Abstract
The development of mouse genetic tools has made a significant contribution to the understanding of skeletal and hematopoietic stem cell niches in bone marrow (BM). However, many experimental designs (e.g., selections of marker genes, target vector constructions, and choices of reporter murine strains) have unavoidable technological limitations and bias, which lead to experimental discrepancies, data reproducibility issues, and frequent data misinterpretation. Consequently, there are a number of conflicting views relating to fundamental biological questions, including origins and locations of skeletal and hematopoietic stem cells in the BM. In this report, we systematically unravel complicated data interpretations via comprehensive analyses of technological benefits, pitfalls, and challenges in frequently used mouse models and discuss their translational relevance to human stem cell biology. Particularly, we emphasize the important roles of using large human genomic data-informatics in facilitating genetic analyses of mouse models and resolving existing controversies in mouse and human BM stem cell biology.
Keywords: stem cell niches, cell identity, bone marrow stromal cells, skeletal stem cells, hematopoietic stem cells, mouse genetics, genomics, epigenomics, nestin, leptin receptor
In this article, Chen and colleagues discuss technological challenges for genetic analysis of bone marrow stem cell niches. This article consists of a concise review, considerable perspectives, and in-depth data analysis. Importantly, the authors provide translational relevance through interrogating mouse genetic models with human genomic datasets. This review would aid to resolve data inconsistencies and their associated stem cell controversies.
Main Text
Introduction
Genetically modified mouse models have been extensively used to trace stem cell niches, evaluate stem cell identities, and provide translational insights into human stem cell biology. Currently, we still face considerable experimental discrepancies and data reproducibility issues related to the use of mouse genetic models, which have led to several major controversies in fundamental biological questions in the bone marrow (BM) stem cell field. For example, the precise locations of hematopoietic stem cell (HSC) niches, which are predominantly determined by the use of various mouse reporter genes, are currently under debate (Acar et al., 2015, Asada et al., 2017, Kunisaki et al., 2013, Oguro et al., 2013). Moreover, “mesenchymal stem cells” are a vague and confusing concept, which was primarily based on “bone marrow stromal cells” (Friedenstein et al., 1966, Owen and Friedenstein, 1988) and on multipotent skeletal stem cells (SSCs), without the use of definitive markers (Bianco and Robey, 2015). Furthermore, it is unclear whether local neural crest cells could directly contribute to SSCs in BM (Isern et al., 2014, Morikawa et al., 2009, Zhou et al., 2014). Thus, BM stem cells represent a more diffuse-and-complex biological area, having many unidentified variables that are responsible for existing experimental discrepancies and data irreproducibility (Morrison, 2014). Nonetheless, it appears that all the above controversies are, at least, associated with one common methodological basis; i.e., the differential use of mouse reporter strains.
During the past two decades, marker gene identifications combined with cell lineage tracing using reporter mouse strains have had a major impact on understanding the complexity of cellular dynamics and commitments of murine BM stem cell niches at various developmental stages. However, choices of marker genes, constructions of reporter murine strains, and even experimental designs have unavoidable bias, which have limited our understanding of BM stem cell biology (reviewed in Bianco and Robey, 2015, Kfoury and Scadden, 2015, Mendez-Ferrer et al., 2015, Morrison and Scadden, 2014). Current technologies used to identify BM stem cells mainly rely on various mouse reporter strains based on limited numbers of marker genes (e.g., nestin [Nes], leptin receptor [Lepr], Cspg4/NG2, and Wnt-1). Perplexingly, these marker genes usually have high levels of expression in non-BM tissues or organs, thus having limited specificity in BM. For instance, in mouse embryos, Nes, Lepr, Cspg4, and Wnt-1 all have higher levels of expression in the brain than in the BM. At present, many unmanageable variables in mouse experiments stem from genetically engineered reporter genes in mouse strains. Therefore, optimizing murine models to resolve existing controversies and to translate the information from animal models into human BM biology has been challenging.
To accurately define diverse BM cell lineages and differentiation, in this review, we systematically untangle the complicated data interpretation using various mouse genetic models. We aim to do the following: (1) briefly discuss the advantages of mouse genetic models and try to resolve inconsistencies, (2) shed light on the technological advantages, pitfalls, and challenges in the development of BM stem cell lineages, and (3) examine the translational relevance of murine models, and utilize existing large human genomic datasets to facilitate data interpretation. Technically, we present this review as a dedicated resource, in which our detailed analyses of the pros and cons of different mouse strains (in the main text and in Tables 1 and S1) would enable scientists to efficiently grasp principles of designing mouse genetic models and of choosing appropriate mouse strains of interest. The genomic and molecular analyses, available in Figures 1, 2, 3, 4, and 5, would help researchers to prospectively understand the translational process based on existing genomic databases. Hence, this resource review may be suitable for a broad range of investigators, scientists, biologists, and trainees in different stem cell fields, particularly for scientists working on the hematological and skeletal systems.
Table 1.
Representative Analyses of Marker Genes Used for Bone Marrow and Skeletal Stem Cell Identities
| Mouse Strains | Major Descriptions | Authors' Comments | References |
|---|---|---|---|
| Col2.3-GFP transgenic mice | express GFP in osteoblasts and osteocytes under the control of the 2.3-kb rat Col 1a1 (procollagen, type 1, alpha 1) promoter | useful for studying bone development and osteoblast lineage tracing; wary of rat subspecies sequence effects | Kalajzic et al., 2002 |
| Cxcl12-dsRed |
|
useful for identifying Cxcl12-expressing perivascular stromal cells and endothelial cells in the bone marrow | Ding and Morrison, 2013 |
|
Cxcl12-GFP knockin mice |
highly enriched in Cxcl12-abundant reticular (CAR) cells within the intra-trabecular space in the bone marrow | endothelial cells and the endosteal surface osteoblasts show faint or undetectable GFP signals | Ara et al., 2003, Sugiyama et al., 2006 |
| Gt(ROSA)26Sortm1(HBEGF)Awai |
|
suitable for ablation of cells that express DTR following diphtheria toxin treatment | Buch et al., 2005 |
| Leprfl/fl |
|
|
Cohen et al., 2001 http://www.jax.org/ |
| Lepr-Cre | L B6.129-Leprtm2(Cre)Rck/J (Lepr-Cre); the targeting vector contains an IRES-NLS-Cre and a neo (flanked by frt sites) inserted immediately 3′ of the stop codon in the last exon of the Lepr gene | transcripts may terminate in many Lepr transcript variants that do not contain the last exon of the canonical Lepr isoform (Lepr-B) | DeFalco et al., 2001 |
|
Mx-1-Cre, transgenic mice |
B6.Cg-Tg(Mx1-cre)1Cgn/J, also known as Mx-Cre and Mx1-Cre (BALB/c): the Mx-1-Cre transgene contains Cre recombinase under the control of the Mx-1 promoter that is silent in healthy mice |
|
Kuhn et al., 1995 http://www.jax.org/ |
| Nes-Cre | Cre recombinase is expressed under the control of the 5.8-kb rat Nes promoter and the 1.8-kb intron 2 enhancer element |
|
Tronche et al., 1999 |
|
Nes-CreERT2 transgenic mice |
|
|
Balordi and Fishell, 2007, Chagin et al., 2007, Feil et al., 1997, Karimian et al., 2008, Lagace et al., 2007, Zimmerman et al., 1994 |
| Nes-GFP | Tg(Nes-EGFP)33Enik: a Nes-GFP reporter in transgenic mice, driven by the 5.8 promoter and 1.8-kb intron 2 enhancer of the rat Nes gene |
|
Lendahl et al., 1990, Mignone et al., 2004, Zimmerman et al., 1994 |
| NG2-CreER™ |
|
useful for inducible Cre recombinase expression in NG2-expressing glia and other cell types | Zhu et al., 2011; http://www.jax.org/ |
| P0-Cre | transgenic mice expressing Cre recombinase directed by the myelin protein zero (P0) gene promoter | genetic tools for labeling neural crest cell lineages such as Schwann cells | Feltri et al., 1999, Yamauchi et al., 1999 |
| Prx1-Cre | B6.Cg-Tg(Prrx1-cre)1Cjt/J: expresses Cre under the control of a Prrx1-derived enhancer | useful for studying limb bud development and patterning | Logan et al., 2002 |
| Wnt1-Cre |
|
|
Danielian et al., 1998, Lewis et al., 2013 |
| Wnt1-Cre2 | Cre expression under the control by 1.3-kb 5′ promoter and 5.5-kb 3′ enhancer |
|
Lewis et al., 2013 |
Cre, Cre recombinase; Cxcl12, chemokine (C-X-C motif) ligand 12; GFP, green fluorescent protein; HSV, herpes simplex virus; IRES, internal ribosome entry site; Neo, neomycin resistance gene; NLS, nuclear localization signal; RFP, red fluorescent protein; TK, thymidine kinase.
