Abstract
We report, herein, a new class of RNAi trigger molecules based on the unconventional parallel hybridization of two oligonucleotide chains. We have prepared and studied several parallel stranded (ps) duplexes, in which the parallel orientation is achieved through incorporation of isoguanine and isocytosine to form reverse Watson-Crick base pairs in ps-DNA:DNA, ps-DNA:RNA, ps-(DNA-2′F-ANA):RNA, and ps-DNA:2′F-RNA duplexes. The formation of these duplexes was confirmed by UV melting experiments, FRET and CD studies. In addition, NMR structural studies were conducted on a ps-DNA:RNA hybrid for the first time. Finally, we provide evidence for the unprecedented finding that ps-DNA:RNA and ps-DNA:2′F-RNA hybrids can engage the RNAi pathway to silence gene expression in vitro.
INTRODUCTION
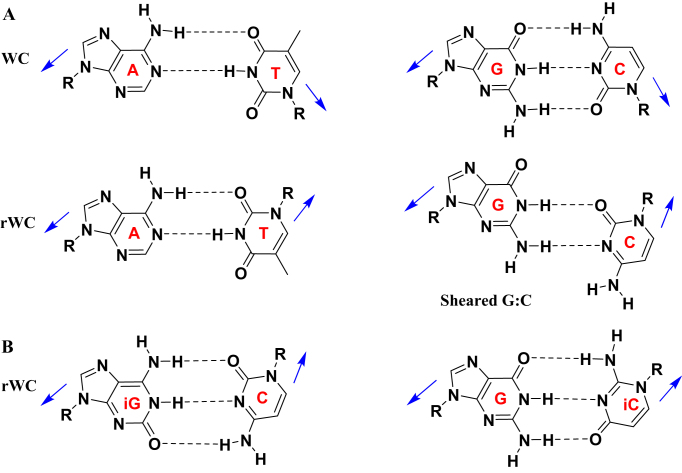
Nucleic acid double-helices typically adopt an antiparallel-stranded (aps) conformation with one strand in 5′-3′ and the other in 3′-5′ orientation, paired via canonical G:C and A:T (U) Watson–Crick (WC) hydrogen bonds (1). However, nucleic acids can also adopt the less common parallel-stranded (ps) arrangement in duplexes (2–7) and higher order structures, such as in triplexes, (8–12) G-quadruplexes (13–17) and i-motifs (18,19). Unlike the antiparallel arrangement, at neutral pH the nucleotides in parallel systems can have their glycosidic bonds oriented in trans, relative to the axis of interaction (so-called reverse WC configuration (rWC), Figure 1A) (8). Interest in parallel stranded duplexes has grown much in recent years, mainly because of their possible roles in biological function (20,21), as an entirely new base pairing system in synthetic biology (22), and in the design of both nucleic acid nanostructures (23) and hybridization probes. However, research has been mainly restricted to ps-DNA:DNA, and our understanding of the formation of other types of parallel duplexes, such as ps-RNA:RNA or ps-DNA:RNA remains quite limited and mostly at the level of thermal stability investigations (24–28). Furthermore, whether these structures have sequence-dependent conformations, or how they interact with small ligands or proteins are areas that are largely unexplored.
Figure 1.
(A) Schematic diagrams of canonical WC and rWC A:T and C:G with cis (top) and trans (bottom) glycosidic bond orientations (shown with arrow). (B) rWC hydrogen bonding in iG:C and G:iC base pairs.
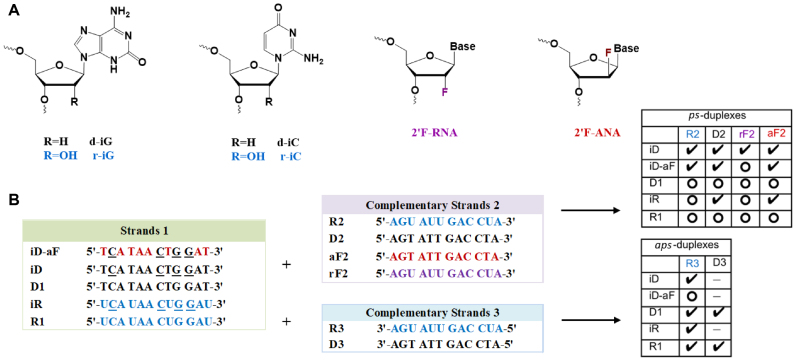
In general, the construction of parallel double-helices of mixed base composition is quite difficult as it must outcompete the formation of the more stable antiparallel helices, mostly requiring the modification of the nucleobases. Isoguanine (iG) and isocytosine (iC) nucleobases have been shown to facilitate the hybridization of parallel DNA:DNA duplexes, resulting in iG:C and G:iC base pairs with restored reverse WC (rWC) hydrogen bonding (Figure 1B) (26,27,29,30). To expand upon the current knowledge in this area, our work explores how sugar and base modifications affect ps-duplex formation and stability, and further investigates whether such ps-duplexes can engage in the RNA interference (RNAi) process. With these aims in mind, we first synthesized several 12-nt oligomers incorporating iG and iC, as well as sugar modifications known to stabilize duplexes in the antiparallel orientation, namely 2′-deoxy-2′-fluoro-ribose (2′F-RNA) and 2′-deoxy-2′-fluoro-arabinose (2′F-ANA) (Figure 2A) (31). This study was followed by the preparation of a few 21-bp ps-duplexes of mixed base composition to investigate their structure and assess, in some cases, their ability to inhibit gene expression via RNAi.
Figure 2.
(A) Structures of chemically modified nucleotides incorporated in ps- and aps-duplexes. (B) Sequence of the synthesized dodecamers for parallel and antiparallel strand pairings. Underlined residues (N) contain the isonucleobase modifications. Check marks indicate duplexes with significant melting transitions; open circles refer to combinations that did not yield a melting transition (see Table 1). “-" means “not measured".
