Summary
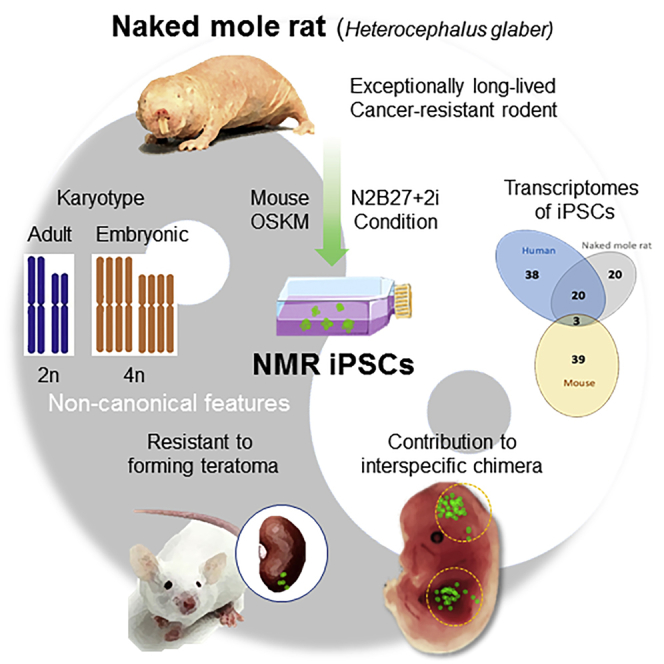
Naked mole rats (NMRs) are exceptionally long-lived, cancer-resistant rodents. Identifying the defining characteristics of these traits may shed light on aging and cancer mechanisms. Here, we report the generation of induced pluripotent stem cells (iPSCs) from NMR fibroblasts and their contribution to mouse-NMR chimeric embryos. Efficient reprogramming could be observed under N2B27+2i conditions. The iPSCs displayed a characteristic morphology, expressed pluripotent markers, formed embryoid bodies, and showed typical differentiation patterns. Interestingly, NMR embryonic fibroblasts and the derived iPSCs had propensity for a tetraploid karyotype and were resistant to forming teratomas, but within mouse blastocysts they contributed to both interspecific placenta and fetus. Gene expression patterns of NMR iPSCs were more similar to those of human than mouse iPSCs. Overall, we uncovered unique features of NMR iPSCs and report a mouse-NMR chimeric model. The iPSCs and associated cell culture systems can be used for a variety of biological and biomedical applications.
Keywords: naked mole rat, induced pluripotent stem cells, aging, lifespan, interspecific chimera, reprogramming
Graphical Abstract
Highlights
-
•
Generation of iPSCs from NMR embryonic and adult fibroblasts with high efficiency
-
•
NMR iPSCs display non-canonical features and are resistant to teratoma formation
-
•
The transcriptome of NMR iPSCs is more similar to that of human than mouse iPSCs
-
•
NMR iPSCs contribute to interspecific mouse-NMR chimera
In this article, Gladyshev and colleagues demonstrate that naked mole rat (NMR) embryonic and adult fibroblasts can be efficiently reprogrammed into induced pluripotent stem cells (iPSCs). NMR iPSCs showed unique features, were resistant to teratoma formation, and contributed to interspecific chimeric embryos.
Introduction
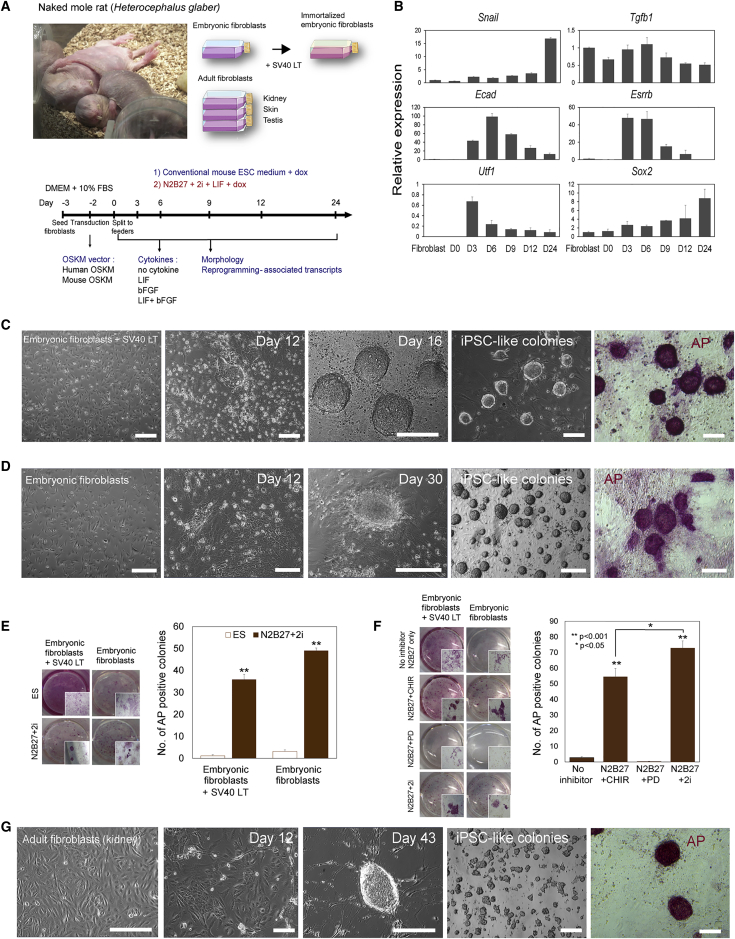
The naked mole rat (NMR; Heterocephalus glaber) (Figure 1A, top) is an emerging model organism for studies on aging and cancer. While NMRs and mice have a similar body size, the former live 10 times longer than the latter (∼30 versus ∼3 years) (Edrey et al., 2011). NMRs are also highly resistant to both spontaneous and experimentally induced cancer (Liang et al., 2010, Seluanov et al., 2009). Moreover, NMRs exhibit the absence of pain sensitization due to hypofunctional TrkA receptor (Omerbasic et al., 2016) and extreme resistance to hypoxia through fructose metabolism to avoid tissue damage (Park et al., 2017). In addition, these animals do not maintain stable body temperature, can live at low oxygen and high carbon dioxide concentrations in the atmosphere, and show other features that are useful for biomedical research (Edrey et al., 2011, Kim et al., 2011).
Figure 1.
Generation of Naked Mole Rat iPSCs
(A) Naked mole rat (Heterocephalus glaber) and fibroblasts used in this study (top). Schematic diagram of the conventional (blue) and NMR (red) reprogramming protocols (bottom).
(B) Real-time PCR analyses of reprogramming-associated gene expression up to day 24 (e.g., D1, day 1; D3, day 3, etc.). All values are mean ± SEM from three independent experiments. Relative expression was normalized to the expression of Gapdh.
(C) Generation of NMR iPSCs from embryonic fibroblasts transfected with SV40 LT. Immortalized cells, cells at day 12 after transduction, and cells at day 16 after transduction, showing iPSC-like morphology and AP staining.
(D) Generation of NMR iPSCs from embryonic fibroblasts. Embryonic fibroblasts, cells at day 12 after transduction, and cells at day 30 after transduction, showing iPSC-like morphology and AP staining.
(E) AP-positive colonies derived from NMR embryonic fibroblasts maintained in ESC and N2B27 media (upper panel). Quantification is in the lower panel. Values represent means ± SD for three independent experiments. ∗∗p < 0.001.
(F) AP-positive colonies derived from NMR embryonic fibroblasts maintained in the indicated medium in combination with inhibitors (left panel). Quantification is in the right panel. Values represent means ± SD for three independent experiments. ∗p < 0.05, ∗∗p < 0.001.
(G) Generation of NMR iPSCs from adult fibroblasts. Adult fibroblasts, cells at day 12 after transduction, and cells at day 43 after transduction, showing iPSC-like morphology and AP staining.
Scale bars, 100 μm. See also Figures S1 and S2.