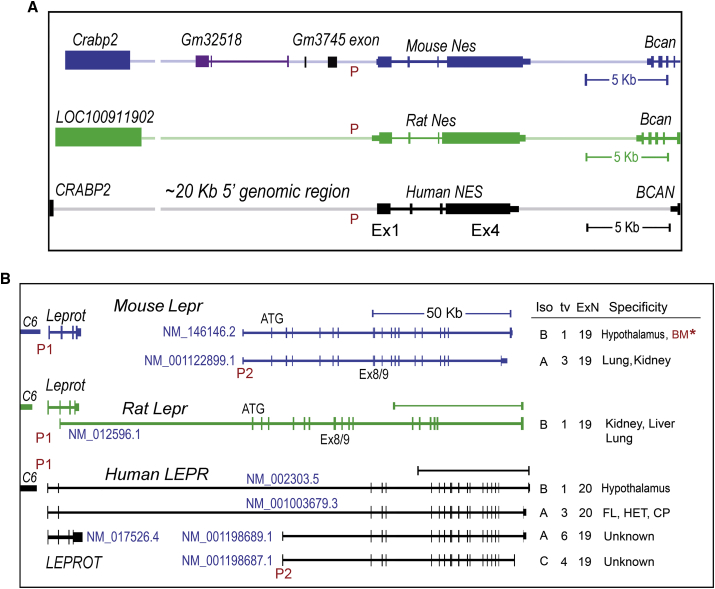
Figure 1.
Genomic Organization of the Nestin and Leptin Receptor Genes
(A) Nestin and (B) leptin receptor genes in mice, rats, and humans. The graphs were created based on recent data from both the National Center for Biotechnology Information (NCBI) (www.ncbi.nlm.nih.gov) and the UCSC Genome Browser (genome.ucsc.edu). The accession numbers for leptin receptor isoforms are: NM_146146.2 (mouse Lepr-B isoform, transcript variant 1, 19 exons), NM_001122899.1 (mouse Lepr-A, transcript variant 3, 19 exons), NM_012596.1 (rat Lepr-B, 19 exons), and NM_002303.5 (human LEPR-B, transcript variant 1, 20 exons), NM_001003679.3 (human LEPR-A, transcript variant 3, 20 exons), NM_001198689.1 (human LEPR-A, transcript variant 6, 19 exons), and NM_001198687.1 (human LEPR-C, transcript variant 4, 19 exons). Representative leptin receptor isoforms, transcript variant identification numbers, exon numbers, and tissue-specific expression patterns were briefly indicated in the right panel. Of note, the asterisk sign (∗) indicates that mouse bone marrow (BM) stromal cells express the Lepr isoform b based on the existence of Lepr-Cre+ cells around sinusoids of the BM (Zhou et al., 2014) and the NLS-Cre cassette (in the Lepr-Cre transgene), which is inserted into the 3′ of the stop codon at exon 19 of the transcript variant 1 (DeFalco et al., 2001).
E or Ex, exon; ExN, exon numbers; FL, fetal liver; HET, hematopoietic tissues; I, intron; Iso, leptin receptor protein isoform; Lepr/LEPR, the leptin receptor gene; Leprot/LEPROT, leptin receptor overlapping transcript; Nes/NES, the gene coding for nestin; tv, transcript variant; P, promoter.
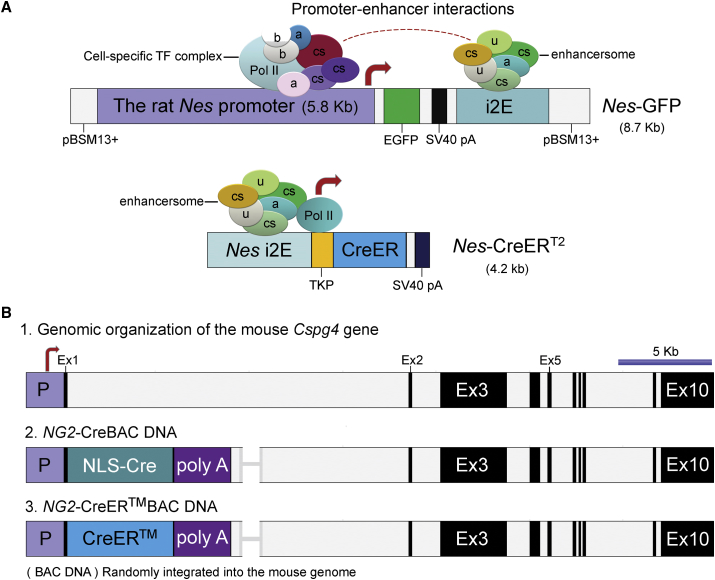
Figure 2.
DNA Transgene Expression Vectors and Regulatory Mechanisms
(A) Transgene expression vectors based on the rat Nes gene. Top panel: Nes-GFP, subcloned into the pBSM13 vector, contains the 5.8-kb rat Nes promoter and the 1.8-kb neural-specific intron-2 enhancer fragment (i2E), which flanked the enhanced version of GFP (EGFP). The 8.7-kb final construct, mimicking the arrangement of the regulatory sequences of the Nes or NES found in the rat, mice, and humans, was used for the pronuclear injections of the fertilized oocytes (Mignone et al., 2004). Lower panel: Nes-CreERT2 comprises the T2 mutant form of a Cre recombinase-estrogen receptor fusion (Cre-ERT2) (Feil et al., 1997) under the control of a thymidine kinase promoter (TKP) driven by the 1.8-kb i2E as described in the top panel (Balordi and Fishell, 2007). In Nes-GFP transgenic mice, a cell-specific transcriptional complex at the promoter might interact with the neural-specific intron-2 enhancersome, thereby mediating different gene expression patterns in miscellaneous cell types including BM cells. However, in the case of Nes-CreERT2 mice, the transgene is largely driven by the intron 2 enhancersome.
(B) Transgene expression vectors based on the chondroitin sulfate proteoglycan four gene (Cspg4), also known as NG2 (neural/glial gene). (1) Genomic organization of the Cspg4 gene is based on the recent genomic information from the NCBI sequence (NM_1390012) with a scale bar (5 kb). (2 and 3) NG2-CreBAC (Zhu et al., 2008) and NG2-CreER™BAC (Zhu et al., 2011) DNAs were used for generating NG2-Cre and NG2-CreER™ transgenic mice, respectively. In brief, a 208-kb mouse bacterial artificial chromosome (BAC) containing the entire Cspg4 gene was modified by introducing a Cre recombinase cDNA with a nuclear localization signal (NLS) or a CreER™ cDNA (Danielian et al., 1993, Littlewood et al., 1995) into exon 1 of the Cspg4 gene, followed by a rabbit β-globin polyadenylation sequence, poly(A). These two transgenes were microinjected into the pronucleus of fertilized oocytes from C57BL/6J mice to generate the transgenic lines of interest.
a, adaptor protein(s); b, basal transcriptional factor(s); Cre-ERT2, Cre recombinase fused to the human estrogen receptor ligand-binding domain with a triple mutation (i.e., G400V/M543A/L544A), which does not bind its natural ligand (17β-estradiol); Cre-ER™, Cre recombinase fused to a G525R mutant form of the mouse estrogen receptor ligand-binding domain; cs, cell-specific, Ex, exon; i2E, the intron 2 enhancer fragment of the rat Nes gene; P, promoter; Pol II, RNA polymerase II; SV40 pA, the polyadenylation sequences from the simian virus 40; TF, transcriptional factor; TKP, a 160-bp herpes simplex virus (HSV) thymidine kinase (TK) promoter; u, unidentified factor(s).
