Abstract
The increasing prevalence of multidrug-resistant Klebsiella pneumoniae strains isolated from hospitals shows the limitation of recent antibiotics used for bacterial eradication. In this study, 81 K. pneumoniae isolates were collected from three hospitals in Tehran. Antibiotic susceptibility test showed the highest rates of resistance to cefotaxim (85.5%) and ceftazidime (78.3%), and the lowest rates of resistance were detected for colistin (16.9%), streptomycin (16.8%), and chloroamphenicol (21.7%). Eleven different resistance patterns were observed. Sixty-six out of 81 isolates (81.5%) were found to be multidrug resistant (MDR), and 35.8% of them belonged to A3 resistance pattern. 7.4% and 66.7% were KPC enzyme and armA gene positive, respectively. RAPD PCR assay of these bacteria showed 5 clusters, 16 single types, and 14 common types, and there was not any correlation between genetic patterns of the isolates and presence of resistance agents. Simultaneous detection of resistance-creating agents could be an important challenge for combination therapy of MDR K. pneumoniae-caused infections.
1. Introduction
In the 1980s, gram-negative pathogens were successfully defeated using cephalosporins, carbapenems, and fluoroquinolones [1]. K. pneumoniae is one of the most widespread nosocomial pathogens causing urinary tract infections, bacteremia, and pneumonia in different parts of the world [2, 3]. Extensive use of antibiotics and thereby the development of various resistance mechanisms have led to the emergence of MDR (multidrug resistance) K. pneumoniae [4]. KPC- (Klebsiella pneumoniae carbapenemase-) producing K. pneumoniae strains are the most common carbapenamase-producing pathogens worldwide [5] that have also been reported in Iran in many studies [6–8]. Seventeen different types in the KPC family have been reported to date [9], among which one family is located on a conjugative plasmid that encodes resistance to gentamicin and tobramycin [10]. The armA gene, which is responsible for resistance to the majority of the aminoglycosides and is the most prevalent in Asia [11], was found to be located on the same plasmid of KPC-producing strains reported formerly from Italy [12], China [13], and Poland [14]. Coexistence of the resistance-inducing agents can result in the emerging of MDR K. pneumoniae strains. Epidemiological characterization of MDR K. pneumoniae can help to prevent the spread of resistant strains [15]. RAPD PCR (random amplification of polymorphic DNA PCR) is a simple, rapid, inexpensive, and widely used typing method which does not require advanced knowledge of DNA sequences of the target organism. RAPD typing has been successfully employed for epidemiological analysis of many bacteria [16] as well as clinical isolates of K. pneumonia [17].
This study aimed to detect the RAPD PCR fingerprint, antibiotic resistance patterns, and the presence of armA gene in clinical KPC-producing isolates and nonproducing isolates of K. pneumoniae.
2. Materials and Methods
2.1. Bacterial Collection
From September 2015 to March 2016, clinical samples were collected from both inpatients and outpatients attending three hospitals in Tehran, Iran. The samples were transferred to the microbiology laboratory, and biochemical tests were performed to identify K. pneumoniae. The pure isolates were stored at −20°C in Trypticase soy broth containing 20% glycerol.
2.2. Susceptibility Testing and Phenotypic Detection of KPC
The antimicrobial susceptibility test was conducted using the Kirby–Bauer disk diffusion method on Mueller–Hinton agar (Merck, Germany). Also, for colistin-resistant isolates, MIC was determined through a broth microdilution method, according to the Clinical and Laboratory Standard Institute (CLSI) guidelines. The isolates were tested for susceptibility to the following 16 antimicrobial agents (MAST Group, Merseyside, UK): ofloxacin (OFX, 5 μg), ciprofloxacin (CIP, 5 μg), norfloxacin (NOR, 10 μg), gentamicin (GM, 10 μg), amikacin (AK, 30 μg), streptomycin (HLS, 10 μg), tobramycin (TOB, 10 μg), kanamycin (K, 30 μg), ticarcillin (TIC, 75 μg), cefotaxim (CTX, 30 μg), ceftazidime (CAZ, 30 μg), azetreonam (ATM, 30 μg), imipenem (IMI, 10 μg), trimethoprim (TR, 5 μg), chloramphenicol (C, 30 μg), and colistin (CO, 10 μg). Escherichia coli ATCC 25922 was used as a control strain. Also, detection of KPC enzyme was performed by the modified Hodge test [18].
