Abstract
Pluripotent stem cells have the property of long-term self-renewal and the potential to give rise to descendants of the three germ layers and hence all mature cells in the human body. Therefore, they hold the promise of offering insight not only into human development but also for human disease modeling and regenerative medicine. However, the generation of mature differentiated cells that closely resemble their in vivo counterparts remains challenging. Recent advances in single-cell transcriptomics and computational modeling of gene regulatory networks are revealing a better understanding of lineage commitment and are driving modern genome editing approaches. Additional modification of the chemical microenvironment, as well as the use of bioengineering tools to recreate the cellular, extracellular matrix, and physical characteristics of the niche wherein progenitors and mature cells reside, is now being used to further improve the maturation and functionality of stem cell progeny.
Keywords: Stem cells, disease modeling, regenerative medicine, genome editing
Introduction
Stem cells have the remarkable property of long-term self-renewal, and at the same time they can give rise to progressively more lineage-committed cells. Pluripotent stem cells (PSCs) are multipotent as they can generate all mature cells of the body. Although murine PSCs were already isolated in the 1980s 1, it was not until 1998 that human PSCs (hPSCs) were first isolated from human blastocysts, termed embryonic stem cells (ESCs) 2. This accomplishment, and, even more so, the creation of so-called ‘induced pluripotent stem cells’, or iPSCs, from mouse 3 and then human 4 fibroblasts in 2006–2007, opened up the possibility of generating any cell type for regenerative medicine. In addition, the availability of human ESCs (hESCs) and iPSCs (collectively termed PSCs) now provides us with tools to better understand human development as well as create models to study human diseases. Fully exploiting that potential, however, remains challenging. Although new stem cell differentiation protocols are published on a weekly basis, many hurdles remain regarding how to create mature stem cell progeny. In this review, we will discuss the current state of the art of stem cell culture and lineage differentiation from stem cells. Current limitations in creating mature PSC progeny are steering investigators to explore novel avenues in stem cell research. We discuss the roads that are being taken and need to be taken to improve the functionality of PSC-derived human tissue cells and ultimately exploit the full potential of hPSCs.
Pluripotency matters
The derivation and culture of hESCs and human iPSCs (hiPSCs) use conditions that differ from those used to isolate their murine counterparts. This results in the capture of cells at different stages of embryonic development. Murine PSCs can be isolated by using several culture methods from mouse embryos. These include culture on mouse embryonic fibroblasts or using mTeSR and LIF, resulting in a population of cells containing chiefly ESCs but also extra-embryonic endoderm progenitors 5. More recently, it has been demonstrated that murine ESCs can be isolated and maintained in a ‘naïve’ ground state (termed 2i/LIF conditions) that resembles pre-implantation pluripotency wherein fate allocation in specific cell populations has not yet occurred. Unlike their murine counterparts, human PSCs in standard culture conditions resemble post-implantation, ‘primed’ epiblast stem cells, wherein initial fate allocation has occurred 6. When post-implantation murine or rat embryos are used, a similar epiblast-like cell type can be isolated as well 7. Such primed human and mouse ESCs have already undergone X-chromosome inactivation. Moreover, the generation of hiPSCs does not lead to reactivation of the X-chromosome. This X-chromosome inactivation state, however, is unstable, as long-term culture of female ‘primed’ hESCs or hiPSCs has been shown to cause ‘X-chromosome erosion’; that is, the inactivation of the silenced X-chromosome is progressively lost. X-chromosome erosion is associated with decreased differentiation potential, and this is inherited by the differentiated progeny 8– 11. In addition, excessive X-chromosome skewing is frequently seen 12– 15, yielding skewed progeny that could manifest or lose disease phenotypes. For example, in the case of Duchenne muscular dystrophy, female carriers of a dystrophin gene mutation rarely manifest with muscular dystrophy because the random nature of X-chromosome inactivation leads to a mixture of cells expressing either the wild-type or the disease gene. In this case, in vitro X-chromosome skewing could misrepresent the in vivo situation 16. Together with the fact that primed cells are not lineage-neutral, this may be a major contributing factor to the variability observed within and between different cell lines 17.
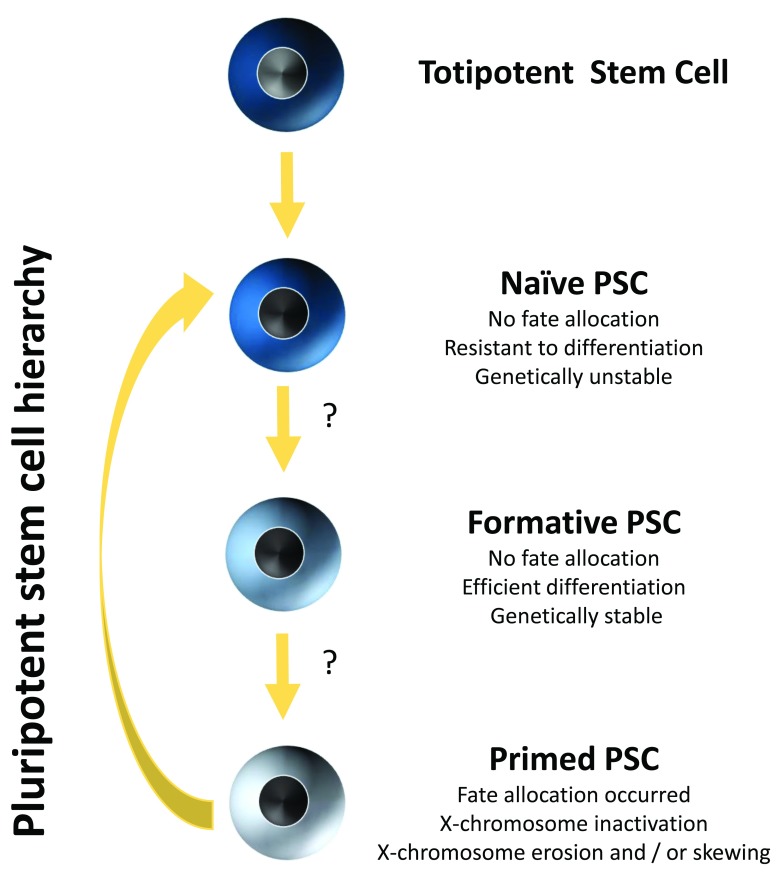
During the last 3 years, a number of protocols have been described in which primed hPSCs can be reset to a naïve state or which allow reprogramming of somatic cells to naïve hPSCs (reviewed in 18). However, such human naïve PSCs are more resistant to differentiation, and a step wherein naïve PSCs are committed to an intermediate primed state is required, at least in some studies 19– 21. For instance, transitioning through a more naïve state proved to be especially beneficial in the case of germ line cell differentiation. The initial fate allocation of regular primed hPSCs significantly dampens germ line competence, but converting hESCs to a more naïve state, using a protocol developed by Gafni et al. 22, drastically improved the efficiency of generating primordial germ cells 23. However, recent reports sound a cautionary note by reporting that naïve hPSCs are genetically unstable 11, 24. Hence, they are not good candidates to start lineage differentiation for either disease modeling or regenerative medicine. However, recent insights into the hierarchy of pluripotency during development have led to the identification of an additional cell population, to which the term ‘formative PSCs’ was assigned, that appears to be located in between naïve and primed PSCs 25 ( Figure 1). Such putative formative PSCs 26 are, like naïve PSCs, lineage-neutral; however, unlike naïve PSCs, they are hypothesized to be able to differentiate efficiently into all different cell lineages. Furthermore, they also appear to be genetically more stable than naïve hPSCs and, because they have higher levels of methyltransferase activities than primed PSCs 27, also may not be subject to X-chromosome erosion. Therefore, developing methods that allow capturing and maintaining hPSCs in this formative state will likely be of great importance to enable robust and controlled lineage differentiation.
