Abstract
Metabolism of 25-hydroxyvitamin D3 (25OHD3) plays a central role in regulating the biologic effects of vitamin D in the body. Although cytochrome P450–dependent hydroxylation of 25OHD3 has been extensively investigated, limited information is available on the conjugation of 25OHD3. In this study, we report that 25OHD3 is selectively conjugated to 25OHD3-3-O-sulfate by human sulfotransferase 2A1 (SULT2A1) and that the liver is a primary site of metabolite formation. At a low (50 nM) concentration of 25OHD3, 25OHD3-3-O-sulfate was the most abundant metabolite, with an intrinsic clearance approximately 8-fold higher than the next most efficient metabolic route. In addition, 25OHD3 sulfonation was not inducible by the potent human pregnane X receptor agonist, rifampicin. The 25OHD3 sulfonation rates in a bank of 258 different human liver cytosols were highly variable but correlated with the rates of dehydroepiandrosterone sulfonation. Further analysis revealed a significant association between a common single nucleotide variant within intron 1 of SULT2A1 (rs296361; minor allele frequency = 15% in whites) and liver cytosolic SULT2A1 content as well as 25OHD3-3-O-sulfate formation rate, suggesting that variation in the SULT2A1 gene contributes importantly to interindividual differences in vitamin D homeostasis. Finally, 25OHD3-3-O-sulfate exhibited high affinity for the vitamin D binding protein and was detectable in human plasma and bile but not in urine samples. Thus, circulating concentrations of 25OHD3-3-O-sulfate appear to be protected from rapid renal elimination, raising the possibility that the sulfate metabolite may serve as a reservoir of 25OHD3 in vivo, and contribute indirectly to the biologic effects of vitamin D.
Introduction
Vitamin D deficiency has been associated with a wide range of adverse health outcomes, such as rickets, osteoporosis, diabetes, multiple sclerosis, and colon cancer (Holick, 2007), although evidence for nonskeletal effects is more limited than that for skeletal disease (Theodoratou et al., 2014). Vitamin D is found in nature in the forms of cholecalciferol (vitamin D3) and ergocalciferol (vitamin D2), with predominately vitamin D3 in plasma from untreated humans. 25-Hydroxyvitamin D3 (25OHD3), a major circulating metabolite of vitamin D3, is used as a clinical biomarker for assessment of vitamin D status; concentrations < 20 ng/ml (50 nM) are considered insufficient for bone health by the Institute of Medicine (Hossein-nezhad and Holick, 2013) and deficient by others (Holick et al., 2011). Multiple factors affect vitamin D sufficiency, including the synthesis of vitamin D3 in the skin, the absorption of vitamin D3 and D2 from food in the intestine, vitamin D 25-hydroxylation activity in the liver, and a combination of 25OHD oxidation and conjugation reactions catalyzed principally in the kidney, liver, and small intestine (DeLuca, 1988; Jones et al., 2014).
Variation in the diet and exposure to sunlight contribute importantly to intra- and interindividual differences in serum 25OHD3 concentration, but much of the observed variance remains unexplained (Fuleihan et al., 2015). Results from genome-wide and candidate gene association studies have implicated CYP2R1, CYP24, GC [plasma vitamin D binding protein (DBP)], and 7-dehydrocholesterol reductase (DHCR7) gene variation in serum 25OHD3 variability, but the extent of their contribution appears low (Ahn et al., 2010; Bu et al., 2010; Berry and Hyppönen, 2011) despite 25OHD3 heritability estimates of 20%–85% (Hunter et al., 2001; Arguelles et al., 2009; Shea et al., 2009).
25OHD3-3-O-sulfate is reported to be a major circulating metabolite of 25OHD3 in humans, with an average circulating concentration comparable to that of 25OHD3 (Axelson, 1985; Shimada et al., 1995; Higashi et al., 2014), and thus, the sulfonation metabolic pathway might contribute importantly to vitamin D homeostasis. Surprisingly little is known about how and where 25OHD3-3-O-sulfate is formed. This information is crucial if the contribution from the sulfonation pathway to vitamin D homeostasis is to be fully evaluated. In this study, we examined the sulfonation of 25OHD3 in humans using recombinant sulfotransferase (SULT) enzymes, liver cytosols, primary hepatocytes, renal tubular epithelial cells, and immortalized intestinal epithelial cells to identify which of the human SULTs is responsible for the sulfonation reaction and in what tissues the reaction occurs, as well as potential genetic and environmental sources of interindividual variability in hepatic sulfonation activity.
The formation of 25OHD3-3-O-sulfate from 25OHD3 could be considered a catabolic process, but some investigators have hypothesized that it also represents an alternative 25OHD3 storage form in the body (Higashi et al., 2010). This is quite plausible because other endogenous steroid sulfate conjugates, such as estradiol-sulfate and dehydroepiandrosterone (DHEA)-sulfate, circulate at relatively high levels, are deconjugated in target tissues, and contribute to certain physiologic functions (Axelson, 1987; Banerjee et al., 2013; Sánchez-Guijo et al., 2015). Similarly, 25OHD3-3-O-sulfate might be retained in the circulation and distributed to different tissues of the body where it could be hydrolyzed to 25OHD3, replenishing the 25OHD3 pool, as needed. With this in mind, we also tested the binding affinity of 25OHD3-3-O-sulfate for DBP and its presence in human urine and bile.
Materials and Methods
Materials
Chemicals.
DHEA, 3-phosphoadenosine-5-phosphosulfate (PAPS), 4-phenyl-1,2,4-triazoline-3,5-dione (PTAD), and rifampicin were purchased from Sigma-Aldrich (St. Louis, MO). The precursors of 4-(4′-dimethylaminophenyl)-1,2,4-triazoline-3,5-dione (DAPTAD) were obtained as follows: 4-(4′-dimethylaminophenyl)-1,2,4-triazolidine-3,5-dione (Santa Cruz Biotechnology, Dallas, TX) and iodobenzene diacetate (Sigma-Aldrich). 25OHD3 was obtained from Calbiochem (La Jolla, CA) and its deuterated form, d6-25OHD3 (containing six deuterium atoms at C-26 and C-27), was purchased from Medical Isotope Inc. (Pelham, NH). Chemically synthesized 25OHD3-3-O-sulfate and its deuterated form, d6-25OHD3-3-O-sulfate, were purchased from Toronto Research Chemicals (Toronto, ON, Canada). 25OHD3-25-O-glucuronide and its deuterated form were generated by enzymatic reactions, isolated, and purified as described (Gao et al., 2017). All other buffers and chemicals were of the highest grade commercially available.
Sequencing-grade trypsin, iodoacetamide, dithiothreitol, and bovine serum albumin (BSA) were purchased from Pierce Biotechnology (Rockford, IL). Two synthetic surrogate peptides (Supplemental Table 1) for both SULT2A1 and BSA were identified using the published protocol (Prasad and Unadkat, 2014a,b) and procured from Thermo Fisher Scientific (Rockford, IL). Chloroform, high-performance liquid chromatography (LC)–grade acetonitrile, methanol, and formic acid were purchased from Fischer Scientific (Fair Lawn, NJ). Ammonium bicarbonate (98% purity) and sodium deoxycholate (98% purity) were obtained from Thermo Fisher Scientific and MP Biomedicals (Santa Ana, CA), respectively.
Cell culture medium, insulin-transferrin-selenium A, penicillin-streptomycin, and amphotericin B were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum was obtained from Atlanta Biologicals (Lawrenceville, GA). Sodium pyruvate was purchased from Cellgro (Herndon, VA). Hydrocortisone and cell dissociation solution was purchased from Sigma-Aldrich. Mouse collagen IV was obtained from Corning (Corning, NY).
Human Recombinant Enzymes and Cells.
For the SULT isoform activity screening experiments, human recombinantly expressed SULT isoforms SULT1A1, SULT1B1, SULT1E1, SULT2A1, and SULT2B1 were purchased from Genway Biotech (San Diego, CA). For kinetic experiments, purified recombinant SULT2A1 was purchased from R&D Systems (Minneapolis, MN). This same product was used as an authentic standard to determine the SULT2A1-specific content of pooled human liver cytosol. Cryopreserved primary human hepatocytes from two different human liver donors were obtained from either Triangle Research Laboratories (Durham, NC) (lot no. GC4008; Caucasian man, aged 69 years; no alcohol, tobacco, or drug use) or Invitrogen (lot no. Hu4237; Caucasian woman, aged 57 years; no alcohol, tobacco, or drug use) and cultured as previously published (Wang et al., 2013a). The human intestinal epithelial LS180 cell line was obtained from American Type Culture Collection (Manassas, VA). Human renal cortical tissue was obtained during the surgical resection of a renal cell carcinoma. Renal tubular epithelial cells were isolated from a noncancerous section of tissue from three donors using established methods (Weber et al., 2016). The surgery was performed at the University of Washington Medical Center and the human subjects protocol was approved by the University of Washington Human Subjects Review Board.
Tissue Samples.
A total of 258 human livers from the University of Washington School of Pharmacy Human Tissue Bank (a resource established by National Institutes of Health National Institute of General Medical Sciences) and the St. Jude Liver Resource (obtained through the Liver Tissue Cell Distribution System, Minneapolis, MN and Pittsburgh, PA, and funded by the Department of Health and Human Services; Shirasaka et al., 2016) were used for this investigation. The collection and use of these tissues for research purposes was approved by the Human Subjects Institutional Review Boards at the University of Washington and St. Jude Children’s Research Hospital. In both cases, all links between archived tissues and the original donors were destroyed to preserve anonymity and facilitate research. Demographic characteristics of the liver bank donors were as follows: 44% female and 56% male; age <1–81 years (mean 43 years); and 93% white European ancestry, 4% black, 1% Hispanic, and 2% unknown.
Outdated human plasma was from the Puget Sound Blood Center (Seattle, WA); anonymous, pooled human bile was kindly provided by Dr. Evan D. Kharasch (Washington University in St. Louis, St. Louis, MO); and anonymous pooled human urine was collected at the University of Washington. These outdated human samples were analyzed under an institution exempt approval from the University of Washington Human Subjects Review Board.
DNA and RNA for DNA sequencing and RNA sequencing (RNA-Seq) analysis were isolated from liver tissue, as described below. Cytosolic fractions were isolated, as previously described (Shirasaka et al., 2016). Sulfonation activity measurements are described below. Total protein concentration in each cytosol preparation was measured by bicinchoninic acid assay (Thermo Fisher Scientific).
Formation of 25OHD3-3-O-Sulfate by Pooled Human Liver Cytosol and Recombinant SULT Isozymes
The following procedures were conducted under low light conditions to avoid potential degradation of vitamin D metabolites. Initial experiments with liver cytosols or SULTs (SULT1A1, SULT1B1, SULT1E1, SULT2A1, and SULT2B1) were carried out in 50 mM Tris-HCl (pH 7.5) solution containing 1 mg/ml pooled liver cytosols or 0.025 mg/ml recombinant SULT, 5 µM 25OHD3, and 0.1 mM PAPS, with a 0.5 ml final volume in glass tubes. Briefly, pooled liver cytosols (n = 10, randomly selected from 258 individual human liver cytosol preparations) or SULT enzymes were mixed with 25OHD3 and prewarmed at 37°C for 5 minutes. The reaction was initiated by adding PAPS, was maintained at 37°C for various times as indicated, and was terminated with an equal volume of ice-cold acetonitrile. Incubations without PAPS served as negative controls.
Kinetic studies were conducted in 50 mM Tris-HCl (pH 7.5) solution containing pooled liver cytosol (1 mg/ml; 0.2 mg in 0.2 ml final volume) or recombinant SULT2A1 (0.025 mg/ml purified SULT2A1 (2.5 µg SULT2A1 in 0.1 ml final volume) under conditions (worked out in pilot studies) that provided linear product formation with respect to protein concentration and time. The range of substrate concentration was 0.25–40 µM for 25OHD3 and each incubation was conducted at 37°C for 30 minutes. Reactions were initiated with the addition of 0.1 mM PAPS and terminated with the addition of two volumes of ice-cold acetonitrile (0.4 ml for cytosol and 0.2 ml for SULT2A1 incubations) containing internal standard d6-25OHD3-3-O-sulfate (100 nM) and the mixtures were transferred to Eppendorf tubes and centrifuged for 5 minutes (12,000g). The supernatants were collected, concentrated under a nitrogen stream, and reconstituted in 20% acetonitrile in mobile phase for LC–mass spectrometry (MS) analysis (see below). Incubations without PAPS served as negative controls.
The rates of product formation were normalized for SULT2A1 or cytosolic protein concentration. We also determined the specific SULT2A1 content of the pooled human liver cytosol preparation (see below) and normalized the cytosolic rates for the SULT2A1-specific content. A simple Michaelis–Menten kinetics model was fit to each 25OHD3 data set using nonlinear regression data analysis (GraphPad Prism software, version 5; GraphPad Software Inc., La Jolla, CA). Each experimental reaction condition was conducted in duplicate (technical replicate) and repeated three times independently (different days) to generate the mean and variance for the kinetic parameters (Km and Vmax).
