Abstract
Synthetic genetic sensors and circuits enable programmable control over timing and conditions of gene expression and, as a result, are increasingly incorporated into the control of complex and multi-gene pathways. Size and complexity of genetic circuits are growing, but stay limited by a shortage of regulatory parts that can be used without interference. Therefore, orthogonal expression and regulation systems are needed to minimize undesired crosstalk and allow for dynamic control of separate modules. This work presents a set of orthogonal expression systems for use in Escherichia coli based on heterologous sigma factors from Bacillus subtilis that recognize specific promoter sequences. Up to four of the analyzed sigma factors can be combined to function orthogonally between each other and toward the host. Additionally, the toolbox is expanded by creating promoter libraries for three sigma factors without loss of their orthogonal nature. As this set covers a wide range of transcription initiation frequencies, it enables tuning of multiple outputs of the circuit in response to different sensory signals in an orthogonal manner. This sigma factor toolbox constitutes an interesting expansion of the synthetic biology toolbox and may contribute to the assembly of more complex synthetic genetic systems in the future.
INTRODUCTION
Over the past decades, following advances in metabolic engineering and synthetic biology, microbial production aroused increasing interest as valuable alternative for the production of a vast number of diverse (bio)chemicals. Efforts have focused on improving performance of industrial strains for synthesis of both native and new-to-the-host products (1–7). Metabolic engineering, with its top-down approach, focuses on altering the activity of enzymatic reactions through the overexpression of rate-limiting steps in combination with the deletion of competing pathways to drive the flux to the product of interest (8–10). This strategy has proven successful in improving yield and productivity of engineered strains. In contrast to metabolic engineering, synthetic biology uses a bottom-up approach making use of engineering principles, whereby complex systems are built by combining separate well-defined parts, to design and construct biological systems. With the concomitant development of DNA parts such as promoter (especially for housekeeping sigma factor 70, σ70, from Escherichia coli) and ribosome binding site (RBS) libraries (11–13), and new (high-throughput) DNA assembly techniques (14–19), combinatorial engineering of pathways emerged (4,20,21). In this way, expression of the various genes of interest can be optimized simultaneously irrespective of prior and/or profound knowledge and whole biological systems can be driven toward the desired phenotype. The use of aforementioned static control elements precludes, however, the adjustment of the expression in response to encountered transient conditions.
In nature, biological systems make use of complex regulatory networks to dynamically control metabolic fluxes in the cell in order to efficiently use cellular resources in transient conditions. One example is the control by transcriptional regulators that sense the concentration of metabolic intermediates and can accordingly repress or activate enzyme synthesis (22). Dynamic pathway control, for example to prevent the accumulation of toxic intermediates, can be implemented during pathway optimization to minimize negative effects on the host (23). In addition, to optimally respond to changing conditions observed by the cell, gene expression should be controllable independently from the host’s native regulatory control. To fulfill these requirements and to ensure optimal production and strain robustness, pathway expression has to occur: (i) in an orthogonal manner and, (ii) each of its steps should be maximally and dynamically fine-tuned. In order to achieve these goals there is a need for orthogonal gene expression elements and regulatory systems that operate without undesired crosstalk. Additionally, the availability of different orthogonal sets of parts would allow for the assembly of more complex genetic circuits. A pathway could then be divided in separate modules, which can be regulated independently in response to varying concentrations of intermediates or byproducts, thereby enabling integration of dynamic control of parts in a pathway. For these reasons, newly developed parts should be specific, orthogonal (strong interactions solely with cognate partner) and compatible in order to be used together in a multiple input multiple output genetic circuit (24,25). When fulfilling all these requirements, parts can be used for modular control of pathways in which orthogonal expression enables maximal independency toward the host and/or between output and the regulation used to control it.
In the last years, tools have become available for orthogonal control of gene expression both on the transcriptional (activators and repressors) and translational level with orthogonal ribosomes, riboswitches, sRNAs and toehold switches as examples for the latter (26–31). An advantage of transcriptional control in gene circuitry is that energy and resources are not wasted on RNA synthesis and, furthermore, more information is available about the underlying mechanisms compared to the regulation of downstream steps in the flow of information processing. Many bacterial regulatory mechanisms control the first step of transcription, which is initiated by binding of the unique RNA polymerase (RNAP) to specific promoter sequences (32–35). A different strategy relies on the use of bacteriophage encoded RNA polymerases of which T7 polymerase is well characterized and functions in an orthogonal manner in bacteria. Mutants of this polymerase have been engineered to obtain orthogonal polymerase–promoter pairs that allow for modular control (36,37). Furthermore, fragmented variants of the T7 polymerase have been used to create transcriptional and gates for integration in genetic circuitry (38) as well as a ‘resource allocator’ (39), where the assembly of different polymerase fragments in various combinations mimics the strategy of the bacterial RNA polymerase with its sigma factors to change cellular gene expression in response to stimuli. Still, for each one of these orthogonal sets there is currently only a limited number of promoters available (36,40). Moreover, phage-derived RNA polymerases generally remain to some extent toxic to the cell when used in bacteria. Conversely, the use of bacterial RNA polymerase or subunits thereof might generate less interference with the host.
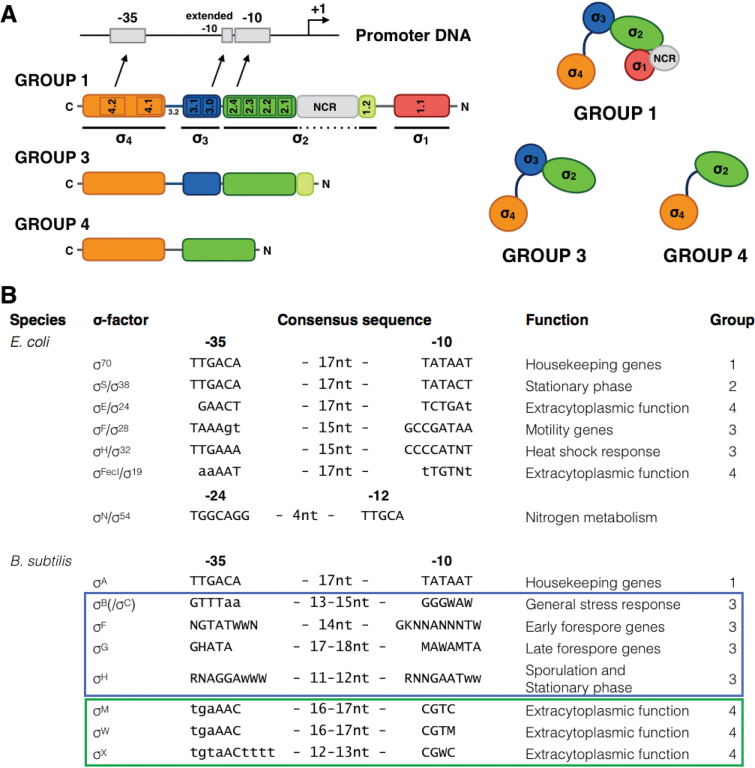
Bacterial RNA polymerases are multi-subunit enzymes, composed of a core enzyme (α2ββ’ω) associated with a sigma subunit (σ). The latter is responsible for promoter selectivity through recognition of specific DNA sequences in the promoter region. In addition to the housekeeping sigma factor (σ70 in E. coli; σA in Bacillus subtilis) that transcribes genes essential for growth, most bacteria have a variable number of alternative sigma factors, which bind competitively to the core enzyme and target the holoenzyme to distinct classes of promoters. This enables the cell to quickly change its genetic expression program in response to stress conditions or other environmental signals. Hence, sigma factors function as a form of global switches. This feature has been utilized by Rhodius et al. (41), who analyzed extracytoplasmic function (ECF) type sigma factors from different species in E. coli for orthogonality toward each other showing their potential for use in complex genetic circuits. Sigma factors are small modular proteins, comprising two to four domains, which differ greatly in sequence and size (Figure 1A). In addition to promoter specificity, they have a large dynamic output ranging from a very low OFF state to high levels of expression when turned ON (42–44). As a result, they are the first important type of selective transcriptional regulators and therefore an attractive starting point to create orthogonal expression systems.