2.3. Detection of armA Gene
Bacteria were cultured in Luria–Bertani (LB) broth medium and were incubated overnight at 37°C. DNA was extracted from K. pneumoniae isolates by using Genomic DNA Isolation Kit (Global Gene Network Bio, cat. no. k-3000, South Korea) according to the manufacturer's instruction. The presence of armA gene was evaluated using PCR technique with the armA-F: TCGGAACTTAAAGACGACGA and armA-R: CCATTCCCTTCTCCTTTCCA (designed in this study) sequences. The PCR product was analyzed by electrophoresis in a 1% (W/V) agarose gel in TBE buffer at 95 V for 45 min, stained with ethidium bromide, and observed under UV lighting using Gel Doc. Then, DNA sequencing was performed by Bioneer Company (Korea), blasted in NCBI, and analyzed by Finch TV software.
2.4. RAPD PCR
Molecular typing of K. pneumoniae isolates was performed using RAPD PCR analysis with the primer 640 (CGTGGGGCCT) (Faza Biotech, Tehran, Iran). Reaction mixtures (25 µl) contained: 12.5 µl (Exprime Taq Premix (2X) Mastermix (Global Gene Network Bio, cat. no. G-5000)), 7.5 µl Distilled water, 200 pmol of primer, and 3 µl DNA template. The program used was as follows: 5 min at 94°C followed by 36 cycles of 1 min at 94°C, 1 min at 36°C, and 2 min at 72°C followed by a final extension at 72°C for 9 min. PCR products were then electrophoresed on 1% agarose gels and visualized by ethidium bromide staining. To determine the similarity rate among the isolates, they were analyzed by the unweighted pair-group method with arithmetic mean (UPGMA) using GelClust software.
3. Results
3.1. Bacterial Isolation
Eighty-one K. pneumoniae were isolated from the patients (44 males and 37 females). Samples were isolated from different clinical specimens including urine, 49 (60.5%); blood cultures, 16 (19.7%); wounds, 5 (6.2%); sputum, 4 (5%); intra-abdominal, 3 (3.7%), and others 4 (5%). The hospital ward distribution was as follows: pediatric ward, 24 (29.6%); outpatient, 18 (22.2%); intensive care unit, 10 (12.3%); surgical unit, 6 (7.4%); infection unit, 2 (2.5%); and others, 21 (26%).
3.2. Determination of Antibiotic Resistance and KPC Enzyme Detection
Antibiogram showed that the resistance rates of the isolates were as follows: ofloxacin 65%, ciprofloxacin 68.7%, norfloxacin 66.3%, gentamicin 66.3%, amikacin 51.8%, tobramycin 56.7%, kanamycin 79.5%, ticarcillin 82%, streptomycin 16.9%, cefotaxim 85.5%, ceftazidime 78.3%, azetreonam 79.5%, imipenem 45.8%, trimethoprim 74.7%, chloramphenicol 21.7%, and colistin 16.9%. One colistin-resistant isolate had a MIC of 8 µg/ml, and the other all had MIC values of 4 µg/ml. All the colistin-resistant isolates except one of them belonged to one hospital. Five colistin-resistant K. pneumoniae exhibited resistance to imipenem of which one was positive for KPC production. The antibiotic resistance pattern of the isolates is shown in Table 1. The highest rate of the resistance belonged to the A9 pattern (64.2%) which found to be resistant to three flouroquinolons. Also, the lowest rate of resistance belonged to the A4 pattern (4.9%) which was resistant to ciprofloxacin, ofloxacin, norfloxacin, imipenem, amikacin, and colistin. MDR is generally determined as resistance to three or more classes of antibiotics [19]. In the present study, 66 out of 81 isolates were resistant to 3 or more classes of antibiotics; thereby, 81.5% of the isolates were found to be MDR. Also, the modified Hodge test showed that 6 isolates (7.4%) were positive for KPC production with 3 resistance patterns (Table 2).
Table 1.
