ABSTRACT
The human MOF (hMOF) protein belongs to the MYST family of histone acetyltransferases and plays a critical role in transcription and the DNA damage response. MOF is essential for cell proliferation; however, its role during replication and replicative stress is unknown. Here we demonstrate that cells depleted of MOF and under replicative stress induced by cisplatin, hydroxyurea, or camptothecin have reduced survival, a higher frequency of S-phase-specific chromosome damage, and increased R-loop formation. MOF depletion decreased replication fork speed and, when combined with replicative stress, also increased stalled replication forks as well as new origin firing. MOF interacted with PCNA, a key coordinator of replication and repair machinery at replication forks, and affected its ubiquitination and recruitment to the DNA damage site. Depletion of MOF, therefore, compromised the DNA damage repair response as evidenced by decreased Mre11, RPA70, Rad51, and PCNA focus formation, reduced DNA end resection, and decreased CHK1 phosphorylation in cells after exposure to hydroxyurea or cisplatin. These results support the argument that MOF plays an important role in suppressing replication stress induced by genotoxic agents at several stages during the DNA damage response.
KEYWORDS: MOF, replication stress, R loop, homologous recombination, PCNA
INTRODUCTION
DNA replication is a fundamental cell process that ensures accurate duplication of the genetic material and subsequent transfer to daughter cells. Intra- as well as extracellular factors constantly challenge the integrity of the replicating DNA, which can result in replication stress. Replication stress is a complex phenomenon that can interfere with the progression, stability, and proper recovery of DNA replication forks after stalling (1, 2). Several factors that can lead to replication stress have been identified, including DNA polymerase stalling, DNA fragile sites, topoisomerase trapping, R-loop formation, oncogenic stress, or DNA secondary structures such as G4 quadruplexes, etc. If these obstacles are not properly resolved, they can result in chromosomal instability and oncogene activation leading to carcinogenesis (3).
DNA replication, as well as the cellular response to replication stress, occurs in the context of the chromatin protein-DNA complex. During S phase of the cell cycle, the chromatin landscape undergoes dramatic changes as the entire genome is duplicated (4), and any failure to maintain chromatin structural integrity and organization can promote replication stress (5, 6). Deregulation of DNA replication, including replication fork stalling and fork collapse, induces DNA damage due to formation of single-stranded DNA (ssDNA) and ultimately double-strand breaks (DSBs) at stalled or collapsed forks, respectively (7, 8). Replicative stress, therefore, can trigger the cellular DNA damage response (DDR), and this response is tightly regulated by the changes in the surrounding chromatin environment.
The accessibility to DNA during various DNA metabolic processes, such as replication, transcription, and the DSB damage response, is tightly regulated by chromatin modifications and chromatin-remodeling factors (9). Chromatin-remodeling factors such as INO80, SMARCAD1, and ATRX also play important roles in stabilization of the replication fork and fork progression (10–13). Histone acetylation is a particularly important posttranslational modification that regulates chromatin folding and compactness, which are crucial for numerous DNA transactions. After completion of DNA repair, modification and reincorporation of histones are necessary to reestablish the functional chromatin environment essential for replication fork progression and stability. Defects in acetylated histone H3 lysine 56 (H3K56ac)-mediated chromatin reassembly in yeast, for example, cause replication fork collapse, hyperrecombination, and large chromosomal rearrangements (14–16). The acetylation of H4K16 (H4K16ac) is also critical for DNA damage repair, as well as gene activation (17, 18), through its influence on both chromatin structure and internucleosome interactions mediated by the histone H4 tail (19–21). Reduced H4K16ac levels have been observed in some cancer cells (22).
Human MOF (hMOF) (males absent on the first), encoded by the KAT8 gene, is a member of the MYST family of histone acetyltransferase (HATs) and specifically acetylates histone H4 at lysine 16 (H4K16ac). MOF is an evolutionarily conserved protein that is associated with transcription regulation, the DNA damage response, and other critical cellular functions (23–26). MOF-mediated H4K16 acetylation alters chromatin-protein interactions as well as the local chromatin structure and impacts DNA transcription and repair (19, 27, 28). Apart from its histone acetylation function, MOF can also interact with and modulate the function of proteins involved in the DNA damage response, such as ATM, 53BP1, etc. (25, 29). Through its interactions with 53BP1, MOF is a key regulator of the pathway choice between homologous recombination (HR) and nonhomologous end joining (NHEJ) for DSB repair (25). In addition, MOF is essential for the survival of postmitotic cells (24, 30, 31). Given the role of MOF and H4K16ac in a wide range of critical cellular processes, MOF silencing leads to multiple defects, including altered gene expression, defective DNA damage repair, genomic instability, embryonic lethality, and defective embryonic stem cell differentiation (23, 29, 32, 33).
In this study, we characterized the role of MOF during replicative stress. Taken together, our results indicate novel functions for MOF in modulating the DNA damage response induced by replication stress and the resolution of stalled DNA replication forks.
RESULTS
Replicative stress-induced cell death is increased by MOF depletion.
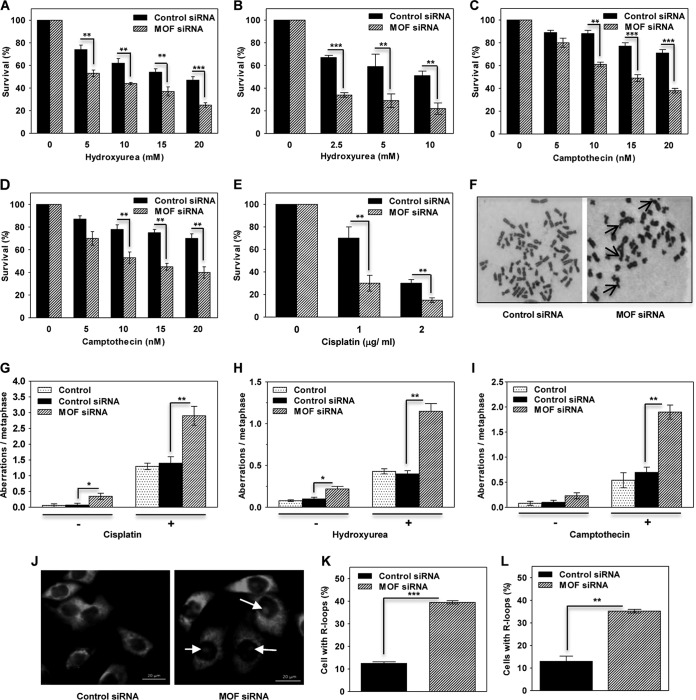
To evaluate the role of MOF in the replication stress response, we examined whether MOF depletion decreased clonogenic cell survival after treatment with three different replicative stress-inducing agents: cisplatin, hydroxyurea (HU), and camptothecin (CPT). Cellular treatment with cisplatin induces DNA inter- as well as intrastrand cross-links, HU induces replication fork stalling by depleting the nucleotide pool available for the DNA polymerases, and camptothecin is a well-known inhibitor of human topoisomerase I. Depletion of MOF with small interfering RNA (siRNA), in either HeLa or U2OS cells, significantly reduced survival of cells after treatment with any of the three aforementioned genotoxic agents (Fig. 1A to E). Increased sensitivity to genotoxic agents is often the result of defective DNA repair and is reflected in an increase in chromosomal aberrations observed at metaphase (34). Analysis of chromosomal aberrations at metaphase in HeLa cells, with or without treatment with either cisplatin, HU, or CPT, indicated that MOF depletion significantly increased aberration levels (Fig. 1F to I). This suggests that MOF functionally contributes to genomic stability in replicating nonstressed cells as well as in cells undergoing replicative stress.
FIG 1.