Figure 1. Pluripotent stem cell (PSC) hierarchy.
Hypothesized hierarchy of human PSCs and their properties.
Lineage differentiation: current state of the art and shortcomings
To induce lineage differentiation from PSCs, investigators have used insights gained from development. Initially, this comprised the generation of embryoid bodies, wherein differentiation occurs spontaneously as a result of signals emanating from the different cell populations that spontaneously develop 25. To control the differentiation process better, subsequent studies have used step-wise addition of growth factors and cytokines, or inhibitors thereof, known to play a role during certain steps of differentiation, combined in the majority in cases with monolayer culture systems. This has enabled the generation of a number of cell types, including cells with features of neural subpopulations (for example, glutamatergic cortical neurons 28, cholinergic neurons 29, dopaminergic neurons 30, oligodendrocytes 31, and astrocytes 32), cardiac muscle 33– 36, or hepatocytes 37– 39, among many others. However, these cells resemble fetal tissue more than adult tissue in the majority of cases. Reasons for this are numerous and will be discussed below.
First, although we have some insight into the progressive maturation of cells in a tissue, this knowledge is based on quantitative reverse transcription polymerase chain reaction or genome-wide transcriptome studies throughout development, assessing gene expression in bulk populations of cells. However, as was already suggested by immunofluorescence staining for a limited number of marker proteins, not all cells in a tissue are equal. This is likely also true for developing cell populations, although such studies in human embryos are, for obvious reasons, not readily feasible. Nevertheless, thanks to the advent of single-cell RNAseq, single-cell variability in mature and developing tissues (the latter chiefly in mouse) can now be further elucidated (reviewed in 40, 41). For instance, it is now clear that neurons and astrocytes are brain region-specific, and transcriptional signatures for these are becoming available (reviewed in 42). Therefore, as has been done for neuronal differentiation protocols, different approaches will likely be needed to generate regionally specific astrocytes.
Second, although the mature phenotype of certain cells, such as hepatocytes, has been well established and can be used to define the maturation state of these cells, this is not the case for all cell types 43. Using astrocytes as an example, the definition of a ‘mature’ astrocyte remains unclear (reviewed in 44). Although in general the presence of glial fibrillary acidic protein or S100β or both has been used to define astrocytes, this is insufficient, as there are other neural precursors, such as radial glia, that could, on the basis of these markers alone, be classified as astrocytes 45. Therefore, markers or assays that reveal the functional properties of these cells, including propagation of calcium waves, glutamate handling, and inflammatory responses, should be used to define astrocytes 46– 48.
Third, standard culture conditions constitute a major roadblock in lineage differentiation, as they do not provide all of the necessary environmental cues. During embryogenesis, cells from different germ layers co-develop in response not only to graded and continuously changing concentrations of chemical factors generated by neighboring cells and cells at some distance 49 but also to cell–cell- and cell–extracellular matrix (ECM)-mediated signals. Although growth factor addition, in what we would call a ‘purist’ approach to a single-cell-type-at-a-time differentiation, tries to recreate the in vivo chemical signals, it is very crude and fails to recreate the subtle progressively changing levels of growth factors and morphogens present in vivo. Obviously, by attempting to make pure populations of differentiated cells, we preclude the influence on cell commitment and maturation of neighboring cells derived from the same or even a distinct germlayer. In addition, differentiation performed in 2D culture is generally in culture plastic wells coated with an ECM such as collagen, laminin, or matrigel. This does not start to recreate the complex and changing ECM or the physical characteristics of developing ‘soft’ organs. In addition, numerous other environmental cues affect cell differentiation—such as electrophysiological activity that regulates cardiac muscle or neural cell maturation 50; mechanical stimulation inducing cardiac differentiation 51; flow/shear force that affects endothelial differentiation 52; or nutrient composition of the blood that affects zonation of cells in the liver 53—all of which are only now starting to be evaluated to enhance lineage maturation.
Finally, the duration of many differentiation protocols does not reflect human gestation. It may not be surprising that differentiated progeny resembles fetal tissue more than adult tissue. For instance, during development, neurogenesis switches to gliogenesis late during gestation 54, which underlies the fact that very lengthy cultures are required to derive astrocytes from PSCs.
Another example is that PSC-derived hepatocytes usually harvested after 3–5 weeks continue to express the fetal hepatocyte gene, alpha-fetoprotein 55, and do not express markers associated with mature hepatic function, such as the detoxifying enzymes of the cytochrome P450 family (CYPs) 56. Although the fetal phenotype of PSC-derived hepatocytes allows disease modeling of, for instance, hepatotropic viruses such as dengue, hepatitis B, and hepatitis C 57– 60, their fetal metabolic signature limits their usefulness in drug toxicity studies 43. In contrast, in the field of regenerative medicine, the immaturity and plasticity of progenitor cells can be advantageous as, for example, dopaminergic neuronal progenitors still hold the ability to migrate and integrate when transplanted in Parkinson’s disease 61. Several clinical trials, conducted between 1990 and 2003, have tested the feasibility and clinical benefit of transplanting fetal mesencephalic grafts in the basal ganglia of patients with Parkinson’s disease 61, 62. Though not all equally successful, these trials served as a proof of principle and paved the way for trials using hPSC-derived progeny 63. Nevertheless, the use of incomplete differentiated progeny from PSCs may hold the risk of causing unwanted excessive proliferation 64 or even teratoma formation 65.