Formation of 25OHD3-3-Sulfate and DHEA-Sulfate by a Panel of Human Liver Cytosols
To characterize and compare interindividual differences in hepatic 25OHD3 and DHEA sulfonation activity, we incubated 258 individual human liver cytosol preparations (0.25 mg/ml) in 50 mM Tris-HCl (pH 7.5), 5 µM 25OHD3 or DHEA (established probe substrate for SULT2A1), and 0.1 mM PAPS, with a 0.1 ml final volume (0.025 mg protein) in glass tubes. After the mixture was prewarmed at 37°C for 5 minutes, the reaction was initiated by adding PAPS and maintained at 37°C for 30 minutes and terminated with addition of an equal volume of ice-cold acetonitrile. Once again, incubations without PAPS served as negative controls. Rates of product formation were normalized for cytosolic protein concentration.
Formation of 25OHD3-3-O-Sulfate and Other 25OHD3 Metabolites in Human Cell Cultures
Cryopreserved human hepatocytes from two donors were thawed and cultured, as previously published (Wang et al., 2013a). After viable cells were plated and cultured in collagen I–coated 96-well plates for 24 hours, the cells were treated with 10 µM rifampicin or vehicle, 0.1% dimethylsulfoxide, in 100 µl solution per well. After 48 hours, the treated cells were washed with saline solution twice and then incubated with 50 nM 25OHD3 for various incubation times (t = 2, 4, 8, or 24 hours). At the end of the treatment period, culture medium was collected from 12 wells and pooled, if necessary.
For quantitation of 25OHD3 oxidative metabolites, cell medium samples (0.5 ml) were subjected to liquid-liquid extraction with the addition of 5 ml ethyl acetate, followed by PTAD derivatization and reconstitution in 40% acetonitrile containing 0.1% formic acid. Formation of both 4β,25(OH)2D3 and 4α,25(OH)2D3 was quantified after treatment of the cell culture medium with β-glucuronidase (1500 IU/ml) prior to the liquid-liquid extraction step to account for significant secondary metabolism, as previously described (Wang et al., 2013a). For quantification of 25OHD3-glucuronides, cell medium (0.25 ml) was quenched with two volumes of ice-cold acetonitrile, and the mixtures were centrifuged at 12,000g for 4 minutes in Eppendorf tubes (Wang et al., 2014). The supernatants were collected, concentrated under a nitrogen stream, and reconstituted in 50 µl mobile phase (20% acetonitrile in 5 mM ammonium acetate) for LC-MS analysis as described below.
For quantitation of 25OHD3-3-O-sulfate, cell medium samples (0.3 ml) were buffered with an equal volume of 0.1 M sodium acetate (pH 4.0) and subjected to solid-phase extraction (SPE) using Waters Oasis WAX anion exchange cartridges (30 mg, 1 cc; Waters, Milford, MA), under conditions suggested by the manufacturer. Briefly, the Oasis cartridges were preconditioned with methanol and 2% acetic acid before loading of diluted samples. After loading, the cartridges were washed with 2% acetic acid and 100% methanol sequentially. 25OHD3-3-O-sulfate was eluted by 3 ml buffer containing a mixture of 5:95 (v/v) ammonium hydroxide (28%) to methanol. The eluates were dried, pooled if necessary, and reconstituted in 50 µl mobile phase solution (20% acetonitrile in 5 mM ammonium acetate) for LC-MS analysis as described below.
To assess the conversion of 25OHD3 to 25OHD3-3-O-sulfate at extrahepatic sites that are important in maintaining systemic vitamin D homeostasis, additional experiments were performed using human renal tubular epithelial cells from three different tissue donors and the LS180 colon carcinoma cell line. LS180 cells (passage 30) and human renal tubule epithelial cells (passage 4) were cultured on uncoated and collagen IV–coated six-well plates, respectively, for 1 week. The cells were then washed with saline solution twice and incubated with 50 nM 25OHD3. Culture medium was collected after 24 hours and concentrations of 25OHD3-3-O-sulfate were quantified using the same LC-MS method, as described for hepatocytes.
Quantitation of 25OHD3-3-O-Sulfate in Recombinant SULT Enzyme, Human Cytosol, and Cell Incubates
Samples reconstituted in mobile phase (20% acetonitrile in 5 mM ammonium acetate, pH 4.6; 15 µl injected on column) were subjected to LC-UV/MS analysis using an Agilent MSD mass spectrometer coupled with an Agilent 1100 series high-performance LC system (Agilent Technologies, Santa Clara, CA). Chromatographic separation was achieved on a Symmetry C18 column (2.1 × 150 mm, 3.5 µm; Waters) and a mobile phase consisting of 5 mM ammonium acetate (A) (pH 4.6) and acetonitrile (B) at 45°C. A linear gradient from 20% B (0–2 minutes) to 90% B (17–22 minutes) in 15 minutes at a flow rate of 0.25 ml/min was employed. UV detection was performed at 265 nm. The mass spectrometer was operated in the negative ionization mode. The interface was maintained at 350°C with a nitrogen nebulization pressure of 25 psi, resulting in a flow of 10 l/min. 25OHD3-3-O-sulfate and d6-25OHD3-3-O-sulfate were detected by selective ion monitoring at a mass-to-charge ratio (m/z) of 479 and 485, respectively, after 110 V ion fragmentation. 25OHD3-3-O-sulfate eluted from the column at approximately 14.4 minutes. Calibration curves were constructed by plotting the peak area ratios of 25OHD3-3-O-sulfate and d6-25OHD3-3-O-sulfate versus the corresponding 25OHD3-3-O-sulfate concentrations and fitting a linear regression equation to the data. Formation of 25OHD3-3-O-sulfate was then quantified by fitting the peak area ratios (metabolites/internal standard) from incubations to the calibration curve.
Quantitation of 25OHD3 Monohydroxy and Glucuronide Metabolites in Human Hepatocyte Incubations
Quantification of 25OHD3 monohydroxy metabolites and primary glucuronides was performed using previously published LC–tandem mass spectrometry (MS/MS) (Wang et al., 2013a) and LC-MS (Wang et al., 2014) methods, respectively. Briefly, LC-MS/MS was performed using an Agilent 1290 series ultra-performance liquid chromatography (UPLC) system and an Agilent 6410 triple quadrupole tandem mass spectrometer equipped with an electrospray ionization (ESI) source (Pala Alto, CA). Separation was achieved on a Hypersil Gold column (2.1 × 100 mm, 1.9 µm; Thermo Fisher Scientific), and multiple reaction monitoring of the transitions m/z 574→314, 574→298, 558→298, 564→298, and 580→314 was employed to detect 1α (4α or 4β),25(OH)2D3, 24,25(OH)2D3, 25OHD3, d6-25OHD3, and d6-1α,25(OH)2D3, respectively (Wang et al., 2013a). Calibration curves were constructed by plotting the peak area ratio for each vitamin D3 metabolite and its internal standard, versus the corresponding concentration, and fitting a linear regression equation to the data. LC-MS analysis was performed using an Agilent MSD mass spectrometer coupled with an Agilent 1100 series LC system (Wang et al., 2014). Chromatographic separation was achieved on a Symmetry C18 column (2.1 × 150 mm, 3.5 µm; Waters). Separations were achieved after a linear gradient from 30% B (2 minutes) to 45% B (25 minutes), held at 45% B for 1 minute and increased to 60% B (35 minutes), increased to 90% in 1 minute and held for 3 minutes (39 minutes), and then equilibrated back to 30% B in 1 minute. 25OHD3 glucuronides and d6-25OHD3-25-glucuronide as the internal standard were detected by selective ion monitoring at m/z 575 and 581, respectively, after 110 V ion fragmentation. Calibration curves were constructed by plotting the peak area ratio of glucuronide metabolites and internal standard versus the corresponding standard 25OHD3-25-glucuronide concentration and fitting a linear regression equation to the data.
Quantitation of 25OHD3-3-O-Sulfate in Human Plasma, Bile, and Urine
Human plasma (0.1 ml) and urine (0.5 ml) were first subjected to protein precipitation using two volumes of acetonitrile followed by dilution with two volumes of 0.1 M sodium acetate (pH 4.0). For human bile (1 ml), the pooled (from different adult donors) sample was first diluted with 0.5 ml water, subjected to liquid-liquid extraction using an equal volume of hexane, twice. The aqueous solution was separated, pooled if necessary, and buffered with an equal volume of 0.1 M sodium acetate (pH 4.0) and subjected to solid-phase anion exchange extraction of 25OHD3-3-O-sulfate, using the Waters Oasis WAX (60 mg, 3 cc) anion exchange cartridges, as described above (cell culture experiments). After SPE, the eluate was transferred to a new glass tube, dried, and reconstituted in 50 µl 20% acetonitrile in 5 mM ammonium acetate. Reconstituted samples were analyzed for 25OHD3-3-O-sulfate concentration, as described above for quantitation of 25OHD3-3-O-sulfate produced during in vitro incubations. The limit in plasma and urine was estimated at 1.0 ng/ml with intra- and interday errors of ≤15%.
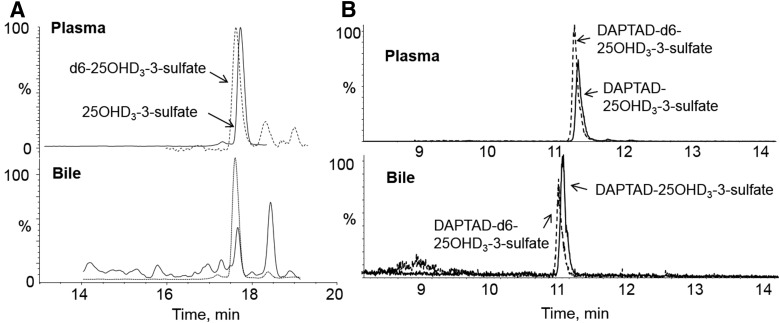
For better detection of 25OHD3-3-O-sulfate in bile samples, a derivatization step was included as part of the sample treatment following a previously reported method (Gao et al., 2017). Briefly, pooled human bile (100 µl) was precipitated with 200 µl acetonitrile and buffered with 1 ml 0.1 M sodium acetate (pH 3.2) and subjected to SPE using Waters Oasis WAX anion exchange cartridges (60 mg, 3 cc). The eluates from SPE were then dried and reconstituted in 10 µl methanol and derivatized with 200 µl 4.5 mM DAPTAD solution for 1 hour at room temperature in the dark (Ogawa et al., 2013). The reaction mixture was then evaporated under N2 flow and reconstituted in 100 µl mobile phase. A centrifugation step was included to remove an insoluble substance before LC-MS/MS analysis. DAPTAD-25OHD3-3-O-sulfate was analyzed using an AB Sciex QTRAP 6500 LC-MS/MS system (Concord, ON, Canada). Chromatographic separation was achieved on a Hypersil Gold column (2.1 × 100 mm, 1.9 μm; Thermo Fisher Scientific) and a mobile phase consisting of 5 mM ammonium acetate (A) (pH 4.6) and acetonitrile (B) at 45°C. Linear gradients were applied with B% increased from 25% to 60% B (1–13 minutes) and from 60% to 90% (13–16 minutes) at a flow rate of 0.25 l/min. The autosampler was maintained at 4°C and the injection volume was 15 µl. The ESI source of the mass spectrometer was operated in the positive ion mode. Detection was by multiple reaction monitoring of the transitions at m/z 699.5→323 for DAPTAD-25OHD3-3-O-sulfate and m/z 705.5→323 for the internal standard DAPTAD-d6-25OHD3-3-O-sulfate. Optimal MS parameters were as follows: curtain gas, 20 psi; nebulizer gas (GS1) and turbo gas (GS2), 40 psi; ion spray voltage, 5500 V; and source temperature, 400°C. Collision energy was 45 eV and declustering potential was 40 V for both analytes. Due to the lack of blank bile matrix, calibration curves were plotted with corrected peak area ratios of DAPTAD-25OHD3-3-O-sulfate and DAPTAD-d6-25OHD3-3-O-sulfate by subtracting “blank” samples without spiking with 25OHD3-3-O-sulfate solution from the ones spiked with serial solutions.
