Abstract
RNase III is a ribonuclease that recognizes and cleaves double-stranded RNA. Across bacteria, RNase III is involved in rRNA maturation, CRISPR RNA maturation, controlling gene expression, and turnover of messenger RNAs. Many organisms have only one RNase III while others have both a full-length RNase III and another version that lacks a double-stranded RNA binding domain (mini-III). The genome of the cyanobacterium Synechococcus sp. strain PCC 7002 (PCC 7002) encodes three homologs of RNase III, two full-length and one mini-III, that are not essential even when deleted in combination. To discern if each enzyme had distinct responsibilities, we collected and sequenced global RNA samples from the wild type strain, the single, double, and triple RNase III mutants. Approximately 20% of genes were differentially expressed in various mutants with some operons and regulons showing complex changes in expression levels between mutants. Two RNase III’s had a role in 23S rRNA maturation and the third was involved in copy number regulation one of six native plasmids. In vitro, purified RNase III enzymes were capable of cleaving some of the known Escherichia coli RNase III target sequences, highlighting the remarkably conserved substrate specificity between organisms yet complex regulation of gene expression.
INTRODUCTION
Transcript degradation plays a pivotal role in determining steady-state transcript levels. Regulation of transcript degradation is inherently critical to coordinate and control gene expression and is a central component of cellular physiology. To accurately control protein levels within a cell, detailed information on rates of transcription, transcript degradation, translation, and protein degradation must be integrated to produce models that can both precisely predict and explain protein levels. Of these four areas one is both vastly understudied and underutilized as a method to control gene expression: transcript degradation. There are many tools to control gene expression at the level of transcription (promoters, induction and repression systems, transcriptional bioswitches (1–3)), translation (ribosome binding sites, translational bioswitches (4,5)) and protein degradation (proteolytic tags (6)). These tools have been applied to bacterial chemical factories and produced significant increases in titers and yields (7–9). Contrastingly, very few examples of controlling gene expression via engineered transcript stability exist and those that do are restricted to proof of concept in select organisms (10–12).
The lack of tools for controlling gene expression on the level of transcript stability most likely stems from the lack of knowledge of how these processes work in living cells. The lag in descriptive and mechanistic knowledge of transcript stability behind those of transcription, translation, and protein turnover may stem from the specific difficulties and challenges in studying RNA turnover such as the short lifetime of transcripts, the many distinct enzymes of which some form complexes, and the disparate activity of these enzymes in vitro and in vivo.
Transcript degradation is facilitated by ribonucleases (RNases) that have distinct characteristics. Models of transcript turnover involve initial cleavage facilitated by endoribonucleases that cleave RNA internally (RNase E, G, III, Y) followed by subsequent cleavage by exoribonucleases that processively remove nucleotides from the ends (RNase II, R, J, PNPase). Models of transcript degradation have been developed primarily based on Gram-negative Escherichia coli and Gram-positive Bacillus subtilis.
Many factors and RNase activities seem to be conserved in these two models, but the specific enzymes can be different. Escherichia coli transcript degradation is facilitated by RNase E and partially by RNase G while B. subtilis possess a somewhat similar RNase Y. Bacillus subtilis has two 5′ to 3′ exoribonucleases (RNase J1 and J2) that facilitate degradation from the 5′ end. Many bacterial groups possess a mixture of these RNases which leads to questions about how transcripts are being efficiently turned over and regulated.
We became interested in a cyanobacterial strain, Synechococcus sp. strain PCC 7002 (hereafter PCC 7002), for both its industrial applications in chemical production and its unique array of RNases. PCC 7002 contains both a homolog of RNase E (A0788) and RNase J (A1273) as well as three homologs of RNase III (A0061, A2542, A0384); the first two being full length RNase III’s while the third being a Mini-RNase III that lacks the double-stranded binding domain (13). These three RNase III homologs are not essential under standard growth conditions and have been deleted in all combinations (13). Here, we aimed to elucidate the function of the three homologs of RNase III to determine if they played distinct or redundant roles in transcript turnover, what structures and sequences they target, and what genes and cellular activities are affected by their action.
MATERIALS AND METHODS
Sample collection
All strains used in this study are listed in Supplemental Table S1. A detailed description of the creation of the RNase III mutant cyanobacterial strains is outlined in (13), but in summary, antibiotic resistance markers flanked by 500 bp homology arms were introduced via natural transformation. After passaging on the appropriate antibiotic(s), the segregation of the strains was verified. Cyanobacteria were cultured on media A+ agar (1.5%) plates with the appropriate antibiotic or antibiotics (gentamicin 30 μg/ml, kanamycin 100 μg/ml and streptomycin 100 μg/ml). Pre-cultures of 20 ml A+ media were bubbled with air overnight. The OD730 nm was measured and 1 L pyrex bottles filled with 900 ml media A+ were inoculated with pre-culture for an OD730 nm = 0.01. Cultures were bubbled with air for 24 h before harvesting 40 ml and adding 10 ml of cold stop solution (10% phenol in ethanol). Samples were centrifuged at 10,000 × g at 4°C for 10 min in a Beckman Coulter Avanti J-E Centrifuge. The resulting cell pellets were frozen in liquid nitrogen and stored at –80°C until further processing.
RNA isolation and RNA-sequencing
RNA was extracted as described previously (14) and submitted to the University of Wisconsin Biotechnology Gene Expression Center for RNA-sequencing. rRNA was removed with a Ribo-Zero Magnetic Kit. A cDNA library was created using a TruSeq Stranded Total RNA Library Kit. DNA libraries were sequenced using an Illumina HiSeq 2500 (1 × 100). Samples had between 5 and 22 million reads, sufficient coverage for bacterial differential gene expression analysis (15).
Differential gene expression analysis
Sequencing files were aligned to the PCC 7002 chromosome (NC_010475), pAQ1 (NC_010476), pAQ3 (NC_010477), pAQ4 (NC_010478), pAQ5 (NC_010479), pAQ6 (NC_010480) and pAQ7 (NC_010474) using Bowtie2 (v 2.2.6) and samtools (v 1.2). Counts for each feature in the general feature format files were obtained with HTSeq (v 0.6.1) (16) using the htseq-count script set on union mode and stranded set to reverse (HTseq v 0.6.1). edgeR (v 3.16.5) was used to test for differential gene expression (17). Specifically, feature counts were loaded and filtered for all genes that had at least one read per million in three samples and tested for differential expression with exactTest using tagwise dispersion. P-values were adjusted with the Benjamini and Hochberg algorithm (18) to estimate the false discovery rate (FDR) and genes with a FDR < 0.005 were classified as differentially expressed. Raw sequence files, feature counts, and differential expression fold change data was deposited in the Gene Expression Omnibus (GSE99279). Raw data files have also been uploaded to the NCBI Sequence Read Archive (accession number SRP107964).
Expression and purification of E. coli RNase III and PCC 7002 RNase III’s
Escherichia coli RNase III (Rnc) and three PCC 7002 RNase III homologs were amplified with primers (Supplemental Table S2) and inserted into pET28b using Gibson assembly (19). Plasmids (Supplemental Table S3) were verified by sequencing and transformed into chemically competent E. coli BL21 (DE3) cells and selected on LB kanamycin plates. Cultures of E. coli BL21 strains containing plasmids (5 ml LB with 50 μg/ml kanamycin) grown for 16 hours were used to inoculate 600 ml of LB (50 μg/ml kanamycin) in a 2 L flask. Cultures were grown at 37°C (250 rpm) until at OD600 nm ∼0.3–0.4 and induced with 1 mM IPTG. 1 ml of culture was collected 0, 1, 2, 3 and 4 h after induction to check overexpression on a SDS-PAGE gel. After 4 h, cultures were spun down at 6,000 × g for 20 min at 4°C (Beckham Coulter Avanti J-E Centrifuge), aspirated, and stored at –80°C. Purification was performed according to published methods (20), with the following deviations. Benzonase (2 μl) was added to the cells resuspended in lysis buffer and sonication was done in short bursts using a Fisher Scientific Sonic Dismembrater Model 500. Cell debris was removed by centrifugation in a Beckman Coulter Avanti J-E Centrifuge at 12,000 rpm at 4°C for 30 min and filtered through a 0.45 μm filter. RNase III variants were purified using an Äkta start purification system (GE) with a 1 ml HisTrapTM HP column (GE Healthcare). Briefly, the column was equilibrated with 10 volumes of buffer A + 5 mM imidazole and the clarified cell lysate was added (1mL/minute). The column was washed with 15 column volumes of buffer A+ 5mM imidazole and then 10 column volumes with buffer A + 60 mM imidazole. Protein was eluted with a gradient (10 column volumes) and collected in 0.5 ml fractions. A SDS-PAGE gel was run to visualize the fractions and the five most concentrated and pure fractions were combined. A PD-10 Desalting Column (GE Healthcare Life Sciences) was used according to manufacturer's instructions to transfer the protein into storage buffer containing 50% glycerol.
