 and pursued further characterization of niclosamide against H. pylori. The MIC of niclosamide against H. pylori was 0.25 μg/mL. Niclosamide was stable in acidic pH and demonstrated partial synergy with metronidazole and proton pump inhibitors, such as omeprazole and pantoprazole. Niclosamide administration at 1 × MIC concentration, eliminated 3-log10 CFU of H. pylori adhesion/invasion to AGS cells. Interestingly, no resistance developed even after exposure of H. pylori bacteria to niclosamide for 30 days. The cytotoxic assay demonstrated that niclosamide is not hemolytic and has an IC50 of 4 μg/mL in hepatic and gastric cell lines. Niclosamide administration decreased transmembrane pH as determined by DiSC3(5) assay indicating that the mechanism of action of the anti-H. pylori activity of niclosamide was the disruption of H. pylori proton motive force. Niclosamide was effective in the Galleria mellonella-H. pylori infection model (p = 0.0001) and it can be develop further to combat H. pylori infection. However, results need to be confirmed with other H. pylori and clinical strains.
Introduction
Helicobacter pylori is a Gram-negative, helically shaped, stomach pathogen associated with human gastric mucosa. This bacillus can cause chronic gastritis, peptic ulcer, gastric mucosa-associated lymphoid tissue (MALT) lymphoma and gastric carcinoma1. About 2.9% of H. pylori infected individuals develop gastric cancer2 and eradication of H. pylori infection decreased the risk of gastric cancer2,3. Moreover, even though the association between the H. pylori infection and colorectal cancer is inconclusive4, meta-analysis by Wu et al., demonstrated that the H. pylori infection was associated with increased occurrence of colorectal cancer5. H. pylori infection is often initiated in childhood6. This results in an ongoing H. pylori infection in about half of the global population7. The World Health Organization (WHO) declared H. pylori a class I gastric carcinogen8 with worldwide prevalence, and increased impact in low socio-economic countries9. Successful antimicrobial treatment for H. pylori infection is extremely challenging due to its survival in a hostile acidic environment and association within the gastric mucosa10. As such, the antimicrobial agents must penetrate thick mucus, remain active in low pH and a successful therapy requires sequential administration of two antibiotics, such as amoxicillin and clarithromycin, along with a proton pump inhibitor (PPI)10. However, traditional antibiotics are failing to clear H. pylori infection at ever increasing rates, primarily due to the emergence of bacterial drug resistance11. For example, H. pylori resistance to clarithromycin among male U.S. veterans increased from 16% to 24% during 2009 to 2013 and resistance to metronidazole is high11.
Similar to the difficulties with antimicrobial drug discovery in general, development of novel anti-H. pylori agents has been challenging due to regulatory guidelines, depressed financial returns, and difficulties determining the mechanism of action of novel compounds12. Repurposing is a powerful approach for identifying potential antimicrobial characteristics of existing clinical molecules that have been prescribed for other therapies in healthcare. Research from others, as well as our group, previously reported the antibacterial activities of anthelmintics on MRSA13,14, Clostridium difficile15 and Pseudomonas aeruginosa quorum sensing16. Anthelmintics are used to treat protozoan-helminth infections in humans and they are among the most used and lowest-priced medicines world-wide17. Niclosamide is primarily used to treat cestodes (tapeworms) such as Taenia saginata, Taenia solium, Hymenolepsis nana18 and it inhibits production of energy derived from anaerobic metabolism19. Niclosamide, is a known anthelmintic that is listed in the WHO essential medicines20, and we reported that the drug was active against H. pylori infection. This manuscript further investigated whether they possess activity against H. pylori strain 60190, both in vitro and in vivo.
Results and Discussion
Antibacterial susceptibility
We conducted a pilot screening for anti- H. pylori activity by a broth microdilution assay using the hit molecules that was identified from our previous HTS14,21, which can be found in the Supplementary Table. S2. Interestingly, we found that the anthelmintics (niclosamide, oxyclozanide, rafoxanide, and closantel) inhibited the growth of H. pylori strain 60190. More specifically, the MICs of niclosamide, oxyclozanide, rafoxanide, and closantel were 0.25, 2.0, 4.0 and 16 μg/mL, respectively (Table 1) and the MBC of niclosamide, oxyclozanide, rafoxanide, and closantel against H. pylori were 0.5, 4.0, 8.0, 32 μg/mL, respectively (Table 1).
Table 1.
Antimicrobial properties of anthelmintics against H. pylori.
| Antimicrobial agent | MIC μg/mL | MBC μg/mL |
|---|---|---|
| Niclosamide | 0.25 | 0.5 |
| Oxyclonazide | 2.0 | 4.0 |
| Rafoxanide | 4.0 | 8.0 |
| Closantel | 16 | 16.0 |
| Amoxicillin | 0.01 | 0.5 |
| Clarithromycin | 0.0025 | 0.25 |
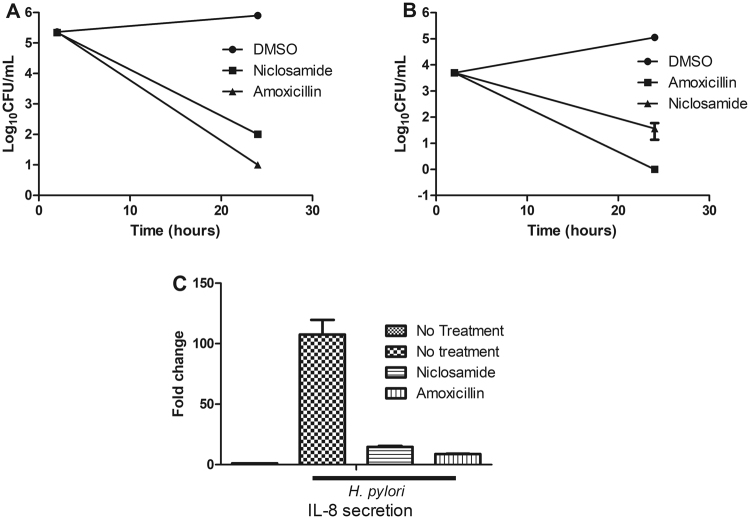
The MIC of niclosamide was the lowest of the anthelmintics tested (0.25 μg/mL, Table 1), therefore we focused on this agent and time to kill assays were used to confirm the killing properties of niclosamide against H. pylori. Bacterial cells (109/mL) exposed to the compounds at 4× MIC showed potent inhibition of the bacterial cell division relative to DMSO controls (Fig. 1). The finding that niclosamide is bacteriostatic against H. pylori is consistent with previous data reported by our research group claiming that niclosamide is bacteriostatic against MRSA13. Of note, clarithromycin, a commonly prescribed antibiotic for H. pylori treatment, is also bacteriostatic22.
Figure 1.
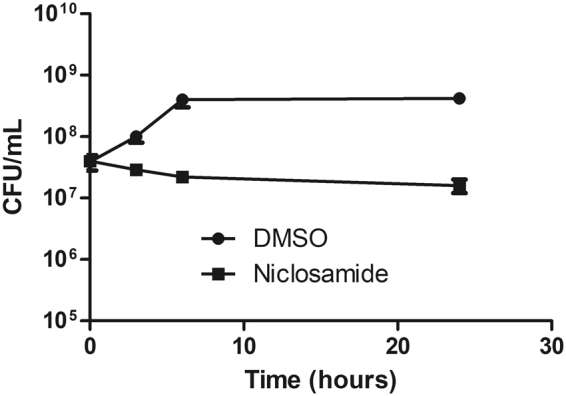
Time to kill assay. Killing kinetics of niclosamide on H. pylori at 4× MIC (1 μg/mL) was determined by a time to kill assay and was found to be bacteriostatic.