Lineage differentiation: ways forward
Genome editing
As stated above, the incomplete lineage differentiation from PSCs for many cell types is caused by our incomplete understanding of developmental processes. However, the newly available high-throughput single-cell transcriptomics and computational modeling of gene regulatory networks now allow in-depth characterization of cellular identity and lineage commitment, not only in adult tissues but also throughout development (the latter at least in mouse development) 66– 68. These studies start to identify previously underestimated heterogeneity in cell populations, in the adult as well as during development 69, 70, and in the meantime provide key targets and knowledge of transcriptional dynamics 68.
Such single-cell RNAseq data combined with novel genome engineering approaches, especially the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) technology (reviewed in 71), allow investigators to ‘engineer’ PSC progeny toward specific lineages. CRISPR technology significantly enhances the frequency of homologous recombination in hPSCs. This is all the more important to create reporter cell lines, enabling the enrichment of precursors or mature cells from mixed populations. However, even more important for lineage differentiation approaches is the ability to exploit the CRISPR system to activate or interfere with gene expression, using CRIPSR-activation (CRISPRa) and CRIPSR-interfering (CRISPRi) approaches. For CRISPRa, a dead Cas (dCas), unable to cause double-strand breaks, is linked to a transactivator sequence, such as multiple herpes simplex virus protein VP16 repeats (VP64) 72, or VP64 fused with p65, a subunit of the ubiquitous NF-κB transcription factor complex (p65) and the Epstein-Barr virus R transactivator (Rta) (so-called VPR) 73, among others. The VPR transactivator, for instance, allows the activation of endogenous gene transcription by a median of 150-fold. Alternatively, the dCas can be fused with a gene repressor such as the Krüppel-associated box (KRAB) domain of Kox1 74 or four copies of the mSin3 domain (termed SID4X) 71 to inhibit gene transcription by up to 15-fold. The CRISPR–dCas-activator/repressor can be integrated into a safe harbor locus 75 and be driven by different inducible promoters to enable the expression of either the transactivator or the repressor. In addition, the use of diverse dCas9 orthologues from different bacteria and their respective single-guide RNA 76 further enhances the possibilities for sequential or combinatorial induction (or both) of transcription factors (TFs) that are insufficiently expressed in PSC progeny while inhibiting TFs that are incorrectly expressed in differentiated progeny. The latter could be either pluripotency TFs but also TFs for lineages other than the desired lineage that are incorrectly activated during the in vitro differentiation process 43, 72, 77– 81.
Chemical engineering of the culture medium
A number of studies have started to test libraries of small molecules to identify factors that enhance differentiation. A good example is pancreatic-beta cell differentiation from PSCs, wherein more than 20 different molecules have been used to create insulin-responsive cells 82. Similar examples can be found in the literature for other cell types, including hepatic, neuronal, or cardiac progeny 36, 83, 84. However, an often-overlooked characteristic of the culture medium is the nutrient microenvironment. It is, however, well known that medium composition can greatly affect cellular behavior in vitro. Several studies demonstrated that nutrient metabolism is one of the major regulators of stem cell fate. For instance, central carbon metabolism plays an important role in PSC maintenance, proliferation, differentiation, and lineage specialization (reviewed in 85). Indeed, PSCs have a glycolytic phenotype 86, and failure to induce glycolysis prevents iPSC generation 87. By contrast, many differentiated progeny—especially, neural cells 88, cardiomyocytes 89, and hepatocytes 90—are dependent on oxidative phosphorylation for energy production. As the latter is fueled by fatty acids or amino acids or both, it will be paramount to analyze the nutrient requirements of precursor populations generated from PSCs and ultimately develop culture media that support these nutrient needs 91 to allow generation of mature lineage-specific cells and simultaneously avoid alternate cell faiths. An example of the latter approach is the eradication of hESCs/iPSCs from cultures by the transient depletion of methionine 92.
Recreation of the stem cell, precursor, and mature cell niche
As discussed above, the ‘purist’ approach to lineage differentiation from PSCs has been to develop differentiation protocols based on monolayer systems coated with a generic ECM layer supplemented with growth factors, morphogens, and cytokines. However, culture in 2D of numerous mature primary cells—such as hepatocytes 93 or glia 46, to name a few—is incompatible with maintaining a mature, differentiated phenotype in vitro. Therefore, it is not surprising that such culture conditions are not suitable for the generation of mature PSC-derived progeny. Hence, researchers are investigating approaches such as co-culture of one or more cell types in 3D culture systems or PSC-derived organoid cultures to better mimic the environment, also named ‘niche’, wherein cell fating and differentiation occur during development and wherein mature cells reside within an organ.
Although organoids from undifferentiated PSCs allow evaluation of the initial steps of cell fating and the spontaneously developing interactions between neighboring cells and their niches, as is thought to occur in vivo, it remains difficult to control these steps, and the creation of mature functional cells from stem cell organoids is not easily achieved. A very good example is the self-organizing brain organoid, initially described by Lancaster and Knoblich, that allows the generation of a variety of brain regional identities, including hindbrain, midbrain, forebrain, and even retinal tissue 94. However, even if this allows the creation of some of the complexity of the brain, inherent to spontaneous development is that the organoids are highly variable within a single experiment and between experiments, and cells do not achieve a terminally differentiated state.
As an alternative, organoids can be generated from different PSC-derived lineage precursors embedded or not in functionalized hydrogels, in combination with chemical signals (growth factors) to recreate the physicochemical properties of the organ wherein the maturing cells reside. This enables greater control of the cell types present in the 3D culture compared with the spontaneous, less controlled differentiation occurring in undifferentiated PSC organoid cultures 95, 96. Until recently, 3D organoid cultures were exclusively performed in matrigel, a poorly defined and largely proteinaceous mixture whose properties cannot be readily modulated and therefore do not allow the evaluation of cell–ECM interactions or do not allow one to understand the role of mechanical forces in cell fate decisions. While many cell types are not intrinsically mechanically active, they encounter and respond to a range of mechanical forces such as shear stress, matrix topography, and rigidity. While we intuitively accept that differentiation induces changes in cell shape, controlling cell shape also affects differentiation 97, 98. Mechano-sensation affects cell morphology, cytoskeletal structure, adhesion, and function, emphasizing the importance of cell–ECM interactions 99, 100. The advent of highly tunable hydrogels based on poly(ethylene glycol) (PEG) 101 or natural gellan gum (GG) 102 has significantly increased the flexibility of probing the role of the microenvironment in specifying stem cell fate. A number of teams have developed ECM component libraries wherein matrix stiffness, matrix degradability, and soluble and ECM peptide factors can be combinatorially and systematically tested in high-throughput approaches. This has resulted in the definition of improved PSC culture and differentiation conditions 103, 104.