In vitro RNA cleavage assays and cleavage site mapping
RNA cleavage assays and 5′ RACE were performed as previously described (21). All cleavage reactions were performed as time courses with aliquots collected without enzyme, without magnesium chloride, and 5, 10, 20, 30 and 60 min after magnesium chloride addition. Some purified 5′ RACE products were subcloned into pGEM-T Easy vectors (Promega) before sequencing.
Plasmid copy number quantification
PCC 7002 (WT and ΔA2542) were cultured overnight in 20 ml media A+ in bubble tubes and diluted to an OD730 nm = 0.01 in 150 ml media A+ in 250 ml pyrex bottle with glass rod for bubbling (in quintuple). Cultures were grown for 24 h before 40 ml was collected and centrifuged for 20 min at 10,000 × g in a Beckham Coulter Avanti J-E Centrifuge at room temperature. Supernatant was removed, and samples were stored at –80°C until processing (10 days). A linear DNA spike-in was made by amplifying ΦX174 Virion DNA (NEB, N3023S) with primer JCC425 and primer JCC426 and purifying the product via ethanol precipitation. The spike-in was added immediately after addition of the TE (with lysozyme) to the frozen cell pellets at a concentration of 10 copies per cell. Cell counts in each sample were determined by measuring OD730 nm and converting through a standard curve of cells/mL (hemocytometer counts) versus OD730 nm. DNA was isolated from four replicates as in (14), except that the pH of the phenol solution was adjusted to 8 using Tris alkaline buffer and purified DNA was diluted 1:1000. The relative copy number was determined by qPCR using an AriaMx Real-Time PCR System. Reactions consisted of 5 μl iQ SYBR Supermix (Bio-Rad, #1708880), 0.3 μl 10 μM primer mix, 3.7 μl of water and 1 μl of template or standard. Cycling conditions consisted of 95°C for 3 min, 95°C for 15 s, 55°C for 30 s, 72°C for 1 min, repeat to step 2 for a total of 35 times, followed by a melt curve from 55°C to 95°C. All samples and standards were run in technical duplicate. Analysis was done using Agilent Aria Software v1.3 and Cq values were determined with an absolute threshold. Sample Cq values were compared to standard curve of five 10-fold dilutions of purified DNA templates (pAQ3: R2 = 0.998, efficiency = 109.9%, slope = –3.11, y-intercept = 1.46; gene F: R2 = 0.999, efficiency = 101.4%, slope = –3.29, y-intercept = 4.64). Absolute quantification was calculated by dividing the ratio of pAQ3 to gene F spike-in. No template controls were well above standard curves (23 and 29 for pAQ3 and gene F respectively). Primer GG607 and primer GG608 were used to detect pAQ3 (116 bp product) and primer JCC437 and primer JCC438 were used to detect control spike-in gene F (98 bp product). Primers were designed with Primer-Blast (NCBI).
Amplifying 23S rRNA extensions and verification of ctRNA expression
1 μg of total RNA extracted from WT, Δ0384, and Δ0061Δ2542Δ0384 was used to create cDNA with the iScriptTM cDNA Synthesis Kit (Bio-Rad #1708891) using random primers. Control reactions lacking reverse transcriptase were simultaneously performed. PCR reactions were set up using 5 μl GoTaq Green Master Mix (Promega #M7123), 1 μl each of forward and reverse primer (10 μM), 2 μl of water, and 1 μl of cDNA template. Reactions were run on a ProFlex PCR system with the following cycling conditions: 95°C for 3 min, 95°C for 30 s, 55°C for 30 s, 72°C for 1 min, repeat to step 2 for a total of 25 times, 72°C for 10 min, and storage at 4°C. PCR products were visualized on a 1% agarose gel with ethidium bromide.
For verification of ctRNA expression, aliquots were taken from WT and ΔA2542 cultures mentioned above, mixed with cold stop solution (10% phenol in ethanol), and stored for 2 weeks at –80°C until RNA was extracted using the Trizol 95 method (22). RNA was quantified using the Qubit RNA high sensitivity assay kit, and 1 μg of RNA and primer GG660 was used to create cDNA using the GoScript™ reverse transcriptase kit (A5000, Promega) according to manufacturer instructions. The presence of the 86 bp product was detected using the same PCR reaction and conditions as mentioned above using primer GG660 and primer GG661.
RESULTS
rRNA processing by RNase III homologs
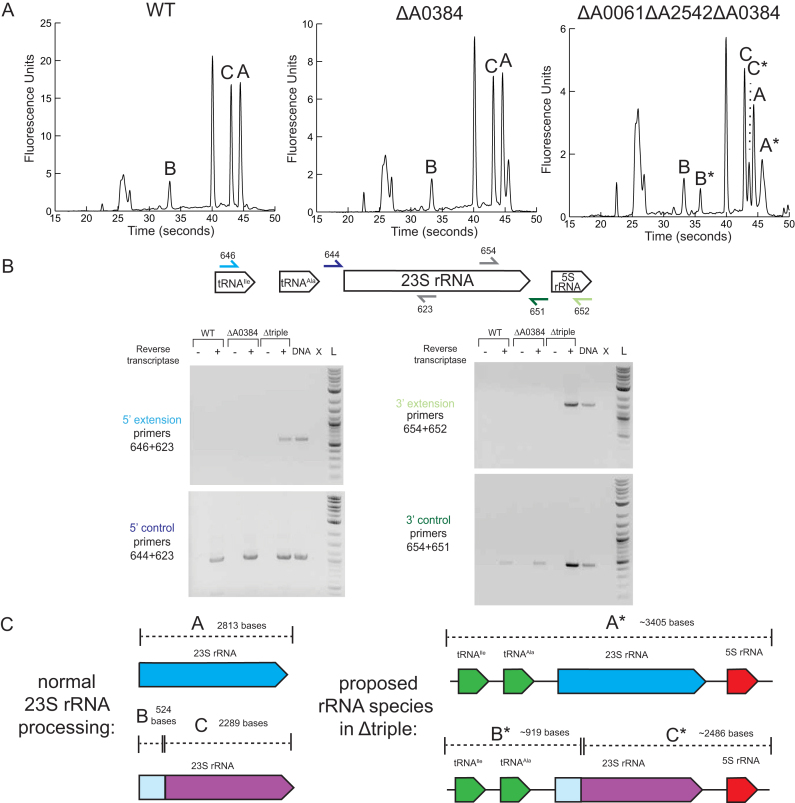
RNase III has been shown to be involved in rRNA processing in many bacterial species (23–25), yeast (26), and in plant chloroplasts (27), so we examined the rRNA of RNase III mutants. We extracted RNA from WT and strains harboring all combinations of RNase III deletions. The distribution of rRNA fragments was evaluated using nanoscale electrophoresis (BioAnalyzer) according to manufacturer instructions. We observed no differences in rRNA profiles in ΔA0061, ΔA2542 or ΔA0061ΔA2542 compared to WT (Supplemental Figure S1). When A0384 was deleted we saw an additional peak slightly larger than the 23S peak (Figure 1A). This resembled traces of rRNA isolated from E. coli Δrnc where the 23S rRNA has ∼25 and ∼50 bases extensions at the 5′ and 3′ end (23) as well as the case of rRNA isolated from B. subtilis mini-III deletion where 23S rRNA had extensions at the 5′ and 3′ end ranging from 2–64 nucleotides (25). We were unable to pinpoint the exact location of these proposed 23S rRNA extensions in ΔA0384 because the 5′ RACE products sequenced corresponded to the mature 5′ end of the 23S rRNA. There were additional peaks in ΔA0061ΔA0384 and ΔA0061ΔA2542ΔA0384, suggesting that A0061 and A0384 perform a redundant function that is altered when both are deleted. Based on the retention times, it appears that in addition to the normal rRNA species ΔA0061ΔA0384 and ΔA0061ΔA2542ΔA0384 also contain a larger rRNA fragment (A*) as well as two other fragments (B* and C*) that appear longer that corresponding fragments in the WT rRNA pool.
Figure 1.