Niclosamide partially synergized with clinical molecules
Use of multiple antimicrobial agents can decrease bacterial resistance and even re-establish the clinical efficacy of certain antibiotics23. Checkerboard assays were carried out to determine whether niclosamide can act synergistically against H. pylori when paired with clinical antibiotics and PPIs. Paired combinations of compounds and their observed fractional inhibitory concentration indices (FICI) are listed in Table 2. Synergistic activity, where the combined antibacterial effect of the 2 antimicrobial agents is more than the sum of their effects alone, is defined by FICI ≤ 0.5, antagonism by FICI > 4.0 and ‘no interaction’ by FICI > 0.5–4.024. In our assays, antagonism was not observed for any of the combinations (amoxicillin, clarithromycin, metronidazole, omeprazole, and pantoprazole) with niclosamide. Niclosamide was partially synergistic with metronidazole, and the PPIs omeprazole and pantoprazole (Table 2). Notably, PPIs are known for their role in decreasing acid secretion, but they do exhibit mild anti-H. pylori activity25,26. The main purpose of PPIs in triple therapy (two antibiotics with one PPI) is to potentiate the antibacterial activity of antibiotics in a hostile gastric acid environment27. Taken together, our results indicate that niclosamide is highly active in acidic pH and can also partially synergize with omeprazole and pantoprazole (Table 2).
Table 2.
FICI value of niclosamide with other test compounds.
| Test compounds | FICI value Niclosamide |
|---|---|
| Amoxicillin | 2.0 |
| Clarithromycin | 1.0 |
| Omeprazole | 0.625 |
| Metronidazole | 0.75 |
| Pantoprazole | 0.75 |
Synergy FICI ≤ 0.5, partial synergy 0.5 < FICI ≤ 1.0, no interaction 1.0 > FICI ≤ 4.0, antagonism FICI > 4.0.
Inhibitory role on adhesion/invasion
Adhesion is the initial step in the H. pylori infection process28. In order to determine the role of niclosamide on adhesion, we allowed H. pylori to adhere to AGS cells for 2 h then administrated niclosamide for 24 h (2× MIC, 0.5 μg/mL). After incubation, we observed that the administrated concentration eliminated the adhered bacteria and reduced CFU/mL counts by 3-log10 (Fig. 2A). H. pylori is also reported as a facultative intracellular bacterium29 and it can survive in the plasma membrane of infected epithelial cells30. To determine the role of niclosamide on H. pylori invasion, a gentamicin protection assay was performed and it revealed that niclosamide (2× MIC, 0.5 μg/mL) reduced 3-log10 CFU in AGS cell lines (Fig. 2B). We found a contradiction between the killing kinetics and adhesion/invasion assays indicating that niclosamide appeared bacteriostatic during the killing kinetics and bacteriocidal during adhesion/invasion assays. We hypothesized that the variance in the efficacy of niclosamide could be due to a difference in bacterial cell concentration. The density of bacterial inoculum in the adhesion/invasion assays was lesser than the time-to-kill assay. A previously reported molecule, omeprazole, acted in a similar manner31 and it appeared bacteriostatic in high density and bactericidal in a low density of bacteria. In comparison, amoxicillin treatment (0.1 μg/mL) eliminated bacterial adhesion and invasion completely.
Figure 2.
In vitro infection assay. (A) Adhesion assay. Niclosamide removed adhered H. pylori on AGS at a concentration of 2× MIC (0.5 μg/mL). (B) Invasion assay Niclosamide prevented H. pylori invasion into AGS at a concentration of 2× MIC (0.5 μg/mL). Data represent the mean ± SEM (n = 3). (C) Real-Time PCR. Niclosamide (at 1.0 μg/mL that is 4× MIC) inhibited H. pylori-induced IL-8 secretion in MKN-28 cells.
Niclosamide treatment inhibits IL-8 secretion
H. pylori infection induces IL-8 secretion32-which, in turn, initiates neutrophil chemotaxis and activation, ultimately causing mucosal damage due to the production of reactive oxygen radicals33 and IL-8 secretion is an important factor in the immunopathogenesis of peptic ulcers and its relevant role in gastric carcinogenesis34. We carried out Real-Time (RT-PCR) to determine whether niclosamide has an inhibitory role in IL-8 secretion. As shown in Fig. 2C, niclosamide treatment (4× MIC, 1.0 μg/mL) during 24 h incubation period inhibited IL-8 secretion in H. pylori (MOI = 50) infected MKN-28 cells.
Stability of niclosamide in acidic pH
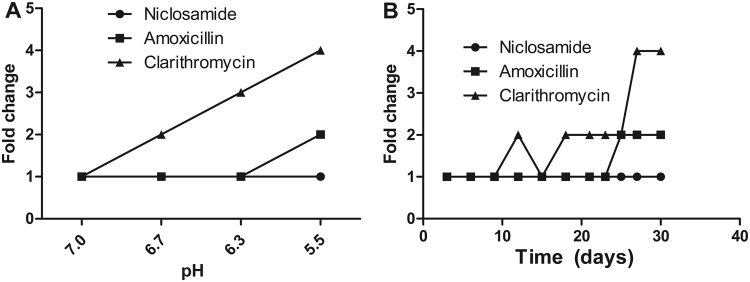
In H. pylori therapy, antibiotics need to be active in acidic pH35,36. However, certain antibiotics lose their potency in low pH37–39. Broth microdilution assays in various acidic pHs were carried out to determine the stability of niclosamide. We found that antibacterial potential of niclosamide did not change in acidic pH (tested between 5.0 to 6.5) (Fig. 3A) and the MIC of niclosamide in low pH remained the same as in neutral pH (MIC 0.25 μg/mL).
Figure 3.
Acid stability of niclosamide and H. pylori resistance to antibiotics. (A) Niclosamide inhibit H. pylori growth at acidic pH. Growth at various pH indicated that MIC of standard antibiotics has increased, but not with niclosamide. (B) Mutation frequency of H. pylori to niclosamide. H. pylori were cultured with clinical antibiotics or niclosamide for 30 days and the emergence of resistance against clinical antibiotics in H. pylori was observed, but not with niclosamide. Fold change defined as change in MIC.
Multi-step mutation frequency
H. pylori resistance to standard antibiotic treatment is increasing and the resistance rate to clarithromycin is highest in North America and amoxicillin resistance is highest in Africa40,41. To determine the potential for resistance, we passaged H. pylori treated with either niclosamide or a standard clinical antibiotic (amoxicillin and clarithromycin) for 30 days. The MIC increased 2-fold for clarithromycin and amoxicillin, while that of niclosamide remained constant through at least 10 passages (Fig. 3B). Similarly, the MIC to amoxicillin and clarithromycin increased when H. pylori bacteria were exposed to these agents for after 15 days and 24 days, respectively. More specifically, during continuous passage with amoxicillin and clarithromycin, the MIC increased by 2-fold and 4-fold, respectively (Fig. 3B), while MIC to niclosamide remained unchanged, indicating a smaller chance of emerging resistance to niclosamide during H. pylori treatment.
