Abstract
Background
F-box protein 32 (FBXO32) (also known as atrogin-1), a member of the F-box protein family, was recently shown to be a transforming growth factor beta (TGF-β)/Smad4 target gene involved in regulating cell survival. It can be transcriptionally silenced by epigenetic mechanisms in some cancers, but its role in colorectal carcinoma (CRC) is unclear. We investigated the role of FBXO32 in CRC and determined its prognostic significance.
Material/Methods
We used real-time quantitative PCR, Western blot, and immunohistochemistry to elucidate the role of FBXO32 in clinical specimens and primary CRC cell lines. Differences in patient survival were determined by the Kaplan-Meier method and log-rank test.
Results
We found that the FBXO32 and SMAD4 levels were higher in normal tissues than in CRC tissues, but PAI-1 and VEGF levels showed the opposite pattern. The expressions of FBXO32 and SMAD4 were related to clinicopathological parameters in CRC. Kaplan-Meier analyses showed that the 5-year overall survival of the low-FBXO32 expression group was significantly shorter than that of the high-FBXO32 expression group (p=0.010).
Conclusions
The fbxo32 gene is a novel tumor suppressor that inhibits CRC progression by inducing differentiation. Elevated expression of FBXO32 predicts longer survival in CRC patients.
MeSH Keywords: Cell Survival, Colorectal Neoplasms, F-Box Proteins, Smad4 Protein, Transforming Growth Factor beta1
Background
Colorectal carcinoma (CRC) is the third most common cancer and the fourth most common cause of cancer mortality worldwide [1]. From a genetic perspective, CRC is one of the best understood neoplasms, but it is still the second most common cause of cancer-related death. The diagnosis and treatment of CRC have been significantly improved in recent years, leading to a substantial reduction in cancer-related mortality [2]. However, CRC is still a major worldwide public health problem. Several biomarkers have been shown to exhibit tumor suppressor functions in CRC [3].
FBXO32 (F-box protein 32) is a member of the F-box protein family, which constitutes 1 of the 4 subunits of the ubiquitin protein ligase complex involved in muscle atrophy [4,5]. FBXO32 is very strongly induced in many catabolic states and plays an important role in the generation of muscle atrophy [6]. Moreover, reports have shown that FBXO32 contributes to 3-deazaneplanocin A-induced apoptosis in breast cancer cells, and thus, it may also regulate cell survival [7]. Recent findings have demonstrated that fbxo32 may regulate cell survival and act as a potential tumor suppressor [8]. A previous study identified fbxo32 as a transforming growth factor-β (TGF-β)/Smad4 signalling pathway target gene, and it was transcriptionally silenced by epigenetic mechanisms in some types of carcinomas [8]. The TGF-β signalling pathway plays a key role in the regulation of cell proliferation, migration, and survival, which affect multiple biological processes, including carcinogenesis, fibrosis and wound healing [9]. As a previous study showed that fbxo32 has a key role in regulating apoptosis in ovarian cancer [10], we investigated whether fbxo32 induces apoptosis in CRC. However, few studies have examined the consequences of TGF-β signalling in CRC. Recently, we identified a TGF-β/Smad4 target fbxo32, which was expressed in normal colorectal cells but was epigenetically silenced in CRC cells showing dysregulated TGF-β signalling. These and several other results have suggested that dysregulation of a signalling pathway may cause epigenetic silencing of downstream targets [11]. Smad4, which is a tumor suppressor gene, is a central mediator in the signalling pathways of the TGF-β superfamily. Smad4 acts as a common partner of activated Smads to help execute their function by interacting with nuclear-pore complexes. Inactivation of the smad4 gene through mutation occurs frequently in pancreatic and CRCs and is associated with malignant progression of cancers [12].
In this study, we focused on the FBXO32 expression in CRC and established the relationship between FBXO32 expression and clinicopathologic features. We also investigated the role of fbxo32 in the regulation of CRC progression in vitro and evaluated the prognostic role of fbxo32 in CRC patients.
Material and Methods
Clinical and tissue samples
We chose primary tumor samples (n=122) and matched corresponding normal mucosa (n=43) from patients between July 2006 and June 2017 in the Linyi Peoples’ Hospital and Qilu Hospital of Shandong University (Table 1). Follow-up was conducted by telephone visit. The patients who died of diseases not directly related to their CRC or due to unexpected events were excluded from this study. The pathological diagnosis was conducted before surgery and validated after surgery. CRC samples for use in scientific research were collected with permission from the patients. All patients provided written informed consent, and our study was approved by the institutional review boards of all institutions involved.
Table 1.
