Abstract
i-Motif (iM) is a four stranded DNA structure formed by cytosine-rich sequences, which are often present in functionally important parts of the genome such as promoters of genes and telomeres. Using electronic circular dichroism and UV absorption spectroscopies and electrophoretic methods, we examined the effect of four naturally occurring DNA base lesions on the folding and stability of the iM formed by the human telomere DNA sequence (C3TAA)3C3T. The results demonstrate that the TAA loop lesions, the apurinic site and 8-oxoadenine substituting for adenine, and the 5-hydroxymethyluracil substituting for thymine only marginally disturb the formation of iM. The presence of uracil, which is formed by enzymatic or spontaneous deamination of cytosine, shifts iM formation towards substantially more acidic pH values and simultaneously distinctly reduces iM stability. This effect depends on the position of the damage sites in the sequence. The results have enabled us to formulate additional rules for iM formation.
INTRODUCTION
Endogenous biochemical reactions and exogenous effects can trigger the formation of reactive oxygen species (ROS) in cells, which react with various molecules in cells including biomacromolecules. Of these, nucleic acids are among the main macromolecular targets of ROS induced damage (1). The physical and biochemical effects of this damage has been thoroughly studied with double helical (2) and a few non-B DNA models, including the four-stranded G-quadruplexes (3–5). Another four-stranded non-canonical nucleic acid architecture is the i-motif (iM), which can also be targeted by intracellular ROS reactions.
Oligo- and polynucleotides with repeating cytosine (C)-rich sequences tend to form iM structures in slightly acidic conditions (6). The iM consists of two parallel stranded duplexes that are held together by hemi-protonated cytosine base pairs (C·C+) (Figure 1A) and are mutually intercalated in antiparallel orientation (Figure 1B). C-rich sequences of the human genome also form iM structures facilitated by negative superhelicity and crowding under physiological conditions. This implicates a biological role of this architecture (7–15). Due to the pH-dependence of iM formation, nanotechnology has been using iM-forming sequences as pH-switches, DNA-nanomotors and other DNA nano-devices (16,17). Earlier (7) as well as recent reviews (8,16,18) have evaluated many aspects of the sequence requirements, kinetics, topology and stability of iMs, their significance in gene expression, and in iM–ligand complex formation. Structural features of iMs have also been studied with nucleotide analogues, similarly to the other four-stranded non-B fold, the G-quadruplexes (4). However, the spectrum of the natural and synthetic building blocks used in iM studies is less comprehensive than with the G-quadruplexes. Primarily, 5-methylcytosine (5mC), an epigenetic modification has been studied in detail (13,19–24). Publications on ROS-induced damage include derivatives of 5mC: 5-hydroxymethylcytosine (5hmC), 5-formylcytosine and 5-carboxycytosine. These oxidized derivatives are not only intermediates of enzymatic demethylation of mC but also serve as epigenetic marks in mammals (13,20). The 5mC, except in the case of hypermethylation, stabilized (20), whereas 5hmC destabilized the iM (13). iMs fold near pH 6, but replacing C with 5mC shifts this pH towards neutral values, while replacing C with 5-bromocytosine (5BrC) shifts the pH to more acidic values (25). Regarding loop modifications, loop length and sequence have been found to significantly impact the thermodynamics and kinetics of the folding of iM DNA (26–31). The effect of substitutions of loop nucleotides of iMs has barely been studied (24,32–34).
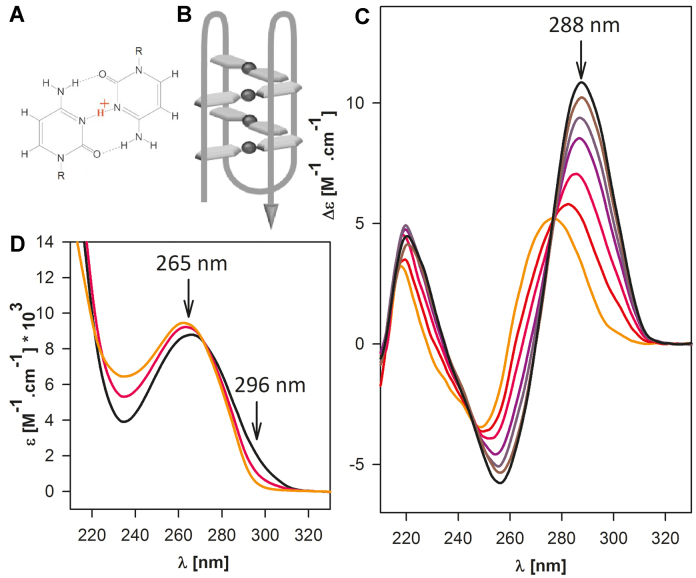
Figure 1.
(A) A hemiprotonized C·C+ base pair; (B) a schematic structure of an intramolecular iM; (C) CD and (D) UV absorption spectra reflecting iM formation induced by decreasing pH from 7.5 (orange) to 5.0 (black). The course of iM formation and unfolding is monitored by changes in Δϵ at 288 nm and ϵ at 296 nm, respectively, as indicated in the figure.
We have long been interested in investigating the effect of natural base lesions on the folding topology of G-quadruplexes of the human telomere DNA sequence. We substituted G bases of the G-tetrads by derivatives of G, such as 8-oxoguanine (35) and an abasic site (36), and recently, by incorporating 8-oxoadenine (8oxoA), adenine abasic sites and 5-hydroxymethyluracil into the TTA loops of the human telomere G-strand quadruplex (3). However, the effects of natural lesions in the complementary C-rich human telomere (htel) sequence have not been studied yet. Here, we present the results of the effects of the most frequently occurring natural base lesions within the loops and the core cytosines of the iM formed by the htel C-strand 22mer sequence, 5′-(CCCTAA)3CCCT-3′. The study includes the influence of the 8oxoA, and the abasic site replacing 2′-deoxyadenosines, and 5hmU (37) substituting for thymine of the TAA loops, and of the uracil, the deamination product of core cytosines (38,39), on the formation and stability of the htel iM.
MATERIALS AND METHODS
Oligonucleotides
Loop-modified DNA sequences (Supplementary Table S1) were synthesized by the Department of Functional Genomics and Proteomics, Masaryk University, Brno (36), and the sequences with substitutions of U for C by Sigma-Aldrich (Haverhill, UK). The lyophilized samples were dissolved in 1 mM sodium phosphate (pH 8.0) with 0.3 mM EDTA. The purity of the samples in terms of length homogeneity was checked by denaturing gel electrophoresis. Before measurements, the DNA samples were thermally denatured in 1 mM sodium phosphate at 90°C for 5 min. Precise concentration of the studied sequences was determined based on UV absorption measured at 260 nm at 90°C using molar absorption coefficient of 8900 M−1·cm−1 calculated for the wild type sequence of (C3TAA)3C3T according to Gray (40). The same coefficient was used for the loop-modified sequences. The extinction coefficients of 9000 and 9050 M−1·cm−1 were used for one and two U/C core substitutions, respectively. UV absorption was measured on a UNICAM 5625 UV/VIS spectrometer (Cambridge, UK).
Circular dichroism
Circular dichroism spectra were measured on a Chirascan-plus CD spectrometer (Applied Photophysics, UK) in the region of 210–330 nm with 0.5 nm step and 0.5 s time-per-point. CD was expressed as a difference in molar absorption of left- and right-handed circularly polarized light (Δϵ) related to base. The measurement was performed at 23°C in 1 cm path-length quartz Hellma cells placed in a thermostated holder.