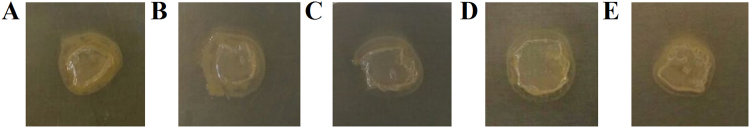
Niclosamide inhibits H. pylori motility at sub-MIC level
Initial colonization of the stomach mucosa by H. pylori is associated with the motility of bacteria42. In order to evaluate the effects, if any, of niclosamide on H. pylori motility, we performed a motility assay and found that motility decreased under niclosamide treatment at a dose-dependent manner (from 0.10 to 0.20 μg/mL). The swarming movement of H. pylori bacteria decreased gradually depending on the concentration of niclosamide. At sub-MIC (0.2 μg/mL) concentrations, the motility was abated (Fig. 4A–E).
Figure 4.
Niclosamide decreased H. pylori motility. (A–E) H. pylori was cultured on soft top agar in the presence or absence of niclosamide. Niclosamide treatment impedes H. pylori swarming movement in a dose-dependent manner. (A) DMSO; (B–E) niclosamide. (B) 75 ng/mL; (C) 100 ng/mL; (D) 150 ng/mL; (E) 200 ng/mL.
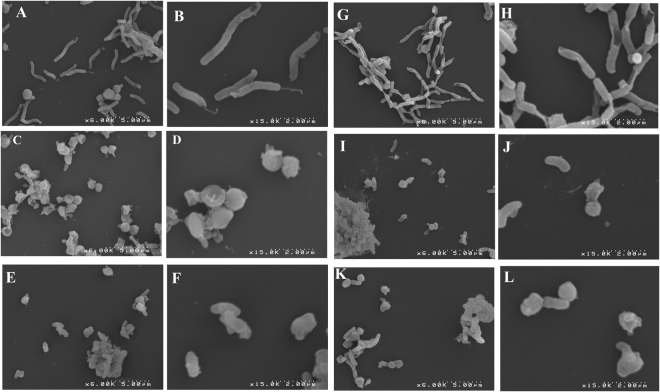
Scanning electron microscopic observations
To study further the decreased motility, we evaluated the morphology of H. pylori bacteria under a scanning electron microscope (SEM) in the presence or absence of niclosamide. The untreated H. pylori cells were helically shaped, had intact membrane surface, and possessed well-formed bacterial flagella. However, niclosamide-treated H. pylori cells became short bacilli and the morphology changed in a dose-dependent manner (Fig. 5C–H). The helical shape of the bacterium was shortened at 1× MIC (Fig. 5C,D) and appeared to become Cocco-bacillary (Fig. 5E–H) at 4× and 8× MIC. The amoxicillin (0.1 μg/mL) and CCCP (10 μM) groups also exhibited morphological changes and decreased cell size. Interestingly, treatment with niclosamide caused flagellar deformation, similar to that reported by Zhang et al.43 after exposure to the antimicrobial peptide cathelicidin. This effect in the bacterial flagella may explain the effects of niclosamide in H. pylori motility.
Figure 5.
Electron microscopic observations. (A–L) H. pylori was cultured with or without test drugs for 3 h in a microaerophilic chamber with agitation and morphological changes were observed under the SEM. Niclosamide treatment (1× , 4× , 8× MIC) caused morphological changes such as a shortened length of curved bacilli (C–H) compared to no treatment (A,B). Amoxicillin (0.1 μg/mL) and CCCP (10 μM) served as control drugs.
Light microscopy imaging for vacuolation
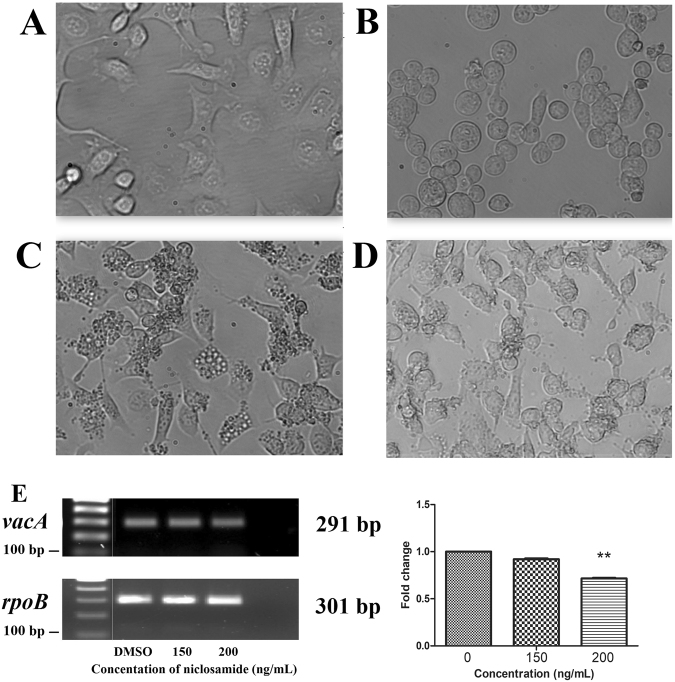
H. pylori bacteria secrete VacA toxin via a type V secretion system44. This toxin binds to host gastric epithelial cells and its internalization leads to vacuolation, characterized by the accumulation of large vacuoles in gastric epithelial cells during the infection process44,45. To demonstrate whether sub-MIC (0.2 μg/mL) concentrations of niclosamide influences toxin expression in H. pylori, RT-PCR was performed. Results indicated that at the sub-MIC level, niclosamide downregulates VacA expression (Fig. 6E). This was further confirmed by the impairment of vacuolation when niclosamide was administered to AGS cocultured with H. pylori (Fig. 6A–D).
Figure 6.
Niclosamide inhibits vacuolation. (A–D) AGS cells were infected with H. pylori (MOI-100) in the presence or absence of niclosamide and vacuolation were visualized by light microscopy imaging. Results revealed that sub-MIC (0.2 μg/mL) of niclosamide prevented vacuolation. (A) AGS alone; (B) AGS treated with niclosamide; (C) AGS cells infection with H. pylori; (D) AGS cells cocultured with H. pylori and niclosamide. (E) Niclosamide down-regulated vacA gene (291 bp) expression at sub-MIC (0.2 μg/mL), which is compared to expression of the housekeeping gene rpoB (301 bp). The grouping of gels cropped from different parts of the same gel demonstrated by the yellow line drawn on spliced position. Full-length gels are presented in Supplementary Figure S3. ***p < 0.001, students t-test comparing DMSO control.