MATERIALS AND METHODS
Synthesis and purification of oligonucleotides
All fully DNA, RNA, 2′-FRNA or 2′-FANA oligonucleotides were readily synthesized using the standard solid phase synthesis and deprotection. All phosphoramidites used were commercially available (Supplementary Figure S20) and were purchased from ChemGenes (Wilmington, MA). For oligonucleotides containing iso modifications, detritylation was performed using 3% dichloroacetic acid in toluene. Phosphoramidites coupling times were 600 s for iso-DNA, iso-RNA, RNA, 2′F-RNA and 2′F-ANA phosphoramidites, except for their guanosine phosphoramidites, which were allowed to couple for 900 s. The removal of CPG support, base protecting groups and the cyanoethyl protecting groups of the iso-containing oligonucleotides occurred best with ammonium hydroxide added to the CPG-bound oligo, and the tightly sealed solution was shaken at 25°C for 72 h. The duration of this deprotection step was proportional to the number of iC and iG modifications in the strand, and was determined by following the reaction with IE-HPLC. Note that minimal exposure to basic deprotection conditions is crucial to avoid iso-C degradation. Deprotection and cleavage from the solid support for those oligos that did not contain iso-nucleobases was accomplished with ammonia:ethanol (3:1, v/v) for 48 h at room temperature.
Oligonucleotides containing iso-RNA and RNA were synthesized with standard 2′-TBDMS phosphoramidites, and desilylation was achieved with neat TEA·3HF for 48 h at room temperature. Oligonucleotides were precipitated by adding 3 M sodium acetate (25 μl) followed by addition of cold butanol (1 ml). All oligonucleotides (except for the iso-modified oligos, which were PAGE purified) were purified by reverse phase HPLC on an Agilent 1200 series using a Waters semipreparative C18 column. An increasing gradient of 5% acetonitrile in 0.1 M triethylammonium acetate, pH 7.0 was used as the mobile phase to separate the oligonucleotides (solution A: 0.1 M TEAA, pH 7.0, solution B: acetonitrile). Purified oligonucleotides were then lyophilized to dryness, and were characterized by ESI-mass spectrometry in negative mode (Supporting Tables S1–S3).
Thermal denaturation experiments and circular dichroism studies
UV thermal denaturation data were obtained on a UV–VIS spectrophotometer equipped with a Peltier temperature controller. Duplex concentration, for both series (12mers and 21mers), was 2 μM (4 μM total concentration of single strands) in 140 mM KCl, 1 mM MgCl2 and 5 mM NaHPO4 (pH 7.2). The temperature was increased at a rate of 0.4°C/min from 7°C to 75°C for 12-bp duplexes and from 7°C to 85°C for 21-bp duplexes. Absorbance values were recorded each minute. Samples were kept under nitrogen flow when below 15°C. Tm values were calculated using the baseline method.
CD spectra were obtained at a duplex concentration of 2 μM (4 μM total concentration of strands) at a range of temperatures. Samples were slowly annealed (90–4°C over several hours) in the same buffer used for thermal denaturation studies prior to recording their CD spectra.
NMR experiments
Samples for NMR studies were dissolved in 500 μl of D2O or H2O/D2O 9:1 (buffer conditions 25 mM Na2HPO4, 100 mM NaCl, pH 7). NMR spectra were acquired in Bruker spectrometers operating at 600 and 800 MHz, and processed with Topspin software. NMR melting experiments were recorded from 5 to 45°C. Two dimensional TOCSY, DQF-COSY and NOESY experiments were recorded in D2O and H2O/D2O at 5 and 25°C (0.7–0.9 mM duplex concentration). The NOESY spectra were acquired at 100 and 250 ms mixing time. For 2D experiments in H2O, water suppression was achieved including WATERGATE module in the pulse prior to acquisition. The spectral analysis program Sparky was used for semiautomatic assignment of the NOESY cross-peaks and quantitative evaluation of the NOE intensities.
Characterization of duplex formation by gel electrophoresis
Samples were prepared to contain 7.5 pmol of oligonucleotides in 10 μl of PBS (Invitrogen) and mixed with 1 μl of DNA loading dye (Thermo Fisher Scientific). Samples were then loaded onto 20% (w/v) acrylamide running gel prepared in a Tris-acetate-EDTA buffer (TAE buffer: 40 mM Tris-acetate, 1 mM EDTA, pH 8.0). Gel was then immersed in TAE buffer and electrophoresed at constant voltage of 60 V for 15 min, followed by 60 min at 150 V. Oligonucleotides were revealed following manufacture's protocol for SYBR Gold nucleic acid gel stain.
Luciferase reporter plasmid cloning and transfection
PsiCHECK-2 dual-luciferase reporter plasmid (Promega) was digested with NotI and XhoI and the insert bearing three fully complementary target sites for siRenilla (Supplementary Table S9) was cloned in the 3′-untranslated region (3′- UTR) of the Renilla gene. The plasmid was confirmed by sequencing and transfected at 20 ng/well using jetPEI (Polyplus) according to the manufacturer's instructions.
Renilla luciferase assay
HEK293T cells were seeded in white 96-well plates at 10 000 viable cells/well and duplexes were transfected after ca. 12–16 h, whereas the reporter plasmid ca. 24 h post-seeding. 48 h later, supernatant was removed, and luminescence was measured on a microtiter plate reader (Mithras LB940, Berthold Technologies) using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer's instructions. Values were normalized against firefly luminescence and the corresponding mock control whose relative luciferase activity was set to 100%. The experiment was repeated in three biological replicates.
Statistical analysis
Statistical analysis was performed with GraphPad Prism. An independent Student's t-test was used to compare the statistical significance of two groups, whereas two-way ANOVA was applied to compare more than two groups.
AGO2 knockdown experiment
siAGO2 siRNA pool was transfected at 50 and 100 nM using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions in HEK293T cells. Following the siAGO2 transfection, duplexes targeting Renilla luciferase mRNA were transfected and Renilla assay was conducted according to the instructions above. The experiment was repeated in two biological replicates.