Clinicopathological characteristics of patients with CRC (n=122).
| Characteristic | Value | % |
|---|---|---|
| Age median (range), years | 63 (21–85) | |
| Gender | ||
| Male | 73 | 59.84 |
| Female | 49 | 40.16 |
| Tumour Nodes Metastases category | ||
| T1 | 2 | 1.64 |
| T2 | 11 | 9.02 |
| T3 | 108 | 88.52 |
| T4 | 1 | 0.82 |
| N0 | 72 | 59.02 |
| N1 | 34 | 27.87 |
| N2 | 16 | 13.11 |
| M0 | 121 | 99.18 |
| M1 | 1 | 0.82 |
| American Joint Committee on Cancer category | ||
| I | 8 | 6.55 |
| II | 64 | 52.46 |
| III | 49 | 40.16 |
| IV | 1 | 0.82 |
| Differentiation | ||
| Low differentiation | 21 | 17.21 |
| Moderate differentiation | 97 | 79.51 |
| High differentiation | 4 | 3.28 |
Histology of all tumour specimens and regional lymph nodes was confirmed with haematoxylin–eosin staining according to the International Union against Cancer Tumour Nodes Metastases classification. Union for International Cancer Control released the eighth edition of the TNM staging of malignancies. AJCC is a classification system developed by the American Joint Committee on Cancer for describing the extent of disease progression in cancer patients.
Materials
RpMI-1640, DMEM (high glucose), McCoy’s 5A and fetal bovine serum (FBS) were obtained from Invitrogen (Gibco, USA). RNeasy RNA kits were purchased from TIANGEN Biotechnology Co., Ltd. (Beijing, China). The neutralizing anti-human FBXO32 monoclonal antibody and anti-GAPDH antibody were from Abcam, Inc. (CA, USA). An immunohistochemistry (IHC) kit was obtained from Maixin Biological Co. (Fuzhou, China).
Immunohistochemistry
FBXO32 and SMAD4 were stained with the two-step staining method of EnVision™, and the major steps are described as follows. Tissue sections were dewaxed in xylene, rehydrated in alcohol, and immersed in 3% hydrogen peroxide for 10 min to suppress endogenous peroxidase activity. Antigen retrieval was conducted by heating each section at 100°C for 30 min in 0.01 mol/l sodium citrate buffer (pH 6.0). After three 5-min rinses in phosphate-buffered saline (PBS), the sections were incubated for 1 h at room temperature with a mouse polyclonal anti-FBXO32 antibody (Epitomics, UK) diluted 1: 50 in PBS, and the bound antibodies were detected with a streptavidin-biotin-peroxidase system (Dako, USA) and 3,3′-diaminobenzidine substrate-chromogen solution (Dako, USA). The slides were counterstained with hematoxylin and inspected by an experienced pathologist. The staining area was scored as 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%), or 4 (76–100%) based on the percentage of positively stained cells [13]. The images were captured with an inverted Nikon Eclipse TE2000-S microscope (Tokyo, Japan) with 200× magnification, and then, the presence of green fluorescence indicative of a-SMA was qualitatively analyzed by 2 investigators blinded to the study. The degree and intensity of staining were independently assessed by 2 pathologists in a blinded manner.
Cell Culture
We purchased the human CRC cell lines HT-29, HCT-116 and Caco-2 from the Institute of Cell and Biochemistry, Chinese Academy of Sciences (Shanghai, China), and the normal human cell line NCM-460 from the American Type Culture Collection (ATCC). We plated HCT-116 and NCM-460 cells in culture plates with McCoy’s 5A medium (Gibco, USA), which was supplemented by 10% FBS (Gibco, USA) and penicillin/streptomycin (Invitrogen) at 100 U/ml; Caco-2 was propagated DMEM (high glucose) medium (Gibco, USA) containing 20% FBS; and HT-29 was cultured in RPMI-1640 medium (Gibco, USA) with 10% FBS. All cells were incubated in a humidified incubator at 37°C in 5% CO2.
Quantitative real-time PCR (qRT-PCR) assay
QRT-PCR assays were performed to detect the expression of fbxo32, smad4, pai-1, and vegf mRNA in NCM460, HCT116, HT-29, and Caco-2 cells. Total RNA was separated from cells by the centrifugal column method (TIANGEN Biotech, Beijing, China), reverse transcribed into cDNA using the Superscript first strand synthesis kit (Invitrogen) in accordance with the manufacturer’s protocol, and amplified by qRT-PCR using a Mini-Opticon real-time PCR detection system (ABI, CA, USA) in SYBR Green II master mix (GeneCopoeia, Guangzhou, China) as described by the manufacturer’s instructions. All data were analyzed by Opticon Monitor software (ABI; 7500). All primer sequences are listed in Table 2.