Until now, tissue samples have been the main source for comparative studies involving NMRs. However, phenotypes of these animals have been difficult to assess, mainly due to the limited access to embryonic material and cell culture models. Live cell models are preferred over tissue samples in assessing and manipulating the unique features of these animals. Pluripotent stem cells could be an important biological resource for such studies. Embryonic stem cells (ESCs) exhibit pluripotency and can form any cell type (Evans and Kaufman, 1981, Martin, 1981, Brook and Gardner, 1997). However, NMRs are eusocial rodents that live in large colonies with only one breeding female. Such reproductive behavior is a major limitation for generating ESCs from blastocysts.
Induced pluripotent stem cells (iPSCs) have similar biological characteristics to ESCs, including morphology, capacity for infinite proliferation, gene expression profiles, the ability to form the derivatives of all three germ layers, and the utility for germline transmission (Takahashi and Yamanaka, 2006, Chin et al., 2009). iPSCs also offer several advantages over ESCs because they can be generated without using embryos, e.g., by manipulating adult cells. The first iPSCs were generated 11 years ago using four pluripotency factors, OCT4, SOX2, KLF4, and c-MYC (OSKM), reprogramming mouse fibroblasts (Takahashi and Yamanaka, 2006). Since then, iPSCs were generated from several mammals, including humans (Takahashi et al., 2007, Yu et al., 2007), rats (Liao et al., 2009), pigs (Esteban et al., 2009, Ezashi et al., 2009), cows (Huang et al., 2011), dogs (Luo et al., 2011), rabbits (Honda et al., 2010), horses (Nagy et al., 2011), buffalos (Deng et al., 2012), sheep (Bao et al., 2011), and bats (Mo et al., 2014). In many studies, iPSCs have served as a substitute for ESCs and a source of somatic cells. The successful generation of NMR iPSCs could bring a much-needed resource for characterizing relevant phenotypes.
Recently, Miyawaki et al. (2016) reported the generation of NMR iPSCs, found resistance to teratoma formation and attributed this feature to elevated Arf expression and mutation in the Eras oncogene. They used a conventional human culture condition to derive NMR iPSCs and found that they could be generated at high oxygen and with a low efficiency of reprogramming in the case of adult fibroblasts. Moreover, chimeric contribution of NMR iPSCs has not been examined that would further support their pluripotency.
Here, we report the development of NMR iPSCs from embryonic and adult fibroblasts using drug-inducible expression of OSKM with high efficiency. The iPSCs displayed the pluripotency and some non-canonical features such as a propensity for a tetraploid karyotype and resistance to forming teratomas. Interestingly, these iPSCs contributed to interspecific chimera despite differences in physiological temperature and phylogenetic distance. Moreover, the transcriptomes of NMR iPSCs were more similar to those of human than mouse iPSCs. These cells and the associated protocols should pave the way for generation of gene-targeted NMR models for biomedical research and provide much-needed cell culture systems to facilitate aging and cancer-related research at the cellular and molecular levels.
Results
Conventional Protocols that Support Preparation of Mouse iPSCs Do Not Favor NMR Cell Reprogramming
To reprogram NMR cells, we employed a doxycycline-inducible lentiviral system, in which mouse or human OSKM were inserted downstream of a tetracycline operator (Carey et al., 2009, Hockemeyer et al., 2008). We first used NMR embryonic fibroblasts (∼45 days postcoitum) (Figure 1A, top) and maintained them in a conventional mouse ESC medium following transduction (Figure 1A, bottom, blue letters). Reprogramming of somatic cells toward iPSCs is thought to proceed through three phases: initiation, maturation, and stabilization (Plath and Lowry, 2011). The initiation phase is marked by the mesenchymal-to-epithelial transition (MET) and bone morphogenic protein signaling (Li et al., 2010, Samavarchi-Tehrani et al., 2010), and E-cadherin impeding reprogramming (Chen et al., 2010). Esrrb and Utf1 represent early markers that predict an eventual reprogramming event, whereas endogenous Sox2 is a late-phase reprogramming factor (Buganim et al., 2012).
To gain insights into MET and NMR cell reprogramming, we quantified NMR-specific E-cadherin (Ecad), Esrrb, Utf1, and Sox2 transcripts on day 6 following transduction (Figure 1A; bottom). Mouse OSKM-transduced NMR cells showed higher expression of these transcripts compared with cells transduced with human OSKM, although both approaches induced expression of the marker genes. In particular, the Ecad levels were 30-fold higher than in control fibroblasts (Figure S1A). We further found that cytokine treatment increased the expression of reprogramming-related genes, with LIF (leukemia inhibitory factor) being a stronger inducer than basic fibroblast growth factor (Figure S1B). Hence, mOSKM and LIF were chosen for further experiments. We screened for changes in marker gene expression until day 24 and also analyzed the initial reprogramming genes Snail and Tgfb1 (Figure 1B). Snail expression increased starting from day 3 and was maximal at day 24. Tgfb1 increased at day 6 and gradually decreased until day 24. Ecad, Esrrb, and Utf1 were dramatically increased from days 3–6 and gradually decreased to day 24. Sox2 was gradually increased to day 24. Thus, transcription factors and cytokines could alter reprogramming-associated gene expression in NMRs. With the same method, we generated mouse iPSCs from embryonic and adult fibroblasts (Figures S1C and S1D). However, NMR cells showed no visible morphological changes until day 24 (Figure S1E), when we detected OCT4-expressed cells (Figure S1F). Nevertheless, this approach did not result in viable ESC-like NMR colonies, suggesting that the conventional protocols, which readily support preparation of mouse iPSCs, are unsatisfactory for deriving NMR iPSCs.
Development of Optimal Protocols to Support Generation of NMR iPSCs
SV40 large T antigen has been reported to improve the efficiency of iPSC generation (Park et al., 2008). Reducing p53 expression can also improve this process (Mali et al., 2008, Utikal et al., 2009, Hong et al., 2009, Hanna et al., 2009a). In fact, SV40 large T antigen may support iPSC generation by inhibiting p53 expression (Bao et al., 2011). Also, unlike mouse cells, rat ESCs and iPSCs require specific culture conditions, such as serum-free defined culture medium (N2B27) with inhibition of the MEK (mitogen-activated protein kinase)/ERK (extracellular signal regulated kinases 1 and 2) pathway and glycogen synthase kinase 3 (GSK3) by small synthetic drugs PD0325901 (PD) and CHIR99021 (CHIR), respectively. These culture conditions (N2B27+2i) were also beneficial for establishing and maintaining ground state pluripotent cells from other species, including mouse and human (Ying et al., 2008, Hanna et al., 2009b, Buehr et al., 2008).
We applied the N2B27+2i culture model to NMR embryonic fibroblasts and immortalized embryonic fibroblasts by inserted SV40 large T antigen in combination with the lentiviral system (Figure 1A, bottom). Remarkably, 16 days after transduction, typical ESC colony-like cells were observed in immortalized embryonic fibroblasts (Figure 1C). These colonies showed strong alkaline phosphatase (AP) activity, suggesting pluripotency (Figure 1C). Interestingly, in the case of normal embryonic fibroblasts, the first colonies were visible at day 30 (Figure 1D). NMR iPSCs could be routinely passaged on feeder systems every 5 days. These colonies exhibited round, tightly packed morphology characterized by a high ratio of nucleus to cytoplasm and prominent nucleoli (Figures 1C and 1D), similar to rat iPSCs. To optimize the protocol, we cultured OSKM-transduced cells in various culture conditions and performed AP staining. These analyses confirmed the difference between conventional ESC and N2B27+2i media (Figure 1E). Clearly, N2B27+2i media dramatically induced AP-positive colonies with typical ESC-like morphology, suggesting that this culture condition is particularly effective for reprogramming NMR cells.
