ABSTRACT
A tightly controlled cellular deoxyribonucleotide (deoxynucleoside triphosphate [dNTP]) pool is critical for maintenance of genome integrity. One mode of dNTP pool regulation is through subcellular localization of ribonucleotide reductase (RNR), the enzyme that catalyzes the rate-limiting step of dNTP biosynthesis. In Saccharomyces cerevisiae, the RNR small subunit, Rnr2-Rnr4, is localized to the nucleus, whereas the large subunit, Rnr1, is cytoplasmic. As cells enter S phase or encounter DNA damage, Rnr2-Rnr4 relocalizes to the cytoplasm to form an active holoenzyme complex with Rnr1. Although the DNA damage-induced relocalization requires the checkpoint kinases Mec1-Rad53-Dun1, the S-phase-specific redistribution does not. Here, we report that the S-phase cyclin–cyclin-dependent kinase (CDK) complex Clb6-Cdc28 controls Rnr2-Rnr4 relocalization in S phase. Rnr2 contains a consensus CDK site and exhibits Clb6-dependent phosphorylation in S phase. Deletion of CLB6 or removal of the CDK site results in an increased association of Rnr2 with its nuclear anchor Wtm1, nuclear retention of Rnr2-Rnr4, and an enhanced sensitivity to the RNR inhibitor hydroxyurea. Thus, we propose that Rnr2-Rnr4 redistribution in S phase is triggered by Clb6-Cdc28-mediated phosphorylation of Rnr2, which disrupts the Rnr2-Wtm1 interaction and promotes the release of Rnr2-Rnr4 from the nucleus.
KEYWORDS: S phase, cyclin-dependent kinases, cyclins, deoxyribonucleotides, nucleocytoplasmic transport, ribonucleotide reductase
INTRODUCTION
Faithful replication of the genome depends on an adequate and balanced deoxyribonucleotide (deoxynucleoside triphosphate [dNTP]) pool (1). Ribonucleotide reductase (RNR) catalyzes the rate-limiting step in de novo dNTP synthesis and, thus, is largely responsible for controlling intracellular dNTP pools (2). All eukaryotic RNRs comprise a large subunit R1 that contains the catalytic and allosteric sites and a small subunit R2 that houses a diferric-tyrosyl radical cofactor that is essential to initiate nucleotide reduction (3, 4). In the budding yeast, Saccharomyces cerevisiae, the R1 subunit is a homodimer encoded by the RNR1 gene, while the R2 subunit is a heterodimer encoded by two paralogous genes, RNR2 and RNR4 (5–7).
Adjusting the dNTP pools to meet the demand of DNA synthesis under different conditions is critical, as levels that are too high or too low can cause increased spontaneous mutagenesis and genomic instability, a hallmark of cancer and aging (8–10). Cells have evolved multiple strategies to tightly control the level and activity of RNR, including allostery, transcription, and inhibitor protein association, as well as subcellular localization (2, 11). Allosteric regulation occurs through the large subunit R1, which alters substrate specificity based on sensing individual dNTP concentrations and overall activity through a dATP-mediated negative-feedback mechanism (2).
In response to DNA damage and replication stress, such as low dNTP levels caused by the RNR inhibitor hydroxyurea (HU), transcription of the RNR genes is rapidly induced by activation of the DNA damage checkpoint kinase cascade Med1-Rad53-Dun1, which inactivates the Crt1-Ssn6-tup1 transcriptional repressor complex (12, 13). The Mec1-Rad53-Dun1 kinase cascade is also responsible for phosphorylation-mediated proteolysis of the Rnr1 inhibitor protein Sml1, thus allowing further RNR activation (14).
RNR subcellular localization is an additional level of regulation (15–18). In the budding yeast, Rnr1 resides in the cytoplasm, whereas Rnr2-Rnr4 is confined to the nucleus except when cells enter S phase or encounter genotoxic stress. Nuclear sequestration of Rnr2-Rnr4 is controlled by two negative regulators of RNR: Dif1, which facilitates its nuclear import, and Wtm1, which binds to and retains it within the nucleus (19–22). DNA damage and replication stress elicit the Mec1-Rad53-Dun1 kinase cascade-mediated phosphorylation and degradation of Dif1, which leads to cytoplasmic enrichment of Rnr2-Rnr4, allowing it to form an active holoenzyme with Rnr1 (21, 22). The physiological importance of tight RNR regulation is manifested by the fact that the lethality of the mec1Δ and rad53Δ checkpoint kinase mutants can be rescued by activation of the RNR pathway, either via overexpression of RNR or via the removal of one of its negative regulators, SML1, WTM1, or DIF1 (13, 19, 21–24).
Rnr2-Rnr4 also relocalizes from the nucleus to the cytoplasm as cells enter S phase, coinciding with an increased demand of dNTP production for DNA replication (17). However, it is unclear whether this S-phase-specific redistribution is controlled by the Mec1-Rad53-Dun1 checkpoint kinase cascade or through a yet-unidentified mechanism.
The G1-to-S-phase transition in yeast cells is coordinated by two conserved kinase complexes, cyclin-dependent kinase (CDK) and Dbf4-dependent kinase (DDK) (25), each comprising a constitutive catalytic subunit, Cdc28 and Cdc7, respectively, and a cell cycle-controlled regulatory subunit, cyclins and Dbf4, respectively (25, 26). CDK and DDK act cooperatively to phosphorylate specific subunits of the prereplication complex, which triggers conformational changes that allow polymerase recruitment and origin firing in S phase. The S-phase CDK activity is governed by sequential association of Cdc28 with six B-type cyclins, Clb1 to -6, which peak in three successive waves, in the order of the pair of S-phase cyclins Clb5/Clb6 and the two pairs of mitotic cyclins Clb3/Clb4 and Clb1/Clb2, during the S-phase pregression (27). The Clb5/Clb6-dependent CDK activity peaks at the G1-to-S transition and is primarily responsible for cooperation with DDK to activate the firing of replication origins.
In this study, we uncover the mechanism that regulates Rnr2-Rnr4 relocalization during S phase. We show that the S-phase-specific redistribution is independent of the Mec1-Rad53-Dun1 kinase cascade and, instead, is controlled by Clb6-Cdc28. Furthermore, we demonstrate that a consensus CDK phosphorylation site, TPSK, at the N terminus of Rnr2 controls relocalization of Rnr2-Rnr4 through modulating the Wtm1-Rnr2 association. A phosphorylation-deficient substitution in Rnr2 enhances both in vivo Rnr2-Wtm1 association and cellular sensitivity to HU. We present evidence indicating that Clb6-Cdc28-mediated phosphorylation of Rnr2 at the consensus TPSK motif plays a critical role in the nucleus-to-cytoplasm redistribution of Rnr2-Rnr4 in S phase.
RESULTS
The nucleus-to-cytoplasm redistribution of the R2 subunit in S phase does not require the DNA damage checkpoint.
