Abstract
Evolutionary thinking continues to challenge our views on health and disease. Yet, there is a communication gap between evolutionary biologists and toxicologists in recognizing the connections among developmental pathways, high-throughput screening, and birth defects in humans. To increase our capability in identifying potential developmental toxicants in humans, we propose to apply evolutionary genetics to improve the experimental design and data interpretation with various in vitro and whole-organism models. We review five molecular systems of stress response and update 18 consensual cell-cell signaling pathways that are the hallmark for early development, organogenesis, and differentiation; and revisit the principles of teratology in light of recent advances in high-throughput screening, big data techniques, and systems toxicology. Multiscale systems modeling plays an integral role in the evolutionary approach to cross-species extrapolation. Phylogenetic analysis and comparative bioinformatics are both valuable tools in identifying and validating the molecular initiating events that account for adverse developmental outcomes in humans. The discordance of susceptibility between test species and humans (ontogeny) reflects their differences in evolutionary history (phylogeny). This synthesis not only can lead to novel applications in developmental toxicity and risk assessment, but also can pave the way for applying an evo-devo perspective to the study of developmental origins of health and disease.
Introduction
Environmental exposures are associated with morbidity and mortality in children’s health globally (WHO 2005; Suk et al. 2016). About 3–4 % of newborn infants are identified as having a structural malformation at birth, including cleft palate, missing limbs (amelia), missing or extra digits (adactyly and polydactyly), no or small eyes (anophthalmia and microphthalmia), hearts that have a leaking septum (ventricular septal defect), spinal cords that close incompletely (spina bifida), and so forth (Thorogood 1997; Epstein 2008). Most developmental defects have a complex etiology, with genetic, environmental, and social contributing factors. Maternal exposures to pharmaceuticals, illicit drugs, and some environmental chemicals are thought to account for 6% of human birth defects (Nelson and Holmes 1989). More than 90,000 manufactured chemicals have been captured in the U.S. Environmental Protection Agency’s (EPA) Toxic Substances Control Act (TSCA) inventory, and, each year, the EPA receives notification of the manufacture of 500 to 1,000 more (U.S. EPA 2015). Most of these chemicals have not been screened for developmental toxicity, and the wide blend of chemical structures elevates the challenge of assessing their risks to human health and the environment (Judson et al. 2009). Even where some data are available, the mechanisms of developmental toxicity for environmental chemicals are not understood in enough detail or depth for risk assessment purposes.
To assess the potential toxicological effects of an ever-increasing number of chemicals, 21st-century toxicology approaches are building large datasets of high-throughput screening (HTS) data to profile in vitro bioactivity of chemical libraries (NRC 2007; Tice et al. 2013). This in vitro bioactivity information covers chemical-biological interactions across a broad suite of experimental platforms and biological scales, from molecular lesions, subcellular events, and cellular disruption, to tissue dysfunction, and is generated from an array of in vitro experimental systems, including human cell lines, stem cells, small model organisms and, most recently, engineered microscale and microphysiological systems (Collins et al. 2008; Tice et al. 2013; Settivari et al. 2015). Guideline testing for reproductive toxicity and prenatal developmental toxicity traditionally has relied on lower throughput approaches in whole-animal studies utilizing vertebrate species [clawed frogs, mice, rats, and rabbits (Carney et al. 2008; Mouche et al. 2011; Robinson et al. 2012; Kalaskar et al. 2014)] and higher throughput, more evolutionarily distant alternatives [zebrafish and invertebrate species, such as hydra, roundworms, water fleas, and fruit flies (Dang et al. 2012; Padilla et al. 2012; Glauber et al. 2013; Liu et al. 2014; Boyd et al. 2016)]. The vast collections of in vitro data now available from 21st-century toxicology approaches, coupled with the diverse and distributed nature of these data and an ever-increasing knowledgebase for embryonic development across diverse species and biological systems, create a major challenge for data access, curation, integration, and interpretation.
Cross-species extrapolation is a challenging and continuing aspect of developmental toxicology and risk assessment. In the 1950s and 60s, use of thalidomide―a sedative used as a morning sickness pill―caused limb deformities (phocomelia) in over 10,000 children (Ito and Handa 2012; Vargesson 2015). Although thalidomide tested negative for limb teratogenesis in rodent species (rats and mice), the phocomelia commonly ascribed to human thalidomide embryopathy has been observed with this drug during embryonic development for rabbits and monkeys, as well as avian and zebrafish species (Hansen et al. 2002; Therapontos et al. 2009; Ema et al. 2010; Ito et al. 2010; see review by Vargesson [2015]; Figure 1). The discordance of susceptibility across species presents a challenge in extrapolating human risk based on data from various model organisms. Another example of species-specific differences can be seen in the Testicular Dysgenesis Syndrome (TDS), the observed clinical linkages of developmental defects in the male reproductive tract to genetic and environmental factors underlying congenital undescended testes (cryptorchidism) and malformations of the genital tubercle [hypospadias (Skakkebaek et al. 2016)]. In the experimental studies by Heger et al. (2012) and HTS modeling by Leung et al. (2016b), rats were shown to be more susceptible than mice to male reproductive toxicants. Some notable examples of chemicals that can induce one or more common endpoints of TDS include bisphenol A, flutamide, phthalates, and vinclozolin (see Leung et al. 2016b, references therein). Only about 20% of male reproductive toxicants reported in rat studies were shown also to be male reproductive toxicants in mouse; those chemicals accounted for about 67% of male reproductive toxicants reported in the mouse studies. To address the discordance of susceptibility in cross-species extrapolation, a novel organizing principle is needed to make the massive amount of toxicity data across multiple experimental systems and species accessible and useful for predictive understanding of developmental toxicity in humans.
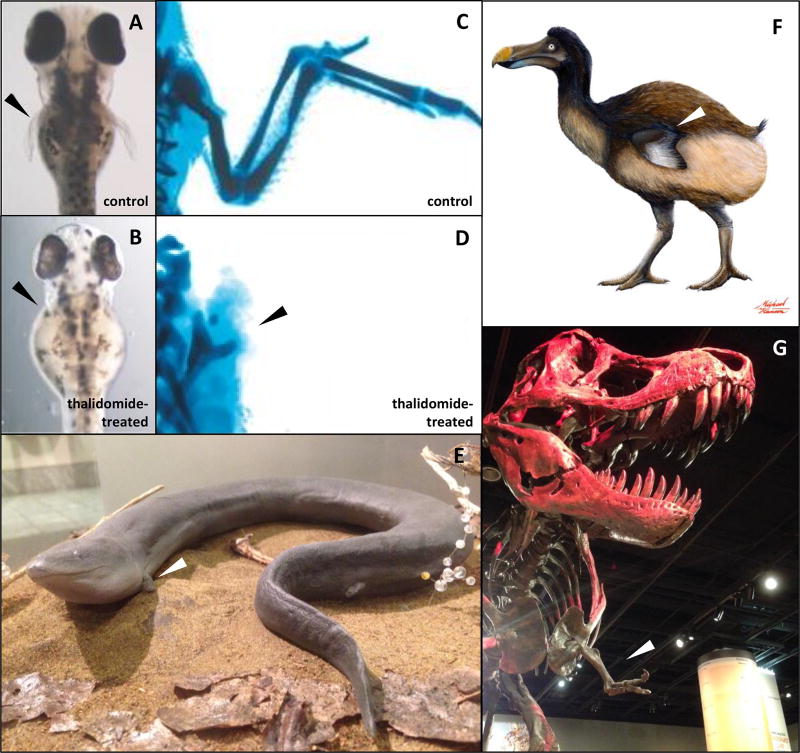
Figure 1. Discovery from Natural Selection: Thalidomide-induced limb deformities in D. rerio and G. gallus in comparison with forelimb atrophy in A. means, R. cucullatus, and T. rex.
Limb deformities (phocomelia) can be observed with exposure to thalidomide in (A, B) D. rerio, (C, D) G. gallus, and humans. In the course of evolution, a similar developmental outcome (forelimb atrophy) is stabilized to better adapt to ecological niches in (E) two-toed amphiuma (Amphiuma means), a native species of salamanders in North Carolina; (F) flightless birds, such as the dodos (Raphus cucullatus); and (G) the family Tyrannosauridae and subfamily Carnotaurinae of theropod dinosaurs, such as Tyrannosaurus rex (Guinard 2015). The images were used with permission of (A–D) the American Association for the Advancement of Science (Ito et al. 2010), and (F) Michael Hanson (Yale University); and taken in (E) the North Carolina Museum of Natural Sciences, Raleigh, NC, and (G) the National Museum of Natural History, Smithsonian Institution, Washington, D.C.
The challenge of integrating and extrapolating toxicity information across species can be met from a phylogenetic perspective. Evolutionary genetics is the study of how genetic variation leads to evolutionary change (Dobzhansky 1937). It includes topics such as the genetic basis of speciation and adaptation, genetic change in response to selection within populations, the origins and patterns of biodiversity, and the processes that maintain biodiversity. These topics have been exploited in diverse fields, such as infectious disease (Ebert 1998; Bull and Lauring 2014), cancer therapeutics (Burrell et al. 2013; Das Thakur and Stuart 2013), and ecotoxicology (Meyer et al. 2002; Wirgin et al. 2011; Whitehead et al. 2012; Reitzel et al. 2014a). Here, we explore ways that evolutionary genetics can be exploited to integrate our understanding of how developmental susceptibility varies between individual species―from small model organisms to humans―with the goals of focusing experimental design and reducing data dimensionality for the in vitro and in vivo assessment of developmental toxicity in general and teratogenesis in particular. Toward this end, we first review five molecular systems of stress response and update 18 consensual cell-cell signaling pathways that are the hallmark for early development, organogenesis, and differentiation. We then revisit the principles of teratology in light of recent advances in HTS technology, big data techniques, and systems toxicology. Finally, we discuss how to use multiscale systems modeling, phylogenetic analysis, and comparative bioinformatics in developmental toxicology and risk assessment.
Developmental Pathways and Toxicity Assessment
Embryonic development is a highly coordinated process that involves division, differentiation, migration, and destruction of cells at specific times and places. Although developmental processes and strategies differ markedly across diverse phyla, much developmental patterning is controlled by cell-cell signaling pathways that are highly conserved. In fact, the same types of molecules evolved into modular signaling pathways and gene regulatory networks, whereas conserved modules are used in different ways, not only across species but the life cycle as well. The 2000 National Research Council report, Scientific Frontiers in Developmental Toxicology and Risk Assessment, listed 17 consensual cell-cell signaling pathways that were then known to be functional in early development (6 pathways), organogenesis (4), and postdifferentiation (7) (NRC 2000; Abbott 2008). Here, we added the Per-ARNT-Sim (PAS) pathway as another relevant, early developmental pathway (Reitzel et al. 2014b; Brown 2014) and listed in Table 1 the intercellular ligands, specific receptors on or within the cell and a set of molecular intermediates that transmit signals to components of the transcription machinery within the cell. From a comparative perspective, conservation of cell signaling implies a fundamental strategy of how molecular information is used by the embryo, where so-called “toolkit genes” appear to play the same role across phyla, and in all vertebrate species from zebrafish to humans. Some of the best examples are transcription factors: Pax6 for eye development (Shaham et al. 2012), Nkx/tinman for heart development (Clowes et al. 2014), Hox genes for axial patterning (Casaca et al. 2014), and Hes-1 for molecular clocks (Harima et al. 2014).