Cells depleted of MOF are sensitive to replication stress induced by cisplatin, hydroxyurea, or camptothecin. (A and B) Clonogenic survival of MOF-depleted HeLa (A) and U2OS (B) cells treated with the indicated concentrations of HU. (C and D) Clonogenic survival of MOF-depleted HeLa (C) and U2OS (D) cells treated with indicated concentrations of CPT. (E) Clonogenic survival of MOF-depleted HeLa cells treated with the indicated concentrations of cisplatin. The experiments were performed in triplicate, and the error bars represent the standard deviation from three independent experiments. P values: **, <0.01; ***, <0.001. (F) Representative images of metaphase spread of cells with control siRNA (untreated) and MOF-depleted (cisplatin-treated) HeLa cells showing various kinds of chromosomal aberrations. Arrows indicate aberrations. (G to I) Bar graphs show the chromosomal aberrations in control and MOF-depleted HeLa cells treated with either 50 μM cisplatin for 1 h (G), 2 mM HU for 24 h (H), or 250 nM CPT for 6 h (I). “Control” refers to cells that were not transfected with siRNA. The error bars represent the standard deviation from three independent experiments. P values: *, <0.05; **, <0.01. (J) Representative images of cells with R loops in control and MOF-depleted cells. Arrows indicate endogenous R loops detected by S9.6 antibody staining, which specifically detects DNA-RNA hybrids. (K and L) Bar graphs show the quantification of the percentages of cells showing endogenous R loops in control and MOF-depleted HeLa (K) and U2OS (L) cells. The error bars represent the standard deviation from three independent experiments. P values: **, <0.01; ***, <0.001.
MOF is critical for transcription, and RNA transcripts can form R loops, three-stranded nucleic acid structures composed of a DNA-RNA hybrid and the associated nontemplate single-stranded DNA (ssDNA), which are prone to DNA damage. Formation of R loops is also favored by collision of replication forks with transcribing DNA (35, 36). In both MOF-depleted HeLa and U2OS cells, we observed an approximately 3-fold-higher level of cells with endogenous R loops than in control cells (Fig. 1J to L). Taken together, the results from the clonogenic survival assay, chromosomal aberration analysis, and R-loop accumulation assay suggest that MOF plays a role in the repair of DNA damage caused by replication stress, both endogenous and in response to stress-inducing agents, in order to maintain genomic stability.
MOF depletion impairs DNA replication fork progression.
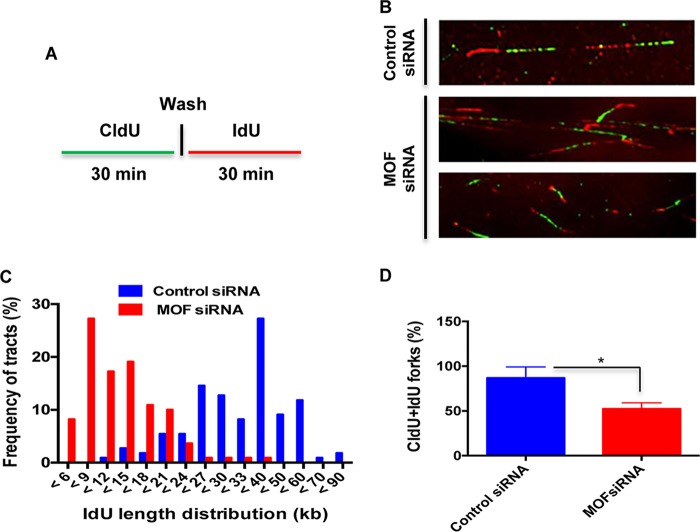
To further determine whether MOF affects DNA replication in cells under normal growth or replicative stress conditions, we employed a DNA fiber assay to evaluate replication fork dynamics at single-molecule resolution. Cells were pulse-labeled with two different thymidine analogs: a 5-chlorodeoxyuridine (CldU) pulse to identify cells with ongoing DNA synthesis, followed by 5-iododeoxyuridine (IdU) pulse-labeling. Individual DNA fibers, which incorporated the CldU and IdU pulses, were detected with fluorescent antibodies against those analogs. Measurement of the IdU tract (red in Fig. 2A) length distribution in DNA fibers with contiguous CldU labeling indicated much shorter lengths in MOF-depleted cells than in control cells (Fig. 2B and C). The frequency of DNA fibers with both CldU and IdU labels at the fork was also decreased in MOF-depleted cells (Fig. 2D). This suggests that nonstressed MOF-depleted cells have defective replication fork progression, which may also account for the slower growth of MOF-depleted cells (23, 37).
FIG 2.
Replication fork progression is impaired in MOF-depleted cells. (A) Scheme of the single DNA fiber analysis. The control and MOF-depleted HeLa cells were labeled with CldU for 30 min, rinsed, and then labeled with IdU for 30 min before being subjected to DNA fiber analysis. (B) Representative images of DNA fiber tracts in control and MOF-depleted cells. The CldU and IdU tracts were visualized in green and red, respectively. (C) Graph representing the frequency of IdU tract length distributions in control and MOF-depleted cells. (D) Bar graph showing the frequency of DNA fiber tracts with contiguous CldU and IdU labels at the replication fork. The experiments were performed three or four times, and for each time approximately 300 fibers were counted. Error bars represent the standard deviation from three independent experiments, with a P value of <0.05 (*) suggesting that the data have statistical significance.
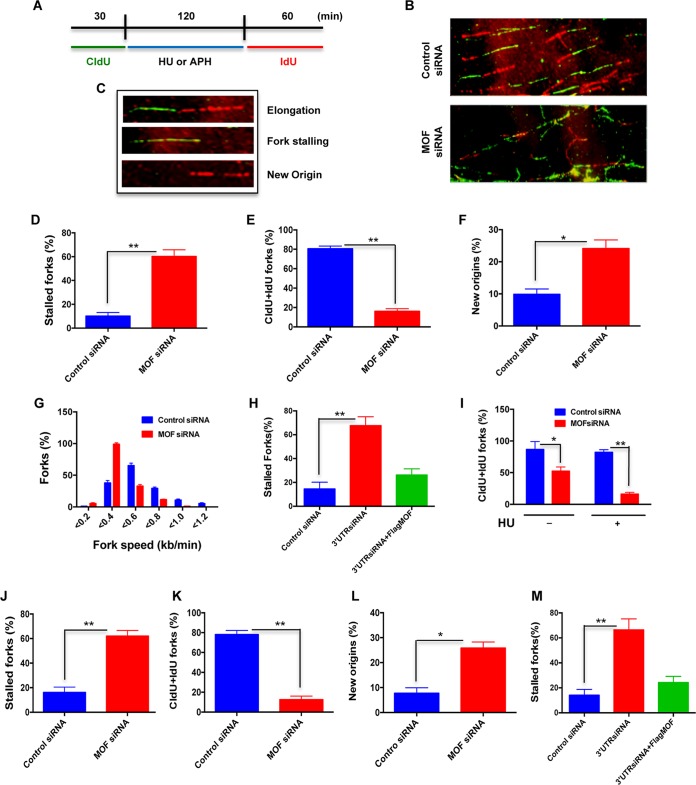
Replication forks encounter numerous DNA template obstacles, which can lead to fork stalling or fork collapse that results in DNA double-strand breaks (DSBs). To understand the role of MOF in replication fork dynamics following transient genotoxic stress-induced replication blockage, we analyzed the restart/recovery of replication forks in MOF-depleted cells after HU or aphidicolin (APH) treatment using the DNA fiber analysis (Fig. 3A and B). We observed primarily three different types of DNA fiber tracts, i.e., those with continuing elongation forks, stalled forks, and fibers in which a new DNA replication origin had fired (Fig. 3C). In cells treated with HU, depletion of MOF enhanced the frequencies of both stalled forks and new origins by approximately 4-fold and 2.5-fold, respectively, in comparison to those in HU-treated control cells (Fig. 3D and F), while replication tracts with CldU and IdU colabeling were significantly reduced (Fig. 3E). A significant decrease in the fork speed was also observed in MOF-depleted cells (Fig. 3G). Fork stalling was rescued by ectopic MOF expression in cells depleted of endogenous MOF (Fig. 3H). A comparison of the frequency of tracts with both CldU- and IdU-labeled forks in MOF-depleted cells showed that the frequency, which was decreased in MOF-depleted cells, was further reduced upon HU treatment (Fig. 3I).
FIG 3.