UV melting
Denaturation/renaturation curves were measured on a Varian Cary 4000 spectrophotometer in a K-RB buffer, pH 5 (specified below). The spectra were collected from 210 to 330 nm in the temperature range 1–85°C in four cycles (up, down, up and down) with 1°C step and 2 min of waiting at each temperature. The final temperature change rate was around 0.25°C·min−1. Melting curves were monitored by absorbance at 296 nm, Tm values were determined from 0 to 1 normalized dependences (the bottom and upper linear baseline-corrected melting curves) as the temperature where half of the sample was folded (41). The error range of Tm determination was ±0.5°C.
pH measurements
pH dependences were measured on pH meter Mettler Toledo (with an In Lab Micro electrode) in Robinson-Britton (K-RB) buffer, which is a mixture of 0.04 M H3BO3, 0.04 M CH3COOH and 0.04 M H3PO4 titrated with 0.2 M KOH to desired pH. The K+ concentration was 0.07 M for pH 7. The oligonucleotides were titrated directly in cells by 0.2 M HCl and pH dependent changes were monitored by a set of values of Δϵ at 288 nm assembled from twenty spectra on average. The pKa values were determined based on the 4-parameter Hill equation used to automatically fit the experimental points by a SigmaPlot software (Systat Software, USA). The error range of pKa determination was ±0.05.
Native PAGE
The molecularity of oligonucleotides was determined from native polyacrylamide gel electrophoresis (PAGE) using an electrophoretic instrument (SE-600; Hoefer Scientific, San Francisco, CA, USA). Gel concentration was 16% and both the gel and the running buffer contained K-RB buffer, pH 5. About 2 μg of DNA was loaded on the gel. Electrophoresis ran at 30 V and 23°C for 18 h for samples with damaged loops and at 50 V and 4°C for 19 h for samples with damaged cytosines. Gels were stained with Stains All dye (Sigma-Aldrich, St. Louis, MO, USA) and scanned using a Personal Densitometer SI, model 375-A (Molecular Dynamics, Sunnyvale, CA, USA).
RESULTS
Formation of an i-motif (iM) is sensitively reflected in the spectra of the electronic circular dichroism (Figure 1C) and in UV absorption spectra (Figure 1D). A single-stranded unordered C-rich human telomere (htel) DNA fragment yields a CD spectrum with a positive maximum at 275 nm and a negative one around 245 nm. With decreasing pH below pH 7, the sequence forms an iM, which is reflected by an increase of the positive CD band and a simultaneous decrease of the negative one, while the whole spectrum shifts towards the red. CD spectrum with a positive maximum at 288 nm and a negative one close to 260 nm is characteristic of the iM (Figure 1C). Cytosine protonation is reflected by an increase in the UV absorption spectrum around 300 nm and the formation of the ordered iM is accompanied by a decrease of the 265 nm maximum and a slight bathochromic shift (Figure 1D) (11,42).
Lesions in iM loops
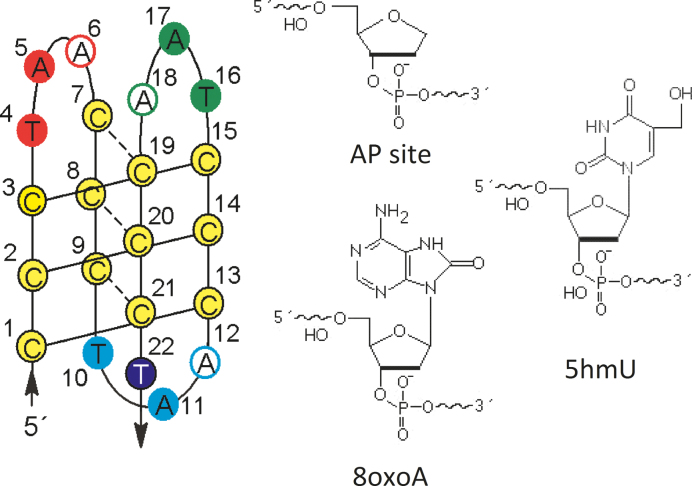
We examined three types of damage which frequently occur in T and A bases forming TAA loops in the human telomere C-rich DNA sequence (C3TAA)3C3T. Adenines were replaced by the abasic (AP) sites or 8-oxoadenines (8oxoA), and thymines were replaced by 5-hydroxymethyluracils (5hmU). The studied sequences are called apAX, 8oxoAX or 5hmUX, where X corresponds to the order of the substituted nucleobase in the studied htel 22-mer. All the studied sequences are listed in Supplementary Table S1. A scheme of the iM structure with the numbering of particular nucleobases is shown in Figure 2.
Figure 2.
Schematic presentation of the htel iM with numbering and colors used for particular nucleotides referenced in the text and the figures in this paper; chemical structures of studied lesions.
Adenine abasic (AP/A) sites
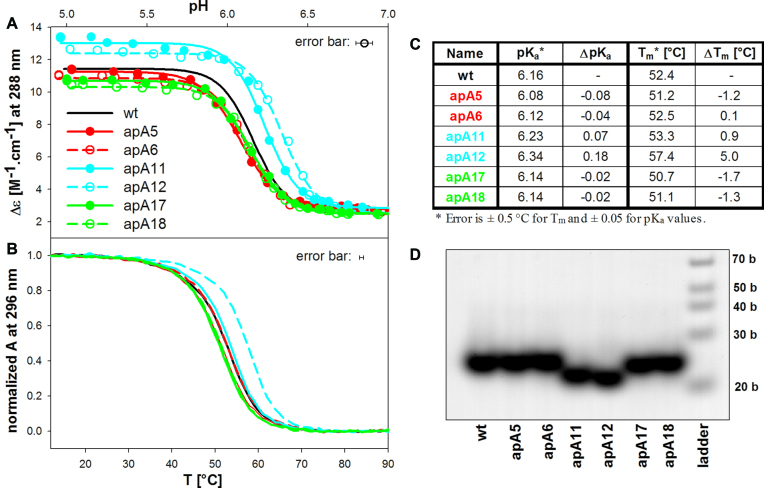
Figure 3A shows a pH-induced formation of iM by htel sequences containing AP lesions instead of particular loop A (AP/A). The iM formation is monitored by changes in CD values of its characteristic positive band (Figure 1C). The presence of AP/A lesions in the first and the third loops has virtually no effect on the formation of iM: Δϵ at 288 nm as a function of pH follows very closely the curve of the htel wild type (wt) sequence. The transition takes place in the region of pH 6.5–5.8 (Figure 3A), with the midpoint of pH ∼6.12 (Figure 3C). Only the AP/A sites in the middle loop affect iM formation significantly. The transition midpoint of apA12 is shifted by 0.18 towards neutral pH. Changes in pKa of other sequences are below the accuracy of their determination. The resulting iMs of apA11 and apA12 reach higher Δϵ values than the iM of the wt (Figure 3A). The AP sites in the middle loop increase the thermostability of the htel iM structure by 0.9 and 5°C for apA11 and apA12, respectively compared with the wt. In contrast, AP sites in the third, 3′end loop, slightly destabilize the iM (Figure 3B and C). The temperature dependences are fully reversible.
Figure 3.
(A) pH dependent iM formation of the AP/A site containing htel sequences monitored by Δϵ at 288 nm; (B) normalized melting curves monitored by UV absorption at 296 nm, (C) table of pKa and Tm values, (D) the 16% native PAGE. The measurements were carried out in K-RB, pH 5 for melting and electrophoresis, and at 23°C for the pH dependence and electrophoresis as stated in the M+M section.
The presence of the AP sites does not alter the molecularity of the iM. All the structures are intramolecular. The missing bases in the middle loop slightly accelerate the migration of the structure (Figure 3D), probably indicating increased compactness of the structures.