Antibiotic resistance patterns.
| Resistance pattern number | Number of isolates (%) | Antibiotics |
|---|---|---|
| A1 | 11 (13.6) | OFX, NOR, CIP, IMI, K, GM, AK, TOB, HLS |
| A2 | 19 (23.4) | OFX, NOR, CIP, IMI, GM, AK, TOB, K |
| A3 | 29 (35.8) | OFX, NOR, CIP, IMI, K, GM |
| A4 | 4 (4.9) | CIP, OFX, NOR, IMI, AK, CO |
| A5 | 12 (14.8) | AK, GM, TOB, K, HLS |
| A6 | 25 (30.9) | CIP, OFX, NOR, IMI, AK |
| A7 | 33 (40.7) | CIP, OFX, NOR, IMI |
| A8 | 30 (37) | CIP, OFX, NOR, AK |
| A9 | 52 (64.2) | CIP, OFX, NOR |
| A10 | 11 (13.6) | TR, TIC, C |
| A11 | 25 (30.9) | IMI, AK |
OFX: ofloxacin; NOR: norfloxacin; CIP: ciprofloxacin; IMI: imipenem; GM: gentamicin; AK: amikacin; TOB: tobramycin; HLS: streptomycin; K: kanamycin; CO: colistin; TR: trimethoprim; TIC: ticarcillin; C: chloramphenicol.
Table 2.
Antibiotic resistance patterns and presence of armA gene.
| Isolate name | Resistance pattern | armA |
|---|---|---|
| 1 | OFX, NOR, CIP, IMI, K, GM, AK, TOB, HLS | + |
| 2 | OFX, NOR, CIP, IMI, K, GM, AK, TOB | + |
| 3 | OFX, NOR, CIP, IMI, K, GM, AK, TOB | − |
| 4 | OFX, NOR, CIP, IMI, K, GM | − |
| 5 | OFX, NOR, CIP, IMI, K, GM | + |
| 6 | OFX, NOR, CIP, IMI, K, GM | + |
OFX: ofloxacin; NOR: norfloxacin; CIP: ciprofloxacin; IMI: imipenem; K: kanamycin; GM: gentamicin; AK: amikacin; TOB: tobramycin; HLS: streptomycin.
3.3. Molecular Detection
The PCR showed that 54 isolates (66.7%) of the isolates had armA gene. Aminoglycoside resistance patterns in armA positive isolates were presented in Table 3.
Table 3.
Aminoglycoside resistance in armA positive isolates.
| Aminoglycosides | Resistant number (%) | armA + number (%) |
|---|---|---|
| Kanamycin | 66 (79.5) | 46 (69.7) |
| Tobramycin | 47 (56.7) | 32 (68) |
| Gentamicin | 55 (66.3) | 38 (69) |
| Amikacin | 43 (51.8) | 32 (74.4) |
| Streptomycin | 14 (16.9) | 12 (87.7) |
3.4. RAPD PCR
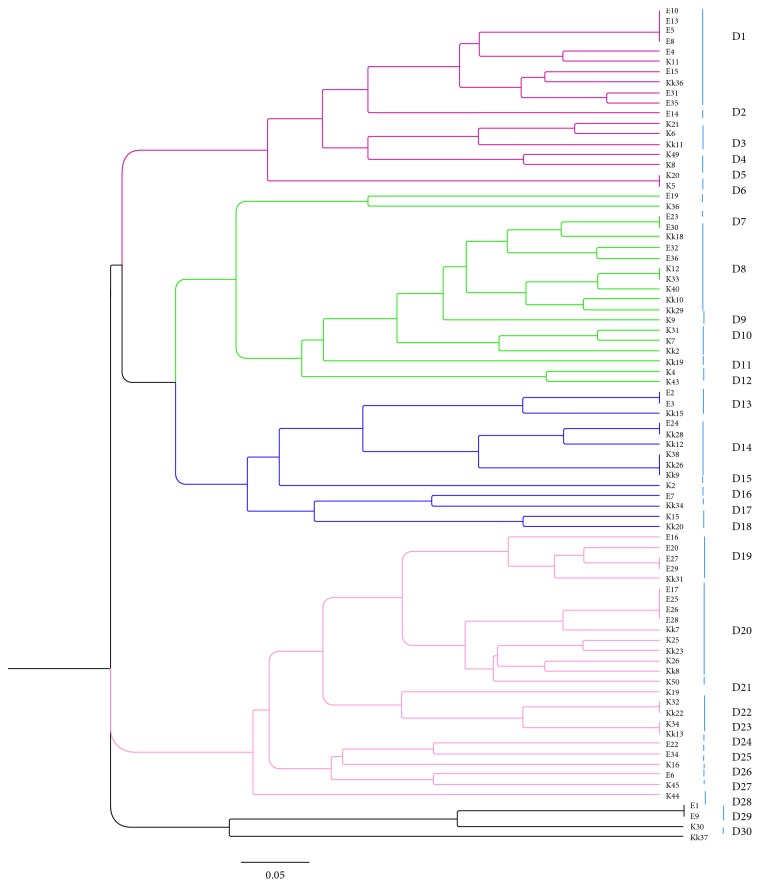
Genetic analysis of RAPD showed 30 distinct patterns from D1 to D30, as 5 distinct clusters (Figure 1). The information of isolates is shown in Table 4. The isolates were considered as the same pattern if the level of similarity was ≥85%. The numbers of the isolates in each cluster (from cluster 1 to 5) were 18, 19, 14, 26, and 4, respectively. The highest redundancy belonged to pattern D1, D8, and D20. There was not any correlation between genetic patterns of the isolates and presence of armA gene or KPC production. The isolates which showed 100% similarity, belonging to different clusters, all found to have armA gene.
Figure 1.
Cluster analysis of Klebsiella pneumoniae based on RAPD typing.
Table 4.
Analysis of strains with their RAPD profiles.
| Isolate number | armA | KPC | Date | Origin | Department | Hospital | RAPD pattern |
|---|---|---|---|---|---|---|---|
| 1 | − | − | February 2016 | Urine | Gynecology and obstetrician | A | D1 |
| 2 | − | − | February 2016 | Urine | Outpatient | A | D1 |
| 3 | − | − | February 2016 | Urine | Outpatient | A | D1 |
| 4 | − | − | February 2016 | Urine | Laparoscopy | A | D1 |
| 5 | − | − | February 2016 | Urine | Outpatient | A | D1 |
| 6 | + | − | September 2015 | Wound | Others | B | D1 |