Table 1. Developmental Pathways.
The 2000 National Research Council report, Scientific Frontiers in Developmental Toxicology and Risk Assessment, proposed 17 consensual cell-cell signaling pathways that are hallmarks of morphogenesis (NRC 2000; Abbott 2008). Here, we add the Per-ARNT-Sim (PAS) pathway, which consists of the hypoxia, circadian, and aryl hydrocarbon receptor (AHR) pathways (McIntosh et al. 2010; Hahn and Karchner 2012; Brown 2014). Some signaling intermediates are shared by different developmental pathways, such as mitogen-activated protein kinases (MAPK) and activator protein 1 (AP-1) by both small G-protein (Ras)-linked receptor tyrosine kinase and receptor guanylate cyclase pathways.
| Embryonic Stages | Developmental Pathways | Ligands, Receptors, and Signaling Intermediates |
|---|---|---|
|
| ||
| Early development and later | 1a and 1b. Wingless-int (Wnt) pathway (canonical and noncanonical) | Wnt proteins, β catenin, and jun N-terminal kinase (JNK) |
| 2. Receptor serine-threonine kinase pathway | Transforming growth factor β (TGFβ), bone morphogenetic proteins (BMPs), and Smad transcription factors | |
| 3. Sonic hedgehog (Shh) pathway | Shh, patched receptor (Ptc), and smoothened (Smo) | |
| 4. Small G-protein (Ras)-linked receptor tyrosine kinase pathway | Epidermal growth factor (EGF), vascular endothelial growth factor (VEGF), platelet-derived growth factor (PDGF), fibroblast growth factor (FGF), insulin-like growth factor, ephrins, protein kinase C (PKC), Ras, Rho, extracellular signal-regulated kinase (ERK), mitogen-activated protein kinase (MAPK), and jun N-terminal kinase (JNK/p38) | |
| 5. Notch-Delta pathway | Notch, Delta, Jagged, and Serrate | |
| 6. Cytokine receptor pathway | Growth hormone, erythropoietin, prolactin, thrombopoietin, interleukins, interferons, and Janus kinase (JAK)/signal transducer and activator of transcription protein (STAT) | |
| 7. Per-ARNT-Sim (PAS) pathway | Hypoxia-inducible factor (HIF), aryl hydrocarbon receptor (AHR), RAR-related orphan receptor A (RORA), heat shock protein 90 (HSP90), AHR repressor, nuclear translocator, and nuclear translocator-like (AHRR ARNT, and ARNTL), clock circadian regulator (CLOCK), and period and cryptochrome circadian clock (PER and CRY) | |
|
| ||
| Organogenesis and later | 8. Interleukin-1 receptor pathway | Nuclear factor-Kappa B (NFκB) and inhibitors (IκB) |
| 9. Nuclear hormone receptor pathway | Estrogen receptor (ER), glucocorticoid receptor (GR), mineralocorticoid receptor (MR), androgen receptor (AR), prostaglandin receptor (PR), thyroid hormone receptor (TR), vitamin D3 receptor (VDR), retinoic acid receptor (RAR), retinoid X receptor (RXR), and peroxisome proliferator-activated receptor (PPAR) | |
| 10. Apoptosis pathway | Caspase proteolytic enzymes, tumor necrosis factor, Fas, BAX, Bcl2, FADD, and TRADD | |
| 11. Receptor phosphotyrosine phosphatase pathway | Dephosphorylation of receptors and intermediates of other pathways | |
|
| ||
| Postdifferentiation | 12. Receptor guanylate cyclase pathway | c-Fos, JunB, cyclic AMP response element-binding protein (CREB), activator protein 1 (AP-1), and ion channels |
| 13. Nitric oxide receptor pathway | A cytoplasmic enzyme that binds NO at a heme group converting GTP to cyclic GMP and affecting transcription via c-Fos | |
| 14. G-protein-coupled receptor (large G proteins) pathway | A very broad range of ligands (proteins, peptides, and small molecules) that bind cell-surface receptors and affect a broad range of events (transcription, metabolism, motility, secretion, and activity of other kinase pathways) | |
| 15, 16, and 17. Integrin, cadherin, and gap pathways | Cell-to-cell signaling and cell-environment signaling that affect adhesion, motility, and passage of ions, metabolites and signaling molecules between cells | |
| 18. Ligand-gated cation channel pathways | Several receptors and ligands (acetylcholine, glutamate, NMDA, and GABA) and affect membrane potentials and calcium-dependent events | |
An adverse developmental outcome often involves (a) direct perturbation of the synthesis or metabolism of the relevant ligands, competitive binding of receptors, or signal transduction of developmental pathways (NRC 2000, Table 1); and/or (b) indirect perturbation by means of disrupting intracellular homoeostasis that results in necrosis or apoptosis of cells that organize and maintain developmental patterning (Gohlke et al. 2007; Simmons et al. 2009). Because developmental pathways are well conserved, a central tenet is that human developmental toxicity can be predicted for environmental chemicals with appropriate in vitro data and in silico models (NRC, 2010; Knudsen et al. 2011; Bouhifd et al. 2014). In vitro HTS bioactivity data are now publicly available in ToxCast/Tox21 (NRC, 2007; Collins et al. 2008; Kavlock et al. 2012; Tice et al. 2013; Sturla et al. 2014), as are relevant data aggregation tools, such as the Adverse Outcome Pathway wiki (AOP; http://aopwiki.org/) and Comparative Toxicogenomics Database (CTD; http://ctdbase.org/). These resources are currently being explored as new approaches to predict potential human health risk of chemical exposure in developmental toxicity in general and teratogenesis in particular (Sipes et al. 2011a; Wu et al. 2013; Leung et al. 2016b). Cell-cell signaling pathways are highly connected to stress response at a molecular level. For instance, programmed cell death (apoptosis) is a consequence of normal signaling or induced stress (Simmons et al. 2009). Intracellular homoeostasis in more general terms is linked critically to stress response that accounts for up to 70% of the measured in vitro bioactivity caused by chemical exposure based on the EPA’s ToxCast HTS data (Judson et al. submitted). Stress response is mediated by at least five molecular systems (Table 2). These have both evolved and diversified in response to environmental stressors, such as ultraviolet radiation; heavy metals; oxidative atmosphere; dietary exposure to secondary metabolites in plant and fungal species; and, possibly, environmental pollutants (Monosson 2012, 2015, Figure 2). In some instances, stress response can lead to adverse outcomes, such as the metabolic activation of mutagenic polycyclic aromatic hydrocarbons (PAHs).
Table 2. Molecular Systems of Stress Response.
Stress response is mediated by at least five highly interconnected molecular systems. Both severe stress and contradictory growth signals can activate programmed cell death [i.e., apoptosis; see Table 1; Kroemer et al. 2010].
| Molecular Systems | Functions |
|---|---|
|
| |
| 1. Antioxidant defenses | Regulate effects of reactive species and redox signaling |
| 2. Xenobiotic metabolism | Increases the solubility of an exogenous chemical to facilitate its removal |
| 3a and 3b. DNA repair and cell cycle pathway | Prevent disruption of normal cell function and development caused by chromatin denaturation and delay certain synthetic processes in the cell cycle until other processes, such as DNA repair, are complete |
| 4. Unfolded protein response | Prevents an accumulation of misfolded and unfolded proteins in the endoplasmic reticulum lumen |
| 5. Autophagy | Degrades cytoplasmic components (including mitochondria) through the lysosome for quality-control |
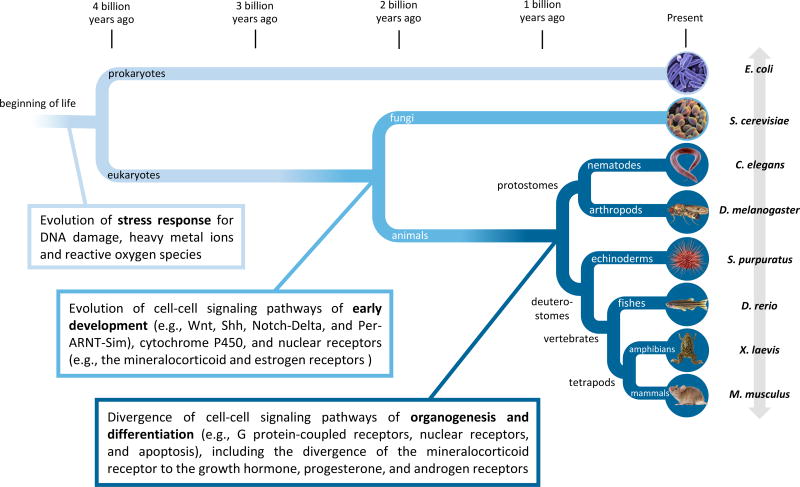
Figure 2. Evolutionary origins of stress response and developmental pathways.
Stress response and developmental pathways (Table 1 and 2) are highly interconnected, and both have diversified in the course of evolution at different time points. For example, the estrogen receptor evolved before the diversification of animals; the androgen receptor evolved at a much later time point (Reitzel and Tarrant 2010; Kassahn et al. 2011). The species divergence times were calculated using the TimeTree knowledgebase (Hedges et al. 2006, 2015).