The above-mentioned hydrogel technology not only can be used to embed spontaneous assembly of different cell types but also can be combined with bioprinting, allowing the creation of stem cell niches. Bioprinting, such as bioextrusion 105, inkjet bioprinting 106, and microvalve-based bioprinting 107, 108, has already been used to create 3D tissue and organs from stem cells. However, the spatial resolution of these approaches is in the 50-μm range, not allowing the precise recreation of cell niches. However, this may be feasible when using laser-guided printing 109, 110, wherein laser settings and droplet size allow very high-precision printing to the single-cell level. This theoretically should allow one to precisely recreate the cellular niches and to probe the effect of the microenvironment (cell–ECM and cell–cell contact) on cellular differentiation.
As cells also respond differentially to the organization and ultrastructure of the ECM, topographical cues embedded in ECM can further guide tissue development. For instance, hPSC-derived cardiomyocytes grown on microgrooves, that topographically resemble aligned collagen fibers in the developing heart, aligned to the substrate, which resulted in improved sarcomere length 111, 112. Therefore, synthetic nanopatterned substrates with well-defined properties are also assessed for their effect on differentiation 113, 114.
Mechanical and electrical engineering
The controlled addition of growth factors and small molecules in conjunction with tailored hydrogels can mimic the physicochemical properties of the immediate in vivo environment. However, developing cells are also subjected to electromechanical forces exerted by the organ in which they develop (reviewed in 115). Therefore, a number of studies have started to test the additional effect of electrical or mechanical stimulation (or both) on cardiac and neural differentiation, as electrical activity is a fundamental property of these cell types 50, 51, 116. For instance, stimulation paradigms have been well described for both rat and mouse primary cultures 117, 118, suggesting that integrating electrical stimulation to enhance the maturation of hPSC-derived neurons is feasible. However, such an approach has not yet been widely applied to human culture systems. Continuous electrical stimulation also improved cardiac differentiation, significantly enhanced connexin expression and sarcomeric structure, and instructed cardiomyocytes to adapt their beating rate 51, a sign of electrophysiological maturity 119. As electrical activity is coupled to contraction in cardiomyocytes, the role of cyclic stress in cardiac muscle maturation is also being tested. Although applying mechanical stimulation improved the transcriptional and functional profile of hPSC-derived cardiomyocytes 120, less is known about the influence of mechanical stimulation on the maturation of other cell types.
Engineering perfusion
The final and perhaps most arduous addition to the 3D-derived models is to integrate the vascular network, to allow the delivery of nutrients and oxygen to the microtissue and to allow re-circulation of endogenous factors and hence aid in specifying cells within a tissue (for example, zonation in a liver sinusoid). Vascular networks have, for instance, already been printed, integrated into skin tissue or on a cardiac patch 109, 121, or incorporated in self-assembling 3D tissues 96. When implanted in rodents, the murine blood vessels of the implant connected with the vascular network, integrating blood vessels into the vascular system of the host 96. In addition, microfluidic systems have been generated allowing the continuous manipulation of, for instance, pre-defined oxygen gradients, the delivery/removal of specific factors or functional components to/from the microtissue, and the alteration of mechanical forces exerted on the tissue 122. For example, Giobbe et al. differentiated hPSCs directly on a chip under perfusion to hepatocytes and cardiomyocytes, which presented with significantly increased functionality as compared with unperfused control cells 123. Even though perfusion is of such importance for the differentiation of PSCs and their derivatives, a major challenge remains to connect very small (micrometer-diameter) capillaries 109, 121 to microfluidic channels that are commonly 5 to 10 times wider than capillaries.
Conclusions
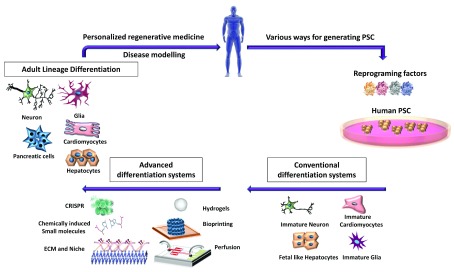
The generation of mature PSC-derived progeny, resembling their in vivo counterpart, is desperately needed in the field of disease modeling and regenerative medicine. The availability of high-throughput single-cell transcriptomics, computational modeling of gene regulatory networks, and new genome editing tools now allows in-depth characterization of cellular identity and lineage commitment. These advances highlight a previously underestimated heterogeneity in cell populations but in the meantime provide key targets and knowledge of cellular state and function. This increased understanding has been one of the driving forces behind the continuous generation of new differentiation protocols. In addition, progress made in the field of (bio)engineering now allows the creation of hydrogels with mechanical stiffness similar to that of a given organ. It also allows one to decorate the gels with ECM components as well as chemical guides, aside from electromechanical stimuli, to drive differentiation in a 3D configuration. In addition, bioprinting techniques have evolved such that specific ECM topologies that instruct differentiation can be recreated, specific cell-to-cell interactions can be incorporated at the (near) single-cell level, and capillary networks can be co-embedded between different cells to enable continuous nutrient support and removal of breakdown products. All of these tools are starting to recreate different aspects of the niches wherein progenitors and mature cells reside, although much is still to be learned about these niches before we will be able to fully recreate them ( Figure 2).
Figure 2. Schematic overview for the advances in lineage differentiation.
Induced pluripotent stem cells (PSCs), for studies of human disease or to create differentiated cells for regenerative medicine, can be generated from any somatic cell, from which the desired cell can be differentiated. However, current differentiation systems generate immature progeny. Recent advances in genome editing (CRISPRa/i), chemical screens, and bioengineering—extracellular matrix (ECM) functionalized hydrogels, bioprinting, and microfluidics—are being used to allow the derivation of more mature and functional PSC progeny, which resemble their in vivo counterparts better, and can be used for personalized and regenerative medicines. CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats.
As we are starting to move away from standard ‘petri dish’ culture systems to high-complexity systems, not only will it be important to ensure reproducibility but also the main challenge will be to adapt these advanced culture systems to medium- and high-throughput formats in a cost-effective way. Thus, even if great strides have been made toward lineage differentiation of stem cells, many challenges are still ahead before it will be possible to generate fully mature and functional cell types.