| 7 | − | − | February 2016 | Urine | Outpatient | A | D1 |
| 8 | + | − | October 2015 | Blood | Pediatric | C | D1 |
| 9 | − | − | February 2016 | Urine | Rheumatology | A | D1 |
| 10 | + | − | March 2016 | Urine | Nephrology | A | D1 |
| 11 | − | − | February 2016 | Urine | Others | A | D2 |
| 12 | + | − | December 2015 | Blood | Hematology | B | D3 |
| 13 | + | − | January 2015 | Wound | Surgery | B | D3 |
| 14 | + | − | January 2015 | Urine | Pediatric | C | D3 |
| 15 | − | + | November 2015 | Urine | Outpatient | B | D4 |
| 16 | − | − | November 2015 | Blood | Infectious | B | D4 |
| 17 | + | − | September 2015 | Intraabdominal | Gastrointestinal | B | D5 |
| 18 | + | + | September 2015 | Sputum | ICU | B | D5 |
| 19 | − | − | February 2016 | Urine | Cancer | A | D6 |
| 20 | + | − | October 2015 | Sputum | Bone marrow transplantation | B | D7 |
| 21 | − | − | February 2016 | Urine | Dermatology | A | D8 |
| 22 | − | + | February 2016 | Urine | Cardiology | A | D8 |
| 23 | + | − | January 2015 | Blood | Pediatric | C | D8 |
| 24 | − | − | March 2016 | Urine | Outpatient | A | D8 |
| 25 | − | − | March 2016 | Urine | Outpatient | A | D8 |
| 26 | + | − | January 2015 | Blood | ICU | B | D8 |
| 27 | − | − | January 2015 | Sputum | ICU | B | D8 |
| 28 | + | − | December 2015 | Urine | Outpatient | B | D8 |
| 29 | + | − | December 2015 | Blood | Pediatric | C | D8 |
| 30 | + | − | December 2015 | Blood | Pediatric | C | D8 |
| 31 | + | − | October 2015 | Wound | Surgery | B | D9 |
| 32 | + | − | November 2015 | Intraabdominal | Cancer | B | D10 |
| 33 | − | − | October 2015 | Others | NICU | B | D10 |
| 34 | + | − | November 2015 | Blood | Pediatric | C | D10 |
| 35 | + | − | December 2015 | Urine | Pediatric | C | D11 |
| 36 | + | − | September 2015 | Sputum | ICU | B | D12 |
| 37 | + | − | October 2015 | Wound | Surgery | B | D12 |
| 38 | + | − | March 2016 | Urine | ICU | A | D13 |
| 39 | + | + | March 2016 | Urine | Others | A | D13 |
| 40 | + | − | January 2015 | Blood | Pediatric | C | D13 |
| 41 | + | − | February 2016 | Urine | Outpatient | A | D14 |
| 42 | + | − | January 2015 | Others | Pediatric | C | D14 |
| 43 | + | − | January 2015 | Urine | Pediatric | C | D14 |
| 44 | + | − | December 2015 | Others | Outpatient | B | D14 |
| 45 | + | + | December 2015 | Urine | Pediatric | C | D14 |
| 46 | + | − | December 2015 | Blood | Pediatric | C | D14 |
| 47 | − | − | September 2015 | Urine | Surgery | B | D15 |
| 48 | − | − | February 2016 | Urine | Outpatient | A | D16 |
| 49 | + | − | October 2015 | Urine | Pediatric | C | D17 |
| 50 | + | − | November 2015 | Urine | Endocrinology | B | D18 |
| 51 | + | − | November 2015 | Blood | Pediatric | C | D18 |
| 52 | − | − | March 2016 | Urine | Cardiology | A | D19 |
| 53 | + | − | March 2016 | Urine | Others | A | D19 |
| 54 | − | − | March 2016 | Urine | Pediatric | A | D19 |
| 55 | − | − | March 2016 | Urine | Outpatient | A | D19 |
| 56 | − | − | January 2015 | Urine | Pediatric | C | D19 |
| 57 | − | − | February 2016 | Urine | Pediatric | A | D20 |
| 58 | − | − | February 2016 | Urine | Surgery | A | D20 |
| 59 | − | − | February 2016 | Urine | ICU | A | D20 |