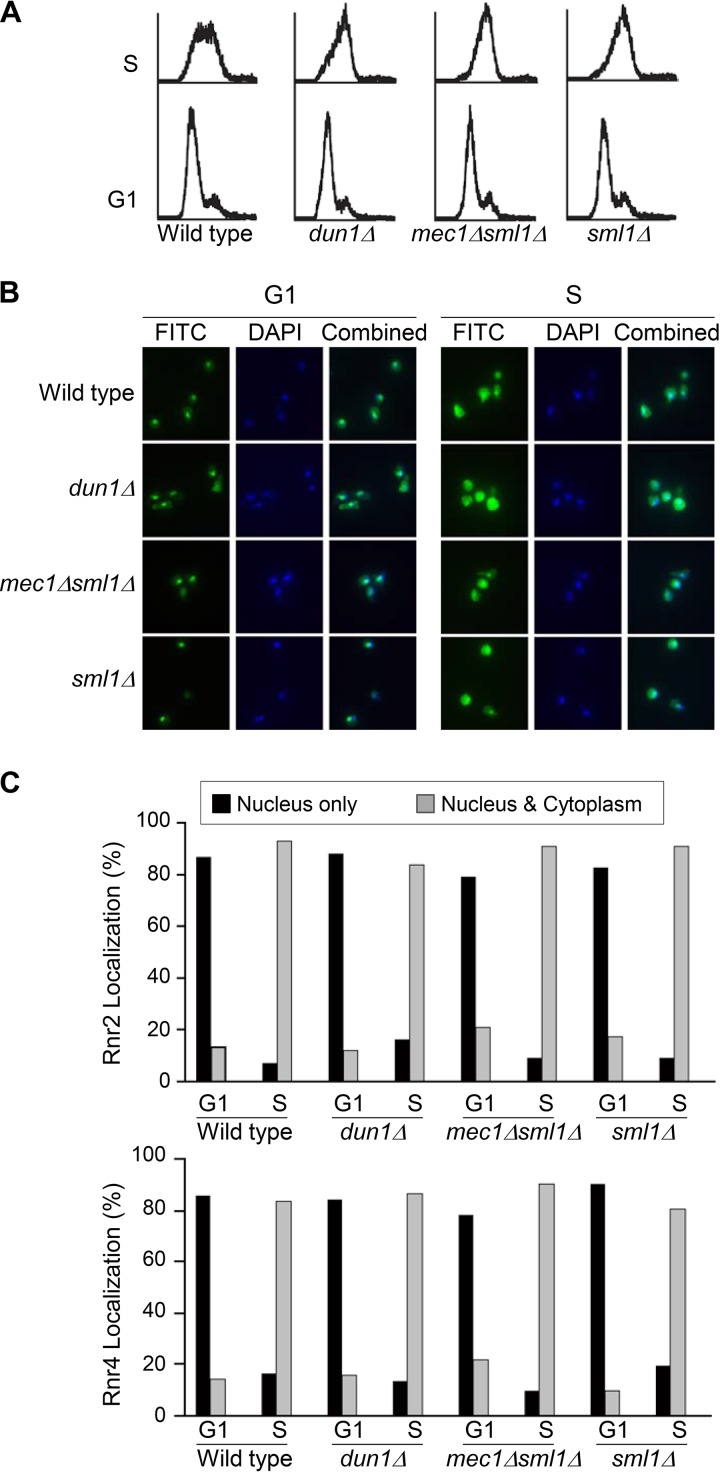
The R2 subunit of yeast RNR is an Rnr2-Rnr4 heterodimer that is cotransported between the nucleus and the cytoplasm (7). We have previously shown that the subcellular localization of the R2 subunit shifts from a predominantly nuclear pattern in cells arrested in the G1 phase to a ubiquitously distributed pattern (i.e., similar signal intensities in the nucleus and the cytoplasm) as cells enter the S phase (17). To determine whether S-phase-specific redistribution of R2 is controlled by the DNA damage checkpoint, we compared Rnr2 and Rnr4 localization in wild-type, dun1Δ, mec1Δ sml1Δ, and sml1Δ cells (sml1Δ suppresses the lethality of mec1Δ) after being released from G1 arrest to S phase. Cells were synchronized in G1 phase by using α-factor-mediated arrest; the majority of the cells were in the middle of the S phase 30 min after being released from the G1 arrest (Fig. 1A). The subcellular localization of Rnr2 and Rnr4 was determined by immunofluorescence analysis using specific polyclonal anti-Rnr2 and anti-Rnr4 antibodies (Fig. 1B). In all strains, the Rnr2 and Rnr4 signals changed from a predominantly nuclear pattern (>80%) in G1-arrested cells to a more ubiquitous pattern as cells entered S phase, with comparable signals (>80%) in the nucleus and the cytoplasm in the majority of the cells (Fig. 1C). The indistinguishable subcellular R2 localization patterns in the wild type and the checkpoint mutants indicate that the S-phase-specific R2 redistribution is not controlled by the DNA damage checkpoint kinase cascade.
FIG 1.
Nucleus-to-cytoplasm redistribution of the R2 subunit in S phase is independent of the DNA damage checkpoint. Wild-type, dun1Δ, mec1Δ sml1Δ, and sml1Δ cells from early-log-phase cultures grown at 30°C were arrested in G1 phase by using α-factor. Half of the cells were harvested at time zero, and the other half were released from the G1 arrest by washing off the α-factor with fresh medium and harvested 30 min later (t = 30 min), when the majority of the cells were in the middle of the S phase. (A) Flow cytometry analysis of DNA content of collected cells. (B) Representative images of cells from the G1 and S phases with DAPI (blue) and anti-Rnr2 and anti-Rnr4 antibody indirect immunofluorescence (FITC, green) staining. Combined panels show superimposed images of DAPI and FITC staining. (C) Quantitative analysis of subcellular localization patterns of Rnr2 and Rnr4 proteins. For each experiment, >150 cells were counted for each sample. The indirect immunofluorescence analyses were repeated three times, and a representative result is shown. Black bars represent percentages of cells with a predominantly nuclear signal, and gray bars represent percentages of cells with equal signal intensities between the nucleus and the cytoplasm.
The S-phase-specific R2 redistribution requires CDK.
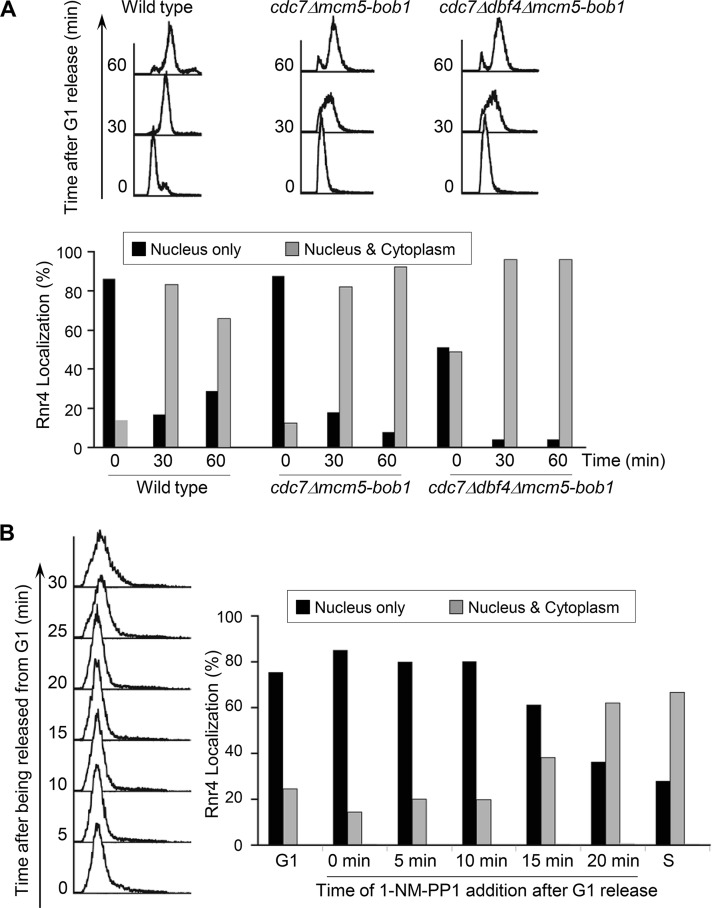
We next asked whether R2 redistribution in S phase is controlled by the two cell cycle-regulated kinases involved in DNA replication initiation, DDK and CDK. The two subunits of DDK, Dbf4 and Cdc7, are essential for mitotic survival. To obtain a viable DDK-deficient mutant, we chose the mcm5-bob1 allele that bypasses the essential function of Cdc7 and Dbf4 (28). Although the cdc7Δ mcm5-bob1 and cdc7Δ dbf4Δ mcm5-bob1 mutants appeared to progress through S phase at a lower rate than the wild-type strain based on fluorescence-activated cell sorting (FACS) analyses of DNA content, both exhibited nucleus-to-cytoplasm redistribution of R2 as cells entered S phase that was similar to that in the wild-type cells (Fig. 2A). The percentage of cells with a predominantly nuclear R2 signal dropped from >80% to <20% as cdc7Δ mcm5-bob1 cells progressed from G1 to S phase. The cdc7Δ dbf4Δ mcm5-bob1 mutant cells exhibited a less-predominantly nuclear localization pattern (50% to 65%) under G1 arrest, which may reflect difficulties in achieving cell cycle synchronization of the triple mutant. Nevertheless, R2 became redistributed in the majority (>90%) of the cdc7Δ dbf4Δ mcm5-bob1 cells after they entered S phase, as in the wild-type cells. Thus, we concluded that DDK is not required for S-phase-specific redistribution of R2.