Niclosamide role on H. pylori proton motive force (PMF)
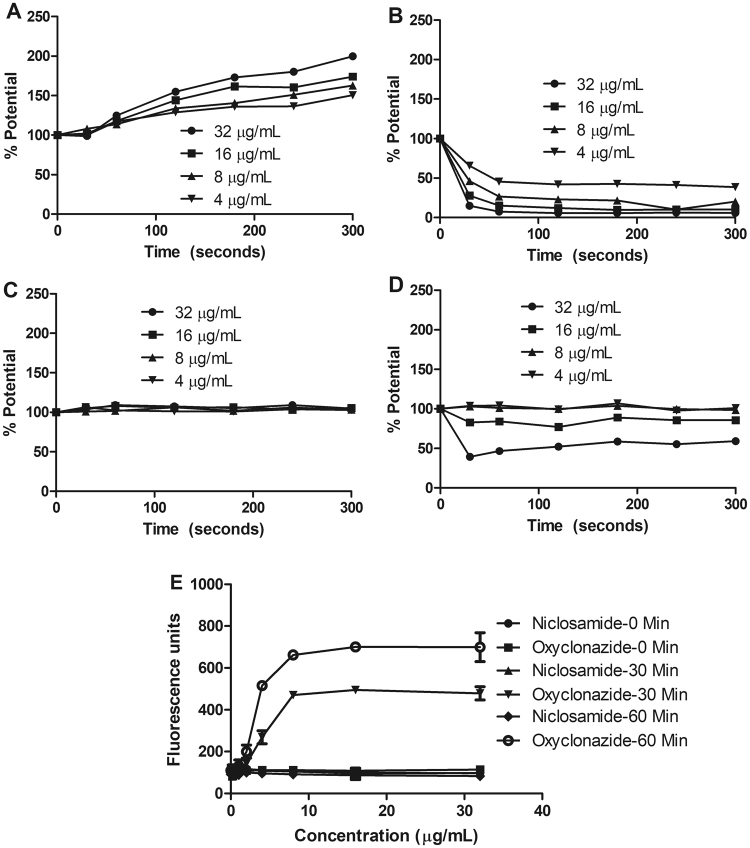
Many antimicrobial agents are known to act on bacterial membranes46 and exposure to niclosamide can disrupt the membrane potential47. We hypothesized that the mechanism of action of niclosamide is likely to disrupt H. pylori PMF. The fluorescent probe DiSC3(5) was loaded with H. pylori bacteria, which accumulates in the cytoplasmic membrane. The dissipation of PMF can be determined by either increase in fluorescence (disruption of membrane potential) or decrease in fluorescence (dissipation of transmembrane pH)48. We found that niclosamide treatment decreased the transmembrane pH of H. pylori (Fig. 7B), as determined by decreased fluorescence. In this series of experiments, we used the known protonophore, CCCP, that disrupts PMF by decreasing transmembrane pH49 as a positive control (Fig. 7D). Valinomycin (Fig. 7A), that can disrupt membrane potential, and amoxicillin (Fig. 7C), that has effects on the membrane, were also included in this study. Finally, the inner-membrane permeabilization was also monitored by uptake of the membrane-impermeable DNA-binding dye Sytox Green into H. pylori by measuring the increase in fluorescence due to drug administration. Exposure of cells to niclosamide (64 μg/mL) did not show any changes in cellular fluorescence, indicating that niclosamide does not cause any physical damage to H. pylori membranes (Fig. 7E).
Figure 7.
Membrane activity of niclosamide. (A–D) Membrane potential. Fluorescence probe Disc3(5) loaded H. pylori was treated with (A) Valinomycin; (B) Niclosamide; (C) Amoxicillin; (D) CCCP and the fluorescence was recorded before and after treatment. Niclosamide caused decreased transmembrane pH, whereas positive control agent valinomycin perturbed membrane potential. Data depicts three independent experiments. (E) Membrane permeability. H. pylori was loaded with Sytox green fluorescence probe and 0.5 h later, treated with niclosamide and the cellular fluorescence was measured. Niclosamide did not cause membrane damage of H. pylori but the oxyclozanide (positive control) cause membrane damage. Data represent the mean ± SEM (n = 3).
Cytotoxicity
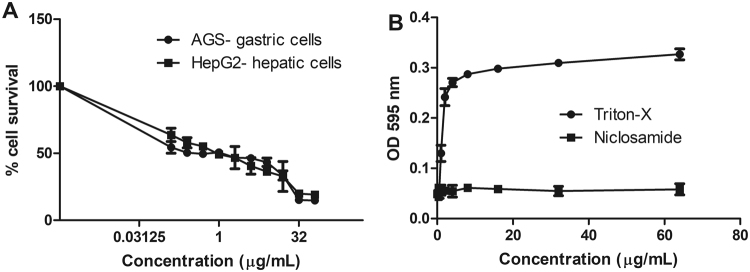
Eventhough niclosamide is approved and widely used agent, we evaluated the cytotoxicity of niclosamide against a gastric cell line (AGS), as well as human red blood cells (h-RBC) and hepatic HepG2 cells. Serially diluted Triton-X (0.001 to 1%) was included as a positive control. As expected, niclosamide caused no hemolysis, while the IC50 against the gastric cell line, was similar to that of hepatic cells (Fig. 8A–C). Notably, niclosamide is not absorbed through the intestinal wall of the host and in murine models, animals survived even after a dose of 5 mg/kg50. The U. S. Environmental Protection Agency has reported that the human oral LD50 of niclosamide is 1 gm/kg and based on the IC50 values of niclosamide in vitro51, it seems to be cytotoxic. However, according to US EPA, the human oral LD50 is much higher51 and niclosamide could eradicate H. pylori from the gastric region with low cytotoxic level.
Figure 8.
Cytotoxicity and hemolysis of niclosamide. (A) Cytotoxicity of niclosamide was assessed in HepG2 and AGS cells IC50 and was found to be 4 μg/mL. (B) Hemolysis of niclosamide was tested with h-RBC and we did not observe hemolysis. Data represent the mean ± SEM (n = 3).
The in vivo efficacy of niclosamide in the Galleria mellonella model
Giannouli et al., developed G. mellonella as an alternative model host for the evaluation of antimicrobial agents against H. pylori infection52. We monitored the surviving larvae every 24 hours, considering them dead if they lacked response to external stimuli. The niclosamide treatment significantly rescued larvae from H. pylori infection, with a survival rate up to 70% compared to the no treatment group (p < 0.0001) (Fig. 9). The clarithromycin injected group served as the positive control (up to 80% survival at 5 days).
Figure 9.
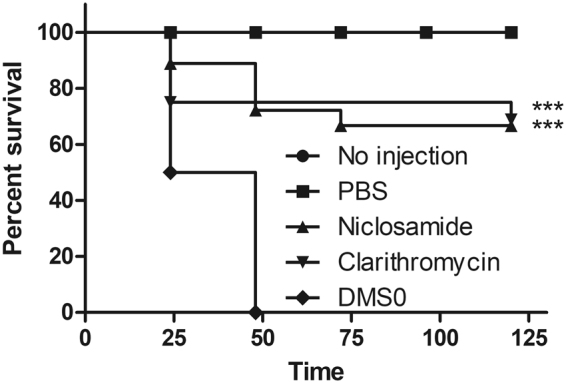
In vivo efficacy of niclosamide in the G. mellonella model. H. pylori were suspended in PBS (OD600 = 0.25) and 10 μL was injected into last left pro-leg of wax-moth. Niclosamide was administered at a concentration of 25 mg/kg and about 60% of wax-moth survived up to 120 h. Data depicts from two independent assays.