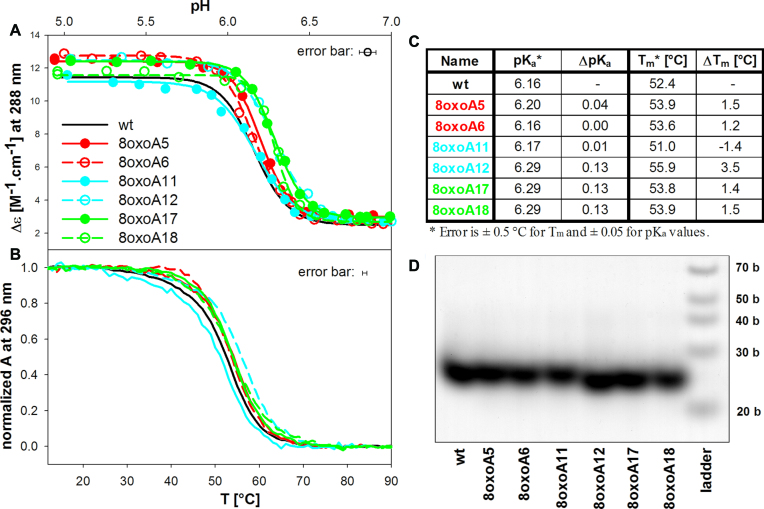
8-oxoadenine (8oxoA) substitution for adenine
pH induced iM formation of 8oxoA htel analogues is shown in Figure 4A. With the exception of 8oxoA11, the presence of the oxo group stabilizes iM formation (Figure 4B and C) and shifts its formation towards neutral pH values for 8oxoA12 and the sequences substituted in the third loop. The effect of 8oxoA substitution in the middle loop is distinct for 8oxoA11 and 8oxoA12. 8oxoA12 forms the most stable and 8oxoA11 the least stable iM. The midpoint of the 8oxoA11 transition to iM is comparable with that of the wt, but it proceeds with lower cooperativity and the CD spectrum of the final iM reaches a lower Δϵ value (Figure 4A). The temperature dependences of all sequences are fully reversible. The 8oxoA/A substitution does not change the molecularity and all iMs remain intramolecular (Figure 4D).
Figure 4.
(A) pH dependent iM formation of the 8oxoA containing htel sequences monitored by Δϵ at 288 nm; (B) normalized melting curves monitored by UV absorption at 296 nm, (C) table of pKa and Tm values, (D) the 16% native PAGE. The conditions are as in Figure 3.
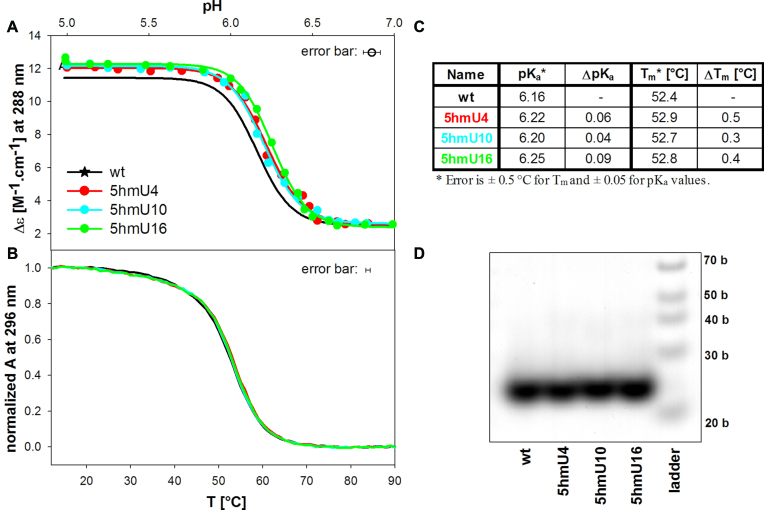
5-hydroxymethyluracil (5hmU) substitution for thymine
The presence of the 5hmU influences only slightly iM formation (Figure 5) and the changes in Tm and pKa are within or close to the accuracy of their determination. Final iMs of the substituted sequences yield slightly higher Δϵ values than the wt (Figure 5A). The modification influences neither the molecularity of the iMs nor affects distinctly their thermostability (Figure 5B, C and D).
Figure 5.
(A) pH dependent iM formation of the 5hmU containing htel sequences monitored by Δϵ at 288 nm; (B) normalized melting curves monitored by UV absorption at 296 nm, (C) table of pKa and Tm values, (D) the 16% native PAGE. The conditions are as in Figure 3.
Substitution of core cytosines by uracil
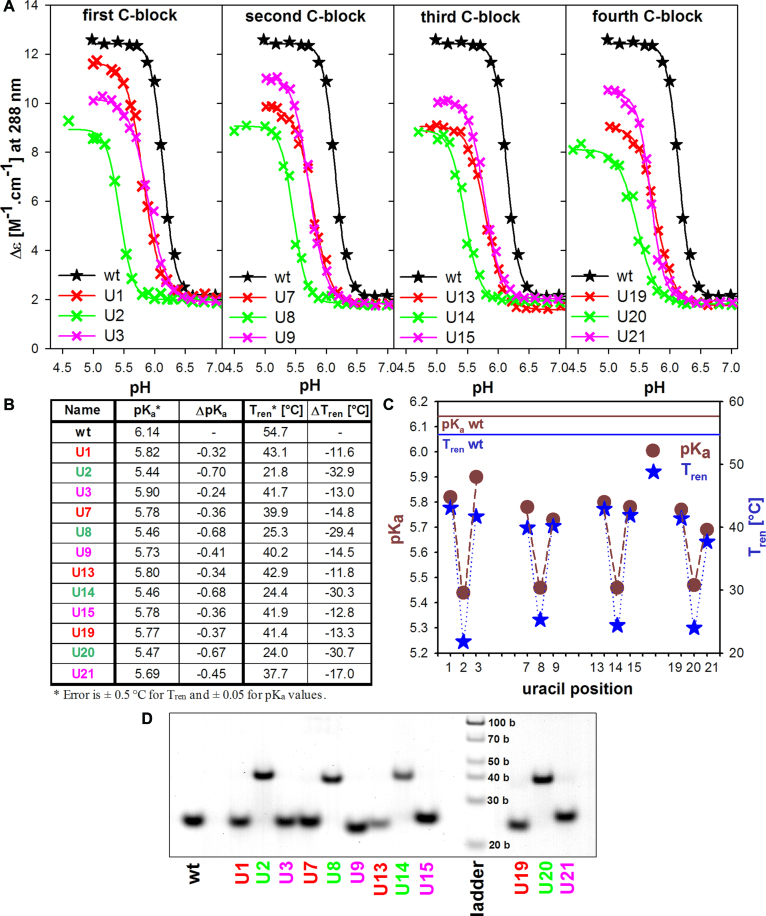
The htel sequences with particular deoxycytosines (Cs) substituted by deoxyuracil (U) are called UX in analogy with the loop-modified sequences (X corresponds to the order of the nucleobase in the (C3TAA)3C3T DNA sequence). Their primary structures are given in Supplementary Table S1. The pH-induced formation of iMs of the sequences monitored by Δϵ at 288 nm is shown in Figure 6A. The modifications have negative influence on iM formation. The transition shifts to more acidic values and simultaneously samples are thermally destabilized compared to the non-modified wt sequence (Figure 6B and C).
Figure 6.
(A) pH dependent iM formation of the uracil containing htel sequences monitored by Δϵ at 288 nm, (B) Table of pKa and Tren (renaturation temperature) values, (C) dependence of pKa (brown circles) and Tren (blue stars) values on uracil position, (D) the 16% native PAGE. The measurements were carried out as shown in Figure 3 but the electrophoresis ran at 4°C.