FIG 2.
S-phase-specific R2 redistribution requires Cdc28 (CDK) but not Cdc7 (DDK). (A) Wild-type, cdc7Δ mcm5-bob1, and cdc7Δ dbf4Δ mcm5-bob1 cells were synchronized in G1 phase before being released into the first S phase. Cells were harvested at 30 min and 60 min after the release and processed for flow cytometry (top) and immunofluorescence and quantitative analyses of Rnr4 subcellular localization patterns (bottom) as described in the legend to Fig. 1. (B) Asynchronously grown cdc28-as1 cells were synchronized in G1 and split into seven equal parts. One was harvested in G1, and another was collected 30 min after being released from G1, when cells entered S phase. For the remaining five cultures, 1-NM-PP1 was added at the indicated time points post-G1 release, and cells were collected 45 min post-G1 release for flow cytometry (left) and immunofluorescence and quantitative analyses of Rnr4 subcellular localization patterns (right) as described in the legend to Fig. 1.
To determine the role of CDK in R2 redistribution during S phase, we took advantage of the analog-sensitive allele cdc28-as1 that encodes a Cdc28 kinase with an enlarged ATP-binding pocket, allowing it to bind the nonhydrolyzable ATP analogue 1-NM-PP1 {4-amino-1-tert-butyl-3-(1′-naphthylmethyl)pyrazolo[3,4-d]pyrimidine}. Treatment of cells with 1-NM-PP1 triggers rapid and highly specific downregulation of Cdc28 kinase activity in vivo (29). The cdc28-as1 mutant cells were released from G1 arrest and the Cdc28 kinase was inhibited at different time points by the addition of 1-NM-PP1. All cells were collected 45 min after G1 release for Rnr4 immunofluorescence analysis. In the absence of 1-NM-PP1, the majority of the cdc28-as1 cells exhibited loss of a predominantly nuclear Rnr4 signal (from 75% to 30%) as they moved from G1 to S phase (Fig. 2B). Treatment with 1-NM-PP at earlier time points after G1 release effectively blocked the loss of Rnr4 from the nucleus (Fig. 2B, 0, 5, and 10 min). In contrast, adding 1-NM-PP1 at later time points (20 min after G1 release) had little effect on Rnr4 redistribution. Taken together, the results strongly indicate that Cdc28 kinase activity is required for R2 redistribution as cells move from G1 to S phase and that the execution point of Cdc28 is in early S phase, within 10 min of G1 release.
Clb6 but not Clb5 is required for R2 redistribution in S phase.
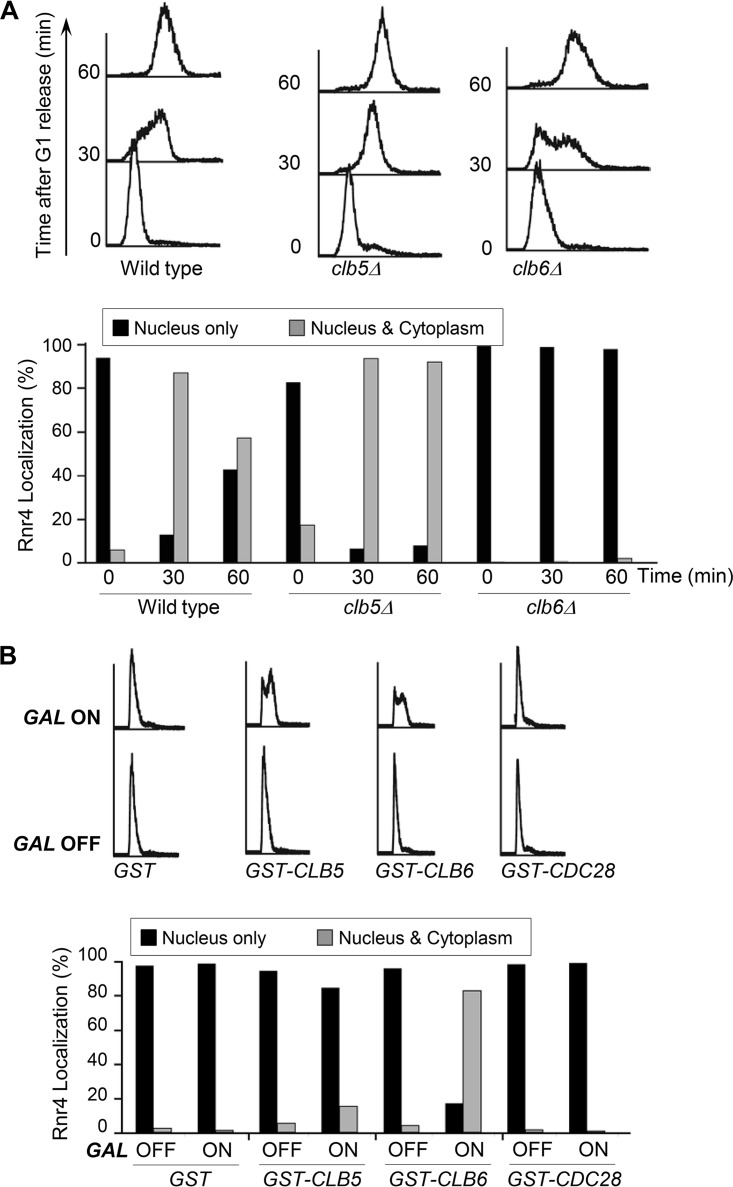
The transition from G1 into S phase is triggered by two early S-phase-specific cyclins, Clb5 and Clb6. To determine whether these two cyclins are involved in R2 redistribution, we compared the R2 subcellular localization patterns between clb5Δ, clb6Δ, and wild-type cells as they entered S phase. The clb5Δ mutant and the wild-type cells exhibited similar patterns of nucleus-to-cytoplasm redistribution of R2 (Fig. 3A). In contrast, the clb6Δ mutant clearly had a deficiency in R2 redistribution, with the majority (>90%) of cells still retaining R2 in the nucleus 30 to 60 min after being released from G1 arrest (Fig. 3A). Thus, we concluded that Clb6-Cdc28 but not Clb5-Cdc28 cyclin-dependent kinase activity is specifically required for R2 redistribution in S phase.
FIG 3.
S-phase-specific R2 redistribution is controlled by Clb6. (A) R2 redistribution in S phase is deficient in clb6Δ but not in clb5Δ mutant cells. Wild-type, clb5Δ, and clb6Δ cells were synchronized in G1 and released into S phase. Cells were collected at G1 and 30 min and 60 min after G1 release for flow cytometry (top) and indirect immunofluorescence (bottom) analyses as described in the legend to Fig. 1. (B) Overexpression of Clb6 but not Clb5 leads to nucleus-to-cytoplasm redistribution of R2 in α-factor-arrested G1 cells. Wild-type cells harboring plasmids expressing PGAL-GST, PGAL-GST-CLB5, PGAL-GST-CLB6, and PGAL-GST-CDC28 were grown to early-log phase in medium containing raffinose as the only carbon source (GAL OFF) and arrested in G1 by using α-factor. The PGAL promoter was turned on by the addition of 2% galactose (GAL ON) and further incubation for 1 h while maintaining α-factor in the medium. Cells were harvested for indirect immunofluorescence (top) and flow cytometry (bottom) analyses as described in the legend to Fig. 1.