Western blot
HEK293T cells were lysed in RIPA buffer (Sigma-Aldrich). Protein concentrations were determined using the BCA assay (Thermo Fisher Scientific). 20–30 μg of total protein were mixed with Laemmli loading buffer (BioRad), heated at 95°C for 5 min and separated on SDS gel and transferred to PVDF western blotting membrane. Non-specific membrane binding was blocked for 60 min at room temperature with 5% milk in phosphate-buffered saline containing 0.05% Tween-20. Membranes were incubated overnight at 4°C with primary antibodies against AGO2 (C34C6; Cell Signaling Technology), or Gapdh (OTI2D9; OriGene). After washing, membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies for 2 h at room temperature in blocking buffer and washed again. Signals generated by the chemiluminescent substrate (GE Healthcare) were captured by a cooled CCD camera (Bio-Rad, Hercules).
RESULTS AND DISCUSSION
Synthesis and structural analysis of 12-bp parallel duplexes
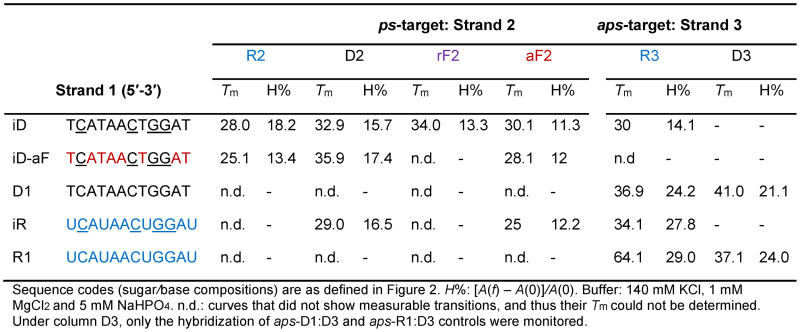
The ps-DNA:DNA dodecamer duplex previously characterized by Seela et al. (27) was selected for exploring the impact of both ribose (2′OH, 2′F) and arabinose (2′F) substitutions on ps-duplex stability (D1:D3, Figure 2B, Table 1). Control oligomers D1 and R1, containing canonical C and G bases, were prepared to assess the requirement of the iso-base modification (iso deoxy and iso ribo G/C monomers) in both DNA and RNA strands (Figure 2B). The entire series (‘Strands 1’) were hybridized to complementary oligomers (‘Strands 2’: D2, R2, aF2, rF2; and ‘Strands 3’: D3, R3) designed to form either parallel or antiparallel duplexes of the same base composition (Figure 2B). Duplex formation was monitored by UV thermal melting experiments, circular dichroism (CD) and NMR spectroscopy.
Table 1. Melting temperatures (Tm) and percent hyperchromicity (H%) for parallel and antiparallel duplexes.
|
As expected, neither a mixture of unmodified D1 + D2, or R1 + D2 exhibited any thermally induced transitions. However, the corresponding iso-modified DNA strand (iD) when combined with DNA (D2) yielded a complex with a Tm of 32.9°C. Also, pairing iD with RNA (R2) and 2′F-RNA (rF2) strands afforded complexes of similar or higher stability compared to iD:D2 (Table 1). The hybrids iD:R2 and iR:D2 shared similar Tm values (Table 1). Overall, these experiments demonstrated that iC and iG incorporations in either DNA or RNA strands are essential for parallel hybridization of these sequences (28,30). Replacing a few DNA residues with 2′F-ANA residues in the iD strand (to give iD-aF) was found to be slightly stabilizing, as assessed by comparing iD-aF:D2 with iD:D2 (ΔTm + 3°C; Table 1). Interestingly, the opposite effect was observed when iD-aF was mixed with RNA (R2); in this case, iD-aF:R2 had lower Tm values relative to iD:R2. This behavior is in contrast to what has been observed with antiparallel DNA:RNA hybrids, where DNA substitution with 2′F-ANA is generally stabilizing (32–34). All attempts to form parallel duplexes comprising RNA and/or 2′F-RNA residues were unsuccessful. This was evident by the lack of any clear UV melting transition when trying any of the following parallel combinations: i.e. duplexes iR + R2, R1 + R2 or iR + rF2. By contrast, the antiparallel control combination R1 + R3 resulted in the formation of the expected antiparallel RNA:RNA duplex (Tm = 64.1°C). Hence, we concluded that for this sequence, at least, duplex formation comprising RNA and 2′F-RNA is favored only in the antiparallel orientation. This observation is in full agreement with the results reported by Seela on a different 12-bp system (28).
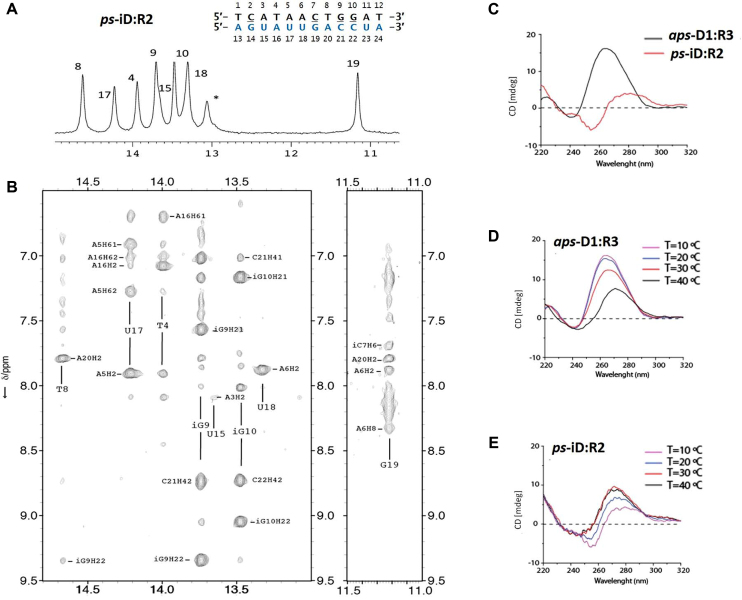
CD spectra of parallel and antiparallel duplexes are shown in Figure 3, Supplementary Figures S1 and S2. Antiparallel duplexes displayed the characteristic strong positive band centered between 260 and 280 nm, and a negative band at 240 nm. However, for the iso-modified duplexes, a negative band centered at 255 nm was observed. Furthermore, ps-duplexes exhibited a rise in the magnitude of the positive CD band upon raising the temperature, whereas an opposite (i.e. hypochromic) effect was seen for the aps-duplexes (Figure 3D, Supplementary Figures S1 and S2). These differences are not surprising given the parallel nature of the duplexes, as well as the altered spectroscopic properties of the iC/iG nucleobases (27). The 1H NMR imino peaks of the 12-bp ps and aps-duplexes at 10°C were very well resolved (Supplementary Figures S3 and S4) and were used to confirm the hybridization of the paired bases in ps- and aps-duplexes. While some imino signals of the paired bases in ps-duplexes were not observed (likely due to the enhanced fraying characteristic of terminal A:T base pairs), the observed signals confirmed the hybridization in all cases (Figure 3A, Supplementary Figures S3 and S4).