Table 2.
Primers for real-time PCR.
| Gene | Accession number | Primer sequence | Product length (bp) |
|---|---|---|---|
| FBXO32 | NM_017617.3 | F: 5′-CCTTTGTGCTTCTGTTCTTCG-3′ | 212 |
| R: 5′-CCACTCATTCTGGTTGTCGTC-3′ | |||
| VEGF | NM_003376.5 | F: 5′-CGCAAGAAATCCCGGTATAA-3′ | 68 |
| R: 5′-AAATGCTTTCTCCGCTCTGA-3′ | |||
| Smad4 | NM_005359.5 | F: 5′-GGTTGCACATAGGCAAAGGT-3′ | 122 |
| R: 5′-ACGCCCAGCTTCTCTGTCTA-3′ | |||
| PAI-1 | NM_033049.3 | F: 5′-CACCCTCAGCATGTTCATTG-3′ | 571 |
| R: 5′-GGTCATGTTGCCTTTCCAGT-3′ | |||
| GAPDH | NM_002046.4 | F: 5′-AGGTCGGAGTCAACGGATTTG-3′ | 532 |
| R: 5′-GTGATGGCATGGACTGTGGT-3′ |
The PCR conditions were as follows: 10 min at 95°C, 40 cycles of 95°C for 10 s, 60°C for 20 s, and 15 s at 72°C. The mRNA level of each sample normalized to the level of GAPDH mRNA and is presented as the unit values of 2^[Ct(gapdh)–Ct(fbxo32)]. The amplified PCR products were analyzed by agarose gel electrophoresis with a 1.5% gel. Each experiment was performed in triplicate.
Western blot analysis
Cells were harvested and lysed using immunoprecipitation assay buffer containing protease inhibitors and phosphatase inhibitors for 30 min on ice. Protein concentrations of the cell lysates were determined by the BCA method (Beyotime). The proteins were placed on 10% gradient gels and then separated by SDS-PAGE (Bio-Rad, Hercules, CA). Resolved proteins were electrophoretically transferred onto PVDF membranes (Invitrogen) and then blocked for 1 h at room temperature with 5% non-fat dry milk in TBST containing 0.1% Tween 20. The membranes were probed with rabbit anti-GAPDH (mAb) (1: 1000) or rabbit anti-FBXO32 monoclonal antibody (mAb) (1: 1000) overnight at 4°C. Then, they were washed with 1×TBST and incubated in secondary antibodies diluted 1: 2000 that were conjugated to horseradish peroxidase (Santa Cruz, USA) for 2 h at room temperature with enhanced chemiluminescence reagents to detect the proteins (Boster, Wuhan, China). The membrane protein bands were imaged using X-film (Kodak Co.) and scanned. All Western blots were performed 3 times at least.
Statistical analysis
The SPSS17.0 statistical software was used analyze experimental data. For statistical analysis, Fisher’s exact test was conducted for any 2×2 tables, Pearson χ2 test was conducted for non-2×2 tables, and the chi-square trend test was conducted for ordinal date. Kaplan-Meier method and log-rank test to determine patient survival, p<0.05 was defined as statistically significant.
Results
FBXO32 and SMAD4 expression in CRC and normal colorectal tissues
FBXO32 and SMAD4 expression was low in CRC samples but high in normal colorectal tissues. Heterogeneous FBXO32 immunostaining was observed in 43 samples of normal tissues and 122 specimens of CRC, as shown in Figure 1.
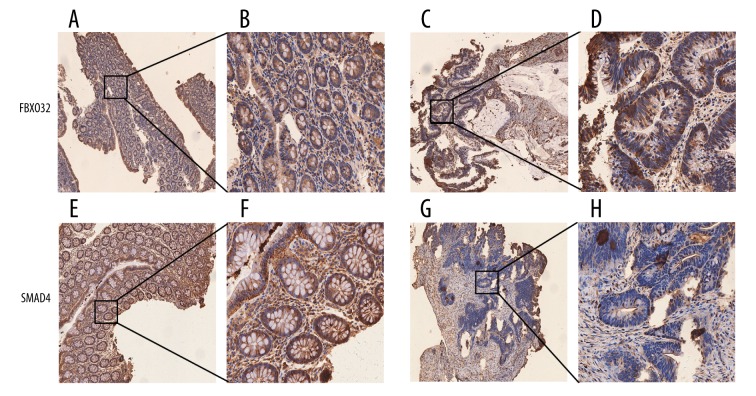
Figure 1.