Depletion of MOF decreases replication restart after replicative stress. (A) Scheme for dual labeling of DNA fibers to evaluate replication restart or recovery following HU- or APH-induced replication fork stalling. (B) Representative images of DNA fiber tracts in control and MOF-depleted cells after HU treatment. CldU tracts were visualized in green and IdU was visualized in red in control and MOF-depleted cells. (C) Examples of DNA fiber tract types representing elongation, stalled fork, and new origins. (D to G) Quantitative analysis of the DNA fiber replication restart assay after HU treatment. Bar graphs show the quantification of the percentage of stalled forks, total tracts with both CldU and IdU labels at the fork (CldU+IdU), percentage of new origins, and fork speed in control and MOF-depleted cells in panels D to G, respectively. The experiment was performed three times, and for each time point about 300 fibers were counted. Error bars represent the standard deviation from three independent experiments. P values are <0.05 (*) and <0.01 (**), showing that the values are statistically significant. (H) Quantitative analysis of replication restart after HU treatment in HeLa cells transfected with siRNA specific to the 3′ untranslated region (UTR) of the mof gene and ectopic expression of FLAG-MOF in these cells. (I) Bar graph showing the frequency of replication tracts with both CldU and IdU labels in the absence or presence of HU treatment in control and MOF-depleted cells. (J to L) Quantitative analysis of the DNA fiber assay following APH treatment. Bar graphs show the measurement of percentage of stalled forks, total tracts with both CldU and IdU labels at the fork (CldU+IdU), and percentage of new origins in control and MOF-depleted cells in panels J to L, respectively. (M) Quantitative analysis of the replication restart assay after APH treatment in cells transfected with siRNA specific to the 3′ UTR of the mof gene followed by ectopic FLAG-MOF expression.
In order to eliminate the possibility of an HU-specific effect, we tested the response of MOF-depleted cells to another replication stress-inducing agent. Aphidicolin (APH) is a DNA polymerase α inhibitor that blocks DNA replication, resulting in replication fork stalling. As with HU-induced replicative stress in MOF-depleted cells, APH treatment of MOF-depleted cells increased fork stalling and new origin firing (Fig. 3J to L), and this effect was rescued by ectopic MOF expression (Fig. 3M). Together, the DNA fiber analysis results demonstrate that MOF is an important factor in replication fork recovery once fork movement is blocked.
MOF depletion delays S-phase progression.
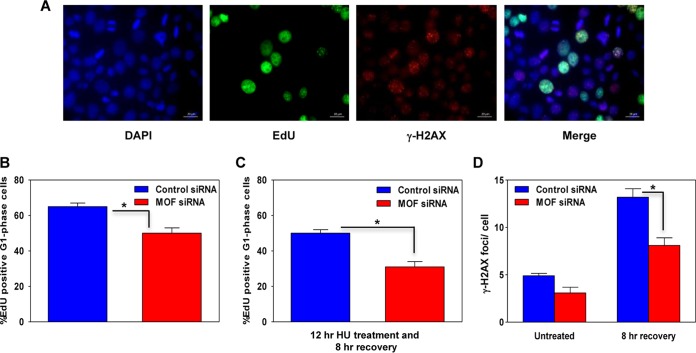
The DNA fiber analysis suggested that MOF-depleted cells would arrest in S/G2 phase. To determine the impact of stalled replication forks on S-phase progression, we employed a 5′-ethynyl-2′deoxyuridine (EdU) labeling/staining technique. The thymidine analog EdU is readily incorporated into nascent DNA during S-phase DNA replication and can subsequently be detected by Cu(I)-catalyzed azide coupling to a fluorescent label (Fig. 4A). An initial flow cytometric analysis based solely on DNA content indicated that under normal growth conditions, the S-phase cell fraction increased slightly after MOF depletion, while the corresponding G1-phase cell fraction declined (data not shown). When we specifically measured the movement of EdU-labeled S/G2-phase cells into G1, a delay in S-phase transit was detected in MOF-depleted cells (Fig. 4A and B).
FIG 4.
Depletion of MOF delays S-phase progression. (A) Representative images of control and MOF-depleted HeLa cells coimmunostained for EdU (green) and γ-H2AX (red), with DAPI-stained nuclei in blue. (B) Graph showing the percentage of EdU-positive G1-phase cells in the asynchronous HeLa cell population in control and MOF-depleted cells after labeling with EdU for 30 min followed by growth of the cells under normal culture conditions for 8 h. (C) Quantification of the percentage of EdU-positive G1-phase cells in control and MOF-depleted cells pretreated with 1 mM HU for 12 h, followed by 8 h of recovery. (D) Numbers of large γ-H2AX foci, representing collapsed forks, in EdU-positive S-phase cells in control and MOF-depleted cells. Foci were counted in approximately 200-EdU positive cells per data set. The error bars represent the standard deviation from three independent experiments. The P value is <0.05 (*), indicating that the difference is statistically significant.
Prolonged exposure to replicative stress usually leads to replication fork collapse, which is a necessary step for initiating DNA repair and resumption of replication. To measure collapsed fork formation, we labeled S-phase cells with a short EdU pulse, followed by incubation in 1 mM HU for 12 h. The HU was washed out, and the cells were then allowed to grow for an additional 8 h under normal culture conditions before analysis of EdU and γ-H2AX focus-positive cells. First, we scored the percentage of EdU-positive cells which had migrated into G1 phase during the 8-h time period after HU treatment and found that fewer cells had progressed from S and G2 phase into G1 phase in MOF-depleted cells (Fig. 4C). Second, we scored cells for large γ-H2AX foci, which arise only after long HU treatment and are thus thought to represent collapsed replication forks. Surprisingly, MOF-depleted cells had a reduced frequency of collapsed forks compared to control cells (Fig. 4D). Overall, the data suggest that MOF-depleted cells have a delayed S/G2-phase transition under normal growth conditions and that under replicative stress, this may result in decreased accumulation of collapsed forks after replication stalling.
MOF depletion impairs DNA repair by HR.