Conclusion
Repurposing is a potentially powerful approach to reveal unexplored antibacterial properties of existing clinical molecules. We found that niclosamide inhibits growth of H. pylori strain 60190 at low MIC and is effective against H. pylori at low pH. Combinatorial activity with clinical antibiotics showed an absence of antagonism. Niclosamide eliminated adhered/invaded H. pylori on gastric epithelial cells and decrease motility and IL-8 secretion. Interestingly, the MOA is mediated through the disruption of PMF and decrease of transmembrane pH. The in vitro and in vivo activity of niclosamide warrants future evaluation with other clinical strains and, eventually, evaluation in clinical trails.
Methods
Bacterial cultures
The H. pylori reference strain 60190 (ATCC 49503) was purchased from American Type Culture Collection (ATCC). Bacteria were cultured on Brucella agar (Becton Dickinson, Braintree, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Long Island, NY, USA) and H. pylori selective supplement (vancomycin – 10.0 mg/l, cefsulodin – 5.0 mg/l, trimethoprim – 5.0 mg/l, amphotericin B – 5.0 mg/l) (Oxoid, Hampshire, UK) and maintained in humidified incubators at 37 °C under an atmosphere of 5% CO2.
Antibacterial susceptibility assays
In vitro antibacterial activity was tested using the broth microdilution assay53. Experiments were carried out in triplicate using Müller-Hinton broth (BD Biosciences, Franklin Lakes, NJ, USA) supplemented with 10% FBS in 96-well plates (BD Biosciences) at a total assay volume of 100 μL. Antimicrobials used in the pilot study was selected from our previous HTS assay14,21,54 and tested against H. pylori. Anthelmentics (niclosamide, oxyclozanide, closantel, rafoxanide) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Two-fold serial dilutions were prepared between the concentration range 0.01–64 μg/mL. An initial bacterial inoculum was adjusted to OD600 = 0.06 and incubated with test compounds at 37 °C for 3 days in humidified incubators under the 5% CO2 atmosphere. OD600 was measured and the lowest concentration of compound that inhibited bacterial growth was reported as the MIC55. The broth microdilution assay was used to demonstrate the stability of the niclosamide using MHB supplemented with 10% FBS. The pH was adjusted to acidic condition with 1 N HCl. The minimal bactericidal concentration (MBC) was determined by plating 5 μL of broth culture from the MIC assay onto Müller-Hinton agar (BD Biosciences) supplemented with 10% FBS. After 72 h, the lowest concentration at which colonies were not observed was reported as the MBC.
Killing kinetic assays
The antibacterial properties of niclosamide against H. pylori were further examined using killing kinetic assays, as previously described13. Briefly, agar grown H. pylori bacteria were suspended in fresh MHB with 10% FBS to a density of 109 cells/mL and placed into 10 mL tubes (BD Biosciences). Test compounds were then added at the 4× MIC and incubated with agitation at 37 °C under microaerophilic conditions. Aliquots were periodically drawn from the tubes, serially diluted and plated onto Brucella agar (BD Biosciences) supplemented with 10% FBS. CFUs were counted after a 3-day incubation and assays were carried out in duplicate.
Checkerboard assays
Antibacterial synergy was tested using the checkerboard assay56. Niclosamide was combined with antibiotics and proton pump inhibitors (PPIs). In this series of experiments cultures of H. pylori were adjusted to OD600 = 0.06 and added to compound pairs that had been serially diluted in the same 96-well plates, vertically for one compound and horizontally for the other. Assays were carried out in triplicate as described in the antibacterial susceptibility assay sub-section. The combinatorial inhibitory concentration was indicated by the fractional inhibitory concentration index (FICI): FICI = MICA combination/MICA alone + MICB combination/MICB alone56.
Adhesion and invasion assays
The AGS (Gastric adenocarcinoma cell lines) cell line was used to examine inhibition of adhesion and invasion of H. pylori by niclosamide, as described by Schmitt et al.57. The AGS cells were grown in DMEM (Gibco), supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin (Gibco) and maintained at 37 °C in 5% CO258,59. Then, 5 × 105 cells in antibiotic and serum-free DMEM were seeded in 6-well plates 24 h prior to infection. H. pylori bacteria at a multiplicity of infection (MOI) = 100 were added and allowed to adhere to the surface of the AGS cells. Planktonic bacteria were removed after 2 h and DMEM with niclosamide (1× MIC) was then added to the wells containing the AGS. The mammalian cells were lysed by 0.1% saponin after 20 h of incubation and the adhered bacteria were then serially diluted, plated in Brucella agar supplemented with 10% FBS, and incubated as described earlier. To determine the bacterial invasion inhibition, AGS cells were treated with DMEM supplemented with 200 μg/mL gentamicin and incubated for 1.30 h to eliminate extracellular bacteria. Antibiotic and serum-free DMEM with and without test compounds were added and incubated in 5% CO2 for 20 h. Cell lysate preparation, plating, and incubation were carried out as we described in the adhesion assay. Assays were carried out in triplicate21,49.
Gene expression assays
To determine the effect of niclosamide in H. pylori VacA toxin expression, bacteria grown on agar were suspended in Brucella broth supplemented with 10% FBS (OD600 = 0.06) in dilutions of niclosamide (0, 100, 150, or 200 ng/mL) and incubated for 3 days60. After 3 days, the bacteria were washed and treated with TRIZOL (Invitrogen, Carlsbad, CA, USA), and the RNA concentration was determined using a NanoVue Plus spectrophotometer (GE Healthcare, Fairfield, CT, USA). PCR amplifications were performed and the products were analyzed by gel electrophoresis (1.0% agarose) containing SYBR® Safe (Invitrogen). Gel images were captured and analyzed using the Quantity One System. To test whether niclosamide treatment inhibits H. pylori-induced IL-8 secretion, MKN-28 cells were co-cultured with H. pylori (MOI = 100) in the presence or absence of niclosamide. At 24 hours, RNA was isolated from the infected host cells. cDNA was synthesized using the Verso cDNA (Invitrogen) synthesis kit according to manufacturer instructions. Quantitative real-time reactions were carried out in a Bio-Rad iTaq universal SYBR (Bio-Rad, Hercules, CA USA) one-step kit and a CFX98 real-time PCR cycler61. The relative gene expression was calculated from Cq values using a ΔΔCq method. The primer sequences and PCR conditions are listed in Supplementary Table S2.
Multi-step emergence of resistance
To determine whether H. pylori can develop resistance against niclosamide, bacterial cells were suspended in MHB with 10% FBS at OD600 = 0.06 in the presence of serially diluted niclosamide and incubated, as described in the antibiotic susceptibility assays sub-section. From the highest drug concentration allowing visible growth at sub-MIC concentration, aliquots were diluted (1:40) into fresh medium. These were used to inoculate the second set of serial drug dilutions as described by Dalhoff et al.62. After a 3 day incubation, H. pylori bacteria were passaged sequentially over 30 days and the OD was recorded.
Motility assays
The motility assay was performed as described previously63. In brief, Brucella agar with 10% FBS was prepared in 2 layers. The bottom layer contained pre-casted 1.5% agar and the softer top layer was comprised of 0.4% agar and niclosamide (75, 100, 150 or 200 ng/mL). Agar-grown H. pylori cells were sliced and the densely grown H. pylori agar slice was placed facing up towards the soft layer and incubated as described in bacteria and mammalian cell culture subsection64.