Abbreviations
CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; CRISPRa, Clustered Regularly Interspaced Short Palindromic Repeats-activation; dCas, dead Cas; ECM, extracellular matrix; ESC, embryonic stem cell; hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cell; hPSC, human pluripotent stem cell; iPSC, induced pluripotent stem cell; PSC, pluripotent stem cell; TF, transcription factor; VP64, virus protein VP16 repeats (4 times repeat); VPR, VP64 fused with p65, a subunit of the ubiquitous NF-κB transcription factor complex (p65) and the Epstein-Barr virus reverse transactivator.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Gareth J Sullivan, Department of Molecular Medicine, University of Oslo, Oslo, Norway
Amander T Clark, Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research, Los Angeles, USA
Funding Statement
This research was supported by the Agency for Innovation by Science and Technology (IWT SBO iPSCAF - 150031). Joke Terryn is a recipient of the IWT SB (141228), and Tine Tricot is a recipient of the FWO (1185918N) grant.
[version 1; referees: 2 approved]
References
- 1. Martin GR: Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78(12):7634–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. : Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–7. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- 3. Takahashi K, Yamanaka S: Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 4. Takahashi K, Tanabe K, Ohnuki M, et al. : Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–72. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 5. Lo Nigro A, de Jaime-Soguero A, Khoueiry R, et al. : PDGFRα + Cells in Embryonic Stem Cell Cultures Represent the In Vitro Equivalent of the Pre-implantation Primitive Endoderm Precursors. Stem Cell Reports. 2017;8(2):318–33. 10.1016/j.stemcr.2016.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nichols J, Smith A: Naive and primed pluripotent states. Cell Stem Cell. 2009;4(6):487–92. 10.1016/j.stem.2009.05.015 [DOI] [PubMed] [Google Scholar]
- 7. Brons IG, Smithers LE, Trotter MW, et al. : Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448(7150):191–5. 10.1038/nature05950 [DOI] [PubMed] [Google Scholar]
- 8. Mekhoubad S, Bock C, de Boer AS, et al. : Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10(5):595–609. 10.1016/j.stem.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 9. Nazor KL, Altun G, Lynch C, et al. : Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10(5):620–34. 10.1016/j.stem.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patel S, Bonora G, Sahakyan A, et al. : Human Embryonic Stem Cells Do Not Change Their X Inactivation Status during Differentiation. Cell Rep. 2017;18(1):54–67. 10.1016/j.celrep.2016.11.054 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 11. Sahakyan A, Kim R, Chronis C, et al. : Human Naive Pluripotent Stem Cells Model X Chromosome Dampening and X Inactivation. Cell Stem Cell. 2017;20(1):87–101. 10.1016/j.stem.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 12. Dhara SK, Benvenisty N: Gene trap as a tool for genome annotation and analysis of X chromosome inactivation in human embryonic stem cells. Nucleic Acids Res. 2004;32(13):3995–4002. 10.1093/nar/gkh746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geens M, Seriola A, Barbé L, et al. : Female human pluripotent stem cells rapidly lose X chromosome inactivation marks and progress to a skewed methylation pattern during culture. Mol Hum Reprod. 2016;22(4):285–98. 10.1093/molehr/gaw004 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Lengner CJ, Gimelbrant AA, Erwin JA, et al. : Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141(5):872–83. 10.1016/j.cell.2010.04.010 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 15. Liu W, Yin Y, Jiang Y, et al. : Genetic and epigenetic X-chromosome variations in a parthenogenetic human embryonic stem cell line. J Assist Reprod Genet. 2011;28(4):303–13. 10.1007/s10815-010-9517-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miyagoe-Suzuki Y, Nishiyama T, Nakamura M, et al. : Induction of Pluripotent Stem Cells from a Manifesting Carrier of Duchenne Muscular Dystrophy and Characterization of Their X-Inactivation Status. Stem Cells Int. 2017;2017: 7906843. 10.1155/2017/7906843 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 17. Osafune K, Caron L, Borowiak M, et al. : Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26(3):313–5. 10.1038/nbt1383 [DOI] [PubMed] [Google Scholar]
- 18. Davidson KC, Mason EA, Pera MF: The pluripotent state in mouse and human. Development. 2015;142(18):3090–9. 10.1242/dev.116061 [DOI] [PubMed] [Google Scholar]
- 19. Guo G, von Meyenn F, Santos F, et al. : Naive Pluripotent Stem Cells Derived Directly from Isolated Cells of the Human Inner Cell Mass. Stem Cell Reports. 2016;6(4):437–46. 10.1016/j.stemcr.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 20. Guo G, von Meyenn F, Rostovskaya M, et al. : Epigenetic resetting of human pluripotency. Development. 2017;144(15):2748–63. 10.1242/dev.146811 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Takashima Y, Guo G, Loos R, et al. : Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158(6):1254–69. 10.1016/j.cell.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gafni O, Weinberger L, Mansour AA, et al. : Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504(7479):282–6. 10.1038/nature12745 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 23. Irie N, Weinberger L, Tang WW, et al. : SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160(1–2):253–68. 10.1016/j.cell.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 24. Pastor WA, Chen D, Liu W, et al. : Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory. Cell Stem Cell. 2016;18(3):323–9. 10.1016/j.stem.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Hough SR, Thornton M, Mason E, et al. : Single-cell gene expression profiles define self-renewing, pluripotent, and lineage primed states of human pluripotent stem cells. Stem Cell Reports. 2014;2(6):881–95. 10.1016/j.stemcr.2014.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith A: Formative pluripotency: the executive phase in a developmental continuum. Development. 2017;144(3):365–73. 10.1242/dev.142679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kalkan T, Olova N, Roode M, et al. : Tracking the embryonic stem cell transition from ground state pluripotency. Development. 2017;144(7):1221–1234. 10.1242/dev.142711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shi Y, Kirwan P, Smith J, et al. : Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15(3):477–86, S1. 10.1038/nn.3041 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 29. Maury Y, Côme J, Piskorowski RA, et al. : Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat Biotechnol. 2015;33(1):89–96. 10.1038/nbt.3049 [DOI] [PubMed] [Google Scholar]