Immunohistochemical staining for FBXO32 and SMAD4 in differentiated tissues (original magnification ×200). (A, B) FBXO32 staining in normal tissues; (C, D) FBXO32 staining in tumor tissues; (E, F) SMAD4 staining in normal tissues; (G, H) SMAD4 staining in tumor tissues.
Although weak staining was observed in several stromal cells, scattered areas of weak heterogeneous FBXO32 immunostaining in epithelial cell membranes were found in 18/43 (41.8%) normal tissue specimens, while 25/43 (58.2%) tissues were positive for FBXO32 (score 1–3). For FBXO32-positive primary CRC samples, heterogeneous weak-moderate (score 1–2), and strong staining (score 3) was respectively found in 86/122 (70.5%) and 36/122 (29.5%) samples, and SMAD4 strong staining in normal tissue 20/43 (46.5%), but only strong staining in normal tissue 41/122 (33.6%).
Association of FBXO32 and Smad4 with clinicopathological characteristics of CRC
The relationship between FBXO32 and SMAD4 with pathological features of cancer patients is shown in Table 3. High expression of FBXO32 tended to be related to the tumor stage (p=0.016) and lymphatic vessel invasion (p=0.031). The incidence of positive FBXO32 expression was increased based on histologic grade (p=0.012). However, SMAD4 expression was only related to tumor grade (p=0.005) and lymphatic vessel invasion (p=0.006).
Table 3.
The expression of FBXO32 and SMAD4 with clinicopathological parameters in CRC.
| Variable | No. of patients | FBXO32 expression levels | P value | SMAD4 expression levels | P value | ||
|---|---|---|---|---|---|---|---|
| Negative (%) | Positive (%) | Negative (%) | Positive (%) | ||||
| Age (years) | |||||||
| <50 | 101 | 78 | 23 | 0.570 | 67 | 34 | 0.697 |
| ≥50 | 21 | 15 | 6 | 13 | 8 | ||
| Gender | 0.057 | ||||||
| Male | 73 | 63 | 10 | 0.927 | 59 | 14 | |
| Female | 49 | 42 | 7 | 32 | 17 | ||
| Tumour stage | 0.016* | 0.869 | |||||
| A–B | 79 | 79 | 3 | 69 | 10 | ||
| C–D | 43 | 36 | 7 | 38 | 5 | ||
| Grade | 0.012* | 0.005* | |||||
| LMP | 21 | 11 | 10 | 13 | 8 | ||
| LG | 97 | 80 | 17 | 85 | 12 | ||
| G | 4 | 3 | 1 | 2 | 2 | ||
| Lymph node metastasis | 0.031* | ||||||
| Negative | 86 | 79 | 7 | 77 | 9 | 0.006* | |
| Positive | 36 | 28 | 8 | 25 | 11 | ||
1. Immunostaining grades observed using light microscopy. The criteria used for assessment were previously reported. 2. Pearson χ2 test for non-2×2 tables was used.
P<0.05 was considered as statistically significant.
fbxo32, smad4, vegf, and pai-1 mRNA and protein levels in primary CRC cell lines
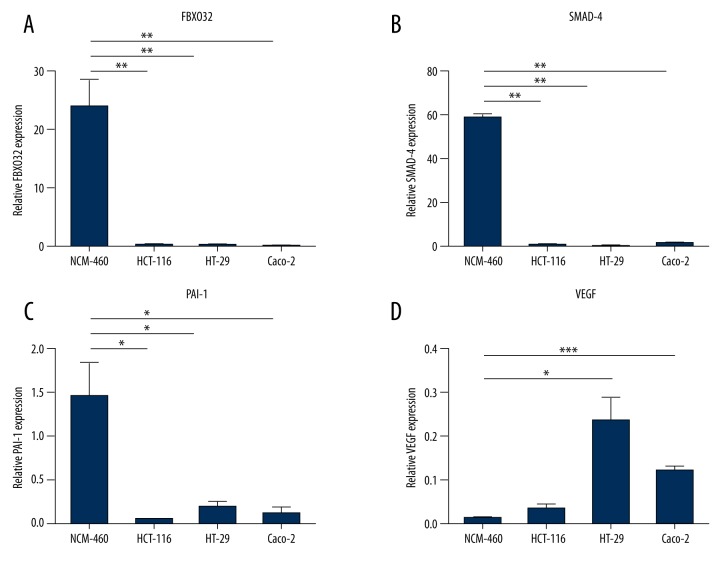
We examined mRNA expression in CRC cell lines and normal colorectal cells using qRT-PCR. The results showed a statistically higher elevation of fbxo32 mRNA expression in non-tumor cell than tumor cells (p<0.01, Figure 2A). Smad4 and pai-1 mRNA levels were also higher in non-tumor cells than in tumor cells (p<0.01, p<0.05, Figure 2B, 2C). However, vegf mRNA expression showed the opposite pattern, notably in HCT-116 and Caco-2 cells (p<0.05, Figure 2D). To investigate whether fbxo32 was elevated at the protein level, we assessed the CRC cell lines were by Western blot. We found that FBXO32 protein levels in tumor cells were much lower than those in non-tumor cells, as shown in (Figure 3), which confirmed the qRT-PCR results. The HT-29, HCT-116 and Caco-2 cell lines showed lower fbxo32 transcript levels than the NCM460 cell line (p<0.05, Figure 3). Similarly, the SMAD4 and PAI-1 protein levels were lower in those CRC cell lines compared to the NCM460 cell line, and vegf also showed an opposite expression pattern (Figure 3).