Overexpression of Clb6 in G1-arrested cells is sufficient to drive R2 redistribution.
To further investigate the role of Clb6 in R2 relocalization, we transformed wild-type cells with plasmids harboring glutathione S-transferase (GST) fusions of CLB5, CLB6, and CDC28 that were under the control of the GAL1,10 promoter. The transformants were kept in the G1 phase by α-factor-mediated arrest in a medium containing raffinose as the sole carbon source (Fig. 3B, GAL OFF), and expression of the GST fusion proteins was induced by the addition of galactose to the medium (Fig. 3B, GAL ON). Within 1 h of GST-Clb6 induction, R2 shifted from a mostly nuclear signal to a more ubiquitous localization pattern (Fig. 3B). In contrast, R2 remained in the nucleus in cells overexpressing GST-Clb5 and GST-Cdc28. Importantly, although the induction of either GST-Clb6 or GST-Clb5 was sufficient to initiate DNA replication in the α-factor-arrested cells (Fig. 3B, top), only GST-Clb6 caused R2 exit from the nucleus, further confirming that R2 redistribution is specifically controlled by Clb6.
Clb6 is required for Rnr2 phosphorylation in S phase.
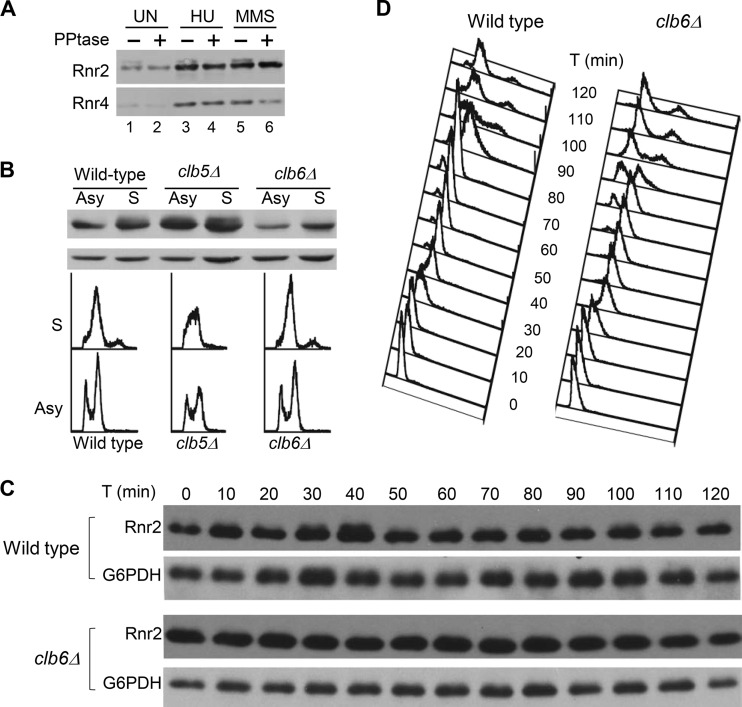
We noticed that a fraction of Rnr2 protein exhibited slower mobility on protein blots of yeast cell extract from asynchronous culture (Fig. 4A, lane 1). No different mobility species of Rnr4 protein were observed under the same experimental conditions. Treatment of cells with the RNR inhibitor hydroxyurea (HU) and the DNA-alkylating reagent methyl methanesulfonate (MMS) resulted in increased Rnr2 and Rnr4 protein levels, consistent with DNA damage checkpoint-mediated transcriptional induction of RNR2 and RNR4 (30). However, the relative ratio between the faster- and more-slowly migrating species of Rnr2 remained unchanged (Fig. 4A, lanes 3 and 5). The more-slowly migrating Rnr2 band was diminished by phosphatase treatment, indicating that it contained phosphorylated Rnr2 (Fig. 4A, lanes 2, 4, and 6). The lack of change in Rnr2 phosphorylation in HU- and MMS-treated cells suggests that the phosphorylation is not mediated by the DNA damage checkpoint kinase cascade Mec1-Rad53-Dun1.
FIG 4.
Rnr2 is phosphorylated in S phase in a Clb6-dependent manner. (A) Rnr2 is a phosphoprotein. Wild-type cells in early log phase were incubated with 125 mM HU or 0.025% of MMS or left untreated (UN) for 1 h before being harvested. Protein extracts were prepared and treated with lambda phosphatase (PPtase, +) or mock treated (−) at 37°C for 30 min before being resolved by SDS-PAGE, and the protein blot was probed with anti-Rnr2 and anti-Rnr4 antibodies. (B) Comparison of Rnr2 phosphorylation between wild-type, clb5Δ, and clb6Δ cells. Cells from early-log-phase culture were split into two halves; one was kept growing asynchronously (Asy), and the other was synchronized in S phase by collecting cells 30 min after being released from an α-factor-mediated G1 arrest. DNA content was analyzed by flow cytometry. Protein extracts were prepared and then resolved by SDS-PAGE, and the protein blot was probed with antibodies against Rnr2 (top) and glucose-6-phosphate dehydrogenase (G6PDH, bottom) as a loading control. (C) Comparison of Rnr2 phosphorylation between wild-type and clb6Δ cells during cell cycle progression. Wild-type and clb6Δ cells from early log-phase cultures were synchronized in G1 by using α-factor and released back into the cell cycle by washing out the α-factor at time zero. Cells were collected at 10-min interval throughout the first 2 h for flow cytometry analysis and Western blotting using anti-Rnr2 and anti-G6PDH antibodies.
To determine whether Rnr2 phosphorylation is cell cycle regulated, we compared Rnr2 protein blotting between G1- and S-phase-synchronized cells. The phosphorylation-dependent slower-mobility form of Rnr2 increased in S-phase cells in the wild-type and clb5Δ mutant strains but diminished in the clb6Δ mutant cells (Fig. 4B), suggesting that it is Clb6 regulated. To further confirm the role of Clb6, wild-type and clb6Δ mutant cells were released from α-factor-mediated G1 arrest and the levels of Rnr2 protein were monitored through the first cell cycle at 10-min intervals. Phosphorylated Rnr2 is enriched in the wild-type cells during the first S phase (30 to 40 min) and, to a lesser degree, during the second S phase (90 to 100 min). In contrast, no phosphorylated Rnr2 species was observed in the clb6Δ mutant at any time point (Fig. 4C). Taking these results together, we concluded that Rnr2 is phosphorylated in S phase in a Clb6-dependent manner.
Mutation resulting in phosphorylation-defective consensus CDK site in Rnr2 leads to nuclear retention of R2 subunit in S phase.
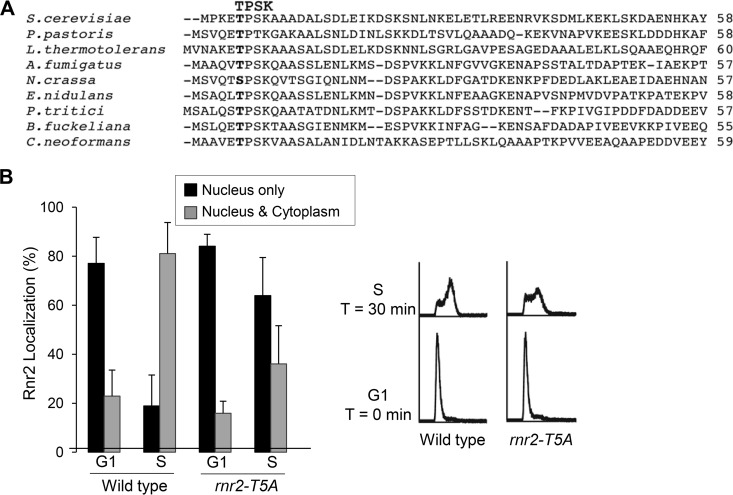
The consensus sequence for Cdc28 (Cdk1)-type CDK phosphorylation is S/T-P-X-K/R or S/T-P (31, 32). Rnr2 contains a single Cdc28 phosphorylation motif, TPSK, at its N terminus, which is conserved in many fungal relatives of S. cerevisiae (Fig. 5A). To determine whether the threonine-5 within this TPSK motif is involved in regulating Rnr2 redistribution in S phase, we generated a phosphorylation-deficient mutant allele, rnr2-T5A, and compared the subcellular localization patterns of Rnr2 wild-type and Rnr2-T5A mutant proteins as cells moved from G1 to S phase. In G1-arrested cells, Rnr2-T5A was primarily localized to the nucleus, just like the wild-type protein. However, as cells were released from G1 to S phase, Rnr2-T5A was much slower than the wild type in leaving the nucleus. At the 30-min time point after release from G1, Rnr2-T5A was still retained in the nucleus in >60% of the cells, while only ∼20% of the wild-type cells had a strong nuclear Rnr2 signal (Fig. 5B). Anti-Rnr4 antibody immunofluorescence staining revealed similar nuclear retention of Rnr4 in the rnr2-T5A mutant (data not shown). Thus, the consensus CDK phosphorylation site in Rnr2 is required for the nucleus-to-cytoplasm redistribution of Rnr2-Rnr4 in S phase.