Role of RNase III in rRNA processing. (A) BioAnalyzer traces of RNA extracted from WT and select RNase III mutants. (B) Extensions of 23S rRNA at the 5′ and 3′ ends in ΔA0061ΔA2542ΔA0384. A diagram of the rRNA operon shows primers used to amplify cDNA made from WT and ΔA0384 and Δtriple strains. Controls using WT PCC 7002 DNA as a template (DNA), no template (X), and 2 log DNA ladder (L) are also shown. (C) Proposed rRNA species seen in the BioAnalyzer traces.The 23S peak (labeled as A) breaks into two fragments B (524 bases) and C (2289 bases). rRNA species from the triple mutant with 5′ and 3′ extensions are labeled with an asterisk.
The 23S rRNA from PCC 7002 and other cyanobacteria is known to fragment into two pieces, but remain functional in the ribosome. The exact location of this fragmentation has not been reported, but similar fragment sizes have been observed in several species (28,29). The mechanism of this 23S rRNA processing is unknown and has been described as ‘slow’ and more rapid in the light than in the dark (28). Through 5′ RACE, we located a 5′ end that corresponded to base 524 of the annotated 23S rRNA suggesting that the full-length rRNA transcript is processed at this location. This site is located in a large loop between helix 24 and helix 2 in E. coli’s 23S rRNA secondary structure (30).
Based on the approximate peak sizes observed in the BioAnalyzer traces, we hypothesized that the larger species when both A0061 and A0384 were absent corresponded to 23S rRNA with extra 5′ and 3′ extensions. We created cDNA using random hexamers and ran PCR with a series of primers to see if we could detect 23S rRNA with extended 5′ or 3′ regions. We were able to amplify products that corresponded to 5′ and 3′ extensions of the 23S rRNA only with cDNA created from the triple mutant when reverse transcriptase was added (Figure 1B). We were able to amplify fragments using primers as far away as located in the intergenic tRNA in the 5′ direction and located in 5S rRNA in the 3′ direction. These 5′ and 3′ extensions suggest a role of A0061 and A0384 in processing the pre-23S rRNA, which contains a long region of complementary sequences (Supplemental Figure S2).
We attempted to complement E. coli Δrnc RNase III mutant strain with the three homologs from PCC 7002, but were unable to complement this rRNA phenotype (data not shown) perhaps because E. coli’s RNase III is known to bind the ribosome and it's specificity is altered by the presence of r-proteins (31).
Differential gene expression in RNase III mutants
To explore further roles of RNase III in PCC 7002, we isolated total RNA from WT, the single, double, and triple mutants and sequenced the rRNA-subtracted transcriptome of each. To explore the impact of each RNase III, we aligned the RNA-sequencing reads to the genome and six plasmids, counted the reads that aligned to each feature, and determined differentially expressed genes compared to WT with a false discovery rate cutoff rate of 0.005. With these parameters, ∼6–18% of genes were up-regulated, and 8–18% of genes were down-regulated when each mutant was compared with WT. We did not observe any substantial changes expression of any of the RNase III genes across any of the mutants (Supplemental Table S4). The number of counts aligning to each feature in each strain can be found in Supplemental Tables S5–S12, and a full list of differentially expressed genes is available in Supplemental Tables S13–S19.
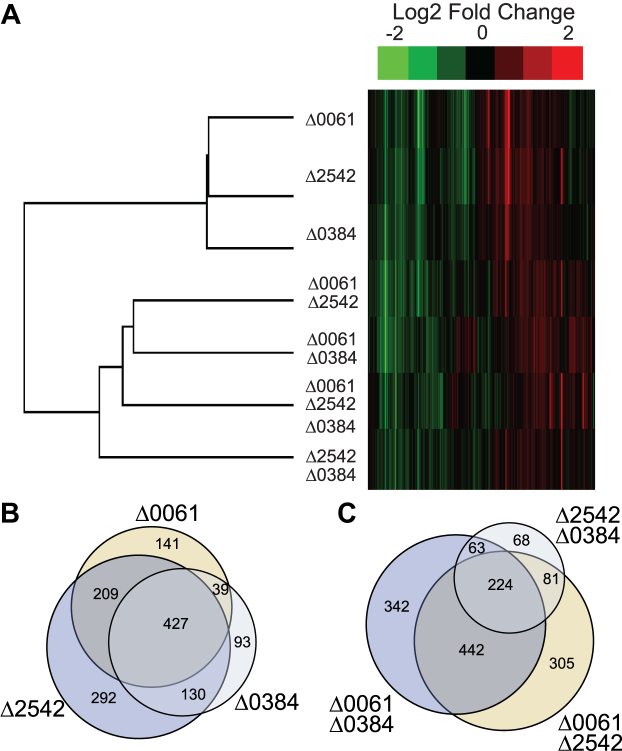
We clustered the log 2-fold change data for each gene and each sample type and found that the gene expression patterns of the single RNase III mutants most resembled each other and that the double and triple mutants clustered farther away (Figure 2A). Of all the differentially expressed genes in the three single mutants, many of them are differentially regulated in all three mutants (Figure 2B). Additionally, there seems to be a significant overlap in differentially expressed genes between the Δ0061Δ0384 and Δ0061Δ2542 double mutants (Figure 2C). These results were initially surprising because we expected to see expression patterns where we could logically conclude specific functions to each homolog. Instead, this data highlights the complexities associated with gene expression regulation mediated by RNases. The following sections describe specific roles that were identified.
Figure 2.
Overall expression patterns in RNase III mutants compared to WT. (A) Heatmap of clustered log2 fold changes of all genes. Venn diagram of overlapping genes that were differentially expressed in the single RNase III mutants (B) and double RNase III mutants (C) (FDR < 0.005).
Up-regulation of pAQ3 copy number in ΔA2542
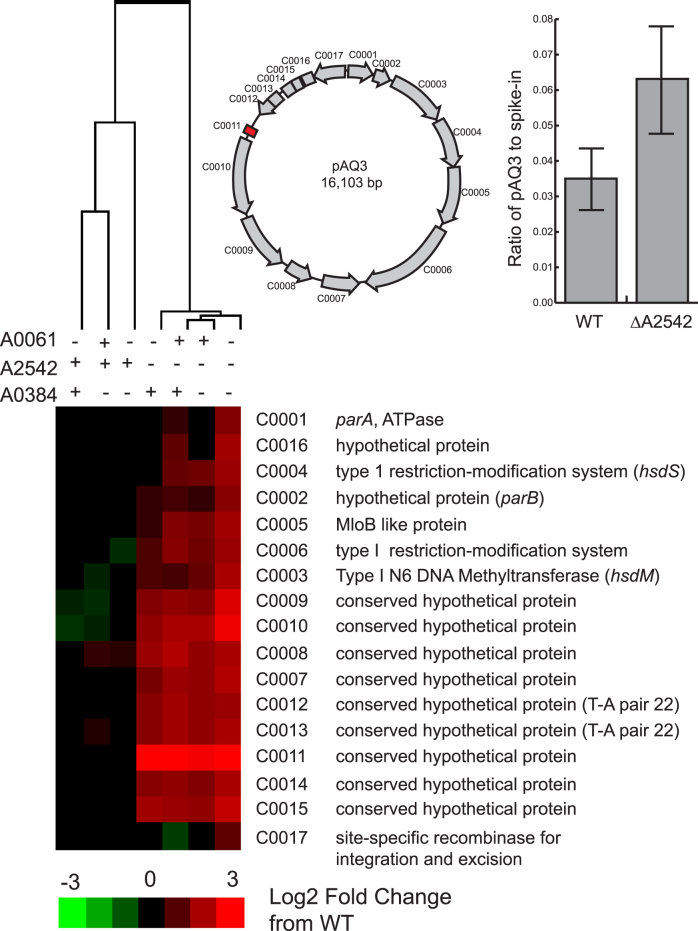
When A2542 was absent (single, double, and triple mutants), we observed significant up-regulation of many genes on pAQ3, one of the six plasmids in PCC 7002. Most pAQ3 genes in strains lacking A2542 had 2–3 times higher expression level than in strains with A2542 present, except for one gene, C0011, that was approximately 16 times more highly expressed (Figure 3). Due to the up-regulation of all of the genes on this plasmid, we hypothesized that we were observing an increase in copy number of this plasmid when A2542 was absent. We used qPCR to determine the ratio of copies of pAQ3 to a control DNA spike-in and found 1.8±0.4 times more copies of pAQ3 in ΔA2542 than WT (Figure 3). We saw similar values when comparing pAQ3 to a control gene on the chromosome (data not shown).
Figure 3.