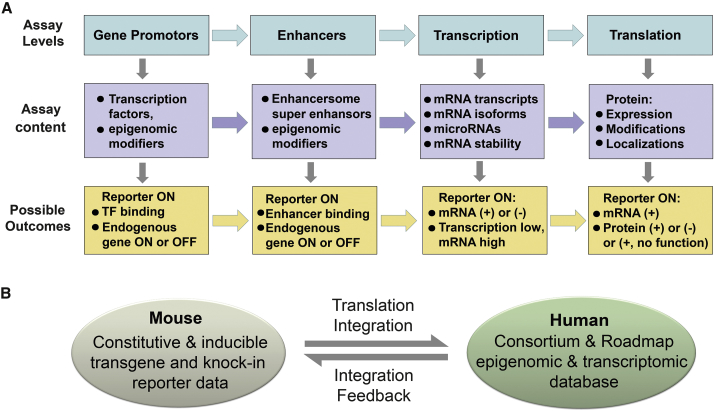
Figure 3.
Gene Regulation, Data Interpretation, and Integration
(A) Regulation of transgene at different molecular levels. Transgene reporter expression may or may not overlap with endogenous gene expression patterns. With regard to a transgene reporter activation, various experimental outcomes may be possible, which need to be confirmed by additional downstream assays (e.g., mRNA and protein expression).
(B) A scheme of data integration between mouse transgene reporter data and human epigenomic databases. Data from mouse genetic models may be directly translated and integrated into human BM biology given that they shared highly similar genomic structures and regulatory elements. Existing genomic and epigenomic databases can be also used to facilitate mouse data interpretation and help design humanized mouse models.
Figure 4.
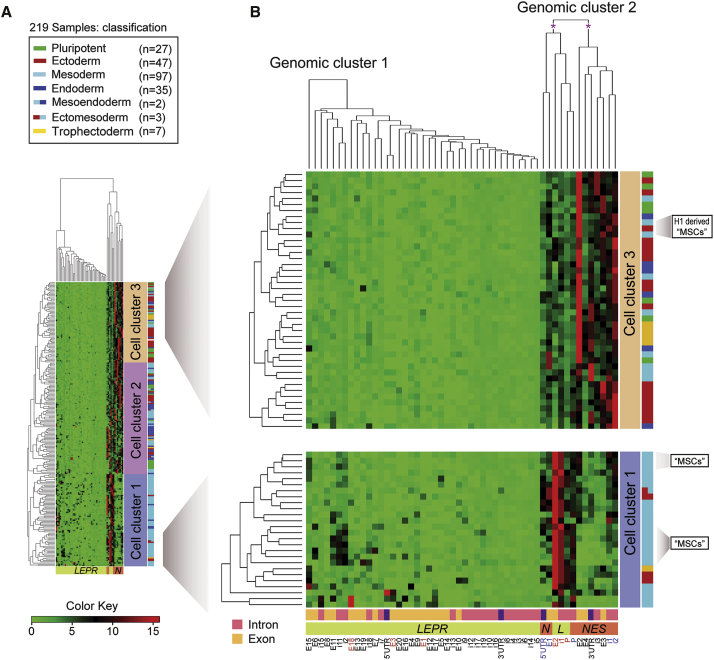
Representative Analysis of the Epigenetic Marker H3K4me1 at the NES and LEPR Loci
(A) Clustering analysis of deposited H3K4me1 ChiP-seq data (www.genboree.org) in 219 samples that comprise cell types from three germ layers and trophectoderm (Table S2, Figure S1). H3K4me1 data for 219 human samples (GEO accession number: GGSM621418) were imported into the Genboree Workbench from Release 9 of the Human Epigenome Atlas (www.genboree.org). Human genome assembly GRCh37/hg19 (February 2009) was used for this analysis. The normalized values for NES and LEPR were exported for cluster analysis and visualization in R (www.cran.r-project.org) using the heatmap.2 function.
(B) The enlarged views of the regions of interest are presented on the right panel. Asterisks indicate the views of truncated dendrograms. Detailed information for these dendrograms is available from Figure S1. Of note, the genomic localization of exon 20 of the LEPR gene is currently not available from the Human Epigenome Atlas (www.genboree.org). Hence, the epigenomic data of exon 20 should be interpreted with caution.
H1, human embryonic stem cell line H1 (WA01); H3K4me1, monomethylated histone H3 lysine 4; I, intron; LEPR or L, the leptin receptor gene; LEPROT or Leprot, leptin receptor overlapping transcript; “MSC,” “mesenchymal stem cells”; NES or N, the gene coding for nestin; UTR, untranslated region.
Figure 5.
Deciphering Molecular Cell-Identity Codes through Integration of Data-Informatics Cascades (from Epigenomics, Transcriptomics, and Chromatin Proteomics) into Regulatory Signatures
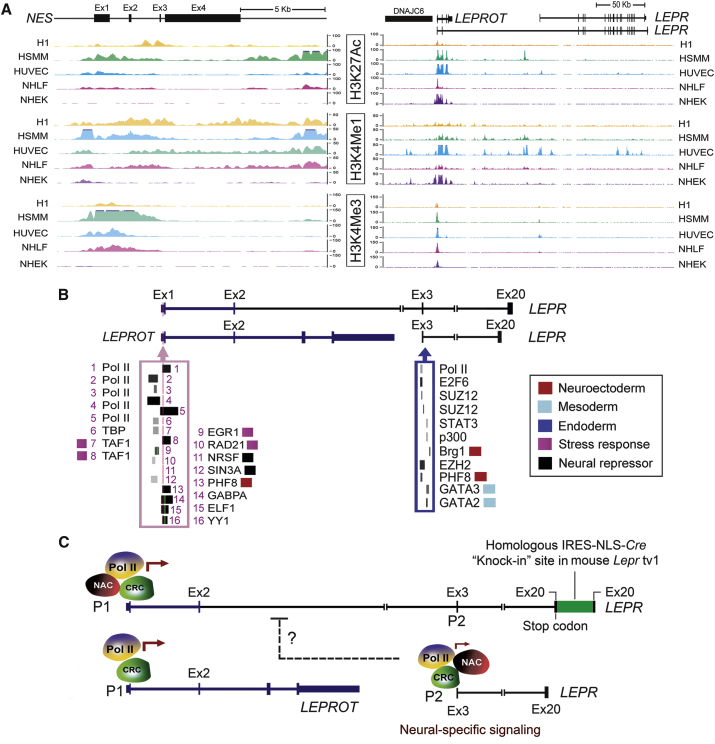
(A) Analysis of the epigenomic markers (H3K27ac, H3K4me1, and H3K4me3 at the NES and LEPR loci) was based on the Encyclopedia of DNA Elements (ENCODE) at the UCSC (genome.ucsc.edu). Human genome assembly GRCh38/hg38 (December 2013) was used for this analysis. The ChiP-seq data are arranged to correspond precisely to their genomic locations. Five (H1, human skeletal muscle cells and myoblasts [HSMM], HUVEC, normal human lung fibroblasts [NHLF], and normal human epidermal keratinocytes [NHEK]) out of seven cell lines are shown.
(B) Mapping of transcriptional regulators on the chromatin at the LEPR and/or LEPROT loci: LEPR (uc001dci.4) is located at chr1:65420652-65641559, based on the orientation of the transcript variant 1 from the RefSeq NM_002303. The enriched transcriptional factors (TFs) on the LEPR/LEPROT gene promoter as well as the LEPR exon 3 regions are shown. Some of these TFs are color-highlighted based on their role in cellular response and in lineage differentiation.