We next examined the effect of inhibitors on AP-positive colony formation using N2B27+ LIF with or without PD or CHIR. The number of AP-positive colonies in the presence of CHIR and 2i was much higher than under other conditions, supporting a role of CHIR in reprogramming (Figure 1F). We also examined the effect of cytokines, with LIF representing the most effective factor (Figure S2A). On the other hand, the reprogramming efficiency was influenced by neither feeder/feeder-free monolayer culture (Figure S2B) nor ascorbic acid (Figure S2C). After serial passaging of ESC-like colonies, extensive spontaneous differentiation was observed. An inhibitor of type 1 transforming growth factor β receptor ALK5 (A83-01) can help maintain rat iPSCs (Li et al., 2009), and we found that it supported homogeneous population and prevented spontaneous differentiation of NMR iPSCs (Figure S2D). We also observed that the expression of SV40 large T accelerated reprogramming of NMR iPSCs, e.g., the reprogramming time was reduced in immortalized fibroblasts compared with normal fibroblasts (16 and 30 days, respectively).
We further applied these culture conditions to reprogram adult somatic fibroblasts isolated from the kidney of a 1-year-old male NMR. In this case, the first colony was visible on day 43, suggesting that the reprogramming period was extended compared with embryonic fibroblasts (Figure 1G). The reprogramming efficiency was 0.17% ± 0.01% for immortalized embryonic fibroblasts, 0.24% ± 0.01% for normal embryonic fibroblasts (embryonic), and 0.085% ± 0.0132% for adult somatic fibroblasts. We also succeeded in generating iPSCs from adult skin and testis cells (Figure S2E), suggesting that the procedure we developed represents a robust protocol for derivation of stable NMR iPSCs. Real-time PCR analysis using primers specific for OSKM transgenes showed no expression of exogenous Oct4 and c-Myc after removal of doxycycline from culture medium (Figure S2F). Taken together, we established effective conditions for cell reprogramming and for maintaining NMR iPSCs.
Pluripotent Characteristics of NMR iPSCs
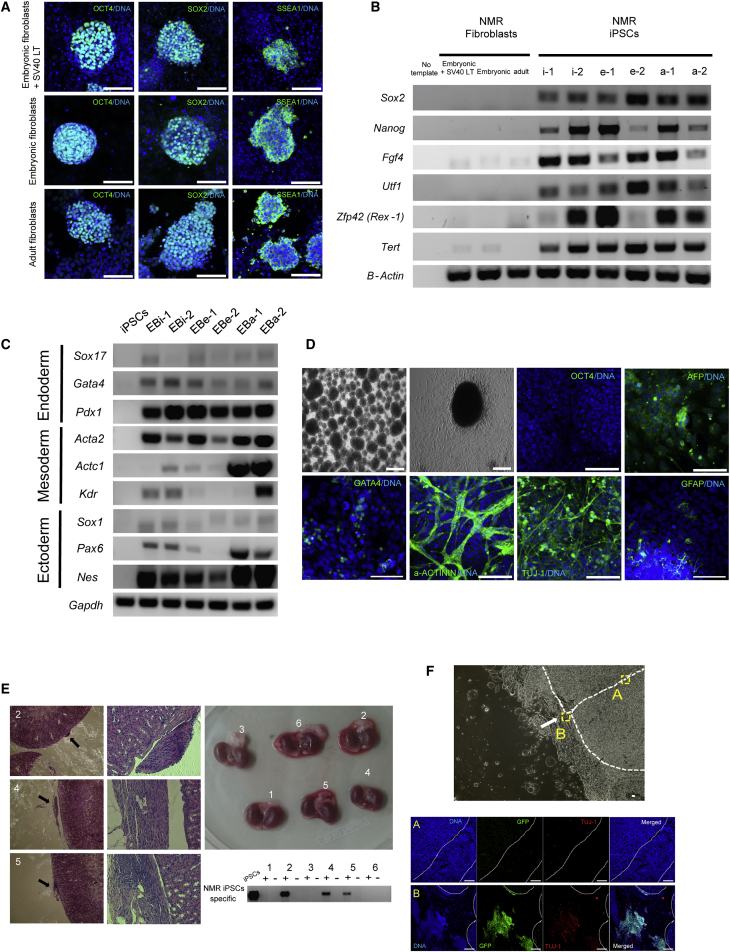
The generated NMR iPSCs could be stably passaged for more than 30 generations. They exhibited pluripotent cell markers, including OCT4, SOX2, and SSEA1 (Figure 2A). RT-PCR analysis revealed upregulation of several endogenous pluripotent genes (Sox2, Nanog, Fgf4, Utf1, Zfp42, and Tert) in NMR iPSCs (Figure 2B). To test for differentiation of NMR iPSCs in vitro, we allowed them to form embryonic bodies (EBs) and undergo differentiation for 7 days, and then analyzed for the presence of markers characterizing each germ layer. NMR iPSCs were able to differentiate into all three germ layers, as demonstrated by high expression of Sox17 (endoderm), Gata4 (endoderm), Pdx1 (endoderm), Acta2 (mesoderm), Actc1 (mesoderm), Kdr (ectoderm), Sox1 (ectoderm), Pax6 (ectoderm), and Nes (Nestin; ectoderm) (Figure 2C). We further examined in vitro differentiation of NMR iPSCs by immunocytochemistry. NMR EBs were transferred onto gelatin-coated plates for continued cultivation, and the attached cells exhibited different morphologies. Immunocytochemistry analyses detected cells positive for alpha-fetoprotein (endoderm), GATA4 (endoderm), alpha-actinin (a-ACTININ; mesoderm), βIII-tubulin (TUJ-1; ectoderm), and glial fibrillary acidic protein (ectoderm) (Figure 2D).
Figure 2.
Characterization of NMR iPSCs
(A) Representative images of NMR iPSC colonies from cells of indicated origin following immunostaining for OCT4, SOX2, and SSEA-1. Scale bars, 100 μm.
(B) RT-PCR analysis of endogenous pluripotent genes expressed in NMR iPSCs. (i) iPSCs from immortalized fibroblasts; (e) iPSCs from embryonic fibroblasts; (a) iPSCs from adult fibroblasts; origin, two independently established clones.
(C) RT-PCR analysis of genes expressed in EBs derived from NMR iPSCs (Immortalized; EBi, embryonic; EBe, adult; EBa origin: two independently established NMR iPSC lines; iPSCs: undifferentiated iPSC line).
(D) In vitro differentiation of NMR iPSCs. EB formation. Immunocytochemical analyses of EB expression: pluripotency marker (OCT4; negative control), endoderm (alpha-fetoprotein [AFP] and GATA4), mesoderm (a-ACTININ), and ectoderm (TUJ-1 and glial fibrillary acidic protein [GFAP]) markers. Scale bars, 100 μm.
(E) Teratoma assay involving NMR iPSCs. Sections of kidney capsules of mice transplanted with NMR iPSCs (five tetraploid and one diploid iPSC; no. 2 kidney was injected with the diploid iPSCs) after 10 weeks (left panel; upper right). Some nerve-like cells were observed on the cortex (left panels, shown by arrows; magnified in the middle panels). H&E staining is also shown. Numbers represent individual kidney samples. Genomic PCR analyses show that the nerve-like cells were of the NMR origin (lower right panel). +, injected kidney capsule; −, non-injected kidney capsule in this panel.
(F) Teratoma assay involving EGFP-expressed NMR iPSCs. The white dashed line shows the border between the kidney capsule and the site of the injection scar. The arrow indicates the injection site of NMR iPSCs into kidney capsules. A, regions of inner cortex; B, regions of injection site on the cortex. Immunocytochemical analyses of EGFP cell expression: neuronal marker (TUJ-1). Scale bars, 100 μm.
NMRs exhibit an unusual mode of thermoregulation in that they are endothermic and capable of employing non-shivering thermogenesis (Buffenstein and Yahav, 1991). NMR fibroblasts grow poorly at the standard 37°C conditions, perhaps because the animals have a body temperature several degrees lower in their natural environment (Lewis et al., 2012). Prior to teratoma assays and chimera production, we examined the effect of high temperature on maintenance of NMR iPSCs and the process of differentiation in vitro. The number of viable NMR iPSCs significantly decreased at 37°C after culturing them for 10 or 20 days (Figure S3A). When we cultured NMR iPSCs in a normal fibroblast medium without pluripotent environment (LIF and 2i), the number of viable cells decreased at day 30, but no significant differences were observed in comparison with the 32°C environment (Figure S3B). We also compared the formation of EBs between 32°C and 37°C, but there was no difference in morphology (Figure S3C) and the number of EBs (Figure S3D). These EBs were differentiated to three germ layers as revealed by immunocytochemistry (Figure S3E). Together, we found that elevated temperature (37°C) led to a reduced number of NMR iPSCs and their derivatives during differentiation in vitro. Nevertheless, some surviving cells could differentiate into all three germ layers even at elevated temperature.