Figure 1.
The bacterial sigma factor is responsible for promoter selectivity through recognition of specific DNA sequences in the promoter region. (A) Sigma factors of the sig70-family are divided in different groups based on their protein domain composition, consisting of two to four domains (42). Subregions 4.2 and 2.4 are of utmost importance in contacting the −35 and −10 promoter elements, respectively. Subregion 3.0 can contact the extended −10 element. (NCR = non-coding region). (B) List of different sigma factors, relevant for this work, originating from Escherichia coli and Bacillus subtilis with their consensus promoter sequences, design (group) and function in the cell.
Here, we present such orthogonal expression systems based on heterologous sigma factors from B. subtilis, a Gram-positive bacterium, which enable independent expression of different sets of genes. Additionally, promoter libraries for three of these sigma factors are built, thus obtaining a wide range of transcription initiation frequencies (TIF) for each sigma factor without loss of their orthogonal character toward each other and toward the host. Used in combination these heterologous sigma factors and their respective promoter libraries allow the assembly of modular circuits in which expression of each and every step of the pathway can be optimized independently.
MATERIALS AND METHODS
For additional information, see Supplementary Data Materials and Methods.
Media and bacterial strains
Lysogeny broth (LB) was used for cloning purposes. Complex medium (853) was used for all further experiments. Strains B. subtilis subsp. subtilis wild-type (LMG 7135), E. coli K12 MG1655 and E. coli Top10 were used for cloning purposes. Escherichia coli MG1655 and knock-in derivatives bearing a heterologous sigma factor integrated in the genome (made through homologous recombination according to the method of Datsenko and Wanner (45) with modifications) were used for all further experiments.
An overview of resulting strains is listed in Supplementary Table S3.
Cloning, growth and fluorescence reporters
Plasmid pTrc99a was used to express sigma factors from B. subtilis and E. coli under control of an isopropyl-β-D-1-thiogalactopyranoside (IPTG) inducible promoter. Plasmid pSC101-mKate2 was used as fluorescent reporter construct to measure the activity of B. subtilis and E. coli promoters. All derivatives of these plasmids were constructed in a seamless ligation reaction, protocol from Zhang et al. (17) with modifications. Composition of the B. subtilis promoters with their sequences are given in Supplementary Table S1. All primers used and resulting constructs are listed in Supplementary Tables S2 and 3. Constructs were verified by DNA sequencing. Competent cells for DNA transformation were prepared by the standard CaCl2 treatment (46).
Growth and fluorescence analysis
All strains (E. coli MG1655 knock-in derivatives and MG1655 transformants bearing a pTrc99a derivative) were tested in triplicate (different colonies = biological triplicates) on microtiter plates and experiments were repeated at least twice (independent plates). For growth analysis optical density (OD) at 600 nm was measured every 10 min and growth curve data were analyzed with the Grofit package in R (47). For fluorescence (FL) analysis: OD and FL were measured every 10 min and values were corrected for media blank. Further, the ratio of FL to OD was calculated and corrected for autofluorescence of the cells.
Promoter library construction
Libraries were constructed using standard molecular cloning protocols. Randomized DNA sequences, depicted as ‘N’ base pairs were included in polymerase chain reaction (PCR) primers to result in the promoter sequences shown in Table 1. Our cloning vector pLibrary is based on the pSC101 plasmid and additionally contains a constitutively expressed sfGFP gene (Supplementary Figure S2). The assembled plasmids were transformed in electrocompetent Top10 cells and subsequently grown overnight in 10 ml selective LB medium at 37°C. We obtained library sizes between 82 000 and 774 000 colony forming units (CFU) (Table 1). Plasmid DNA was extracted for all individual libraries with a Qiagen Plasmid Mini Kit and stored at −20°C.
Table 1. Library sequences overview and the maximum practically obtained coverage.
| Library | Sequence (5′ → 3′) | # transformants (×10−3) | Max. library coverage (%) |
|---|---|---|---|
| UP_B | TGTTTAAAAAAATGTCGGAGAACGT | N.A. | N.A. |
| UP_F | GTAAAGATGCGTCCTGTTCTGCGAT | N.A. | N.A. |
| UP_W | TGATAAACTTATTTTATAAAAAAAT | N.A. | N.A. |
| UP_D | GGTCTATGAGTGGTTGCTGGATAAC | N.A. | N.A. |
| UTR_D | AGGGAGAGCACAACGGTTTCCCTCTACAAATAATTTTGTTTAACTTT | N.A. | N.A. |
| PB2 | UP_B - GTTTAT TTTTTTGAAAAA GGGTAT GTAACTTGTA | N.A. | N.A. |
| LB2–1 | UP_B - GTTTATNNNNNNNNNNNNGGGTAT GTAACTTGTA | 475 | 2.83 |
| LB2–2 | UP_B - GTTTAT TTTTTTNNNNNNGGGTAT GTAACTTGTA | 298 | 100 |
| LB2–3 | UP_B - GTTTATNNNNNNGAAAAA GGGTAT GTAACTTGTA | 82 | 100 |
| PF3 | UP_F - GTTTA AAAACGATCTTTTTT TCTCATAAT AGTAGAAACA | N.A. | N.A. |
| LF3–1 | UP_F - GTTTANNNNNNNNNNNNNNN KCTCATAAT AGTAGAAACA | 751 | 0.035 |
| LF3–2 | UP_F - GTTTA AAAACGATNNNNNNN KCTCATAAT AGTAGAAACA | 762 | 100 |
| LF3–3 | UP_F - GTTTANNNNNNNNCTTTTTT TCTCATAAT AGTAGAAACA | 687 | 100 |
| PW2 | UP_W - TGAAAC CTTTTGAAACGAAGCT CGTA TACATACAGA | N.A. | N.A. |
| LW2–1 | UP_W - TGAAACNNNNNNNNNNNNNNNNCGTA TACATACAGA | 554 | 0.013 |
| LW2–2 | UP_W - TGAAAC CTTTTGAANNNNNNNNCGTA TACATACAGA | 666 | 100 |
| LW2–3 | UP_W - TGAAACNNNNNNNNACGAAGCT CGTA TACATACAGA | 774 | 100 |
| PproD* | UP_D - TTTACG GGCATGCATAAGGCTCG TATAAT ATATTC - UTR_D | N.A. | N.A. |
| LproD-1* | UP_D - TTTACGNNNNNNNNNNNNNNTCG TATAAT ATATTC - UTR_D | 391 | 0.15 |
| LproD-2* | UP_D - TTTACGNNNNNNNNNNNNNNNNNTATAAT ATATTC - UTR_D | 542 | 0.0032 |
Sequences represent the ‘promoter library site’ in Supplementary Figure S1.
Flow cytometry analysis
Each of the 12 B. subtilis promoter libraries and original promoters were transformed by electroporation into the E. coli K12 MG1655 strains containing either the genes for heterologous sigma factor B, W or F and in the wild-type strain, resulting in 48 new strains or pools. Furthermore, the control constructs containing the Phigh libraries and the construct containing only sfGFP were transformed in the WT strain giving a total of 52 strains. All strains were cultured overnight (12 h) at 30°C in 5 ml selective 853 medium. Cultures were then resuspended and diluted in filtered phosphate-buffered saline for flow cytometry analysis. All data were processed with a custom-written R script mainly using flowCore, flowPeaks and ggplot2 packages. Automated gating was based on the sfGFP reference FL and forward scatter (Supplementary Figure S4). The data reported in this study are generated by asinh transformation of the red over green fluorescence (RFP/sfGFP) and subtraction of the mean value of the negative control harboring no RFP gene (FluorescenceCT).