Figure 2.
The fbxo32, smad-4, pai-1 and vegf mRNA levels determine in cancer cell lines by qRT-PCR. (* p<0.05, ** p<0.01, *** p<0.001).
Figure 3.
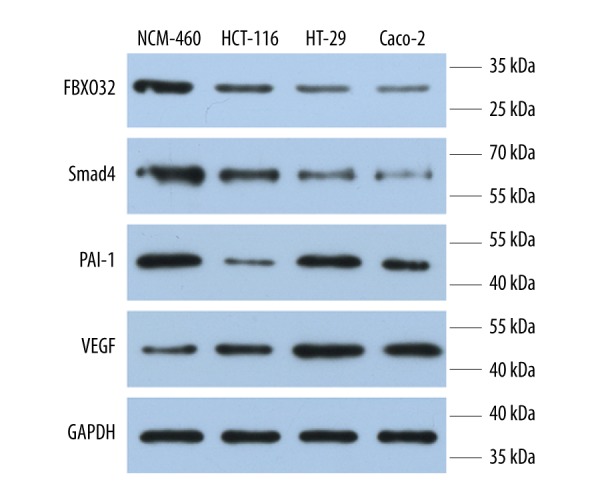
Evaluation of FBXO32, SMAD-4, PAI-1 and VEGF protein levels in cancer cell lines by Western blot.
Correlations of FBXO32 expression with survival of CRC
To further investigate the correlation of FBXO32 expression with the survival of CRC, Kaplan-Meier analyses were performed. As shown in (Figure 4), the 5-year overall survival of low-FBXO32 expression group was significantly shorter than that of high-FBXO32 expression group (p=0.010), indicating that down-regulation of FBXO32 might be correlated with poor survival of CRC.
Figure 4.
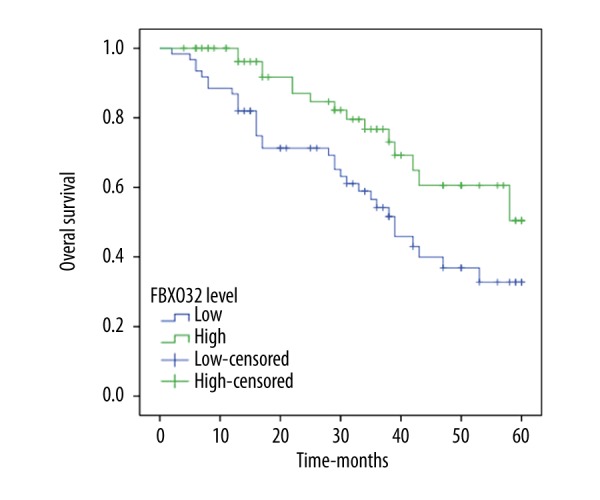
Overall survival analysis according to the Kaplan-Meier method for FBXO32 expression (log-rank test, p=0.010).
Discussion
Tumor metastasis and invasion consist of disruption of the basement membrane, stromal infiltration, extravasation and tumor cell invasion into a target organ. Our investigation of the TGF-β/Smad4 downstream target fbxo32 further supports this hypothesis. Fbxo32 is a downstream gene of smad4. Nuclear proteins are synthesized in the cytoplasm and need to be imported via the nuclear-pore complexes into the nucleus [14]. As a key gene of the TGF-β/Smad4 pathway, smad4 nuclear translocation may play a crucial role in regulating target gene expression [15]. Smad4 was originally isolated as a tumor suppressor gene on chromosome 18q21.1 in pancreatic ductal adenocarcinomas [13]. Inactivation of smad4 at the gene or protein level has been shown to be essential for the progression of several tumors [16,17]. Smad4 inactivation in cancers can be attributed to several factors, including homozygous deletion or loss of heterozygosity [12,18], and further studies should be performed to elucidate the exact mechanism of smad4 inactivation in CRC. The TGF-β signalling pathway is central in cell differentiation and stem cell maintenance [19]. Disruption of the TGF-β signal transduction pathway has been observed in a significant subset of human cancers. In recent reports, disruption of TGF-β signalling was shown to promote the formation of cancer stem/progenitor cells in liver and breast cancer [20,21]. Dysregulation of the TGF-β/Smad4 signalling pathway is a common event leading to loss of growth inhibition in CRC [22]. However, the mechanism underlying this dysregulation is still unclear.