FIG 5.
A consensus Cdk1 phosphorylation site at the N terminus of Rnr2 is required for S-phase-specific R2 redistribution. (A) Alignment of the N-terminal 55 to 60 residues of the Rnr2 orthologs from Saccharomyces cerevisiae, Pichia pastoris, Lactobacillus thermotolerans, Aspergillus fumigatus, Neurospora crassa, Emericella nidulans, Pyrenophora triticirepentis, Botryotinia fuckeliana, and Cryptococcus neoformans. The conserved TPSK motif is indicated by boldface. (B) Comparison of Rnr2 subcellular localization patterns in G1- and S-phase-synchronized wild-type and rnr2-T5A mutant cells. Cell cycle synchronization, flow cytometry, and indirect immunofluorescence were as described in the legend to Fig. 1A. Three independent samples of each strain were processed for anti-Rnr2 antibody staining, with >150 cells examined for each sample. The error bars represent standard deviations.
Both clb6Δ and phosphorylation-defective rnr2-T5A mutants have enhanced Rnr2-Wtm1 interaction in vivo.
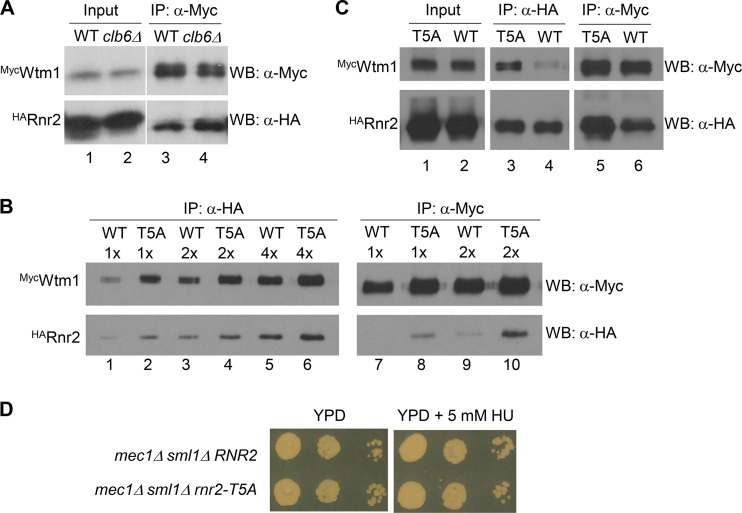
The increased nuclear localization of R2 in clb6Δ mutant cells may result from increased Dif1-facilitated nuclear import, enhanced Wtm1-mediated nuclear retention, or a combination of both. Since we did not observe any increase in Dif1 protein levels in clb6Δ cells relative to the levels in wild-type cells (data not shown), we surmised that the clb6Δ mutant may have an increased Wtm1-Rnr2 association. To test this hypothesis, we tagged the genomic copies of WTM1 and RNR2 with an N-terminal MYC and a hemagglutinin (HA) epitope, respectively, and probed the in vivo Wtm1-Rnr2 interaction by coimmunoprecipitation. We found that more HARnr2 was brought down with the same amount of MycWtm1 in the anti-Myc antibody immunocomplexes from the asynchronously grown clb6Δ mutant than from the wild-type cells (Fig. 6A, lanes 3 and 4), which is consistent with a stronger Wtm1-Rnr2 association in the clb6Δ mutant.
FIG 6.
The Rnr2-Wtm1 interaction is weakened by clb6Δ and phosphorylation-defective mutation of the consensus CDK site in Rnr2. (A) The clb6Δ mutant has increased Wtm1-Rnr2 interaction relative to the level in the wild type. MycWTM1 HARNR2 wild-type (WT) and MycWTM1 HARNR2 clb6Δ mutant (clb6Δ) cells were grown to early log phase before being harvested for protein extract preparation. For each sample, 500 μg of total protein extract was subjected to immunoprecipitation (IP) using a monoclonal anti-Myc (9E10) antibody. Immunocomplexes were resolved by SDS-PAGE, and the protein blot was probed with rabbit polyclonal anti-Myc and anti-HA antibodies. WB, Western blotting. (B) T5A substitution in Rnr2 increases the Wtm1-Rnr2 interaction in vivo. Protein extracts were prepared from asynchronously grown wild-type (WT, MycWTM1 HARNR2) and T5A mutant (MycWTM1 HArnr2-T5A) cells. Amounts of 500 μg each of protein extracts were subjected to immunoprecipitation using monoclonal anti-HA (12CA5) and anti-Myc (9E10) antibodies. Immunocomplexes were serially diluted, resolved by SDS-PAGE, and probed with rabbit polyclonal anti-Myc and anti-HA antibodies, respectively. (C) Comparison of Wtm1-Rnr2 and Wtm1–Rnr2-T5A interactions in S phase cells is shown. The MycWTM1, HARNR2 (WT) and MycWTM1, HArnr2-T5A (T5A) cells were synchronized in S phase by being released from α-factor-mediated G1 arrest and collected 30 min later. Protein extraction, immunoprecipitation, and Western blotting were as described in the legend to panel B. For Input lanes, 10 μg of total protein extract was loaded for each sample. (D) The rnr2-T5A mutant allele enhances sensitivity to HU in the mec1Δ sml1Δ background. Tenfold serial dilutions of mec1Δ sml1Δ and mec1Δ sml1Δ rnr2-T5A mutant cells from asynchronously growing cultures were plated on YPD plates without or with 5 mM hydroxyurea (HU). Images were taken after incubation at 30°C for 2 days.
We then asked whether (phosphorylation of) threonine-5 of the TPSK motif in Rnr2 is involved in regulation of the Wtm1-Rnr2 interaction by comparing reciprocal Wtm1-Rnr2 coimmunoprecipitation between wild-type and rnr2-T5A mutant cells. We first performed immunoprecipitation in cells from asynchronously grown cultures. Serial dilution and protein blotting revealed that more MycWtm1 was brought down with the same amount of HARnr2 in the anti-HA antibody immunocomplexes (Fig. 6A, lane 3 versus lane 4), and more HARnr2 was brought down with the same amount of MycWtm1 in the anti-Myc antibody immunocomplexes (Fig. 6B, lane 8 versus lane 9). The increased Wtm1–Rnr2-T5A interaction is more obvious in cells that were synchronized in S phase (Fig. 6C, lane 3 versus lane 4 and lane 5 versus lane 6). Together, our data demonstrate that the phosphorylation-deficient Rnr2-T5A mutant protein has a strong association with Wtm1 in vivo.
rnr2-T5A mutation increases HU sensitivity in a mec1Δ sml1Δ background.