Library promoter selection and characterization
The three libraries constructed for each sigma factor were assembled (e.g. LB2-1, L B2-2 and L B2-3 together) prior to transformation. These assemblies were transformed by electroporation in strains bearing the gene for the respective cognate sigma factor. Next, samples for FACS were cultured and prepared as described above (flow cytometry analysis). Each library was sorted in twelve bins, with equal boundaries for the three libraries. Hence, the portion of each library sorted in a particular bin varies, strongly dependent on the expression profile, from 23% to under 0.1% of the population. Bins were plated and four colonies were picked from each bin (48 in total) for a FL-based selection (n = 1) of 9 or 10 promoters for each library representing the acquired expression range. sfGFP expression was also taken into account for this selection and colonies with aberrant expression were excluded. DNA was extracted from microtiter plate (MTP) cultures of the selected strains with an adapted protocol using a Qiagen Plasmid Mini Kit for lysis and neutralization, and ethanol precipitation for purification. Subsequent retransformation in strains with or without heterologous sigma factor was performed with fresh, in transformation and storage solution (TSS) buffer, chemically competent prepared cells. Four biological replicates were analyzed on microtiter plates by measuring FL after reaching the stationary growth phase. The reported values were obtained by first correcting mKate2 and sfGFP FL for media blank and subsequently, calculating the ratio of mKate2 over sfGFP FL (FluorescenceC).
RESULTS
Selection of heterologous sigma factors and promoters
The sequence and structure of core RNAP is highly conserved among Bacteria, which is not the case for the associating sigma factors. Although the latter vary greatly in size (from 20 to more than 70 kDa), they are functionally similar as indicated by the generation of functional hybrid holoenzymes composed of a sigma factor from one bacterium with the core enzyme from a distantly related one (41). In general, organisms that encounter varied environments, need frequent adjustments of their metabolism and/or must respond to many stresses contain a larger number of sigma factors. For this work with E. coli as host, 6 out of the 19 known/predicted sigma factors from the distantly related Gram-positive model organism B. subtilis were selected (Figure 1B). This selection is primarily based on the predicted promoter consensus sequences that differ substantially from the E. coli σ70 consensus to achieve specific recognition. All of the selected sigma factors (B, F, G, M, W, X) belong to the sig70-type, but with different domain compositions (Figure 1A) characteristic for the different groups of the sig70-family.
SigB from Gram-positive bacteria is functionally similar to RpoS in Gram-negative bacteria as they are both responsible for stationary-phase and stress response gene expression. However, they exhibit a dissimilar build (belonging to group 3 and group 2, respectively) and differ in genetic organization and regulation of expression and activity (44,48,49). Furthermore, their consensus promoter sequences are different, rendering sigma factor B a promising first choice for orthogonal gene expression in E. coli. In contrast, sigma factor D of B. subtilis was not taken into consideration as it is a functional ortholog of FliA from E. coli, with conserved promoter recognition and function, as indicated by the functional complementation of an E. coli fliA mutant with sigD (50). Sigma factors E, F, G, H and K are all involved in the sporulation cascade in which each one has a specific role. From these sigma factors E and K were dismissed because of their functional dependence on post-translational modifications (51,52). Sigma factors M, W and X are three out of the seven sigma factors from B. subtilis that belong to the ECF subfamily of sig70-type sigma factors. ECFs are among the smallest sigma factors bearing only domain 2 and 4, which are the most conserved of the four regions of bacterial sigma factors and correspond to domains mostly involved in binding core RNAP and DNA recognition (Figure 1A). Because of their minimalistic build which lacks domain 3, they are expected to have lower affinity for core RNAP compared to other sigma factors and no (specific) recognition of an extended −10 promoter element (42,53–55). Additionally, they have low non-specific DNA binding activity compared to σ70. The ECF family constitutes the largest group of alternative sigma factors and its members usually regulate functions related to cell envelope homeostasis.
For each of the six investigated sigma factors from B. subtilis, three to four cognate naturally occurring promoters (−60 to +2) expected to have different TIF were selected (Supplementary Table S1) (56–62).
Plasmid and genome-based expression of sigma factors
Sigma factors and mutant derivatives thereof were inserted either in a mid-copy number plasmid (≈40 copies) under control of an IPTG inducible promoter (pTrc99a derivative) or introduced into the chromosome (single copy) downstream of rpoS, while the corresponding promoters were placed upstream of a red fluorescent reporter gene on a low copy number plasmid (pSC101 derivative). The obtained sigma factor mutants bearing one or two amino acid substitutions (B S201R/R240H, M E73K, M A130V, W L94F, X T152P) were not created purposefully, but acquired during cloning. These mutations could be the result of standard PCR errors as is probably the case for sigma factor W of which both a WT and a mutant version were attained. For sigma factor M, we did not succeed in cloning the wild-type gene despite several attempts but instead obtained multiple variants each bearing a single mutation, resulting in a variety of (partially) functional proteins and non-functional ones with an out-of-frame mutation or early stop codon. These results suggest a possible growth inhibiting effect of the heterologous wild-type protein on the E. coli host in the conditions used. However, the heterologous sigma factors and mutants thereof (either on plasmid or genome) that were used further in this study showed either no or only minor effects on growth of the host (Supplementary Figure S1). To reduce some inherent problems of working with combinations of different plasmids and as expression from the chromosome is generally more stable and proved to be sufficient for efficient initiation at cognate promoters (63), all further experiments were conducted with genome-based expression of heterologous sigma factors.
Functionality and orthogonality of heterologous sigma factors and promoters in E. coli
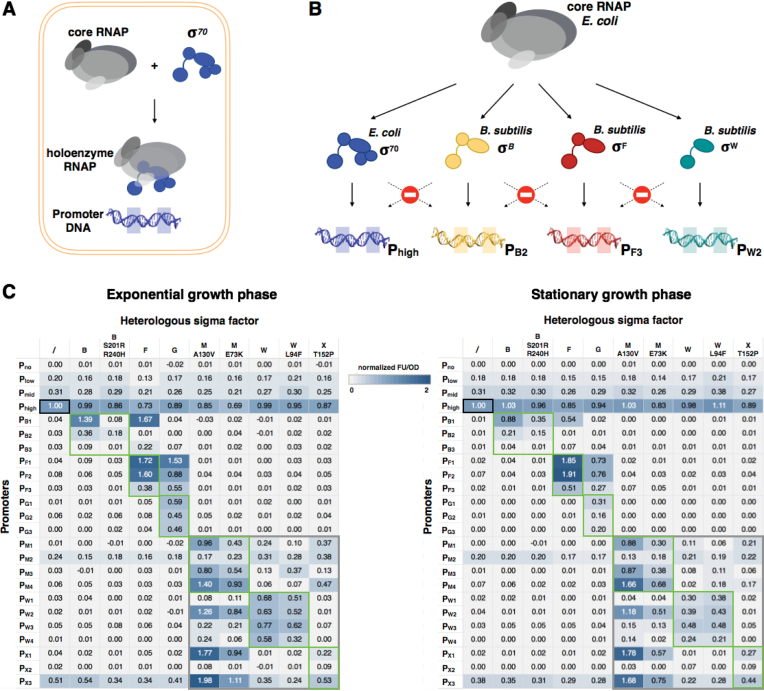
During exponential growth σ70 is most abundantly present in E. coli cells and binds with core RNAP to form a holoenzyme in order to initiate transcription from its promoter sequences (Figure 2A). Three σ70-specific promoters with different TIF were used as positive control and reference during FL measurements (12), whereas a promoter-less plasmid construct was used as negative control. In E. coli MG1655, the FL signal produced by these constructs corresponds to a relative expression level of 0.20, 0.31 and 1, respectively for the promoter with low, medium and high TIF (relative to promoter Phigh with the high TIF as reference and equal to 1) both in the exponential and stationary growth phase, while the negative control showed no measurable expression above background (Figure 2C). When the four reference plasmids were introduced in MG1655 recombinant strains bearing a heterologous sigma factor integrated in the chromosome, FL dropped to 0.7 in the presence of sigma factors F and M E73K and more moderately (0.85–0.9) in the presence of sigma factors B S201R/R240H, G, M A130V and X T152P. This result may be expected as these recombinant strains produce an additional sigma factor that will compete with the native sigma factors for binding to the limiting amount of core RNAP (Figure 2B and C). In contrast, we measured a small increase in FL for the recombinant strain bearing mutant sigma factor W L94F (in stationary growth phase), which could indicate some recognition of σ70-specific promoters by this heterologous sigma factor (Figure 2C).