(C) Multiple regulatory models for the LEPR/LEPROT locus: the full-length human LEPR gene (containing 20 exons) is transcribed from the P1 promoter. In the Lepr-IRES-NLS-Cre targeting construct (containing the neo gene, flanked by the FRT sites) was introduced by homologous recombination immediately after the mouse Lepr stop codon at exon 19 (human exon 20 counterpart) (DeFalco et al., 2001). This Lepr-Cre knockin mouse model has been widely used to monitor transcriptional activity of the full-length mouse Lepr gene that encodes the Lepr-B protein isoform (Kunisaki et al., 2013, Mizoguchi et al., 2014, Ono et al., 2014, Zhou et al., 2014).
Brg1, SMARCA4 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4); CRC, chromatin-remodeling complexes including histone modifications enzymes; E2F6, E2F transcription factor 6; EGR1, early growth response 1; ELF1, E74-Like factor 1 (Ets domain transcription factor); Ex, exon; EZH2, enhancer of Zeste 2 polycomb repressive complex 2 subunit; GABPA, GA binding protein transcription factor, alpha subunit 60 kDa; GATA2, GATA binding protein 2; GATA3, GATA binding protein 3; H1, H1 (WA01) human embryonic stem cell line; HSMM, human skeletal muscle myoblasts (mesoderm); HUVEC, human umbilical vein endothelial cells (mesoderm) from blood vessels; LEPROT, leptin receptor overlapping transcript; NAC: neuronal lineage activator complexes that include either BRG1 or PHF8 or both; NHEK, normal human epidermal keratinocytes (ectoderm); NHLF, normal human lung fibroblasts; NRSF, known as REST (RE1-silencing transcription factor); P1, the promoter of the canonical LEPR gene; P2, an alternative promoter at the 5′ of exon 3 of LEPR; p300, EP300 (E1A binding protein P300); PHF8, PHD finger protein 8, a histone lysine demethylase that preferentially acts on histones in the monomethyl or dimethyl states; Pol II, RNA polymerase II; RAD21, RAD21 cohesin complex component; SIN3A, SIN3 transcription regulator family member A; STAT3, signal transducer and activator of transcription 3 (acute-phase response factor); SUZ12, SUZ12 polycomb repressive complex 2 subunit; TAF1, TAF1 RNA polymerase II, TATA-box binding protein (TBP)-associated factor, 250 kDa; TBP, TATA-box binding protein; YY1, YY1 transcription factor.
Mouse Genetic Models: Advantages and Problems Solved
Mouse genetic models have dramatically advanced our understanding of many fundamental developmental processes in both the skeletal and hematological systems, thereby accelerating the processes of translational medicine (Bianco et al., 2013, Frenette et al., 2013, Morrison and Scadden, 2014). These mouse models offer cell lineage mapping in vivo, a powerful approach to study specific cell types, numbers, physiological and pathological states, and particularly cell signaling pathways in stem cell niches (Tables 1 and S1).
Stem cell niches can be briefly defined as a specific microenvironment that contains and sustains stem cells in an undifferentiated state. The basic components of a BM stem cell niche comprise BM stroma, extracellular matrices, HSCs, SCCs, Cxcl12-abundant reticular (CAR) cells, adipocytes, endothelial cells, and different types of stromal cells not fully defined to date. These niche-supporting cells secrete specific niche factors encoded by many HSC niche maintenance genes (such as Cxcl12, KitL, Angpt1, and Lepr) at restricted regions and mediate many intercellular interactions (Isern et al., 2014). Several known niche-supporting cells include CAR cells (Sugiyama et al., 2006), NG2+/Nes-GFPhigh cells (Kunisaki et al., 2013), and Lepr-Cre+/Nes-GFPlow cells (Zhou et al., 2014). Technically, niche-associated gene promoter or enhancer activity as well as mRNA expression can be monitored and targeted by different fluorescent reporter proteins such as GFP. Thus far, mouse genetic models combined with imaging analysis have been the most widely used tool to successfully answer long-standing questions in developmental biology, which include the origins, identities, and locations of postnatal SSCs and HSCs. It is clear now that the major source of SCCs in human BM is tightly associated with CD146+/CD45−/Ter119− reticular pericytes (Sacchetti et al., 2016) and Lepr-Cre+/Nes-GFPlow cells near perisinusoids (Zhou et al., 2014) in mice. The major HSC niche has also been confidently localized at BM perivascular regions containing specific types of stromal cells (Acar et al., 2015, Kunisaki et al., 2013).
Tables 1 and S1 summarize a significant amount of data, with point-to-point interpretations and comments of each reporter mouse strain, related to BM cell lineage development, perivascular stromal cells, and neural crest cells. However, to better understand the pros and cons of genetically modified strains, we comprehensively analyzed two frequently used transgenes (i.e., Nes and Lepr) at different developmental stages (Table S1; Figure 1). These two individual genes are chosen, not only for their frequent use in BM niche studies, but also for their transcriptional activities that have empowered us to mark several important niche-supportive cell populations (i.e., Nes-GFPhigh, Nes-GFPlow, and Lepr-Cre+ BM stromal cells) (Kunisaki et al., 2013, Zhou et al., 2014). Accordingly, there is an increasing body of data generated from using these mouse models (Table S1). For example, combined with other transgene reporters such as NG2-CreER™, a tamoxifen-inducible Cre recombinase-estrogen fusion protein driven by the NG2 promoter-enhancer, scientists were able to identify two important BM cell populations. These two distinct cell populations; i.e., NG2-CreERTM+/Nes-GFPhigh and Lepr-Cre+/Nes-GFPlow cells, likely constitute a distinct HSC niche at the periarteriolar and a major SSC source at the perisinusoidal regions (Kunisaki et al., 2013, Zhou et al., 2014). The existence of distinct HSC niches, presumably with different functions, is currently an important topic under debate (Acar et al., 2015, Asada et al., 2017, Kunisaki et al., 2013, Oguro et al., 2013). Despite the enthusiasm of applying transgene-based models for in vivo cell-fate mapping, there are emerging controversial concepts, inconsistent data, and inappropriate data interpretation due to the limitations of mouse genetic systems.
Mouse Genetic Models: Disadvantages, Pitfalls, and Experimental Discrepancies
Noticeably, there are numerous limitations of mouse genetic models, which can be introduced by the experimental design of generating genetically engineered mice, to experimental data collection and interpretation. In general, the causes of experimental variability could be classified into the following four major categories, which include the following: (1) the designs of transgenes or targeting vectors used for generating transgenic mice; (2) random chromosomal integrations of genetically identical transgenes or similar transgenes; (3) methods of gene expression (e.g., constitutive versus inducible gene expression systems) and associated cellular cytotoxicity; and (4) complicated dynamic changes of cellular and molecular states of cells in BM throughout development. In the following sections, we will use some representative examples to highlight the above-mentioned major causes of experimental variability and discrepancies.
Mouse Genetic Model Designs: Genomic Elements, Orientations, and Gene Reporter Data Interpretation
It is worth noting that the choice of promoter sequences and enhancer elements in a reporter construct might have a major impact on reporter activity in mouse models. As far as Nes-GFP and Nes-CreERT2 mouse models are concerned, both transgenes certainly report transcriptional activation of the Nes gene. However, their transcriptional activities are apparently controlled by two different genetic systems (Figure 2A). Nes-GFP expression is driven essentially by the 5′ 5.8-kb promoter and the 1.8-kb intron 2 enhancer (i2E) (Table S1; Figure 2A). However, in Nes-CreERT2 transgenic mice, a tamoxifen-inducible Cre-estrogen fusion cassette (i.e., CreERT2) is driven by a thymidine kinase promoter under the control of the 5′ i2E element (Figure 2A). Thus, the 4.2-kb transgene fragment excludes the majority of the rat 5′ Nes promoter sequence, which has a different genomic orientation from that of the Nes-GFP construct (Mignone et al., 2004).