Figure 3.
1D 1H-NMR spectrum (A) and 2D-NOESY (B) of 12-bp ps-iD:R2 duplex. Star sign (*) indicates the not assigned imino resonance. CD spectra comparison of 12-bp anti parallel (D1:R3) versus parallel (iD:R2) hybrids at 10°C (C) and their CD melting profiles (D1:R3 (D) and iD:R2 (E)) at 10, 20, 30 and 40°C. Sequences of constituent strands are shown in Figure 2B.
To better understand the effect of the iso-modifications, the 12-bp ps-iD:R2 duplex was further analyzed by 2D-NMR spectroscopy (Figure 3A, B and Supplementary Figures S3, S5, S6). The intra-residual H1′-H8 NOE cross-peaks intensity indicated that all bases are in the anti conformation. The sequential H2′/H2″-H6/8 and H1′-H6/H8 cross-peaks observed were consistent with a right-handed duplex structure (Supplementary Figure S6). Multiple NOEs associated with base stacking were observed in both strands. Base pair formation was detected from A3:U5 to A11:U23, with the NOE cross-peaks at H2A-H3T/U, consistent with rWC pairing (35,36). Analysis of the exchangeable protons showed that amino protons were clearly visible in iG:C base pairs, but were not observed in G:iC base pairs. This effect has been previously described to be likely due to the different rotation rates of amino groups in natural nucleobases and their isomers, suggesting the slower rotation rates in iG and faster in iC, compared to the canonical G and C bases, respectively (37). Accordingly, the imino region of ps-iD:R2 duplex showed fewer imino signals than the number of possible base pairs in the dodecamer (Figure 3A). Also, the imino protons of the base pairs at 5′- and 3′-termini were absent in the spectra, likely due to the multiple A:T/U and iC:G pairs at these positions. Exchangeable protons of iG:C base pairs appeared at comparable chemical shifts to A:U/T base pairs (Figure 3A). On the contrary, the guanine imino protons of the iC7:G19 base pair appeared at significantly higher field (Figure 3A and B, signal at 11.2 ppm).
Temperature-dependent 1D experiments showed higher stability of some base pairs. Most imino protons disappeared at around 25°C, while imino resonances for the segment iC7 to iG10 could be observed at higher temperatures, suggesting that these central base pairs are the most stable in this duplex (Supplementary Figure S5). Overall, these data show that hybridization of iD with R2 occurs though rWC base pairing. Full NMR peak assignment, together with a complete structural determination of this parallel duplex is in progress.
While our efforts to induce the formation of ps-RNA:RNA duplexes failed, formation of stable ps-DNA:RNA (iD:R2) and ps-DNA:2′F-RNA (iD:rF2) hybrids motivated us to test these constructs in gene silencing experiments. This seemed reasonable as siRNA duplexes with systematic substitution of DNA in their passenger strand have been shown to be effective triggers of RNA interference (RNAi) with significantly reduced off-target effects (38–41).
Synthesis and structural analysis of 21-bp parallel siRNAs
Design strategy
Short interfering RNA (siRNA) duplexes are typically 21-bp in length and are used as exogenous triggers of the RNAi gene regulation pathway (42). In classical antiparallel siRNA designs, the ‘guide’ strand shares full WC complementarity with target mRNA, while the ‘passenger’ strand has the same sequence as the target region on mRNA. Consequently, three strategies seemed possible for designing parallel duplexes as RNAi triggers (Figure 4). The first strategy involved the incorporation of iG and iC nucleotides into the DNA passenger strand of a ps-DNA:RNA duplex, with its constituent RNA guide strand designed to bind to the target mRNA in normal antiparallel direction (Figure 4A). The second and third strategies (Figure 4B and C) would require placing the iso modifications in the RNA guide strand. Given that formation of ps-RNA:RNA was not feasible, and that iG:C and iC:G base pairings are destabilizing when incorporated in aps-duplexes, the first strategy was pursued (Figure 4A).
Figure 4.
Strategies to design rWC parallel si-duplexes targeting Bcl-2 incorporating iG and iC, with guide strand antiparallel (A and C) and parallel (B) to the mRNA. In all cases the si-duplex, itself, is parallel stranded. Codes: DNA residues shown in black (d-iC underlined), RNA residues shown in blue (r-iC underlined).
Guidelines to achieve stable ps-hybrids
Thermal stabilities of the various 21-nt long parallel hybrids targeting the P53, CCND1, Renilla luciferase, and Bcl-2 genes were evaluated in a buffer representative of intracellular condition (details in the supporting information). Of these, only the hybrids with Renilla and Bcl-2 sequences afforded stable ps-duplexes. Based on these results and those obtained with the 12-bp duplexes, five criteria (guidelines) emerged for obtaining stable ps-hybrids: (i) In general, sequences with high GC content (50% or more) are desirable, as they allow for a higher number of stabilizing iG:C and iC:G base pairs in the parallel orientation. (ii) Sequences with ‘GG’ repeats should be avoided as they promote formation of competing higher order structures like G-quadruplexes. (iii) Blunt-ended hybrids afford a larger number of base pairs (21-bp versus 19-bp), and hence, greater stabilization (albeit not necessarily better cellular activity; see below). (iv) Sequences with high iG content in the DNA strand are preferred over high iC content, in agreement with an earlier report pointing to the greater stabilization provided by iG:C over iC:G base pairs (27). Finally, (v) similar to what is observed with aps-duplexes, incorporation of 2′F-RNA modifications into the RNA strand provides higher duplex stabilization.