Quantitation of DHEA-Sulfate in Recombinant SULT Enzyme and Human Cytosol Incubates
Incubation samples quenched with 50% acetonitrile were centrifuged at 12,000g for 5 minutes. The supernatant was transferred to a clean glass tube and the solvent was removed under a nitrogen stream. The sample residues were reconstituted in mobile phase (30% acetonitrile in 5 mM ammonium acetate, pH 4.6) then subjected to LC-MS analysis using an Agilent MSD mass spectrometer coupled with an Agilent 1100 series LC system. Sample injection volumes were 20 µl on column and chromatographic separation of the analytes was achieved on a Symmetry C18 column (2.1 × 150 mm, 3.5 µm; Waters), with a mobile phase consisting of 5 mM ammonium acetate (A) (pH 4.6) and acetonitrile (B) at 40°C. A linear gradient from 30% B (0–2 minutes) to 50% B (11–11.5 minutes) over 15 minutes at a flow rate of 0.25 ml/min was employed. DHEA-sulfate and d6-DHEA-sulfate eluted from the column at approximately 6.5 minutes. The mass spectrometer was operated in the negative ionization mode. The interface was maintained at 350°C with a nitrogen nebulization pressure of 35 psi, resulting in a flow of 10 l/min. DHEA-sulfate and d6-DHEA-sulfate were detected by selective ion monitoring at m/z 367 and 373, respectively, after 120 V ion fragmentation. Calibration standards were prepared in incubation buffer and processed as described for the incubation samples. Calibration curves were constructed by plotting the peak area ratios of DHEA-sulfate and d6-DHEA-sulfate versus the corresponding DHEA-sulfate concentrations and fitting a linear regression equation to the data. Formation of DHEA-sulfate was then quantified by fitting the peak area ratios (metabolites/internal standard) from incubations to the calibration curve.
Hepatic RNA-Seq Analysis
RNA Isolation.
Human liver RNA was isolated from tissue samples and purified using a NucleoSpin miRNA kit (Macherey-Nagel, Duren, Germany; Clontech Laboratories, Mountain View, CA) according to the manufacturer’s protocol. Briefly, approximately 30 mg liver tissue was combined with 4°C lysis buffer, homogenized using a TissueLyser LT (Qiagen, Valencia, CA), and allowed to sit at room temperature for 5 minutes. The solution was then added to the column. After centrifugation to bind the large RNA to the column, the column was treated with an rDNase solution at room temperature for at least 15 minutes. The flow-through containing the small RNA was treated, to precipitate out the protein. The small RNA was then bound to a new column. After three wash steps of each column, the resulting large and small RNA were each eluted, quantitated, and bioanalyzed for quality control using a Bioanalyzer 2100 (Agilent). Only RNA with an RNA integrity number (RIN) ≥ 7.0 was submitted for sequencing.
cDNA Library Preparation.
Next-generation sequencing libraries were prepared from 1.25 µg total RNA using the TruSeq Stranded mRNA kit (Illumina, San Diego, CA). All of the steps required for sequence library construction have been automated and were performed on a Sciclone NGSx Workstation (Perkin Elmer, Waltham, MA). Ribosomal RNA was depleted by poly(A) enrichment and first- and second-strand cDNA syntheses were performed. Each library was uniquely barcoded using the Illumina adapters and amplified using a total of 13 cycles of polymerase chain reaction. After amplification and cleanup, library concentrations were quantified using the Quant-it dsDNA Assay (Life Technologies, Carlsbad, CA). Libraries were normalized and pooled based on Agilent 2100 Bioanalyzer results (Agilent Technologies). Pooled libraries were size selected using a Pippin Prep (Sage Science, Beverly, MA) and then balanced by mass and pooled in batches of 96 with a final pool concentration of 2 to 3 nM for sequencing on the HiSeq 2500 instrument (Illumina, San Diego, CA).
Read Processing and Final Analysis Pipeline.
The read processing pipeline included the following steps. First, base calls were generated in real time on the HiSeq instrument (Illumina). Second, Illumina RTA-generated BCL files were converted to FASTQ files. Third, custom scripts developed in house were used to process the FASTQ files and to output demultiplexed FASTQ files by lane and index sequence. Fourth, sequence read and base quality were checked using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) and FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Fifth, sequences were aligned to hg19 with reference transcriptome Ensembl version 67 (ftp://ftp.ensembl.org/pub/release-67/embl/homo_sapiens/) using Tophat (Kim et al., 2013). Finally, custom scripts for quality assessment generated metrics.
All aligned read data were subject to the following steps. First, lane-level bam data files were merged using the Picard MergeSamFiles tool and suspected polymerase chain reaction duplicates were marked, but not removed, in the alignment files using the Picard MarkDuplicates tool (http://broadinstitute.github.io/picard/). Second, local realignment was performed around indels, and base quality score recalibration was run using GATK tools (McKenna et al., 2010). Third, variant detection was performed with the GATK Unified Genotyper version 2.6.5 (DePristo et al., 2011). Fourth, aligned data were used for isoform assembly and quantitation with Cufflinks (Kim et al., 2013; Trapnell et al., 2013); genomic features (e.g., SULT2A1) were quantitated with featureCounts (Liao et al., 2014). Finally, sample-level QC scripts were executed to generate final sample-level statistics. Differential expression analysis was performed concurrently with the Cufflinks suite of tools and DESeq (Anders and Huber, 2010).
DNA Sequence Analysis
DNA was isolated from liver samples, as described previously (Shirasaka et al., 2016), and candidate genes were sequenced using the PGRN-Seq platform developed for the Pharmacogenomics Research Network by Gordon et al. (2016). For SULT2A1, sequence coverage included exons 1, 5, and 6, exon-intron boundaries, and parts of the 3′ and 5′ untranslated regions, which were all aligned to the hg19 human reference genome. Sequence data were filtered using VCFtools (Danecek et al., 2011). The average locus read depth was 155. All single nucleotide variants (SNVs) passed a Phred quality score > 50, and for the SULT2A1 locus, the average Phred quality score for all variant positions was 389,614. SNVs were annotated with SeattleSeq annotation, briefly described by Ng et al., (2009).
Quantitation of SULT2A1 and SULT1A1 Proteins in Hepatic Cytosol
Sample Preparation for SULT2A1, SULT1A1, and BSA Protein Quantification.
BSA was used as a protein internal standard to address intersample and interday variability in the trypsin digestion, whereas heavy labeled peptides were used as a control to address LC-MS/MS instrument variability. Ten microliters of BSA (100 μg total) was spiked into 80 μl liver cytosolic samples (2.0 mg/ml), and the protein mixture was denatured, reduced, and alkylated as per published protocol (Prasad and Unadkat, 2014a; Prasad et al., 2016). Similarly, the denatured/alkylated protein was then desalted and precipitated using methanol/chloroform/water precipitation followed by resuspension and digestion, as discussed previously (Prasad et al., 2016). The digestion reaction was quenched by 20 µl labeled peptide internal standard cocktail containing the two heavy peptides listed in Supplemental Table 1 (prepared in 50% acetonitrile in water containing 0.1% formic acid) plus 10 μl blank solvent (50% acetonitrile in water containing 0.1% formic acid). The samples were centrifuged at 5000g for 5 minutes at 4°C, and 5 µl of the supernatants was injected onto the LC-MS/MS system.
LC-MS/MS Peptide Analysis.
Surrogate peptides of SULT2A1, SULT1A1, and BSA were quantified using optimized LC-MS/MS parameters listed in Supplemental Tables 1 and 2 using an AB Sciex 6500 tandem mass spectrometer coupled to Waters Acquity UPLC system (Waters, Hertfordshire, UK) with few modifications, mainly in the LC parameters. Briefly, a UPLC column (Acquity UPLC HSS T3 1.8 µm, 2.1 × 100 mm; Waters) with a Security Guard column (C18, 4 × 2.0 mm) from Phenomenex (Torrance, CA) was eluted (0.3 ml/min) with a gradient mobile phase consisting of water and acetonitrile (with 0.1% formic acid; see below). The injection volume was 5 µl (approximately 10 μg total protein). The parent to product ion transitions for the analyte peptides and their respective stable isotope labeled (SIL) peptides were monitored using optimized LC-MS/MS parameters (Supplemental Tables 1 and 2) in ESI positive ionization mode. Peak areas of each fragment were calculated, multiple fragments of each peptide parent ion were averaged, and the area ratios of light versus heavy were calculated. Finally, SULT2A1 and SULT1A1 area ratios were normalized by the BSA ratios. Each sample was analyzed twice on 2 different days and average of the 2 days was considered for the data analysis. For absolute quantification of SULT2A1 in the pooled cytosol sample, nine serial dilutions of the purified protein were digested and processed similar to the cytosol samples. The on-column amounts of SULT2A1 standard ranged from 0.15 (lower limit of quantification) to 38 fmol.
Binding of 25OHD3-3-O-Sulfate and 25OHD3 to the DBP
The binding assays were performed as described previously (Horst et al., 1981). Briefly, rat plasma was diluted 1:5000 (v/v) in 0.05 M phosphate buffer (pH 7.5) containing 0.01% gelatin and 0.01% merthiolate (PBG buffer). Each assay mixture was placed in a borosilicate glass tube and consisted of the following: 1) 0.5 ml 1:5000 dilution of vitamin D–deficient rat plasma in PBG buffer, 2) 6000–8000 cpm [23,24-3H]-25OHD3 in 20 µl 100% ethanol, and 3) vitamin D metabolites in 25 µl ethanol. After 1 to 2 hours of incubation at 4°C, the bound vitamin D metabolites were separated from free vitamin D metabolites by adding 0.2 ml of a mixture of cold 1.0% Norit A Charcoal and 0.1% Dextran T-70 (Sigma-Aldrich, St Louis, MO) in PBG buffer to each tube. After 30 minutes at 4°C, the tubes were spun at 1000g for 10 minutes in a refrigerated centrifuge. A portion (0.5 ml) of the supernatant was removed for quantitation of the bound [3H]-25OHD3. The binding experiment was conducted with rat plasma so that we could directly compare new results to historical data generated by our laboratory using rat plasma (Wang et al., 2014). Previously, Bouillon et al. (1980) concluded that the heterogeneity between the genes encoding for DBP in rats and humans, and the subsequent structural difference between the two proteins, has no effect on their affinity or capacity to bind vitamin D metabolites.
Statistical Analysis
All data are expressed as the mean ± S.D. unless stated otherwise. Group comparisons were made using one-way analysis of variance with Dunnett’s multiple-comparisons test. For all cell- and recombinant enzyme-based experiments, we used GraphPad Prism software (version 5) for the statistical analyses, and a P value < 0.05 was considered statistically significant. For results from experiments with human livers, we used a linear trend test (one-way analysis of variance) to assess SULT2A1 genotype-phenotype associations, and a P value < 0.05 was considered to be statistically significant. The regression coefficient (r2) was calculated in Microsoft Office Excel 2010 (Microsoft Corporation, Redmond, WA).
A multivariate linear regression analysis of genetic, demographic, and other variables for the liver donors was performed in R software (version 3.3.3; R: A language and environment for statistical computing. R Core Team, 2017. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.) to elucidate major determinants of interindividual variability in 25OHD3 sulfonation activity. Full details are given in the Supplemental Methods. Regression analyses were performed with the log-transformed activity data, because of its non-normal (skewed) distribution. A P value < 0.05 was considered to be statistically significant for all tests.
Results
Formation of 25OHD3-3-O-Sulfate by Human Liver Cytosols and Recombinant SULTs
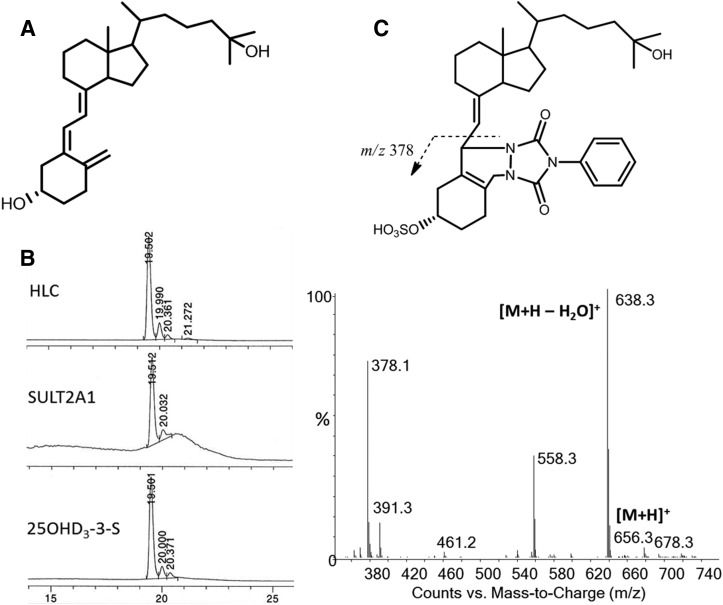
Sulfonation of 25OHD3 is catalyzed by cytosolic SULT isozymes. 25OHD3 contains two hydroxyl groups and presumably two different sulfate conjugate isomers either at the C-3 or C-25 positions could be produced (Fig. 1A). Incubation of 25OHD3 with pooled human liver cytosols and PAPS cofactor generated a single major sulfate product (retention time: 19.5 minutes) with selective mass ion monitoring at m/z 479, which corresponds to [M ‒ H]‒ under the negative ion mode (Fig. 1B). An identical peak was also detected with recombinant SULT2A1 and SULT1A1 incubations, but not SULT1B1, SULT1E1, and SULT2B1, under the incubation conditions that were employed. The activity of SULT2A1 greatly exceeded that of SULT1A1, which was seen in only one of three replicate experiments. The LC-MS characteristics of the predominant cytosolic metabolite were identical. Its LC retention time matched with that of the synthesized standard 25OHD3-3-O-sulfate, indicating conjugation with a sulfate group at the C-3 position (Fig. 1B). This was further confirmed by the ion fragmentation pattern after PTAD derivation. As shown in Fig. 1C, an identical daughter ion at m/z 378 indicated a direct conjugation at the C-3 position and cleavage between C-6 and C-7, which is in agreement with the fragmentation pattern of standard 25OHD3-3-O-sulfate. Based on these data, we conclude that the predominant product of SULT2A1 and human liver cytosol is 25OHD3-3-O-sulfate. A minor peak (retention time: 20.0 minutes) with selective mass ion at m/z 479 was also observed in incubations with either SULT2A1 or human liver cytosol; however, whether this peak is 25OHD3-25-O-sulfate or a 25OHD3-3-O-sulfate stereoisomer remains unknown (Fig. 1B).