Not surprisingly, some experimental discrepancies have been observed from these two genetically different transgenes. Nes-CreERT2+ and Nes-GFP+ cells were not co-localized, but differentially presented at the prenatal stage in BM (Table S1). Nes-CreERT2+ cells are likely involved in fetal bone development based on their locations near the osteochondral junction and trabecular bone at the prenatal stage, but not in committing to neonatal and postnatal bone development (Isern et al., 2014). It appears that Nes-CreERT2+ cells co-localize with Nes-GFP+ pericytes at the neonatal stage (i.e., P0 to P14) (Isern et al., 2014). Seemingly, Nes-GFP+ stromal cells did not contribute to fetal endochondrogenesis (Isern et al., 2014), but subsequently initiated their role in specifying osteoblasts at the neonatal stage (i.e., P0 to P10) (Ono et al., 2014). Consistently, a Lepr-Cre+/Nes-GFPlow/+ cell population, without Nes-CreERT2 expression, was shown to be a major SSC source in the mouse BM at a postnatal stage (Zhou et al., 2014). Thus, Nes-CreERT2+ and Nes-GFP+ cells have distinct functions in specifying SSC lineage development in BM. Still, the underlying molecular basis for the above discrepancies between Nes-GFP and Nes-CreERT2 mouse strains remains to be elucidated (Isern et al., 2014, Ono et al., 2014).
We speculate that the above discrepancies could be partially explained by the different orientation of their genomic and vector elements (Figure 2A). The Nes-CreERT2 transgene, containing the i2E, likely functions as a weaker reporter of neuronal enhancer complexes (due to its orientation). Nes-GFP, containing the Nes promoter and i2E and mimicking the orientation of the endogenous Nes gene, reports a wide-range of transcriptional activities, ranging from weak to strong GFP signals, at different developmental stages (Figure 2A). Likewise, there are two transcriptional activities that regulate Nes-GFP, which are differentially associated with specification of the SSC lineage (defined by Lepr-Cre+/Nes-GFPlow/+) and with the periarteriolar HSC niche (defined by NG2-CreERTM+/Nes-GFPhigh cells).
In the case of Lepr regulation, one potential misinterpretation of transcriptomic data might also be due to the orientation of reporter genes (e.g., GFP) that are used to depict transcriptional activity of different transcript variants. The Lepr has a large and complicated genomic organization, which transcribes multiple mRNA variants from a promoter (designated as P2), which is different from that of humans (P1) (Figure 1B). Interestingly, the mouse P2 promoter-initiated transcripts are often terminated at different exons that are proximal to mouse exon 19 (exon 20 in the human counterpart) of the canonical Lepr gene (Figure 1B). Therefore, a knockin reporter immediately after exon 19 in mice only depicts the canonical Lepr transcriptional activities (DeFalco et al., 2001), likely masking or underscoring the contribution of different Lepr isoforms or alternative transcripts (terminated at a different exon) to lineage differentiation (Kunisaki et al., 2013, Zhou et al., 2014). In general, Nes- or Lepr-driven transgene or knockin gene expression seems far more complicated than their endogenous mRNA and protein expression at different developmental stages (Table S1). Therefore, genomic elements and their orientations must be taken into consideration when designing or choosing desired genetic models, and interpretation of transgene reporter data.
Random Chromosomal Integration of Transgenes
Besides the interference from genomic elements and their orientations, random chromosomal integrations of the same transgene could be another major factor that explains different transgene expression patterns in mouse BM. Theoretically, no two transgenic lines are created equal when the genomic elements are randomly integrated in the mouse genome. The variability of transgenic lines has created some confusion and misinterpretation of data when using transgenic reporter as markers. In the case of the Nes gene, there are approximately nine Nes-CreERT2 transgenic lines that have been designed and used to study mouse neural stem cells and progenitors (Sun et al., 2014). However, the expression patterns of these Nes-CreERT2 lines vary greatly in the mouse brain, apparently because of random chromosomal integration. Only very small subset lines expressed Nes-CreER at the neurogenic zones of the adult brain, like that of the endogenous Nes gene (Sun et al., 2014). Thus, each individual line should be fully characterized prior to use for a specific need.
Gene Expression Methods: Constitutive Versus Inducible Expression
It is conceivable that different gene manipulations would also have a significant impact on their expression patterns. As already discussed above, Nes-GFP and Nes-CreERT2 have different transgene expression patterns in the BM (Isern et al., 2014, Ono et al., 2014, Zhou et al., 2014), which may be partially explained by their differences in regulatory elements and their orientations in the expression vectors (Figure 2A). Moreover, these two transgenes also differ in the regulatory elements that control their expression. Nes-GFP has a constitutively active Nes promoter and i2E in neurogenic cells (Figure 2A). Nes-CreERT2 contains a tamoxifen-inducible CreERT2. Nevertheless, the definite role of CreERT2 in the contribution to experimental discrepancies remains to be determined for the complexity of the two transgene systems. Moreover, it also remains to be established whether a constitutive versus inducible modification would lead to a significant experimental difference or discrepancy.
Fortunately, a pairwise comparison between another two transgene expression systems (i.e., NG2-Cre and NG2-CreER™), in which the genetic elements (i.e., the nuclear localization signal [NLS] and CreER™) that control constitutive and inducible expression, respectively, are the only difference (Figure 2B). This comparison presents a convincingly positive answer to the above question (Asada et al., 2017). NG2-Cre, with a constitutively active NLS, marks BM stromal cells at both periarteriolar and sinusoidal areas; whereas NG2-CreER™, with a tamoxifen-inducible Cre-ER™, preferentially marks periarteriolar cells, presumably presenting distinct HSC niche-supporting function (Asada et al., 2017). Thus, NG2-Cre- and NG2-creER™-marked cells showed differential HSC niche-supporting functions, in which NG2-Cre+, but not NG2-creERTM+, cells are the source of the major HSC niche factor, Scf, whose deletion in NG2-Cre mice led to a defect in multi-lineage reconstitution in the BM (Asada et al., 2017). Clearly, these results provide insights into how different genetic approaches can impact on experimental conclusions, thereby presenting, at least partial resolution, of the current debate between the existence of distinct and uniform HSC niches in BM (Acar et al., 2015, Kunisaki et al., 2013).
Despite the inducible systems that enable a spatial-temporal activation of marker genes for single-cell lineage tracing, some gene-inducible systems are particularly leaky in terms of their system specificity. Moreover, the side effects of inducible reagents on a particular cell type should be taken into account. This could be exemplified by the intriguing ligand-dependent Cre recombinase that is inducible by administration of tamoxifen. Tamoxifen blocks the actions of estrogen, a female hormone, and is used to treat several types of breast cancer in clinics. It has been shown that tamoxifen has mixed estrogen-agonist effects and may alter bone and chondrocyte growth, and signal transduction between endothelial cells and pericytes (Table 1) (Chagin et al., 2007, Karimian et al., 2008). Furthermore, cellular cytotoxicity may be encountered in both constitutive and inducible systems. Such cytotoxicity has been observed in Cre-ER activation in induced hematological disorders (Higashi et al., 2009), thus rendering non-specific phenotypes to BM stromal cells.
It was also shown that high levels of Cre recombinase expression in mouse embryo fibroblasts induced DNA damage and inhibited cell growth in a Cre-ER activity-dependent manner (Loonstra et al., 2001), and, in neuronal stem and progenitor cells, led to increased aneuploidy, cell death, and brain developmental defects (Forni et al., 2006). These studies highlight the potential problems for developmental studies of BM cells, especially Nes-CreERT2- and NG2-CreER™-expressing cells with a high neurogenic promoter or enhancer activity. Consequently, it is unknown whether there are significant amounts of NG2-CreERhigh and Nes-CreERhigh cells in previous studies (Acar et al., 2015, Kunisaki et al., 2013), which may be eliminated due to a high nuclear Cre activity. Hence, it is important to titrate Cre activity in each individual transgenic line, to use low levels of Cre-ER that permit for desired recombination without cell cytotoxicity, and to have tight tamoxifen-inducible controls when these Cre-based systems are used to study complicated dynamic changes of SSC and HSC niches.