Gene regulatory networks are the biological circuitry that ultimately drive morphogenesis and differentiation (Davidson et al. 2002; Longabaugh et al. 2005). Because the molecular systems of stress response are highly interconnected to developmental pathways in the course of evolution (Figure 2), severe stress can influence the cell fate/death decisions in embryonic development. Understanding mechanisms of developmental toxicity thus requires tools, models, and approaches to analyze network structure and state dynamics (Knudsen and Kavlock 2008; Sturla et al. 2014). For example, the induction of eye defects associated with early developmental exposure to alcohol in mice is foreshadowed by substrain-dependent (C57BL/6N versus C57BL/6J) reprogramming of gene regulatory networks (Green et al. 2007). Mapping ethanol-responsive KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways revealed strain-dependent downregulation of ribosomal proteins and proteasome and upregulation of glycolysis and the pentose phosphate pathway in C57BL/6N embryos that are resistant to alcohol-induced microphthalmia and significant upregulation of tight-junction, focal adhesion; adherens junction; and regulation of the actin cytoskeleton (and near-significant upregulation of Wnt signaling and apoptosis) pathways in both sensitive (C57BL/6J) and resistant substrains. Expression networks constructed computationally from these altered genes identified entry points for ethanol at several gene hubs in the network. These findings are consistent with the view that developmental exposure to ethanol alters common signaling pathways linking receptor activation to cytoskeletal reorganization. One hypothesis is that oxidative stress can disrupt the Wnt, Notch-Delta, and apoptosis pathways that determine the patterning and outcome of eye development (Smith et al. 2005; Yamaguchi et al. 2005, Table 1). Although humans have a greater biological complexity than most model organisms (Figure 3), the gene regulatory networks that connect stress response and developmental pathways in humans may be conserved in model organisms, such as C. elegans, Drosophila, and zebrafish. If so, these small model organisms can provide a useful HTS platform to assess potential hazard of developmental toxicity and teratogenesis in humans (Sipes et al. 2011b; Rand et al. 2014; Beekhuijzen et al. 2015; Boyd et al. 2016).
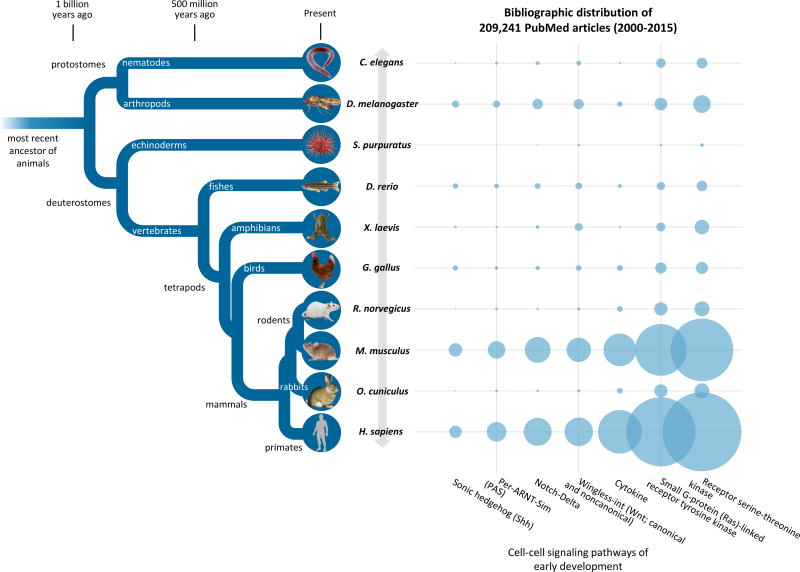
Figure 3. Number of PubMed Articles Between 2000 and 2015 for nine model organisms and humans for seven consensual cell-cell signaling pathways in early development.
Rats, mouse, and rabbits―the three conventional models in teratology―share the last common ancestor with humans approximately 90 million years ago (TimeTree knowledgebase; Hedges et al. 2006, 2015). Citations of all 209,241 articles are provided in Supplementary Material, Excel Workbook S1.
Evolutionary Synthesis in Experimental Teratology
Although today’s birth defects research mostly constitutes experimental studies, the field is deeply rooted in observational studies. Historically, observations of embryos and embryonic stages were made and recorded by Aristotle in the 4th century BC. In the 18th and early 19th century, Johann Friedrich Meckel (1812), Etienne Geoffroy Saint-Hilaire (1822), and Charles Féré (1909), among others, began to study abnormal development using the scientific method. In the early 20th century, experimental teratology began to link developmental defects to hypoxia (Fantel 1996; Ornoy 2007), infection (Elliott 2001; Avgil and Ornoy 2006; De Santis et al. 2006), radiation (Heynick and Merritt 2003; De Santis et al. 2007), nutritional disturbances (Brent et al. 1990; Collins and Mao 1999), alcohol consumption (Jones 2011; Murawski et al. 2015), and exposure to chemicals (Wilson 1959, 1973, 1977; Barrow 1977). These observations were summarized in the Wilson’s Principles of Teratology (Wilson 1973).
Susceptibility to teratogenesis depends on the genotype of the conceptus and the manner in which this interacts with adverse environmental factors.
Susceptibility to teratogenesis varies with the developmental stage at the time of exposure to an adverse influence.
Teratogenic agents act in specific ways (mechanisms) on developing cells and tissues to initiate sequences of abnormal developmental events (pathogenesis).
The access of adverse influences to developing tissues depends on the nature of the influence (agent).
The four manifestations of deviant development are death, malformation, growth retardation, and functional deficit.
Manifestations of deviant development increase in frequency and degree as dosage increases, from the no-effect to the totally lethal level.
The Wilson’s Principles introduced the concepts of gene-environment interactions (Principle I), windows of susceptibility (II), mechanisms of action (III), and dose-response relationships (VI) to experimental teratology. Although the original six principles are still basically valid today (Jelínek 2005; Friedman 2010), recent advances in HTS technology, big data techniques, and systems toxicology now are changing the nature of cross-species extrapolation (Knudsen and Kavlock 2008; Sturla et al. 2014). For instance, AOP is an analytical framework proposed by the Organization for Economic Co-operation and Development (OECD) and EPA for integrating a large volume of research literature and HTS data in risk assessment (Ankley et al. 2010; OECD 2013; Sturla et al. 2014). In the AOP framework, the molecular, biochemical, and histological data are organized across different levels of biological organization. Such a massive amount of toxicity data across multiple experimental systems and species requires a new organizing principle to make it accessible and useful for predicting developmental toxicity in humans.
The discordance of susceptibility between test species and humans (ontogeny) reflects their differences in evolutionary history (phylogeny).
This organizing principle represents a synthesis between experimental teratology and evolutionary genetics. Originally, the discordance of susceptibility among mammalian species (including humans) was curated by Wilson in Environment and Birth Defects (1973). Such discordance reflects the morphogenetic and gene regulatory underpinnings of individual species’ biological development (i.e., ontogeny); it also reflects species differences in evolutionary history (i.e., phylogeny) (Haeckel, 1866). Here, we propose to apply the concept of “ontogeny recapitulates phylogeny” as a testable hypothesis to connect disparate pieces of toxicity data under the integrative AOP framework and to apply evolutionary genetics to evaluate the roles of different molecular targets in developmental toxicity and risk assessment. In the next three sections, we will discuss how to apply this concept in multiscale systems modeling, phylogenetic analysis, and comparative bioinformatics (Figure 4).
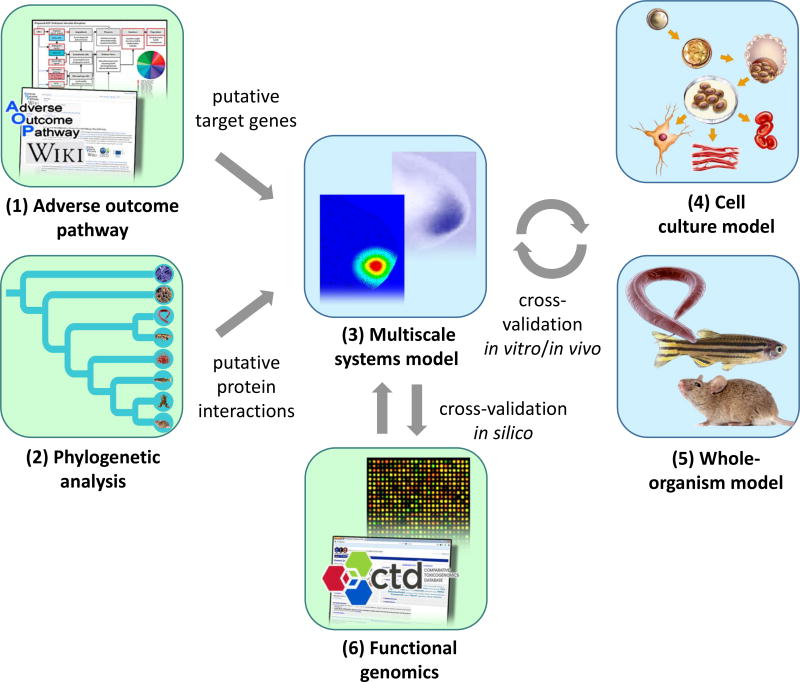
Figure 4. Evolutionary approach to cross-species extrapolation.
Adverse Outcome Pathway (AOP) is a useful framework to integrate a large volume of toxicity data from research literature and high-throughput screening for predicting developmental toxicity in humans. An adverse outcome can be linked to a molecular initiating event (MIE) that involves a developmental toxicant and a molecular target. Risk assessment can be started with the bioinformatics analysis (green) of (1) an AOP to predict MIEs and target genes and (2) phylogenetic analysis of putative protein interactions. Multiscale systems models (3) can be built to examine the spatio-temporal dynamics of developmental patterning. The results of computer simulations can be cross-validated in vitro, in vivo, and in silico using (4) cell culture models, (5) whole-organism models, and (6) functional genomics. This approach can improve the design and data interpretation with different experimental models (blue), thereby increasing the capability in identifying potential developmental toxicants in humans.
Multiscale Systems Modeling
Multiscale systems modeling is an important approach for understanding the spatio-temporal dynamics of embryonic development (Knudsen and Kavlock 2008; Morelli et al. 2012; Sturla et al. 2014). An embryo is composed of many interacting parts (molecules, cells, and tissues) in an intricate arrangement; cell differentiation and developmental patterning are precisely orchestrated by genetic pathways and cellular processes. Networks of individual interactions ultimately govern how the system behaves in response to chemical-induced perturbation. Multiscale modeling and simulation are thus an important approach for discovery and synthesis of biological design principles underlying the response of complex adaptive systems to developmental toxicity. This approach has been used to unravel complex series of multicellular events in neocortical neurogenesis (Gohlke et al. 2007), angiogenesis (Kleinstreuer et al. 2013), circadian rhythms (DeWoskin et al. 2014), limb bud formation (Uzkudun et al. 2015), and urethral tube closure (Leung et al. 2016a). It can serve as a focal point for cross-species validation of the spatio-temporal dynamics of developmental patterning in vitro and in vivo, thereby providing an essential tool for data integration in teratogenicity assessment [i.e., in vitro to in vivo extrapolation (IVIVE) (Gohlke et al. 2008; Tal et al. (submitted), Figure 4)].