Fig. 1.
Formation of 25OHD-3-O-sulfate by human liver cytosols and SULT2A1. (A and B) Considering the structure of 25OHD3 shown in (A), sulfonation occurs preferentially at the 3-OH position as seen from its formation by human liver cytosol and SULT2A1 (B). (C) Derivatization with PTAD yields an adduct that, after ionization, gives a diagnostic ion at m/z 378 that includes the site of sulfonation. HLC, human liver cytosol.
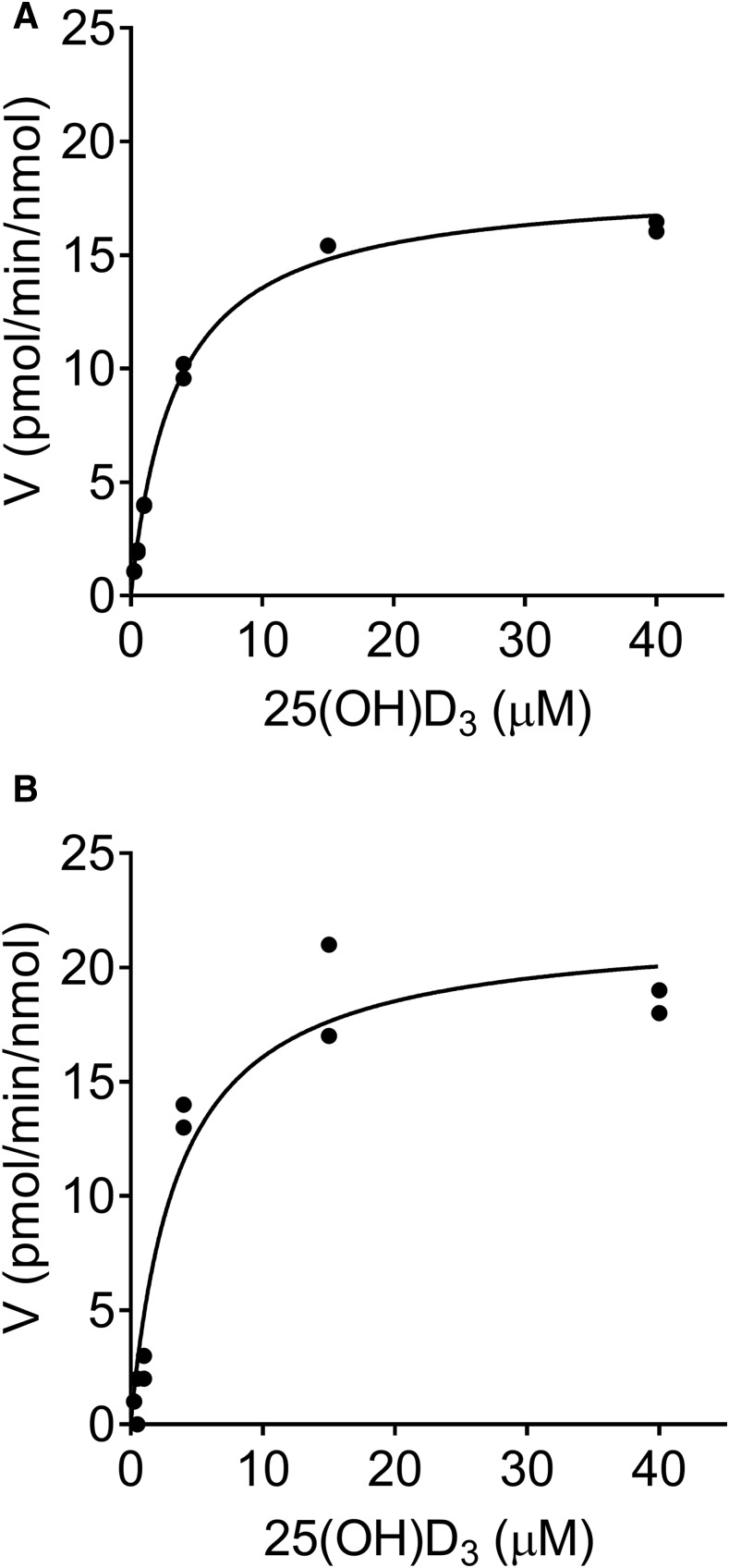
Kinetic studies of 25OHD3-3-O-sulfate formation were performed in vitro using pooled human liver cytosols and recombinantly expressed SULT2A1 (Fig. 2). The Km and Vmax values for 25OHD3-3-O-sulfate formation estimated using a simple Michaelis–Menten kinetics model are presented in Table 1. Incubations with human liver cytosol and SULT2A1 yielded nearly identical mean apparent Km values (3.43 vs. 3.36 µM, respectively). In addition, when normalized to the measured specific SULT2A1 content (1360 ± 106 pmol/mg protein) of the pooled human liver cytosol used in these experiments, the mean Vmax value was comparable to that of the pooled recombinant human SULT2A1 (17.7 and 21.3 pmol/min per nanomole SULT2A1, respectively). The corresponding intrinsic sulfonation clearances were also similar (5.15 and 6.59 µl/min per nanomole SULT2A1, respectively). These results suggest that SULT2A1 is the predominant source of sulfonation activity in the human liver and could represent an important metabolic route for 25OHD3 clearance.
Fig. 2.
Kinetics of 25OHD3-3-O-sulfate formation. (A and B) Concentration-dependent formation of 25OHD3-3-O-sulfate by pooled human liver cytosol (A) and recombinant SULT2A1 (B) are shown. Individual data points from duplicate incubations at each substrate concentration are shown. The solid line represents the result of fitting a simple Michaelis–Menten equation to the substrate concentration-rate data. Mean parameter (Km and Vmax) estimates from three separate experiments for each enzyme system are shown in Table 1. The Vmax values were normalized to the SULT2A1 protein content.
TABLE 1.
Kinetic parameters for 25OHD3-3-O-sulfate formation in vitro
Values are the mean ± S.D. of three independent experiments conducted on different days.
| Parameter | Human Liver Cytosol | SULT2A1 |
|---|---|---|
| Vmax (pmol/min per milligram) | 24.0 ± 1.88 | |
| Vmax (pmol/min per nanomole) | 17.7 ± 1.38 | 21.3 ± 3.88 |
| Km (µM) | 3.43 ± 0.02 | 3.36 ± 1.10 |
| CLint (µl/min per milligram) | 7.00 ± 0.54 | |
| CLint (µl/min per nanomole) | 5.15 ± 0.40 | 6.59 ± 1.13 |
A simple Michaelis–Menten model was fitted to the data. Pooled human liver cytosol was prepared from different randomly selected livers (n = 10). Cytosolic Vmax and CLint values normalized for total protein (in milligrams) and specific SULT2A1 content (in nanomoles) are presented. SULT2A1 protein was a recombinantly expressed, purified product obtained from R&D Systems. CLint, intrinsic clearance.
25OHD3-3-O-Sulfate Is the Major Metabolite Formed from 25OHD3 by Human Hepatocytes
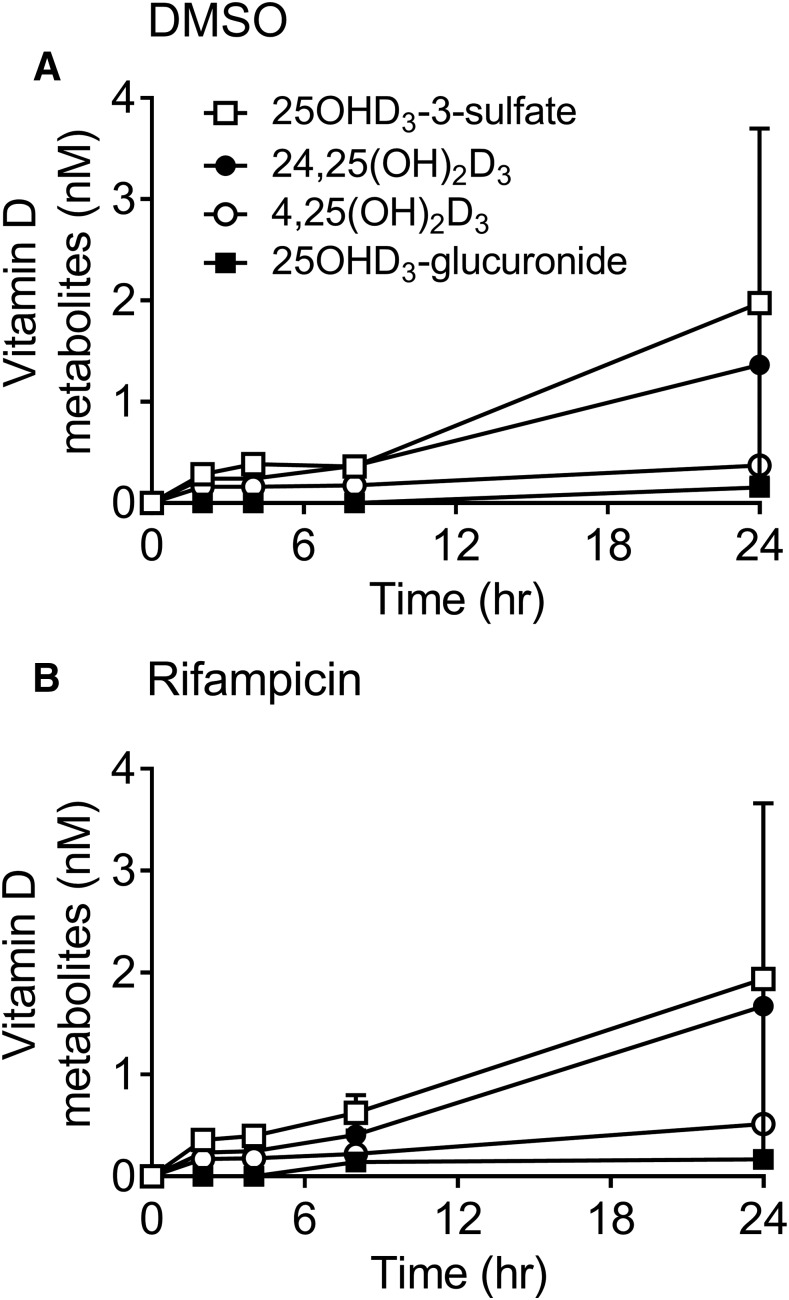
Previous studies have demonstrated that at nonphysiologic concentrations (1–5 µM), 25OHD3 is metabolized to 1α,25(OH)2D3, 24R,25(OH)2D3, 4α,25(OH)2D3, 4β,25(OH)2D3, 25OHD3-3-O-glucuronide, 25OHD3-25-O-glucuronide, and putative 5,6-trans-25OHD3-25-O-glucuronide when incubated with human hepatocytes (Wang et al., 2014). However, formation of 25OHD3-3-O-sulfate in human hepatocytes was not determined. For this investigation, a more physiologic concentration of 25OHD3 (50 nM) was applied to cultured human hepatocytes to generate a metabolite profile. As shown in Fig. 3A, all major metabolites of 25OHD3, except 1α,25(OH)2D3, were detected in the incubations. 25OHD3-3-O-sulfate was the most abundant product observed, followed by 24R,25(OH)2D3, 4,25(OH)2D3 [4α,25(OH)2D3 and 4β,25(OH)2D3], and 25OHD3-glucuronides. Formation of these 25OHD3 metabolites occurred in a linear, time-dependent manner, except for 4β,25(OH)2D3 and 4α,25(OH)2D3 formation, which underwent extensive sequential glucuronidation, as previously reported (Wang et al., 2013a).
Fig. 3.