Complexity of Dynamic Changes of Cellular and Molecular States in Development
Currently, the big challenge is to deeply understand the complexities of cellular and molecular states of BM cells at different developmental stages, which are thought to be tightly co-regulated by largely unknown mechanisms. At the cellular level, some cell identities present only in a transient state at a specific stage, which sometimes are too dynamic to be identified. For example, we have discussed that Nes-GFP+ and Nes-CreERT2+ cells are differentially presented in both prenatal and postnatal BMs. Nes-CreERT2 transcription may be repressed before the formation of the primary ossification center, but de-repressed after the development of the primary ossification center and the marrow cavity (Ono et al., 2014). Under the condition of tamoxifen induction at P0 and chase to P7, Isern et al. (2014) found that Nes-CreERT2+ cells were highly co-localized with Nes-GFP+ cells in BM. These data suggest that the Nes i2E expression is dominant at the neonatal stage (i.e., P0 to P7), which might have coupled with one core transcriptional mechanism that regulates BM stem cell niches in a developmental stage-specific manner. Of note, promoter/enhancer activity, mRNA expression, and protein expression of the marker gene of interest may be consistent or inconsistent at different developmental stages (Table S1). Thus, specific developmental windows used for induction experiments and retrieving data are required to be consistent or specifically identified for comparative studies.
Regardless of the existence of complicated cell types in BM, the underlying mechanisms that regulate their cell identities involve gene regulation not only at the transcriptional level, but also at many different molecular levels (e.g., epigenomic, post-transcriptional, and translational modifications) (Figure 3A). These complicated gene regulatory mechanisms might make transgenic data interpretation even more difficult. Therefore, we should be aware that reporter gene expression is not always consistent with its mRNA and protein expression patterns (Figure 3A). One could not assume that Nes-GFPhigh cells must have high levels of endogenous Nes mRNAs or nestin protein expression. In general, each individual mouse strain (e.g., Nes-GFP or Nes-CreERT2) should be considered as an independent assay tool for in vivo cell fate, functional analysis, and translational studies of mouse BM biology. In the following sections, we will further discuss the translational implication, potential challenges, and future considerations of mouse genetic models, mainly based on integrating existing genetic data and genomic informatics from both mouse and human studies (Figure 3B).
Mouse Genetic Models versus Human Resource Databases: Translational Relevance, Challenges, and Prospective Considerations
In the biomedical field, the ultimate goals of using diverse animal models are to provide translatable biomedical information to understand the etiology of human diseases and to derive effective clinical treatments for patients. On the one hand, the success of this translational approach relies on the understanding of the interplay of datasets between murine models and the human cells. On the other hand, the availability of genome-wide datasets in the post human genome era offers the possibility to optimize mouse genetic models through existing coherent human datasets (Birney et al., 2007, Kundaje et al., 2015) (Figure 3B). However, the above interplay approaches have not been well integrated to guide stem cell research. Here, we will focus on the interpretation of mouse model data based on the transcriptomic complexity of marker gene transcripts in both humans and mice. Furthermore, we will shed light on how “generic” epigenomic markers from redundant human epigenomic databases could provide prospective molecular cell identities for facilitating translational biology.
Transcriptomic Complexity of Marker Gene Transcripts
To better integrate human genomic data with mouse models, we initially analyzed the genomic organization of the human NES and LEPR loci, due to the availability of datasets, and because the two reporters, Nes-GFP and Lepr-Cre, have been extensively used to categorize stem cell identities in animal models as discussed above (Tables 1 and S1). Figure 1A illustrates the genomic organization of these two genes from mice, rats, and humans. In the case of the homolog genes that encode the nestin protein, there are significantly conserved intronic and exonic structures, but with some variations found in the 20- to 25-kb 5′ genomic regions (Figure 1A). Likely, the similar genomic organizations of Nes and NES among mice, rats, and humans would make in vivo animal studies more relevant to clinical sittings.
However, with respect to the leptin receptor genes, genomic sequence data reveal significant differences between the species in terms of gene structures, function, transcription start sites, alternative transcripts, and the locations of the last exon in each individual transcript (Figure 1B). During embryonic development, LEPR isoform A (LEPR-A) is expressed in fetal liver, hematopoietic tissues, and the choroid plexus. In adults, LEPR-A is highly expressed in mesoendoderm-derived tissues (such as heart, liver, small intestine, prostate, and ovary) (www.SWISS_Prot). However, LEPR-B (the canonical isoform) is highly expressed in neuroectodermal tissues, including the choroid plexus and hypothalamus, in adult humans and mice (Figure 1B). Notably, in humans, there are at least five LEPR protein isoforms derived from six mRNA transcripts, which are expressed in a tissue-specific manner (Figure 1B). Thus, this genomic or proteomic information is particularly useful for us to design GFP- or Cre-based Lepr constructs in murine models to study tissue-specific regulation of cellular states.
Furthermore, the LEPR and LEPROT (leptin receptor overlapping transcripts) genes, which encode two distinct proteins, share the same promoter and the first two exons (Figure 1B). The orientation of the two genes are similar in both human and rat genomes, but different from that of mice (Figure 1B). The mouse Leprot is approximately 50 kb away from Lepr (Figure 1B). Importantly, we need to determine where the reporter activity is initiated. The differential activation of LEPR and LEPROT promoters or enhancers may render opposite interpretations of the results.
Generic Epigenomic Markers and Molecular Cell Identities
Redundant epigenomic databases represent a valuable tool for defining various epigenomic and transcriptional states during development. It is unknown whether we could also accurately define molecular cell identities using a panel of “generic” epigenomic markers, which are currently available in miscellaneous human epigenomic databases. We evaluated the presence or absence of the monomethylated histone H3 lysine four epigenomic marker (H3K4me1), at the human NES and LEPR loci, enabled by the availability of a large H3K4me1 chromatin immunoprecipitation sequencing (ChiP-seq) dataset in 219 human cell samples (www.genboree.org). The 219 samples comprise cell types from all three germ layers and the trophectoderm (Table S2; Figure S1). H3K4me1 usually pre-marks the enhancers that are not active, but primed for activation, in the absence of external stimuli or signals (Shlyueva et al., 2014). As shown in Figure 4, the dendrogram reveals two genomic clusters that separate the majority of marked introns and exons of LEPR from those of NES (Figures 4A and 4B). Moreover, H3K4me1 segregates the previously well-characterized regulatory regions (i.e., intron 1 and 2 enhancers, denoted as i1E and i2E, respectively) of the NES (or Nes) gene, validating the reliability of using H3K4me1 for cell-identity classification in this analysis. Therefore, H3K4me1 segregates all samples into three major cell clusters, in which cell clusters 1 and 3 are clearly different (Figures 4A and 4B). Cell cluster 1 (containing predominantly mesodermal derivatives) is apparently regulated by H3K4me1 on the promoter region, intron 1, and exon 2 of the LEPR gene (Figure 4B, lower panel). Interestingly, H3K4me1 marks cell cluster 3, containing predominantly neural and epidermal/ectodermal derivatives (e.g., brain and foreskin tissues), on introns 1 and 2 of the NES promoter (Figure 4B, upper panel). The inclusion of pluripotent stem cells and their differentiated cell types in the cell cluster 3 merely reflects the developmental proximity between the neuroectoderm and embryonic epiblasts. Cell cluster 2, which partially overlaps with the cell clusters 1 and 3, requires additional markers to identify their cell identities. Nevertheless, these data suggest that even a generic marker (such as H3K4me1) on limited genomic loci (e.g., LEPR and NES) could bear remarkable epigenetic information to classify mesodermal and ectodermal disparities.