Heatmap of gene expression of coding sequences on pAQ3 in RNase III mutants compared to WT. Log2 fold change data for each gene was clustered using Cluster 3.0 (hierarchical clustering, correlation uncentered, centroid linkage). Genes not meeting the FDR cutoff (<0.005) were given a value of zero. A schematic of the coding sequences on pAQ3 and their locations and orientations is shown. There was a significant difference in pAQ3 levels compared to a DNA spike-in are shown for WT and ΔA2542 (P < 0.05, Student's t-test, two-sided, equal variance, n = 4). Averages are shown with the error bars showing the standard deviation.
We hypothesized that C0011 and A2542 may be involved in copy number control because C0011 was the only coding sequence differentially expressed in ΔA2542 after correcting for DNA copy number. Based on the approximate start location of the C0011 mRNA as visualized in the RNA-seq data, we hypothesized that the start codon was mis-annotated, and the protein is actually 50 amino acids (coordinates 13,101–13,253 in NC_010477) instead of the original 63 amino acids. Protein homology searches (BLASTP) showed weak homology to CopG family proteins (Thermus sp. WG and Subdoligranulum varaiable DSM 15176). This protein has been characterized in prototype rolling circle plasmid pMV158 and its derivative pLS1, relatively small multicopy streptococcal plasmids that can replicate in both Gram-positive and Gram-negative hosts (32). Copy number control in pMV158 and pLS1 is controlled by two components: (i) CopG, a small ribbon-helix-helix transcriptional repressor protein that binds a 50 bp operator sequence and autorepresses the CopG-replication protein transcript and (ii) a small countertranscribed RNA (ctRNA) that binds to a region of the replication protein (33). The replication protein, RepB, in these plasmids is the rate-limiting factor that initiates plasmid replication by nicking the double-stranded origin (dso) and it's synthesis is controlled by a ctRNA that blocks an atypical ribosome binding site (34,35).
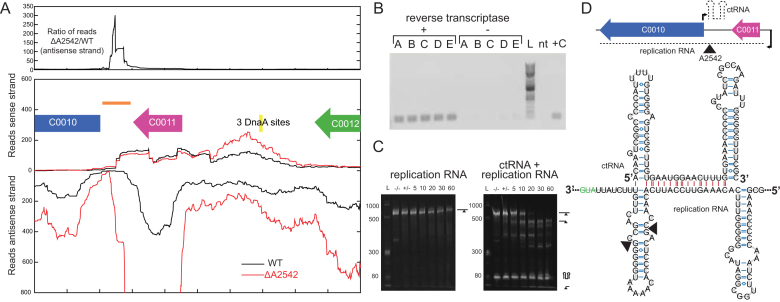
C0011 is predicted to share a similar ribbon-helix-helix structure (as predicted by Phyre2 (36)) and also has the highly conserved glycine in the turn between the two α-helices (37). The gene downstream of C0011 (C0010) is in a similar position as RepB from pMV158 but has little homology. We were unable to detect the presence of a small ctRNA on the sense strand in the RNA-seq data in WT or ΔA2542 (Figure 4A), but we did see a large increase in the transcript on the antisense strand between C0011 and C0010. There is a large increase in the ratio of reads of the antisense strand when comparing ΔA2542 to WT, and this region corresponds to the location of the ctRNA in other systems. Folding this region using mfold (38) reveals a 86-bp structure very similar to ctRNA’s from other plasmids that consists of two hairpins surrounding a 12 bp single stranded region. We hypothesize that we could not see this ctRNA in the RNA-sequencing data because the library preparation was not designed to target small RNAs (<100 nucleotides). We showed that it is expressed by amplifying this ctRNA from RNA samples treated with reverse transcriptase (Figure 4B). We compared ctRNA expression levels between WT and ΔA2542 but there was no statistically significant difference (data not shown).
Figure 4.
Identification of ctRNA on pAQ3. (A) RNA-seq coverage of portion of pAQ3. An example replicate is shown for WT and ΔA2542 with reads aligning to the sense strand displayed at the top and reads aligning to the antisense strand displayed in the opposite direction. The top panel shows the ratio of ΔA2542 to WT of the antisense strand. (B) Verification of expression of ctRNA by amplification off of cDNA (86 bp product). Replicates A-E made from WT RNA are shown along with a reference 2 log ladder (L), a no-template (nt), and positive control (+C). (C) In vitro cleavage assays of the replication RNA and the replication RNA and ctRNA with A2542. Cleavage reactions containing 5 μg of RNA were initiated by addition of MgCl2 and samples were removed at 5, 10, 20, 30 and 60 minutes and run on a urea polyacrylamide gel. Initial samples were also taken before the addition of enzyme (–/–) and MgCl2 (+/-). (D) A diagram of the location and orientations of these two RNAs and the predicted structure of the ctRNA as predicted by mfold (38). A section of the replication RNA is shown with the marked sites of A2542 cleavage in the in vitro cleavage reaction. Intramolecular bonds are shown in blue and intermolecular interactions are shown in red. The structured RNA is shown, but these RNAs are perfectly complementary (being transcribed from the same DNA segment), and could exist as a perfectly double-stranded 86 bp region.
We hypothesized that because RNase III enzymes cleave double stranded RNA, A2542 could be cutting the duplex of ctRNA and replication RNA. We used in vitro transcription to make these two species and tested purified A2542’s ability to cut both the replication RNA alone as well as the annealed duplex. These reactions were initiated by the addition of magnesium chloride and samples were taken and quenched with EDTA at a series of time points. A2542 was unable to cleave replication RNA alone even after 60 minutes, but was able to cleave the duplex of the ctRNA and replication RNA (Figure 4C). The ctRNA was not cleaved, but we identified the cleavage site within the replication RNA by purifying RNA from the reaction and performing 5′ RACE. A structure of the annealed duplexes is shown with the cleavage sites (Figure 4D). These sites do not have the traditional polar 2 nucleotide overhangs and it's possible one was created through a subsequent cleavage event. These regions could also form a 86 bp perfectly doubled-stranded region. A0384 could not cleave the duplex of ctRNA and replication RNA in vitro, but interestingly A0061 could despite no observed differences in pAQ3 gene expression (Supplemental Figure S3).
Global gene expression changes in RNase III mutants
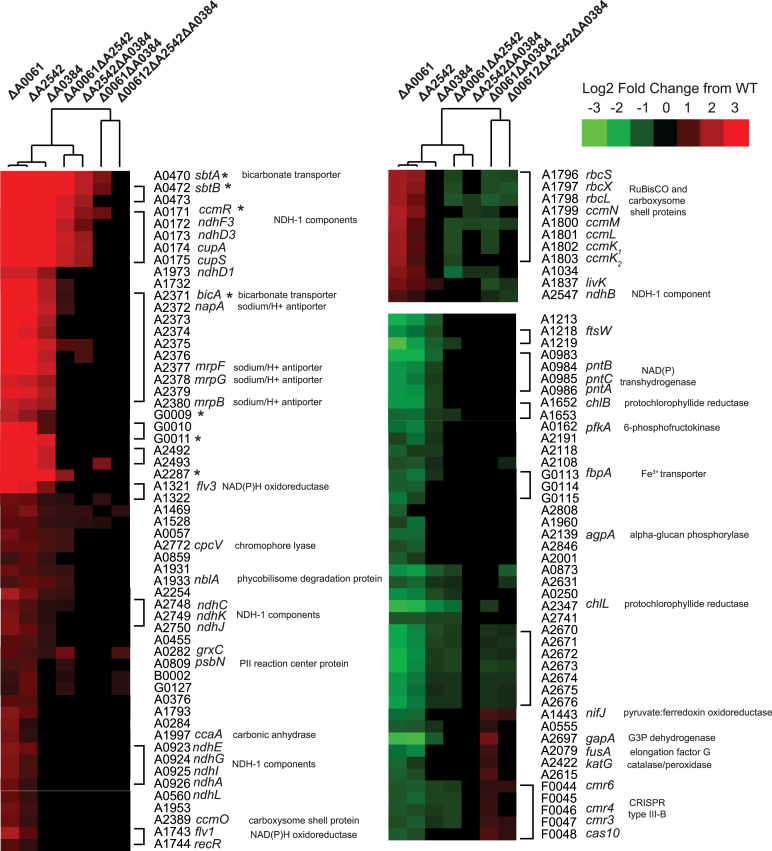
A set of genes, including bicarbonate transporters and NADH dehydrogenase components, were very strongly upregulated, including sbtA, bicA, ndhF3, ndhD3, ndhD1 (Figure 5) in each of the single RNase III mutants. Many of these genes are regulated by a transcriptional repressor ccmR in cyanobacteria (39). CcmR has been characterized in other model cyanobacteria and shown to repress itself as well as structural components of inorganic carbon transport (40). The CcmR binding motif was determined for PCC 7002 (41) and we observed up-regulation in the single RNase III mutants of all genes predicted to contain this motif (Figure 5 marked with *). Interestingly, the same transcripts in the double and triple mutants are not as significantly changed from WT if changed at all (Figure 5, right columns of heat map) indicating that there is more complex regulation involved.