Time-dependent formation of 25OHD3 metabolites by human hepatocytes. (A and B) Primary human hepatocytes pretreated for 24 hours with DMSO (A) or 10 µM rifampicin in DMSO (B) were incubated with 50 nM 25OHD3 for an additional 0–24 hours and the products were analyzed for 25OHD3-3-O-sulfate (open squares), 24R,25(OH)2D3 (closed circles), 25OHD3-3-O-glucuronide (closed squares), and 4,25(OH)2D3 (open circles) by LC-MS/MS, as described in the Materials and Methods. Each data point represents the mean ± S.D. of three replicate hepatocyte incubations per donor. Two donors were used in the study. The mean rates of formation of each metabolite in hepatocytes (or rifampicin-treated hepatocytes) were as follows: 0.16 (or 0.16) pmol/h per 106 cells (25OHD3-3-O-sulfate), 0.11 (or 0.13) pmol/h per 106 cells [24R,25(OH)2D3], 0.01 (or 0.01) pmol/h per 106 cells (25OHD3-O-glucuronide), and 0.03 (0.04) pmol/h per 106 cells (4,25(OH)2D3); measured after β-glucuronidase treatment. DMSO, dimethylsulfoxide.
By comparison, renal tubule epithelial cells and LS180 intestinal epithelial cells showed no detectable formation of 25OHD3-3-O-sulfate during a 24-hour incubation with 50 nM 25OHD3 (data not shown). These cells have previously been shown to catalyze the 24-hydroxylation of 25OHD3 under similar culture conditions (Zheng et al., 2012; Weber et al., 2016). Based on the assay limit of detection, culture conditions, and the approximate number of cells per well, it was estimated that 25OHD3 sulfonation activity per renal tubule epithelial cell or LS180 cell was <10% that of cryopreserved human hepatocytes.
Chronic exposure to pregnane X receptor (PXR) agonists, such as rifampicin, can cause a reduction in plasma 25OHD3 blood levels (Brodie et al., 1980) by a mechanism that is not completely understood. In a previous study, we showed that treatment of human hepatocytes with rifampicin significantly increased the 25OHD3 4-hydroxylation pathway by induction of hepatic CYP3A4 (Wang et al., 2013a). The effect of rifampicin on 25OHD3 sulfonation is unclear (Echchgadda et al., 2007; Fang et al., 2007) and thus was explored. After incubation of hepatocytes with 10 µM rifampicin or vehicle (0.1% dimethylsulfoxide) for 48 hours, the cells were washed and incubated with 50 nM 25OHD3 for 2, 4, 8, and 24 hours. There was no apparent difference in 25OHD3-3-O-sulfate formation between rifampicin-treated and control hepatocytes, whereas as expected, 4,25(OH)2D3 formation was significantly (37.6% after 24 hours) induced by rifampicin treatment (Fig. 3B).
Associations between SULT2A1 Gene Variants and Hepatic SULT2A1 mRNA and Protein Abundances and Cytosolic Sulfonation Activities
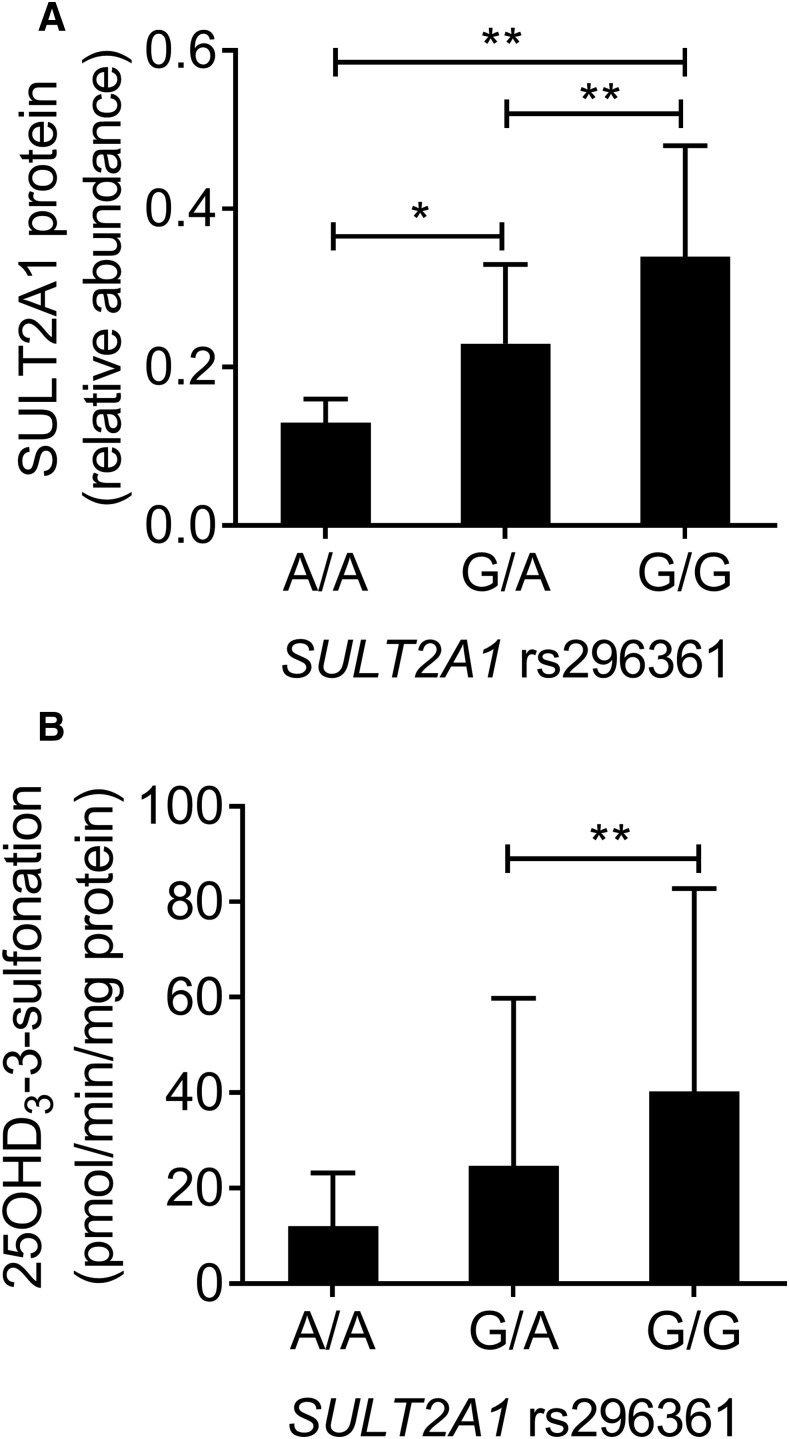
With the identification of SULT2A1 as the principal catalyst of 25OHD3-3-O-sulfate formation, we sought to test whether variation in the SULT2A1 gene sequence might contribute to interindividual differences in hepatic sulfonation activity. To that end, we accessed preexisting sequence data (Gordon et al., 2016) for parts of the SULT2A1 gene (coverage included exons 1, 5, and 6; intron-exon boundaries; and parts of the 3′ and 5′ untranslated regions) in DNA isolated from 258 human livers. Relative to the reference human genome, a total of 12 different SNVs were identified; eight in untranslated regions, one intronic, one synonymous coding, and two nonsynonymous coding variants (Supplemental Fig. 1). Of these, eight SNVs had minor allele frequencies > 0.6%. Three variants were common (rs296366, T>C; rs296365, C>G; and rs296361, G>A) and detected at an allele frequency of 80%, 27%, and 15%, respectively, and all were in Hardy-Weinberg equilibrium. Of the three common SNVs, two (rs296366 and rs296365) were located in the 3′ region of the gene and one was near the intron 1/exon 1 boundary (rs296361). All three exhibited a high degree of linkage disequilibrium (data not shown). An assessment of SULT2A1 mRNA content of the same livers was obtained by RNA-Seq analysis and revealed significant associations between rs296361, but not rs296366 and rs296365, and the relative level of hepatic SULT2A1 mRNA content, with a 70% lower mean level of mRNA in livers homozygous for the rs296361 A allele, compared with the reference G allele (Supplemental Fig. 2). Accordingly, we focused further analysis on rs296361 because of its association with mRNA level and potential of the intron 1 SNV to disrupt mRNA splicing and message accumulation.
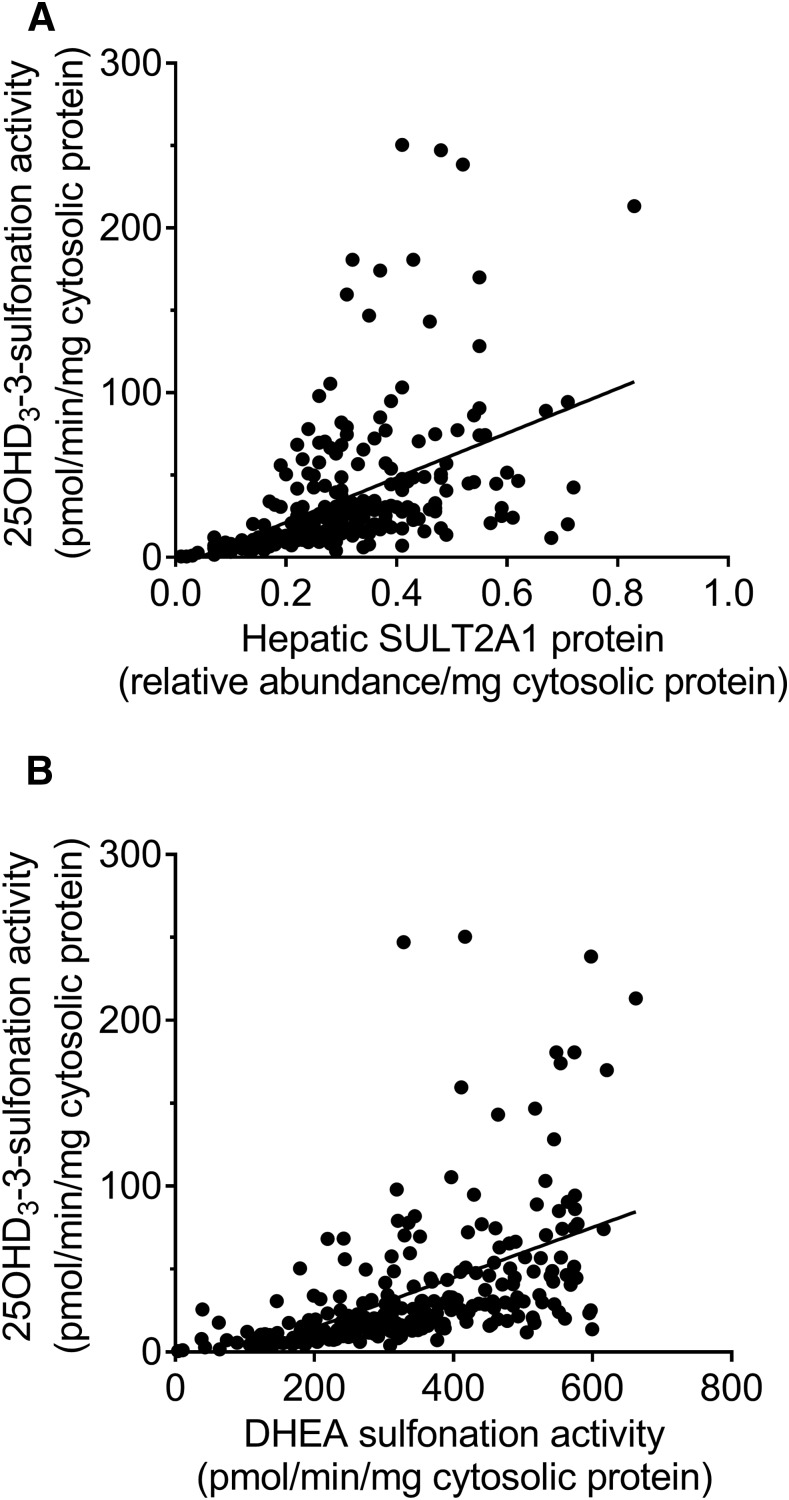
Cytosol isolated from the same bank of human livers was subjected to quantitative analysis of SULT2A1 protein content and 25OHD3 sulfonation activity at a substrate concentration of 5 µM. There were significant associations between SULT2A1 rs296361 and total SULT2A1 protein (Fig. 4A) and the cytosolic 25OHD3-3-O-sulfate formation rate (Fig. 4B). Consistent with the mRNA and protein results, the mean rate of 25OHD3-3-O-sulfate formation in cytosol from livers homozygous for the variant allele was only 30% of that found for the livers homozygous for the reference allele. Not surprisingly, there was a statistically significant correlation between SULT2A1 protein content and the 25OHD3-3-O-sulfate formation rate, where differences in the protein level explained 22% of the unadjusted variance in sulfonation activity (Fig. 5A).
Fig. 4.
Association between SULT2A1 rs296361 and SULT2A1 protein content and 25OHD3 sulfonation rate in human liver. (A and B) Cytosol was isolated from 258 human livers for quantitation of SULT2A1 protein content (A) and 25OHD3 3-O-sulfonation rate (B). Mean ± S.D. data for rs296361 G>A genotypes are shown. Group results were compared by ANOVA, followed by pairwise comparisons if the ANOVA result was significant. *P < 0.05; **P < 0.01. Of additional note, the comparison of the 25OHD3 3-O-sulfonation rate for the homozygous rs296361 GG and AA groups reached near significance (P = 0.052 with post hoc analysis). ANOVA, analysis of variance.