- 30. Cajánek L, Ribeiro D, Liste I, et al. : Wnt/beta-catenin signaling blockade promotes neuronal induction and dopaminergic differentiation in embryonic stem cells. Stem Cells. 2009;27(12):2917–27. 10.1002/stem.210 [DOI] [PubMed] [Google Scholar]
- 31. Douvaras P, Fossati V: Generation and isolation of oligodendrocyte progenitor cells from human pluripotent stem cells. Nat Protoc. 2015;10(8):1143–54. 10.1038/nprot.2015.075 [DOI] [PubMed] [Google Scholar]
- 32. Shaltouki A, Peng J, Liu Q, et al. : Efficient generation of astrocytes from human pluripotent stem cells in defined conditions. Stem Cells. 2013;31(5):941–52. 10.1002/stem.1334 [DOI] [PubMed] [Google Scholar]
- 33. Laflamme MA, Chen KY, Naumova AV, et al. : Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25(9):1015–24. 10.1038/nbt1327 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 34. Mummery C, Ward-van Oostwaard D, Doevendans P, et al. : Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107(21):2733–40. 10.1161/01.CIR.0000068356.38592.68 [DOI] [PubMed] [Google Scholar]
- 35. Kehat I, Kenyagin-Karsenti D, Snir M, et al. : Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108(3):407–14. 10.1172/JCI12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Burridge PW, Matsa E, Shukla P, et al. : Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11(8):855–60. 10.1038/nmeth.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roelandt P, Vanhove J, Verfaillie C: Directed differentiation of pluripotent stem cells to functional hepatocytes. Methods Mol Biol. 2013;997:141–7. 10.1007/978-1-62703-348-0_11 [DOI] [PubMed] [Google Scholar]
- 38. Schwartz RE, Linehan JL, Painschab MS, et al. : Defined conditions for development of functional hepatic cells from human embryonic stem cells. Stem Cells Dev. 2005;14(6):643–55. 10.1089/scd.2005.14.643 [DOI] [PubMed] [Google Scholar]
- 39. Basma H, Soto-Gutiérrez A, Yannam GR, et al. : Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology. 2009;136(3):990–9. 10.1053/j.gastro.2008.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kumar P, Tan Y, Cahan P: Understanding development and stem cells using single cell-based analyses of gene expression. Development. 2017;144(1):17–32. 10.1242/dev.133058 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 41. Wen L, Tang F: Single-cell sequencing in stem cell biology. Genome Biol. 2016;17:71. 10.1186/s13059-016-0941-0 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 42. Poulin J, Tasic B, Hjerling-Leffler J, et al. : Disentangling neural cell diversity using single-cell transcriptomics. Nat Neurosci. 2016;19:1131–41. 10.1038/nn.4366 [DOI] [PubMed] [Google Scholar]
- 43. Godoy P, Schmidt-Heck W, Natarajan K, et al. : Gene networks and transcription factor motifs defining the differentiation of stem cells into hepatocyte-like cells. J Hepatol. 2015;63(4):934–42. 10.1016/j.jhep.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Engel M, Do-Ha D, Muñoz SS, et al. : Common pitfalls of stem cell differentiation: a guide to improving protocols for neurodegenerative disease models and research. Cell Mol Life Sci. 2016;73(19):3693–709. 10.1007/s00018-016-2265-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bramanti V, Tomassoni D, Avitabile M, et al. : Biomarkers of glial cell proliferation and differentiation in culture. Front Biosci (Schol Ed). 2010;2:558–70. 10.2741/s85 [DOI] [PubMed] [Google Scholar]
- 46. Krencik R, Weick JP, Liu Y, et al. : Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat Biotechnol. 2011;29(6):528–34. 10.1038/nbt.1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Serio A, Bilican B, Barmada SJ, et al. : Astrocyte pathology and the absence of non-cell autonomy in an induced pluripotent stem cell model of TDP-43 proteinopathy. Proc Natl Acad Sci U S A. 2013;110(12):4697–702. 10.1073/pnas.1300398110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roybon L, Lamas NJ, Garcia AD, et al. : Human stem cell-derived spinal cord astrocytes with defined mature or reactive phenotypes. Cell Rep. 2013;4(5):1035–48. 10.1016/j.celrep.2013.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Watt FM, Hogan BL: Out of Eden: stem cells and their niches. Science. 2000;287(5457):1427–30. 10.1126/science.287.5457.1427 [DOI] [PubMed] [Google Scholar]
- 50. Park SY, Park J, Sim SH, et al. : Enhanced differentiation of human neural stem cells into neurons on graphene. Adv Mater. 2011;23(36):H263–7. 10.1002/adma.201101503 [DOI] [PubMed] [Google Scholar]
- 51. Eng G, Lee BW, Protas L, et al. : Autonomous beating rate adaptation in human stem cell-derived cardiomyocytes. Nat Commun. 2016;7: 10312. 10.1038/ncomms10312 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 52. Adamo L, Naveiras O, Wenzel PL, et al. : Biomechanical forces promote embryonic haematopoiesis. Nature. 2009;459(7250):1131–5. 10.1038/nature08073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kietzmann T: Metabolic zonation of the liver: The oxygen gradient revisited. Redox Biol. 2017;11:622–30. 10.1016/j.redox.2017.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 54. Yamaguchi M, Seki T, Imayoshi I, et al. : Neural stem cells and neuro/gliogenesis in the central nervous system: understanding the structural and functional plasticity of the developing, mature, and diseased brain. J Physiol Sci. 2016;66(3):197–206. 10.1007/s12576-015-0421-4 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Vanhove J, Pistoni M, Welters M, et al. : H3K27me3 Does Not Orchestrate the Expression of Lineage-Specific Markers in hESC-Derived Hepatocytes In Vitro. Stem Cell Reports. 2016;7(2):192–206. 10.1016/j.stemcr.2016.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Baxter M, Withey S, Harrison S, et al. : Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes. J Hepatol. 2015;62(3):581–9. 10.1016/j.jhep.2014.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lang J, Vera D, Cheng Y, et al. : Modeling Dengue Virus-Hepatic Cell Interactions Using Human Pluripotent Stem Cell-Derived Hepatocyte-like Cells. Stem Cell Reports. 2016;7(3):341–54. 10.1016/j.stemcr.2016.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 58. Xia Y, Carpentier A, Cheng X, et al. : Human stem cell-derived hepatocytes as a model for hepatitis B virus infection, spreading and virus-host interactions. J Hepatol. 2017;66(3):494–503. 10.1016/j.jhep.2016.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 59. Shlomai A, Schwartz RE, Ramanan V, et al. : Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc Natl Acad Sci U S A. 2014;111(33):12193–8. 10.1073/pnas.1412631111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Roelandt P, Obeid S, Paeshuyse J, et al. : Human pluripotent stem cell-derived hepatocytes support complete replication of hepatitis C virus. J Hepatol. 2012;57(2):246–51. 10.1016/j.jhep.2012.03.030 [DOI] [PubMed] [Google Scholar]
- 61. Kordower JH, Chu Y, Hauser RA, et al. : Lewy body-like pathology in long-term embryonic nigral transplants in Parkinson's disease. Nat Med. 2008;14(5):504–6. 10.1038/nm1747 [DOI] [PubMed] [Google Scholar]