Structural analysis and validation of ps-hybrid formation
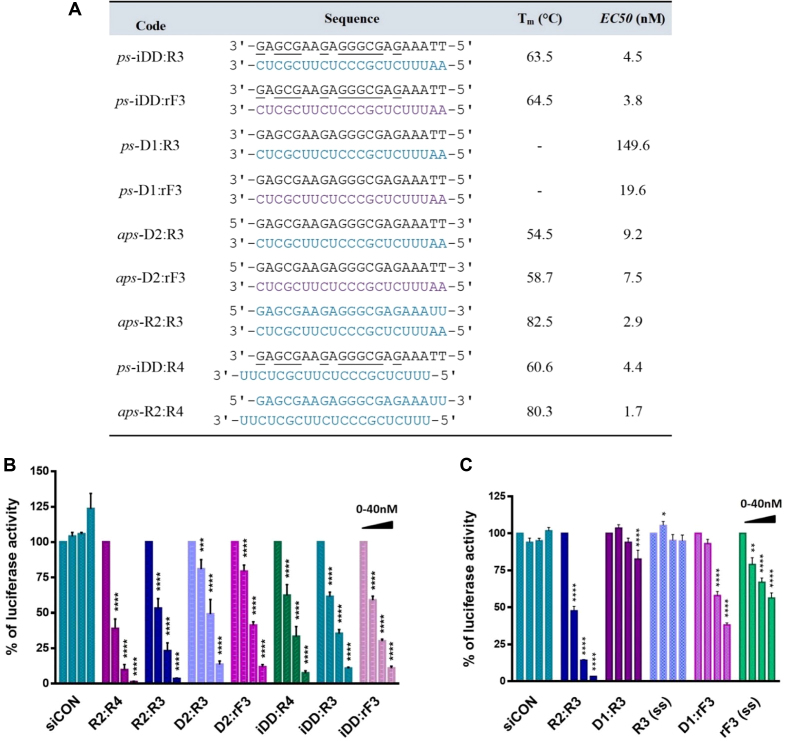
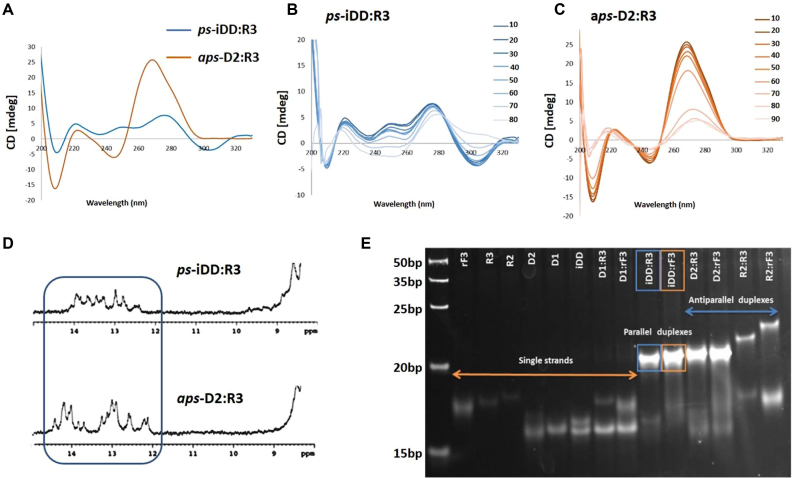
Duplexes targeting the Renilla luciferase (21-bp) and Bcl-2 (19-bp) mRNAs were synthesized (iDD:R3 and iDD:rF3; Supplementary Tables S2 and S3) and their properties were assessed by UV thermal denaturation, CD, gel electrophoresis and 1H NMR. The Tm values for iDD:R3 and iDD:rF3 parallel hybrids (targeting the Bcl-2) were exceptionally high (Supplementary Table S4, Figures S7 and S8). Notably, for the Renilla mRNA-targeting system, these parallel duplexes exhibited even higher Tm values than the corresponding antiparallel duplexes (D2:R3 and D2:rF3) (Figure 6A; Supplementary Figures S13 and S14). Gel electrophoresis under native conditions showed the presence of a single duplex species for iDD:R3 and iDD:rF3, as well as for the control (antiparallel) duplexes targeting Renilla luciferase and Bcl-2 mRNAs (Figure 5E and Supplementary Figure S9 respectively).
Figure 6.
(A) Sequences, melting temperature, and half maximal effective concentration (EC50) values of duplexes targeting Renilla luciferase mRNA. Legend: RNA (R3 and R4) and 2′F-RNA (rF3) sequences are colored blue and dark blue, respectively; DNA sequences (iDD, D1 and D2) are colored black. G and C are d-iG and d-iC, respectively. All duplexes are composed of the passenger (sense) strand on top and the guide (antisense) strand at the bottom. (B) Knockdown of Renilla luciferase in HEK293T cells by ps- and aps-siRNAs. (C) Mixtures (D1+R3) and (D1+rF3) as well as single stranded guide strands (R3 and rF3) were separately tested against Renilla luciferase mRNA with siCON as negative control. All samples were tested at 0, 2.5, 10 and 40 nM concentrations. Error bars represent standard error of the mean (SEM) of three biological replicates (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
Figure 5.