Figure 5.
Heatmap of differentially expressed genes that showed different expression patterns in the single RNase III and double or triple RNase III mutants. Genes that were differentially expressed compared to WT with a fold-change greater than 2 or less –2 were clustered and the genes that had different expression patterns in the single mutants compared to the double or triple mutants were extracted. Genes were re-ordered to be next to those shown to or be predicted to be in the same operon. Those known or predicted to be a part of the CcmR regulon are marked (*). Genes in the same operon are grouped with brackets.
Additional genes were also regulated in a similar manner to those containing CcmR binding motifs. This group, which has not been linked to the CcmR regulon, include carbonic anhydrase that converts bicarbonate to CO2 inside of the carboxysome, NDH-1 components thought to be involved in CO2 transport, NAD(P)H oxidoreductases, as well as the subunits of RuBisCO (rbcS, rbcL) and carboxysome-encoding operon (ccmK2K1LMN). Additionally, in the single mutants we observed down-regulation of several groups of genes: transhydrogenases (pntCBA), glycolysis genes (pfkA, gapA), protochlorophyllide reductases (chlB, chlL) and pyruvate ferredoxin oxidoreductase (nifJ). These patterns indicate that RNase III plays a role in regulating the key genes in carbon fixation.
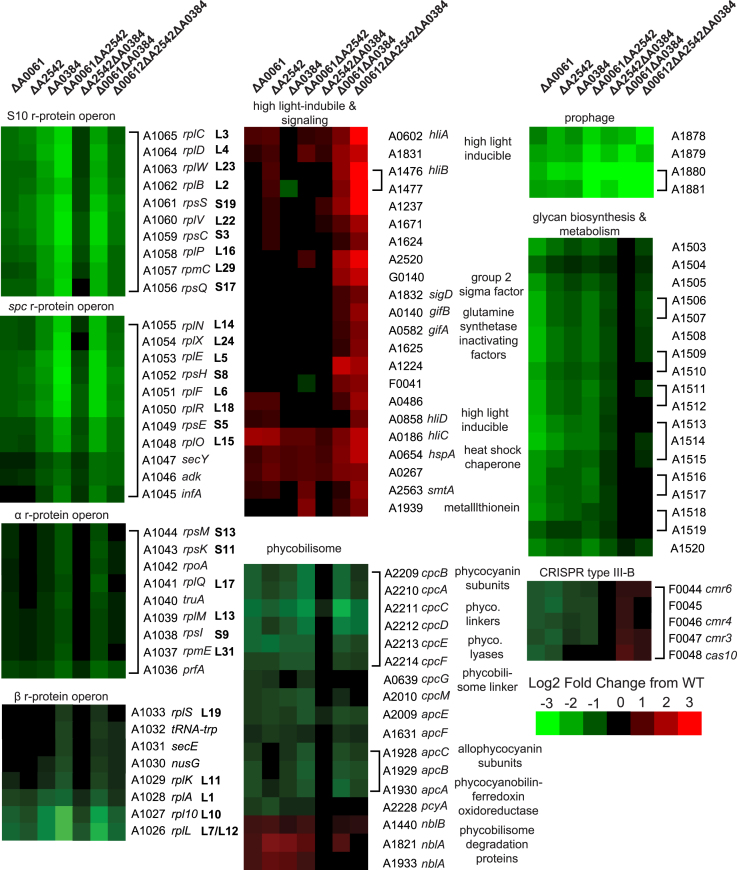
Other major changes in RNase III mutants include down-regulation of prophage genes, down-regulation of r-protein genes, and down-regulation of genes involved in glycan biosynthesis and metabolism (Figure 6). We saw down-regulation of many components of the phycobilisome and saw up-regulation of phycobilisome degradation proteins. Interestingly we found differential regulation of several genes that are part of the CRISPR type III RAMP module on pAQ6 (F0044–F0047). RNase III has been shown to be involved in processing of the CRISPR precursor transcript into the mature crRNA in type II CRISPR systems (42), but has not been shown to be involved in type III RNA-targeting CRISPR systems. We were unable to detect any differences in expression of the crRNA from the type III array on pAQ6 perhaps because the RNA-sequencing did not efficiently capture RNA < 100 nucleotides, but this potential involvement of RNase III regulating the protein components of the CRISPR type III-B system has not been reported before. We saw many global shifts in related-genes, indicating a role for RNase III’s in maintaining proper gene expression.
Figure 6.
Heatmaps of clustered groups of differentially expressed genes in all samples compared to WT. Genes were re-ordered to be next to those shown to or be predicted to be in the same operon. Genes in the same operon are grouped with brackets.
In vitro cleavage assays with purified RNase III homologs
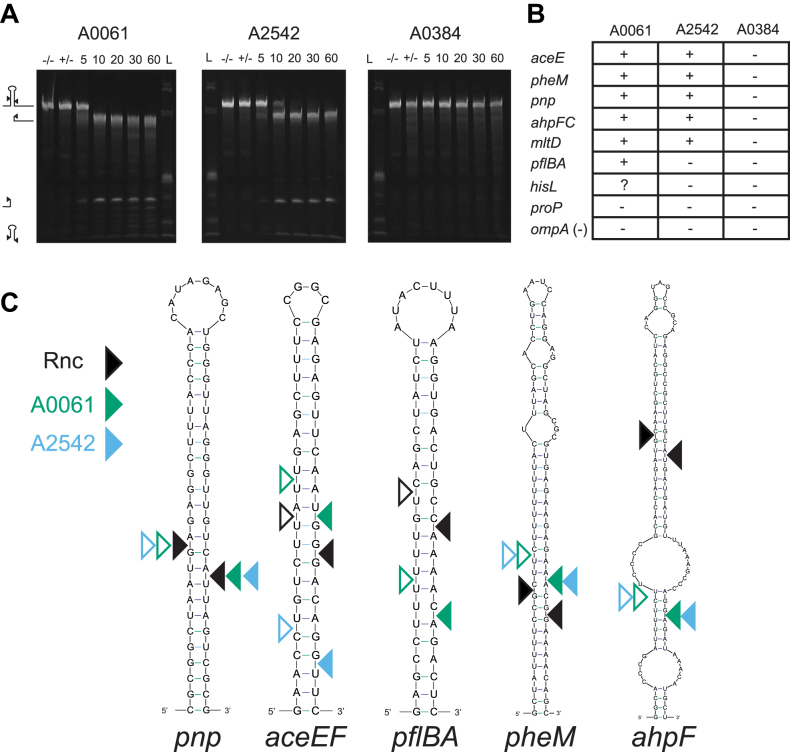
We were intrigued with the somewhat contradictory observations involving the specificities and redundancies of the three RNase III’s and decided to test their ability to cleave known RNase III targets from E. coli. We created RNA transcripts via in vitro transcription and tested the ability of purified cyanobacteria RNase III proteins to cut each transcript. Reactions were initiated with the addition of magnesium chloride and quenched with excess EDTA at a series of time points. Reaction products were run on a denaturing polyacrylamide gel and visualized with the addition of ethidium bromide. We observed cleavage of these transcripts as evidence by the disappearance of the large full-length band or the inability to cut these targets as evidence by the presence of the band after the 60-minute time course (Figure 7A). Both A0061 and A2542 could cleave many of the E. coli RNase III targets, but we never observed cleavage by A0384 (Figure 7B), the mini-III. One E. coli RNase III target, proP, could not be cut by any of these three enzymes. For a select few targets, we performed 5′ RACE on cleavage reactions to pinpoint the exact location of these cleavage events. A0061 and A2542 cut the pnp transcript at the same location as the E. coli RNase III enzyme, but for several others A0061 and A2542 cleaved the same stem at neighbouring bases (range -21 to +3) (Figure 7C). We tested the oligomerization of these three purified proteins by incubating them with the cross-linker disuccinimidyl suberate and running them on a polyacrylamide gel. Like E. coli’s RNase III and all other known RNase III’s, we saw the formation of dimers of A0061 and A0384 but we did not observe dimers for A2542 (Supplemental Figure S4). We hypothesize that our lack of observation of a dimer for A2542 may be due to structural differences or lack of closely located primary amines in the dimer. The ability of these RNase III’s to cleave some non-native targets precisely and some non-precisely highlights the intricate specificities of these enzymes and supports the idea that these enzymes are more finely-tuned to regulate gene expression than is currently appreciated.