Accordingly, we further analyzed three major epigenomic markers, H3K27ac, H3K4me1, and H3K4me3, at the NES and LEPR loci, based on the Encyclopedia of DNA Elements at UCSC (2003–2012) (genome.ucsc.edu). Unlike H3K4me1, H3K27ac marks active enhancers at transcriptional factor-accessible genomic loci (Creyghton et al., 2010), whereas H3K4me3 marks gene promoters that are active or poised to be active (Benayoun et al., 2014, Lauberth et al., 2013). In brief, we were able to integrate data-informatics cascades from epigenomics, transcriptomics, and chromatin proteomics into regulatory complexes for monitoring cell identity in human embryonic stem cell line H1 (WA01) and other mesodermal or ectodermal cell lines (i.e., human skeletal muscle cells and myoblasts, HUVECs [human umbilical vein endothelial cells], normal human lung fibroblasts, and normal human epidermal keratinocytes) (Figure 5A). Mapping of potential transcriptional regulators on chromatin at the NES and LEPR loci (e.g., at BM stromal cell cluster 1, Figure 4B) would provide new insights into Nes-GFP and Lepr-Cre transcriptome activities that are commonly monitored in mouse models.
Indeed, LEPR represents a complicated regulation due to the presence of multiple alternative transcripts and the co-regulated LEPROT gene (Figure 1B). The three histone markers are increased on the promoter region adjacent to exons 1 and 2 among the four cell lines (except H1) (Figure 5A, right panel). Interestingly, H3K4me1 was located at multiple regions in intron 2 and in two 3′ exonic areas in HUVECs (Figure 5A, right panel). The biological consequences of these sites remain unclear. However, they might be associated with alternative transcription start sites of the gene, therefore potentially interfering with Lepr-Cre transcriptome interpretation in endothelial cells. Based on the recruitment of RNA polymerase II, we identified at least two promoters (i.e., P1 and P2) on the full-length LEPR. The two promoters appear to be consistent with their epigenetic states (Figures 5A, right panel, 5B, and 5C). These data confirm the presence of alternative transcripts due to differential initiation of transcription under diverse cellular contexts.
Interestingly, a neuronal repressor complex (that involves both NRSF and SIN3A) was drastically downregulated at the P1 promoter. Concomitantly, there is an increase in neural activation complex (NAC) that contains PFH8, GABPA, and ELF1 at the both P1 and P2 promoters (Figures 5B and 5C). The P2 promoter (located at the 5′ end of exon 3) seems to be a weaker promoter compared with P1. Moreover, P2 is apparently regulated by the NAC that includes either BRG1 or PHF8 (or both), polycomb group repressive complex proteins (e.g., EZH2 and SUZ12), and GATA binding proteins (e.g., GATA2 and GATA3) (Figure 5B). Thus, a neural-specific regulation of the P2 promoter has been implicated in the human LEPR gene (Satoh et al., 2009).
Taken together, transcription factor profiling of the LEPR promoters reveals a potential molecular switch between neuroectodermal and mesodermal regulation, suggesting a possible coupling mechanism between sequential de-repression and activation, which controls cell-type-specific transcriptional activity at the P1 and P2 promoters (Figure 5C). Of note, we need to be aware of using eipgenomic data from human cell lines, which might increase the possibility of altered epigenetic marker expression under certain cell culture conditions. In the future, these analyses should include large-scale epigenomic data from human tissues and purified human cell populations. It would be also desirable to have a side-by-side comparative analysis between mouse and human epigenomic datasets. Ultimately, we would be able to make humanized mouse models by integrating partial human genomic or epigenomic information into a transgenic mouse model for translational studies.
Concluding Remarks
Constitutive and inducible expression based on various types of transgenes have identified a plethora of functionally important stem cell and progenitor populations in BM. Various experimental discrepancies, data irreproducibility, and misinterpretations could be explained, minimized, and circumvented if we have a better understanding of these mouse genetic systems. Ideally, we should develop and apply non-toxic, cell-type-specific, regulatable, and humanized mouse genetic systems, combined with other technological approaches for in vivo cell-fate analysis. Theoretically, molecular signatures of cell identities could be evident at multiple levels of gene regulation, resulting in activation of transcriptional complexes, mRNA transcription, and protein translation at different developmental stages. Practically, we need to be aware of these differences when we interpret data based on mouse reporter activity (e.g., from Nes-GFP, Nes-CreER, and Lepr-Cre), mRNA transcripts, and protein expression. Each gene regulation or expression mechanism should be considered as an independent assay for lineage analysis. Importantly, all genetic and epigenetic assays should be combined with definitive surface marker analysis and genome-wide “clusterome” to accurately define specific cellular states and cell identities. Precise understanding of the regulation of reporter transcriptomes in murine models would enable us to accurately decipher diverse cell fates in the BM.
Acknowledgments
This work was supported by the Intramural Research Program of the NIH at the National Institute of Neurological Disorders and Stroke (to K.G.C. and K.R.J.) and the National Institute of Dental and Craniofacial Research (to P.G.R.). We thank Dr. Ronald D. McKay for discussion and support of this work.
Footnotes
Supplemental Information includes one figure and two tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.09.014.
Contributor Information
Kevin G. Chen, Email: cheng@mail.nih.gov.
Pamela G. Robey, Email: probey@dir.nidcr.nih.gov.
Supplemental Information
References
- Acar M., Kocherlakota K.S., Murphy M.M., Peyer J.G., Oguro H., Inra C.N., Jaiyeola C., Zhao Z., Luby-Phelps K., Morrison S.J. Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature. 2015;526:126–130. doi: 10.1038/nature15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ara T., Tokoyoda K., Sugiyama T., Egawa T., Kawabata K., Nagasawa T. Long-term hematopoietic stem cells require stromal cell-derived factor-1 for colonizing bone marrow during ontogeny. Immunity. 2003;19:257–267. doi: 10.1016/s1074-7613(03)00201-2. [DOI] [PubMed] [Google Scholar]
- Asada N., Kunisaki Y., Pierce H., Wang Z., Fernandez N.F., Birbrair A., Ma'ayan A., Frenette P.S. Differential cytokine contributions of perivascular haematopoietic stem cell niches. Nat. Cell Biol. 2017;19:214–223. doi: 10.1038/ncb3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balordi F., Fishell G. Mosaic removal of hedgehog signaling in the adult SVZ reveals that the residual wild-type stem cells have a limited capacity for self-renewal. J. Neurosci. 2007;27:14248–14259. doi: 10.1523/JNEUROSCI.4531-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayoun B.A., Pollina E.A., Ucar D., Mahmoudi S., Karra K., Wong E.D., Devarajan K., Daugherty A.C., Kundaje A.B., Mancini E. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158:673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P., Robey P.G. Skeletal stem cells. Development. 2015;142:1023–1027. doi: 10.1242/dev.102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P., Cao X., Frenette P.S., Mao J.J., Robey P.G., Simmons P.J., Wang C.Y. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat. Med. 2013;19:35–42. doi: 10.1038/nm.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch T., Heppner F.L., Tertilt C., Heinen T.J., Kremer M., Wunderlich F.T., Jung S., Waisman A. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- Chagin A.S., Karimian E., Zaman F., Takigawa M., Chrysis D., Savendahl L. Tamoxifen induces permanent growth arrest through selective induction of apoptosis in growth plate chondrocytes in cultured rat metatarsal bones. Bone. 2007;40:1415–1424. doi: 10.1016/j.bone.2006.12.066. [DOI] [PubMed] [Google Scholar]
- Cohen P., Zhao C., Cai X., Montez J.M., Rohani S.C., Feinstein P., Mombaerts P., Friedman J.M. Selective deletion of leptin receptor in neurons leads to obesity. J. Clin. Invest. 2001;108:1113–1121. doi: 10.1172/JCI13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian P.S., White R., Hoare S.A., Fawell S.E., Parker M.G. Identification of residues in the estrogen receptor that confer differential sensitivity to estrogen and hydroxytamoxifen. Mol. Endocrinol. 1993;7:232–240. doi: 10.1210/mend.7.2.8469236. [DOI] [PubMed] [Google Scholar]