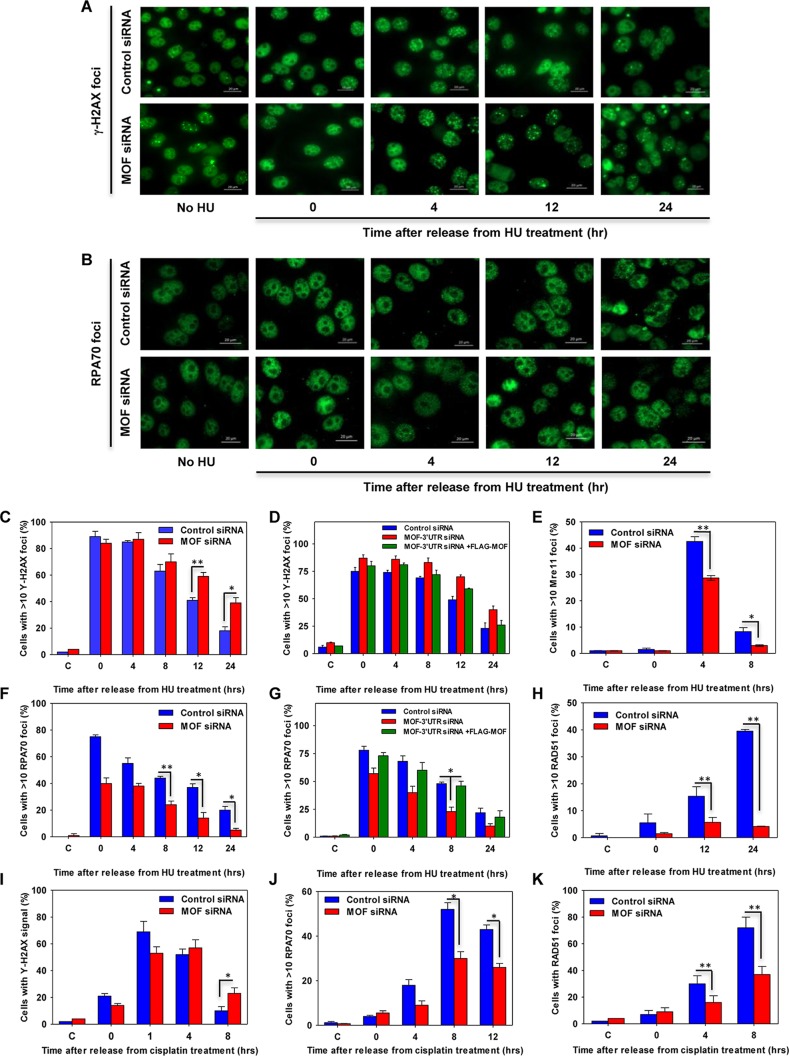
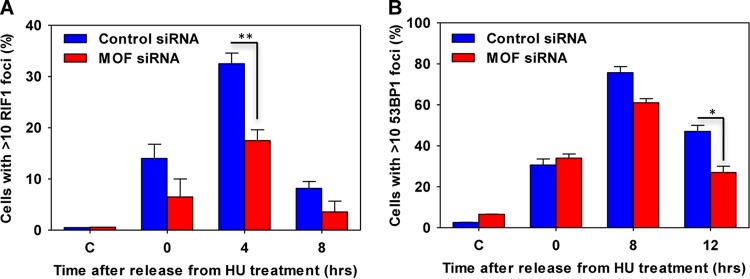
Collapsed forks generated during prolonged exposure to replication stress are repaired via the homologous recombination (HR)-mediated DSB repair pathway. To evaluate the role of MOF in HR, we measured the recruitment of HR-related proteins to HU-induced DNA damage sites in MOF-depleted cells by immunofluorescence focus formation. Measured over the course of 24 h, there was an increased persistence of γ-H2AX foci in MOF-depleted cells, which was rescued by FLAG-tagged MOF expression, suggesting that there was a defect in the processing of DNA damage (Fig. 5A, C, and D). The appearance of Mre11 foci, an early step in the DNA damage response, was also significantly reduced in MOF-depleted cells, suggesting that MOF might be an important factor in the early events of DNA end resection (Fig. 5E). The appearance of other downstream HR proteins, such as RPA and RAD51, at DNA damage sites was also decreased in MOF-depleted cells (Fig. 5B and F to H). These repair defects were not unique to HU-induced fork collapse, as similar results were observed when MOF-depleted cells were treated with cisplatin (Fig. 5I to K). Interestingly, a significant decrease in the appearance of NHEJ-related factors, such as RIF1 and 53BP1, was also detected, which is consistent with previous findings that MOF can influence the NHEJ pathway (Fig. 6) (18, 25).
FIG 5.
Homologous-recombination-mediated DNA repair is impaired in MOF-depleted cells. (A) Representative images of γ-H2AX focus formation at different time points after the release from HU treatment in control and MOF-depleted cells. (B) Representative images of RPA70 focus formation in control and MOF-depleted cells after HU treatment and release for different time points. (C to K) The control and MOF-depleted HeLa cells were treated with either 2 mM HU for 24 h or 50 μM cisplatin for 1 h and analyzed for focus formation of different HR proteins at various time intervals after the release from stress. (C, E, F, and H) Quantification of time-dependent γH2AX, Mre11, RPA70, and Rad51 focus formation, respectively, in control and MOF-depleted cells after treatment with HU. The experiments were performed two times for each protein, and for every time point approximately 150 cells were counted. P values: *, <0.05; **, <0.01. (D and G) Quantification of time-dependent γH2AX (D) and RPA70 (G) focus formation after HU treatment and release for different time points in control cells and cells transfected with 3′ UTR-specific MOF siRNA and depletion followed by ectopic FLAG-MOF expression. (I to K) Quantification of time-dependent γH2AX, RPA70, and Rad51 focus formation, respectively, in control and MOF-depleted cells after cisplatin treatment and release for different time points.
FIG 6.
Nonhomologous end joining-mediated DNA repair is impaired in MOF-depleted cells. The bar graphs represent the RIF1 (A) and 53BP1 (B) focus formation after HU treatment and release for the indicated time points under normal growth conditions.
MOF depletion inhibits DNA end resection.
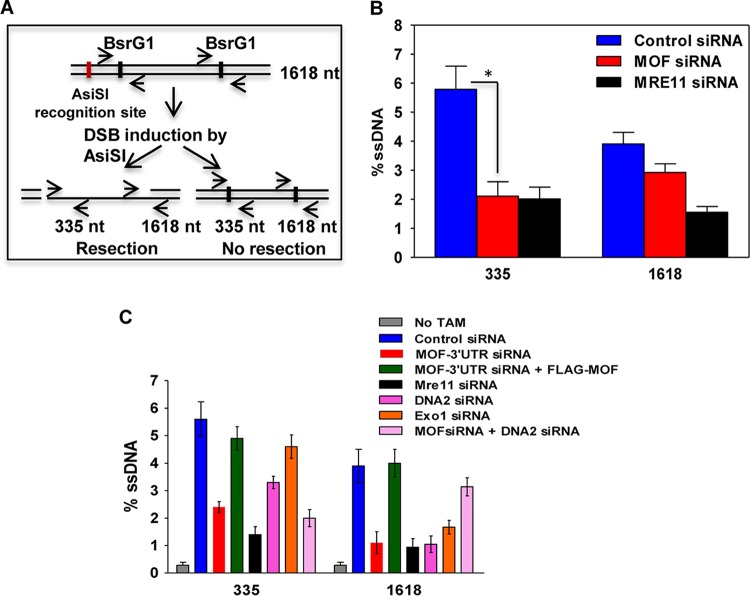
Since MOF-depleted cells had reduced Mre11, RPA, and Rad51 focus formation, we reasoned that MOF might influence the early DNA end resection step of DSB repair. Therefore, DNA end resection was measured using an established estrogen receptor (ER)-AsiSI assay (38) that quantitates the amount of ssDNA generated during resection by quantitative PCR (qPCR). A significant decrease in ssDNA production was observed in MOF-depleted cells (Fig. 7B), suggesting that MOF contributes to efficient DNA end resection at DSBs. This may be one factor explaining why the absence of MOF reduces the recruitment of downstream factors required for HR-mediated repair. To further investigate the role of MOF in DNA end resection, the effect of MOF depletion was compared with that of Mre11, DNA2, or Exo1 depletion and also with that of simultaneous MOF and Mre11, DNA2 or Exo1 depletion. Depletion of endogenous MOF decreased resection, which was rescued by transfection with siRNA-insensitive FLAG-MOF. Individual depletion of Mre11, DNA2, or Exo1 all resulted in decreased resection; however, attempts to simultaneously deplete MOF and Mre11 or Exo1 resulted in cell lethality (data not shown). Simultaneous MOF and DNA2 depletion produced no additive decrease in DNA end resection at the nucleotide 335 site, suggesting that these two proteins act in the same pathway (Fig. 7C).
FIG 7.
End resection at a DNA DSB site is inhibited in MOF-depleted cells. (A) Scheme of the experimental design. After site-specific DSB induction by ER-AsiSI, the amount of intermediate ssDNA generated in the cells was measured by quantitative PCR using site-specific primers flanking nucleotide positions 335 and 1618 downstream of the DSB induction site. (B) Quantification of the percent ssDNA in control and MOF-depleted cells by quantitative PCR. Mre11-depleted ER-AsiSI U2OS cells were used as a positive control for the experiment. The experiments were performed three times, and the error bars represent the standard deviation from three independent experiments. The P value is < 0.05 (*), indicating that the difference is statistically significant. (C) Quantification of the percent ssDNA in cells transfected with different siRNA as indicated.
MOF interacts with PCNA and affects its ubiquitylation and recruitment to DNA damage sites.