Convergent evolution and developmental system drift
Multiscale systems modeling is essential for prediction of a human adverse outcome based on observations in various species. In the course of evolution, a certain set of developmental outcomes are stabilized in various species by natural selection, conferring better adaption to ecological niches (Figure 1). As such, some degree of similarity in morphogenesis and tissue organization often is seen between test species and humans. A similar developmental outcome can evolve independently in different species (i.e., convergent evolution). For instance, the single-lens eyes of cephalopods and vertebrates are similar in structure but different in embryonic development. Alternatively, as a developmental outcome is conserved (thus preserving the species’ ecological niches), the underlying pathways can undergo genetic drift without evolutionary penalty. Such pathway divergence without phenotypic change is known as developmental system drift (True and Haag 2001). C. elegans and P. pacificus share a common ancestor that lived approximately 250 million years ago and a similar vulvar structure that arises from the same set of precursor cells. Yet, the vulvar precursor cells are induced by the small G-protein (Ras)-linked receptor tyrosine kinase pathway in C. elegans, as compared with the Wnt pathway in P. pacificus (Wang and Sommer, 2011).
Because convergent evolution and developmental system drift both can contribute to species-specific differences in developmental mechanisms, the molecular initiating events (MIEs) and key events can be different for the same adverse outcome in different species, even if the MIEs are initiated by the same class of chemical teratogens. This is best exemplified by the fetal male reproductive system response to phthalate exposure in rats, mice, rabbits, and humans (Johnson et al. 2012; Albert and Jégou 2014; Cunha et al. 2015). Mice share a more recent common ancestor with rats (approximately 20 million years ago) than with rabbits and humans [approximately 90 million years ago (Hedges et al. 2006, 2015, Figure 3)], but mice and humans are both less susceptible than rabbits and rats to phthalate exposure. This likely results from species-specific differences in both toxicokinetics and developmental system drift. A low-dose effect (e.g., endocrine disruption in wildlife populations) often can be linked to a specific MIE in different species and is thus predictable by comparing the phylogenetic congruence of a specific receptor (Karchner et al. 2006; Lalone et al. 2013). In contrast, a high-dose effect (e.g., idiosyncratic toxicity of pharmaceuticals in humans) often is linked to different MIEs in different species, thereby requiring multiscale systems modeling to identify key events at the multicellular level for cross-species extrapolation.
Systems models as a focal point of cross-species extrapolation
Multiscale systems modeling plays an integral role in the evolutionary approach to cross-species extrapolation (Figure 4). The AOP Wiki and phylogenetic analysis both can provide useful information for developing a conceptual model for an adverse developmental outcome based on research literature, HTS data, and sequence information (as discussed in the next section). As an additional line of evidence, the conceptual model can be validated using databases such as CTD. Afterward, a computer simulation can be run to examine the spatio-temporal dynamics of the adverse developmental outcome, which can be further validated using cell culture and whole-organism HTS models. This approach can both improve the interpretation of HTS data and guide the development of new HTS assays.
Phylogenetic Analysis
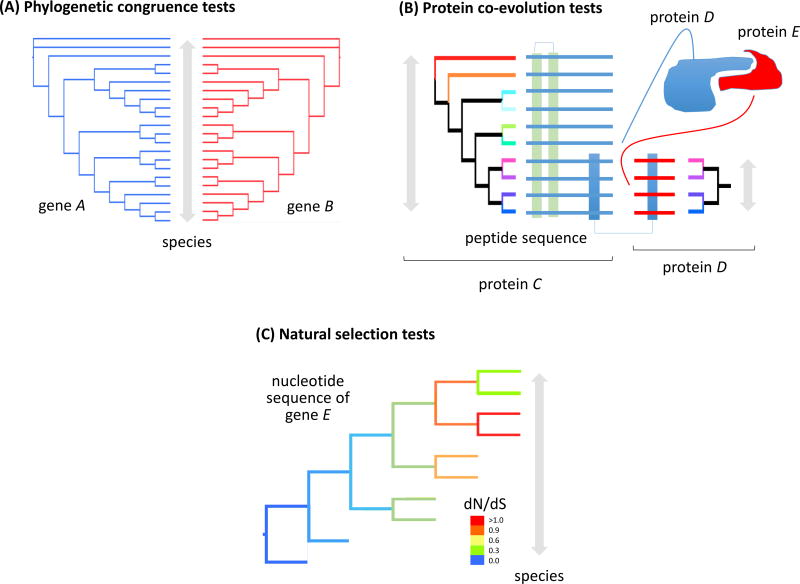
Phylogenetics is the study of the evolutionary relationships between DNA sequences by making comparisons between those sequences (Yang and Rannala 2012). It can be divided into two basic approaches. The first approach involves detecting evolutionary congruence of potentially interacting proteins, which can be examined in two ways: (1) simple assessment of phylogenetic congruence of two genes (Figure 5A); and (2) detection of co-evolution of whole proteins, as well as specific residues (Figure 5B). The second approach involves the detection of natural selection on genes, on individual residues of proteins, and on specific branches of the tree of life (Figure 5C), which enables inferences about selection or protein interactions within an evolutionary context.
Figure 5. Statistical tests for phylogenetic congruence, protein co-evolution, and natural selection.
(A) Phylogenetic congruence tests measure and visualize the extent to which tree topologies differ. These tests can be used to provide evidence for differing patterns of evolution among genes, as well as gene duplication, horizontal gene transfer, or other nonvertical inheritance. Phylogenetic congruence tests may be classified as character congruence tests, of which the incongruence length difference test is most well-known, and topological congruence tests, including the tanglegram shown here (the gene A and B topologies are congruent). (B) Tests for protein co-evolution measure the coordinated changes that occur in pairs of proteins or protein residues, typically to maintain or refine catalytic or ligand-binding interactions (between protein C and D; modified from de Juan et al. 2013). (C) dN/dS is an indicator of natural selection acting on a genetic locus. It is the ratio of mutations that change amino acids (nonsynonymous mutations) to those that do not (synonymous mutations). If selection is absent, and mutations are caused by random genetic drift, dN/dS = 1. dN/dS > 1 suggests positive directional selection, indicating adaptation of gene E in those species (red lineages in the tree). dN/dS < 1 suggests balancing selection, such that mutations of gene E are reducing the fitness of those species (orange through blue lineages).
Phylogenetic congruence and protein co-evolution tests
Congruence is a measurement of similarity between two objects. In evolutionary biology, different genes are thought to have different evolutionary histories because of lineage sorting effects (Page and Charleston 1998) and differences in selective pressures on genes (Oleksyk et al. 2010). Similarity in evolutionary history can be reflected in phylogenetic congruence, which is used to hypothesize potential relationships between genes, such as gene duplication, horizontal gene transfer, nonvertical inheritance, and differing rates of evolution among genes. Phylogenetic congruence tests measure character information (i.e., character congruence tests, such as incongruence length difference tests, maximum likelihood tests, and Bayesian analyses) and tree shape [i.e., topological congruence tests, such as consensus-based measurements and tree distances (Planet 2006)]. Although congruence of two genes does not necessarily indicate that the two gene products are interacting or co-evolving, this approach provides an estimate of potential interaction prior to biochemical examination.
Phylogenetic congruence tests have been used to examine nuclear receptors, cytochrome P450 reductases, the basic helix-loop-helix (bHLH) family of transcription factors (Nelson et al. 2013; McRobb et al. 2014; Reitzel et al. 2014b). For example, the aryl hydrocarbon receptor (AhR), a member of the bHLH and PAS families, accounts for the developmental toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin [TCDD (Schmidt and Bradfield 1996)]. Multiple AhR homologs have been identified in some taxa because of whole genome duplication (Hahn and Karchner 2012). In addition to differences in the DNA sequences of single genes, whole genome duplication contributes to species-specific differences of TCDD, polychlorinated biphenyl (PCB), and PAH toxicity. Also, maximum likelihood and Bayesian analyses were used to identify the putative ancestral gene lineages of cytochrome P450 reductases and mineralocorticoid receptors [Figure 1 (Kassahn et al. 2011; Nelson et al. 2013)].
Recently, LaLone et al. (2013, 2014) described a new tool (SeqAPASS) based on an algorithm that aligns the DNA sequences of molecular targets and assesses a similarity cutoff for cross-species extrapolation, which also takes into consideration conserved functional protein domains and gene ontology. SeqAPASS can be used to assess the degree of conservation of molecular targets between species, such as zebrafish and humans, as well as other commonly used model organisms in biomedical and environmental toxicology research. This approach has been used to describe how key nodes in the ontogenetic regulation of developmental angiogenesis have evolved phylogenetically across the diverse species of interest (Tal et al. submitted). For example, homology in a signature predicting embryonic vascular disruption appears first in the receptor tyrosine kinase signaling systems, followed by the urokinase plasminogen activating receptor system and chemokines/G-protein-coupled receptor system. Zebrafish, in particular, display a strong conservation of the angiogenic and vessel remodeling pathways relative to the extracellular matrix- and cytokine-mediated pathways.
Co-evolution refers to the coordinated changes that occur in pairs of organisms or biomolecules (de Juan et al. 2013). Co-evolution of proteins typically occurs to maintain or refine functional interactions. Much progress has been made in the development of computational tools for the detection of protein co-evolution (Fares and McNally 2006; Tillier and Charlebois 2009; Ochoa and Pazos 2010). These tools can predict (1) functional properties of a protein family, such as substrate-binding specificity of a given enzyme; (2) interactions of entangled protein residues, such as catalytic and binding sites; and (3) interprotein interactions, such as partners for ligand-receptor pairs. Hypotheses of protein co-evolution can be examined by means of ancestral reconstruction and synthesis of proteins (Bridgham et al. 2010; Harms and Thornton 2013). Because stress response closely interacts with developmental pathways at the molecular level, tests for protein co-evolution can be useful in understanding the mechanism of developmental toxicity caused by exposure to reactive oxygen species and DNA-damaging agents (Hales 2005; Wells et al. 2005).
Tests for natural selection
Natural selection on proteins can be categorized as neutral (drift), negative (purifying), or positive (Darwinian); these three evolutionary states can be determined for an individual gene or protein domain. One test for selection is the ratio of nonsynonymous (dN) to synonymous (dS) substitutions (Kryazhimskiy and Plotkin 2008; Mugal et al. 2014)]. The dN/dS ratio describes the relationship of mutations that change amino acids (dN mutations) to mutations that are “silent” and do not change amino acids (dS mutations). Although the dN/dS ratio often is used in population genetics, it is also useful in molecular biology and evolution to determine whether the common ancestor of a group of organisms experienced an increase in natural selection at a genetic locus. If selection is absent and mutations are caused by random genetic drift, then dN/dS = 1. When dN/dS > 1, this suggests positive selection, indicating adaptation in that gene, and, when dN/dS < 1, this suggests purifying selection, indicating that mutations of the gene are reducing the fitness of the organism. Additional statistical assessments of protein functional divergence include site-specific tests of altered amino acid property or evolutionary rate (Gu, 2006). Natural selection can be assessed at the whole-genome level using robust statistical methods (Pentony et al. 2012).