| 60 | − | − | February 2016 | Urine | Others | A | D20 |
| 61 | + | − | November 2015 | Urine | Pediatric | C | D20 |
| 62 | + | − | November 2015 | Urine | Outpatient | B | D20 |
| 63 | + | − | January 2015 | Urine | Pediatric | C | D20 |
| 64 | + | − | November 2015 | Blood | Outpatient | B | D20 |
| 65 | + | − | January 2015 | Urine | Pediatric | C | D20 |
| 66 | + | − | January 2015 | Others | Outpatient | B | D20 |
| 67 | + | − | September 2015 | Wound | Others | B | D21 |
| 68 | + | − | November 2015 | Blood | Hematology | B | D22 |
| 69 | + | − | November 2015 | Blood | Pediatric | C | D22 |
| 70 | + | − | September 2015 | Urine | NICU | B | D22 |
| 71 | + | − | September 2015 | Urine | Pediatric | C | D22 |
| 72 | + | − | March 2016 | Urine | Surgery | A | D23 |
| 73 | + | − | March 2016 | Urine | Cardiology | A | D24 |
| 74 | + | − | December 2015 | Blood | NICU | B | D25 |
| 75 | − | − | February 2016 | Urine | Pediatric | A | D26 |
| 76 | + | − | October 2015 | Urine | Outpatient | B | D27 |
| 77 | + | − | December 2015 | Urine | ICU | B | D28 |
| 78 | − | − | February 2016 | Urine | Outpatient | A | D29 |
| 79 | − | − | February 2016 | Urine | Outpatient | A | D29 |
| 80 | + | + | January 2015 | Intraabdominal | Infectious | B | D29 |
| 81 | + | − | November 2015 | Blood | Pediatric | C | D30 |
4. Discussion
In the present study, 81 K. pneumoniae were isolated from clinical samples. Antibiotic resistance patterns and their relationships among different clonal isolates were investigated. Eleven antibiotic patterns were found (A1–A11), showing much diversity in the resistance patterns. In the study by Hassan et al. [16] and Ben-Hamouda et al. [17], different antibiotic resistance patterns were reported, too [20]. In the present study, 81.5% were MDR. The presence of multidrug resistance community acquired K. pneumoniae highlights the need for accurate planning to control and prevent of the dissemination of MDR strains. Most of the KPC-producing isolates harbored armA gene and were resistant to carbapenems, aminoglycosides, and fluoroquinolones. According to our previous study, Real Time PCR showed an increased expression level of OqxAB and AcrAB efflux pumps in fluoroquinolone-resistant isolates in comparison with the sensitive ones (data not shown). So, the role of efflux pump in creating fluoroquinolone-resistant strains could be identified in these isolates [21]. Also, the aminoglycoside resistance rates suggested 16S rRNA methylase activity. In the present study, approximately 70% of the aminoglycoside-resistant strains carried the armA gene as Zhou et al. reported [22]. Therefore, some of these K. pneumoniae isolates have three features of resistance: KPC, efflux pumps, and armA gene. So they can be resistant to fluoroquinolones, cephalosporins, carbapenems and a spectrum of aminoglycosides as well. These strains can turn out to an important challenge for community and hospital officials by disseminating among the patients in hospitals and making the treatment process more difficult. Coexistence of the active efflux pump, armA gene, and KPC enzymes in K. pneumoniae can help to resist against the combination therapy. This hypothesis is also recommended by Zacharczuk et al. [14] and Jiang et al. [13] who observed KPC production and armA gene in clinical isolates of K. pneumoniae.