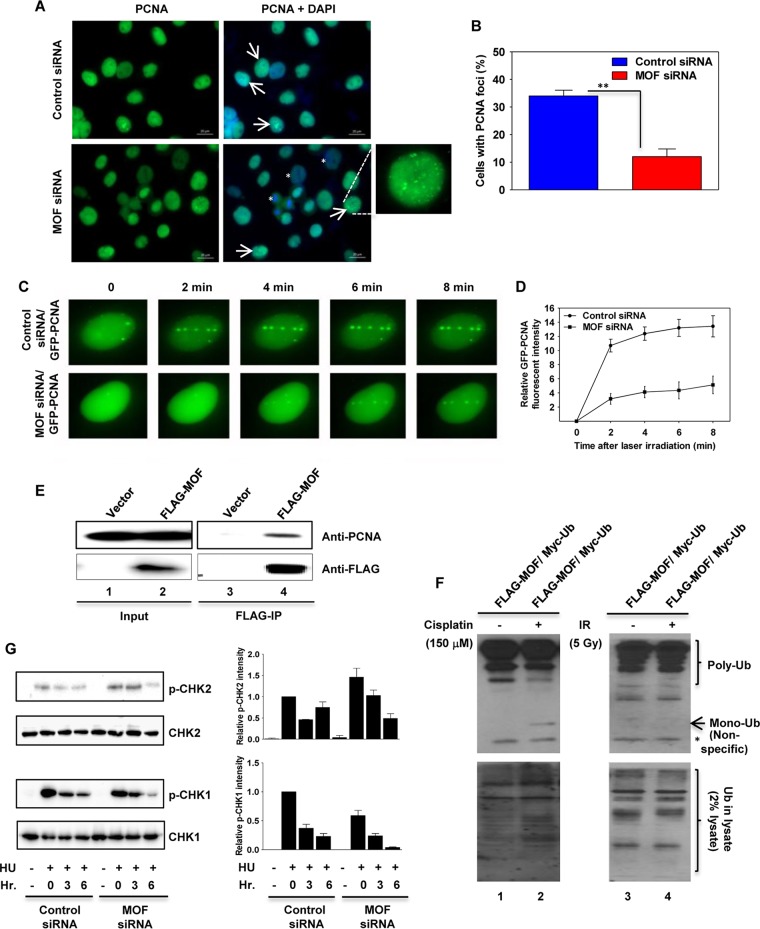
PCNA is an important factor in coordinating DNA replication with repair protein function (39, 40). In order to understand MOF involvement in the replication stress response, we determined whether MOF affected the recruitment of PCNA to DNA damage sites. We measured PCNA focus formation after HU treatment by immunostaining and found that 12 h after release from HU treatment, the MOF-depleted cells had a reduced number of cells with PCNA foci compared to control cells (Fig. 8A and B). MOF depletion results in loss of H4K16ac, which enhances chromatin condensation and thereby reduces protein factor access to DNA. Consistent with these observations, we observed a significant number of cells with decreased PCNA staining in MOF-depleted cells (Fig. 8A). A laser microirradiation technique was then used to determine whether MOF depletion alters the dynamics of PCNA recruitment to DNA damage sites along the damage track. The results showed that in control cells, green fluorescent protein (GFP)-PCNA recruitment to the DNA damage track was readily detected within 2 min and was undiminished through 8 min postirradiation (Fig. 8C and D). In MOF-depleted cells, there was no visible PCNA recruitment within the first 2 min of the DNA damage and only very slow recruitment after that compared to the case for control cells (Fig. 8C and D), suggesting that PCNA recruitment was delayed in the absence of MOF. Furthermore, Western blot analysis of FLAG immunoprecipitates prepared from HeLa cells transfected with FLAG-tagged MOF detected coimmunoprecipitating PCNA with MOF (Fig. 8E). In response to DNA damage, PCNA is either mono- or polyubiquitylated to promote translesion DNA synthesis (TLS). We measured the ubiquitylation of PCNA in cells overexpressing MOF and treated with cisplatin or ionizing radiation (IR). Interestingly, PCNA ubiquitylation was increased in cisplatin-treated cells and not in response to IR (Fig. 8F). These results suggest that MOF is important for PCNA ubiquitylation and therefore affects PCNA recruitment to DNA damage sites associated with replicative stress.
FIG 8.
MOF depletion reduces PCNA ubiquitination, delays PCNA recruitment to DNA damage sites, and inhibits CHK1 phosphorylation. (A) Recruitment of PCNA to DNA damage sites is decreased in MOF-depleted cells. Control and MOF-depleted HeLa cells were treated with HU for 24 h, and 12 h after release from HU treatment, the cells were immunostained with PCNA antibody. The cells with PCNA foci are marked with arrows. The enlarged image in the bottom right panel shows a representative image of the cells with PCNA foci. The less-PCNA-stained cells in MOF-depleted cells are marked with an asterisk. (B) Quantification of the percentage of cells with PCNA foci in control and MOF-depleted cells. Error bars represent the standard deviation from three independent experiments. P value: **, <0.01. (C) Recruitment of PCNA onto laser-induced DNA damage sites. Exponentially growing control and MOF-depleted HeLa cells were transfected with cDNA coding for EGFP-PCNA and then microirradiated, and time-lapse images were captured at different time intervals. (D) Kinetics of GFP-PCNA relative fluorescence intensity at the DNA damage sites measured in control and MOF-depleted HeLa cells at different time intervals after microirradiation. The experiments were performed two times, and each time about 50 cells was targeted with the laser and PCNA recruitment kinetics were measured. (E) FLAG-tagged MOF was used to coimmunoprecipitate endogenous PCNA (lane 4) from extracts prepared from transfected cells. The input and the IgG controls are shown in lanes 1 and 3, respectively. (F) FLAG-MOF and Myc-ubiquitin expression vectors were cotransfected into HeLa cells, followed by treatment with either cisplatin (150 μM) or IR (5 Gy). The increased PCNA monoubiquitylation induced by cisplatin is shown in lane 2. The monoubiquitinated band is marked with an arrow. The lowermost band, which is marked with an asterisk, corresponds to a nonspecific band. (G) The control and MOF-depleted HeLa cells were treated with HU and released from inhibition for the indicated times before being subjected to Western blotting with p-CHK1 and p-CHK2 antibodies. As a loading control, the same blots were reprobed with CHK1 and CHK2 antibodies. The bar graphs on the right show the Western blot quantification for the three independent experiments.
Generation of aberrant replication fork structures containing ssDNA-bound RPA activates the replication stress response, which is mediated by ATR kinase. Activated ATR kinase then phosphorylates downstream substrates such as CHK1 and RPA32 (41, 42). Phosphorylation of CHK1 by ATR helps stabilize the replication fork and delays cell cycle progression and late origin firing (43). We examined the CHK1 phosphorylation status in cells after HU treatment by Western blotting and found that MOF depletion significantly reduced CHK1 phosphorylation in HU-treated cells (Fig. 8G), while CHK2 phosphorylation, which is mediated by ATM in response to DNA damage, was increased.
Taken together, these results suggest that MOF suppresses replication stress by modulating PCNA function and abrogating the signaling pathway normally induced by replication stress.
DISCUSSION
The histone acetyltransferase MOF acetylates lysine 16 of histone H4 (H4K16ac), a histone modification that is unique in its ability to control chromatin structure and protein interactions. Experimental evidence suggests that histone H4K16ac can modulate both higher-order chromatin structure and functional interactions between nonhistone proteins and the chromatin fiber (19, 28). The H4K16ac modification poses a structural constraint on the formation of higher-order chromatin, suggesting the modification may contribute to the DDR by forcing chromatin into a more open, accessible configuration, and/or it could also potentially serve as a structural element or signal to facilitate assembly of DDR components. We demonstrated previously that H4K16 acetylation impacts DNA DSB repair (18), and it has also been shown that H4K16ac limits 53BP1 association with damaged chromatin to promote repair by the HR pathway (44). Here we describe a previously unrecognized role of MOF in replication and the replicative stress response. Our results provide strong evidence that MOF is involved in suppressing replication stress arising under normal cell growth conditions as well as the increased stress induced by exogenous factors.