We have shown previously that Rnr1 is constitutively cytoplasmic, while Rnr2-Rnr4 resides predominantly in the nucleus except during replication or after DNA damage (17). Given that the rnr2-T5A mutant exhibited increased/prolonged nuclear retention of Rnr2-Rnr4, we predicted that these cells should be more sensitive than the wild type to the effect of the RNR inhibitor HU. To test this, we introduced RNR2 and rnr2-T5A alleles into a mec1Δ sml1Δ background to avoid checkpoint activation and, therefore, to separate the effect of transcriptional induction of the RNR genes from the localization of Rnr2-Rnr4. As shown by the results in Fig. 6D, the rnr2-T5A mutation showed no effect on growth in the absence of HU, but it significantly compromised the viability of the mec1Δ sml1Δ mutant on a 5 mM HU plate compared to the effect of the wild-type RNR2 allele. The increased HU sensitivity of the rnr2-T5A mutant was not due to a decreased level of Rnr2 protein, as we saw no difference in Rnr2 levels between RNR2 and rnr2-T5A strains (data not shown).
DISCUSSION
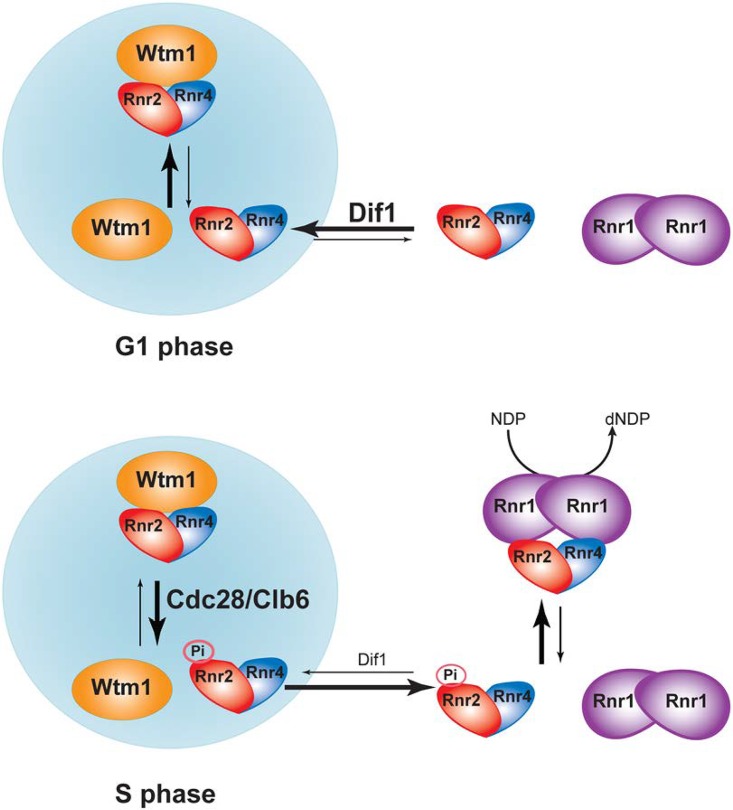
Optimization of cellular dNTP concentrations is important for high-fidelity DNA replication and repair. A main prerequisite for cells to enter S phase is to enlarge their dNTP pools in order to meet the demand of genome duplication, which is achieved largely through upregulation of RNR. In addition to the increase of RNR gene transcription and proteolysis of the RNR inhibitor Sml1 (24, 33) at entry into S phase, the budding yeast further enhances RNR activity by promoting nuclear release of the heterodimeric R2 subunit Rnr2-Rnr4 so it can form the active holoenzyme with the cytoplasmic R1. In this study, we investigated the mechanism underlying the checkpoint-independent, S-phase-specific R2 redistribution. The amount of R2 in the nucleus is a net outcome of its nuclear import, facilitated by Dif1, nuclear retention by Wtm1, and nuclear export. Our results support a model in which the S-phase cyclin-CDK complex Clb6-Cdc28 phosphorylates Rnr2 at its N-terminal TPSK motif, which weakens the Rnr2-Wtm1 interaction and facilitates the release of preexisting Rnr2-Rnr4 from the nucleus (Fig. 7). The nucleus-to-cytoplasm R2 redistribution is further facilitated by S-phase-specific proteolysis of Dif1 (21), leading to decreased nuclear import of newly translated R2. Consistent with this model, a previous study has shown that deletion of CLB6 makes the mec1Δ sml1Δ mutant more sensitive to HU (34), similar to the effect of the phosphorylation-deficient T5A mutant of Rnr2 (Fig. 6D). On the other hand, overexpression of CLB6 can partially suppress the HU sensitivity of the chk1Δ dun1Δ checkpoint double mutant (35), which may be partially accounted for by an increase in cytoplasmic localization of R2.
FIG 7.
A model illustrating Cdc28/Clb6-mediated nucleus-to-cytoplasm redistribution of R2 subunit during G1-to-S-phase transition.
Intriguingly, our results demonstrate that the redistribution of R2 in S phase is specifically controlled by Clb6 but not its paralog Clb5, with which it shares 50% primary sequence identity (36). CLB5 and CLB6 are named S-phase cyclins because they both peak at S-phase entry and play major roles in initiating DNA replication. The Clb5 protein level remains elevated throughout the S phase, whereas Clb6 is rapidly degraded in early unperturbed S phase via an SCFCdc4- and anaphase promoter complex (APC)-dependent pathway during normal cell cycle progression (37, 38). Consistent with the different expression patterns, Clb5 is required for the firing of both early and late replication origins, while Clb6 is only involved in the firing of the early origins (37). Although the clb6Δ mutant shows no apparent S-phase defect, as opposed to the clb5Δ mutant, the clb5Δ clb6Δ double mutant exhibits a more severe S-phase delay and an increased HU sensitivity relative to those of each single mutant (39, 40), suggesting overlapping albeit distinct roles of the two cyclins (39). It is likely that the Clb5-Cdc28 and Clb6-Cdc28 kinase complexes differ in their specificities toward a subset of target substrates while sharing most others. A previous study has shown that Clb6-Cdc28 but not Clb5-Cdc28 triggers nuclear export of transcription factor Swi6 in early S phase by specifically phosphorylating Swi6 at serine-160, thus altering Swi4/Swi6 (SBF) and Mbp1/Swi6 (MBF)-mediated transcription at the G1/S transition (41). Our results indicate that the phosphorylation of Rnr2, like that of Swi6, is specifically dependent on Clb6-Cdc28 and triggers nuclear release of the R2 subunit.
Regulation of the mammalian RNR by phosphorylation of its two R2 subunits, RRM2 and p53R2, has also been reported. RRM2, the main R2 subunit in proliferating cells, peaks in S phase and is degraded in G2 upon completion of DNA replication. The G2 proteolysis of RRM2 is triggered by CDK-mediated phosphorylation of Thr-33 (42). RRM2 contains an additional CDK phosphorylation site, Ser-20 (43), although the physiological significance of this phosphorylation is unclear. The DNA damage-inducible p53R2 is phosphorylated by ATM at Ser-72 in response to UV irradiation, which increases p53R2's stability and cellular survival under genotoxic stress (44). p53R2 has also been shown to interact with kinase MEK2 (extracellular signal-regulated kinase [ERK] kinase 2/mitogen-activated protein [MAP] kinase kinase 2), which is required for serum-stimulated increase in RNR activity (45). It would thus appear that phosphorylation of an RNR subunit is a common theme of RNR regulation both during normal cell cycle progression and in response to DNA damage.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth conditions.
The yeast strains and plasmids used in this study are listed in Table 1. The growth of yeast strains and genetic manipulations were performed as previously described (46). The rich yeast extract-peptone-dextrose (YPD) medium contained 1% Bacto yeast extract, 2% Bacto peptone, and 2% glucose. The synthetic complete (SC) medium contained 0.17% yeast nitrogen base without amino acids and (NH4)2SO4 (catalog number Y20060; Research Products International), 0.5% (NH4)2SO4, and all 20 amino acids at concentrations as described previously (46); the carbon source used was 2% glucose, raffinose, or galactose. Selective (i.e., dropout) media were SC omitting one or multiple amino acids. For solid media, 2% Bacto agar was added before autoclaving. Hydroxyurea (HU) (product number H8627; Sigma-Aldrich) and methyl methanesulfonate (MMS) (product number M4016; Sigma-Aldrich) were added to the media at final concentrations of 125 mM and 0.025%, respectively. 1-NM-PP1 (catalog number A603003; Toronto Research Chemicals, Inc.) was added to the cdc28-as1 culture at a final concentration of 30 μM. For cell cycle synchronization experiments, α-factor (catalog number RP01002; GenScript) was used at a final concentration of 10 μg/ml.