Scanning electron microscopy
H. pylori bacteria were suspended in Brucella broth with 10% FBS in the presence or absence or niclosamide (1x ,4x ,8x MIC) for 2 h under microaerophilic conditions with agitation. After 2 h, the bacterial cells were harvested and adhered to coverslips using a 1% poly-L-lysine (Sigma-Aldrich) solution. Then, cells were fixed in a 5% glutaraldehyde (Sigma-Aldrich), 4% paraformaldehyde (Sigma-Aldrich), 0.1 M sodium cacodylate (Sigma-Aldrich) solution. After fixation, the cells were washed in a 0.1 M sodium cacodylate buffer and dehydrated by immersion in increasing concentration of ethanol (from 30 to 100%). The critical point drying method (CPD) was applied to dehydrate the samples, the coverslips were mounted on aluminum stubs, and then sputter coated with gold (Emitech K550, Ashford, Kent, UK). Images were taken on a Hitachi 2700 Scanning electron microscope.
Light microscopy imaging for vacuolation
AGS cells were harvested and seeded into a 6 well plate at a concentration of 5 × 105 cells, 24 h before experimentation. H. pylori bacteria were harvested and washed with sterile PBS and co-cultured with AGS at a MOI of 100. After incubation for 24 h, the cells were washed, magnified under a microscope, and examined for vacuolation.
Membrane permeabilization assays
Sytox Green (Life Technologies, Carlsbad, CA, USA) was used to probe the effects of niclosamide on H. pylori membrane permeabilization, as previously described65. Assays were carried out in duplicate in 96-well plates (Corning). Bacterial cells were re-suspended in phosphate buffered saline (PBS, pH 7.4) to OD595 nm = 0.2. Sytox Green was added at a final concentration of 5 μM and cells were incubated in the dark for 30 minutes. Cell suspensions (50 μL) were added to 50 μL of compound (64 μg/mL in PBS), and the fluorescence intensity was measured (excitation 485 nm, emission 530 nm) periodically over 60 minutes. DMSO was included as the vehicle control. Membrane disruption effects of compounds were indicated by an increase in cellular fluorescence caused by the enhanced permeability of the membrane impermeable DNA staining dye.
Membrane potential
Membrane potential measurements were performed as described by Scott et al.66. In brief, H. pylori cells (OD595 nm = 0.2) were washed with a buffer (PBS, 100 mM KCl, 20 mM Glucose) and loaded with 5.0 μM of DISC3 (5) (Molecular Probes/Thermo Fisher Scientific, ON, Canada) dissolved in DMSO. The fluorescence was recorded (excitation 610 nm, emission 660 nm) periodically and valinomycin, amoxicillin, carbonyl cyanide m-chlorophenyl hydrazone (CCCP - a protonophore) were used as controls. The percentage of membrane potential change was calculated by comparing drug-treated cells with the untreated control, over all time points.
Mammalian cell cytotoxicity assays
HepG2 and AGS cells were used to test the cytotoxicity of niclosamide, as described by Kwon et al.67. Cells were grown in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin/streptomycin (Gibco) and maintained at 37 °C in 5% CO2. 5 × 104 cells in 100 μL were added to wells of 96-well plates. Compounds were serially diluted in serum and antibiotic-free DMEM and added to the monolayer and incubated at 37 °C in 5% CO2 for 24 h. At 4 h prior to the end of the incubation period, 10 μL of 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2, 4-disulfophenyl)-2H-tetrazolium (WST-1) solution (Roche, Mannheim, Germany) was added to each well. The WST-1 reduction was monitored at 450 nm using a Vmax microplate reader. Assays were performed in triplicate and the percentage survival was calculated by comparing the compound-treated wells to the DMSO-treated vehicle controls.
Human blood cell (RBC) hemolysis assays
Human erythrocytes (Rockland Immunochemicals, Limerick, PA, USA) were used to measure the hemolytic activity of the compounds, as described by Isnansetyo et al.68 Human erythrocytes (4%, in PBS, 50 μL) were added to 50 μL of serially diluted test compounds in PBS in 96-well plates. After incubating at 37 °C for 1 h, the plates were centrifuged at 500 × g for 5 min and 50 μL of the supernatant from each well was transferred to a second 96-well plate. Absorbance (540 nm) was used as a measure of hemolytic activity. Assays were carried in triplicate.
G. mellonella MRSA infection assay
Assays were performed as described by Giannouli et al.52. Twelve randomly selected G. mellonella larvae (Vanderhorst, Inc., St. Mary’s, OH, USA) between 300–350 mg were used for each group in the experiment. H. pylori cells were washed with PBS and diluted to OD600 nm = 0.3, before inoculation into G. mellonella larvae. A 10 μL inoculum was injected into the last left proleg using a 10 μL Hamilton syringe. After 2 h, compounds were administered at into the last right proleg and the wax moths were incubate at 37 °C. Three control groups - (1) injected with PBS only, (2) inoculated with H. pylori but treated with sham injections, (3) no manipulation - were included. G. mellonella survival was evaluated up to 120 h and considered dead if unresponsive to touch. Killing curves and differences in survival were analyzed by the Kaplan-Meier method using GraphPad Prism version 6.04 (GraphPad Software, La Jolla, CA, USA). Statistical analysis (Kruskal-Wallis test) was carried out using the same program.
Statistical analysis
Statistical analysis (Two-way ANOVA followed by Bonfererroni post-test) was carried out using GraphPad Prism version 6.04 (GraphPad Software, La Jolla CA, USA) and p values of < 0.05 were considered significant.
Data availability statement
No datasets were generated or analysed during the current study.
Electronic supplementary material
Acknowledgements
This study was supported by NIH grant P01 AI083214 to EM. We thank to Dr. Steven F Moss (Brown University, Providence, USA) for provided the MKN-28, AGS (Gastric adenocarcinoma cell line).