Figure 7.
PCC 7002 RNase III’s are capable of cleaving E. coli RNase III targets. (A) Cleavage assays of in vitro transcribed E. coli RNase III target (aceEF) incubated with purified cyanobacteria RNase III’s. Cleavage reactions containing 5 μg of RNA were initiated by addition of MgCl2 and samples were removed at 5, 10, 20, 30 and 60 min and run on a urea polyacrylamide gel. Initial samples were also taken before the addition of enzyme (–/–) and MgCl2 (+/-). (B) Summary of cleavage reactions of sequences known to be cleaved by E. coli’s RNase III. A highly structured RNA but not known to be cleaved by E. coli’s RNase III was included as a negative control (ompA). (C) Locations of cyanobacteria RNase III cleavage of known E. coli RNase III targets. Structures and locations of E. coli RNase III cleavage are shown as in (21). Filled in triangles indicate 5′ ends found with 5′ RACE of in vitro cleavage reactions while open triangles indicate position of the other cleavage event where the inferred other cleavage event would be to leave the characteristic two nucleotide 3′-overhangs.
DISCUSSION
rRNA processing
By analyzing the rRNA traces of these RNase III mutants we were able to determine the role and approximate location of cleavage of A0061 and A0384 on the 23S rRNA. Without A0384, there are slight extensions of the 23S rRNA at the 5′ and 3′ ends indicating A0384 cleavage in the paired region proximal to the mature 23S rRNA ends. A0061 seems to cleave more distal in this paired region and without A0061 or A0384, the 23S rRNA has large 5′ and 3′ extensions and includes both tRNAs and the 5S rRNA. From these samples, we were unable to tell if these RNase IIIs play a role in 16S rRNA processing, but in many organisms RNase III cleaves the stem surrounding the 16S rRNA but other enzymes functionally complement absence of RNase III. These results coincide with the role mini-III in 23S rRNA maturation in B. subtilis and the full-length RNase III in 23S rRNA processing as found in many organisms. The presence of a third homolog of RNase III in some cyanobacteria (13) may indicate a specialized role beyond rRNA processing.
The analysis was initially not as straight forward due to the known rRNA fragmentation of the 23S rRNA in cyanobacteria. Using 5′ RACE we were able to pinpoint the fragmentation site and show that it occurs within a predicted loop region. rRNA fragmentation is widespread in bacteria (43), but still there is little known about its consequences on physiology. Some organisms have intervening sequences within their rRNA, such as Rhodobacter capsulatus, Salmonella and Campylobacter, which are removed through RNase III cleavage and subsequent processing by RNase E (44). Here, we saw no evidence of RNase III involvement in 23S rRNA fragmentation in PCC 7002 and postulate that it is caused by RNase E, other endonucleases, or be naturally labile in PCC 7002.
RNase III plasmid copy number regulation
Our RNA-seq data demonstrated a significant upregulation of genes on the native plasmid pAQ3. We hypothesize that A2542 controls the copy number of pAQ3 by recognizing and cleaving the complex of a short counter-transcribed RNA (ctRNA) and a coding transcript, as we demonstrated in vitro. Initially, we saw a build-up of the coding transcript only, but the similarities in gene organization with rolling-circle plasmid replication plasmids (pMV158 and pLS1) led us to detect the ctRNA that was not visible in the RNA-sequencing data. In pMV158 and pLS1 this ctRNA blocks translation of the downstream replication protein, which is the rate-limiting factor for plasmid replication initiation. No one has reported if RNase III plays a role in degrading the ctRNA and coding transcript complex in pMV158 and pLS1. We propose that in PCC 7002 A2542 is just one part of copy number regulation because in its absence copy number is only up-regulated 2-fold. Interestingly, A0061 could also cleave the complex in vitro despite seeing no differences in vivo. This could indicate differential specificities or spatial localizations in vivo (A0061 processes rRNA while A2542 does not). A2542 could ensure proper turnover of the blocked ctRNA, coding transcript complex. This mode of regulation is unique to pAQ3 (the other five native plasmids do not contain any similar elements) and does not appear to be widely conserved in cyanobacterial plasmids.
Global mRNA expression changes in RNase III mutants
We saw many coordinated changes in gene expression within groups of genes involved in related processes. One such intriguing group included all known members of the CcmR regulon (involved in bicarbonate transport and NDH-1 components thought to be involved in CO2 transport) as well as other NDH-1 components, RuBisCO and carboxysome genes, carbonic anhydrase, NAD(P)H oxidoreductases, and other uncharacterized proteins. These genes were only up-regulated in the single RNase III mutants and not the double or triple mutants. For another group of genes we saw down-regulation in the single mutants only. This included an operon encoding NAD(P)H dehydrogenases, glycolytic enzymes, a pyruvate:ferredoxin oxidoreductase, and protochlorophyllide reductases. Taken together we hypothesize that in the mutant strains there may be issues with the redox status or sensing of the redox status of the cell. The up-regulation of genes in the CcmR regulon was the most striking and in PCC 6803 α-ketoglutarate (αKG) and NADP+ have been shown to be co-repressors of CcmR (45). Changes in αKG or NADP+ could be causing derepression of the CcmR regulon, but it seems that there is also CcmR-independent regulation of many genes with related functions. This intriguing pattern of differential gene expression between single and double and triple mutants could suggest that when a single RNase III is absent, cells can compensate with coordinated changes involving CO2 fixation and redox balance. However, when two or all of the RNase III’s are absent cellular objectives change resulting in different gene expression patterns.
We noticed coordinated regulation of the r-proteins in almost most the mutants (Figure 6). Due to the role of RNase III in rRNA processing, it might make sense to down-regulate r-protein synthesis when there is mis-processing of the rRNA. Interestingly, we saw down-regulation even in samples where we did not observe any defects in rRNA processing (ΔA0061, ΔA2542, ΔA0061ΔA2542). This is the first example of coordinated regulation of r-protein transcript levels due to the absence of RNase III. Additional regulation of r-protein synthesis is not surprising because r-protein synthesis is highly regulated both through translational feedback (46) and at the level of transcription initiation (47).
We observed extreme down-regulation of several genes located in a prophage. We saw down-regulation of four collinear genes (A1878-A1881) that look like they could be encoded on the same transcript. There are many examples of RNase III processing sites within phage (lambda, T7, T3), and the mini-III in B. subtilis was shown to be essential for the degradation of toxin genes that are part of a cryptic prophage (48). Additionally, in E. coli RNase III is shown to process a prophage encoded transcript into the DicF sRNA that affects cell division and other processes (49).
There were several other groups of genes where we saw coordinated changes in gene expression across RNase III mutants. We saw up-regulation of many high light inducible proteins especially in the triple RNase III mutant as well as other proteins involved in signalling including sigD and glutamine synthetase inactivating factors. We saw down-regulation of many genes involved in glycan biosynthesis and metabolism (A1503-A1520). Similarly, we saw down-regulation of many phycobilisome components in all samples except ΔA2542ΔA0384. In this same set of samples we saw up-regulation of several distantly-located phycobilisome degradation proteins (nblA’s and nblB).
Based on prior work in E. coli, we expected to identify distinct processing sites within specific RNAs when comparing samples from mutants to the WT. Instead, we observed significant changes in expression for many genes but no clear spikes in RNA levels that would indicate RNase III target sites. This observation is demonstrated in Supplemental Table S5 which highlights the difference in distribution of comparing sequencing coverage between RNase III mutants and WT in both E. coli and cyanobacteria. In the cyanobacterial RNase III dataset we observed global, coordinated shifts in expression of genes of related functions involve genes that are located at different locations in the chromosome, and we hypothesize that some regulator or group of regulators are responsible for this synchronized movement (transcription factor, sRNA, or other). When we see perturbations of these groups of genes in RNase III mutants, RNase III function could be directly involved in these regulators or in the sensing mechanism, or these perturbations are due to secondary effects of the lack of RNase III cleavage.