Furthermore, about 17% of the isolates were resistant to colistin, among which all but one isolate had MIC 4 µg/ml. Although colistin-resistant isolates were related to five different clusters, 42.9% of them belonged only to the fourth cluster from one hospital, indicating a genetically specific circulating cluster. Colistin is considered as effective treatment against MDR and carbapenem-resistant bacteria, such as K. pneumoniae, but resistance to this agent has begun to emerge. So, more studies to determine the best treatment for infections caused by resistant K. pneumoniae are needed. In the present study, genotyping analysis showed different genetic patterns among pathogenic K. pneumoniae isolates. Lim et al. [23], Ben-Hamouda et al. [17], Pai et al. [24], and Eftekhar and Nouri [25] performed separately the genotyping of clinical K. pneumoniae strains and showed that they were genetically diverse and heterogeneous. Therefore, this tool has got the ability to identify related and unrelated isolates. The correlation between the antibiotic resistance patterns and RAPD analysis demonstrated that different genetic patterns had different antibiotyping profiles. Also, KPC-producing K. pneumoniae were found to belong to different clusters, and the results of RAPD PCR implicated that there is no correlation between the genetic patterns and presence of armA gene or KPC-producing K. pneumoniae, as Ma et al. [26] reported that nonclonally spread of ESBL-producing K. pneumoniae strains with armA or rmtB mediates aminoglycoside resistance. It seems that because most resistance genes are carried by the mobile genetic elements, they can easily transmit among the bacteria.
Of importance, the low number of the isolates was one of our limitations. Collecting more clinical isolates, using more powerful discriminating typing methods such as PFGE and analysis of armA and blakpc genes' expression level may improve the quality of our results in the following studies.
5. Conclusion
Although fluoroquinolones seem to be a good choice for treating Klebsiella infections, several studies have shown increasing resistance of K. pneumoniae isolates to these agents. Aminoglycosides have been considered as an adequate therapeutic against both gram-negative and gram-positive pathogens and also combination therapy with β-lactams and aminoglycosides is well accepted for the treatment of the systemic infections caused by K. pneumoniae, so simultaneous detection of resistance creating agents to fluoroquinolones, β-lactams, and aminoglycosides in these strains is of clinical importance. Emphasis on the suitable use of antibiotics, effective infection control measures, and identification of antibiotic resistance mechanisms by molecular procedures are necessary to reduce the incidence of infections caused by antibiotic-resistant organisms.
Acknowledgments
This work was supported by the Karaj Azad University and Shahid Beheshti University.