Figure 2.
Functionality and orthogonality of heterologous sigma factors and promoters in Escherichia coli. (A) Schematic view of core RNA polymerase consisting of 5 subunits (α2ββ’ω)—given in different gray shades) that can bind the housekeeping sigma factor σ70 (given in blue) to form a holoenzyme able to transcribe from a σ70-specific promoter (also given in blue) in an E. coli cell. (B) Concept overview: an ideal orthogonal expression system combining several heterologous Bacillus subtilis sigma factors (namely sigma factor B, F and W given in yellow, red and green respectively) and cognate promoters introduced in the E. coli host. All sigma factors are able to bind the host core RNAP and form a holoenzyme that is able to recognize/transcribe specifically from its cognate promoter and not from the other promoters to yield a perfect orthogonal system without cross-talk. (C) Cross reactivity of sigma factor–promoter pairs measured in both exponential and stationary growth phase. Colors indicate activity (relative FL/OD) defined as the activity of the promoter-sigma pair divided by the activity of the reference (black square), a strong σ70 promoter (Phigh (12)) in wild-type E. coli MG1655. A gray square encloses the ECF sigma factors (M, W, X) in which some cross-talk is observed. Functionality of heterologous sigma factors from B. subtilis in E. coli strains bearing a cognate plasmid-borne promoter–reporter gene construct is shown by green rectangles. Data are the means of at least three biological replicates.
To determine potential recognition of the different B. subtilis promoters by the host transcription machinery, the various plasmid-borne reporter constructs were introduced in wild-type E. coli MG1655. Promoters PM2 and PX3 led to a rather high expression level (above 0.20), while all other promoters generated a signal of 0.08 or below compared to the reference signal. As the aforementioned promoters are recognized and transcribed by one or more E. coli sigma factor(s), (which is not desirable in this study in view of the targeted orthogonality) FL will be produced by all MG1655 derivatives. All other promoters appeared to be fully orthogonal toward native E. coli sigma factors (Figure 2C).
Subsequently, combinations of Bacillus sigma factors and their cognate promoters were tested to determine whether the different heterologous sigma factors are functionally expressed in E. coli (ability to bind the host core RNAP and initiate transcription at their cognate Bacillus promoters) (Figure 2B). With the exception of sigma factor B and its two amino acid substitutions derivative (B S201R/R240H) that showed only minor expression initiated from their cognate promoter PB3 and X T152P with PX2, FL signals were measured for all sigma-promoter pairs covering a wide range from 0.17 up to almost 2 (Figure 2C).
To verify orthogonality between the distinct sigma factors from B. subtilis all possible combinations of sigma factor–promoter pairs were generated in E. coli and relative FL signals were measured during both the exponential and stationary growth phase. The results indicate that sigma factor B and its mutant derivative only recognize their cognate promoters and prove to be orthogonal. Sigma factor F is orthogonal toward all promoters but promoter PB1 and PB3 of sigma factor B, while sigma factor G recognizes both its cognate promoters and those of sigma factor F. The ECF-type sigma factors (M, W, X) on the other hand show crosstalk among each other yet remain orthogonal toward the promoters of all other sigma factors tested. W (or its derivative) and X T152P are fully orthogonal toward each other and the host, but not with M (Figure 2).
Hence, the combination of sigma factors B, F, W and X of B. subtilis appears to be suited for the creation of an orthogonal set, which can be used to independently regulate gene expression in a modular pathway.
Orthogonality of B. subtilis promoters to native E. coli sigma factors
The concentration of alternative sigma factors in the cell can vary in response to different stress conditions. Even between exponential and stationary growth phase the intracellular concentrations of some alternative sigma factors differ. Shimada et al. (64) observed no change in protein concentrations between both growth phases for σ70, σ28 and σ54, while σ38 (/σS) is abundantly more present in stationary phase, and concentrations of σ24, σ32 and σ19 are only slightly higher during stationary phase. In spite of potential differences between exponential and stationary growth phase, the same trend of recognition was observed for all tested sigma factor–promoter pairs (Figure 2C).
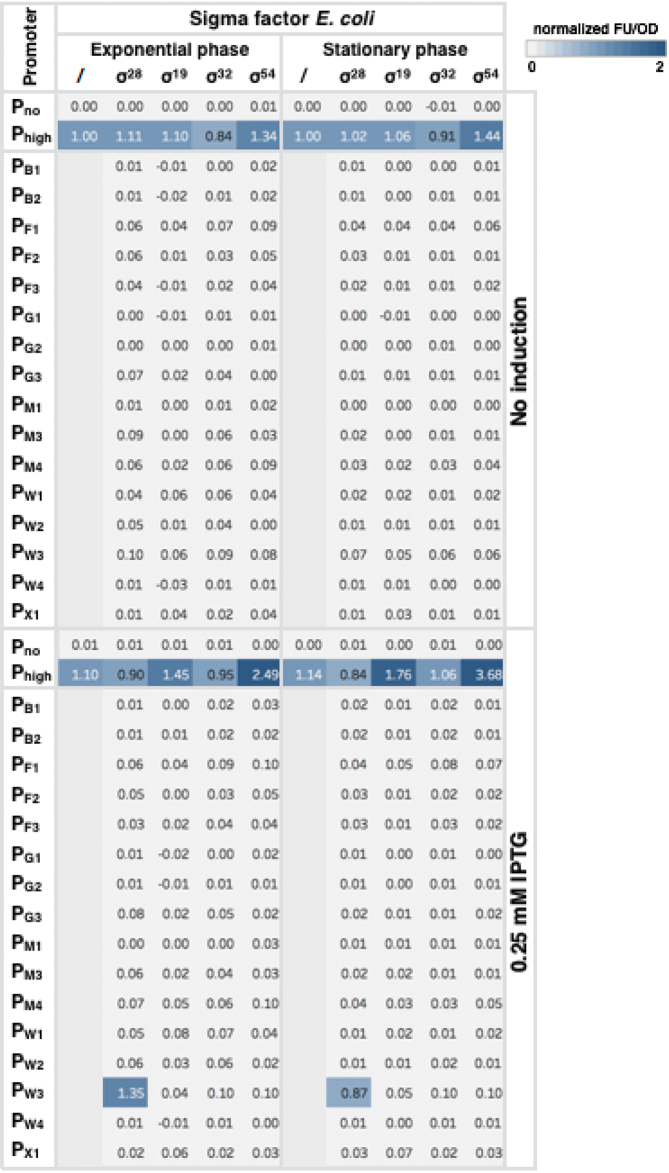
Still, to further evaluate orthogonality of the heterologous promoters to the native E. coli sigma factors, E. coli sigma factors (σ24, σ28, σ19, σ32 and σ54) were placed under control of an IPTG inducible promoter on a mid-copy number plasmid (≈40 copies). Unfortunately, sigma factor σ24 could not be cloned successfully in the conditions and system used in this study (early stop codon). Combinations of the other four sigma factor constructs together with the promoter reporter plasmids were transformed in E. coli and expression was measured with and without induction by IPTG. The results show that there is only recognition of promoter PW3 by σ28 upon induction with IPTG (Figure 3).
Figure 3.