Fig. 5.
Correlation between cytosolic 25OHD3-3-sulfonation rate and SULT2A1 protein content and DHEA sulfonation rate in human liver. Cytosol was isolated from 258 human livers for quantitation of SULT2A1 protein content, 25OHD3 3-O-sulfonation rate, and DHEA 3-sulfonation rate. (A and B) Linear regressions between 25OHD3 3-O-sulfonation rate and SULT2A1 protein content (A; r2 = 0.22) and between 25OHD3 3-O-sulfonation rate and DHEA 3-sulfonation rate (B; r2 = 0.27) are shown.
Correlation between 25OHD3-3-O-Sulfate Formation and DHEA-Sulfate Formation by Human Liver Cytosol
DHEA is considered to be a selective probe substrate for SULT2A1 (Mueller et al., 2015) and we thus determined the rate of formation of DHEA-3-O-sulfate in liver cytosolic preparations incubated with 5 µM DHEA. As seen in Supplemental Fig. 3, there was a significant association between the SULT2A1 rs296361 variant and the DHEA-3-O-sulfate formation rate, which was also strongly correlated with cytosolic SULT2A1 protein content (r2 = 0.71; Supplemental Fig. 4). There was also a significant correlation between the rates of 25OHD3-3-O-sulfate and DHEA-3-O-sulfate formation among the panel of human livers tested ( (r2 = 0.27, P < 0.0001; Fig. 5B), indicative of a common enzyme catalyzing the two reactions. However, an examination of the 25OHD3 sulfonation rate regression curves suggested the possibility of a second enzyme contributing to 25OHD3-3-O-sulfate formation at the substrate concentration tested. Because we had observed some activity with the recombinantly expressed SULT1A1 enzyme, we quantified SULT1A1 protein levels in the panel of human liver cytosols. There was no association between cytosolic SULT1A1 protein content and 25OHD3-3-O-sulfate formation rate (data not shown).
Stratification of the 25OHD3 sulfonation activity by the liver bank source revealed higher average sulfonation activity in the University of Washington livers, compared with the St. Jude livers (data not shown). This was not the case for DHEA sulfonation activity. A multivariate analysis that considered SULT2A1 genotype and donor sex, age, ethnicity, liver repository (St. Jude or University of Washington), and duration of tissue storage at −80°C as covariates revealed significant contributions from SULT2A1 rs296361 genotype, liver repository, and storage duration to the variance in the log 25OHD3-3-O-sulfate formation rate (Table 2). The full regression model explained 47% of the variance in the log 25OHD3 sulfonation rate.
TABLE 2.
Multivariate regression analysis of human cytosol 25OHD3 3-sulfonation activity
| Characteristic | β-Coefficient in Full Modela | Significance in Full Model, P Value | Variability Explained, R2b |
|---|---|---|---|
| Fully adjusted model | 2.2 × 10−16 | 0.47 | |
| rs296361 (additive effect) | 0.50 ± 0.10 | 4.8 × 10−7 | |
| Sex (reference: male) | −0.11 ± 0.10 | 0.27 | |
| Age (years) | −0.004 ± 0.003 | 0.10 | |
| Ethnicity (reference: white) | −0.28 ± 0.32 | 0.37 | |
| Liver bank (reference: University of Washington) | −1.59 ± 0.14 | 2.0 × 10−16 | |
| Storage time (months) | −0.03 ± 0.01 | 7.1 × 10−3 |
Variability in the log liver cytosolic 25OHD3 sulfonation rate explained by predictors in a multiple linear regression is shown. A subset (n = 226) of the overall dataset (n = 258) was included; 32 samples were excluded because of missing age. The significance of the association of each variable with the log 25OHD3 sulfonation rate is indicated by the P value and was determined using a likelihood ratio test. Values are β-coefficients ± S.E. from a multiple linear regression model. β-coefficients were determined as the change in the log 25(OH)D3 sulfonation rate (in picomoles per minute per milligram) per unit change in the predictor variable, when all other predictor variables were held constant.
The goodness of fit of a multiple linear regression, on a scale of 0–1, as reflected in the R2 value for the fully adjusted model of the log 25(OH)D3 sulfonation rate as the outcome variable and all predictors in the table (rs296361, sex, age in years, ethnicity, liver bank, and duration of tissue storage in months) is shown. Reference groups for categorical variables are shown in parentheses.
Detection of 25OHD3-3-O-Sulfate in Pooled Human Plasma, Bile, and Urine
The presence or absence of 25OHD3-3-O-sulfate in human plasma, bile, and urine was evaluated. Due to the complexity of these biologic matrices, an optimized extraction method was developed and employed to partially purify the biosamples individually. An anion exchange cartridge was used to extract 25OHD3-3-O-sulfate from plasma, bile, and urine samples. Deuterium labeled sulfate standard was applied to identify the potential sulfate peak and was used to estimate and then correct for the total recovery during SPE and subsequent steps. As shown in Fig. 6A, a representative chromatogram derived from plasma extracts clearly showed the presence of 25OHD3-3-O-sulfate in human plasma. An analysis of plasma from 21 healthy adults produced mean concentrations of 130.8 ± 70.8, 96.0 ± 44.0, 12.0 ± 7.7, and 0.26 ± 0.14 nM for 25OHD3, 25OHD3-3-O-sulfate, 24R,25(OH)2D3, and 4β,25(OH)2D3, respectively.
Fig. 6.
Detection of 25OHD3-3-O-sulfate in human plasma and bile by LC-MS/MS. (A and B) Biologic samples were subjected to SPE and LC-MS/MS analysis (A) or SPE and derivatization with DAPTAD and LC-MS/MS analysis (B), as described in the Materials and Methods. Peaks corresponding to the d6-25OHD3-3-O-sulfate and 25OHD3-3-O-sulfate analytes are shown.
The presence of 25OHD3-3-O-sulfate in human bile was suggested by detection after SPE of a peak with a mass ion of m/z 479, which corresponds to [M ‒ H]‒ under the negative ion mode, that coeluted with standard 25OHD3-3-O-sulfate (Fig. 6A). Reacting the solid-phase extract with the vitamin D derivatizing agent, PTAD, resulted in a loss of the ion at m/z 479 (data not shown). To confirm this finding, we reacted the extracted material with another derivatizing reagent (DAPTAD), which provided greater analytical sensitivity. As seen in Fig. 6B, a single product coeluting with the derivatized deuterated internal standard was observed. Comparing the signal to that of reference 25OHD3-3-O-sulfate standards yielded an estimated pooled bile concentration of 2.55 nM (mean of two replicate determinations).
25OHD3-3-O-sulfate was not detected in 0.5 ml human urine (n = 3) under underivatized or derivatized conditions.
Binding of 25OHD3-3-O-Sulfate to Rat Plasma DBP
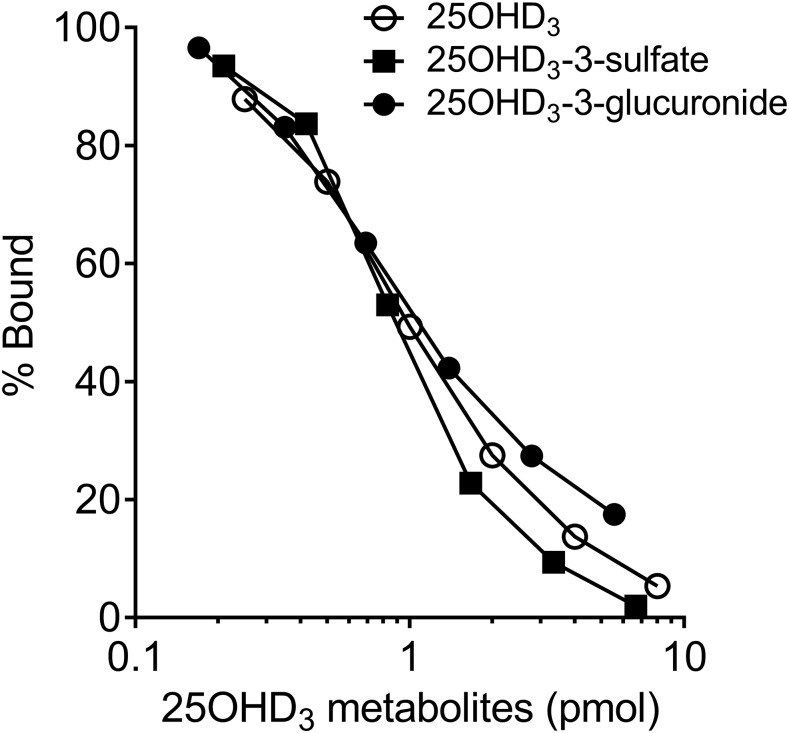
The presence of 25OHD3-3-O-sulfate in plasma, but not urine, suggested the possibility that it might have significant binding affinity for the DBP, restricting its excretion by the kidney, as occurs for 25OHD3. Previous studies have shown that 25OHD3-3-O-glucuronide, but not 25OHD3-25-O-glucuronide, binds tightly to DBP (Wang et al., 2014). Due to the structure similarity, we surmised that 25OHD3-3-O-sulfate could also bind to DBP. To test this hypothesis, the concentration-dependent binding of 25OHD3-3-O-sulfate, 25OHD3-3-O-glucuronide, and 25OHD3 to rat plasma DBP was measured by radioligand binding assay (Horst et al., 1981). As seen in Fig 7. the binding affinity of 25OHD3-3-O-sulfate for DBP was essentially identical to that of 25OHD3 and 25OHD3-3-O-glucuronide. The mean EC50 values for 25OHD3, 25OHD3-3-O-glucruonide, and 25OHD3-3-O-sulfate binding to DBP under current incubation conditions were 0.82, 0.74, and 0.72 pmol, respectively.
Fig. 7.
Binding of 25OHD-3-O-sulfate to rat DBP. A competitive binding assay was performed as described in the Materials and Methods. A range of amounts of 25OHD3, 25OHD3-3-O-sulfate, and 25OHD3-3-O-glucuronide were incubated with 3H-25OHD3 and rat DBP and the percent 3H-25OHD3 bound was calculated for each amount of unlabeled 25OHD3-3-O-sulfate. EC50 values from a single binding site model were as follows: 0.82 pmol (25OHD3), 0.72 pmol (25OHD3-3-O-sulfate), and 0.74 pmol (25OHD3-3-O-glucuronide).
Discussion
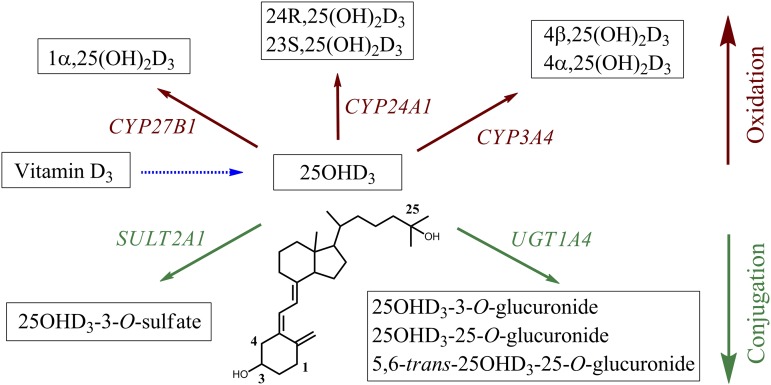
Previous studies from our research group demonstrated that glucuronidation of 25OHD3 occurs in humans and is catalyzed primarily by UGT1A3 and UGT1A4 (Wang et al., 2014). Herein, we confirm that 25OHD3-3-O-sulfate is also a quantitatively important circulating product of 25OHD3 (Axelson, 1985; Shimada et al., 1995) and we report for the first time that it is generated primarily in the liver by the enzyme SULT2A1. The sulfate conjugate was the dominant metabolic product of 25OHD3 produced by primary human hepatocytes when incubated with a physiologically relevant 50 nM concentration of 25OHD3. Thus, interindividual differences in sulfonation activity may be a major source of variation in circulating blood 25OHD3 concentrations (Fig 8)., assuming that hepatic clearance of 25OHD3 is an important determinant of 25OHD3 accumulation in the body.
Fig. 8.
Metabolic pathways (oxidation and conjugation) of 25OHD3 and their corresponding enzymes in human liver.
Results from our kinetic experiments, revealing comparable Km and Vmax values when normalized for SULT2A1 content, suggest that this isozyme is the predominant catalyst of the 25OHD3 sulfonation reaction in the human liver. This conclusion is supported in part by the positive regression of liver cytosolic sulfonation activity against SULT2A1 protein content and the strong association between SULT2A1 genetic variation and liver cytosolic sulfonation activity. However, we note that approximately one-half of the variance in liver cytosolic 25OHD3 sulfonation activity remained unexplained and that variation in cytosolic SULT2A1 protein content explained only 21% of the unadjusted metabolic activity toward this substrate, in contrast to a much stronger correlation between cytosolic DHEA sulfonation activity and SULT2A1 protein content. Nonspecific factors associated with the liver procurement site explained a substantial fraction of the observed variance in liver cytosolic 25OHD3 sulfonation activity and variable storage time and possible loss of activity but not measured protein was also contributory (Table 2). We speculate that these and possibly other factors (e.g., uncontrolled substrate-selective protein-SULT2A1 interactions) masked what would otherwise have been a stronger association between SULT2A1 protein level and sulfonation activity.