- 62. Olanow CW, Gracies JM, Goetz CG, et al. : Clinical pattern and risk factors for dyskinesias following fetal nigral transplantation in Parkinson's disease: a double blind video-based analysis. Mov Disord. 2009;24(3):336–43. 10.1002/mds.22208 [DOI] [PubMed] [Google Scholar]
- 63. Barker RA, Studer L, Cattaneo E, et al. : G-Force PD: a global initiative in coordinating stem cell-based dopamine treatments for Parkinson's disease. NPJ Parkinsons Dis. 2015;1: 15017. 10.1038/npjparkd.2015.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Roy NS, Cleren C, Singh SK, et al. : Functional engraftment of human ES cell-derived dopaminergic neurons enriched by coculture with telomerase-immortalized midbrain astrocytes. Nat Med. 2006;12(11):1259–68. 10.1038/nm1495 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 65. Lee AS, Tang C, Rao MS, et al. : Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med. 2013;19(8):998–1004. 10.1038/nm.3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Semrau S, Goldmann JE, Soumillon M, et al. : Dynamics of lineage commitment revealed by single-cell transcriptomics of differentiating embryonic stem cells. Nat Commun. 2017;8(1): 1096. 10.1038/s41467-017-01076-4 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 67. Collombet S, van Oevelen C, Sardina Ortega JL, et al. : Logical modeling of lymphoid and myeloid cell specification and transdifferentiation. Proc Natl Acad Sci U S A. 2017;114(23):5792–9. 10.1073/pnas.1610622114 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 68. Smith ZD, Sindhu C, Meissner A: Molecular features of cellular reprogramming and development. Nat Rev Mol Cell Biol. 2016;17(3):139–54. 10.1038/nrm.2016.6 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Dulken BW, Leeman DS, Boutet SC, et al. : Single-Cell Transcriptomic Analysis Defines Heterogeneity and Transcriptional Dynamics in the Adult Neural Stem Cell Lineage. Cell Rep. 2017;18(3):777–90. 10.1016/j.celrep.2016.12.060 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 70. Yang L, Wang WH, Qiu WL, et al. : A single-cell transcriptomic analysis reveals precise pathways and regulatory mechanisms underlying hepatoblast differentiation. Hepatology. 2017;66(5):1387–401. 10.1002/hep.29353 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 71. Dominguez AA, Lim WA, Qi LS: Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol. 2016;17(1):5–15. 10.1038/nrm.2015.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Maeder ML, Linder SJ, Cascio VM, et al. : CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10(10):977–9. 10.1038/nmeth.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chavez A, Scheiman J, Vora S, et al. : Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015;12(4):326–8. 10.1038/nmeth.3312 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. Gilbert LA, Larson MH, Morsut L, et al. : CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–51. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 75. Ordovás L, Boon R, Pistoni M, et al. : Efficient Recombinase-Mediated Cassette Exchange in hPSCs to Study the Hepatocyte Lineage Reveals AAVS1 Locus-Mediated Transgene Inhibition. Stem Cell Reports. 2015;5(5):918–31. 10.1016/j.stemcr.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Esvelt KM, Mali P, Braff JL, et al. : Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10(11):1116–21. 10.1038/nmeth.2681 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 77. Shalem O, Sanjana NE, Zhang F: High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16(5):299–311. 10.1038/nrg3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Larson MH, Gilbert LA, Wang X, et al. : CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8(11):2180–96. 10.1038/nprot.2013.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Konermann S, Brigham MD, Trevino AE, et al. : Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583–8. 10.1038/nature14136 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 80. Gilbert LA, Horlbeck MA, Adamson B, et al. : Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159(3):647–61. 10.1016/j.cell.2014.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 81. Mandegar MA, Huebsch N, Frolov EB, et al. : CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell. 2016;18(4):541–53. 10.1016/j.stem.2016.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 82. Pagliuca FW, Millman JR, Gürtler M, et al. : Generation of functional human pancreatic β cells in vitro. Cell. 2014;159(2):428–39. 10.1016/j.cell.2014.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 83. Siller R, Greenhough S, Naumovska E, et al. : Small-molecule-driven hepatocyte differentiation of human pluripotent stem cells. Stem Cell Reports. 2015;4(5):939–52. 10.1016/j.stemcr.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Charbord J, Poydenot P, Bonnefond C, et al. : High throughput screening for inhibitors of REST in neural derivatives of human embryonic stem cells reveals a chemical compound that promotes expression of neuronal genes. Stem Cells. 2013;31(9):1816–28. 10.1002/stem.1430 [DOI] [PubMed] [Google Scholar]
- 85. Mathieu J, Ruohola-Baker H: Metabolic remodeling during the loss and acquisition of pluripotency. Development. 2017;144(4):541–51. 10.1242/dev.128389 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 86. Varum S, Rodrigues AS, Moura MB, et al. : Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One. 2011;6(6):e20914. 10.1371/journal.pone.0020914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Folmes CD, Nelson TJ, Martinez-Fernandez A, et al. : Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14(2):264–71. 10.1016/j.cmet.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wong-Riley MT: Cytochrome oxidase: an endogenous metabolic marker for neuronal activity. Trends Neurosci. 1989;12(3):94–101. 10.1016/0166-2236(89)90165-3 [DOI] [PubMed] [Google Scholar]
- 89. Lopaschuk GD, Jaswal JS: Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J Cardiovasc Pharmacol. 2010;56(2):130–40. 10.1097/FJC.0b013e3181e74a14 [DOI] [PubMed] [Google Scholar]
- 90. Rui L: Energy metabolism in the liver. Compr Physiol. 2014;4(1):177–97. 10.1002/cphy.c130024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Monterey MD, Szerlip NJ, Mathupala SP: Low-cost media formulation for culture of brain tumor spheroids (neurospheres). Biotechniques. 2013;55(2):83–8. 10.2144/000114066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shiraki N, Shiraki Y, Tsuyama T, et al. : Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014;19(5):780–94. 10.1016/j.cmet.2014.03.017 [DOI] [PubMed] [Google Scholar]