Orthogonality of Bacillus subtilis promoters to native Escherichia coli sigma factors. Cross reactivity of sigma factor–promoter pairs measured in both exponential and stationary phase; and with and without addition of IPTG to induce expression of the E. coli sigma factors. Colors indicate activity (relative FL/OD) defined as the activity of the promoter–sigma pair divided by the activity of the reference, a σ70 promoter (Phigh (12)) in wild-type E. coli MG1655. B. subtilis promoters (PM2 and PX3) that were non-orthogonal toward E. coli (Figure 2C), and promoters with low TIF (<0.10) with their cognate sigma factor (Figure 2C, PB3 and PX2) were dismissed from this analysis.
Promoter libraries, construction and characterization
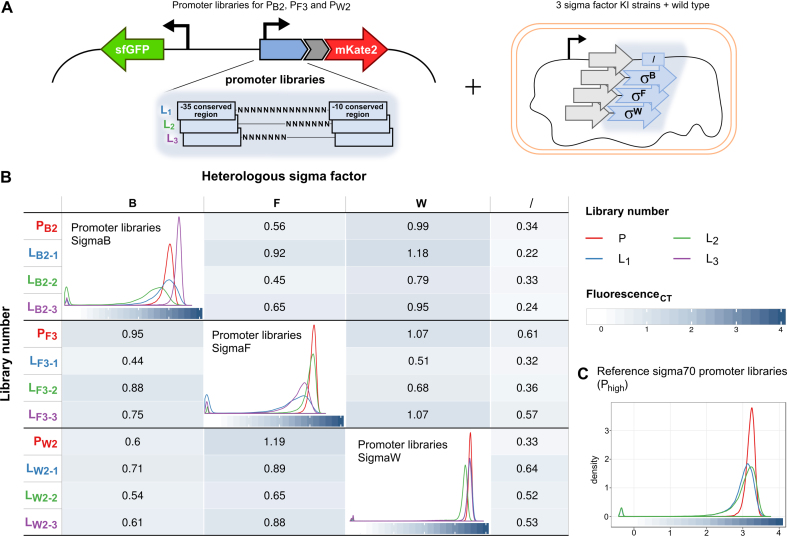
Besides orthogonality, tunability is another required characteristic of biological parts to enable predictable and scalable genetic designs (65). To meet this requirement, three orthogonal heterologous sigma factor–promoter pairs, toward each other and the host, were selected for the construction of promoter libraries. These are sigma factors B, F and W with their promoters PB2, PF3 and PW2, respectively. As sigma factor specificity is primarily determined by the conserved −10 and −35 promoter elements, these regions were not altered in the design with a view to preserve orthogonality (66). Instead, previous work demonstrated that sequences surrounding the −10 and −35 promoter elements are excellent targets to generate variability in expression levels (13,15). Aiming for enhanced tunability, suitable for the application of interest, and simultaneously restricting the potential loss of orthogonality to a minimum, three distinct libraries were designed for each promoter, targeting different regions of the spacer that separates the −10 and −35 promoter elements (Table 1). For the various selected B. subtilis promoters, library 1 (Lσ-1) consists of a completely randomized spacer sequence. For the construction of libraries 2 and 3 (Lσ-2, Lσ-3), half of the spacer, situated adjacent to either the −10 or the −35 conserved region of the promoter, was randomized. The −10 conserved regions include the extended −10 box for sigma factor B and F promoters, which is therefore constant over all libraries, with exception of the −16 position of F promoter libraries L1 and L2. Here a T/G (K) nucleotide was incorporated since G-16 is conserved and important for recognition by sigma factor F, but happens to be a T in promoter PF3 (58,67,68). Promoters for sigma factor W do not have the extension to their −10 consensus region since ECF sigma factors do not have an extended −10 interacting domain. In addition to the B. subtilis promoter libraries, as a control, promoter libraries were created for an E. coli σ70 promoter Phigh (12). Lhigh-1 has a randomized spacer analogous to the other L1 libraries. With Lhigh-2, the influence on the expression range of including the extended −10 region in the randomization is explored. Libraries (Lσ-n) and original promoters (Pσ) were all cloned in the pLibrary vector, which is a derivative of the pSC101-mKate2 vector used in this study (Figure 4A and Supplementary Figure S2). pLibrary additionally contains a constitutive promoter driving stable sfGFP expression to adjust for extrinsic factors influencing gene expression. In the negative control vector ‘sfGFP-only’, the promoter library cloning site and mKate2 were deleted. All 48 possible combinations consisting of the original promoter and pLibrary constructs transformed in strains bearing a cognate, a non-cognate or no heterologous sigma factor (MG1655) (Figure 4A) were grown overnight and characterized by flow cytometry. Furthermore, this series of strains was supplemented with MG1655 transformants bearing the Phigh libraries and the negative control containing only sfGFP. Figure 4B shows the libraries’ FL distribution for cognate pairs and population means for non-cognate pairs. The libraries (for both heterologous promoters as the E. coli-specific promoter) show distinct expression profiles for the different spacer randomizations characterized by a broadening of the expression range and/or a shift relative to the respective unmodified promoter. Generally, it appears that the orthogonal character of the generated libraries is well conserved. This is indicated by the mean FL of the distributions (for non-cognate pairs) that are in the same range or even lower, when comparing the distributions mean FL of the libraries (Lσ-n) with their original promoter (Pσ) within the same strain (Figure 4B). However, randomizing promoter DNA sequences has the potential to result in the creation of an undesirable subpopulation of inactive promoters and can therefore contribute to the lowering of the distribution means. Nevertheless, libraries tested in presence of their cognate sigma factor show that these subpopulations only account for a small percentage of the respective population (7.04% or less), except in two instances (LB2–2: 17.51% and LF3–1: 21.50%) (Supplementary Figure S5). On the other hand, in the presence of a non-cognate sigma factor, few libraries show an increase in number of active promoters indicating that a small subpopulation of promoters lost its orthogonal character, but this effect is limited to a maximum of 13.16% of the total population.
Figure 4.
Promoter libraries setup and characterization. (A) Original promoters PB2, PF3 and PW2 were selected to create three types of libraries by randomizing parts of the spacer sequence between the −10 and −35 conserved regions of the promoters. mKate2 expression is used as reporter to characterize library promoters and constitutively expressed sfGFP to correct for extrinsic factors. Vectors containing original promoters and their libraries were transformed in strains containing each of the cognate and non-cognate sigma factors and wild-type Escherichia coli. (B) All created strains were subjected to flow cytometry analysis. The presented data have been processed by asinh transformation of the mKate2/sfGFP ratio and corrected with the negative control (Supplementary Data Materials and Methods). Population means are displayed for non-cognate promoter–sigma factor pairs and whole library TIF distributions for cognate pairs. (C) Additionally, created σ70 promoter libraries act as a reference for native E. coli promoter library distributions.
To allow for (semi-)rational engineering using a defined set of promoters, with discrete expression levels within the achievable expression range, and to further investigate to which extent individual promoters retain their orthogonal properties, a selection of promoters was made for each sigma factor and characterized in detail. To obtain the representative sets of promoters, all three libraries (Lσ-1,2,3) generated for each sigma factor (B, F and W) were combined and transformed into the strain bearing the respective cognate sigma factor gene in the genome. Cell sorting (FACS) was used to split each mixed library into 12 separate bins, covering the entire expression range. The same bin layout/division was used for the three assembled libraries resulting in bin counts ranging from 23% to under 0.1% of the population. This was done to ensure the selection of promoters with well underrepresented TIF for subsequent screening. From each bin, four colonies were randomly picked and FL was measured for the cognate pairs before selecting 9 or 10 promoters for each library that cover a broad expression range. The resulting promoter set for each sigma factor was further characterized for orthogonality by making all combinations of promoters and strains bearing a cognate, a non-cognate or no heterologous sigma factor (wild-type MG1655).