Tests for natural selection are useful in understanding the functional properties of individual molecular components in a developmental pathway. For instance, the dN/dS ratio was used to examine the functional divergence of the hedgehog gene family in vertebrates (Pereira et al. 2014). The hedgehog proteins are composed of two main domains: (1) the signaling Hedge (N-terminal) and (2) cholesterol-binding Hog (C-terminal) domains (Burglin 2008). There is a strong positive selection at the amino acid isoelectric point property of the Hedge domain, suggesting a hotspot of functional divergence at the vertebrate lineage. Yet, the amino acid sequences of both Hedge and Hog are mostly under purifying selection, which stabilizes the three-dimensional structure and functions of the hedgehog proteins. In fetal alcohol syndrome, sonic hedgehog (SHH) signaling is disrupted by early developmental exposure to ethanol via indirect perturbation of cholesterol metabolism (Sulik 2005).
Comparative Bioinformatics
The application of comparative bioinformatics in birth defects research has been reviewed elsewhere (Singh et al. 2007; Knudsen and Kavlock 2008). Here, we focus on the bioinformatics tools relevant to evolutionary genetics. The Comparative Toxicogenomics Database (CTD) provides manually curated data describing chemical-gene-disease interactions from diverse species (vertebrates and invertebrates) and experimental systems [in vitro and in vivo (Davis et al. 2015)]. Transcriptomic databases with similar capability include Chemical Effects in Biological Systems (CEBS, http://www.niehs.nih.gov/research/resources/databases/cebs/) and the Toxicogenomics Project [TGP, http://toxico.nibiohn.go.jp/english/ (McHale et al. 2010; Igarashi et al. 2015)]. The use of comparative toxicogenomics can complement molecular phylogenetics in deciphering the spatio-temporal dynamics of gene expression that account for phenotypic plasticity and the critical susceptibility window in embryonic development, both of which can be further examined in silico using multiscale systems models (Figure 3).
Novel invertebrate models, such as A. queenslandica (sponge) and N. vectensis (sea anemone), have provided important insights into developmental mechanisms (Degnan et al. 2009; Nakanishi et al. 2014; Hashimshony et al. 2015). Biological information of many invertebrate species is now publicly available through educational and reference databases, such as Encyclopedia of Life, Open Tree of Life, and Tree of Life Knowledge and Information Network (TOLKIN; Beaman et al. 2012; Parr et al. 2014; Hinchliff et al. 2015). The National Center for Biotechnology Information (NCBI) Taxonomy Browser organizes DNA sequences from multiple databases into a taxonomy tree, covering about 10% of all known eukaryotic species (Acland et al. 2014). If the species of interest has not been covered in NCBI, then gene identification and annotation can be done using next-generation sequencing and sequence alignment software (Allen et al. 2015). These bioinformatics tools are invaluable in developing novel models of morphogenesis and developmental toxicity.
Comparisons of developmental outcomes across species require consistent ontology and diagnostic criteria. A number of comparative diagnostic databases have been developed to provide assistance in visual recognition with the aid of photographs of abnormal development in laboratory animals. For instance, the DevTox database (http://www.devtox.org/) currently consists of more than 2,500 images, featuring examples of external, skeletal, soft tissue, and maternal-fetal effects for prenatal exposures in rats mice, rabbits, hamsters, primates, guinea pigs, minipigs, dogs, and birds (Solecki et al. 2010). Other similar initiatives, such as the Comparative Atlas of External Malformations in Laboratory Animals and Humans (Roux et al. 2003) and the Laboratory Animal Congenital Anomaly Database (https://center5.umin.ac.jp/cadb/nsearch.cgi?lang=1), provide essential in vivo references for characterizing phenotypic plasticity in different species.
Conclusion
Conservation of cell signaling across diverse species is an important organizing principle in developmental biology and the genomic sciences. New tools and models in bioinformatics and systems biology have expanded the boundaries of traditional evolutionary genetics, as well as those of traditional toxicology. Together, these concepts are now changing the wider perception of chemical safety assessment and toxicology by contributing to a framework developed around AOPs. A formal connection between evolutionary genetics and developmental pathways is a lynchpin for their integration into toxicity assessment and, specifically, toward improving our understanding of the role of environmental factors underlying human birth defects. Here, we demonstrate (1) how to connect disparate pieces of toxicity data under the concept of “ontogeny recapitulates phylogeny”; and (2) how to apply evolutionary genetics to evaluate the roles of different molecular targets. This synthesis not only can lead to novel applications in developmental toxicity and risk assessment, but also can pave the way for applying an evo-devo perspective to the study of developmental origins of health and disease.
Supplementary Material
The seven cell-cell signaling pathways of early development (Table 1) were mapped to 312 MeSH terms, 209,241 PubMed articles (2000–2015), and nine model organisms and humans (C. elegans, D. melanogaster, S. purpuratus, D. rerio, X. laevis, G. gallus, M. musculus, R. norvegicus, O. cuniculus, and H. sapiens). WNT, wingless-int pathway (canonical and noncanonical; 13 MeSH terms); RSTK, receptor serine-threonine kinase pathway (146); SHH, sonic hedgehog pathway (7); RAS, small G-protein-linked receptor tyrosine kinase pathway (77); ND, Notch-Delta pathway (17); CTK, cytokine receptor pathway (20); and PAS, Per-ARNT-Sim pathway (32).
Highlights.
Here, we review five molecular systems of stress response and update 18 consensual cell-cell signaling pathways that are the hallmark for early development, organogenesis, and differentiation
We revisit the principles of teratology in light of recent advances in high-throughput screening, big data techniques, and systems toxicology.
This synthesis paves the way for applying an evo-devo perspective to the study of developmental origins of health and disease to improve experimental design and data interpretation with various in vitro and whole-organism models.
Acknowledgments
We would like to thank Nancy C. Baker, Bevin P. Engelward, Mark E. Hahn, E. Sidney Hunter III, Thomas B. Knudsen, Joel N. Meyer, Angela M. Zoss, and the Comparative and Veterinary Specialty Section, the Molecular and Systems Biology Specialty Section, and the Scientific Liaison Coalition of the Society of Toxicology for helpful comments.
Footnotes
Conflict of interest: The authors declare they have no actual or potential competing financial interests.
References
- Abbott BD. Signal transduction pathways as targets for teratogens. In: Hansen DK, Abbott BD, editors. Target Organ Toxicology Series, Developmental Toxicology. 3. Boca Raton, FL: Taylor & Francis; 2008. pp. 43–67. [Google Scholar]
- Acland A, Agarwala R, Barrett T, Beck J, Benson DA, Bollin C, Bolton E, Bryant SH, Canese K, Church DM, Clark K, DiCuccio M, Dondoshansky I, Federhen S, Feolo M, Geer LY, Gorelenkov V, Hoeppner M, Johnson M, Kelly C, Khotomlianski V, Kimchi A, Kimelman M, Kitts P, Krasnov S, Kuznetsov A, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Karsch-Mizrachi I, Murphy T, Ostell J, O'Sullivan C, Panchenko A, Phan L, Pruitt DP, Rubinstein W, Sayers EW, Schneider V, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Siyan K, Slotta D, Soboleva A, Soussov V, Starchenko G, Tatusova TA, Trawick BW, Vakatov D, Wang Y, Ward M, John Wilbur W, Yaschenko E, Zbicz K. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014;42:D7–D17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert O, Jégou B. A critical assessment of the endocrine susceptibility of the human testis to phthalates from fetal life to adulthood. Hum Reprod Update. 2014;20:231–249. doi: 10.1093/humupd/dmt050. [DOI] [PubMed] [Google Scholar]
- Allen JM, Huang DI, Cronk QC, Johnson KP. aTRAM - automated target restricted assembly method: a fast method for assembling loci across divergent taxa from next-generation sequencing data. BMC Bioinformatics. 2015;16:98. doi: 10.1186/s12859-015-0515-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, Mount DR, Nichols JW, Russom CL, Schmieder PK, Serrrano JA, Tietge JE, Villeneuve DL. Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ Toxicol Chem. 2010;29:730–741. doi: 10.1002/etc.34. [DOI] [PubMed] [Google Scholar]
- Avgil M, Ornoy A. Herpes simplex virus and Epstein-Barr virus infections in pregnancy: consequences of neonatal or intrauterine infection. Reprod Toxicol. 2006;21:436–445. doi: 10.1016/j.reprotox.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Barrow MV. A Brief History of Teratology to the Early 20th Century. Teratology. 1977;4:119–130. [Google Scholar]
- Beaman RS, Traub GH, Dell CA, Santiago N, Koh J, Cellinese N. TOLKIN—Tree of Life Knowledge and Information Network: filling a gap for collaborative research in biological systematics. PLoS One. 2012;7:e39352. doi: 10.1371/journal.pone.0039352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekhuijzen M, de Koning C, Flores-Guillén ME, de Vries-Buitenweg S, Tobor-Kaplon M, van de Waart B, Emmen H. From cutting edge to guideline: A first step in harmonization of the zebrafish embryotoxicity test (ZET) by describing the most optimal test conditions and morphology scoring system. Reprod Toxicol. 2015;56:64–76. doi: 10.1016/j.reprotox.2015.06.050. [DOI] [PubMed] [Google Scholar]
- Bouhifd M, Hogberg HT, Kleensang A, Maertens A, Zhao L, Hartung T. Mapping the human toxome by systems toxicology. Basic Clin Pharmacol Toxicol. 2014;115:24–31. doi: 10.1111/bcpt.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd WA, Smith MV, Co CA, Pirone JR, Rice JR, Shockley KR, Freedman JH. Developmental effects of the ToxCast™ phase I and II chemicals in Caenorhabditis elegans and corresponding responses in zebrafish, rats, and rabbits. Environ Health Perspect. 2016;124:586–593. doi: 10.1289/ehp.1409645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent RL, Beckman DA, Jensen M, Koszalka TR. Experimental yolk sac dysfunction as a model for studying nutritional disturbances in the embryo during early organogenesis. Teratology. 1990;41:405–413. doi: 10.1002/tera.1420410406. [DOI] [PubMed] [Google Scholar]