In the CCC trimer, the substitution of the external cytosines (with odd number positions) by U (U/C) maintains intramolecular folding of the iM but shifts its formation by 0.36 pH units on average to more acidic pH (Figure 6A and B). Their final iM structures reach lower values of Δϵ than the wt. U1 displays the highest Δϵ of all modified sequences, which indicates that its iM is least affected by the loss of the 5′ end C (C1) and it is (together with its counterpart U13) the most stable of the modified sequences. In contrast, U21 with a substituted 3′end C (C21), is the least stable structure (Figure 6B and C). U1 transforms into iM at a lower pKa value than the odd number U3 in the same block. This first C block is, however, an exception. In all other blocks, the sequences with higher position numbers (U9, U15 and U21) display higher Δϵ than the preceding odd number sequences (U7, U13 and U19) (Figure 6A). Destabilization of the iM by the loss of cytosine-forming pair is similar for U1 and U13 (43.1°C, 42.9°C), and for U3 and U15 (41.7°C, 41.9°C), and becomes different towards the molecule end, i.e. Tm of U7 and U19 (39.9°C and 41.4°C, respectively), and of U9 and U21 (40.2°C and 37.7°C, respectively). The kinetics of iM formation of all odd sequences is rapid and their melting and renaturation proceed along the same curve (an example is shown in Supplementary Figure S1A).
The substitution of the inner (middle) cytosines in the CCC blocks (in even positions) is accompanied by much more extensive iM destabilization than the substitution of odd positions. Mainly, the U/C substitution in the even positions hinders intramolecular folding of the sequence so that all four inner U/C sequences form bimolecular iMs (Figure 6D). The electrophoresis shown in Figure 6D ran at low temperature. On the electrophoresis running at room temperature, all the odd-number sequences remain intramolecular but the even-number sequences provide, in addition to bimolecular species, a slight band in the middle between intramolecular and bimolecular species corresponding to low content of unstructured forms (Supplementary Figure S2). The bimolecular iMs display lower Δϵ values than the odd-number sequences (Figure 6A), their formation proceeds at much lower pH values, and the kinetics of the process are slow. The slow kinetics results in a distinct hysteresis between melting and iM reformation (Supplementary Figure S1B). Moreover, the denaturation profile corresponding to intermolecular iMs could not be fitted by a single sigmoidal curve (denaturation probably consists of multiple steps) so that the temperature of the midpoint of the renaturation curves (Tren) was used for the comparison of iM thermostabilities of all the U/C substituted sequences (Tren is identical with Tm for intramolecular motifs). The ΔTren of the bimolecular iMs is about 30°C lower and pKa is shifted on average by nearly 0.7 pH units toward acid pH compared to the iM of the wt.
Clustered U/C substitutions
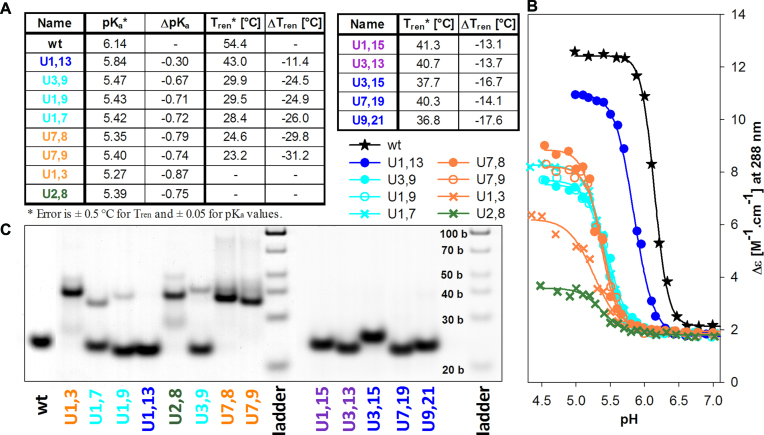
In the following study, we combined U/C substitutions in various htel sequence positions. The pKa and Tren values of the sequences are given in Figure 7A. Changes in pKa and Tren values well correlate in the whole set of U/C substituted sequences (Supplementary Figure S3). iMs were not properly formed for some U/C combinations so that Tren could not be determined.
Figure 7.
(A) Table of pKa and Tren values of the two uracils containing htel sequences (colors differentiate particular types of mutual U positions); (B) pH dependent iM formation monitored by Δϵ at 288 nm and room temperature; (C) the 16% native PAGE at pH 5 and 4°C. Conditions as stated in M+M section.
Sequences with two substituted Us for the outside (odd number) Cs localized in distinct C blocks (Figure 7, cyan, blue and violet) preferentially fold into intramolecular iMs. Those in opposite (parallel oriented) blocks form intramolecular iMs exclusively (Figure 7C, blue and violet). iM of U1,13 with a substituted first C·C+ pair from the 5′end, is the least destabilized of all double U/C combinations - by 11.4°C (Figure 7A), which corresponds well with the destabilization of single substituted U1 or U13 (11.6 and 11.8°C). iMs of further sequences with two outside U/C substitutions replacing whole C·C+ pair (Figure 7, blue), U7,19, U3,15 or U9,21 are destabilized by 14.1, 16.7 and 17.6°C, respectively (Figure 7A, right) and those with U/C substitutions replacing Cs of two different pairs (Figure 7, violet), U1,15 and U3,13 are destabilized by 13.1°C and 13.7°C, respectively.
U/C substitutions in neighbouring (antiparallel-oriented) C blocks (Figure 7, cyan), U1,7, U1,9 and U3,9, are destabilized by 26.0, 24.9 and 24.5°C compared to wt, i.e. significantly more than the previous sequences and more than the two respective single mutants. They predominantly fold intramolecularly but, in addition, they provide a band on the electrophoresis (Figure 7C) corresponding to a slight population of bimolecular iMs. The sequences are unstructured at room temperature as follows from the electrophoresis shown in Supplementary Figure S2.
Sequences containing both substitutions in the same block (U1,3, U7,8 and U7,9) (Figure 7, orange) are strongly destabilized: They form only intermolecular iMs and only partly at room temperature and at still lower pH values (Figure 7C and Supplementary Figure S2). U7,8 and U7,9 provide, in addition to bimolecular, very little populated four molecular structures at low temperature (Figure 7C).
iMs of sequences with substituted two inner (even number) cytosines by U are destabilized heavily so that Tren of their formation cannot be determined. U2,8 (Figure 7, green) forms iM only partly at low temperatures and/or at very acidic pH around 4.5. A band corresponding to the presence of unstructured form is visible on the electrophoresis even at low temperature (Figure 7C). Similarly, U2,20 and U8,14 form iM only intermolecularly at low temperature (Supplementary Figure S4, green). U8,14 assembles into four-molecular iM in addition to bimolecular one. Interestingly, the substitution of two inner cytosines forming C·C+ pair, as it is with U2,14 and U8,20, preserves, in addition to bimolecular, an intramolecular iM folding (Supplementary Figure S4, dark green). The intramolecular folding even prevails for U2,14.
Other sequences with still more U/C substitutions are listed in Supplementary Tables S1 and S2 and their electrophoresis at low temperature is shown in Supplementary Figure S4. At room temperature, these sequences provide one clear electrophoretic band corresponding to unstructured form (not shown). Substitutions of all three Cs in the blocks for U have a similar effect to two U/C substitutions in the same C-block. Bimolecular iM is observed for U1,2,3 and bimolecular and a smear of higher order forms for U7,8,9 at low temperature running electrophoresis (Supplementary Figure S4). U1,2,3,19,20,21 with two fully substituted C blocks, forms bimolecular iM, in addition to unstructured forms, and U7,8,9,13,14,15 forms a slightly faster running bimolecular iM (Supplementary Figure S4).