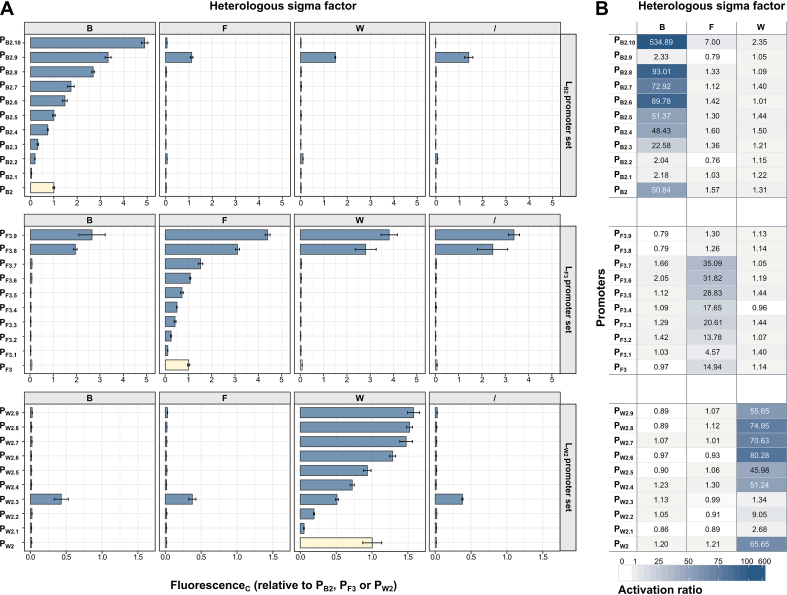
Figure 5A shows the FL measurements for the promoter sets relative to their respective original promoter as reference (PB2, PF3 and PW2, with cognate sigma factor, put equal to 1). The corresponding promoter DNA sequences are displayed in Supplementary Table S4. Expression for sigma factor B promoters ranges from 0.05 to 4.89, for sigma factor F promoters from 0.11 to 4.39 and for sigma factor W promoters from 0.05 to 1.57, all relative to their respective original promoter. The promoter with lowest TIF in presence of its cognate sigma factor, PB2.1, still shows an activation ratio of 2.18 over its expression in WT E. coli K12 (Figure 5B). The highest activation ratio is observed for PB2.10 in presence of sigma B, where expression is 535 times higher than in absence of a heterologous sigma factor. It is noticeable that in some cases the activation ratios are lower than 1, most pronounced for PF3.8 and PF3.9 in presence of sigma B or PB2.9 in presence of sigma F, which is conform the results shown in Figure 2, where promoters recognized by endogenous sigma factor(s) can result in lower expression by production of an additional sigma factor, possibly due to competition for core RNAP. When examining the sequences (Supplementary Table S4), it appears that for some promoters the spacer length differs from the respective original promoter and consequentially alters the promoters’ properties. The spacer from PW2.2 and PW2.3 is 1 bp shorter while PW2.1 misses 5 bp. Conversely, PB2.10 has an additional 2 bp. The occurrence of shorter sequences can be explained by an inaccurate DNA oligonucleotide synthesis process, whereby ∼60% of the ordered oligonucleotides only have the expected length due to imperfect base coupling efficiency (as reported by the supplier).
Figure 5.
Representative set of individual promoters for libraries LB2, LF3 and LW2. A selection of 9 or 10 promoters was made for each library. The three sets were analyzed in presence of their cognate sigma factor, a non-cognate sigma factor or in absence of a heterologous sigma factor (wild-type MG1655). (A) For each promoter–sigma factor couple, the media blank corrected mKate to sfGFP ratio is calculated (FluorescenceC). Depicted values are relative to the respective original promoter as reference (PB2, PF3 and PW2, with cognate sigma factor, put equal to 1). (B) Activation ratios are calculated as the ratio of the activity of a promoter in presence of a heterologous sigma factor to the activity of that promoter in absence of a heterologous sigma factor (wild-type MG1655).
DISCUSSION
With increasing interest and current successes in microbial production, the synthesis of more complex molecules is undertaken, which often involve long metabolic pathways (1,3,10,69,70). Recently, production of compounds such as taxol, fatty acids and many other products utilized a modular metabolic engineering approach, in which a pathway is divided in several modules on different vectors that bear different characteristics (copy number, promoters, RBS) (71–75). Synthetic biology aids to deal with these complex pathways by implementing genetic circuitry whereby well-defined parts are used to control separate steps and/or modules of the pathway. Therefore, a toolbox for orthogonal expression with systems that can be used together in the host to independently direct and regulate transcription of different genes is an important asset. In addition, tunability or the ability to optimize expression of independent genes remains a major benefit. In this work, this was achieved by using bacterial sigma subunits to create an orthogonal expression system in combination with the corresponding promoter libraries.
The effect of the heterologous sigma factors on growth in the conditions tested here was only minor to non-existent (Supplementary Figure S1). This is probably due, in part, to the low expression levels of sigma factors required to exert their function. It was shown that most of the assayed naturally occurring promoters specific for sigma factors from B. subtilis are orthogonal toward the host, as they are not recognized by E. coli sigma factors (Figures 2C and 3). This outcome was anticipated as σA of B. subtilis recognizes the same promoter consensus sequence as σ70 of E. coli and alternative sigma factors are often orthogonal toward their housekeeping sigma factor (Figure 1B) (42,44). Further, successful expression and functionality of the different heterologous sigma factors in E. coli was shown and their orthogonality toward the host and each other was evaluated (Figures 2C and 3). No crosstalk was observed between heterologous ECF sigma factors (M, W and X) and non-ECF factors (B, F, G). Most of the latter show also no interference between each other except sigma factor G, which recognizes promoters specific for F. This is physiologically relevant in B. subtilis as sigma factor G is expressed in a subsequent step of the sporulation cascade as compared to F (51,52). In contrast within the ECF-type sigma factors presented here, sigma factor M shows crosstalk with all, but W (and its derivative) and X T152P are orthogonal toward each other (Figure 2C). The −10 region of the promoter is the key determinant for this specificity with sequence CGAC and CGTA for sigma factor X and W respectively (76).
According to the obtained results, up to four of the analyzed heterologous sigma factors can be combined to independently express genes. As not all combinations of sigma factors are suited, this results in two possibilities to combine four orthogonal sigma factors, nine combinations with three and fifteen with two. However, besides orthogonality it remains important to be able to tune expression (65). In this work, a set of promoter libraries for orthogonal sigma factors B, F and W with an expanded expression range was successfully created while orthogonality is largely conserved. Randomization of the spacer sequence between −10 and −35 conserved regions is confirmed as a reliable method to introduce expression variability (13,15) and, as shown here, has the ability to simultaneously conserve specificity toward specific sigma factors. In depth characterization of a representative set of individual promoters for each sigma factor shows this conserved orthogonality is absolute for the majority of promoters (Figure 5). However, some promoters experience (a partial) loss of orthogonality, especially toward the host. Different regions in the spacer were randomized (corresponding to different libraries) but no correlation could be established between randomization pattern and resulting expression profile as effects are opposite for sigma factor B and F. However, there is a clear difference in expression profile (broad versus narrow range; low versus high TIF) between the different libraries generated per sigma factor. This variation can be of use depending on the application of interest. In contrast to the libraries for promoters PB2 and PF3, the different libraries for PW2 show a quite narrow range and similar TIF (Figure 4B). This might be due to the fact that sigma factor W belongs to group 4 of sig70-type family sigma factors, which comprises the smallest sigma factors consisting of only two domains, 2 and 4, that are primarily involved in interaction with the −10 and −35 promoter elements respectively (Figure 1A). Therefore, altering the spacer sequence might not be sufficient to create a broad expression range for this type of sigma factors. One possibility could be to shorten the spacer region to lower expression, as observed for the final selected sigma factor W promoters with lowest TIF (PW2.1, PW2.2 and PW2.3) that all have a shorter spacer than in the original promoter. Additionally, for PB2.10 the introduction of an extra 2 bp lengthened the spacer from 12 to 14 bp, which is more closely in line with the consensus (Figure 1B) and therefore could drive the relatively high TIF. Spacer length is a known expression determinant and can therefore be considered to be included in library designs together with the randomization of the spacer. Alternatively, one might consider to alter the sequence of the conserved boxes (preferably only in the −35 element to maintain specificity) to introduce larger expression variability for the ECF-type sigma factors, although this may occasion a risk of losing specificity along the way (76). Other possibilities would be changing sequences upstream of the −35 (UP element) or downstream of −10 conserved regions.