- Bridgham JT, Eick GN, Larroux C, Deshpande K, Harms MJ, Gauthier ME, Ortlund EA, Degnan BM, Thornton JW. Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol. 2010;8:e1000497. doi: 10.1371/journal.pbio.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA. Circadian clock-mediated control of stem cell division and differentiation: beyond night and day. Development. 2014;141:3105–3111. doi: 10.1242/dev.104851. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Lauring AS. Theory and empiricism in virulence evolution. PLoS Patho. 2014;10:e1004387. doi: 10.1371/journal.ppat.1004387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin TR. The Hedgehog protein family. Genome Biol. 2008;9:241. doi: 10.1186/gb-2008-9-11-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- Carney EW, Tornesi B, Markham DA, Rasoulpour RJ, Moore N. Species-specificity of ethylene glycol-induced developmental toxicity: toxicokinetic and whole embryo culture studies in the rabbit. Birth Defects Res B Dev Reprod Toxicol. 2008;83:573–581. doi: 10.1002/bdrb.20178. [DOI] [PubMed] [Google Scholar]
- Casaca A, Santos AC, Mallo M. Controlling Hox gene expression and activity to build the vertebrate axial skeleton. Dev Dyn. 2014;243:24–36. doi: 10.1002/dvdy.24007. [DOI] [PubMed] [Google Scholar]
- Clowes C, Boylan MG, Ridge LA, Barnes E, Wright JA, Hentges KE. The functional diversity of essential genes required for mammalian cardiac development. Genesis. 2014;52:713–737. doi: 10.1002/dvg.22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MD, Mao GE. Teratology of retinoids. Annu Rev Pharmacol Toxicol. 1999;39:399–430. doi: 10.1146/annurev.pharmtox.39.1.399. [DOI] [PubMed] [Google Scholar]
- Collins FS, Gray GM, Bucher JR. Toxicology. Transforming environmental health protection. Science. 2008;319:906–907. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha GR, Sinclair A, Risbridger G, Hutson J, Baskin LS. Current understanding of hypospadias: relevance of animal models. Nat Rev Urol. 2015;12:271–280. doi: 10.1038/nrurol.2015.57. [DOI] [PubMed] [Google Scholar]
- Dang Z, Cheng Y, Chen HM, Cui Y, Yin HH, Traas T, Montforts M, Vermeire T. Evaluation of the Daphnia magna reproduction test for detecting endocrine disruptors. Chemosphere. 2012;88:514–523. doi: 10.1016/j.chemosphere.2012.03.012. [DOI] [PubMed] [Google Scholar]
- Das Thakur M, Stuart DD. The evolution of melanoma resistance reveals therapeutic opportunities. Cancer Res. 2013;73:6106–6110. doi: 10.1158/0008-5472.CAN-13-1633. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan Zj, Schilstra MJ, Clarke PJ, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Davis AP, Grondin CJ, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL, Wiegers TC, Mattingly CJ. The Comparative Toxicogenomics Database’s 10th year anniversary: update 2015. Nucleic Acids Res. 2015;43:D914–D920. doi: 10.1093/nar/gku935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Juan D, Pazos F, Valencia A. Emerging methods in protein co-evolution. Nat Rev Genet. 2013;14:249–261. doi: 10.1038/nrg3414. [DOI] [PubMed] [Google Scholar]
- De Santis M, Cavaliere AF, Straface G, Caruso A. Rubella infection in pregnancy. Reprod Toxicol. 2006;21:390–398. doi: 10.1016/j.reprotox.2005.01.014. [DOI] [PubMed] [Google Scholar]
- De Santis M, Cesari E, Nobili E, Straface G, Cavaliere AF, Caruso A. Radiation effects on development. Birth Defects Res C Embryo Today. 2007;81:177–182. doi: 10.1002/bdrc.20099. [DOI] [PubMed] [Google Scholar]
- Degnan BM, Vervoort M, Larroux C, Richards GS. Early evolution of metazoan transcription factors. Curr Opin Genet Dev. 2009;19:591–599. doi: 10.1016/j.gde.2009.09.008. [DOI] [PubMed] [Google Scholar]
- DeWoskin D, Geng W, Stinchcombe AR, Forger DB. It is not the parts, but how they interact that determines the behaviour of circadian rhythms across scales and organisms. Interface Focus. 2014;4:20130076. doi: 10.1098/rsfs.2013.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the Origin of Species. Columbia University Press; New York, NY: 1937. pp. 1–364. [Google Scholar]
- Ebert D. Evolution – Experimental evolution of parasite. Science. 1998;282:1432–1435. doi: 10.1126/science.282.5393.1432. [DOI] [PubMed] [Google Scholar]
- Elliott DJ, Eppes SC, Klein JD. Teratogen update: Lyme disease. Teratology. 2001;64:276–281. doi: 10.1002/tera.1074. [DOI] [PubMed] [Google Scholar]
- Ema M, Ise R, Kato H, Oneda S, Hirose A, Hirata-Koizumi M, Singh AV, Knudsen TB, Ihara T. Fetal malformations and early embryonic gene expression response in cynomolgus monkeys maternally exposed to thalidomide. Reprod Toxicol. 2010;29:49–56. doi: 10.1016/j.reprotox.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Epstein C. Human malformations and their genetic basis. In: Epstein C, Erickson RP, Wynshaw-Boris A, editors. Inborn Errors of Development: The Molecular Basis of Clinical Disorders of Morphogenesis. 2. New York, NY: Oxford University Press; 2008. pp. 3–8. [Google Scholar]
- Fantel AG. Reactive oxygen species in developmental toxicity: review and hypothesis. Teratology. 1996;53:196–217. doi: 10.1002/(SICI)1096-9926(199603)53:3<196::AID-TERA7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Fares MA, McNally D. CAPS: coevolution analysis using protein sequences. Bioinformatics. 2006;22:2821–2822. doi: 10.1093/bioinformatics/btl493. [DOI] [PubMed] [Google Scholar]
- Féré C. Index des travaux de Ch. Féré. Normandie Med. 1909;24:313–314. [Google Scholar]
- Friedman JM. The principles of teratology: are they still true? Birth Defects Res A Clin Mol Teratol. 2010;88:766–768. doi: 10.1002/bdra.20697. [DOI] [PubMed] [Google Scholar]
- Glauber KM, Dana CE, Park SS, Colby DA, Noro Y, Fujisawa T, Chamberlin AR, Steele RE. A small molecule screen identifies a novel compound that induces a homeotic transformation in Hydra. Development. 2013;140:4788–4796. doi: 10.1242/dev.094490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohlke JM, Griffith WC, Faustman EM. Computational models of neocortical neuronogenesis and programmed cell death in the developing mouse, monkey, and human. Cereb Cortex. 2007;17:2433–2442. doi: 10.1093/cercor/bhl151. [DOI] [PubMed] [Google Scholar]
- Gohlke JM, Griffith WC, Faustman EM. Computational models of ethanol-induced neurodevelopmental toxicity across species: Implications for risk assessment. Birth Defects Res B Dev Reprod Toxicol. 2008;83:1–11. doi: 10.1002/bdrb.20137. [DOI] [PubMed] [Google Scholar]
- Green ML, Singh AV, Zhang Y, Nemeth KA, Sulik KK, Knudsen TB. Reprogramming of genetic networks during initiation of the Fetal Alcohol Syndrome. Dev Dyn. 2007;236:613–631. doi: 10.1002/dvdy.21048. [DOI] [PubMed] [Google Scholar]
- Gu X. A simple statistical method for estimating type-II (cluster-specific) functional divergence of protein sequences. Mol Bio Evol. 2006;23:1937–45. doi: 10.1093/molbev/msl056. [DOI] [PubMed] [Google Scholar]
- Guinard G. Introduction to evolutionary teratology, with an application to the forelimbs of Tyrannosauridae and Carnotaurinae (Dinosauria: Theropoda) Evol Biol. 2015;42:20–41. [Google Scholar]
- Haeckel E. Generelle morphologie der organismen. Berlin: G. Reimer; 1866. [General Morphology of the Organisms] [Google Scholar]
- Hahn ME, Karchner SI. The AH Receptor in Biology and Toxicology. Hoboken, NJ: Wiley; 2012. Structural and functional diversification of AHRs during metazoan evolution; pp. 389–403. [Google Scholar]
- Hales BF. DNA repair disorders causing malformations. Curr Opin Genet Dev. 2005;15:234–240. doi: 10.1016/j.gde.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Hansen JM, Harris KK, Philbert MA, Harris C. Thalidomide modulates nuclear redox status and preferentially depletes glutathione in rabbit limb versus rat limb. J Pharmacol Exp Ther. 2002;300:768–776. doi: 10.1124/jpet.300.3.768. [DOI] [PubMed] [Google Scholar]
- Harima Y, Imayoshi I, Shimojo H, Kobayashi T, Kageyama R. The roles and mechanism of ultradian oscillatory expression of the mouse Hes genes. Semin Cell Dev Biol. 2014;34:85–90. doi: 10.1016/j.semcdb.2014.04.038. [DOI] [PubMed] [Google Scholar]
- Harms MJ, Thornton JW. Historical contingency and its biophysical basis in glucocorticoid receptor evolution. Nature. 2014;512:203–207. doi: 10.1038/nature13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T, Feder M, Levin M, Hall BK, Yanai I. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature. 2015;519:219–222. doi: 10.1038/nature13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger NE, Hall SJ, Sandrof MA, McDonnell EV, Hensley JB, McDowell EN, Martin KA, Gaido KW, Johnson KJ, Boekelheide K. Human fetal testis xenografts are resistant to phthalate-induced endocrine disruption. Environ Health Perspect. 2012;120:1137–1143. doi: 10.1289/ehp.1104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heynick LN, Merritt JH. Radiofrequency fields and teratogenesis. Bioelectromagnetics. 2003;6:S174–S186. doi: 10.1002/bem.10127. [DOI] [PubMed] [Google Scholar]
- Hinchliff CE, Smith SA, Allman JF, Burleigh JG, Chaudhary R, Coghill LM, Crandall KA, Deng J, Drew BT, Gazis R, Gude K, Hibbett DS, Katz LA, Laughinghouse HD, 4th, McTavish EJ, Midford PE, Owen CL, Ree RH, Rees JA, Soltis DE, Williams T, Cranston KA. Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proc Natl Acad Sci USA. 2015;112:12764–12769. doi: 10.1073/pnas.1423041112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y, Nakatsu N, Yamashita T, Ono A, Ohno Y, Urushidani T, Yamada H. Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Res. 2015;43:D921–D927. doi: 10.1093/nar/gku955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Handa H. Deciphering the mystery of thalidomide teratogenicity. Congenit Anom (Kyoto) 2012;52:1–7. doi: 10.1111/j.1741-4520.2011.00351.x. [DOI] [PubMed] [Google Scholar]
- Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- Jelínek R. The contribution of new findings and ideas to the old principles of teratology. Reprod Toxicol. 2005;20:295–300. doi: 10.1016/j.reprotox.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Johnson KJ, Heger NE, Boekelheide K. Of mice and men (and rats): phthalate-induced fetal testis endocrine disruption is species-dependent. Toxicol Sci. 2012;129:235–248. doi: 10.1093/toxsci/kfs206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KL. The effects of alcohol on fetal development. Birth Defects Res C Embryo Today. 2011;93:3–11. doi: 10.1002/bdrc.20200. [DOI] [PubMed] [Google Scholar]
- Judson RS, Houck KA, Martin MT, Richard AM, Knudsen TB, Shah I, Little SB, Wambaugh JF, Setzer RW, Kothiya P, Phuong J, Filer DL, Smith DJ, Reif DM, Rotroff DM, Kleinstreuer NC, Sipes NS, Xia M, Huang R, Crofton KM, Thomas RS. Analysis of the effects of cell stress and cytotoxicity on in vitro assay activity in the ToxCast dataset. doi: 10.1093/toxsci/kfw148. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R, Richard A, Dix DJ, Houck K, Martin M, Kavlock R, Dellarco V, Henry T, Holderman T, Sayre P, Tan S, Carpenter T, Smith E. The toxicity data landscape for environmental chemicals. Environ Health Perspect. 2009;117:685–695. doi: 10.1289/ehp.0800168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaskar VK, Lauderdale JD. Mouse embryonic development in a serum-free whole embryo culture system. J Vis Exp. 2014;85:e50803. doi: 10.3791/50803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchner SI, Franks DG, Kennedy SW, Hahn ME. The molecular basis for differential dioxin sensitivity in birds: role of the aryl hydrocarbon receptor. Proc Natl Acad Sci USA. 2006;103:6252–6257. doi: 10.1073/pnas.0509950103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassahn KS, Ragan MA, Funder JW. Mineralocorticoid receptors: evolutionary and pathophysiological considerations. Endocrinology. 2011;152:1883–1890. doi: 10.1210/en.2010-1444. [DOI] [PubMed] [Google Scholar]
- Kavlock R, Chandler K, Houck K, Hunter S, Judson R, Kleinstreuer N, Knudsen T, Martin M, Padilla S, Reif D, Richard A, Rotroff D, Sipes N, Dix D. Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem Res Toxicol. 2012;25:1287–1302. doi: 10.1021/tx3000939. [DOI] [PubMed] [Google Scholar]
- Kleinstreuer N, Dix D, Rountree M, Baker N, Sipes N, Reif D, Spencer R, Knudsen T. A computational model predicting disruption of blood vessel development. PLoS Comput Biol. 2013;9:e1002996. doi: 10.1371/journal.pcbi.1002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen TB, Kavlock RJ. Comparative bioinformatics and computational toxicology. In: Abbott B, Hansen D, editors. Developmental toxicology. 3. New York, NY: Taylor and Francis; 2008. pp. 3–360. [Google Scholar]
- Knudsen TB, Kavlock RJ, Daston GP, Stedman D, Hixon M, Kim JH. Developmental toxicity testing for safety assessment: new approaches and technologies. Birth Defects Res B Dev Reprod Toxicol. 2011;92:413–420. doi: 10.1002/bdrb.20315. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalone CA, Villeneuve DL, Burgoon LD, Russom CL, Helgen HW, Berninger JP, Tietge JE, Severson MN, Cavallin JE, Ankley GT. Molecular target sequence similarity as a basis for species extrapolation to assess the ecological risk of chemicals with known modes of action. Aquat Toxicol. 2013;144–145:141–154. doi: 10.1016/j.aquatox.2013.09.004. [DOI] [PubMed] [Google Scholar]
- LaLone CA, Berninger JP, Villeneuve DL, Ankley GT. Leveraging existing data for prioritization of the ecological risks of human and veterinary pharmaceuticals to aquatic organisms. Philos Trans R Soc Lond B Biol Sci. 2014;369:1656. doi: 10.1098/rstb.2014.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung MCK, Hutson MS, Seifert AW, Spencer RM, Knudsen TB. Computational modeling and simulation of genital tubercle development. Reprod Toxicol. 2016a doi: 10.1016/j.reprotox.2016.05.005. advance publication. [DOI] [PubMed] [Google Scholar]
- Leung MCK, Phuong J, Baker NC, Sipes NS, Klinefelter GR, Martin MT, McLaurin KW, Setzer RW, Darney SP, Judson RS, Knudsen TB. Systems toxicology of male reproductive development: profiling 774 chemicals for molecular targets and adverse outcomes. Environ Health Perspect. 2016b doi: 10.1289/ehp.1510385. advance publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Campo EM, Bossing T. Drosophila embryos as model to assess cellular and developmental toxicity of multi-walled carbon nanotubes (MWCNT) in living organisms. PLoS One. 2014;9:e88681. doi: 10.1371/journal.pone.0088681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longabaugh WJ, Davidson EH, Bolouri H. Computational representation of developmental genetic regulatory networks. Dev Biol. 2005;283:1–16. doi: 10.1016/j.ydbio.2005.04.023. [DOI] [PubMed] [Google Scholar]
- McHale CM, Zhang L, Hubbard AE, Smith MT. Toxicogenomic profiling of chemically exposed humans in risk assessment. Mutat Res. 2010;705:172–183. doi: 10.1016/j.mrrev.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh BE, Hogenesch JB, Bradfield CA. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu Rev Physiol. 2010;72:625–645. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- McRobb FM, Sahagún V, Kufareva I, Abagyan R. In silico analysis of the conservation of human toxicity and endocrine disruption targets in aquatic species. Environ Sci Technol. 2014;48:1964–1972. doi: 10.1021/es404568a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckel JF. Handbuch der Pathologischen Anatomie. Vol. 1. Germany, Leipzig: Reclam; 1812. [Google Scholar]
- Meyer JN, Nacci DE, Di Giulio RT. Cytochrome P4501A (CYP1A) in killifish (Fundulus heteroclitus): heritability of altered expression and relationship to survival in contaminated sediments. Toxicol Sci. 2002;68:69–81. doi: 10.1093/toxsci/68.1.69. [DOI] [PubMed] [Google Scholar]
- Monosson E. Evolution in a Toxic World: How Life Responds to Chemical Threats. Washington, DC: Island Press; 2012. pp. 1–232. [Google Scholar]
- Monosson E. Release: toxics in the wild. In: Monosson E, editor. Unnatural Selection: How We Are Changing Life, Gene by Gene. Washington, DC: Island Press; 2015. pp. 109–126. [Google Scholar]
- Morelli LG, Uriu K, Ares S, Oates AC. Computational approaches to developmental patterning. Science. 2012;336:187–191. doi: 10.1126/science.1215478. [DOI] [PubMed] [Google Scholar]
- Mouche I, Malesic L, Gillardeaux O. FETAX assay for evaluation of developmental toxicity. Methods Mol Biol. 2011;691:257–269. doi: 10.1007/978-1-60761-849-2_15. [DOI] [PubMed] [Google Scholar]
- Mugal CF, Wolf JB, Kaj I. Why time matters: codon evolution and the temporal dynamics of dN/dS. Mol Biol Evol. 2014;31:212–231. doi: 10.1093/molbev/mst192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawski NJ, Moore EM, Thomas JD, Riley EP. Advances in Diagnosis and Treatment of Fetal Alcohol Spectrum Disorders: From Animal Models to Human Studies. Alcohol Res. 2015;37:97–108. doi: 10.35946/arcr.v37.1.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N, Sogabe S, Degnan BM. Evolutionary origin of gastrulation: insights from sponge development. BMC Biol. 2014;12:26. doi: 10.1186/1741-7007-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (NRC) Scientific Frontiers in Developmental Toxicology and Risk Assessment. Washington, DC: National Academies Press; 2000. pp. 1–354. [PubMed] [Google Scholar]
- National Research Council (NRC) Toxicity Testing in the 21st Century. Washington, DC: National Academies Press; 2007. pp. 1–216. [Google Scholar]
- National Research Council (NRC) Toxicity Pathway-Based Risk Assessment. Washington, DC: National Academies Press; 2010. pp. 1–134. [Google Scholar]
- Nelson DR, Goldstone JV, Stegeman JJ. The cytochrome P450 genesis locus: the origin and evolution of animal cytochrome P450s. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120474. doi: 10.1098/rstb.2012.0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson K, Holmes LB. Malformations due to presumed spontaneous mutations in newborn infants. N Engl J Med. 1989;320:19–23. doi: 10.1056/NEJM198901053200104. [DOI] [PubMed] [Google Scholar]
- Ochoa D, Pazos F. Studying the co-evolution of protein families with the Mirrortree web server. Bioinformatics. 2010;26:1370–1371. doi: 10.1093/bioinformatics/btq137. [DOI] [PubMed] [Google Scholar]
- Oleksyk TK, Smith MW, O’Brien SJ. Genome-wide scans for footprints of natural selection. Philos Trans R Soc Lond B Biol Sci. 2010;365:185–205. doi: 10.1098/rstb.2009.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization for Economic Co-operation and Development (OECD) Series on Testing and Assessment, No. 184. Paris, France: OECD; 2013. Guidance Document on Developing and Assessing Adverse Outcome Pathways; pp. 1–45. [Google Scholar]
- Ornoy A. Embryonic oxidative stress as a mechanism of teratogenesis with special emphasis on diabetic embryopathy. Reprod Toxicol. 2007;24:31–41. doi: 10.1016/j.reprotox.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Padilla S, Corum D, Padnos B, Hunter DL, Beam A, Houck KA, Sipes N, Kleinstreuer N, Knudsen T, Dix DJ, Reif DM. Zebrafish developmental screening of the ToxCast™ Phase I chemical library. Reprod Toxicol. 2012;33:174–187. doi: 10.1016/j.reprotox.2011.10.018. [DOI] [PubMed] [Google Scholar]
- Page RD, Charleston MA. Trees within trees: phylogeny and historical associations. Trends Ecol Evol. 1998;13:356–359. doi: 10.1016/s0169-5347(98)01438-4. [DOI] [PubMed] [Google Scholar]
- Parr CS, Wilson N, Leary P, Schulz KS, Lans K, Walley L, Hammock JA, Goddard A, Rice J, Studer M, Holmes JT, Corrigan RJ., Jr The Encyclopedia of Life v2: Providing Global Access to Knowledge About Life on Earth. Biodivers Data J. 2014;2:e1079. doi: 10.3897/BDJ.2.e1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentony MM, Winters P, Penfold-Brown D, Drew K, Narechania A, DeSalle R, Bonneau R, Purugganan MD. The Plant Proteome Folding Project: structure and positive selection in plant protein families. Genome Biol Evol. 2012;4:360–371. doi: 10.1093/gbe/evs015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira J, Johnson WE, O’Brien SJ, Jarvis ED, Zhang G, Gilbert MT, Vasconcelos V, Antunes A. Evolutionary genomics and adaptive evolution of the Hedgehog gene family (Shh, Ihh and Dhh) in vertebrates. PLoS One. 2014;9:e74132. doi: 10.1371/journal.pone.0074132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planet PJ. Tree disagreement: measuring and testing incongruence in phylogenies. J Biomed Inform. 2006;39:86–102. doi: 10.1016/j.jbi.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Rand MD, Montgomery SL, Prince L, Vorojeikina D. Developmental toxicity assays using the Drosophila model. Curr Protoc Toxicol. 2014;59:1.12.1–1.12.20. doi: 10.1002/0471140856.tx0112s59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Tarrant AM. Correlated evolution of androgen receptor and aromatase revisited. Mol Biol Evol. 2010;27:2211–2215. doi: 10.1093/molbev/msq129. [DOI] [PubMed] [Google Scholar]