DISCUSSION
Numerous studies have been performed to uncover the effect of natural lesions of nucleotides on the structure and stability of G-quadruplexes (reviewed in 43). It has been demonstrated with G-quadruplexes of the G-rich htel DNA sequence that the impact of lesions depends on the type and location of the damage in the sequence: Lesions formed in the TTA loops either destabilize, do not affect or in some cases even stabilize the G-quadruplex (3,44–46), and can change the folding topology. For instance, abasic (AP) sites substituting for adenine nucleotides stabilize double-chain-reversal folding of the loops and induce the formation of hybrid quadruplexes (3,45). The presence of AP sites in all three loops transforms the quadruplex into parallel form (47). On the other hand, 8-oxoadenine slightly stabilizes the htel G-quadruplex without affecting its folding type (3,44). Depending on the folding topology, the majority of lesions formed in the core guanines destabilize the G-quadruplex (35,36,48) and, chiefly, restrict its conformational variability (47). Nevertheless, none of the single natural modifications present in G-rich sequences studied so far can hinder their folding into intramolecular G-quadruplexes. Only the clustering lesions prove to be deleterious for the folding or, depending on their position in the molecule, can ruin a G-quadruplex structure. No analogous studies with the same type of natural lesions as those found in htel G-quadruplex models have been conducted with iM structures forming from the C-rich htel sequences. Results presented in this paper are the first such evaluations. Similarly to the results for G-quadruplexes, the impact of lesions in iMs has been found here to be different in the TAA loops and in the core cytosines (Supplementary Table S1).
Loop damage
In contrast to G-quadruplexes, iMs cannot change their topology so that lesions in TAA loops of the C-rich htel sequence cause mostly minor changes in the formation and stability of its iM (overall data in Supplementary Table S1). This holds for all three modifications studied here, namely adenine abasic sites (apA), 8-oxoadenine (8oxoA) substituting adenines and 5-hydroxymethyluracil (5hmU) substituting thymines. None of these loop modifications, as in the case of htel G-quadruplexes, change the molecularity of the htel iMs.
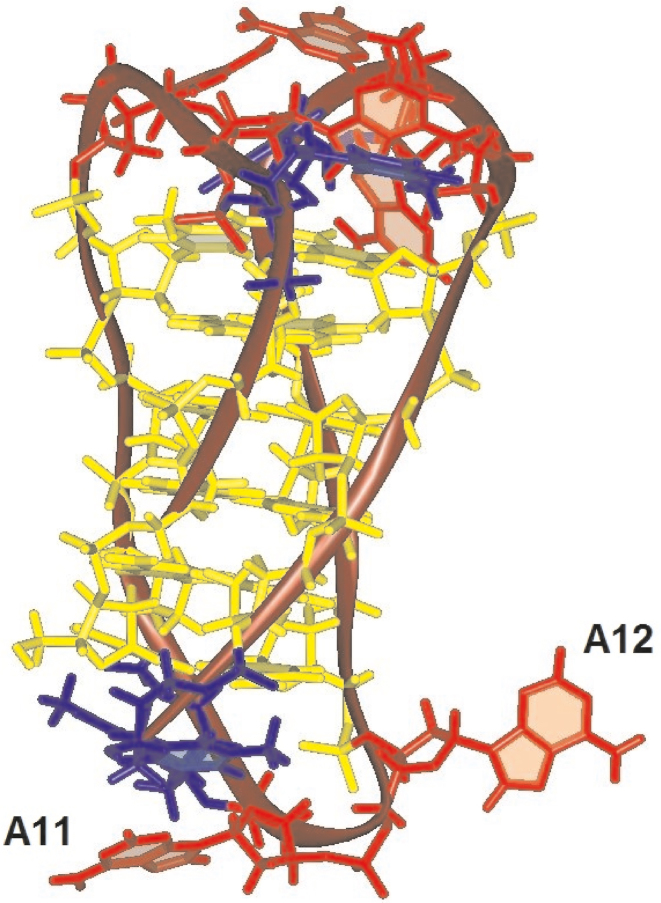
Adenine is a large base, and the two purines probably cannot squeeze into the narrow space so that they protrude out of the iM core as shown in Figure 8. It can be expected that removing this flanking base will contribute to the stability of the iM structure as well as to its compactness. Missing A11 results in only slight iM stabilization and a shift of its formation toward less acidic pH. These changes are more extensive with A12. The released space upon the loss of one adenine may be filled by the other one so that the iM becomes more compact, which is observed on the PAG electrophoresis (Figure 3D). No important base interactions take place with adenines in the major grooves so that their loss does not significantly affect iM formation or stability.
Figure 8.
The 3D structure of human telomere sequence (PDB structure 1ELN). Cytosines are marked with yellow, adenines with red and thymines with blue color.
In the case of the replacement of adenines by 8-oxoadenine, the iM formation is slightly stabilized and its formation is shifted to less acidic pH values. This is obvious with sequences substituted in the third loop, thus at the 3′ end. Similarly to the iMs, the stabilizing effect of the 8oxo group in loop adenines of htel G-quadruplexes was also observed (3,44) and it was argued (44) that it was the first example among the natural DNA lesions that stabilized the htel G-quadruplex DNA. Interestingly, the 8oxoA substitution has a different effect in the case of 8oxoA11, which forms the least stable iM of all modified samples, and for 8oxoA12, the iM of which is the most stable. Oxygen on the carbon C8 of the adenine A11 may cause a steric problem which may be responsible for the observed slight iM destabilization. In contrast, the oxygen on A12 may drag the base closer to the iM core, likely to T22, and prevent its fluctuation. With increasing stability of the iM, the structure becomes more compact and this can be traced on the PAGE electrophoresis of 8oxoA12 (Figure 4D).
Substitution of thymines by 5hmU does not, within the accuracy of the measurement, influence iM stability. The explanation may follow from the PDB structures of iM (1ELN and 1EL2) (49) according to which it seems that thymines do not participate in any stacking or binding interactions.
Substitution of core cytosines by uracil
A substitution of core cytosines by uracil devastates iM structure much more than the damage of loops (Supplementary Table S1), as a single U/C substitution causes the loss of one C·C+ pair and thus an extensive destabilization of the structure. A loss of outer (odd number) cytosines does not hinder intramolecular iM folding but the iMs are formed at lower pH values (ΔpKa 0.24–0.45) and are by 11.6–17.0°C less stable than iM of the wt. It follows from Figure 6C that the readiness of iM formation slightly decreases with the location of the substituted C closer to 3′end. The most stable is the iM of U1 and its pairing counterpart U13, followed by U3 and the counterpart U15, and the least stable of the odd-number sequences is U21. U1 behaves atypically: It displays the highest Δϵ value of all modified structures so that its iM is least deformed by the substitution. Despite its least destabilization, it forms iM at slightly lower pH than U3. The opposite is true for all other odd number U/C substitutions: U7, U13 and U19 display lower Δϵ values than respective U9, U15, and U21. Their structures are thus more affected by the sequence modification, but their iMs are less destabilized. The order of stability of the odd-number substituted sequences is: U1>U3, U7∼U9, U13>U15 and U19>U21.