In conclusion, the ‘sigma factor toolbox’ generated in this study, composed of a selected set of orthogonal sigma factors with cognate promoter libraries exhibiting a wide variety of TIFs, constitutes an interesting tool for the assembly and expression of complex synthetic circuits.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fonds voor Wetenschappelijk Onderzoek Vlaanderen [G.0321.13N to M.D.M., D.C.]; Ghent University Special Research Fund [BOF16/IOP/040 to M.D.M.]; Fonds voor Wetenschappelijk onderzoek Vlaanderen (FWO-Vlaanderen) (to BVH); Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen) Ph.D. Grant (to M.V.B.). Funding for open access charge: Fonds Wetenschappelijk Onderzoek [G.0321.13N].
Conflict of interest statement. None declared.
REFERENCES
- 1. Wargacki A.J., Leonard E., Win M.N., Regitsky D.D., Santos C.N.S., Kim P.B., Cooper S.R., Raisner R.M., Herman A., Sivitz A.B. et al. An engineered microbial platform for direct biofuel production from brown macroalgae. Science. 2012; 335:308–313. [DOI] [PubMed] [Google Scholar]
- 2. Cheon S., Kim H.M., Gustavsson M., Lee S.Y.. Recent trends in metabolic engineering of microorganisms for the production of advanced biofuels. Curr. Opin. Chem. Biol. 2016; 35:10–21. [DOI] [PubMed] [Google Scholar]
- 3. Pandey R.P., Parajuli P., Koffas M.A.G., Sohng J.K.. Microbial production of natural and non-natural flavonoids: Pathway engineering, directed evolution and systems/synthetic biology. Biotechnol. Adv. 2016; 34:634–662. [DOI] [PubMed] [Google Scholar]
- 4. Paddon C.J., Keasling J.D.. Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat. Rev. Microbiol. 2014; 12:355–367. [DOI] [PubMed] [Google Scholar]
- 5. Trantas E.A., Koffas M.A.G., Xu P., Ververidis F.. When plants produce not enough or at all: metabolic engineering of flavonoids in microbial hosts. Front. Plant Sci. 2015; 6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keasling J.D. Manufacturing molecules through metabolic engineering. Science. 2010; 330:1355–1358. [DOI] [PubMed] [Google Scholar]
- 7. Woolston B.M., Edgar S., Stephanopoulos G.. Metabolic engineering: past and future. Annu. Rev. Chem. Biomol. Eng. 2013; 4:259–288. [DOI] [PubMed] [Google Scholar]
- 8. Singh A., Cher Soh K., Hatzimanikatis V., Gill R.T.. Manipulating redox and ATP balancing for improved production of succinate in E. coli. Metab. Eng. 2011; 13:76–81. [DOI] [PubMed] [Google Scholar]
- 9. Tai M., Stephanopoulos G.. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 2013; 15:1–9. [DOI] [PubMed] [Google Scholar]
- 10. Arense P., Bernal V., Charlier D., Iborra J.L., Foulquié-Moreno M.R., Cánovas M.. Metabolic engineering for high yielding L(-)-carnitine production in Escherichia coli. Microb. Cell Fact. 2013; 12:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mutalik V.K., Guimaraes J.C., Cambray G., Lam C., Christoffersen M.J., Mai Q.-A., Tran A.B., Paull M., Keasling J.D., Arkin A.P. et al. Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods. 2013; 10:354–360. [DOI] [PubMed] [Google Scholar]
- 12. Davis J.H., Rubin A.J., Sauer R.T.. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 2011; 39:1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Mey M., Maertens J., Lequeux G.J., Soetaert W.K., Vandamme E.J.. Construction and model-based analysis of a promoter library for E. coli: an indispensable tool for metabolic engineering. BMC Biotechnol. 2007; 7:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trubitsyna M., Michlewski G., Cai Y., Elfick A., French C.E.. PaperClip: rapid multi-part DNA assembly from existing libraries. Nucleic Acids Res. 2014; 42:e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coussement P., Maertens J., Beauprez J., Van Bellegem W., De Mey M.. One step DNA assembly for combinatorial metabolid engineering. Metab. Eng. 2014; 23:70–77. [DOI] [PubMed] [Google Scholar]
- 16. Quan J., Tian J.. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009; 4:e6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Y., Werling U., Edelmann W.. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012; 40:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Werner S., Engler C., Weber E., Gruetzner R., Marillonnet S.. Fast track assembly of multigene constructs using golden gate cloning and the MoClo system. Biobugs. 2012; 3:38–43. [DOI] [PubMed] [Google Scholar]
- 19. Van Hove B., Guidi C., De Wannemaeker L., Maertens J., De Mey M.. Recursive DNA assembly using protected oligonucleotide duplex assisted cloning (PODAC). ACS Synth. Biol. 2017; 6:943–949. [DOI] [PubMed] [Google Scholar]
- 20. De Mey M., Maertens J., Boogmans S., Soetaert W.K., Vandamme E.J., Cunin R., Foulquié-Moreno M.R.. Promoter knock-in: a novel rational method for the fine tuning of genes. BMC Biotechnol. 2010; 10:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zou R., Zhou K., Stephanopoulos G., Too H.P.. Combinatorial engineering of 1-Deoxy-D-Xylulose 5-Phosphate pathway using cross-lapping in vitro assembly (CLIVA) method. PLoS OnE. 2013; 8:e79557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De Paepe B., Peters G., Coussement P., Maertens J., De Mey M.. Tailor-made transcriptional biosensors for optimizing microbial cell factories. J. Ind. Microbiol. Biotechnol. 2017; 44:623–645. [DOI] [PubMed] [Google Scholar]
- 23. Wu G., Yan Q., Jones J.A., Tang Y.J., Fong S.S., Koffas M.A.G.. Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol. 2016; 34:652–664. [DOI] [PubMed] [Google Scholar]
- 24. Bradley R.W., Buck M., Wang B.. Tools and principles for microbial gene circuit engineering. J. Mol. Biol. 2016; 428:862–888. [DOI] [PubMed] [Google Scholar]
- 25. Slusarczyk A.L., Lin A., Weiss R.. Foundations for the design and implementation of synthetic genetic circuits. Nat. Rev. Genet. 2012; 13:406–420. [DOI] [PubMed] [Google Scholar]
- 26. An W., Chin J.W.. Synthesis of orthogonal transcription-translation networks. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:8477–8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Salis H.M., Mirsky E.A., Voigt C.A.. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009; 27:946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li G.-W., Oh E., Weissman J.S.. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012; 484:538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sharma V., Nomura Y., Yokobayashi Y.. Engineering complex riboswitch regulation by dual genetic selection. J. Am. Chem. Soc. 2008; 130:16310–16315. [DOI] [PubMed] [Google Scholar]
- 30. Na D., Yoo S.M., Chung H., Park H., Park J.H., Lee S.Y.. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol. 2013; 31:170–174. [DOI] [PubMed] [Google Scholar]
- 31. Green A.A., Silver P.A., Collins J.J., Yin P.. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014; 159:925–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A.. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013; 152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stanton B.C., Nielsen A.A.K., Tamsir A., Clancy K., Peterson T., Voigt C.A.. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat. Chem. Biol. 2013; 10:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L.A.. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013; 41:7429–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brewster R.C., Jones D.L., Phillips R.. Tuning promoter strength through RNA polymerase binding site design in Escherichia coli. PLoS Comput. Biol. 2012; 8:e1002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Temme K., Hill R., Segall-Shapiro T.H., Moser F., Voigt C.A.. Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 2012; 40:8773–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meyer A.J., Ellefson J.W., Ellington A.D.. Directed evolution of a panel of orthogonal T7 RNA polymerase variants for in vivo or in vitro synthetic circuitry. ACS Synth. Biol. 2015; 4:1070–1076. [DOI] [PubMed] [Google Scholar]