CD spectra (A–C), 1H NMR (D) and native PAGE (E) for 21-bp ps-iDD:R3 and aps-D2:R3 siRNAs targeting Renilla Luciferase mRNA. (A) Comparison of CD spectra for ps-iDD:R3 versusaps-D2:R3 duplexes at 10°C. (B, C) Temperature-dependent CD spectra, recorded at 10 degree intervals (10–90°C range) in a buffer containing 140 mM KCl, 1 mM MgCl2 and 5 mM Na2HPO4, at pH 7.2 (duplex concentration 10 μM). (D) Imino region of the 1D 1H NMR spectra of parallel iDD:R3 vs. antiparallel D2:R3 duplexes at 10°C. Buffer condition: 25 mM sodium phosphate, 100 mM NaCl at pH 7. (E) Bands corresponding to parallel duplexes on gel are specified with a rectangle.
To further confirm the parallel strand orientation of selected duplexes targeting Bcl-2 (Supplementary Table S5), a Fluorescence Resonance Energy Transfer (FRET) parallel probe was designed (Supplementary Table S5 and Figures S10 and S11) in which the iDD and R3 strands of the ps-iDD:R3 duplex were labelled with Cyanine3 (Cy3) and Cyanine5 (Cy5) dyes, respectively. Control probes were designed with aps-D2:R3 duplexes labelled with Cy3 and Cy5 at the 3′-end of D2 and 5′-end of R3 strands (positive control C1; FRET emission due to dyes in close proximity) or 5′-end of both D2 and R3 strands (negative control C2; no FRET emission due to dyes at the opposite ends). Our experiment demonstrated that under annealing conditions, both ps-iDD:R3 and aps-D2:R3 (C1) hybrids allowed for energy transfer to occur with very high efficiency, validating that the dyes are held in close proximity in both probes, and confirming the parallel strand orientation in the iDD:R3 duplex (Supplementary Table S5).
The CD spectra of iDD:R3 and iDD:rF3 parallel duplexes targeting Renilla luciferase mRNA somewhat resembled those exhibited by left-handed helices (Figure 5A–C and Supplementary Figure S16 respectively), with the negative band at 305 nm disappearing upon raising the temperature. The corresponding antiparallel duplexes, D2:R3 and D2:rF3, however, presented CD profiles that were typical of A-form helices (Supplementary Figure S16). In agreement with our 12-nt studies, 1H NMR studies on the Renilla mRNA-targeting ps-iDD:R3 versus aps-D2:R3 duplexes showed that the exchangeable imino protons of iDD:R3 (Figure 5D) appear at chemical shifts comparable to those observed for D2:R3 (12–14 ppm), with no signals detected at regions characteristic of G-quadruplexes (11–12 ppm) (43).
Gene-silencing activity of ps-siRNAs
A library of siRNAs targeting Renilla luciferase mRNA was examined (Figure 6) in order to assess their gene-silencing capabilities. The silencing properties of this series were tested in HEK239T cells using a Renilla dual reporter assay (Figure 6B and C), in which three fully complementary target sites were present in the 3′UTR of Renilla luciferase (Supplementary Table S9). For this series, two configurations were considered: a 19-bp duplex with the classical 2-nt overhang (ps-iDD:R4) and a blunt ended 21-bp duplex (ps-iDD:R3), along with their antiparallel counterparts (aps-R2:R4 and aps-R2:R3, respectively). Renilla luciferase levels following treatment with these duplexes at a range of concentrations are shown in Figure 6. Both R2:R4, and the blunt ended R2:R3 control siRNA duplexes significantly reduced Renilla mRNA levels, showing that both classical and blunt ended configurations performed well for the unmodified antiparallel siRNAs. As expected, the corresponding aps-hybrids, D2:R3 and D2:rF3, were also active, albeit to a lesser extent relative to R2:R3. Remarkably, all of the ps-hybrids tested (i.e. iDD:R3, iDD:R4 and iDD:rF3 duplexes) had superior activity relative to their antiparallel counterparts (i.e. D2:R3 and D2:rF3) (Figure 6B). However, none of them exceeded the activity of the native antiparallel siRNAs (R2:R3 and R2:R4). Neither (D1+R3), R3 or rF3 produced any significant gene knockdown. (D1 + rF3) displayed some activity, however, it was over several-fold lower compared to ps-D1:rF3 (Figure 6C).
A library of siRNAs targeting Bcl-2 mRNA was also examined. Caco-2 cells were transfected with duplexes and single strands and Bcl-2 mRNA expression levels were determined via qRT-PCR. All samples tested exhibited activity, with the native siRNA aps-R2:R3 being the most potent among this series (Supplementary Figure S12). Interestingly, in this case, R3 and rF3 single strands exhibited a dose-dependent reduction of Bcl2-mRNA. As no duplex formation was observed for (D1 + R3) and (D1 + rF3) (Supplementary Figure S9), the silencing activity observed for these mixtures likely arises from the activity of the RNA and 2′F-RNA single strands, as has been observed for ss-siRNAs (44,45).
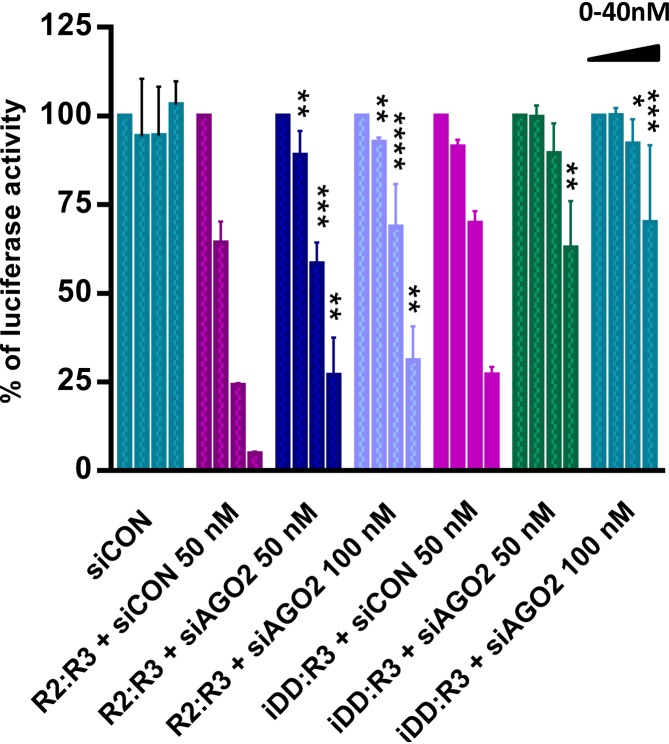
To assess whether the Renilla mRNA-targeting duplexes were operating via the RNAi pathway, we conducted an assay using siRNAs targeting AGO2. Argonaute2 protein (AGO2) is a key component of RNA-induced gene silencing complex, which is crucial for siRNA-mediated repression of target genes. Thus, the potency of the Renilla mRNA-targeting ps-iDD:R3 hybrid and control aps-R2:R3 duplex was again assessed, this time in the presence of increasing concentrations of ‘siAGO2’, a previously characterized siRNA pool targeting the 3′-UTR of AGO2 mRNA (Supplementary Figure S19) (46). The results from this experiment demonstrated that in both ps- and aps- cases, cells treated with siAGO2 led to a significant reduction in luciferase inhibition relative to the cells treated with targeting duplexes without siAGO2 (Figure 7), suggesting that the ps-duplexes are functioning through canonical RNAi pathway. The residual activity of the targeting duplexes (e.g. in the presence of 100 nM ‘siAGO2’) may be due to incomplete knockdown of AGO2, and/or the activity of other Argonaute proteins (e.g. AGO1, 3 and 4), under these conditions.