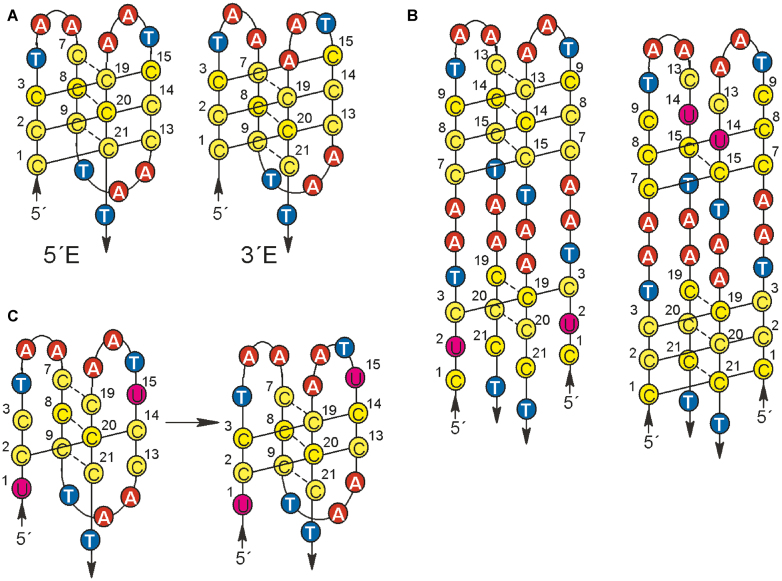
Intramolecular iMs can adopt two distinct intercalation topologies differing in the position of the intercalated pairs (50,51). The structure with the outermost C·C+ pair containing 5′C (C1) (Figure 9A, left) is called 5′E, and the structure, in which the outermost pair contains 3′end C (C21) is called 3′E structure (Figure 9A, right). The above mentioned order in iM destabilization by U/C substitution indicates that C3, C15 and C21 may be situated inside the iM. The substitution of inner pairs is more deleterious for iM stability as it results in the loss of one pair intercalated between two inversely oriented pairs, thus in the loss of the regular alternation of parallel and antiparallel C·C+ pairs. This situation would come up if the iM adopted 5′E arrangement, which is drawn in the schematic presentation in Figure 2.
Figure 9.
(A) Possible 5′E and 3′E topologies of intramolecular wt iM (49,51); (B) suggested structures of bimolecular iM of U2 and U14; (C) a shift in C·C+ pairing of U1,15.
The substitution of the middle, thus even-numbered Cs by U causes much more distinct destabilization than in the case of the odd-number ones. Mainly, these sequences are no longer able to fold intramolecularly. The iM can only fold intermolecularly (Figure 6D). This is a distinction from the effect of modified guanines in the middle tetrad of htel G-quadruplexes where formation of some amount of bimolecular, sometimes also tetramolecular species have been observed (35,36,52) but the intramolecular quadruplexes always dominated. In this respect, the C-rich htel strand is more vulnerable than the G-rich strand as the damage of the middle C of the C triad within a long molecule makes the respective region incapable to form iM.
Bimolecular iMs consist of two units separated by TAA triplets (Figure 9B). The stability of the bimolecular iMs decreases in the order of U8, U14, U20, U2 (Figure 6C). Bimolecular iM is thus more stable if it contains the complete number of six C·C+ pairs at the end of the structure than on the side of the loops (Figure 9B). Thus, the part close to the loop copes with the loss of the middle pair better than the end part. Based on the analogy with the intramolecular iMs, the part with the U in the middle of the sequence would be unable to form iM. It can be thus expected that the TAA loop forms a bulge and the remaining part with the rest of not damaged C·C+ pairs can stack on the complete six pair iM.
Based on simultaneous substitutions of two Cs by Us, we can assess the following simple rules of iM folding. Depending on the mutual position of the substituted C, several variants may occur:
- Substituted outer (odd number) cytosines
- in parallel oriented chains and pairing cytosines: Substitutions of Cs involved in pairing lead to demotion of this C·C+ pair from the motif, while its intramolecular folding is preserved. U1,13 forms the most stable iM. The same thermostability and pKa of the iM formation of U1,13 (blue in Figure 7) as of the single substituted U1 and U13 indicates that with the loss of one of these pairing cytosines the whole pair is lost and, simultaneously, that C with U do not pair at acidic pH. Further examples of this type of damage are U3,15, U7,19 and U9,21 (Figure 7A, right). All lack one C·C+ pair but still form intramolecular iMs (Figure 7C). It follows from the preceding results that the iM stability is more affected by the loss of inner than outer C·C+ pairs that do not disturb alternating C·C+ pairing of parallel and antiparallel oriented chains. A comparison of the distinctly higher Tren of U1,13 (43.0°C) than U3,15 (37.7°C) and, in the same way, higher Tren of U7,19 (40.3°C) than of U9,21 (36.8°C) (Tables A in Figure 7) again supports that the htel iM adopts 5′E motif, in which the less vulnerable C1·C13+ and C7·C19+ pairs are situated outside the iM core (Figure 9A, left). Just the 5′E structure was found by NMR (51) as a major conformation in an equilibrium of htel-21 (C3TAA)3C3 iM conformers.
- in parallel oriented chains and cytosines that do not pair: (Examples are U1,15 and U3,13 (violet in Figure 7A and C) with cytosines situated cornerwise.) Though this damage concerns two pairs, their iMs are relatively little destabilized (by 13.1°C and 13.7°C) and the structure remains predominantly intramolecular (Figure 7C). This indicates that a shift in C pairing takes place so that only one C·C+ pair is lost (Figure 9C).
- in antiparallel oriented chains: (Examples are U1,7, U1,9 or U3,9 (cyan in Figure 7). Destabilization of their iMs is more distinct than in the case of previous sequences, namely 26.0°C, 24.9°C and 24.5°C, respectively. Similar shift in C·C+ pairing may occur in analogous cases (Supplementary Figure S5). Four C pairs remain preserved, which still allows the formation of an intramolecular structure, though a minority presence of bimolecular fraction can be traced on the electrophoresis (Figure 7C).
Substituted inner (even number) cytosines: These substitutions (Figure 7 and Supplementary Figure S4, green) destabilize iM distinctly, so that the sequences form iMs only partly at room temperature and Tren values cannot be determined (Supplementary Table S2). They all form bimolecular iMs at low temperatures but distinctly populated depending on mutual positions of the inner U/C substitutions. Those in the first and third or second and fourth C blocks (light green in Supplementary Figure S4), parallel-oriented in the intramolecular arrangement, occur in the same parts of the bimolecular iMs: In the first case, they are in the part at the molecule ends, in the second case, in the part close to the loop. The latter sequence forms, in addition, tetramolecular iMs. U/C substitutions in neighbouring C blocks (first and second or third and fourth C blocks) occur in both parts of the bimolecular iM so that both its regions are affected. Population of unstructured forms can be found on their low temperature electrophoresis pattern (Figure 7C and Supplementary Figure S4). On the other hand, the substitutions of Cs, which form pairs in intramolecular iM, remain to a large extent intramolecular (Supplementary Table S2 and Supplementary Figure S4, dark green).
Substituted cytosines in the same block (orange in Figure 7 and Supplementary Figure S4): These substitutions also have a large devastating effect on iM formation. None of these sequences form intramolecular iMs. Bimolecular structures are formed at low temperature and a part of molecules remains still unstructured or four-molecular iMs are formed with some sequences (Figure 7C). The structures melt around room temperature. In all these sequences, one part of the bimolecular iMs contains only one and three C·C+ pairs, probably stacked on the second intact iM part. The part with possible one and three C·C+ pairs contributes only marginally to the bimolecular iM stability as the sequences with the whole U/C substituted block (U1,2,3 or U7,8,9) behave similarly on the electrophoresis (Figure 7C and Supplementary Figure S4). Also in this case, the two or three substitutions in the region close to the loop are better tolerated than those situated at the molecule ends. The sequences with U/C in the regions close to the loops are more susceptible to form four-molecular iMs (Supplementary Figure S4).
It can be concluded that for the formation of an intramolecular iM, the presence of two and three or at least two and two regularly alternating parallel and antiparallel C·C+ pairs are required. On violation of the regular alternation of parallel and antiparallel oriented intercalated pairs (by a loss of the middle C), exclusively bimolecular iMs are formed. In the case of a bimolecular iM, two and one alternating parallel antiparallel pairs are tolerated as well, which then probably stack on the intact six C·C+ pairs containing iM of the second part of the molecule. The damage in the region close to the iM loop is then better tolerated than at the end of the molecule.