TABLE 1.
Yeast strains and plasmids used in this study
| Strain or plasmid | Relevant description | Reference or source |
|---|---|---|
| Strains | ||
| Y300 | MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 17 |
| MHY26 | Y300 dun1::HIS3 | This study |
| MHY363 | Y300 sml1::KAN | 22 |
| MHY365 | Y300 sml1::HIS3 mec1::HIS3 | This study |
| AXY2466 | Y300 sml1::HIS3 mec1::HIS3 rnr2::KAN pMH813 | This study |
| AXY2488 | Y300 sml1::HIS3 mec1::HIS3 rnr2::KAN pMH1653 | This study |
| XWY58 | Y300 clb5::KAN | This study |
| XWY61 | Y300 rnr2::KAN pMH1653 | This study |
| XWY65 | Y300 rnr2::KAN pMH800C | This stud |
| XWY77 | Y300 clb6::KAN | This study |
| XWY86 | Y300 wtm1::MYC-WTM1 rnr2::KAN pXW97 | This study |
| XWY88 | Y300 wtm1::MYC-WTM1 rnr2::KAN pMH1387 | This study |
| SLJ1386 | MATa bar1 cdc28-as1 | M. Winey |
| P211 | MATa ura3 lys2 cyh2 his3 leu2 bob1-1 cdc7Δ1::HIS3 | R. Sclafani |
| P235 | MATa ura3 lys2 cyh2 his3 leu2 bob1-1 cdc7Δ1::HIS3 dbf4Δ1::URA3 | R. Sclafani |
| RSY743 | MATa trp1 leu2 ade1 arg4 his3 1 his7 cyh2 | R. Sclafani |
| RSY755 | RSY743 clb5::ARG4 | R. Sclafani |
| RSY756 | RSY743 clb6::ADE1 | R. Sclafani |
| Plasmids | ||
| pRS314 | CEN TRP1 | 47 |
| pRS415 | CEN LEU2 | 47 |
| pRS424 | 2μm TRP1 | 47 |
| pXW80 | pRS424-PGAL1,10-GST-CLB5 | This study |
| pXW81 | pRS424-PGAL1,10-GST-CLB6 | This study |
| pXW82 | pRS424-PGAL1,10-GST-CDC28 | This study |
| pXW97 | pRS415-PRNR2-HA-rnr2(T5A) | This study |
| pMH762 | pRS424-PGAL1,10-GST | This study |
| pMH800C | pRS314-PRNR2-3×Myc-RNR2 | This study |
| pMH813 | pRS415-PRNR2-3×Myc-RNR2 | 48 |
| pMH1387 | pRS415-PRNR2-HA-RNR2 | This study |
| pMH1653 | pRS314-PRNR2-3×Myc-rnr2(T5A) | This study |
Indirect immunofluorescence.
Preparation of yeast spheroplasts, immunofluorescence staining, image acquisition, and quantitative analysis of subcellular localization patterns were performed as previously described (7). DNA was visualized by staining with 1 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI) (product number D9542; Sigma-Aldrich). The polyclonal anti-Rnr2 and anti-Rnr4 antibodies used in immunostaining were described previously (17).
Flow cytometry.
Amounts of 0.5 × 107 to 1.0 × 107 cells of each sample were fixed in 1 ml of 70% ethanol for 30 min. The fixed cells were resuspended in 1 ml of 1× phosphate-buffered saline (PBS), pH 7.4, for 1 h for rehydration. The rehydrated cells were resuspended in 100 μl of FACS buffer (0.2 M Tris-HCl, pH 8.0, 20 mM EDTA, pH 8.0) with the addition of 0.1% RNase A and incubated for 4 h at 37°C. The cells were then resuspended in 100 μl of 1× PBS, pH 7.4, with 50 μg/ml of propidium iodide (PI; Sigma) for DNA staining for 1 h at room temperature. Before flow cytometry, PI-stained cells were diluted with the addition of 900 μl of 1× PBS, pH 7.4, and sonicated briefly (20% output for 10 s) on a sonicator (Sonifier 250) to break up aggregated cells. For each sample, ∼10,000 cells were scanned in a Beckman Coulter Epics XL MCL flow cytometer, and the data were imported into and processed with DeltaGraph (RedRock).
Protein extraction, immunoprecipitation, and phosphatase treatment.
Protein extracts were prepared by using glass bead disruption in a Bullet Blender (Next Advance). Two different extraction solutions were used. For direct immunoblotting detection of steady-state protein levels, trichloroacetic acid (TCA) was employed to extract protein from 1 × 107 to 1 × 108 cells during the cell cycle for each loading as described previously (7). For immunoprecipitation and phosphatase treatment, cells were lysed in a protein lysis buffer of 50 mM Tris-HCl, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, supplemented with 1× protease inhibitor cocktail (Roche Applied Science). Protein extracts were then centrifuged at 13,400 × g for 15 min to remove debris, and protein concentrations were determined by using the Bradford protein assay (Bio-Rad). For phosphatase treatment, 25 to 50 μg of total protein extracts was incubated with 200 units of lambda phosphatase (New England BioLabs) at 37°C for 30 min. All immunoprecipitation steps were performed at 4°C. For each sample, 0.5 to 1 mg of total protein extract was diluted to a final volume of 200 μl with the protein lysis buffer, and the mixture incubated with primary antibodies (1:100 dilution) overnight. The antibody-protein complexes were precipitated by absorption to protein A-Sepharose beads for 4 h and washed twice with a high-salt buffer (50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 1% Triton X-100, 1 mM DTT). Proteins were separated by 8-to-10% SDS-PAGE, transferred to a nitrocellulose membrane, and probed with primary and secondary antibodies. Blots were developed by using an enhanced chemiluminescence substrate (Perkin-Elmer).
Monoclonal anti-Myc (9E10) and anti-HA (12CA5) antibodies were purchased from Covance and Roche Applied Sciences, respectively. Rabbit polyclonal anti-Myc and anti-HA antibodies were from Santa Cruz Biotechnology. Horseradish peroxidase (HRP)- and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse and goat anti-rabbit antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc. Polyclonal anti-GST and anti-Zwf1 (glucose-6-phosphate dehydrogenase [G6PDH]) antibodies were from Sigma.
ACKNOWLEDGMENTS
We thank Robert Sclafani and Mark Winey for their generosity in sharing yeast strains.
This work was supported by National Institutes of Health grant number CA125574 to M.H.