In our study, we used immunohistochemistry, qRT-PCR and Western blot analyses to test FBXO32 expression and found that FBXO32 levels were high in normal colorectal and low in CRC samples. Fbxo32 expression was low in CRC, and it may be a tumor suppressor gene. Our results suggest that dysregulation of the TGF-β/Smad4 signalling pathway may contribute to fbxo32 methylation in CRC. Promoter hypermethylation of fbxo32 was observed in ovarian cancer cells showing constitutive smad4 nuclear translocation [23]. A previous study showed that the AKT signalling pathway negatively regulates fbxo32, which indicates that it may also be involved in regulating cell survival [24]. However, the role of smad4 in the methylation of fbxo32 requires further investigation. The mechanism of fbxo32 silencing in breast cancer cells may be different than that in CRC, and off-target effects of 5azaDC cannot be overlooked [7]. Under conditions of long-term signal disruption, recruitment of repressive factors causes progressive accumulation of DNA methylation at the targeted promoter, which eventually establishes an epigenetic memory for the generation of an inactive heterochromatin state [25]. Aberrant promoter methylation of TGFBR1 was reported to cause TGF-β resistance in gastric cancer, but when the selected CpG sites were different, the methylation frequency was also altered [26,27]. Many studies have shown that multiple growth factors and their receptors, including the vegf system, play roles in tumor invasion and metastasis [28], and the regulation of the vegf signalling axis has become a major focus of current research. Smad4 restoration influenced angiogenesis by decreasing the expression of vegf [29]. Smad4 also inhibited the secretion of vegf-a and vegf-c to promote angiogenesis in tumors by autocrine or paracrine pathways, which may be a mechanism underlying inhibition of tumor growth [30].
Overall, we believed that fbxo32 is downregulated in CRC with impaired TGF-β/Smad4 signalling, and promoter hypermethylation may be one of the mechanisms for inactivation of fbxo32 in CRC, particularly in CRC patients with an UGIC family history of North China [31]. Additionally, decreased expression and hypermethylation of fbxo32, along with a positive UGIC family history and stage III and IV classification, are highly predictive of metastasis and poor prognosis in CRC. Further studies need to be performed to determine whether fbxo32 can be used as a target to improve clinical outcomes of CRC. Above results support the hypothesis that aberrant TGF-β/Smad4 signalling can cause epigenetic silencing of its downstream target fbxo32 in CRC. Plasminogen activator inhibitor-1 (pai-1) is a proteolytic factor that plays a role in invasion and metastatic diffusion by modulating extracellular matrix degradation, cell proliferation and adhesion, which is required for the invasive process, as observed during wound healing and cancer invasion [32,33]. Pai-1 has been suggested as a target for the development of antithrombotic agents and promotion of fibrinolysis [34]. Furthermore, one of the major regulators of pai-1 expression, TGF-β1, was increased in pre-malignant oral leukoplakia and in oral squamous cell carcinoma [35,36].
Thus, fbxo32 is epigenetically silenced in CRC cells with impaired TGF-β/Smad4 signalling and may be a new tumor suppressor in CRC. As high levels of fbxo32 methylation predict survival in CRC patients, we hypothesized that fbxo32 may act as a methylation biomarker for this disease.
There are 2 limitations in our study. First, due to the tumor samples are drawn from the specimen database, normal mucosa samples are obtained from volunteers, there were fewer normal mucosa samples than tumors. Our future study in this area will include more normal mucosa samples. Second, the study lacked information about relative levels of genes expression in CRC cases. In our next study, we will assess genes expression in CRC cases.
Conclusions
We showed that FBXO32 was expressed in CRC and was negatively associated with the primary tumor stage and nuclear grade. Elevated expression of FBXO32 predicts a longer survival in CRC patients. Therefore, targeting fbxo32 may be a potential strategy for the treatment of CRC due to its tumor suppressor function by the process of TGF-β. However, the in-depth mechanism requires further investigation.
Footnotes
Conflict of interest
None.