- 38. Shis D.L., Bennett M.R.. Library of synthetic transcriptional AND gates built with split T7 RNA polymerase mutants. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:5028–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Segall-Shapiro T.H., Meyer A.J., Ellington A.D., Sontag E.D., Voigt C.A.. A ‘resource allocator’ for transcription based on a highly fragmented T7 RNA polymerase. Mol. Syst. Biol. 2014; 10:742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jones J.A., Vernacchio V.R., Lachance D.M., Lebovich M., Fu L., Shirke A.N., Schultz V.L., Cress B., Linhardt R.J., Koffas M.A.G.. ePathOptimize: a combinatorial approach for transcriptional balancing of metabolic pathways. Sci. Rep. 2015; 5:11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rhodius V.A., Segall-Shapiro T.H., Sharon B.D., Ghodasara A., Orlova E., Tabakh H., Burkhardt D.H., Clancy K., Peterson T.C., Gross C.A. et al. Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Mol. Syst. Biol. 2013; 9:702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paget M. Bacterial sigma factors and anti-sigma factors: Structure, function and distribution. Biomolecules. 2015; 5:1245–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kazmierczak M.J., Wiedmann M., Boor K.J.. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 2005; 69:527–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gruber T.M., Gross C.A.. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003; 57:441–466. [DOI] [PubMed] [Google Scholar]
- 45. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dagert M., Ehrlich S.D.. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979; 6:23–28. [DOI] [PubMed] [Google Scholar]
- 47. Kahm M., Hasenbrink G., Lichtenberg-Fraté H., Ludwig J., kschischo M.. Grofit: fitting biological growth curves with R. J. Stat. Softw. 2010; 33:7. [Google Scholar]
- 48. Hengge-Aronis R. Stationary phase gene regulation: what makes an Escherichia coli promoter sigmaS-selective. Curr. Opin. Microbiol. 2002; 5:591–595. [DOI] [PubMed] [Google Scholar]
- 49. Lonetto M., Gribskov M., Gross C.A.. The sigma 70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 1992; 174:3843–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen Y.F., Helmann J.D.. Restoration of motility to an Escherichia coli fliA flagellar mutant by a Bacillus subtilis sigma factor. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:5123–5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Errington J. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 2003; 1:117–126. [DOI] [PubMed] [Google Scholar]
- 52. Piggot P.J., Hilbert D.W.. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 2004; 7:579–586. [DOI] [PubMed] [Google Scholar]
- 53. Maeda H., Fujita N., Ishihama A.. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 2000; 28:3497–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mitchell J.E., Zheng D., Busby S.J.W., Minchin S.D.. Identification and analysis of ‘extended -10’ promoters in Escherichia coli. Nucleic Acids Res. 2003; 31:4689–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grigorova I.L., Phleger N.J., Mutalik V.K., Gross C.A.. Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:5332–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Petersohn A., Bernhardt J., Gerth U., Höper D., Koburger T., Völker U., Hecker M.. Identification of sigma(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J. Bacteriol. 1999; 181:5718–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Amaya E., Khvorova A., Piggot P.J.. Analysis of promoter recognition in vivo directed by F of Bacillus subtilis by using random-sequence oligonucleotides. J. Bacteriol. 2001; 183:3623–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang S.T., Setlow B., Conlon E.M., Lyon J.L., Imamura D., Sato T., Setlow P., Losick R., Eichenberger P.. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 2006; 358:16–37. [DOI] [PubMed] [Google Scholar]
- 59. Britton R.A., Eichenberger P., Gonzalez-Pastor J.E., Fawcett P., Monson R., Losick R., Grossman A.D.. Genome-wide analysis of the stationary-phase sigma factor (Sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 2002; 184:4881–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Eiamphungporn W., Helmann J.D.. The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 2008; 67:830–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cao M., Kobel P.A., Morshedi M.M., Wu M.F.W., Paddon C., Helmann J.D.. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 2002; 316:443–457. [DOI] [PubMed] [Google Scholar]
- 62. Cao M., Helmann J.D.. The Bacillus subtilis extracytoplasmic-function X factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J. Bacteriol. 2004; 186:1136–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tyo K.E.J., Ajikumar P.K., Stephanopoulos G.. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat. Biotechnol. 2009; 27:760–765. [DOI] [PubMed] [Google Scholar]
- 64. Shimada T., Tanaka K., Ishihama A.. The whole set of the constitutive promoters recognized by four minor sigma subunits of Escherichia coli RNA polymerase. PLoS One. 2017; 12:e0179181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lucks J.B., Qi L., Whitaker W.R., Arkin A.P.. Toward scalable parts families for predictable design of biological circuits. Curr. Opin. Microbiol. 2008; 11:567–573. [DOI] [PubMed] [Google Scholar]
- 66. Browning D.F., Busby S.J.W.. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004; 2:57–65. [DOI] [PubMed] [Google Scholar]
- 67. Sun D., Fajardo-Cavazos P., Sussman M.D., Tovar-Rojo F., Cabrera-Martinez R.M., Setlow P.. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by E sigma F: identification of features of good E sigma F-dependent promoters. J. Bacteriol. 1991; 173:7867–7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bagyan I., Casillas-Martinez L., Setlow P.. The katX gene, which codes for the catalase in spores of Bacillus subtilis, is a forespore-specific gene controlled by sigmaF, and KatX is essential for hydrogen peroxide resistance of the germinating spore. J. Bacteriol. 1998; 180:2057–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Paddon C.J., Westfall P.J., Pitera D.J., Benjamin K., Fisher K., McPhee D., Leavell M.D., Tai A., Main A., Eng D. et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013; 496:528–532. [DOI] [PubMed] [Google Scholar]
- 70. Jullesson D., David F., Pfleger B., Nielsen J.. Impact of synthetic biology and metabolic engineering on industrial production of fine chemicals. Biotechnol. Adv. 2015; 33:1395–1402. [DOI] [PubMed] [Google Scholar]
- 71. Ajikumar P.K., Xiao W.H., Tyo K.E.J., Wang Y., Simeon F., Leonard E., Mucha O., Phon T.H., Pfeifer B., Stephanopoulos G.. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science. 2010; 330:70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xu P., Gu Q., Wang W., Wong L., Bower A.G.W., Collins C.H., Koffas M.A.G.. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat. Commun. 2013; 4:1409. [DOI] [PubMed] [Google Scholar]
- 73. Gu L., Zhang J., Du G., Chen J.. Multivariate modular engineering of the protein secretory pathway for production of heterologous glucose oxidase in Pichia pastoris. Enzyme Microb. Technol. 2015; 68:33–42. [DOI] [PubMed] [Google Scholar]
- 74. Jones J.A., Toparlak Ö.D., Koffas M.A.. Metabolic pathway balancing and its role in the production of biofuels and chemicals. Curr. Opin. Biotechnol. 2015; 33:52–59. [DOI] [PubMed] [Google Scholar]
- 75. Wu J., Liu P., Fan Y., Bao H., Du G., Zhou J., Chen J.. Multivariate modular metabolic engineering of Escherichia coli to produce resveratrol from L-tyrosine. J. Biotechnol. 2013; 167:404–411. [DOI] [PubMed] [Google Scholar]
- 76. Qiu J., Helmann J.D.. The -10 region is a key promoter specificity determinant for the Bacillus subtilis extracytoplasmic-function sigma factors sigma(X) and sigma(W). J. Bacteriol. 2001; 183:1921–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.