Although previous studies demonstrated that MOF-depleted cells have proliferation defects, with embryonic lethality in Mof−/− knockout mice, the molecular details were unknown (23, 32, 45). We employed a robust approach using multiple assays to test MOF involvement in the replicative stress response. We found similar sensitivities of MOF-depleted cells to different replication stress-inducing agents which have different modes of action, suggesting that the MOF function in resolving replication stress is quite diverse and might influence different stress response pathways (Fig. 1). For example, the increased accumulation of R loops in MOF-depleted cells also suggests that it contributes to resolving RNA-DNA hybrids. These R loops are formed as a result of RNA transcription-DNA replication collisions and are a major source of DNA break-mediated genome instability (46). As in human cells, the common fragile site FRA3B, which contains the 1.5-Mb-long fragile histidine triad (FHIT) gene, is associated with transcription/replication collisions and R-loop accumulation (46). Therefore, suppression of R-loop-mediated genome instability by MOF is important and could occur through multiple, even simultaneous, mechanisms including gene transcription regulation (47–50), as well as by the altered DNA damage responses and affects on DNA replication observed here.
The DNA fiber analysis detected increased endogenous replication fork stalling in nonstressed MOF-depleted cells, which was further increased to approximately 4-fold after either HU or APH treatment (Fig. 2 and 3). Many factors could contribute to replication fork stalling, including fragile sites, DNA polymerase inhibition, aberrant secondary structures such as G quadruplexes and R-loop formation, etc. The interference with replication progression by R loops has been shown in organisms from bacteria to humans. In Escherichia coli, an rRNA gene operon transcribed head-on into the direction of replication fork progression and a transcribed R-loop-forming IgS region cause replication fork stalling and chromosomal rearrangements (51, 52). In human cells, TOP1 depletion causes transcription to interfere with replication in an R-loop-dependent manner (53). Apart from R loops, additional factors may also contribute to replication stalling. In cells with a deficiency of cotranscriptional protein factors, the resulting RNA polymerase stalling may cause an excess of R loops such that an arrested RNA polymerase II (Pol II) remains at the site, where it contributes to the block of an advancing replisome (54, 55). Thus, our data suggest that MOF could suppress stalled replication forks in three possible ways: first, by removing the DNA structural constraints for replication restart or recovery; second, by removing/preventing R-loop-mediated fork stalling; and third, by recruiting PCNA at the DNA damage site to reactivate replication (discussed below). These observations suggest that fork stalling and the decreased fork speed could contribute to the proliferation defects and S-phase cell arrest in MOF-depleted cells. Surprisingly, we observed a decrease in the total number of collapsed forks per cell in MOF-depleted cells. This could be attributed to the slower fork progression in MOF-depleted cells and is consistent with our previous finding that lamin A/C depletion decreased fork collapse but increased overall DNA damage (56).
Collapsed forks formed during prolonged exposure to replication stress are removed by a DNA double-strand break (DSB) repair pathway. Previous studies have shown that MOF protein is critical for DSB repair in response to ionizing radiation (IR)-induced DNA damage (18, 25, 29, 57). While MOF depletion can inhibit both the HR and NHEJ repair pathways (18), our results are consistent with defective recruitment of HR-related proteins to DNA damage sites produced by replicative stress-inducing agents such as HU and cisplatin (Fig. 5 and 6). More significantly, we found that DNA end resection was reduced in MOF-depleted cells (Fig. 7), indicating that MOF is involved in the initial steps of HR-mediated DSB repair and is thus critical for subsequent stress-induced repairosome complex formation at the DNA damage site.
Our results showed that MOF protein interacts with PCNA and contributes to PCNA ubiquitylation, enabling its recruitment to DNA damage sites (Fig. 8). PCNA is important for both DNA replication and DNA repair. Cells have evolved bypass mechanisms to prevent blocked replication forks from collapse (58). One important means is to switch from accurate replicative polymerases (i.e., Polδ or Polε) to specialized translesion polymerases that can read through lesions, a process termed translesion DNA synthesis (TLS) that is directly controlled through covalent modification of PCNA by ubiquitin (59). In response to DNA damage, PCNA is either mono- or polyubiquitylated to initiate the rescue, but error-prone, TLS pathway. Our findings clearly show that MOF facilitates PCNA recruitment to replicative-stress DNA damage sites through modulation of PCNA ubiquitylation.
The persistence of RPA-bound ssDNA that is adjacent to stalled, newly replicated dsDNA generates a signaling platform for activation and recruitment of a number of replication stress response proteins, such as the ATM and ATR kinases. ATR is a central replication stress response kinase that, once activated, phosphorylates substrates such as RPA and CHK1, which promotes replication fork stabilization and fork restart to faithfully complete DNA replication under stress conditions and ensure cell survival (41, 42, 60, 61). Phosphorylation of CHK1 at serine 317 was significantly decreased in MOF-depleted cells in response to HU treatment (Fig. 8G). This observation is consistent with our findings of decreased single-stranded DNA generation and RPA recruitment to DSBs in MOF-depleted cells, which should decrease ATR activation. Altered CHK1 function is also mechanistically consistent with our observation of decreased replication restart in MOF-depleted cells. Similar results were also seen previously when we depleted the proapoptotic protein MCL-1 (62).
In summary, the present data suggest that MOF has multiple roles in the replication stress response. The decreased recruitment of HR proteins and abrogation of replication stress-induced DNA damage signaling after MOF depletion suggest that MOF influences the replication stress response by acting during the early initial events. This might also point toward its role in repairosome organization at the replication-induced DNA damage sites, which is consistent with its role in modulating chromatin dynamics in response to DNA damage.
MATERIALS AND METHODS
Cell lines and transfection.
HeLa and U2OS cells were purchased from ATCC (Manassas, VA) and cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin at 37°C in a humidified atmosphere with 5% CO2. The cells were transfected with either control siRNA or MOF siRNA using an Amaxa Nucleofector electroporation kit and following the manufacturer's protocol (Lonza, Allendale, NJ).
Clonogenic survival assay.
Forty-eight hours after transfection (HeLa or U2OS cells with either control or MOF siRNA), 600 cells were seeded onto 60-mm dishes in triplicate. The next day, the cells were treated with the indicated concentrations of either cisplatin for 1 h, hydroxyurea (HU) for 20 h, or camptothecin (CPT) for 12 h and then washed thrice with phosphate-buffered saline (PBS) and refed with fresh DMEM. The cells were allowed to grow for 13 days, and the surviving colonies (greater than 50 cells) were fixed, stained with crystal violet, and counted.
DNA replication restart assay.
Exponentially growing cells were pulse-labeled with 50 mM 5-chlorodeoxyuridine (CldU) for 30 min, washed three times with PBS, treated with 2 mM hydroxyurea (HU) for 2 h, washed three times with PBS, incubated again in fresh medium containing 50 mM 5-iododeoxyuridine (IdU) for 60 min, and then washed three times in PBS. DNA fiber spreads were made by a modification of a procedure described previously (56). Briefly, cells labeled with IdU and CldU were mixed with unlabeled cells at a ratio of 1:10, and 2-μl cell suspensions were dropped onto a glass slide and then mixed with 20 μl hypotonic lysis solution (10 mM Tris-HCl [pH 7.4], 2.5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 0.5% Nonidet P-40) for 8 min. Air-dried slides were fixed, washed with 1× PBS, blocked with 5% bovine serum albumin (BSA) for 15 min, and incubated with primary antibodies against IdU and CldU (rat anti-IdU monoclonal antibody [MAb] [1:150 dilution; Abcam] and mouse anti-CldU MAb [1:150 dilution; BD]) and secondary antibodies (anti-rat Alexa Fluor 488-conjugated [1:150 dilution] and anti-mouse Alexa Fluor 568-conjugated [1:200 dilution] antibodies) for 1 h each. Slides were washed with 1× PBS with 0.1% Triton X-100 and mounted with Vectashield mounting medium without 4′,6-diamidino-2-phenylindole (DAPI). ImageJ software was used to analyze the DNA fibers. For each data set, about 300 fibers were counted for stalled forks, new origins, or elongated forks, and the results were summarized in bar graphs.
Measurement of collapsed replication forks.