SULT2A1 is known to metabolize hydroxysteroids, such as estradiol, DHEA and bile acids (Chatterjee et al., 2005). That it also catalyzes 25OHD3 sulfonation is not surprising, considering structural similarities. The regiospecificity of SULT2A1 toward 25OHD3 sulfonation, almost exclusively at the 3-position, is similar to that observed for hydroxysteroids and oxysterols. Interestingly, the related SULT isoform, SULT2B1, showed little activity toward 25OHD3, which is in contrast to its ability to generate sulfate metabolites of hydroxysteroids and oxysterols that are substrates for SULT2A1 (Falany and Rohn-Glowacki, 2013). SULT2B1 is expressed primarily in extrahepatic tissues (Falany and Rohn-Glowacki, 2013), whereas SULT2A1 is highly expressed in the liver and adrenal cortex and less so in the gastrointestinal tract (Chatterjee et al., 2005). This distribution pattern suggests that there will be limited 25OHD3 sulfonation activity outside of the liver. Consistent with these findings, no detectable 25OHD3 sulfonation was observed in either human intestinal LS180 cells or renal epithelial cells incubated with a physiologically relevant concentration of 25OHD3.
We found no evidence that hepatic 25OHD3 sulfonation activity is induced by PXR agonists, based on our negative results with rifampicin and primary human hepatocytes, further supporting a role for enhanced oxidative metabolism in the reduction of plasma 25OHD3 levels after chronic rifampicin treatment (Brodie et al., 1980). This result is concordant with the findings of Fang et al. (2007), who reported inconsistent inductive responses in a relatively large number of primary human hepatocyte preparations and, with additional experiments, suggested that PXR actually represses SULT2A1 expression and that sporadic inductive responses in hepatocytes are mediated by a non-PXR mechanism. However, it is at odds with other published results from PXR-transfected Caco-2 and HepG2 systems that suggest transcriptional enhancement of SULT2A1 by PXR agonists (Echchgadda et al., 2007). Characterization of 25OHD3-3-O-sulfate levels after rifampicin treatment in vivo could help clarify the discrepancy. It is noteworthy that SULT2A1 is thought to be regulated by 1α,25(OH)2D3 (Echchgadda et al., 2004), suggesting an autofeedback mechanism of vitamin D regulation, as described for CYP24 (Haussler et al., 2013) and proposed for CYP3A4 (Xu et al., 2006; Wang et al., 2013a).
Partial sequence analysis of the SULT2A1 gene revealed a strong association between an SNV (rs296361) and cytosolic sulfonation activity and SULT2A1 protein abundance. Accordingly, we hypothesize that this SULT2A1 SNV may contribute to interindividual differences in the circulating plasma 25OHD3-3-O-sulfate and 25OHD3 levels, something that should be tested in future investigations. The minor allele frequency for rs296361, as reported in the National Center for Biotechnology Information dbSNP database, is 16.1%, 0.4%, 0.3%, 6.2%, and 10.6% for Europeans, East Asians, Africans, admixed Americans, and South Asians, respectively. Interestingly, although the variant SNV resides within intron 1, it was also strongly associated with hepatic SULT2A1 mRNA abundance, suggesting that RNA splicing or the accumulation of the mature message is somehow affected. RNA-Seq analysis did not reveal alternative splice variants, suggesting that the efficiency of splicing was altered by the SNV, perhaps by altering the function of a splice enhancer or repressor sequence. It is also possible that the intron 1 SNV is in linkage disequilibrium with another allelic variant not detected by our sequencing analysis (e.g., in unsequenced exonic and intronic regions or an upstream or downstream regulatory region of the SULT2A1 gene) and that rs296361 has no causal effect on the SULT2A1 phenotype. It is important to note that a copy number variation (CNV) in the SULT2A1 gene locus has also been described (Ekström and Rane, 2015) and has been associated with the urinary excretion of DHEA-sulfate and testosterone-sulfate (Schulze et al., 2013). However, the frequency of the CNV appears to be much lower than that of rs296361. Whether there is linkage disequilibrium between the two gene variants remains to be determined, as we did not test for the CNV with the sequencing platform that we employed. Regardless of the mechanism behind the rs296361–SULT2A1 phenotype associations, the data strongly suggest that this common genetic variation could influence the biologic effects of 25OHD3, DHEA, and other important hormone signaling molecules metabolized by SULT2A1.
25OHD3-3-O-sulfate was found to have a high binding affinity for DBP, which explains its relatively high abundance in plasma and absence from urine. The crystal structure of DBP indicates that the vitamin D binding site is a cleft, which can easily accommodate large substituents at the C-3 position of 25OHD3 (e.g., conjugated moieties) (Verboven et al., 2002). High binding affinity to DBP would reduce the renal excretion of 25OHD3-3-O-sulfate into urine. We speculate that the complex of 25OHD3-3-O-sulfate and DBP is filtered and then reabsorbed in renal proximal tubules by megalin/cubilin-mediated endocytosis, as shown for the 25OHD3-DBP complex (Rowling et al., 2006).
Although 25OHD3-3-O-sulfate is a major circulating form of vitamin D3, whether it possesses biologic activity directly or indirectly is unclear. A number of studies have been conducted to understand the biologic activities of vitamin D3-sulfate, a conjugated metabolite of vitamin D3 (Higaki et al., 1965; Sahashi et al., 1967a,b, 1969). Vitamin D3-sulfate was synthesized previously (Reeve et al., 1981) and its biologic activity was determined in a vitamin D–deficient rat model. Activity was observed, but only at doses higher than what can be elicited by vitamin D3 (Nagubandi et al., 1981). Later studies also showed less biologic activity of vitamin D3-sulfate than free vitamin D3 in vivo (Cancela et al., 1985). However, in each of these studies, vitamin D3-sulfate was administered, rather than having the metabolite generated in situ, with the uncertainties of bioavailability and access to cellular sites that complicate quantitative comparisons. Thus, it is possible that 25OHD3-3-O-sulfate might undergo hydrolysis, catalyzed by ubiquitous sulfatases and regenerate 25OHD3. This type of hormone conjugate cycling is observed for estrogen and DHEA (Mueller et al., 2015), where the sulfo-conjugates are the dominant form in blood circulation and are distributed to peripheral tissues where desulfonation can occur. In the case of DHEA-3-O-sulfate, conversion to DHEA is followed by metabolism to androstenedione and downstream androgens and estrogens (Strott, 2002).
Finally, given the detection of 25OHD3-3-O-sulfate in bile, we are intrigued by the possibility that preferential delivery of the hormone conjugate to the duodenum and upper small intestine might explain the preferential expression of vitamin D receptor target genes, such as CYP3A4, transient receptor potential cation channel subfamily V member 6 (TRPV6), and calbindin D9K, in the upper small intestine (Wang et al., 2013b). Results from unpublished studies indicate that 25OHD3-3-O-sulfate is a substrate for the cell uptake transporter, organic anion transporting polypeptide 2B1 (OATP2B1), which is expressed in the intestinal epithelia (Drozdzik et al., 2014). Once absorbed into mucosal epithelial cells, 25OHD3-3-O-sulfate could be hydrolyzed to 25OHD3 and then undergo 1α-hydroxylation to the active hormone and contribute to the regulation of TPRV6, calbindin D9K, and CYP3A4 (Wang et al., 2013b). With regard to the kidney, 25OHD3-3-O-sulfate bound to the DBP in blood could be filtered in the glomerulus and then reabsorbed in the proximal tubular epithelium through the action of megalin/cubilin, similar to what occurs for the 25OHD3-DBP complex (Negri, 2006). Again, intracellular hydrolysis of the conjugate and bioactivation to 1α,25(OH)2D3 could contribute to the known biologic effects of vitamin D in this tissue. Further work is needed to explore these mechanistic hypotheses.
Acknowledgments
We thank Dr. Evan D. Kharasch (Washington University in St. Louis, St. Louis, MO) for generously providing human bile. We also thank Dr. Nicholas J. Koszewski and Dr. Jesse P. Goff (Iowa State University, Ames, IA) for helping with the DBP competitive assay.
Abbreviations
- 25OHD3
25-hydroxyvitamin D3
- BSA
bovine serum albumin
- CNV
copy number variation
- DAPTAD
4-(4′-dimethylaminophenyl)-1,2,4-triazoline-3,5-dione
- DBP
vitamin D binding protein
- DHEA
dehydroepiandrosterone
- ESI
electrospray ionization
- LC
liquid chromatography
- m/z
mass-to-charge ratio
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- PAPS
3-phosphoadenosine-5-phosphosulfate
- PTAD
4-phenyl-1,2,4-triazoline-3,5-dione
- PXR
pregnane X receptor
- RNA-Seq
RNA sequencing
- SNV
single nucleotide variant
- SPE
solid-phase extraction
- SULT
sulfotransferase
- UPLC
ultra-performance liquid chromatography
Authorship Contributions
Participated in research design: Wong, Wang, Foti, Prasad, Chaudhry, Schuetz, Horst, Mao, de Boer, Thummel.
Conducted experiments: Wong, Wang, B. Chapron, Suzuki, Claw, Gao, Foti, Prasad, A. Chapron, Calamia, Horst.
Contributed new reagents or analytic tools: Foti, Schuetz.
Performed data analysis: Wong, Wang, B. Chapron, Suzuki, Claw, Gao, Foti, Prasad, A. Chapron, Calamia, Chaudhry, Schuetz, Horst, Mao, Thornton, Thummel.
Wrote or contributed to the writing of the manuscript: Wong, Wang, B. Chapron, Suzuki, Claw, Gao, Foti, Prasad, A. Chapron, Calamia, Chaudhry, Schuetz, Horst, Mao, de Boer, Thornton, Thummel.