To determine whether NMR iPSCs displayed in vivo pluripotency, six lines of NMR iPSCs were injected into renal capsules of six severe combined immunodeficiency (SCID) mice. No teratomas were detected 12 weeks following injection. However, nerves were found on the cortex in half of the injected kidneys, as revealed by histological examination and NMR-specific sequences (Figure 2E, left panel). We also used EGFP-expressed iPSCs (Figure S4A) for teratoma assay and found that these cells were positive for a neuronal marker (TUJ-1) by immunocytochemistry (Figure 2F). Thus, these nerve cells were derived from the NMR iPSCs rather than from mouse cells, and this happened without tumor formation.
Propensity for a Tetraploid Karyotype in NMR Fibroblasts and iPSCs
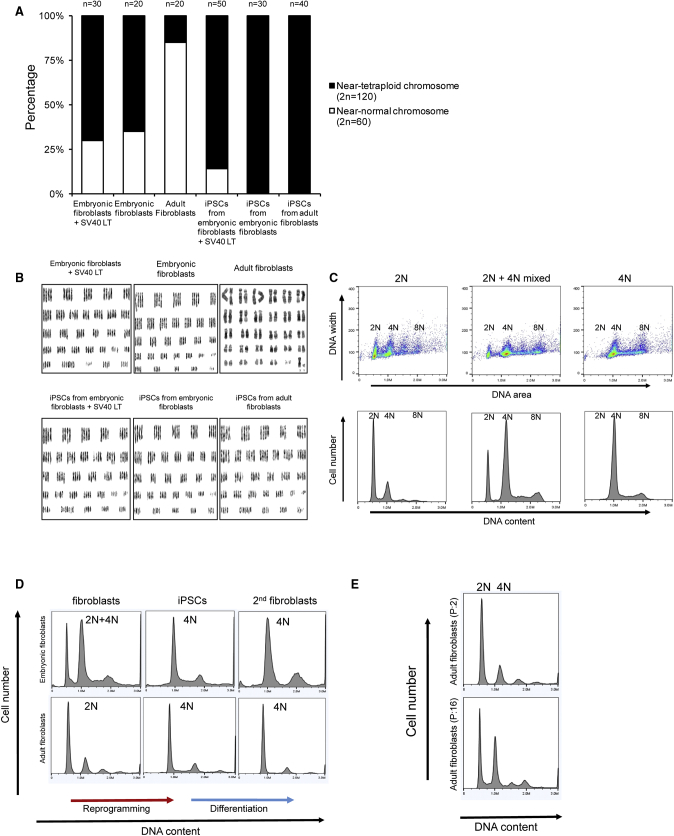
Karyotype abnormality is often associated with quality of iPSCs. We karyotyped 12 NMR iPSCs lines, which were derived from three different fibroblast lines, at passage 10. Except for one line (2N = 60 chromosomes) derived from immortalized fibroblasts, NMR iPSCs lines showed a sign of tetraploidy (2N = 120 chromosomes) (Figure 3A). We also karyotyped the seven fibroblast lines that were used for generation of NMR iPSCs. Interestingly, embryonic fibroblasts also showed the tetraploid karyotype, while adult fibroblasts exhibited primarily normal karyotype (Figures 3A and 3B). To clarify these characteristics of NMR cells, we performed DNA content analysis in fibroblasts and their respective iPSCs by fluorescence-activated cell sorting. Embryonic fibroblasts exhibited diploid (2N), tetraploid (4N), and mixed diploid/tetraploid DNA, while adult fibroblasts were primarily diploid (2N) (Figure 3C). These findings support the idea that NMRs may rely on the increased use of tetraploid cells. We noted that iPSCs derived from embryonic fibroblasts did not change DNA content, whereas iPSCs from adult fibroblasts increased their tetraploidy (4N). The DNA content of each iPSC line did not change during differentiation (Figure 3D). We also found that 4N cells increased during long-term culture of fibroblasts (Figure 3E). We did not observe differences between NMR tetraploid and diploid iPSCs when they were grown at 32°C and 37°C (Figures S3A and S3B) or subjected to the teratoma assay (Figure 2E, left panel). Overall, NMR embryonic fibroblasts and iPSCs showed a high propensity for the tetraploid karyotype.
Figure 3.
Propensity of NMR Fibroblasts and iPSCs for a Tetraploid Karyotype
(A) Percentage of diploid and tetraploid NMR fibroblasts and iPSCs based on karyotype analyses. n, number of cells with chromosome counts. Seven NMR fibroblast and 12 NMR iPSC lines were analyzed.
(B) Karyotype analysis showing tetraploid karyotypes of NMR embryonic fibroblasts and iPSCs. Adult fibroblasts show a mostly diploid karyotype.
(C) DNA content of NMR embryonic fibroblasts and iPSCs as analyzed by fluorescence-activated cell sorting.
(D) Changes in the DNA content during reprogramming and differentiation in embryonic and adult cells.
(E) Changes in the DNA content of NMR adult fibroblasts during long-term culture.
Contribution to Chimera Embryogenesis
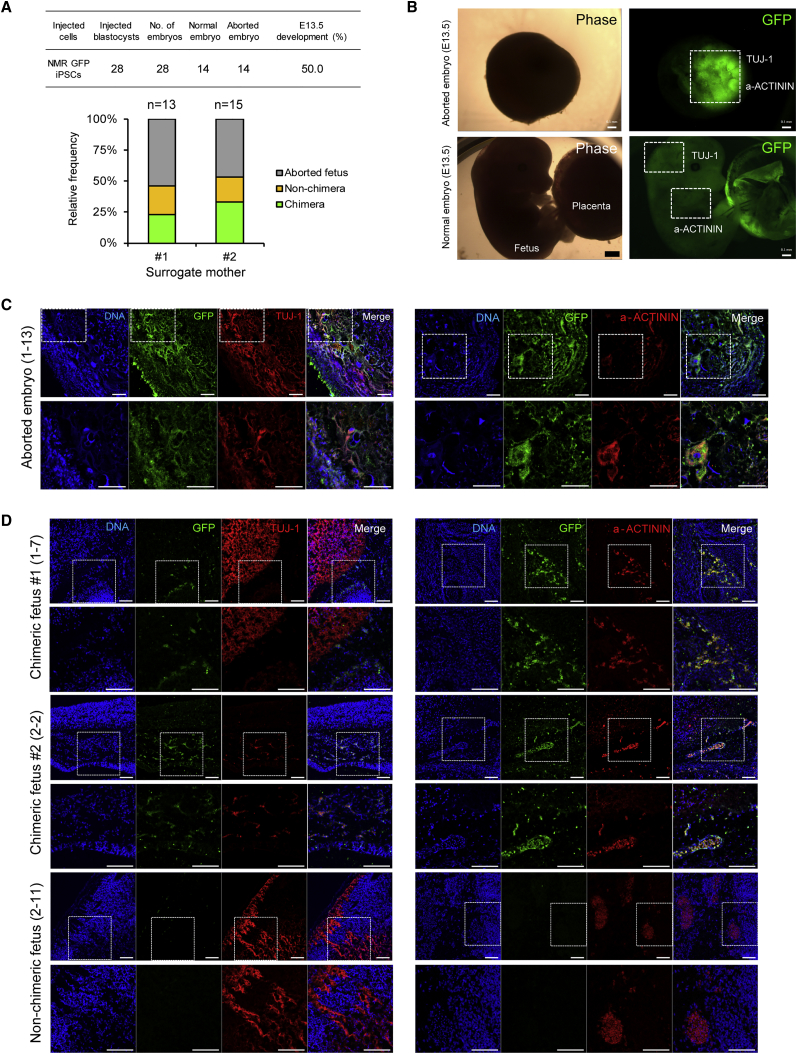
To test for the ability of NMR iPSCs to generate interspecies chimera in vivo, we prepared NMR iPSCs that expressed EGFP (Figure S4A) and exhibited pluripotent cell makers (Figure S4B). These cells were microinjected into embryonic day 3.5 (E3.5) blastocysts of C57BL/6 mice and allowed to develop to the E13.5 (Figure S4C). After dissection, a total 28 embryos were obtained from two surrogate mothers, with an equal number of normal and aborted embryos (Figure 4A; upper). To confirm the interspecies chimerism of NMR iPSCs, genomic DNA extracted from normal and aborted embryos was PCR-amplified using NMR iPSCs-specific primers (Figure S4D). About a quarter of normal embryos from two surrogated mothers were confirmed to be interspecific chimera by genomic PCR (Figure 4A, bottom). Compared with previous data in rat-mouse interspecific chimera production (Kobayashi et al., 2010), the percentage of NMR iPSCs chimerism in mouse fetus was similar, even though there are species-specific developmental patterns of NMRs, rats and mice. Whole-mount immunostaining with anti-GFP antibodies revealed the presence of NMR iPSC-derived cells in chimeric embryos (Figures S4D and S4E). Aborted embryos generally showed higher integration of GFP-expressing cells (Figure S5D). Intriguingly, we found that NMR iPSC-derived cells could integrate into both placenta and normal fetus (Figure S5E), suggesting their contribution to embryonic and extra-embryonic tissues. We further examined the identity of these GFP-positive cells by immunostaining with different lineage markers such as TUJ-1 (ectoderm) and a-ACTININ (mesoderm) (Figure 4B). In several chimeric embryos, we found that GFP-positive cells co-expressed appropriate lineage-specific markers in aborted embryo and normal fetuses (Figures 4C and 4D). Together, these data suggest that NMR iPSC-derived cells could repopulate into the mouse early embryos with further differentiation and contribute to interspecies chimera.