Abbreviations
- RAPD:
Random amplification of polymorphic DNA
- KPC:
Klebsiella pneumoniae carbapenemase
- PCR:
Polymerase chain reaction
- CLSI:
Clinical and Laboratory Standards Institute
- MDR:
Multiple drug resistance
- NCBI:
National Center for Biotechnology Information
- ESBL:
Extended-spectrum beta-lactamases
- PFGE:
Pulsed field gel electrophoresis.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Livermore D. M. Current epidemiology and growing resistance of gram-negative pathogens. The Korean Journal of Internal Medicine. 2012;27(2):128–142. doi: 10.3904/kjim.2012.27.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Padilla E., Llobet E., Domenech-Sanchez A., Martinez-Martinez L., Bengoechea J. A., Alberti S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrobial Agents and Chemotherapy. 2010;54(1):177–183. doi: 10.1128/aac.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdel-Hady H., Hawas S., El-Daker M., El-Kady R. Extended-spectrum β-lactamase producing Klebsiella pneumoniae in neonatal intensive care unit. Journal of Perinatology. 2008;28(10):685–690. doi: 10.1038/jp.2008.73. [DOI] [PubMed] [Google Scholar]
- 4.Souli M., Galani I., Antoniadou A., et al. An outbreak of infection due to β-lactamase Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae in a Greek university hospital: molecular characterization, epidemiology, and outcomes. Clinical Infectious Diseases. 2010;50(3):364–373. doi: 10.1086/649865. [DOI] [PubMed] [Google Scholar]
- 5.Mezzatesta M. L., Gona F., Caio C., et al. Emergence of an extensively drug-resistant ArmA- and KPC-2-producing ST101 Klebsiella pneumoniae clone in Italy. Journal of Antimicrobial Chemotherapy. 2013;68(8):1932–1934. doi: 10.1093/jac/dkt116. [DOI] [PubMed] [Google Scholar]
- 6.Azimi L., Rastegar-Lari A., Talebi M., Ebrahimzadeh-Namvar A., Soleymanzadeh-Moghadam S. Evaluation of phenotypic methods for detection of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae in Tehran. Journal of Medical Bacteriology. 2015;2(3-4):26–31. [Google Scholar]
- 7.Bina M., Pournajaf A., Mirkalantari S., Talebi M., Irajian G. Detection of the Klebsiella pneumoniae carbapenemase (KPC) in K. pneumoniae isolated from the clinical samples by the phenotypic and genotypic methods. Iranian Journal of Pathology. 2015;10(3):199–205. [PMC free article] [PubMed] [Google Scholar]
- 8.Nobari S., Shahcheraghi F., Ghezelgeh F. R., Valizadeh B. Molecular characterization of carbapenem-resistant strains of Klebsiella pneumoniae isolated from Iranian patients: first identification of blaKPC gene in Iran. Microbial Drug Resistance. 2014;20(4):285–293. doi: 10.1089/mdr.2013.0074. [DOI] [PubMed] [Google Scholar]
- 9.Wang D., Chen J., Yang L., Mou Y., Yang Y. Phenotypic and enzymatic comparative analysis of the KPC variants, KPC-2 and its recently discovered variant KPC-15. PLoS One. 2014;9(10) doi: 10.1371/journal.pone.0111491.e111491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sacha P., Ostas A., Jaworowska J., et al. The KPC type beta-lactamases: new enzymes that confer resistance to carbapenems in Gram-negative bacilli. Folia Histochemica et Cytobiologica. 2009;47(4):537–543. doi: 10.2478/v10042-009-0079-y. [DOI] [PubMed] [Google Scholar]
- 11.Fritsche T. R., Castanheira M., Miller G. H., Jones R. N., Armstrong E. S. Detection of methyltransferases conferring high-level resistance to aminoglycosides in Enterobacteriaceae from Europe, North America, and Latin America. Antimicrobial Agents and Chemotherapy. 2008;52(5):1843–1845. doi: 10.1128/aac.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frasson I., Lavezzo E., Franchin E., et al. Antimicrobial treatment and containment measures of an extremely drug-resistant Klebsiella pneumoniae ST101 isolate carrying pKPN101-IT, a novel fully sequenced blaKPC-2 plasmid. Journal of Clinical Microbiology. 2012;50(11):3768–3772. doi: 10.1128/JCM.01892-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang Y., Yu D., Wei Z., Shen P., Zhou Z., Yu Y. Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrobial Agents and Chemotherapy. 2010;54(9):3967–3969. doi: 10.1128/aac.00137-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zacharczuk K., Piekarska K., Szych J., et al. Emergence of Klebsiella pneumoniae coproducing KPC-2 and 16S rRNA methylase ArmA in Poland. Antimicrobial Agents and Chemotherapy. 2011;55(1):443–446. doi: 10.1128/aac.00386-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Souza Lopes A. C., Rodrigues J. F., de Morais Júnior M. A. Molecular typing of Klebsiella pneumoniae isolates from public hospitals in Recife, Brazil. Microbiological Research. 2005;160(1):37–46. doi: 10.1016/j.micres.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Hassan R., Barwa R., Shaaban M. I., Adel L. Random amplified DNA polymorphism of Klebsiella pneumoniae isolates from Mansoura University Hospitals, Egypt. African Journal of Microbiology Research. 2015;9(9):621–630. [Google Scholar]
- 17.Ben-Hamouda T., Foulon T., Ben-Cheikh-Masmoudi A., Fendri C., Belhadj O., Ben-Mahrez K. Molecular epidemiology of an outbreak of multiresistant Klebsiella pneumoniae in a Tunisian neonatal ward. Journal of Medical Microbiology. 2003;52(5):427–433. doi: 10.1099/jmm.0.04981-0. [DOI] [PubMed] [Google Scholar]
- 18.Tsai Y.-K., Fung C. P., Lin J. C., et al. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrobial Agents and Chemotherapy. 2011;55(4):1485–1493. doi: 10.1128/aac.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swick M. C., Morgan-Linnell S. K., Carlson K. M., Zechiedrich L. Expression of multidrug efflux pump genes acrAB-tolC, mdfA, and norE in Escherichia coli clinical isolates as a function of fluoroquinolone and multidrug resistance. Antimicrobial Agents and Chemotherapy. 2011;55(2):921–924. doi: 10.1128/aac.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shahcheraghi F., Moezi H., Feizabadi M. M. Distribution of TEM and SHV beta-lactamase genes among Klebsiella pneumoniae strains isolated from patients in Tehran. Medical Science Monitor. 2007;13(11):BR247–BR250. [PubMed] [Google Scholar]
- 21.Pakzad I., Karin M. Z., Taherikalani M., Boustanshenas M., Lari A. R. Contribution of AcrAB efflux pump to ciprofloxacin resistance in Klebsiella pneumoniae isolated from burn patients. GMS Hygiene and Infection Control. 2013;8(2) doi: 10.3205/dgkh000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Y., Yu H., Guo Q., et al. Distribution of 16S rRNA methylases among different species of gram-negative bacilli with high-level resistance to aminoglycosides. European Journal of Clinical Microbiology & Infectious Diseases. 2010;29(11):1349–1353. doi: 10.1007/s10096-010-1004-1. [DOI] [PubMed] [Google Scholar]
- 23.Lim K. T., Yeo C. C., Md Yasin R., Balan G., Thong K. L. Characterization of multidrug-resistant and extended-spectrum β-lactamase-producing Klebsiella pneumoniae strains from Malaysian hospitals. Journal of Medical Microbiology. 2009;58(11):1463–1469. doi: 10.1099/jmm.0.011114-0. [DOI] [PubMed] [Google Scholar]
- 24.Pai H., Kang C. I., Byeon J. H., et al. Epidemiology and clinical features of bloodstream infections caused by AmpC-type-β-lactamase-producing Klebsiella pneumoniae. Antimicrobial Agents and Chemotherapy. 2004;48(10):3720–3728. doi: 10.1128/aac.48.10.3720-3728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eftekhar F., Nouri P. Correlation of RAPD-PCR profiles with ESBL production in clinical isolates of Klebsiella pneumoniae in Tehran. Journal of Clinical and Diagnostic Research. 2015;9(1):DC01–DC03. doi: 10.7860/JCDR/2015/10651.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma L., Lin C. J., Chen J. H., et al. Widespread dissemination of aminoglycoside resistance genes armA or rmtB in Klebsiella pneumoniae isolates in Taiwan producing CTX-M type extended spectrum β-lactamase. Antimicrobial Agents and Chemotherapy. 2009;53(1):104–111. doi: 10.1128/aac.00852-08. [DOI] [PMC free article] [PubMed] [Google Scholar]