Footnotes
This work was supported in part by the National Institutes of Health (NIH) National Institute of General Medical Sciences [Grants R01-GM63666, U01-GM092676, and T32-GM007750], the NIH National Institute of Environmental Health Sciences [Grant P30-ES007033], the NIH National Center for Advancing Translational Sciences [Grant TL1-TR000422], the NIH National Cancer Institute [Grant P30-CA21765], and the American Lebanese Syrian Associated Charities. Human livers used in this study were from the University of Washington School of Pharmacy Human Tissue Bank, established by the NIH National Institute of General Medical Sciences [Program Project Grant on Drug Interactions P01-GM32165], and the St. Jude Liver Resource, funded by the Department of Health and Human Services [Contract HHSN276201200017C].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, Gallicchio L, Jacobs EJ, Ascherio A, Helzlsouer K, Jacobs KB, et al. (2010) Genome-wide association study of circulating vitamin D levels. Hum Mol Genet 19:2739–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguelles LM, Langman CB, Ariza AJ, Ali FN, Dilley K, Price H, Liu X, Zhang S, Hong X, Wang B, et al. (2009) Heritability and environmental factors affecting vitamin D status in rural Chinese adolescent twins. J Clin Endocrinol Metab 94:3273–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelson M. (1985) 25-Hydroxyvitamin D3 3-sulphate is a major circulating form of vitamin D in man. FEBS Lett 191:171–175. [DOI] [PubMed] [Google Scholar]
- Axelson M. (1987) The cholecalciferol sulphate system in mammals. J Steroid Biochem 26:369–373. [DOI] [PubMed] [Google Scholar]
- Banerjee N, Fonge H, Mikhail A, Reilly RM, Bendayan R, Allen C. (2013) Estrone-3-sulphate, a potential novel ligand for targeting breast cancers. PLoS One 8:e64069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D, Hyppönen E. (2011) Determinants of vitamin D status: focus on genetic variations. Curr Opin Nephrol Hypertens 20:331–336. [DOI] [PubMed] [Google Scholar]
- Bouillon R, van Baelen H, de Moor P. (1980) Comparative study of the affinity of the serum vitamin D-binding protein. J Steroid Biochem 13:1029–1034. [DOI] [PubMed] [Google Scholar]
- Brodie MJ, Boobis AR, Dollery CT, Hillyard CJ, Brown DJ, MacIntyre I, Park BK. (1980) Rifampicin and vitamin D metabolism. Clin Pharmacol Ther 27:810–814. [DOI] [PubMed] [Google Scholar]
- Bu FX, Armas L, Lappe J, Zhou Y, Gao G, Wang HW, Recker R, Zhao LJ. (2010) Comprehensive association analysis of nine candidate genes with serum 25-hydroxy vitamin D levels among healthy Caucasian subjects. Hum Genet 128:549–556. [DOI] [PubMed] [Google Scholar]
- Cancela L, Marie PJ, Le Boulch N, Miravet L. (1985) Vitamin D3 3 beta sulfate has less biological activity than free vitamin D3 during pregnancy in rats. Biol Neonate 48:274–284. [DOI] [PubMed] [Google Scholar]
- Chatterjee B, Echchgadda I, Song CS. (2005) Vitamin D receptor regulation of the steroid/bile acid sulfotransferase SULT2A1. Methods Enzymol 400:165–191. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 1000 Genomes Project Analysis Group (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca HF. (1988) The vitamin D story: a collaborative effort of basic science and clinical medicine. FASEB J 2:224–236. [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdzik M, Gröer C, Penski J, Lapczuk J, Ostrowski M, Lai Y, Prasad B, Unadkat JD, Siegmund W, Oswald S. (2014) Protein abundance of clinically relevant multidrug transporters along the entire length of the human intestine. Mol Pharm 11:3547–3555. [DOI] [PubMed] [Google Scholar]
- Echchgadda I, Song CS, Oh T, Ahmed M, De La Cruz IJ, Chatterjee B. (2007) The xenobiotic-sensing nuclear receptors pregnane X receptor, constitutive androstane receptor, and orphan nuclear receptor hepatocyte nuclear factor 4alpha in the regulation of human steroid-/bile acid-sulfotransferase. Mol Endocrinol 21:2099–2111. [DOI] [PubMed] [Google Scholar]
- Echchgadda I, Song CS, Roy AK, Chatterjee B. (2004) Dehydroepiandrosterone sulfotransferase is a target for transcriptional induction by the vitamin D receptor. Mol Pharmacol 65:720–729. [DOI] [PubMed] [Google Scholar]
- Ekström L, Rane A. (2015) Genetic variation, expression and ontogeny of sulfotransferase SULT2A1 in humans. Pharmacogenomics J 15:293–297. [DOI] [PubMed] [Google Scholar]
- Falany CN, Rohn-Glowacki KJ. (2013) SULT2B1: unique properties and characteristics of a hydroxysteroid sulfotransferase family. Drug Metab Rev 45:388–400. [DOI] [PubMed] [Google Scholar]
- Fang HL, Strom SC, Ellis E, Duanmu Z, Fu J, Duniec-Dmuchowski Z, Falany CN, Falany JL, Kocarek TA, Runge-Morris M. (2007) Positive and negative regulation of human hepatic hydroxysteroid sulfotransferase (SULT2A1) gene transcription by rifampicin: roles of hepatocyte nuclear factor 4alpha and pregnane X receptor. J Pharmacol Exp Ther 323:586–598. [DOI] [PubMed] [Google Scholar]
- Fuleihan Gel-H, Bouillon R, Clarke B, Chakhtoura M, Cooper C, McClung M, Singh RJ. (2015) Serum 25-hydroxyvitamin D levels: variability, knowledge gaps, and the concept of a desirable range. J Bone Miner Res 30:1119–1133. [DOI] [PubMed] [Google Scholar]
- Gao C, Bergagnini-Kolev MC, Liao MZ, Wang Z, Wong T, Calamia JC, Lin YS, Mao Q, Thummel KE. (2017) Simultaneous quantification of 25-hydroxyvitamin D3-3-sulfate and 25-hydroxyvitamin D3-3-glucuronide in human serum and plasma using liquid chromatography-tandem mass spectrometry coupled with DAPTAD-derivatization. J Chromatogr B Analyt Technol Biomed Life Sci 1060:158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AS, Fulton RS, Qin X, Mardis ER, Nickerson DA, Scherer S. (2016) PGRNseq: a targeted capture sequencing panel for pharmacogenetic research and implementation. Pharmacogenet Genomics 26:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussler MR, Whitfield GK, Kaneko I, Haussler CA, Hsieh D, Hsieh JC, Jurutka PW. (2013) Molecular mechanisms of vitamin D action. Calcif Tissue Int 92:77–98. [DOI] [PubMed] [Google Scholar]
- Higaki M, Takahashi M, Suzuki T, Sahashi Y. (1965) Metabolic activities of vitamin D in animals. 3. Biogenesis of vitamin D sulfate in animal tissues. J Vitaminol (Kyoto) 11:261–265. [DOI] [PubMed] [Google Scholar]
- Higashi T, Goto A, Morohashi M, Ogawa S, Komatsu K, Sugiura T, Fukuoka T, Mitamura K. (2014) Development and validation of a method for determination of plasma 25-hydroxyvitamin D3 3-sulfate using liquid chromatography/tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 969:230–234. [DOI] [PubMed] [Google Scholar]
- Higashi T, Shimada K, Toyo’oka T. (2010) Advances in determination of vitamin D related compounds in biological samples using liquid chromatography-mass spectrometry: a review. J Chromatogr B Analyt Technol Biomed Life Sci 878:1654–1661. [DOI] [PubMed] [Google Scholar]
- Holick MF. (2007) Vitamin D deficiency. N Engl J Med 357:266–281. [DOI] [PubMed] [Google Scholar]
- Holick MF, Binkley NC, Bischoff-Ferrari HA, Gordon CM, Hanley DA, Heaney RP, Murad MH, Weaver CM, Endocrine Society (2011) Evaluation, treatment, and prevention of vitamin D deficiency: an Endocrine Society clinical practice guideline. J Clin Endocrinol Metab 96:1911–1930. [DOI] [PubMed] [Google Scholar]
- Horst RL, Reinhardt TA, Beitz DC, Littledike ET. (1981) A sensitive competitive protein binding assay for vitamin D in plasma. Steroids 37:581–591. [DOI] [PubMed] [Google Scholar]
- Hossein-nezhad A, Holick MF. (2013) Vitamin D for health: a global perspective. Mayo Clin Proc 88:720–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter D, De Lange M, Snieder H, MacGregor AJ, Swaminathan R, Thakker RV, Spector TD. (2001) Genetic contribution to bone metabolism, calcium excretion, and vitamin D and parathyroid hormone regulation. J Bone Miner Res 16:371–378. [DOI] [PubMed] [Google Scholar]
- Jones G, Prosser DE, Kaufmann M. (2014) Cytochrome P450-mediated metabolism of vitamin D. J Lipid Res 55:13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JW, Gilligan LC, Idkowiak J, Arlt W, Foster PA. (2015) The regulation of steroid action by sulfation and desulfation. Endocr Rev 36:526–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagubandi S, Londowski JM, Bollman S, Tietz P, Kumar R. (1981) Synthesis and biological activity of vitamin D3 3 beta-sulfate. Role of vitamin D3 sulfates in calcium homeostasis. J Biol Chem 256:5536–5539. [PubMed] [Google Scholar]
- Negri AL. (2006) Proximal tubule endocytic apparatus as the specific renal uptake mechanism for vitamin D-binding protein/25-(OH)D3 complex. Nephrology (Carlton) 11:510–515. [DOI] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, et al. (2009) Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461:272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Ooki S, Morohashi M, Yamagata K, Higashi T. (2013) A novel Cookson-type reagent for enhancing sensitivity and specificity in assessment of infant vitamin D status using liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom 27:2453–2460. [DOI] [PubMed] [Google Scholar]
- Prasad B, Gaedigk A, Vrana M, Gaedigk R, Leeder JS, Salphati L, Chu X, Xiao G, Hop C, Evers R, et al. (2016) Ontogeny of hepatic drug transporters as quantified by LC-MS/MS proteomics. Clin Pharmacol Ther 100:362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B, Unadkat JD. (2014a) Comparison of heavy labeled (SIL) peptide versus SILAC protein internal standards for LC-MS/MS quantification of hepatic drug transporters. Int J Proteomics 2014:451510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B, Unadkat JD. (2014b) Optimized approaches for quantification of drug transporters in tissues and cells by MRM proteomics. AAPS J 16:634–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve LE, DeLuca HF, Schnoes HK. (1981) Synthesis and biological activity of vitamin D3-sulfate. J Biol Chem 256:823–826. [PubMed] [Google Scholar]
- Rowling MJ, Kemmis CM, Taffany DA, Welsh J. (2006) Megalin-mediated endocytosis of vitamin D binding protein correlates with 25-hydroxycholecalciferol actions in human mammary cells. J Nutr 136:2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahashi Y, Suzuki T, Higaki M, Asano T. (1967a) Metabolism of vitamin D in animals. V. Isolation of vitamin D sulfate from mammalian milk. J Vitaminol (Kyoto) 13:33–36. [DOI] [PubMed] [Google Scholar]
- Sahashi Y, Suzuki T, Higaki M, Asano T. (1969) Antirachitic potency of vitamin D sulfate in human milk. J Vitaminol (Kyoto) 15:78–82. [DOI] [PubMed] [Google Scholar]
- Sahashi Y, Suzuki T, Higaki M, Takahashi M, Asano T. (1967b) Metabolic activities of vitamin D in animals. VI. Physiological activities of vitamin D sulfate. J Vitaminol (Kyoto) 13:37–40. [DOI] [PubMed] [Google Scholar]
- Sánchez-Guijo A, Oji V, Hartmann MF, Traupe H, Wudy SA. (2015) Simultaneous quantification of cholesterol sulfate, androgen sulfates, and progestagen sulfates in human serum by LC-MS/MS. J Lipid Res 56:1843–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze J, Johansson M, Thörngren JO, Garle M, Rane A, Ekström L. (2013) SULT2A1 gene copy number variation is associated with urinary excretion rate of steroid sulfates. Front Endocrinol (Lausanne) 4:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea MK, Benjamin EJ, Dupuis J, Massaro JM, Jacques PF, D’Agostino RB, Sr, Ordovas JM, O’Donnell CJ, Dawson-Hughes B, Vasan RS, et al. (2009) Genetic and non-genetic correlates of vitamins K and D. Eur J Clin Nutr 63:458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Mitamura K, Kitama N. (1995) Quantitative determination of 25-hydroxyvitamin D3 3-sulphate in human plasma using high performance liquid chromatography. Biomed Chromatogr 9:229–232. [DOI] [PubMed] [Google Scholar]
- Shirasaka Y, Chaudhry AS, McDonald M, Prasad B, Wong T, Calamia JC, Fohner A, Thornton TA, Isoherranen N, Unadkat JD, et al. (2016) Interindividual variability of CYP2C19-catalyzed drug metabolism due to differences in gene diplotypes and cytochrome P450 oxidoreductase content. Pharmacogenomics J 16:375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strott CA. (2002) Sulfonation and molecular action. Endocr Rev 23:703–732. [DOI] [PubMed] [Google Scholar]
- Theodoratou E, Tzoulaki I, Zgaga L, Ioannidis JP. (2014) Vitamin D and multiple health outcomes: umbrella review of systematic reviews and meta-analyses of observational studies and randomised trials. BMJ 348:g2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboven C, Rabijns A, De Maeyer M, Van Baelen H, Bouillon R, De Ranter C. (2002) A structural basis for the unique binding features of the human vitamin D-binding protein. Nat Struct Biol 9:131–136. [DOI] [PubMed] [Google Scholar]
- Wang Z, Lin YS, Dickmann LJ, Poulton EJ, Eaton DL, Lampe JW, Shen DD, Davis CL, Shuhart MC, Thummel KE. (2013a) Enhancement of hepatic 4-hydroxylation of 25-hydroxyvitamin D3 through CYP3A4 induction in vitro and in vivo: implications for drug-induced osteomalacia. J Bone Miner Res 28:1101–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Schuetz EG, Xu Y, Thummel KE. (2013b) Interplay between vitamin D and the drug metabolizing enzyme CYP3A4. J Steroid Biochem Mol Biol 136:54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wong T, Hashizume T, Dickmann LZ, Scian M, Koszewski NJ, Goff JP, Horst RL, Chaudhry AS, Schuetz EG, et al. (2014) Human UGT1A4 and UGT1A3 conjugate 25-hydroxyvitamin D3: metabolite structure, kinetics, inducibility, and interindividual variability. Endocrinology 155:2052–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber EJ, Chapron A, Chapron BD, Voellinger JL, Lidberg KA, Yeung CK, Wang Z, Yamaura Y, Hailey DW, Neumann T, et al. (2016) Development of a microphysiological model of human kidney proximal tubule function. Kidney Int 90:627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Hashizume T, Shuhart MC, Davis CL, Nelson WL, Sakaki T, Kalhorn TF, Watkins PB, Schuetz EG, Thummel KE. (2006) Intestinal and hepatic CYP3A4 catalyze hydroxylation of 1alpha,25-dihydroxyvitamin D(3): implications for drug-induced osteomalacia. Mol Pharmacol 69:56–65. [DOI] [PubMed] [Google Scholar]
- Zheng XE, Wang Z, Liao MZ, Lin YS, Shuhart MC, Schuetz EG, Thummel KE. (2012) Human PXR-mediated induction of intestinal CYP3A4 attenuates 1α,25-dihydroxyvitamin D3 function in human colon adenocarcinoma LS180 cells. Biochem Pharmacol 84:391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]