Figure 4.
Interspecific Chimera
(A) Summary of interspecies chimera production (E13.5). Relative frequencies of aborted, non-chimeras, and chimeras during embryonic development from two surrogated mothers (bottom panel). Data from genotyping results. See also Figure S4D.
(B) Representative images showing an aborted embryo and a normal embryo (E13.5). White boxes indicate the sagittal section of brain and heart regions in a normal fetus (bottom panel). Scale bars, 100 μm.
(C) Representative images showing the integration of NMR iPSC-derived cells into mouse E13.5 aborted embryos (embryo number; 1–13 from Figure S4D). Anti-GFP antibody was co-stained with anti-TUJ1 (left panels) and anti-a-ACTININ (right panels) antibodies. The bottom panels are enlargements of the white boxes. Scale bars, 100 μm.
(D) Representative images showing the integration of NMR iPSC-derived cells into mouse E13.5 normal fetuses (embryo number; 1-7 and 2-2 from Figure S4D). Non-chimera as control (embryo number; 2-11 from Figure S4D). Anti-GFP antibody was co-stained with anti-TUJ1 (left panels) and anti-a-ACTININ (right panels) antibodies. The bottom panels are enlargements of the white boxes. Scale bars, 100 μm.
Comparative Gene Expression Analyses of Mouse, Human, and NMR iPSCs
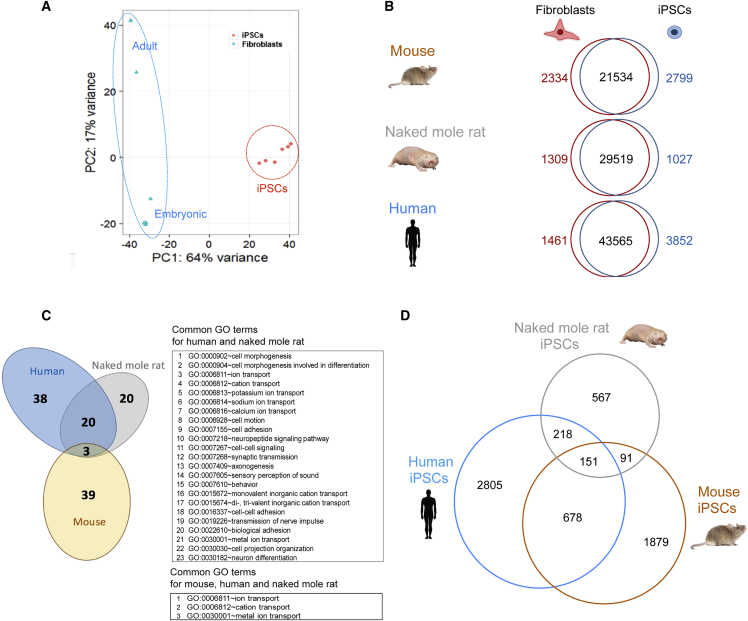
To gain insights into similarities and differences in gene expression among mouse, human, and NMR iPSCs, we performed RNA sequencing (RNA-seq) analyses of six lines of fibroblasts and their respective iPSC lines of the NMR. Principal-component analysis revealed tight clustering of all iPSCs and their split from the cells of origin (Figure 5A). Next, we performed RNA-seq analyses of four mouse fibroblast lines and the respective iPSCs, which were generated in parallel with the NMR iPSCs. We also used a published human fibroblast/iPSC dataset (Choi et al., 2015). First, we identified differentially expressed genes (DEGs) between fibroblasts and iPSCs for each species (Figure 5B), revealing elevated expression of 2,799 mouse, 1,027 NMR, and 3,852 human genes in iPSCs. These corresponded to 42 gene ontology (GO) terms in mouse, 43 in NMR, and 61 in human (Figures S5A–S5C). Interestingly, 23 GO terms overlapped between NMR and human iPSCs, which were related to cell morphogenesis, ion transport, and neuronal function, whereas only 3 GO terms were common to mouse and NMR iPSCs (Figure 5C). Next, we found 151 DEGs overlapped among NMR, human, and mouse iPSCs by gene symbol (Figure 5D). Excluding common 151 DEGs, only 91 DEGs overlapped between NMR and mouse iPSCs, whereas 218 DEGs overlapped between NMR and human iPSCs. The data suggest that the reprogramming-associated transcriptome changes in NMR cells are more similar to those of human cells.
Figure 5.
Gene Expression Analysis of Mouse, NMR, and Human iPSCs
(A) Principal-component analysis of global gene expression patterns of NMR fibroblasts and iPSCs derived from them.
(B) Venn diagrams showing pairwise comparison of gene expression. Red and blue numbers denote differentially expressed genes (false discovery rate < 0.05 and fold-change > 2); black numbers denote expressed genes with no significant differences between indicated samples.
(C) Venn diagram showing common GO terms among NMR, human, and mouse. The set of 23 (NMR/human) and 3 (NMR/mouse or human/mouse) GO terms is shown. See also Figure S5.
(D) Venn diagram showing unique and common differentially expressed genes in NMR, human and mouse iPSCs.
Discussion
We report the development and characterization of NMR iPSCs and describe protocols for robust preparation of these cells. The NMR is a mouse-sized rodent, but viable NMR iPSCs could not be generated by employing a conventional system that supports mouse iPSCs derivation. Although we observed altered expression of reprogramming-associated genes (Ecad, Esrrb, Utf1, and Sox2) and detected OCT4-expressing cells, this procedure did not lead to reprogramming. However, further analyses revealed the importance of N2B27 medium for NMR iPSC derivation. We also found that LIF+2i conditions, which activate LIF/STAT3 and Wnt/β-catenin signaling, inhibit FGF/MEK, and support naive PSCs across species (Ye et al., 2014), were necessary and sufficient for inducing and maintaining pluripotency of NMR cells. Also, N2B27-3i was superior to N2B27-2i, highlighting positive influence of the ALK5 inhibitor A83-01, as previously reported for rat iPSCs (Li et al., 2009). In addition, we examined appropriate conditions for feeder-free monolayer culture and found that NMR iPSCs grown on Matrigel maintained AP expression. This finding opens opportunities for feeder-free iPSC culture, transfection, and cell differentiation without EB formation. We further found that the expression of SV40 large T accelerated reprogramming of NMR iPSCs. Although this factor did not seem to alter the reprogramming efficiency (0.17% versus 0.24%), the reprogramming time was significantly reduced (16 versus 30 days).