The complex changes we see in gene expression could be due to RNA binding activity without cleavage. In bacteriophage λ, RNase III is thought to bind to the upstream region of cIII and exposes it's ribosome binding site, and stimulating translation (50). There is also the possibility that these RNase III’s are post-translationally modified, affecting their activity. When E. coli is infected with the T7 phage, the phage T7 protein kinase phosphorylates the native RNase III which stimulates it's activity up to 4-fold (51). Another possible explanation for these complex gene expression changes is the existence of RNase III inhibitors. A protein inhibitor was found in E. coli (YmdB) that prevents RNase III dimerization and as a consequence, activity (52). We found no homolog of YmdB in PCC 7002, but in E. coli they observed other unknown YmdB independent inhibition of RNase III.
Effects of RNase III on global gene expression has been observed for a few organisms including E. coli and Staphylococcus aureus (53,54). RNase III has been shown to auto-regulate its own expression by cleaving a structure in its 5′ UTR in many organisms including E. coli, S. aureus, and Strepomyces coelicolor (54–56). We did not observe this phenomenon in PCC 7002. RNase III is also known to process the 5′ UTR of PNPase in several organisms (57–59), but we found no evidence of this in PCC 7002.
We recently showed the action of RNase III on transcripts encoding several key metabolic enzymes including pyruvate dehydrogenase in E. coli (21). We did not see any RNase III-mediated regulation of any of these metabolic enzyme homologs in PCC 7002. This raises an interesting point that RNase III is involved in the regulation of key metabolic processes and enzymes in both heterotrophs and autotrophs. Transcriptional and posttranscriptional regulation of photosynthesis has been shown to be extremely important in physiological response and signaling shifts in response to changing conditions, and RNase IIIs may be a very important player influencing these interactions.
Cleavage activity and specificities of RNase III homologs
Without putative PCC 7002 RNase III targets, we tested the ability of cyanobacterial enzymes to cleave known E. coli targets in vitro in effort to investigate conservation of RNase III targets across species. A0061 and A2542, both full-length enzymes, could cleave many of the E. coli targets, but not all. Additionally, A0061 could cleave the pflBA target while A2542 could not. By extracting RNA after the in vitro cleavage reaction and performing 5′ RACE we pinpointed the exact location of these cleavage events. For the E. coli pnp target, both A0061 and A2542 cleaved at the same location where the E. coli RNase III enzyme did (Figure 7). For all other targets we tested, A0061 and A2542 cut in nearby but not identical locations as the E. coli enzyme (ranging from –21 to +3 bases). For one transcript, aceEF, A0061 and A2542 cut at different locations within the same stem loop structure. We were not surprised to see that A0384, the mini-III, could not cleave any of these E. coli targets because the E. coli enzyme is a full-length RNase III. Expanding the list of specific targets of many different RNase III’s may one day reveal the complex substrate enzyme recognition pairing and allow for a more predictive approach to finding RNase III target sequences.
CONCLUSION
We have identified the roles of two homologs of RNase III in 23S rRNA processing, the third homolog's role in copy number control of one plasmid, and highlighted the complex global changes in gene expression involving CO2 fixation, redox status, and light harvesting. We hypothesize that many of these changes are indirect effects of RNase III activity and indicate a role of RNase III’s in either sensing environmental changes or mediating changes. This work highlights the important role of post-transcriptional regulation and its influence on gene expression. We showed both specific and redundant activities of these RNase III’s on in vitro targets and show that while both A0061 and A2542 can cleave the duplex structure controlling plasmid copy number regulation in vitro, only A2542 acts on this duplex in vivo. This work highlights the continued need for in vivo datasets to tease apart specificities and unravel the complex gene regulatory network enabled by post-transcriptional RNA processing.
DATA AVAILABILITY
All software used is open source. Bowtie2 is available through Sourceforge (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). Samtools is hosted by GitHub (https://github.com/samtools/samtools). HTSeq is available from the Python Package Index (https://pypi.python.org/pypi/HTSeq). Mfold is available through the online web server (http://unafold.rna.albany.edu/?q=mfold). BLASTP is available through the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins). Cluster 3.0 can be downloaded from http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm. Phyre2 can be used online (http://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index).
Raw sequence files, feature counts, and differential expression fold change data was deposited in the Gene Expression Omnibus (GSE99279). Raw data files have also been uploaded to the NCBI Sequence Read Archive (accession number SRP107964).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the University of Wisconsin Biotechnology Center Gene Expression Center for providing RNA library preparation and the DNA Sequencing Facility for their sequencing services.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US Department of Energy [DE-SC0010329]; William F. Vilas Trust [Vilas Associates award to B.P.]; National Institutes of Health Biotechnology Training Program Fellowship [NIGMS-5 T32 GM08349 to G.G.]. Funding for open access charge: US Department of Energy [DE-SC0010329].
Conflict of interest statement. None declared.
REFERENCES
- 1. Politz M.C., Copeland M.F., Pflger B.F.. Artificial repressors for controlling gene expresison in bacteria. Chem. Commun. 2013; 49:4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chappell J., Watters K.E., Takahashi M.K., Lucks J.B.. A renaissance in RNA synthetic biology: New mechanisms, applications and tools for the future. Curr. Opin. Chem. Biol. 2015; 28:47–56. [DOI] [PubMed] [Google Scholar]
- 3. Keasling J.D. Gene-expression tools for the metabolic engineering of bacteria. Trends Biotechnol. 1999; 17:452–460. [DOI] [PubMed] [Google Scholar]
- 4. Oesterle S., Gerngross D., Schmitt S., Roberts T.M., Panke S.. Efficient engineering of chromosomal ribosome binding site libraries in mismatch repair proficient Escherichia coli. Sci. Rep. 2017; 7:12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Topp S., Reynoso C.M.K., Seeliger J.C., Goldlust I.S., Desai S.K., Murat D., Shen A., Puri A.W., Komeili A., Bertozzi C.R. et al. Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl. Environ. Microbiol. 2010; 76:7881–7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cameron D.E., Collins J.J.. Tunable protein degradation in bacteria. Nat. Biotechnol. 2014; 32:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peralta-Yahya P.P., Zhang F., del Cardayre S.B., Keasling J.D.. Microbial engineering for the production of advanced biofuels. Nature. 2012; 488:320–328. [DOI] [PubMed] [Google Scholar]
- 8. Adrio J.L., Demain A.L.. Recombinant organisms for production of industrial products. Bioeng. Bugs. 2010; 1:116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pickens L., Tang Y., Chooi Y.H.. Metabolic engineering for the production of natural products. Annu. Rev. Chem. Biomol. Eng. 2014; 2:211–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Babiskin A.H., Smolke C.D.. Synthetic RNA modules for fine-tuning gene expression levels in yeast by modulating RNase III activity. Nucleic Acids Res. 2011; 39:8651–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carrier T.A., Keasling J.D.. Engineering mRNA stability in E. coli by the addition of synthetic hairpins using a 5′ cassette system. Biotechnol. Bioeng. 1997; 55:577–580. [DOI] [PubMed] [Google Scholar]
- 12. Smolke C.D., Carrier T.A., Keasling J.D.. Coordinated, differential expression of two genes through directed mRNA cleavage and stabilization by secondary structures. Appl. Environ. Microbiol. 2000; 66:5399–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cameron J.C., Gordon G.C., Pfleger B.F.. Genetic and genomic analysis of RNases in model cyanobacteria. Photosynth. Res. 2015; 126:171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khodurksy A., Bernstein J., Peter B., Rhodius V., Wendisch V., Zimmer D.. Escherichia coli spotted double-strand DNA microarrays. Methods Mol. Biol. 2003; 224:61–78. [DOI] [PubMed] [Google Scholar]
- 15. Haas B.J., Chin M., Nusbaum C., Birren B.W., Livny J.. How deep is deep enough for RNA-Seq profiling of bacterial transcriptomes. BMC Genomics. 2012; 13:734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anders S., Pyl P.T., Huber W.. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995; 57:289–300. [Google Scholar]
- 19. Gibson D.G., Young L., Chuang R., Venter J.C., Hutchison C.A., Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–345. [DOI] [PubMed] [Google Scholar]