Source of support: This work was supported by a grant from the Shandong Province Natural Science Fund (no. ZR2014HL067)
References
- 1.Abbruzzese C, Diodoro MG, Sperduti I, et al. Detection of phosphorylated insulin receptor in colorectal adenoma and adenocarcinoma: Implications for prognosis and clinical outcome. J Cell Physiol. 2015;230:562–67. doi: 10.1002/jcp.24733. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Ward E, Thun M. Declining death rates reflect progress against cancer. PLoS One. 2010;5:e9584. doi: 10.1371/journal.pone.0009584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li WX, Xiao HW, Hong XQ, et al. Predictive value of CK20 in evaluating the efficacy of treatment and prognosis after surgery for colorectal cancer. Genet Mol Res. 2015;14:5823–29. doi: 10.4238/2015.May.29.14. [DOI] [PubMed] [Google Scholar]
- 4.Hanai J, Cao P, Tanksale P, et al. The muscle-specific ubiquitin ligase atrogin-1/MAFbx mediates statin-induced muscle toxicity. J Clin Invest. 2007;117:3940–51. doi: 10.1172/JCI32741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li HH, Kedar V, Zhang C, et al. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J Clin Invest. 2004;114:1058–71. doi: 10.1172/JCI22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerlinger-Romero F, Yonamine CY, Junior DC, et al. Dysregulation between TRIM63/FBXO32 expression and soleus muscle wasting in diabetic rats: Potential role of miR-1-3p, -29a/b-3p, and -133a/b-3p. Mol Cell Biochem. 2017;427:187–99. doi: 10.1007/s11010-016-2910-z. [DOI] [PubMed] [Google Scholar]
- 7.Tan J, Yang X, Zhuang L, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–63. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo W, Zhang M, Guo Y, et al. FBXO32, a new TGF-beta/Smad signaling pathway target gene, is epigenetically inactivated in gastric cardia adenocarcinoma. Neoplasma. 2015;62:646–57. doi: 10.4149/neo_2015_078. [DOI] [PubMed] [Google Scholar]
- 9.Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science. 2002;296:1646–47. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- 10.Guo Y-H, Li Y-N, Zhao J-R, et al. HBc binds to the CpG islands of HBV cccDNA and promotes an epigenetic permissive state. Epigenetics. 2014;6:720–26. doi: 10.4161/epi.6.6.15815. [DOI] [PubMed] [Google Scholar]
- 11.Leu YW, Yan PS, Fan M, et al. Loss of estrogen receptor signaling triggers epigenetic silencing of downstream targets in breast cancer. Cancer Res. 2004;64:8184–92. doi: 10.1158/0008-5472.CAN-04-2045. [DOI] [PubMed] [Google Scholar]
- 12.Iacobuzio-Donahue CA, Song J, Parmiagiani G, et al. Kern2 missense mutations of MADH4: Characterization of the mutational hot spot and functional consequences in human tumors. Clin Cancer Res. 2004;10:1597–604. doi: 10.1158/1078-0432.ccr-1121-3. [DOI] [PubMed] [Google Scholar]
- 13.Hahn SA, Schutte M, Hoque AT, et al. DPC4, A candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–53. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 14.Michael W, Xiao Y, Chu XD. Functions of mammalian Smad genes as revealed by targeted gene disruption in mice. Cytokine Growth Factor Rev. 2000;11:49–58. doi: 10.1016/s1359-6101(99)00028-3. [DOI] [PubMed] [Google Scholar]
- 15.Chou JL, Su HY, Chen LY, et al. Promoter hypermethylation of FBXO32, a novel TGF-beta/SMAD4 target gene and tumor suppressor, is associated with poor prognosis in human ovarian cancer. Lab Invest. 2010;90:414–25. doi: 10.1038/labinvest.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuchi M, Masuda N, Miyazaki T, et al. Decreased Smad4 expression in the transforming growth factor-beta signaling pathway during progression of esophageal squamous cell carcinoma. Cancer. 2002;95:737–43. doi: 10.1002/cncr.10727. [DOI] [PubMed] [Google Scholar]
- 17.Che XM, Natsugoe S, Sonshin T, et al. Preserved Smad4 expression in the transforming growth factor β signaling pathway is a favorable prognostic factor in patients with advanced gastric cancer. Clin Cancer Res. 2001;7:277–82. [PubMed] [Google Scholar]
- 18.Miyaki M, Kuroki T. Role of Smad4 (DPC4) inactivation in human cancer. Biochem Biophys Res Commun. 2003;306:799–804. doi: 10.1016/s0006-291x(03)01066-0. [DOI] [PubMed] [Google Scholar]