The resulting iPSCs could be maintained for prolonged periods of time, with no notable difference in morphology after 30 passages. These cells expressed typical markers of pluripotency and differentiated into all three germ layers. On the other hand, some phenotypes of the NMR iPSCs were different from those of iPSCs from other species. For example, we found that NMR iPSCs were resistant to forming teratoma when injected into the mouse renal tubule, instead forming nerves, consistent with tumor resistance from early development stages. It is interesting that NMR iPSCs may be able to differentiate in vivo without forming tumors. It is an attractive possibility that these approaches may lead to the development of tumor-free iPSCs for therapeutic applications. Besides, NMR iPSCs showed a propensity for tetraploidy, which was observed in various culture models and was more prevalent in embryonic cells. Although tetraploidy may be associated with abnormality in cells, in the NMR this phenotype appeared regardless of cells and conditions used. Inappropriate tetraploidization is a frequent event in early stages of cancer development (Davoli and de Lange, 2011). However, despite a high percent of tetraploid cells, NMR iPSCs contributed to interspecific chimera and did not give rise to teratoma. In the mouse, tetraploid ESCs possess essential pluripotency, differentiation potency to form teratoma, and show contribution to the inner cell masses in aggregated chimeric blastocysts (Imai et al., 2015). Besides, the viable tetraploid mammal Tympanoctomys barrerae (Octodontidae) displays genomic imprinting and X-chromosome inactivation, which is partially conserved in the tetraploid genome (Bacquet et al., 2008). Future comprehensive epigenetic or genomic analyses are necessary to elucidate these characteristics of embryonic fibroblasts and iPSCs in the NMR. These characteristics may be related to known NMR phenotypes, e.g., early contact inhibition and resistance to cancer.
When our study was prepared for publication, another group reported the development of NMR iPSCs (Miyawaki et al., 2016). They also found resistance to teratoma formation and attributed this feature to elevated Arf expression. We used a different procedure to derive iPSCs and found that they could be generated at low oxygen and with a higher (∼30-fold) efficiency of reprogramming in the case of adult fibroblasts. Most importantly, we demonstrated chimeric contribution of NMR iPSCs for robust evidence of pluripotency. The availability of different iPSC reagents, especially since they differ in their characteristics, should be of great benefit to the research community.
The teratoma phenotype of our iPSCs prompted us to examine the ability of NMR iPSCs to contribute to embryonic development of mice. Interspecific chimera were generated several years ago using mouse blastocysts injected with rat iPSCs and rat blastocysts injected with mouse iPSCs (Kobayashi et al., 2010). Both mouse and rat iPSCs could contribute to various cell types of the embryos and adult animals (Kobayashi et al., 2010). More recently, chimera models were developed involving rhesus monkey and human cells (Fang et al., 2014, Mascetti and Pedersen, 2016). We found that NMR iPSCs could contribute to embryonic development when injected into mouse blastocysts, followed by implantation. Although NMRs are characterized by slower development (∼70-day gestation period) compared with mouse (19–21 days), apparently NMR cells could divide and differentiate within mouse embryos, at least until E13.5. They were also detected in the placenta and contributed to fetus through different lineages such as ectoderm (TUJ-1) and mesoderm (a-ACTININ). To produce interspecific chimera, genetic diversification and evolutionary distance should be considered as they form a “xenobarrier” (Wu et al., 2016). Interestingly, the chimeric contribution of NMR cells to the mouse embryo was successful despite the great evolutionary distance (70 million years). Analysis of chimeric embryos suggested a possibility of naive pluripotency of the NMR iPSCs, and an opportunity of production of fully developed interspecific chimera between mouse and NMR.
We also examined gene expression changes associated with reprogramming of NMR cells, and these were more similar to those of human than mouse iPSCs. Interestingly, despite the fact that the ancestor of mouse and NMR diverged from the ancestor of humans about 93 million years ago, NMR iPSCs share more common GO terms with human iPSCs than with mouse iPSCs. Although NMR is a rodent, NMRs and humans are characterized by extended lifespans and longer gestation periods, especially compared with what would be expected based on their body sizes.
Our study highlights potential uses of the developed NMR iPSCs for various types of biomedical applications. We showed that they could differentiate into various cell types, be implanted into embryos, and be manipulated genetically despite their propensity for tetraploidy. Previously, studies involving NMRs were limited to the analyses of live animals and tissue samples. Very few cell culture studies have been performed, and they were limited to adult fibroblasts. In addition, no cell lines have been reported for NMR, precluding characterization of the remarkable phenotypes associated with this animal. With the development of NMR iPSCs, some of the major hurdles in NMR research may be overcome, accelerating studies on the phenotypes of these extraordinary animals.
Experimental Procedures
Animal Ethics
The animal welfare, use, and care were carried out according to the protocols approved by the Institutional Animal Care and Use Committee of the Brigham and Women's Hospital, Harvard University, and University of Illinois-Chicago.
Isolation and Culture of Embryonic and Adult Fibroblasts
NMRs used in this work were from the facility at University of Illinois in Chicago. The NMR embryos were harvested at the early stage of fetogenesis, ∼45 days postcoitum. Each fetus was harvested and dissected, voided of internal organs and brain, and the remaining part was washed in sterile PBS to remove remaining blood. Each fetus was teased into fine pieces in 5 mL of 0.25% trypsin-EDTA solution, transferred into a 15 mL tube, and digested overnight at 4°C. DMEM with 20% fetal bovine serum (FBS) was added to inactivate trypsin and tissues were pipetted vigorously and repeatedly to break up the digested tissues into a cell suspension. The cells were plated onto cell culture plates and incubated in a humidified incubator at 32°C with 3% O2 and 5% CO2 atmosphere. Adherent cells were NMR embryonic fibroblasts. Adult fibroblasts were established from kidney, testis, and skin of a 1-year-old male. Mouse embryonic and adult fibroblasts were from mouse fetus (E13.5) and lung of a 10-week-old C57BL/6 male, respectively. Both NMR and mouse fibroblasts were maintained in DMEM (DMEM high glucose + GlutaMAX) with 10% (mouse) and 15% (NMR) FBS, 1× antibiotic/antimycotic, 1× non-essential amino acids, 0.1 mM β-mercaptoethanol. All NMR cells were cultured at 32°C or 37°C, 3% O2, and 5% CO2, while mouse cells were cultured at 37°C and 5% CO2. NMR fibroblasts used in this study are listed in Table S1.
Reprogramming of Mouse and NMR Fibroblasts to iPSCs
After 1 day of lentiviral transduction, approximately 100,000 infected fibroblasts were seeded per well of 6-well plate. Three different culture conditions were used to test the ability of cells to reprogram. First, lentiviral infected mouse and NMR fibroblasts were grown onto mytomycin C-inactivated or gamma-irradiated MEF feeder cells in ESC medium in the presence of 2 mg/mL doxycycline (DMEM supplemented with 15% FBS, 1,000 units/mL mouse LIF or 10 ng/mL basic fibroblast growth factor, 0.1 mM β-mercaptoethanol, 1% non-essential amino acids, 2 mM glutamine, and 1% antibiotic/antimycotic). The other conditions also represented the culture of infected fibroblasts grown on MEF feeder cells or Matrigel matrix, but we used N2/B27+2i medium (1:1 mixture of N2 medium [DMEM/F12 supplemented with 1× N2], 1% antibiotic/antimycotic, 0.005% BSA, 0.1 mM β-mercaptoethanol, and 1% non-essential amino acids) and B27 medium (neurobasal medium supplemented with 1× B27 [without retinoic acid], 1% antibiotic/antimycotic, 2 mM L-glutamine, 0.1 mM β-mercaptoethanol, 0.005% BSA, and 1% non-essential amino acids) supplemented with 1,000 U/mL mLIF, 3 μM GSK3b inhibitor (CHIR99021), and 1 μM MEK1/2 inhibitor (PD0325901). In each of two culture conditions, growth media were replaced every 24 hr.