The presented results show that the effect of naturally forming lesions on htel DNA differs depending on the position of the damage. The three different lesions studied occurring in loops did not substantially change intramolecular structure of htel iM, while the C-to-U lesion targeting iM core and especially their clusters, which may be formed by e.g. ionizing irradiation, are more deleterious for the iM structure and stability. Even a single C damage within a three C block is more dangerous than the analogous damage in the complementary G-rich htel sequence as it hinders intramolecular iM folding and thus makes iM formation impossible in the appropriate region of the C-rich DNA molecule. Recently, a considerable amount of experimental evidence has accumulated proving that not only G-quadruplexes but also iMs are important biological elements controlling DNA function, namely gene expression (53–57). In this respect, it is essential to note the magnitude of damage a single or multiple base lesions can cause in the iM structure. The present study can also help to search for potential iM forming regions in genomes, which were reported (56,58) to differ from the potentially G-quadruplex forming complementary regions. The conclusions following from the results may be also useful for designing new sensors and nano-devices.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Czech Science Foundation [15-06785S, 17-12075S, 17-19170Y]; SYMBIT [CZ.02.1.01/0.0/0.0/15 003/0000477] financed by the ERDF. Funding for open access charge: SYMBIT [CZ.02.1.01/0.0/0.0/15 003/0000477] financed by the ERDF.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993; 362:709–715. [DOI] [PubMed] [Google Scholar]
- 2. von Sonntag C. New aspects in the free-radical chemistry of pyrimidine nucleobases. Free Radic. Res. Commun. 1987; 2:217–224. [DOI] [PubMed] [Google Scholar]
- 3. Konvalinova H., Dvorakova Z., Renciuk D., Bednarova K., Kejnovska I., Trantirek L., Vorlickova M., Sagi J.. Diverse effects of naturally occurring base lesions on the structure and stability of the human telomere DNA quadruplex. Biochimie. 2015; 118:15–25. [DOI] [PubMed] [Google Scholar]
- 4. Sagi J. G-quadruplexes incorporating modified constituents: a review. J. Biomol. Struct. Dyn. 2014; 32:477–511. [DOI] [PubMed] [Google Scholar]
- 5. Sagi J. Designing G-quadruplex topologies by focusing on sequence modifications. G-Quadruplex Structures, Formation and Role in Biology. 2016; NY: Nova Science Publishers, Inc. [Google Scholar]
- 6. Gehring K., Leroy J.L., Gueron M.. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature. 1993; 363:561–565. [DOI] [PubMed] [Google Scholar]
- 7. Gueron M., Leroy J.L.. The i-motif in nucleic acids. Curr. Opin. Struct. Biol. 2000; 10:326–331. [DOI] [PubMed] [Google Scholar]
- 8. Day H.A., Pavlou P., Waller Z.A.. i-Motif DNA: structure, stability and targeting with ligands. Bioorg. Med. Chem. 2014; 22:4407–4418. [DOI] [PubMed] [Google Scholar]
- 9. Brazier J.A., Shah A., Brown G.D.. i-Motif formation in gene promoters: unusually stable formation in sequences complementary to known G-quadruplexes. Chem. Commun. (Camb.). 2012; 48:10739–10741. [DOI] [PubMed] [Google Scholar]
- 10. Sun D., Hurley L.H.. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: Implications for drug targeting and control of gene expression. J. Med. Chem. 2009; 52:2863–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simonsson T., Pribylova M., Vorlickova M.. A nuclease hypersensitive element in the human c-myc promoter adopts several distinct i-tetraplex structures. Biochem. Biophys. Res. Commun. 2000; 278:158–166. [DOI] [PubMed] [Google Scholar]
- 12. Skolakova P., Foldynova-Trantirkova S., Bednarova K., Fiala R., Vorlickova M., Trantirek L.. Unique C. elegans telomeric overhang structures reveal the evolutionarily conserved properties of telomeric DNA. Nucleic. Acids. Res. 2015; 43:4733–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bhavsar-Jog Y.P., Van Dornshuld E., Brooks T.A., Tschumper G.S., Wadkins R.M.. Epigenetic modification, dehydration, and molecular crowding effects on the thermodynamics of i-motif structure formation from C-rich DNA. Biochemistry. 2014; 53:1586–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Selvam S., Mandal S., Mao H.. Quantification of chemical and mechanical effects on the formation of the G-quadruplex and i-motif in duplex DNA. Biochemistry. 2017; 56:4616–4625. [DOI] [PubMed] [Google Scholar]
- 15. Cui J., Waltman P., Le V.H., Lewis E.A.. The effect of molecular crowding on the stability of human c-MYC promoter sequence i-motif at neutral pH. Molecules. 2013; 18:12751–12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dong Y.C., Yang Z.Q., Liu D.S.. DNA nanotechnology based on i-motif structures. Acc. Chem. Res. 2014; 47:1853–1860. [DOI] [PubMed] [Google Scholar]
- 17. Nesterova I.V., Nesterov E.E.. Rational design of highly responsive pH sensors based on DNA i-motif. J. Am. Chem. Soc. 2014; 136:8843–8846. [DOI] [PubMed] [Google Scholar]
- 18. Benabou S., Avino A., Eritja R., Gonzalez C., Gargallo R.. Fundamental aspects of the nucleic acid i-motif structures. RSC Adv. 2014; 4:26956–26980. [Google Scholar]
- 19. Yang B., Rodgers M.T.. Base-pairing energies of protonated nucleoside base pairs of dCyd and m(5)dCyd: implications for the stability of DNA i-motif conformations. J. Am. Soc. Mass Spectrom. 2015; 26:1394–1403. [DOI] [PubMed] [Google Scholar]
- 20. Xu B., Devi G., Shao F.. Regulation of telomeric i-motif stability by 5-methylcytosine and 5-hydroxymethylcytosine modification. Org. Biomol. Chem. 2015; 13:5646–5651. [DOI] [PubMed] [Google Scholar]
- 21. Canalia M., Leroy J.L.. Structure, internal motions and association-dissociation kinetics of the i-motif dimer of d(5mCCTCACTCC). Nucleic Acids Res. 2005; 33:5471–5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Han X., Leroy J.L., Gueron M.. An intramolecular i-motif: the solution structure and base-pair opening kinetics of d(5mCCT3CCT3ACCT3CC). J. Mol. Biol. 1998; 278:949–965. [DOI] [PubMed] [Google Scholar]
- 23. Nonin S., Phan A.T., Leroy J.L.. Solution structure and base pair opening kinetics of the i-motif dimer of d(5mCCTTTACC): a noncanonical structure with possible roles in chromosome stability. Structure. 1997; 5:1231–1246. [DOI] [PubMed] [Google Scholar]
- 24. Phan A.T., Leroy J.L.. Intramolecular i-motif structures of telomeric DNA. J. Biomol. Struct. Dyn. 2000; 17(Suppl. 1):245–251. [DOI] [PubMed] [Google Scholar]
- 25. Lannes L., Halder S., Krishnan Y., Schwalbe H.. Tuning the pH response of i-motif DNA oligonucleotides. ChemBioChem. 2015; 16:1647–1656. [DOI] [PubMed] [Google Scholar]
- 26. Reilly S.M., Morgan R.K., Brooks T.A., Wadkins R.M.. Effect of interior loop length on the thermal stability and pK(a) of i-motif DNA. Biochemistry. 2015; 54:1364–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fujii T., Sugimoto N.. Loop nucleotides impact the stability of intrastrand i-motif structures at neutral pH. Phys. Chem. Chem. Phys. 2015; 17:16719–16722. [DOI] [PubMed] [Google Scholar]