- Danielian P.S., Muccino D., Rowitch D.H., Michael S.K., McMahon A.P. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- DeFalco J., Tomishima M., Liu H., Zhao C., Cai X., Marth J.D., Enquist L., Friedman J.M. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608–2613. doi: 10.1126/science.1056602. [DOI] [PubMed] [Google Scholar]
- Ding L., Morrison S.J. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R., Wagner J., Metzger D., Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Feltri M.L., D'Antonio M., Previtali S., Fasolini M., Messing A., Wrabetz L. P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann. N. Y. Acad. Sci. 1999;883:116–123. [PubMed] [Google Scholar]
- Forni P.E., Scuoppo C., Imayoshi I., Taulli R., Dastru W., Sala V., Betz U.A., Muzzi P., Martinuzzi D., Vercelli A.E. High levels of Cre expression in neuronal progenitors cause defects in brain development leading to microencephaly and hydrocephaly. J. Neurosci. 2006;26:9593–9602. doi: 10.1523/JNEUROSCI.2815-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenette P.S., Pinho S., Lucas D., Scheiermann C. Mesenchymal stem cell: keystone of the hematopoietic stem cell niche and a stepping-stone for regenerative medicine. Annu. Rev. Immunol. 2013;31:285–316. doi: 10.1146/annurev-immunol-032712-095919. [DOI] [PubMed] [Google Scholar]
- Friedenstein A.J., Piatetzky S., II, Petrakova K.V. Osteogenesis in transplants of bone marrow cells. J. Embryol. Exp. Morphol. 1966;16:381–390. [PubMed] [Google Scholar]
- Higashi A.Y., Ikawa T., Muramatsu M., Economides A.N., Niwa A., Okuda T., Murphy A.J., Rojas J., Heike T., Nakahata T. Direct hematological toxicity and illegitimate chromosomal recombination caused by the systemic activation of CreERT2. J. Immunol. 2009;182:5633–5640. doi: 10.4049/jimmunol.0802413. [DOI] [PubMed] [Google Scholar]
- Isern J., Garcia-Garcia A., Martin A.M., Arranz L., Martin-Perez D., Torroja C., Sanchez-Cabo F., Mendez-Ferrer S. The neural crest is a source of mesenchymal stem cells with specialized hematopoietic stem cell niche function. Elife. 2014;3:e03696. doi: 10.7554/eLife.03696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalajzic I., Kalajzic Z., Kaliterna M., Gronowicz G., Clark S.H., Lichtler A.C., Rowe D. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J. Bone Miner. Res. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- Karimian E., Chagin A.S., Gjerde J., Heino T., Lien E.A., Ohlsson C., Savendahl L. Tamoxifen impairs both longitudinal and cortical bone growth in young male rats. J. Bone Miner. Res. 2008;23:1267–1277. doi: 10.1359/jbmr.080319. [DOI] [PubMed] [Google Scholar]
- Kfoury Y., Scadden D.T. Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell. 2015;16:239–253. doi: 10.1016/j.stem.2015.02.019. [DOI] [PubMed] [Google Scholar]
- Kuhn R., Schwenk F., Aguet M., Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisaki Y., Bruns I., Scheiermann C., Ahmed J., Pinho S., Zhang D., Mizoguchi T., Wei Q., Lucas D., Ito K. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace D.C., Whitman M.C., Noonan M.A., Ables J.L., DeCarolis N.A., Arguello A.A., Donovan M.H., Fischer S.J., Farnbauch L.A., Beech R.D. Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. J. Neurosci. 2007;27:12623–12629. doi: 10.1523/JNEUROSCI.3812-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauberth S.M., Nakayama T., Wu X., Ferris A.L., Tang Z., Hughes S.H., Roeder R.G. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendahl U., Zimmerman L.B., McKay R.D. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60:585–595. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- Lewis A.E., Vasudevan H.N., O'Neill A.K., Soriano P., Bush J.O. The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev. Biol. 2013;379:229–234. doi: 10.1016/j.ydbio.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood T.D., Hancock D.C., Danielian P.S., Parker M.G., Evan G.I. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M., Martin J.F., Nagy A., Lobe C., Olson E.N., Tabin C.J. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- Loonstra A., Vooijs M., Beverloo H.B., Allak B.A., van Drunen E., Kanaar R., Berns A., Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Ferrer S., Scadden D.T., Sanchez-Aguilera A. Bone marrow stem cells: current and emerging concepts. Ann. N. Y. Acad. Sci. 2015;1335:32–44. doi: 10.1111/nyas.12641. [DOI] [PubMed] [Google Scholar]
- Mignone J.L., Kukekov V., Chiang A.S., Steindler D., Enikolopov G. Neural stem and progenitor cells in nestin-GFP transgenic mice. J. Comp. Neurol. 2004;469:311–324. doi: 10.1002/cne.10964. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T., Pinho S., Ahmed J., Kunisaki Y., Hanoun M., Mendelson A., Ono N., Kronenberg H.M., Frenette P.S. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev. Cell. 2014;29:340–349. doi: 10.1016/j.devcel.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa S., Mabuchi Y., Niibe K., Suzuki S., Nagoshi N., Sunabori T., Shimmura S., Nagai Y., Nakagawa T., Okano H. Development of mesenchymal stem cells partially originate from the neural crest. Biochem. Biophys. Res. Commun. 2009;379:1114–1119. doi: 10.1016/j.bbrc.2009.01.031. [DOI] [PubMed] [Google Scholar]
- Morrison S.J. Time to do something about reproducibility. Elife. 2014;3 doi: 10.7554/eLife.03981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S.J., Scadden D.T. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro H., Ding L., Morrison S.J. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono N., Ono W., Mizoguchi T., Nagasawa T., Frenette P.S., Kronenberg H.M. Vasculature-associated cells expressing nestin in developing bones encompass early cells in the osteoblast and endothelial lineage. Dev. Cell. 2014;29:330–339. doi: 10.1016/j.devcel.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen M., Friedenstein A.J. Stromal stem cells: marrow-derived osteogenic precursors. Ciba Found. Symp. 1988;136:42–60. doi: 10.1002/9780470513637.ch4. [DOI] [PubMed] [Google Scholar]
- Sacchetti B., Funari A., Remoli C., Giannicola G., Kogler G., Liedtke S., Cossu G., Serafini M., Sampaolesi M., Tagliafico E. No identical “mesenchymal stem cells” at different times and sites: human committed progenitors of distinct origin and differentiation potential are incorporated as adventitial cells in microvessels. Stem Cell Reports. 2016;6:897–913. doi: 10.1016/j.stemcr.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T., Yoshino S., Katano A., Ishizuka T., Tomaru T., Shibusawa N., Hashimoto K., Yamada M., Mori M. Isolation of a novel leptin receptor gene promoter preferentially functioning in neuronal cells. Biochem. Biophys. Res. Commun. 2009;389:673–677. doi: 10.1016/j.bbrc.2009.09.056. [DOI] [PubMed] [Google Scholar]
- Shlyueva D., Stampfel G., Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Kohara H., Noda M., Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Sun M.Y., Yetman M.J., Lee T.C., Chen Y., Jankowsky J.L. Specificity and efficiency of reporter expression in adult neural progenitors vary substantially among nestin-CreER(T2) lines. J. Comp. Neurol. 2014;522:1191–1208. doi: 10.1002/cne.23497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P.C., Bock R., Klein R., Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Abe K., Mantani A., Hitoshi Y., Suzuki M., Osuzu F., Kuratani S., Yamamura K. A novel transgenic technique that allows specific marking of the neural crest cell lineage in mice. Dev. Biol. 1999;212:191–203. doi: 10.1006/dbio.1999.9323. [DOI] [PubMed] [Google Scholar]
- Zhou B.O., Yue R., Murphy M.M., Peyer J.G., Morrison S.J. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Bergles D.E., Nishiyama A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development. 2008;135:145–157. doi: 10.1242/dev.004895. [DOI] [PubMed] [Google Scholar]
- Zhu X., Hill R.A., Dietrich D., Komitova M., Suzuki R., Nishiyama A. Age-dependent fate and lineage restriction of single NG2 cells. Development. 2011;138:745–753. doi: 10.1242/dev.047951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman L., Parr B., Lendahl U., Cunningham M., McKay R., Gavin B., Mann J., Vassileva G., McMahon A. Independent regulatory elements in the nestin gene direct transgene expression to neural stem cells or muscle precursors. Neuron. 1994;12:11–24. doi: 10.1016/0896-6273(94)90148-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.