AP Staining and Immunofluorescence
AP staining was performed using the Alkaline Phosphatase Staining Kit (Stemgent) according to manufacturer's instructions. For fluorescence immunocytochemistry, iPSCs were cultured on mitotically inactivated MEFs for 2–3 days in the case of mouse iPSCs and for 6–7 days in the case of NMR iPSCs, and fixed using PBS containing 4% paraformaldehyde (Sigma) for 15 min at room temperature. After rinsing with PBS, fixed cells were blocked and permeabilized for 1 hr in PBS containing 10% (v/v) goat serum (Sigma) in 0.1% Triton X-100-PBS. Primary antibodies were as follows: OCT4 (Santa Cruz Biotech; sc-5279), SOX2 (Millipore; AB5603), NANOG (Millipore; AB5731), and SSEA-1 (Santa Cruz Biotech; sc-21702), which were diluted 1:200 in blocking solution. Fixed cells were incubated with primary antibody overnight at 4°C. After washing out the unbound antibodies, fixed cells were incubated with secondary antibodies (labeled with Alexa 488, 1:200 dilution, Jackson ImmunoResearch; 111-545-144, 115-545-003) for 1 hr at room temperature. Nuclei were counterstained using 1 μg/mL Hoechst 33342 (Life Technologies; H3570). All fluorescence imaging was conducted using an LSM 700 confocal microscope (Zeiss).
Teratoma Formation
NMR iPSCs or EGFP-expressed iPSCs grown on Matrigel were collected by 0.05% trypsin treatment and injected under the kidney capsules into non-obese diabetic (NOD)-SCID (NOD.CB17-Prkdc/J) mice. The kidney capsules were collected 10 weeks after the injection and processed for paraffin embedding and H&E staining or immunofluorescence co-staining with primary polyclonal antibodies against GFP (chicken IgY; Invitrogen; A10262) and TUJ-1(1:500, Covance; MMS-435P). Secondary antibodies were Alexa 488-conjugated anti-chicken IgY (Invitrogen; A-11039) and Alexa Fluor 647 anti-rabbit IgG (1:200 dilution, Jackson ImmunoResearch; 111-607-003). Genomic DNA of kidney capsules was extracted using a DNeasy Blood & Tissue Kit (QIAGEN). PCR with GoTaq polymerase using specific primers to M2rtTA lentiviral vector was performed to confirm the origin of NMR iPSCs in the kidney capsules.
Karyotype Analysis
Karyotyping was performed at the Cytogenetics Core Laboratory in Brigham and Women's Hospital using standard protocols for chromosomal G-banding.
RT-PCR and Real-Time PCR Analysis
To perform qPCR, total RNA from individual samples was extracted using TRIzol reagent (Invitrogen, MA) according to the manufacturer's instructions. cDNA was synthesized using a High Capacity RNA-to-cDNA Kit (Applied Biosystems, CA). PCR primers are listed in Table S2. Relative gene expression was calculated by normalizing the threshold cycle (Ct) values of each gene to that of the GAPDH via the Δ-Ct method (Livak and Schmittgen, 2001).
Interspecies Chimera Production
To generate chimeric mouse embryos containing NMR iPSCs, C57BL/6-derived blastocysts injected with EGFP-expressed NMR iPSCs were transplanted into the uteri of foster mothers (ICR; CD1 mouse). The generated chimera features were analyzed at E13.5. They were fixed with 4% paraformaldehyde, embedded for frozen sectioning in optimum cutting temperature compound, and co-immunostained with primary polyclonal antibodies against GFP (chicken IgY; Invitrogen) and TUJ-1(1:500, Covance; MMS-435P), or a-ACTININ (1:200, Sigma; A7732). Secondary antibodies were Alexa 488-conjugated anti-chicken IgY (Invitrogen; A10262) and Alexa Fluor 647 anti-rabbit or -mouse IgG (1:200 dilution, Jackson ImmunoResearch; 111-607-003, 115-605-206). Genomic DNA of chimeric mouse embryos was extracted using DNeasy Blood & Tissue Kit (QIAGEN). PCR with GoTaq polymerase using the same primers as in the teratoma formation section was performed to confirm the origin of NMR iPSCs in the embryos.
RNA-Seq and Data Analysis
Total RNA was extracted from mouse and NMR samples using an RNeasy Plus Kit (QIAGEN). Short-insert, paired-end libraries were prepared using the Illumina TruSeq Sample Preparation Kit v2, and sequenced bidirectionally (101 bp in each direction) on Illumina HiSeq 2000 (Illumina, San Diego, CA). Mus musculus (GRCm38) and Heterocephalus glaber (hetGla2) reference genomes and annotation files were from the NCBI. Differential gene expression analysis for RNA-seq data was performed as follows: FastQC package (version 0.11.4) was used for quality control, raw data were trimmed using Trim_Galore (version 0.4.0) and aligned to a reference genome using TopHat2 (version 2.0.9); Bowtie 2.1.0.0 was used for aligning reads to genes. Transcripts were assembled using Cufflinks, and feature counts performed using a Python package HTseq. A generated simple-count matrix was used for follow-up analysis based on EdgeR and DESeq. Raw sequencing data for 20 biological samples have been deposited into the Short Read Archive database under accession number SRA: SRP116326. Human fibroblast and iPSC datasets were from GEO: GSE36552 (Choi et al., 2015).
Statistical Analysis
Statistical analyses were performed using a two-tailed t test (∗t test, p < 0.05; ∗∗t test, p < 0.001). Values are shown as the mean ± SEM of multiple independent experiments, not technical replicates.
Author Contributions
S.G.L. and A.E.M. performed iPSC experiments, and S.H.Y., J.K.P., K.H.C., R.T.B., C.K.L., and T.J.P. contributed to their characterization or performed other biological experiments. S.G.L. and A.V.L. analyzed RNA-seq data, and V.N.G. supervised the study. S.G.L. and V.N.G. prepared the manuscript, with contribution from all other authors.
Acknowledgments
We thank Dr. Lin Wu and the Genome Modification Facility, Harvard University, for technical assistance with teratoma and chimera assays. This work was supported by NIH AG047745, CA080946, and AG047200, by the Glenn Foundation, and by the Next-Generation BioGreen 21 Program (no. PJ01130012015), Republic of Korea.
Published: October 26, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.09.013.