Author Contributions
N.T. and E.M. conceived the project. N.T. designed the project, carried out the laboratory experiments, analyzed the data and wrote original manuscript. J.P. performed scanning electron microscopy. D.C. performed in vivo experiments. J.P., E.M. and N.T. edited the manuscript. E.M. supervised this project. All authors reviewed an accepted the final version of this manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-22037-x.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Thung I, et al. The global emergence of Helicobacter pylori antibiotic resistance. Alimentary pharmacology & therapeutics. 2016;43:514–533. doi: 10.1111/apt.13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hagymási K, Tulassay Z. Helicobacter pylori infection: new pathogenetic and clinical aspects. World journal of gastroenterology: WJG. 2014;20:6386. doi: 10.3748/wjg.v20.i21.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham DY. Helicobacter pylori update: gastric cancer, reliable therapy, and possible benefits. Gastroenterology. 2015;148:719–731. e713. doi: 10.1053/j.gastro.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin Y-L, Chiang J-K, Lin S-M, Tseng C-E. Helicobacter pylori infection concomitant with metabolic syndrome further increase risk of colorectal adenomas. World Journal of Gastroenterology: WJG. 2010;16:3841. doi: 10.3748/wjg.v16.i30.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu, Q., Yang, Z. P., Xu, P., Gao, L. C. & Fan, D. M. Association between helicobacter pylori infection and the risk of colorectal neoplasia: a systematic review and meta‐analysis. Colorectal Disease15 (2013). [DOI] [PubMed]
- 6.Weyermann M, Rothenbacher D, Brenner H. Acquisition of Helicobacter pylori infection in early childhood: independent contributions of infected mothers, fathers, and siblings. The American journal of gastroenterology. 2009;104:182. doi: 10.1038/ajg.2008.61. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen TN, Barkun AN, Fallone CA. Host determinants of Helicobacter pylori infection and its clinical outcome. Helicobacter. 1999;4:185–197. doi: 10.1046/j.1523-5378.1999.99294.x. [DOI] [PubMed] [Google Scholar]
- 8.Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nature reviews. Cancer. 2002;2:28. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 9.Eusebi LH, Zagari RM, Bazzoli F. Epidemiology of Helicobacter pylori infection. Helicobacter. 2014;19:1–5. doi: 10.1111/hel.12165. [DOI] [PubMed] [Google Scholar]
- 10.Romano M, Cuomo A. Eradication of Helicobacter pylori: a clinical update. Medscape General Medicine. 2004;6:19. [PMC free article] [PubMed] [Google Scholar]
- 11.Shiota S, Reddy R, Alsarraj A, El-Serag HB, Graham DY. Antibiotic resistance of Helicobacter pylori among male United States veterans. Clinical Gastroenterology and Hepatology. 2015;13:1616–1624. doi: 10.1016/j.cgh.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodvold KA, McConeghy KW. Methicillin-resistant Staphylococcus aureus therapy: past, present, and future. Clinical infectious diseases. 2014;58:S20–S27. doi: 10.1093/cid/cit614. [DOI] [PubMed] [Google Scholar]
- 13.Rajamuthiah R, et al. Repurposing salicylanilide anthelmintic drugs to combat drug resistant Staphylococcus aureus. PLoS One. 2015;10:e0124595. doi: 10.1371/journal.pone.0124595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajamuthiah R, et al. Whole animal automated platform for drug discovery against multi-drug resistant Staphylococcus aureus. PLoS One. 2014;9:e89189. doi: 10.1371/journal.pone.0089189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gooyit M, Janda KD. Reprofiled anthelmintics abate hypervirulent stationary-phase Clostridium difficile. Scientific reports. 2016;6:33642. doi: 10.1038/srep33642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imperi F, et al. New life for an old drug: the anthelmintic drug niclosamide inhibits Pseudomonas aeruginosa quorum sensing. Antimicrobial agents and chemotherapy. 2013;57:996–1005. doi: 10.1128/AAC.01952-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall A. Anthelmintics: drugs for treating worms. Africa health. 1998;20:4–6. [PubMed] [Google Scholar]
- 18.Kappagoda, S., Singh, U. & Blackburn, B. G. In Mayo Clinic Proceedings. 561–583 (Elsevier) (2011). [DOI] [PMC free article] [PubMed]
- 19.Craig, C. R. & Stitzel, R. E. Modern Pharmacology with Clinical Applications. (Lippincott Williams & Wilkins, 2004).
- 20.Organization, W. H. The selection and use of essential medicines: report of the WHO Expert Committee, 2007:(including the 15th model list of essential medicines). (2007).
- 21.Jayamani E, et al. Characterization of a Francisella tularensis-Caenorhabditis elegans Pathosystem for the Evaluation of Therapeutic Compounds. Antimicrobial agents and chemotherapy. 2017;61:e00310–00317. doi: 10.1128/AAC.00310-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Francesco V, et al. Mechanisms of Helicobacter pylori antibiotic resistance: An updated appraisal. World journal of gastrointestinal pathophysiology. 2011;2:35. doi: 10.4291/wjgp.v2.i3.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torella JP, Chait R, Kishony R. Optimal drug synergy in antimicrobial treatments. PLoS Comput Biol. 2010;6:e1000796. doi: 10.1371/journal.pcbi.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Odds, F. C. (Br Soc Antimicrob Chemo, 2003).
- 25.Gatta L, et al. Antimicrobial activity of esomeprazole versus omeprazole against Helicobacter pylori. Journal of Antimicrobial Chemotherapy. 2003;51:439–442. doi: 10.1093/jac/dkg085. [DOI] [PubMed] [Google Scholar]
- 26.Adamek RJ, Szymanski C, Pfaffenbach B. Pantoprazole suppresses Helicobacter pylori without affecting cure. Helicobacter. 1999;4:266–271. doi: 10.1046/j.1523-5378.1999.99991.x. [DOI] [PubMed] [Google Scholar]
- 27.Sugiyama, T. In Proton Pump Inhibitors: A Balanced View Vol. 32, 59–67 (Karger Publishers, 2013).
- 28.Odenbreit S. Adherence properties of Helicobacter pylori: impact on pathogenesis and adaptation to the host. International journal of medical microbiology. 2005;295:317–324. doi: 10.1016/j.ijmm.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Dubois A, Borén T. Helicobacter pylori is invasive and it may be a facultative intracellular organism. Cellular microbiology. 2007;9:1108–1116. doi: 10.1111/j.1462-5822.2007.00921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan S, Tompkins LS, Amieva MR. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS pathogens. 2009;5:e1000407. doi: 10.1371/journal.ppat.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirshahi F, Fowler G, Patel A, Shaw G. Omeprazole may exert both a bacteriostatic and a bacteriocidal effect on the growth of Helicobacter pylori (NCTC 11637) in vitro by inhibiting bacterial urease activity. Journal of clinical pathology. 1998;51:220–224. doi: 10.1136/jcp.51.3.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaoka Y, et al. Induction of various cytokines and development of severe mucosal inflammation by cagA gene positive Helicobacter pylori strains. Gut. 1997;41:442–451. doi: 10.1136/gut.41.4.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, et al. Association of antral mucosal levels of interleukin 8 and reactive oxygen radicals in patients infected with Helicobacter pylori. Clinical Science. 1997;92:69–73. doi: 10.1042/cs0920069. [DOI] [PubMed] [Google Scholar]
- 34.Crabtree JE, Lindley I. Mucosal interleukin-8 and Helicobacter pylori-associated gastroduodenal disease. European journal of gastroenterology & hepatology. 1994;6:S33–38. [PubMed] [Google Scholar]
- 35.Cyphert EL, Wallat JD, Pokorski JK, von Recum HA. Erythromycin Modification That Improves Its Acidic Stability while Optimizing It for Local Drug Delivery. Antibiotics. 2017;6:11. doi: 10.3390/antibiotics6020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daarmaillac, V., Bouchard, S., Lamouliatte, H. & Mégraud, F. In Gastroenterology. A78–A78 (Wb Saunders Co Independence Square West Curtis Center, Ste 300, Philadelphia, Pa 19106–3399).