- 28. Gurung S.P., Schwarz C., Hall J.P., Cardin C.J., Brazier J.A.. The importance of loop length on the stability of i-motif structures. Chem. Commun. (Camb.). 2015; 51:5630–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lieblein A.L., Furtig B., Schwalbe H.. Optimizing the kinetics and thermodynamics of DNA i-motif folding. ChemBioChem. 2013; 14:1226–1230. [DOI] [PubMed] [Google Scholar]
- 30. McKim M., Buxton A., Johnson C., Metz A., Sheardy R.D.. Loop sequence context influences the formation and stability of the i-motif for DNA oligomers of sequence (CCCXXX)4, where X = A and/or T, under slightly acidic conditions. J. Phys. Chem. B. 2016; 120:7652–7661. [DOI] [PubMed] [Google Scholar]
- 31. Benabou S., Garavis M., Lyonnais S., Eritja R., Gonzalez C., Gargallo R.. Understanding the effect of the nature of the nucleobase in the loops on the stability of the i-motif structure. Phys. Chem. Chem. Phys. 2016; 18:7997–8004. [DOI] [PubMed] [Google Scholar]
- 32. Lee I.J., Kim B.H.. Monitoring i-motif transitions through the exciplex emission of a fluorescent probe incorporating two (Py)A units. Chem. Commun. (Camb.). 2012; 48:2074–2076. [DOI] [PubMed] [Google Scholar]
- 33. Pasternak A., Wengel J.. Modulation of i-motif thermodynamic stability by the introduction of UNA (unlocked nucleic acid) monomers. Bioorg. Med. Chem. Lett. 2011; 21:752–755. [DOI] [PubMed] [Google Scholar]
- 34. Perlikova P., Karlsen K.K., Pedersen E.B., Wengel J.. Unlocked nucleic acids with a pyrene-modified uracil: synthesis, hybridization studies, fluorescent properties and i-motif stability. ChemBioChem. 2014; 15:146–156. [DOI] [PubMed] [Google Scholar]
- 35. Vorlickova M., Tomasko M., Sagi A.J., Bednarova K., Sagi J.. 8-Oxoguanine in a quadruplex of the human telomere DNA sequence. FEBS J. 2012; 279:29–39. [DOI] [PubMed] [Google Scholar]
- 36. Skolakova P., Bednarova K., Vorlickova M., Sagi J.. Quadruplexes of human telomere dG(3)(TTAG(3))(3) sequences containing guanine abasic sites. Biochem. Biophys. Res. Commun. 2010; 399:203–208. [DOI] [PubMed] [Google Scholar]
- 37. Pfaffeneder T., Spada F., Wagner M., Brandmayr C., Laube S.K., Eisen D., Truss M., Steinbacher J., Hackner B., Kotljarova O. et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 2014; 10:574–581. [DOI] [PubMed] [Google Scholar]
- 38. Bransteitter R., Pham P., Scharff M.D., Goodman M.F.. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schutsky E.K., Nabel C.S., Davis A.K.F., DeNizio J.E., Kohli R.M.. APOBEC3A efficiently deaminates methylated, but not TET-oxidized, cytosine bases in DNA. Nucleic. Acids. Res. 2017; 45:7655–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gray D.M., Hung S.H., Johnson K.H.. Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol. 1995; 246:19–34. [DOI] [PubMed] [Google Scholar]
- 41. Mergny J.L., Lacroix L.. Analysis of thermal melting curves. Oligonucleotides. 2003; 13:515–537. [DOI] [PubMed] [Google Scholar]
- 42. Manzini G., Yathindra N., Xodo L.E.. Evidence for intramolecularly folded i-DNA structures in biologically relevant CCC-repeat sequences. Nucleic. Acids. Res. 1994; 22:4634–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sagi J. In what ways do synthetic nucleotides and natural base lesions alter the structural stability of G-quadruplex nucleic acids. J. Nucleic. Acids. 2017; 2017:1641845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aggrawal M., Joo H., Liu W.B., Tsai J., Xue L.. 8-Oxo-7,8-dihydrodeoxyadenosine: The first example of a native DNA lesion that stabilizes human telomeric G-quadruplex DNA. Biochem. Biophys. Res. Commun. 2012; 421:671–677. [DOI] [PubMed] [Google Scholar]
- 45. Babinsky M., Fiala R., Kejnovska I., Bednarova K., Marek R., Sagi J., Sklenar V., Vorlickova M.. Loss of loop adenines alters human telomere d AG(3)(TTAG(3))(3) quadruplex folding. Nucleic. Acids. Res. 2014; 42:14031–14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Virgilio A., Esposito V., Mayol L., Giancola C., Petraccone L., Galeone A.. The oxidative damage to the human telomere: effects of 5-hydroxymethyl-2′-deoxyuridine on telomeric G-quadruplex structures. Org. Biomol. Chem. 2015; 13:7421–7429. [DOI] [PubMed] [Google Scholar]
- 47. Kejnovska I., Bednarova K., Renciuk D., Dvorakova Z., Skolakova P., Trantirek L., Fiala R., Vorlickova M., Sagi J.. Clustered abasic lesions profoundly change the structure and stability of human telomeric G-quadruplexes. Nucleic Acids Res. 2017; 45:4294–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Virgilio A., Petraccone L., Esposito V., Citarella G., Giancola C., Galeone A.. The abasic site lesions in the human telomeric sequence d[TA(G(3)T(2)A)(3)G(3)]: a thermodynamic point of view. Biochim. Biophys. Acta. 2012; 1820:2037–2043. [DOI] [PubMed] [Google Scholar]
- 49. Phan A.T., Gueron M., Leroy J.L.. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 2000; 299:123–144. [DOI] [PubMed] [Google Scholar]
- 50. Esmaili N., Leroy J.L.. i-Motif solution structure and dynamics of the d(AACCCC) and d(CCCCAA) tetrahymena telomeric repeats. Nucleic. Acids. Res. 2005; 33:213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lieblein A.L., Buck J., Schlepckow K., Fürtig B., Schwalbe H.. Time‐resolved NMR spectroscopic studies of DNA i‐motif folding reveal kinetic partitioning. Angew. Chem. Int. Ed. Engl. 2012; 51:250–253. [DOI] [PubMed] [Google Scholar]
- 52. Tomasko M., Vorlickova M., Sagi J.. Substitution of adenine for guanine in the quadruplex-forming human telomere DNA sequence G(3)(T(2)AG(3))(3). Biochimie. 2009; 91:171–179. [DOI] [PubMed] [Google Scholar]
- 53. Kang H.J., Kendrick S., Hecht S.M., Hurley L.H.. The transcriptional complex between the BCL2 i-motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J. Am. Chem. Soc. 2014; 136:4172–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brown R.V., Wang T., Chappeta V.R., Wu G., Onel B., Chawla R., Quijada H., Camp S.M., Chiang E.T., Lassiter Q.R. et al. The consequences of overlapping G-quadruplexes and i-motifs in the platelet-derived growth factor receptor beta core promoter nuclease hypersensitive element can explain the unexpected effects of mutations and provide opportunities for selective targeting of both structures by small molecules to downregulate gene expression. J. Am. Chem. Soc. 2017; 139:7456–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Renciuk D., Rynes J., Kejnovska I., Foldynova-Trantirkova S., Andang M., Trantirek L., Vorlickova M.. G-quadruplex formation in the Oct4 promoter positively regulates Oct4 expression. Biochim. Biophys. Acta. 2017; 1860:175–183. [DOI] [PubMed] [Google Scholar]
- 56. Wright E.P., Huppert J.L., Waller Z.A.E.. Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH. Nucleic Acids Res. 2017; 45:2951–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kaiser C.E., Van Ert N.A., Agrawal P., Chawla R., Yang D., Hurley L.H.. Insight into the complexity of the i-motif and G-quadruplex DNA structures formed in the KRAS promoter and subsequent drug-induced gene repression. J. Am. Chem. Soc. 2017; 139:8522–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dhakal S., Yu Z.B., Konik R., Cui Y.X., Koirala D., Mao H.B.. G-quadruplex and i-motif are mutually exclusive in ILPR double-stranded DNA. Biophys. J. 2012; 102:2575–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.