Supplemental Information
References
- Bacquet C., Imamura T., Gonzalez C.A., Conejeros I., Kausel G., Neildez-Nguyen T.M., Paldi A., Gallardo M.H. Epigenetic processes in a tetraploid mammal. Mamm. Genome. 2008;19:439–447. doi: 10.1007/s00335-008-9131-z. [DOI] [PubMed] [Google Scholar]
- Bao L., He L., Chen J., Wu Z., Liao J., Rao L., Ren J., Li H., Zhu H., Qian L. Reprogramming of ovine adult fibroblasts to pluripotency via drug-inducible expression of defined factors. Cell Res. 2011;21:600–608. doi: 10.1038/cr.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffenstein R., Yahav S. Is the naked mole-rat (Heterocephalus glaber) an endothermic yet poikilothermic mammal? J. Therm. Biol. 1991;16:227–232. [Google Scholar]
- Brook F.A., Gardner R.L. The origin and efficient derivation of embryonic stem cells in the mouse. Proc. Natl. Acad. Sci. USA. 1997;94:5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr M., Meek S., Blair K., Yang J., Ure J., Silva J., McLay R., Hall J., Ying Q.L., Smith A. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Buganim Y., Faddah D.A., Cheng A.W., Itskovich E., Markoulaki S., Ganz K., Klemm S.L., van Oudenaarden A., Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Hanna J., Saha K., Gao Q., Mitalipova M., Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc. Natl. Acad. Sci. USA. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Yuan D., Wei B., Jiang J., Kang J., Ling K., Gu Y., Li J., Xiao L., Pei G. E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells. 2010;28:1315–1325. doi: 10.1002/stem.456. [DOI] [PubMed] [Google Scholar]
- Chin M.H., Mason M.J., Xie W., Volinia S., Singer M., Peterson C., Ambartsumyan G., Aimiuwu O., Richter L., Zhang J. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Lee S., Mallard W., Clement K., Tagliazucchi G.M., Lim H., Choi I.Y., Ferrari F., Tsankov A.M., Pop R. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat. Biotechnol. 2015;33:1173–1181. doi: 10.1038/nbt.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T., de Lange T. The causes and consequences of polyploidy in normal development and cancer. Ann. Rev. Cell Dev. Biol. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- Deng Y., Liu Q., Luo C., Chen S., Li X., Wang C., Liu Z., Lei X., Zhang H., Sun H. Generation of induced pluripotent stem cells from buffalo (Bubalus bubalis) fetal fibroblasts with buffalo defined factors. Stem Cells Dev. 2012;21:2485–2494. doi: 10.1089/scd.2012.0018. [DOI] [PubMed] [Google Scholar]
- Edrey Y.H., Park T.J., Kang H., Biney A., Buffenstein R. Endocrine function and neurobiology of the longest-living rodent, the naked mole-rat. Exp. Gerontol. 2011;46:116–123. doi: 10.1016/j.exger.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Esteban M.A., Xu J., Yang J., Peng M., Qin D., Li W., Jiang Z., Chen J., Deng K., Zhong M. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J. Biol. Chem. 2009;284:17634–17640. doi: 10.1074/jbc.M109.008938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Ezashi T., Telugu B.P., Alexenko A.P., Sachdev S., Sinha S., Roberts R.M. Derivation of induced pluripotent stem cells from pig somatic cells. Proc. Natl. Acad. Sci. USA. 2009;106:10993–10998. doi: 10.1073/pnas.0905284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R., Liu K., Zhao Y., Li H., Zhu D., Du Y., Xiang C., Li X., Liu H., Miao Z. Generation of naive induced pluripotent stem cells from rhesus monkey fibroblasts. Cell Stem Cell. 2014;15:488–496. doi: 10.1016/j.stem.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Hanna J., Markoulaki S., Mitalipova M., Cheng A.W., Cassady J.P., Staerk J., Carey B.W., Lengner C.J., Foreman R., Love J. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009;4:513–524. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Saha K., Pando B., van Zon J., Lengner C.J., Creyghton M.P., van Oudenaarden A., Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Soldner F., Cook E.G., Gao Q., Mitalipova M., Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A., Hirose M., Hatori M., Matoba S., Miyoshi H., Inoue K., Ogura A. Generation of induced pluripotent stem cells in rabbits: potential experimental models for human regenerative medicine. J. Biol. Chem. 2010;285:31362–31369. doi: 10.1074/jbc.M110.150540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Li T., Alonso-Gonzalez L., Gorre R., Keatley S., Green A., Turner P., Kallingappa P.K., Verma V., Oback B. A virus-free poly-promoter vector induces pluripotency in quiescent bovine cells under chemically defined conditions of dual kinase inhibition. PLoS One. 2011;6:e24501. doi: 10.1371/journal.pone.0024501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H., Kano K., Fujii W., Takasawa K., Wakitani S., Hiyama M., Nishino K., Kusakabe K.T., Kiso Y. Tetraploid embryonic stem cells maintain pluripotency and differentiation potency into three germ layers. PLoS One. 2015;10:e0130585. doi: 10.1371/journal.pone.0130585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.B., Fang X., Fushan A.A., Huang Z., Lobanov A.V., Han L., Marino S.M., Sun X., Turanov A.A., Yang P. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479:223–227. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Yamaguchi T., Hamanaka S., Kato-Itoh M., Yamazaki Y., Ibata M., Sato H., Lee Y.S., Usui J., Knisely A.S. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell. 2010;142:787–799. doi: 10.1016/j.cell.2010.07.039. [DOI] [PubMed] [Google Scholar]
- Lewis K.N., Mele J., Hornsby P.J., Buffenstein R. Stress resistance in the naked mole-rat: the bare essentials - a mini-review. Gerontology. 2012;58:453–462. doi: 10.1159/000335966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Li W., Wei W., Zhu S., Zhu J., Shi Y., Lin T., Hao E., Hayek A., Deng H., Ding S. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009;4:16–19. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Liang S., Mele J., Wu Y., Buffenstein R., Hornsby P.J. Resistance to experimental tumorigenesis in cells of a long-lived mammal, the naked mole-rat (Heterocephalus glaber) Aging Cell. 2010;9:626–635. doi: 10.1111/j.1474-9726.2010.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Cui C., Chen S., Ren J., Chen J., Gao Y., Li H., Jia N., Cheng L., Xiao H. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell. 2009;4:11–15. doi: 10.1016/j.stem.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo J., Suhr S.T., Chang E.A., Wang K., Ross P.J., Nelson L.L., Venta P.J., Knott J.G., Cibelli J.B. Generation of leukemia inhibitory factor and basic fibroblast growth factor-dependent induced pluripotent stem cells from canine adult somatic cells. Stem Cells Dev. 2011;20:1669–1678. doi: 10.1089/scd.2011.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Ye Z., Hommond H.H., Yu X., Lin J., Chen G., Zou J., Cheng L. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells. 2008;26:1998–2005. doi: 10.1634/stemcells.2008-0346. [DOI] [PubMed] [Google Scholar]
- Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascetti V.L., Pedersen R.A. Human-mouse chimerism validates human stem cell pluripotency. Cell Stem Cell. 2016;18:67–72. doi: 10.1016/j.stem.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki S., Kawamura Y., Oiwa Y., Shimizu A., Hachiya T., Bono H., Koya I., Okada Y., Kimura T., Tsuchiya Y. Tumour resistance in induced pluripotent stem cells derived from naked mole-rats. Nat. Commun. 2016;7:11471. doi: 10.1038/ncomms11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X., Li N., Wu S. Generation and characterization of bat-induced pluripotent stem cells. Theriogenology. 2014;82:283–293. doi: 10.1016/j.theriogenology.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy K., Sung H.K., Zhang P., Laflamme S., Vincent P., Agha-Mohammadi S., Woltjen K., Monetti C., Michael I.P., Smith L.C. Induced pluripotent stem cell lines derived from equine fibroblasts. Stem Cell Rev. 2011;7:693–702. doi: 10.1007/s12015-011-9239-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omerbasic D., Smith E.S., Moroni M., Homfeld J., Eigenbrod O., Bennett N.C., Reznick J., Faulkes C.G., Selbach M., Lewin G.R. Hypofunctional TrkA accounts for the absence of pain sensitization in the African naked mole-rat. Cell Rep. 2016;17:748–758. doi: 10.1016/j.celrep.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Park T.J., Reznick J., Peterson B.L., Blass G., Omerbasic D., Bennett N.C., Kuich P., Zasada C., Browe B.M., Hamann W. Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science. 2017;356:307–311. doi: 10.1126/science.aab3896. [DOI] [PubMed] [Google Scholar]
- Plath K., Lowry W.E. Progress in understanding reprogramming to the induced pluripotent state. Nat. Rev. Genet. 2011;12:253–265. doi: 10.1038/nrg2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Golipour A., David L., Sung H.K., Beyer T.A., Datti A., Woltjen K., Nagy A., Wrana J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Seluanov A., Hine C., Azpurua J., Feigenson M., Bozzella M., Mao Z., Catania K.C., Gorbunova V. Hypersensitivity to contact inhibition provides a clue to cancer resistance of naked mole-rat. Proc. Natl. Acad. Sci. USA. 2009;106:19352–19357. doi: 10.1073/pnas.0905252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Utikal J., Polo J.M., Stadtfeld M., Maherali N., Kulalert W., Walsh R.M., Khalil A., Rheinwald J.G., Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Greely H.T., Jaenisch R., Nakauchi H., Rossant J., Belmonte J.C. Stem cells and interspecies chimaeras. Nature. 2016;540:51–59. doi: 10.1038/nature20573. [DOI] [PubMed] [Google Scholar]
- Ye S., Liu D., Ying Q.L. Signaling pathways in induced naive pluripotency. Curr. Opin. Genet. Dev. 2014;28:10–15. doi: 10.1016/j.gde.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.