Figure 7.
Renilla luciferase dual reporter assay demonstrating the effect of AGO2 knockdown on the potency of parallel and antiparallel siRNAs iDD:R3 and R2:R3 against Renilla mRNA knockdown. siRNAs iDD:R3 and R2:R3 were tested at 0, 2.5, 10 and 40 nM concentrations. siCON represents the non-targeting negative siRNA control. Statistical tests for R2:R3 and iDD:R3 were run separately against ‘R2:R3+siCON’ and ‘iDD:R3+siCON’-treatments respectively. Error bars represent standard error of the mean (SEM) of two biological replicates (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001).
CONCLUSIONS
We demonstrate here that iC/iG-modified DNA sequences can form parallel stranded duplexes with RNA (28) and 2′F-RNA under physiological ionic strength and pH. While the general applicability of these constructs as RNAi triggers remains to be further investigated, our preliminary findings suggest that ps-DNA:RNA and DNA:2′F-RNA hybrids (e.g. iDD:R3, iDD:rF3), when carefully designed, can engage the RNAi pathway to knockdown the expression of a specific gene, with potencies similar to conventional aps-DNA:RNA and DNA:2′F-RNA duplexes. The parallel-stranded RNAi triggers studied here, like conventional siRNA duplexes, have the added advantage of using native RNA (or 2′F-RNA) as guide strands (38,39,41,47,48). Moreover, since the passenger strand of the ps-duplexes could be heavily modified (e.g. DNA, 2′F-ANA sugars) (31), this would reduce its likelihood to act as guide strand, a feature that would in principle minimize off-target, and possibly, immunostimulatory effects (38). While our data suggest that ps-duplexes can undergo gene knockdown via RNAi pathway, the underlying processes contributing to the overall activity of a parallel duplex need to be carefully investigated. In light of the different helical structure of parallel duplexes, it is reasonable to assume that the potency of these systems can be partly influenced by their different protein binding affinity, although the impacts of other parameters, for example cellular transport or other competing mechanisms cannot be yet ruled out.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Sciences and Engineering Research Council of Canada Discovery (to M.J.D.); Drug Development and Training Program (DDTP) grant from The Canadian Institutes of Health Research (to M.H. and M.Y.A.); Swiss National Science Foundation [CRSII3_141942 to J.H.]; Gebert Rüf Foundation [GRS-041/11 to J.C.L.]; MINECO [BFU2014-52864-R to C.G.]; CSIC-JAE contract (to N.M.P.). Funding for open access charge: The Natural Sciences and Engineering Research Council of Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1. Watson J.D., Crick F.H.C.. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953; 171:737–738. [DOI] [PubMed] [Google Scholar]
- 2. Rippe K., Jovin T.M.. David MJ, Lilley JED. Methods Enzymol. 1992; 211:Academic Press; 199–220. [DOI] [PubMed] [Google Scholar]
- 3. Ramsing N.B., Rippe K., Jovin T.M.. Helix-coil transition of parallel-stranded DNA. Thermodynamics of hairpin and linear duplex oligonucleotides. Biochemistry. 1989; 28:9528–9535. [DOI] [PubMed] [Google Scholar]
- 4. Rippe K., Ramsing N.B., Klement R., Jovin T.M.. A parallel stranded linear DNA duplex incorporating dG · dC base pairs. J. Biomol. Struct. Dyn. 1990; 7:1199–1209. [DOI] [PubMed] [Google Scholar]
- 5. van de Sande J., Ramsing N., Germann M., Elhorst W., Kalisch B., von Kitzing E., Pon R., Clegg R., Jovin T.. Parallel stranded DNA. Science. 1988; 241:551–557. [DOI] [PubMed] [Google Scholar]
- 6. Shchyolkina A.K., Lysov Yu P., Il’ichova I.A., Chernyi A.A., Golova Yu B., Chernov B.K., Gottikh B.P., Florentiev V.L.. Parallel stranded DNA with AT base pairing. FEBS Lett. 1989; 244:39–42. [DOI] [PubMed] [Google Scholar]
- 7. Raghunathan G., Miles H.T., Sasisekharan V.. Parallel nucleic acid helices with hoogsteen base pairing: symmetry and structure. Biopolymers. 1994; 34:1573–1581. [DOI] [PubMed] [Google Scholar]
- 8. Frank-Kamenetskii M.D., Mirkin S.M.. Triplex DNA structures. Annu. Rev. Biochem. 1995; 64:65–95. [DOI] [PubMed] [Google Scholar]
- 9. Lavelle L., Fresco J.R.. UV spectroscopic identification and thermodynamic analysis of protonated third strand deoxycytidine residues at neutrality in the triplex d(C+-T)6:[d(A-G)6d(C−-T)6]; evidence for a proton switch. Nucleic Acids Res. 1995; 23:2692–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marfurt J., Leumann C.. Evidence for C−H⋅⋅⋅O hydrogen bond assisted recognition of a pyrimidine base in the parallel DNA triple-helical motif. Angew. Chem. Int. Ed. 1998; 37:175–177. [Google Scholar]