- Reitzel AM, Karchner SI, Franks DG, Evans BR, Nacci D, Champlin D, Vieira VM, Hahn ME. Genetic variation at aryl hydrocarbon receptor (AHR) loci in populations of Atlantic killifish (Fundulus heteroclitus) inhabiting polluted and reference habitats. BMC Evol Biol. 2014a;14:6. doi: 10.1186/1471-2148-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Passamaneck YJ, Karchner SI, Franks DG, Martindale MQ, Tarrant AM, Hahn ME. Aryl hydrocarbon receptor (AHR) in the cnidarian Nematostella vectensis: comparative expression, protein interactions, and ligand binding. Dev Genes Evol. 2014b;224:13–24. doi: 10.1007/s00427-013-0458-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JF, Verhoef A, Pennings JL, Pronk TE, Piersma AH. A comparison of gene expression responses in rat whole embryo culture and in vivo: time-dependent retinoic acid-induced teratogenic response. Toxicol Sci. 2012;126:242–254. doi: 10.1093/toxsci/kfr342. [DOI] [PubMed] [Google Scholar]
- Roux C, Guittin P, Stadler J, Morin A, Boutemy C, Hiss D, Barrow PC. Comparative Atlas of External Malformations in Laboratory Animals and Humans. Louvain-la-Neuve, Belgium: De Boeck Universite; 2003. pp. 1–152. [Google Scholar]
- Saint-Hilaire EG. Philosophie anatomique des monstruositks humaines. France, Paris: Riqnoux; 1822. [Google Scholar]
- Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- Settivari RS, Ball N, Murphy L, Rasoulpour R, Boverhof DR, Carney EW. Predicting the future: opportunities and challenges for the chemical industry to apply 21st-century toxicity testing. J Am Assoc Lab Anim Sci. 2015;54:214–223. [PMC free article] [PubMed] [Google Scholar]
- Shaham O, Menuchin Y, Farhy C, Ashery-Padan R. Pax6: a multi-level regulator of ocular development. Prog Retin Eye Res. 2012;31:351–376. doi: 10.1016/j.preteyeres.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Simmons SO, Fan CY, Ramabhadran R. Cellular stress response pathway system as a sentinel ensemble in toxicological screening. Toxicol Sci. 2009;111:202–225. doi: 10.1093/toxsci/kfp140. [DOI] [PubMed] [Google Scholar]
- Singh AV, Rouchka EC, Rempala GA, Bastian CD, Knudsen TB. Integrative database management for mouse development: systems and concepts. Birth Defects Res C Embryo Today. 2007;81:1–19. doi: 10.1002/bdrc.20089. [DOI] [PubMed] [Google Scholar]
- Sipes NS, Martin MT, Reif DM, Kleinstreuer NC, Judson RS, Singh AV, Chandler KJ, Dix DJ, Kavlock RJ, Knudsen TB. Predictive models of prenatal developmental toxicity from ToxCast high-throughput screening data. Toxicol Sci. 2011a;124:109–127. doi: 10.1093/toxsci/kfr220. [DOI] [PubMed] [Google Scholar]
- Sipes NS, Padilla S, Knudsen TB. Zebrafish: as an integrative model for twenty-first century toxicity testing. Birth Defects Res C Embryo Today. 2011b;93:256–267. doi: 10.1002/bdrc.20214. [DOI] [PubMed] [Google Scholar]
- Skakkebaek NE, Rajpert-De Meyts E, Buck Louis GM, Toppari J, Andersson AM, Eisenberg ML, Jensen TK, Jørgensen N, Swan SH, Sapra KJ, Ziebe S, Priskorn L, Juul A. Male Reproductive Disorders and Fertility Trends: Influences of Environment and Genetic Susceptibility. Physiol Rev. 2016;96:55–97. doi: 10.1152/physrev.00017.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AN, Miller LA, Song N, Taketo MM, Lang RA. The duality of beta-catenin function: a requirement in lens morphogenesis and signaling suppression of lens fate in periocular ectoderm. Dev Biol. 2005;285:477–489. doi: 10.1016/j.ydbio.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Solecki R, Heinrich V, Rauch M, Chahoud I, Grote K, Wölffel B, Buschmann J, Morawietz G, Kellner R, Lingk W. Comprehensive Toxicology. Vol. 12. Oxford: Academic Press; 2010. The DevTox site: harmonized terminology and database; pp. 339–346. [Google Scholar]
- Sturla SJ, Boobis AR, FitzGerald RE, Hoeng J, Kavlock RJ, Schirmer K, Whelan M, Wilks MF, Peitsch MC. Systems toxicology: from basic research to risk assessment. Chem Res Toxicol. 2014;27:314–329. doi: 10.1021/tx400410s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk WA, Ahanchian H, Asante KA, Carpenter DO, Diaz-Barriga F, Ha EH, Huo X, King M, Ruchirawat M, da Silva ER, Sly L, Sly PD, Stein RT, van den Berg M, Zar H, Landrigan PJ. Environmental pollution: an under-recognized threat to children's health, especially in low- and middle-income countries. Environ Health Perspect. 2016;124:A41–45. doi: 10.1289/ehp.1510517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulik KK. Genesis of alcohol-induced craniofacial dysmorphism. Exp Biol Med. 2005;230:366–375. doi: 10.1177/15353702-0323006-04. [DOI] [PubMed] [Google Scholar]
- Tal T, Kilty C, Smith A, LaLone C, Kennedy B, Tennant A, McCollum C, Bondesson M, Knudsen T, Padilla S, Kleinstreuer N. Screening for chemical vascular disruptors in zebrafish to evaluate a predictive model for developmental vascular toxicity. doi: 10.1016/j.reprotox.2016.12.004. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therapontos C, Erskine L, Gardner ER, Figg WD, Vargesson N. Thalidomide induces limb defects by preventing angiogenic outgrowth during early limb formation. Proc Natl Acad Sci USA. 2009;106:8573–8578. doi: 10.1073/pnas.0901505106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorogood P. The relationship between genotype and phenotype: Some basic concepts. In: Thorogood P, editor. Embryos, Genes, and Birth Defects. New York, NY: Wiley; 1997. pp. 1–16. [Google Scholar]
- Tice RR, Austin CP, Kavlock RJ, Bucher JR. Improving the human hazard characterization of chemicals: a Tox21 update. Environ Health Perspect. 2013;121:756–765. doi: 10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillier ER, Charlebois RL. The human protein coevolution network. Genome Res. 2009;19:1861–1871. doi: 10.1101/gr.092452.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, Haag ES. Developmental system drift and flexibility in evolutionary trajectories. Evol Dev. 2001;3:109–119. doi: 10.1046/j.1525-142x.2001.003002109.x. [DOI] [PubMed] [Google Scholar]
- United States Environmental Protection Agency (U.S. EPA) TSCA chemical substance inventory. 2015 http://www.epa.gov/tsca-inventory/about-tsca-chemical-substance-inventory.
- Uzkudun M, Marcon L, Sharpe J. Data-driven modelling of a gene regulatory network for cell fate decisions in the growing limb bud. Mol Syst Biol. 2015;11:815. doi: 10.15252/msb.20145882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargesson N. Thalidomide-induced teratogenesis: history and mechanisms. Birth Defects Res C Embryo Today. 2015;105:140–156. doi: 10.1002/bdrc.21096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sommer RJ. Antagonism of LIN-17/Frizzled and LIN-18/Ryk in nematode vulva induction reveals evolutionary alterations in core developmental pathways. PLoS Biol. 2011;9:e1001110. doi: 10.1371/journal.pbio.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells PG, Bhuller Y, Chen CS, Jeng W, Kasapinovic S, Kennedy JC, Kim PM, Laposa RR, McCallum GP, Nicol CJ, Parman T, Wiley MJ, Wong AW. Molecular and biochemical mechanisms in teratogenesis involving reactive oxygen species. Toxicol Appl Pharmacol. 2005;207:354–366. doi: 10.1016/j.taap.2005.01.061. [DOI] [PubMed] [Google Scholar]
- Whitehead A, Pilcher W, Champlin D, Nacci D. Common mechanism underlies repeated evolution of extreme pollution tolerance. Proc Biol Sci. 2012;279:427–433. doi: 10.1098/rspb.2011.0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JG. Experimental studies on congenital malformations. J Chron Dis. 1959;10:111–130. doi: 10.1016/0021-9681(59)90026-8. [DOI] [PubMed] [Google Scholar]
- Wilson JG. Environment and Birth Defects. New York, NY: Academic Press; 1973. pp. 1–305. [Google Scholar]
- Wilson JG. Current status of teratology: general principles and mechanisms derived from animal studies. In: Wilson J, Fraser F, editors. Handbook of Teratology. New York, NY: Plenum Press; 1977. pp. 47–74. [Google Scholar]
- Wirgin I, Roy NK, Loftus M, Chambers RC, Franks DG, Hahn ME. Mechanistic basis of resistance to PCBs in Atlantic tomcod from the Hudson River. Science. 2011;331:1322–1325. doi: 10.1126/science.1197296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) Children’s Health and the Environment. A Global Perspective. Geneva, Switzerland: WHO; 2005. pp. 1–367. [Google Scholar]
- Wu S, Fisher J, Naciff J, Laufersweiler M, Lester C, Daston G, Blackburn K. Framework for identifying chemicals with structural features associated with the potential to act as developmental or reproductive toxicants. Chem Res Toxicol. 2013;26:1840–1861. doi: 10.1021/tx400226u. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Tonou-Fujimori N, Komori A, Maeda R, Nojima Y, Li H, Okamoto H, Masai I. Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and Notch signaling pathways. Development. 2005;132:3027–3043. doi: 10.1242/dev.01881. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Molecular phylogenetics: principles and practice. Nat Rev Genet. 2012;13:303–314. doi: 10.1038/nrg3186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The seven cell-cell signaling pathways of early development (Table 1) were mapped to 312 MeSH terms, 209,241 PubMed articles (2000–2015), and nine model organisms and humans (C. elegans, D. melanogaster, S. purpuratus, D. rerio, X. laevis, G. gallus, M. musculus, R. norvegicus, O. cuniculus, and H. sapiens). WNT, wingless-int pathway (canonical and noncanonical; 13 MeSH terms); RSTK, receptor serine-threonine kinase pathway (146); SHH, sonic hedgehog pathway (7); RAS, small G-protein-linked receptor tyrosine kinase pathway (77); ND, Notch-Delta pathway (17); CTK, cytokine receptor pathway (20); and PAS, Per-ARNT-Sim pathway (32).