- 19.Watabe T, Miyazono K. Roles of TGF-beta family signaling in stem cell renewal and differentiation. Cell Res. 2009;19:103–15. doi: 10.1038/cr.2008.323. [DOI] [PubMed] [Google Scholar]
- 20.Bierie B, Stover DG, Abel TW, et al. Transforming growth factor-beta regulates mammary carcinoma cell survival and interaction with the adjacent microenvironment. Cancer Res. 2008;68:1809–19. doi: 10.1158/0008-5472.CAN-07-5597. [DOI] [PubMed] [Google Scholar]
- 21.Tang Y, Kitisin K, Jogunoori W, et al. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc Natl Acad Sci USA. 2008;105:2445–50. doi: 10.1073/pnas.0705395105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun C, Wang FJ, Zhang HG, et al. miR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-beta/Smad4 pathway. World J Gastroenterol. 2017;23:1816–27. doi: 10.3748/wjg.v23.i10.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan MW, Huang YW, Hartman-Frey C, et al. Aberrant transforming growth factor β1 signaling and SMAD4 nuclear translocation confer epigenetic repression of ADAM19 in ovarian cancer. Neoplasia. 2008;10:908–19. doi: 10.1593/neo.08540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stitt TN, Drujan D, Clarke BA, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 25.Madakashira BP, Sadler KC. DNA methylation, nuclear organization, and cancer. Front Genet. 2017;8:76. doi: 10.3389/fgene.2017.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Periyasamy S, Ammanamanchi S, Tillekeratne MP, et al. Repression of transforming growth factor-beta receptor type I promoter expression by Sp1 deficiency. Oncogene. 2000;19:4660–67. doi: 10.1038/sj.onc.1203822. [DOI] [PubMed] [Google Scholar]
- 27.Pinto M, Oliveira C, Cirnes L, et al. Promoter methylation of TGFbeta receptor I and mutation of TGFbeta receptor II are frequent events in MSI sporadic gastric carcinomas. J Pathol. 2003;200:32–38. doi: 10.1002/path.1327. [DOI] [PubMed] [Google Scholar]
- 28.Abdul-Aziz MA, Amin AK, El-Rouby DH, et al. Lymphangiogenesis in oral squamous cell carcinoma: Correlation with VEGF-C expression and lymph node metastasis. Int J Dent. 2017;2017:7285656. doi: 10.1155/2017/7285656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarte-Waldhoff I, Volpert OV, Bouck NP, et al. Smad4/DPC4-mediated tumor suppression through suppression of angiogenesis. Proc Natl Acad Sci USA. 2000;97:9624–29. doi: 10.1073/pnas.97.17.9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Lv X, Xiao J, et al. Smad4 Inhibits VEGF-A and VEGF-C Expressions via Enhancing Smad3 Phosphorylation in Colon Cancer. Anat Rec (Hoboken) 2017;300(9):1560–69. doi: 10.1002/ar.23610. [DOI] [PubMed] [Google Scholar]
- 31.Wang LH, Kim SH, Lee JH, et al. Inactivation of SMAD4 tumor suppressor gene during gastric carcinoma progression. Clin Cancer Res. 2007;13:102–10. doi: 10.1158/1078-0432.CCR-06-1467. [DOI] [PubMed] [Google Scholar]
- 32.Coates AS, Winer EP, Goldhirsch A, et al. Tailoring therapies-improving the management of early breast cancer: St GallenInternational Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol. 2015;26(8):1533–46. doi: 10.1093/annonc/mdv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viala M, Alexandre M, Thezenas S, et al. Prognostic impact of the inclusion of uPA/PAI-1 for adjuvant treatment decision-making in ER+/Her2– pN0 early breast cancers. Breast Cancer Res Treat. 2017;165(3):611–21. doi: 10.1007/s10549-017-4373-7. [DOI] [PubMed] [Google Scholar]
- 34.Peng S, Xue G, Gong L, et al. A long-acting PAI-1 inhibitor reduces thrombus formation. Thromb Haemost. 2017;117:1338–47. doi: 10.1160/TH16-11-0891. [DOI] [PubMed] [Google Scholar]
- 35.Angela F, Logullo, Suely N, et al. Transforming growth factor b1 (TGFβ1) expression in head and neck squamous cell carcinoma patients as related to prognosis. J Oral Pathol Med. 2003;32:139–45. doi: 10.1034/j.1600-0714.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- 36.Wagner VP, Cardoso PR, Dos Santos JN. Immunohistochemical study of TGF-β1 in oral leukoplakia and oral squamous cell carcinoma: Correlations between clinicopathologic factors and overall survival. Appl Immunohistochem Mol Morphol. 2017;25(9):651–59. doi: 10.1097/PAI.0000000000000355. [DOI] [PubMed] [Google Scholar]