- 37.Baudoux P, et al. Combined effect of pH and concentration on the activities of gentamicin and oxacillin against Staphylococcus aureus in pharmacodynamic models of extracellular and intracellular infections. J Antimicrob Chemother. 2007;59:246–253. doi: 10.1093/jac/dkl489. [DOI] [PubMed] [Google Scholar]
- 38.Falagas ME, McDermott L, Snydman DR. Effect of pH on in vitro antimicrobial susceptibility of the Bacteroides fragilis group. Antimicrob Agents Chemother. 1997;41:2047–2049. doi: 10.1128/aac.41.9.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemaire S, Van Bambeke F, Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. Role of acidic pH in the susceptibility of intraphagocytic methicillin-resistant Staphylococcus aureus strains to meropenem and cloxacillin. Antimicrob Agents Chemother. 2007;51:1627–1632. doi: 10.1128/AAC.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghotaslou R, Leylabadlo HE, Asl YM. Prevalence of antibiotic resistance in Helicobacter pylori: A recent literature review. World journal of methodology. 2015;5:164. doi: 10.5662/wjm.v5.i3.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsumoto H, et al. Helicobacter pylori eradication with proton pump inhibitors or potassium-competitive acid blockers: the effect of clarithromycin resistance. Digestive diseases and sciences. 2016;61:3215–3220. doi: 10.1007/s10620-016-4305-0. [DOI] [PubMed] [Google Scholar]
- 42.Ottemann KM, Lowenthal AC. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infection and immunity. 2002;70:1984–1990. doi: 10.1128/IAI.70.4.1984-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, et al. Critical role of antimicrobial peptide cathelicidin for controlling helicobacter pylori survival and infection. The Journal of Immunology. 2016;196:1799–1809. doi: 10.4049/jimmunol.1500021. [DOI] [PubMed] [Google Scholar]
- 44.Galmiche A, Rassow J. Targeting of Helicobacter pylori VacA to mitochondria. Gut microbes. 2010;1:392–395. doi: 10.4161/gmic.1.6.13894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seto K, Hayashi-Kuwabara Y, Yoneta T, Suda H, Tamaki H. Vacuolation induced by cytotoxin from Helicobacter pylori is mediated by the EGF receptor in HeLa cells. FEBS letters. 1998;431:347–350. doi: 10.1016/S0014-5793(98)00788-1. [DOI] [PubMed] [Google Scholar]
- 46.Hurdle JG, O’neill AJ, Chopra I, Lee RE. Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nature Reviews Microbiology. 2011;9:62–75. doi: 10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao J, He Q, Gong Z, Chen S, Cui L. Niclosamide suppresses renal cell carcinoma by inhibiting Wnt/β-catenin and inducing mitochondrial dysfunctions. SpringerPlus. 2016;5:1436. doi: 10.1186/s40064-016-3153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farha MA, Verschoor CP, Bowdish D, Brown ED. Collapsing the proton motive force to identify synergistic combinations against Staphylococcus aureus. Chemistry & biology. 2013;20:1168–1178. doi: 10.1016/j.chembiol.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 49.Tharmalingam, N. et al. Activity of a novel protonophore against methicillin-resistant Staphylococcus aureus. Future Med Chem In press (2017). [DOI] [PMC free article] [PubMed]
- 50.Arundel, J. et al. Chemotherapy of gastrointestinal helminths. Vol. 77 (Springer Science & Business Media, 2012).
- 51.Prevention, U. S. Niclosamide EPA (1999).
- 52.Giannouli M, et al. Use of larvae of the wax moth Galleria mellonella as an in vivo model to study the virulence of Helicobacter pylori. BMC microbiology. 2014;14:228. doi: 10.1186/s12866-014-0228-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wikler, M. A. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard. (Clinical and Laboratory Standards Institute, 2006).
- 54.Tharmalingam, N. et al. Antibacterial Properties of Four Novel Hit Compounds from a Methicillin-Resistant Staphylococcus aureus-Caenorhabditis elegans High-Throughput Screen Microbial Drug Resistance (Accepted) (2018). [DOI] [PMC free article] [PubMed]
- 55.Gwisai, T. et al. Repurposing niclosamide as a versatile antimicrobial surface coating against device-associated, hospital-acquired bacterial infections. Biomedical Materials (2017). [DOI] [PMC free article] [PubMed]
- 56.Zheng, Z. et al. Synergistic efficacy of Aedes aegypti antimicrobial peptide cecropin A2 and tetracycline against Pseudomonas aeruginosa. Antimicrob Agents Chemother61 (2017). [DOI] [PMC free article] [PubMed]
- 57.Schmitt DM, et al. The use of resazurin as a novel antimicrobial agent against Francisella tularensis. Front Cell Infect Microbiol. 2013;3:93. doi: 10.3389/fcimb.2013.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tharmalingam N, et al. Piperine treatment suppresses Helicobacter pylori toxin entry in to gastric epithelium and minimizes β-catenin mediated oncogenesis and IL-8 secretion in vitro. American journal of translational research. 2016;8:885. [PMC free article] [PubMed] [Google Scholar]
- 59.Lee MH, et al. Menadione induces G2/M arrest in gastric cancer cells by down-regulation of CDC25C and proteasome mediated degradation of CDK1 and cyclin B1. American journal of translational research. 2016;8:5246. [PMC free article] [PubMed] [Google Scholar]
- 60.De Kimpe SJ, Kengatharan M, Thiemermann C, Vane JR. The cell wall components peptidoglycan and lipoteichoic acid from Staphylococcus aureus act in synergy to cause shock and multiple organ failure. Proceedings of the National Academy of Sciences. 1995;92:10359–10363. doi: 10.1073/pnas.92.22.10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng, Z. et al. Antimicrobial activity of 1,3,4-oxadiazole derivatives against planktonic cells and biofilm of Staphylococcus aureus. Future Medicinal Chemistry Accepted (2017). [DOI] [PMC free article] [PubMed]
- 62.Dalhoff A. Comparative in vitro and in vivo activity of the C-8 methoxy quinolone moxifloxacin and the C-8 chlorine quinolone BAY y 3118. Clinical infectious diseases. 2001;32:S16–S22. doi: 10.1086/319371. [DOI] [PubMed] [Google Scholar]
- 63.Suerbaum S, Josenhans C, Labigne A. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA-and flaB-negative mutants by electroporation-mediated allelic exchange. Journal of bacteriology. 1993;175:3278–3288. doi: 10.1128/jb.175.11.3278-3288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tharmalingam N, et al. Inhibitory effect of piperine on Helicobacter pylori growth and adhesion to gastric adenocarcinoma cells. Infectious agents and cancer. 2014;9:43. doi: 10.1186/1750-9378-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim W, et al. Identification of an Antimicrobial Agent Effective against Methicillin-Resistant Staphylococcus aureus Persisters Using a Fluorescence-Based Screening Strategy. PLoS One. 2015;10:e0127640. doi: 10.1371/journal.pone.0127640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scott DR, et al. Expression of the Helicobacter pylori ureI gene is required for acidic pH activation of cytoplasmic urease. Infection and immunity. 2000;68:470–477. doi: 10.1128/IAI.68.2.470-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kwon B, et al. Aberrant cell cycle reentry in human and experimental inclusion body myositis and polymyositis. Hum Mol Genet. 2014;23:3681–3694. doi: 10.1093/hmg/ddu077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Isnansetyo A, Kamei Y. MC21-A, a bactericidal antibiotic produced by a new marine bacterium, Pseudoalteromonas phenolica sp. nov. O-BC30(T), against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47:480–488. doi: 10.1128/AAC.47.2.480-488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.