- 20. Amarasinghe A.K., Calin-Jageman I., Harmouch A., Sun W., Nicholson A.W.. Escherichia coli ribonuclease III: affinity purification of hexahistidine-tagged enzyme and assays for substrate binding and cleavage. Methods Enzymol. 2001; 342:143–158. [DOI] [PubMed] [Google Scholar]
- 21. Gordon G.C., Cameron J.C., Pfleger B.F.. RNA sequencing identifies new RNase III cleavage sites in Escherichia coli and reveals increased regulation of mRNA. MBio. 2017; 8:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pinto G.L., Thapper A., Sontheim W., Lindblad P.. Analysis of current and alternative phenol based RNA extraction methodologies for cyanobacteria. BMC Mol. Biol. 2009; 10:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. King T.C., Sirdeshmukh R., Schlessinger D.. RNase III cleavage is obligate for maturation but not for function of Escherichia coli pre-23S rRNA. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Young R.A., Steitz J.A.. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc. Natl. Acad. Sci. U.S.A. 1978; 75:3593–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Redko Y., Bechhofer D.H., Condon C.. Mini-III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis. Mol. Microbiol. 2008; 68:1096–1106. [DOI] [PubMed] [Google Scholar]
- 26. Elela S.A., Igel H., Ares M.. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996; 85:115–124. [DOI] [PubMed] [Google Scholar]
- 27. Hotto A.M., Castandet B., Gilet L., Higdon A., Condon C., Stern D.B.. Arabidopsis chloroplast mini-ribonuclease III participates in rRNA maturation and intron recycling. Plant Cell. 2015; 27:724–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doolittle W.F. Postmaturational cleavage of 23S ribosomal ribonucleic-acid and its metabolic control in blue-green-alga Anacystis nidulans. J. Bacteriol. 1973; 113:1256–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shan J., Clokie M.. Preparation of RNA from bacteria infected with bacteriophages: a case study from the marine unicellular Synechococcus sp. WH7803 infected by phage S-PM2. Methods Mol. Biol. 2009; 171–176. [DOI] [PubMed] [Google Scholar]
- 30. Petrov A.S., Bernier C.R., Hershkovits E., Xue Y., Waterbury C.C., Hsiao C., Stepanov V.G., Gaucher E.A., Grover M.A., Harvey S.C. et al. Secondary structure and domain architecture of the 23S and 5S rRNAs. Nucleic Acids Res. 2013; 41:7522–7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Allas U., Liiv A., Remme J.. Functional interaction between RNase III and the Escherichia coli ribosome. BMC Mol. Biol. 2003; 4:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. del Solar G., Diaz R., Espinosa M.. Replication of the streptococcal plasmid pMV158 and derivatives in cell-free extracts of Escherichia coli. MGG Mol. Gen. Genet. 1987; 206:428–435. [DOI] [PubMed] [Google Scholar]
- 33. del Solar G., Acebo P., Espinosa M.. Replication control of plasmid pLS1: efficient regulation of plasmid copy number is exerted by the combined action of two plasmid components, CopG and RNA II. Mol. Microbiol. 1995; 18:913–924. [DOI] [PubMed] [Google Scholar]
- 34. López-Aguilar C., Ruiz-Masó J.A., Rubio-Lepe T.S., Sanz M., del Solar G.. Translation initiation of the replication initiator repB gene of promiscuous plasmid pMV158 is led by an extended non-SD sequence. Plasmid. 2013; 70:69–77. [DOI] [PubMed] [Google Scholar]
- 35. del Solar G., Espinosa M.. The copy number of plasmid pLS1 is regulated by two trans-acting plasmid products: the antisense RNA II and the repressor protein, RepA. Mol. Microbiol. 1992; 6:83–94. [DOI] [PubMed] [Google Scholar]
- 36. Kelly L.A., Mezulis S., Yates C., Wass M., Sternberg M.. The Phyre2 web portal for protein modelling, prediction, and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Acebo P., García De Lacoba M., Rivas G., Andreu J.M., Espinosa M., del Solar G.. Structural features of the plasmid pMV158-encoded transcriptional repressor CopG, a protein sharing similarities with both helix-turn-helix and β-sheet DNA binding proteins. Proteins Struct. Funct. Genet. 1998; 32:248–261. [DOI] [PubMed] [Google Scholar]
- 38. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Woodger F.J., Bryant D.A., Price G.D.. Transcriptional regulation of the CO2-concentrating mechanism in a euryhaline, coastal marine cyanobacterium, Synechococcus sp. Strain PCC 7002: role of NdhR/CcmR. J. Bacteriol. 2007; 189:3335–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang H.L., Postier B.L., Burnap R.L.. Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J. Biol. Chem. 2004; 279:5739–5751. [DOI] [PubMed] [Google Scholar]
- 41. McClure R.S., Overall C.C., Mcdermott J.E., Hill E.A., Markillie L.M., Mccue L.A., Taylor R.C., Ludwig M., Bryant D.A., Beliaev A.S.. Network analysis of transcriptomics expands regulatory landscapes in Synechococcus sp. PCC 7002. Nucleic Acids Res. 2016; 44:8810–8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E.. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011; 471:602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Evguenieva-Hackenberg E. Bacterial ribosomal RNA in pieces. Mol. Microbiol. 2005; 57:318–325. [DOI] [PubMed] [Google Scholar]
- 44. Zahn K., Inui M., Yukawa H.. Divergent mechanisms of 5′ 23S rRNA IVS processing in the alpha-proteobacteria. Nucleic Acids Res. 2000; 28:4623–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Daley S.M.E., Kappell A.D., Carrick M.J., Burnap R.L.. Regulation of the cyanobacterial CO2-concentrating mechanism involves internal sensing of NADP+ and α-ketogutarate levels by transcription factor CcmR. PLoS One. 2012; 7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nomura M., Yates J.L., Dean D., Post L.E.. Feedback regulation of ribosomal protein gene expression in Escherichia coli: structural homology of ribosomal RNA and ribosomal protein mRNA. Proc. Natl. Acad. Sci. U.S.A. 1980; 77:7084–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lemke J.J., Sanchez-Vazquez P., Burgos H.L., Hedberg G., Ross W., Gourse R.L.. Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Durand S., Gilet L., Condon C.. The essential function of B. subtilis RNase III is to silence foreign toxin genes. PLoS Genet. 2012; 8:e1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Balasubramanian D., Ragunathan P.T., Fei J., Vanderpool C.K.. A Prophage-Encoded Small RNA Controls Metabolism and Cell Division in Escherichia coli. mSystems. 2016; 1:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Altuvia S., Locker-Giladi H., Koby S., Ben-Nun O., Oppenheim A.B.. RNase III stimulates the translation of the cIII gene of bacteriophage lambda. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:6511–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mayer J.E., Schweiger M.. RNase III is positively regulated by T7 protein kinase. J. Biol. Chem. 1983; 258:5340–5343. [PubMed] [Google Scholar]
- 52. Kim K.S., Manasherob R., Cohen S.N.. YmdB: A stress-responsive ribonuclease-binding regulator of E. coli RNase III activity. Genes Dev. 2008; 22:3497–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stead M.B., Marshburn S., Mohanty B.K., Mitra J., Castillo L.P., Ray D., Van Bakel H., Hughes T.R., Kushner S.R.. Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res. 2011; 39:3188–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lioliou E., Sharma C.M., Caldelari I., Helfer A.C., Fechter P., Vandenesch F., Vogel J., Romby P.. Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet. 2012; 8:e1002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bardwell J.C., Régnier P., Chen S.M., Nakamura Y., Grunberg-Manago M., Court D.L.. Autoregulation of RNase III operon by mRNA processing. EMBO J. 1989; 8:3401–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xu W., Huang J., Cohen S.N.. Autoregulation of absB (RNase III) expression in Streptomyces coelicolor by endoribonucleolytic cleavage of absB operon transcripts. J. Bacteriol. 2008; 190:5526–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jarrige A.C., Mathy N., Portier C.. PNPase autocontrols its expression by degrading a double-stranded structure in the pnp mRNA leader. EMBO J. 2001; 20:6845–6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gatewood M.L., Bralley P., Jones G.H.. RNase III-dependent expression of the rpsO-pnp operon of Streptomyces coelicolor. J. Bacteriol. 2011; 193:4371–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Haddad N., Saramago M., Matos R.G., Prévost H., Arraiano C.M.. Characterization of the biochemical properties of Campylobacter jejuni RNase III. Biosci. Rep. 2013; 33:300082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All software used is open source. Bowtie2 is available through Sourceforge (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). Samtools is hosted by GitHub (https://github.com/samtools/samtools). HTSeq is available from the Python Package Index (https://pypi.python.org/pypi/HTSeq). Mfold is available through the online web server (http://unafold.rna.albany.edu/?q=mfold). BLASTP is available through the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins). Cluster 3.0 can be downloaded from http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm. Phyre2 can be used online (http://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index).
Raw sequence files, feature counts, and differential expression fold change data was deposited in the Gene Expression Omnibus (GSE99279). Raw data files have also been uploaded to the NCBI Sequence Read Archive (accession number SRP107964).