REFERENCES
- 1.Mathews CK. 2015. Deoxyribonucleotide metabolism, mutagenesis and cancer. Nat Rev Cancer 15:528–539. doi: 10.1038/nrc3981. [DOI] [PubMed] [Google Scholar]
- 2.Nordlund P, Reichard P. 2006. Ribonucleotide reductases. Annu Rev Biochem 75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 3.Cotruvo JA, Stubbe J. 2011. Class I ribonucleotide reductases: metallocofactor assembly and repair in vitro and in vivo. Annu Rev Biochem 80:733–767. doi: 10.1146/annurev-biochem-061408-095817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang M, Parker MJ, Stubbe J. 2014. Choosing the right metal: case studies of class I ribonucleotide reductases. J Biol Chem 289:28104–28111. doi: 10.1074/jbc.R114.596684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang C, Liu G, Huang M. 2014. Ribonucleotide reductase metallocofactor: assembly, maintenance and inhibition. Front Biol (Beijing) 9:104–113. doi: 10.1007/s11515-014-1302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perlstein DL, Ge J, Ortigosa AD, Robblee JH, Zhang Z, Huang M, Stubbe J. 2005. The active form of the Saccharomyces cerevisiae ribonucleotide reductase small subunit is a heterodimer in vitro and in vivo. Biochemistry 44:15366–15377. doi: 10.1021/bi051616+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.An X, Zhang Z, Yang K, Huang M. 2006. Cotransport of the heterodimeric small subunit of the Saccharomyces cerevisiae ribonucleotide reductase between the nucleus and the cytoplasm. Genetics 173:63–73. doi: 10.1534/genetics.105.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chabes A, Stillman B. 2007. Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 104:1183–1188. doi: 10.1073/pnas.0610585104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niida H, Shimada M, Murakami H, Nakanishi M. 2010. Mechanisms of dNTP supply that play an essential role in maintaining genome integrity in eukaryotic cells. Cancer Sci 101:2505–2509. doi: 10.1111/j.1349-7006.2010.01719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabouri N, Viberg J, Goyal DK, Johansson E, Chabes A. 2008. Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic Acids Res 36:5660–5667. doi: 10.1093/nar/gkn555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanvisens N, Bano MC, Huang M, Puig S. 2011. Regulation of ribonucleotide reductase in response to iron deficiency. Mol Cell 44:759–769. doi: 10.1016/j.molcel.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Z, Elledge SJ. 1992. Isolation of crt mutants constitutive for transcription of the DNA damage inducible gene RNR3 in Saccharomyces cerevisiae. Genetics 131:851–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang M, Zhou Z, Elledge SJ. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94:595–605. doi: 10.1016/S0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 14.Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J 20:3544–3553. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T. 2003. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev 17:1130–1140. doi: 10.1101/gad.1090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue L, Zhou B, Liu X, Qiu W, Jin Z, Yen Y. 2003. Wild-type p53 regulates human ribonucleotide reductase by protein-protein interaction with p53R2 as well as hRRM2 subunits. Cancer Res 63:980–986. [PubMed] [Google Scholar]
- 17.Yao R, Zhang Z, An X, Bucci B, Perlstein DL, Stubbe J, Huang M. 2003. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc Natl Acad Sci U S A 100:6628–6633. doi: 10.1073/pnas.1131932100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lincker F, Philipps G, Chaboute ME. 2004. UV-C response of the ribonucleotide reductase large subunit involves both E2F-mediated gene transcriptional regulation and protein subcellular relocalization in tobacco cells. Nucleic Acids Res 32:1430–1438. doi: 10.1093/nar/gkh310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YD, Elledge SJ. 2006. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev 20:334–344. doi: 10.1101/gad.1380506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, An X, Yang K, Perlstein DL, Hicks L, Kelleher N, Stubbe J, Huang M. 2006. Nuclear localization of the Saccharomyces cerevisiae ribonucleotide reductase small subunit requires a karyopherin and a WD40 repeat protein. Proc Natl Acad Sci U S A 103:1422–1427. doi: 10.1073/pnas.0510516103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YD, Wang J, Stubbe J, Elledge SJ. 2008. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol Cell 32:70–80. doi: 10.1016/j.molcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X, Huang M. 2008. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol Cell Biol 28:7156–7167. doi: 10.1128/MCB.01388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev 12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X, Muller EG, Rothstein R. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell 2:329–340. doi: 10.1016/S1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 25.Toone WM, Aerne BL, Morgan BA, Johnston LH. 1997. Getting started: regulating the initiation of DNA replication in yeast. Annu Rev Microbiol 51:125–149. doi: 10.1146/annurev.micro.51.1.125. [DOI] [PubMed] [Google Scholar]
- 26.Labib K. 2010. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev 24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendenhall MD, Hodge AE. 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 62:1191–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sclafani RA, Tecklenburg M, Pierce A. 2002. The mcm5-bob1 bypass of Cdc7p/Dbf4p in DNA replication depends on both Cdk1-independent and Cdk1-dependent steps in Saccharomyces cerevisiae. Genetics 161:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 30.Huang MX, Elledge SJ. 1997. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol Cell Biol 17:6105–6113. doi: 10.1128/MCB.17.10.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennelly PJ, Krebs EG. 1991. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem 266:15555–15558. [PubMed] [Google Scholar]
- 32.Enserink JM, Kolodner RD. 2010. An overview of Cdk1-controlled targets and processes. Cell Div 5:11. doi: 10.1186/1747-1028-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elledge SJ, Davis RW. 1990. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev 4:740–751. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- 34.Manfrini N, Gobbini E, Baldo V, Trovesi C, Lucchini G, Longhese MP. 2012. G(1)/S and G(2)/M cyclin-dependent kinase activities commit cells to death in the absence of the S-phase checkpoint. Mol Cell Biol 32:4971–4985. doi: 10.1128/MCB.00956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caldwell JM, Chen Y, Schollaert KL, Theis JF, Babcock GF, Newlon CS, Sanchez Y. 2008. Orchestration of the S-phase and DNA damage checkpoint pathways by replication forks from early origins. J Cell Biol 180:1073–1086. doi: 10.1083/jcb.200706009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwob E, Nasmyth K. 1993. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev 7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- 37.Jackson LP, Reed SI, Haase SB. 2006. Distinct mechanisms control the stability of the related S-phase cyclins Clb5 and Clb6. Mol Cell Biol 26:2456–2466. doi: 10.1128/MCB.26.6.2456-2466.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu SY, Kuan VJ, Tzeng YW, Schuyler SC, Juang YL. 2016. The anaphase-promoting complex works together with the SCF complex for proteolysis of the S-phase cyclin Clb6 during the transition from G1 to S phase. Fungal Genet Biol 91:6–19. doi: 10.1016/j.fgb.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Kuhne C, Linder P. 1993. A new pair of B-type cyclins from Saccharomyces cerevisiae that function early in the cell cycle. EMBO J 12:3437–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu WS, Erickson SL, Tsai HJ, Andrews CA, Vas AC, Clarke DJ. 2011. S-phase cyclin-dependent kinases promote sister chromatid cohesion in budding yeast. Mol Cell Biol 31:2470–2483. doi: 10.1128/MCB.05323-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geymonat M, Spanos A, Wells GP, Smerdon SJ, Sedgwick SG. 2004. Clb6/Cdc28 and Cdc14 regulate phosphorylation status and cellular localization of Swi6. Mol Cell Biol 24:2277–2285. doi: 10.1128/MCB.24.6.2277-2285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D'Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, Saraf A, Florens L, Washburn MP, Pagano M. 2012. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell 149:1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan AK, Persad S, Litchfield DW, Wright JA. 1999. Ribonucleotide reductase R2 protein is phosphorylated at serine-20 by P34cdc2 kinase. Biochim Biophys Acta 1448:363–371. doi: 10.1016/S0167-4889(98)00115-3. [DOI] [PubMed] [Google Scholar]
- 44.Chang L, Zhou B, Hu S, Guo R, Liu X, Jones SN, Yen Y. 2008. ATM-mediated serine 72 phosphorylation stabilizes ribonucleotide reductase small subunit p53R2 protein against MDM2 to DNA damage. Proc Natl Acad Sci U S A 105:18519–18524. doi: 10.1073/pnas.0803313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piao C, Youn CK, Jin M, Yoon SP, Chang IY, Lee JH, You HJ. 2012. MEK2 regulates ribonucleotide reductase activity through functional interaction with ribonucleotide reductase small subunit p53R2. Cell Cycle 11:3237–3249. doi: 10.4161/cc.21591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burke D, Dawson D, Stearns T. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual, 2000 ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 47.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, An X, Stubbe J, Huang M. 2013. Investigation of in vivo roles of the C-terminal tails of the small subunit (ββ') of Saccharomyces cerevisiae ribonucleotide reductase: contribution to cofactor formation and intersubunit association within the active holoenzyme. J Biol Chem 288:13951–13959. doi: 10.1074/jbc.M113.467001. [DOI] [PMC free article] [PubMed] [Google Scholar]