- 93. Heslop JA, Rowe C, Walsh J, et al. : Mechanistic evaluation of primary human hepatocyte culture using global proteomic analysis reveals a selective dedifferentiation profile. Arch Toxicol. 2017;91(1):439–52. 10.1007/s00204-016-1694-y [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 94. Lancaster MA, Knoblich JA: Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9(10):2329–40. 10.1038/nprot.2014.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Koui Y, Kido T, Ito T, et al. : An In Vitro Human Liver Model by iPSC-Derived Parenchymal and Non-parenchymal Cells. Stem Cell Reports. 2017;9(2):490–8. 10.1016/j.stemcr.2017.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 96. Takebe T, Sekine K, Enomura M, et al. : Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499(7459):481–4. 10.1038/nature12271 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 97. Yang C, Tibbitt MW, Basta L, et al. : Mechanical memory and dosing influence stem cell fate. Nat Mater. 2014;13(6):645–52. 10.1038/nmat3889 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 98. Sun Y, Yong KM, Villa-Diaz LG, et al. : Hippo/YAP-mediated rigidity-dependent motor neuron differentiation of human pluripotent stem cells. Nat Mater. 2014;13(6):599–604. 10.1038/nmat3945 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 99. Engler AJ, Sen S, Sweeney HL, et al. : Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–89. 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 100. Yeung T, Georges PC, Flanagan LA, et al. : Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil Cytoskeleton. 2005;60(1):24–34. 10.1002/cm.20041 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 101. Sala A, Hänseler P, Ranga A, et al. : Engineering 3D cell instructive microenvironments by rational assembly of artificial extracellular matrices and cell patterning. Integr Biol (Camb). 2011;3(11):1102–11. 10.1039/c1ib00045d [DOI] [PubMed] [Google Scholar]
- 102. Cerqueira MT, da Silva LP, Santos TC, et al. : Gellan gum-hyaluronic acid spongy-like hydrogels and cells from adipose tissue synergize promoting neoskin vascularization. ACS Appl Mater Interfaces. 2014;6(22):19668–79. 10.1021/am504520j [DOI] [PubMed] [Google Scholar]
- 103. Caiazzo M, Okawa Y, Ranga A, et al. : Defined three-dimensional microenvironments boost induction of pluripotency. Nat Mater. 2016;15(3):344–52. 10.1038/nmat4536 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 104. Ishihara K, Ranga A, Lutolf MP, et al. : Reconstitution of a Patterned Neural Tube from Single Mouse Embryonic Stem Cells. Methods Mol Biol. 2017;1597:43–55. 10.1007/978-1-4939-6949-4_4 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 105. Domingos M, Dinucci D, Cometa S, et al. : Polycaprolactone Scaffolds Fabricated via Bioextrusion for Tissue Engineering Applications. Int J Biomater. 2009;2009: 239643. 10.1155/2009/239643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gao G, Yonezawa T, Hubbell K, et al. : Inkjet-bioprinted acrylated peptides and PEG hydrogel with human mesenchymal stem cells promote robust bone and cartilage formation with minimal printhead clogging. Biotechnol J. 2015;10(10):1568–77. 10.1002/biot.201400635 [DOI] [PubMed] [Google Scholar]
- 107. Blaeser A, Duarte Campos DF, Puster U, et al. : Controlling Shear Stress in 3D Bioprinting is a Key Factor to Balance Printing Resolution and Stem Cell Integrity. Adv Healthc Mater. 2016;5(3):326–33. 10.1002/adhm.201500677 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 108. Faulkner-Jones A, Fyfe C, Cornelissen DJ, et al. : Bioprinting of human pluripotent stem cells and their directed differentiation into hepatocyte-like cells for the generation of mini-livers in 3D. Biofabrication. 2015;7(4):044102. 10.1088/1758-5090/7/4/044102 [DOI] [PubMed] [Google Scholar]
- 109. Gaebel R, Ma N, Liu J, et al. : Patterning human stem cells and endothelial cells with laser printing for cardiac regeneration. Biomaterials. 2011;32(35):9218–30. 10.1016/j.biomaterials.2011.08.071 [DOI] [PubMed] [Google Scholar]
- 110. Guillotin B, Souquet A, Catros S, et al. : Laser assisted bioprinting of engineered tissue with high cell density and microscale organization. Biomaterials. 2010;31(28):7250–6. 10.1016/j.biomaterials.2010.05.055 [DOI] [PubMed] [Google Scholar]
- 111. Carson D, Hnilova M, Yang X, et al. : Nanotopography-Induced Structural Anisotropy and Sarcomere Development in Human Cardiomyocytes Derived from Induced Pluripotent Stem Cells. ACS Appl Mater Interfaces. 2016;8(34):21923–32. 10.1021/acsami.5b11671 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 112. Atmanli A, Domian IJ: Recreating the Cardiac Microenvironment in Pluripotent Stem Cell Models of Human Physiology and Disease. Trends Cell Biol. 2017;27(5):352–64. 10.1016/j.tcb.2016.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 113. Kshitiz, Park J, Kim P, et al. : Control of stem cell fate and function by engineering physical microenvironments. Integr Biol (Camb). 2012;4(9):1008–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Reimer A, Vasilevich A, Hulshof F, et al. : Scalable topographies to support proliferation and Oct4 expression by human induced pluripotent stem cells. Sci Rep. 2016;6: 18948. 10.1038/srep18948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Discher DE, Janmey P, Wang YL: Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–43. 10.1126/science.1116995 [DOI] [PubMed] [Google Scholar]
- 116. Balikov DA, Fang B, Chun YW, et al. : Directing lineage specification of human mesenchymal stem cells by decoupling electrical stimulation and physical patterning on unmodified graphene. Nanoscale. 2016;8(28):13730–9. 10.1039/c6nr04400j [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 117. Wagenaar DA, Pine J, Potter SM: Searching for plasticity in dissociated cortical cultures on multi-electrode arrays. J Negat Results Biomed. 2006;5:16. 10.1186/1477-5751-5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bologna LL, Nieus T, Tedesco M, et al. : Low-frequency stimulation enhances burst activity in cortical cultures during development. Neuroscience. 2010;165(3):692–704. 10.1016/j.neuroscience.2009.11.018 [DOI] [PubMed] [Google Scholar]
- 119. Denning C, Borgdorff V, Crutchley J, et al. : Cardiomyocytes from human pluripotent stem cells: From laboratory curiosity to industrial biomedical platform. Biochim Biophys Acta. 2016;1863(7 Pt B):1728–48. 10.1016/j.bbamcr.2015.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 120. Ruan JL, Tulloch NL, Saiget M, et al. : Mechanical Stress Promotes Maturation of Human Myocardium From Pluripotent Stem Cell-Derived Progenitors. Stem Cells. 2015;33(7):2148–57. 10.1002/stem.2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Michael S, Sorg H, Peck CT, et al. : Tissue engineered skin substitutes created by laser-assisted bioprinting form skin-like structures in the dorsal skin fold chamber in mice. PLoS One. 2013;8(3):e57741. 10.1371/journal.pone.0057741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. van Duinen V, Trietsch SJ, Joore J, et al. : Microfluidic 3D cell culture: from tools to tissue models. Curr Opin Biotechnol. 2015;35:118–26. 10.1016/j.copbio.2015.05.002 [DOI] [PubMed] [Google Scholar]
- 123. Giobbe GG, Michielin F, Luni C, et al. : Functional differentiation of human pluripotent stem cells on a chip. Nat Methods. 2015;12(7):637–40. 10.1038/nmeth.3411 [DOI] [PubMed] [Google Scholar]