HeLa cells were labeled with 1 μM 5-ethynyl-2′-deoxyuridine (EdU) for 30 min to stain S-phase cells, washed twice with 1× PBS, incubated in medium with 1 mM HU for 12 h, washed again with 1× PBS to remove HU, and incubated in fresh medium without HU for 8 h before being stained for γ-H2AX and EdU. EdU-positive S/G2- and G1-phase cells were identified by their size and DAPI intensity (DNA content). First, we scored only large γ-H2AX foci, which arise only after long HU treatment and thus represent collapsed replication forks. Second, we scored the percentage of EdU-positive cells which migrated into G1 phase during the 8-h time period after HU treatment. γ-H2AX foci were counted in approximately 200 EdU-positive cells per data set.
Immunofluorescence microscopy.
The HeLa cells transfected with control and MOF siRNAs were allowed to grow for 60 to 72 h in chambered slides and then treated with either 2 mM HU or 50 μM cisplatin, followed by recovery for the indicated time intervals. The cells were then fixed with either 4% paraformaldehyde or methanol and immunostained as described previously (56). Fluorescent images of foci were captured with a Zeiss Axio Imager M2 microscope, and the foci were counted by ImageJ software.
Laser microirradiation.
Control and MOF-depleted U2OS cells transfected with GFP-tagged PCNA were grown on a 35-mm glass-bottom cell culture dish for 24 h. Under normal growth conditions, the cells were microirradiated by a 365-nm laser with the optimum intensity on a Carl Zeiss microscope fitted with a focused laser delivery apparatus. Time-lapse images were captured to quantify accumulation of GFP-PCNA at the DNA damage site using Carl Zeiss Zen software as described previously (56).
Quantitative resection assay using the ER-AsiSI system.
A quantitative DNA resection assay was used to measure ssDNA intermediates as described previously (38). The ER-AsiSI U2OS cells were incubated with tamoxifen (300 ng/ml) for 3 h and then mixed with 0.6% low-melting-temperature agarose in PBS at a concentration of 6 × 106 cells per ml. Fifty microliters of cell suspension was solidified as an agar ball by dropping onto Parafilm. The agar ball was serially treated with 1 ml of EDTA-sarcosine-proteinase K (ESP) buffer (0.5 M EDTA, 2% N-lauroylsarcosine, 1 mg/ml of proteinase K, and 1 mM CaCl2 [pH 8.0]) and high-salt (HS) buffer (1.85 M NaCl, 0.15 M KCl, 5 mM MgCl2, 2 mM EDTA, 4 mM Tris, and 0.5% Triton X-100 [pH 7.5]) for 20 h each time at 16°C with rotation, followed by washing with 1 ml of phosphate buffer (8 mM Na2HPO4, 1.5 mM KH2PO4, 133 mM KCl, and 0.8 mM MgCl2 [pH 7.4]) for 1 h at 4°C with rotation. After heating of the agar ball at 70°C for 10 min, it was diluted 15-fold with 70°C double-distilled water (ddH2O) and mixed with 2× NEB restriction enzyme buffer 4. Sixty microliters of genomic DNA sample was digested with 60 U of restriction enzyme BsrGI (New England BioLabs) or mock digested at 37°C overnight, and 3-μl quantities of mock- or BsrGI-digested samples were used as templates in quantitative PCRs (qPCRs) with 20-μl mixtures.
PCNA ubiquitination assay.
The HeLa cells were cotransfected with FLAG-tagged MOF and Myc-tagged ubiquitin expression plasmids. Twenty-four hours later, the cells were treated with 150 μM cisplatin for 6 h or 5 Gy IR, followed by a 2-h recovery in normal medium supplemented with MG-132 proteosome inhibitor before being harvested. The lysates were immunoprecipitated with anti-Myc antibody, and then mono- or poly ubiquitylated PCNA was detected by Western blotting with PCNA antibody (PC10; Santa Cruz, CA).
Statistical analysis.
The standard deviation was calculated from three or four independent experiments. The t test was employed to calculate two-tailed P values and the statistical significance.
ACKNOWLEDGMENTS
This work was supported by funds from the Houston Methodist Research Institute and by National Institutes of Health grants (RO1CA129537 and RO1GM109768).
We have no conflict of interest.
REFERENCES
- 1.Mazouzi A, Velimezi G, Loizou JI. 2014. DNA replication stress: causes, resolution and disease. Exp Cell Res 329:85–93. doi: 10.1016/j.yexcr.2014.09.030. [DOI] [PubMed] [Google Scholar]
- 2.Zeman MK, Cimprich KA. 2014. Causes and consequences of replication stress. Nat Cell Biol 16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halazonetis TD, Gorgoulis VG, Bartek J. 2008. An oncogene-induced DNA damage model for cancer development. Science 319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 4.Groth A, Rocha W, Verreault A, Almouzni G. 2007. Chromatin challenges during DNA replication and repair. Cell 128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 5.Khurana S, Oberdoerffer P. 2015. Replication stress: a lifetime of epigenetic change. Genes (Basel) 6:858–877. doi: 10.3390/genes6030858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alabert C, Groth A. 2012. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol 13:153–167. doi: 10.1038/nrm3288. [DOI] [PubMed] [Google Scholar]
- 7.Blow JJ, Ge XQ, Jackson DA. 2011. How dormant origins promote complete genome replication. Trends Biochem Sci 36:405–414. doi: 10.1016/j.tibs.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branzei D, Foiani M. 2007. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amst) 6:994–1003. doi: 10.1016/j.dnarep.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 9.Hunt CR, Ramnarain D, Horikoshi N, Iyengar P, Pandita RK, Shay JW, Pandita TK. 2013. Histone modifications and DNA double-strand break repair after exposure to ionizing radiations. Radiat Res 179:383–392. doi: 10.1667/RR3308.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papamichos-Chronakis M, Peterson CL. 2008. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol 15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- 11.Rowbotham SP, Barki L, Neves-Costa A, Santos F, Dean W, Hawkes N, Choudhary P, Will WR, Webster J, Oxley D, Green CM, Varga-Weisz P, Mermoud JE. 2011. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol Cell 42:285–296. doi: 10.1016/j.molcel.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 12.Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM. 2008. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol 18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- 13.Leung KH, Abou El Hassan M, Bremner R. 2013. A rapid and efficient method to purify proteins at replication forks under native conditions. Biotechniques 55:204–206. doi: 10.2144/000114089. [DOI] [PubMed] [Google Scholar]
- 14.Driscoll R, Hudson A, Jackson SP. 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clemente-Ruiz M, Gonzalez-Prieto R, Prado F. 2011. Histone H3K56 acetylation, CAF1, and Rtt106 coordinate nucleosome assembly and stability of advancing replication forks. PLoS Genet 7:e1002376. doi: 10.1371/journal.pgen.1002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myung K, Pennaneach V, Kats ES, Kolodner RD. 2003. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc Natl Acad Sci U S A 100:6640–6645. doi: 10.1073/pnas.1232239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krishnan V, Chow MZ, Wang Z, Zhang L, Liu B, Liu X, Zhou Z. 2011. Histone H4 lysine 16 hypoacetylation is associated with defective DNA repair and premature senescence in Zmpste24-deficient mice. Proc Natl Acad Sci U S A 108:12325–12330. doi: 10.1073/pnas.1102789108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, Bhadra U, Pandita RK, Porteus MH, Chen DJ, Cote J, Pandita TK. 2010. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol 30:3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 20.Oppikofer M, Kueng S, Martino F, Soeroes S, Hancock SM, Chin JW, Fischle W, Gasser SM. 2011. A dual role of H4K16 acetylation in the establishment of yeast silent chromatin. EMBO J 30:2610–2621. doi: 10.1038/emboj.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang R, Erler J, Langowski J. 2017. Histone acetylation regulates chromatin accessibility: role of H4K16 in inter-nucleosome interaction. Biophys J 112:450–459. doi: 10.1016/j.bpj.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Perez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. 2005. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 23.Gupta A, Guerin-Peyrou TG, Sharma GG, Park C, Agarwal M, Ganju RK, Pandita S, Choi K, Sukumar S, Pandita RK, Ludwig T, Pandita TK. 2008. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol Cell Biol 28:397–409. doi: 10.1128/MCB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta A, Hunt CR, Pandita RK, Pae J, Komal K, Singh M, Shay JW, Kumar R, Ariizumi K, Horikoshi N, Hittelman WN, Guha C, Ludwig T, Pandita TK. 2013. T-cell-specific deletion of Mof blocks their differentiation and results in genomic instability in mice. Mutagenesis 28:263–270. doi: 10.1093/mutage/ges080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta A, Hunt CR, Hegde ML, Chakraborty S, Udayakumar D, Horikoshi N, Singh M, Ramnarain DB, Hittelman WN, Namjoshi S, Asaithamby A, Hazra TK, Ludwig T, Pandita RK, Tyler JK, Pandita TK. 2014. MOF phosphorylation by ATM regulates 53BP1-mediated double-strand break repair pathway choice. Cell Rep 8:177–189. doi: 10.1016/j.celrep.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhadra MP, Horikoshi N, Pushpavallipvalli SN, Sarkar A, Bag I, Krishnan A, Lucchesi JC, Kumar R, Yang Q, Pandita RK, Singh M, Bhadra U, Eissenberg JC, Pandita TK. 2012. The role of MOF in the ionizing radiation response is conserved in Drosophila melanogaster. Chromosoma 121:79–90. doi: 10.1007/s00412-011-0344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horikoshi N, Kumar P, Sharma GG, Chen M, Hunt CR, Westover K, Chowdhury S, Pandita TK. 2013. Genome-wide distribution of histone H4 Lysine 16 acetylation sites and their relationship to gene expression. Genome Integr 4:3. doi: 10.1186/2041-9414-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandita TK. 2013. Histone H4 lysine 16 acetylated isoform synthesis opens new route to biophysical studies. Proteomics 13:1546–1547. doi: 10.1002/pmic.201300145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, Lucchesi JC, Khanna KK, Ludwig T, Pandita TK. 2005. Involvement of human MOF in ATM function. Mol Cell Biol 25:5292–5305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar R, Hunt CR, Gupta A, Nannepaga S, Pandita RK, Shay JW, Bachoo R, Ludwig T, Burns DK, Pandita TK. 2011. Purkinje cell-specific males absent on the first (mMof) gene deletion results in an ataxia-telangiectasia-like neurological phenotype and backward walking in mice. Proc Natl Acad Sci U S A 108:3636–3641. doi: 10.1073/pnas.1016524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valerio DG, Xu H, Chen CW, Hoshii T, Eisold ME, Delaney C, Cusan M, Deshpande AJ, Huang CH, Lujambio A, Zheng YG, Zuber J, Pandita TK, Lowe SW, Armstrong SA. 2017. Histone acetyltransferase activity of MOF is required for MLL-AF9 leukemogenesis. Cancer Res 77:1753–1762. doi: 10.1158/0008-5472.CAN-16-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas T, Dixon MP, Kueh AJ, Voss AK. 2008. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Mol Cell Biol 28:5093–5105. doi: 10.1128/MCB.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Li L, Pandey R, Byun JS, Gardner K, Qin Z, Dou Y. 2012. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell 11:163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandita RK, Sharma GG, Laszlo A, Hopkins KM, Davey S, Chakhparonian M, Gupta A, Wellinger RJ, Zhang J, Powell SN, Roti Roti JL, Lieberman HB, Pandita TK. 2006. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol Cell Biol 26:1850–1864. doi: 10.1128/MCB.26.5.1850-1864.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy D, Zhang Z, Lu Z, Hsieh CL, Lieber MR. 2010. Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: a nick can serve as a strong R-loop initiation site. Mol Cell Biol 30:146–159. doi: 10.1128/MCB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aguilera A, Garcia-Muse T. 2012. R loops: from transcription byproducts to threats to genome stability. Mol Cell 46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Taipale M, Akhtar A. 2005. Chromatin mechanisms in Drosophila dosage compensation. Prog Mol Subcell Biol 38:123–149. doi: 10.1007/3-540-27310-7_5. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y, Paull TT. 2015. Direct measurement of single-stranded DNA intermediates in mammalian cells by quantitative polymerase chain reaction. Anal Biochem 479:48–50. doi: 10.1016/j.ab.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 39.Moldovan GL, Pfander B, Jentsch S. 2007. PCNA, the maestro of the replication fork. Cell 129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 40.Mailand N, Gibbs-Seymour I, Bekker-Jensen S. 2013. Regulation of PCNA-protein interactions for genome stability. Nat Rev Mol Cell Biol 14:269–282. doi: 10.1038/nrm3562. [DOI] [PubMed] [Google Scholar]
- 41.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev 19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zou L, Elledge SJ. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 43.Lee J, Dunphy WG. 2013. The Mre11-Rad50-Nbs1 (MRN) complex has a specific role in the activation of Chk1 in response to stalled replication forks. Mol Biol Cell 24:1343–1353. doi: 10.1091/mbc.E13-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. 2013. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol 20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, Akhtar A. 2005. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol 25:6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helmrich A, Ballarino M, Tora L. 2011. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell 44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 47.Suka N, Luo K, Grunstein M. 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet 32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 48.Akhtar A, Becker PB. 2000. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell 5:367–375. doi: 10.1016/S1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 49.Fullgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, Joseph B. 2013. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 500:468–471. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheikh BN, Bechtel-Walz W, Lucci J, Karpiuk O, Hild I, Hartleben B, Vornweg J, Helmstädter M, Sahyoun AH, Bhardwaj V, Stehle T, Diehl S, Kretz O, Voss AK, Thomas T, Manke T, Huber TB, Akhtar A. 2016. MOF maintains transcriptional programs regulating cellular stress response. Oncogene 35:2698–2710. doi: 10.1038/onc.2015.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boubakri H, de Septenville AL, Viguera E, Michel B. 2010. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J 29:145–157. doi: 10.1038/emboj.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X. 2011. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev 25:2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, Pommier Y, Tazi J, Coquelle A, Pasero P. 2009. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol 11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drolet M, Phoenix P, Menzel R, Masse E, Liu LF, Crouch RJ. 1995. Overexpression of RNase H partially complements the growth defect of an Escherichia coli delta topA mutant: R-loop formation is a major problem in the absence of DNA topoisomerase I. Proc Natl Acad Sci U S A 92:3526–3530. doi: 10.1073/pnas.92.8.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rondon AG, Jimeno S, Garcia-Rubio M, Aguilera A. 2003. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J Biol Chem 278:39037–39043. doi: 10.1074/jbc.M305718200. [DOI] [PubMed] [Google Scholar]
- 56.Singh M, Hunt CR, Pandita RK, Kumar R, Yang CR, Horikoshi N, Bachoo R, Serag S, Story MD, Shay JW, Powell SN, Gupta A, Jeffery J, Pandita S, Chen BP, Deckbar D, Lobrich M, Yang Q, Khanna KK, Worman HJ, Pandita TK. 2013. Lamin a/c depletion enhances DNA damage-induced stalled replication fork arrest. Mol Cell Biol 33:1210–1222. doi: 10.1128/MCB.01676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, Corsa CA, Pan PW, Wu L, Ferguson D, Yu X, Min J, Dou Y. 2010. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol 30:5335–5347. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barbour L, Xiao W. 2003. Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: a yeast model. Mutat Res 532:137–155. doi: 10.1016/j.mrfmmm.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 59.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 60.Nam EA, Cortez D. 2011. ATR signalling: more than meeting at the fork. Biochem J 436:527–536. doi: 10.1042/BJ20102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. 2007. The structural determinants of checkpoint activation. Genes Dev 21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mattoo AR, Pandita RK, Chakraborty S, Charaka V, Mujoo K, Hunt CR, Pandita TK. 2017. MCL-1 depletion impairs DNA double-strand break repair and reinitiation of stalled DNA replication forks. Mol Cell Biol 37:e00535-16. doi: 10.1128/MCB.00535-16. [DOI] [PMC free article] [PubMed] [Google Scholar]