- 11. Marfurt J., Parel S.P., Leumann C.J.. Strong, specific, monodentate G-C base pair recognition by N7-inosine derivatives in the pyrimidine•purine-pyrimidine triple-helical binding motif. Nucleic Acids Res. 1997; 25:1875–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun J.-s., Carestier T., Hélène C.. Oligonucleotide directed triple helix formation. Curr. Opin. Struct. Biol. 1996; 6:327–333. [DOI] [PubMed] [Google Scholar]
- 13. Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S.. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006; 34:5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blackburn E.H., Szostak J.W.. The molecular structure of centromeres and telomeres. Annu. Rev. Biochem. 1984; 53:163–194. [DOI] [PubMed] [Google Scholar]
- 15. Hurley L.H. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002; 2:188–200. [DOI] [PubMed] [Google Scholar]
- 16. Sundquist W.I., Klug A.. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989; 342:825–829. [DOI] [PubMed] [Google Scholar]
- 17. Sen D., Gilbert W.. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364–366. [DOI] [PubMed] [Google Scholar]
- 18. Guéron M., Leroy J.-L.. The i-motif in nucleic acids. Curr. Opin. Struct. Biol. 2000; 10:326–331. [DOI] [PubMed] [Google Scholar]
- 19. Völker J., Klump H.H., Breslauer K.J.. The energetics of i-DNA tetraplex structures formed intermolecularly by d(TC5) and intramolecularly by d[(C5T3)3C5]. Biopolymers. 2007; 86:136–147. [DOI] [PubMed] [Google Scholar]
- 20. Tchurikov N.A., Chistyakova L.G., Zavilgelsky G.B., Manukhov I.V., Chernov B.K., Golova Y.B.. Gene-specific silencing by expression of parallel complementary RNA in Escherichia coli. J. Biol. Chem. 2000; 275:26523–26529. [DOI] [PubMed] [Google Scholar]
- 21. Liu Y., Zhang Y., Yang L., Yao T., Xiao C.. Gene-specific silencing induced by parallel complementary RNA in Pseudomonas aeruginosa. Biotech. Lett. 2009; 31:1571–1575. [DOI] [PubMed] [Google Scholar]
- 22. Tchurikov N.A. Natural DNA sequences complementary in the same direction: evidence for parallel biosynthesis. Genetica. 1992; 87:113–117. [DOI] [PubMed] [Google Scholar]
- 23. Soyfer V., Potaman V.. Triple-Helical Nucleic Acids. 1996; NY: Springer; 1–46. [Google Scholar]
- 24. Rich A., Davies D.R., Crick F.H.C., Watson J.D.. The molecular structure of polyadenylic acid. J. Mol. Biol. 1961; 3:71–86. [DOI] [PubMed] [Google Scholar]
- 25. Tchurikov N.A., Ponomarenko N.A., Golova Y.B., Chernov B.K.. The formation of parallel RNA-RNA duplexes in vitro. J. Biomol. Struct. Dyn. 1995; 13:507–513. [DOI] [PubMed] [Google Scholar]
- 26. Sugiyama H., Ikeda S., Saito I.. Remarkably stable parallel-stranded oligonucleotides containing 5-methylisocytosine and isoguanine. J. Am. Chem. Soc. 1996; 118:9994–9995. [Google Scholar]
- 27. Seela F., He Y., Wei C.. Parallel-stranded oligonucleotide duplexes containing 5-methylisocytosine-guanine and isoguanine-cytosine base pairs. Tetrahedron. 1999; 55:9481–9500. [Google Scholar]
- 28. Ingale S.A., Leonard P., Tran Q.N., Seela F.. Duplex DNA and DNA–RNA hybrids with parallel strand orientation: 2′-deoxy-2′-fluoroisocytidine, 2′-deoxy-2′-fluoroisoguanosine, and canonical nucleosides with 2′-fluoro substituents cause unexpected changes on the double helix stability. J. Org. Chem. 2015; 80:3124–3138. [DOI] [PubMed] [Google Scholar]
- 29. Seela F., Melenewski A., Wei C.. Parallel-stranded duplex DNA formed by a new base pair between guanine and 5-aza-7-deazaguanine. Bioorg. Med. Chem. Lett. 1997; 7:2173–2176. [Google Scholar]
- 30. Seela F., Wei C., Melenewski A., He Y., Kröschel R., Feiling E.. Parallel-stranded DNA formed by new base pairs related to the isoguanine-cytosine or isocytosine-guanine motifs. Nucleosides Nucleotides. 1999; 18:1543–1548. [Google Scholar]
- 31. Deleavey G.F., Damha M.J.. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012; 19:937–954. [DOI] [PubMed] [Google Scholar]
- 32. Wilds C.J., Damha M.J.. 2′-Deoxy-2′-fluoro-β-d-arabinonucleosides and oligonucleotides (2′F-ANA): synthesis and physicochemical studies. Nucleic Acids Res. 2000; 28:3625–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Watts J.K., Martín-Pintado N., Gómez-Pinto I., Schwartzentruber J., Portella G., Orozco M., González C., Damha M.J.. Differential stability of 2′F-ANA•RNA and ANA•RNA hybrid duplexes: roles of structure, pseudohydrogen bonding, hydration, ion uptake and flexibility. Nucleic Acids Res. 2010; 38:2498–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anzahaee M.Y., Watts J.K., Alla N.R., Nicholson A.W., Damha M.J.. Energetically important C−H···F−C pseudohydrogen bonding in water: evidence and application to rational design of oligonucleotides with high binding affinity. J. Am. Chem. Soc. 2010; 133:728–731. [DOI] [PubMed] [Google Scholar]
- 35. Zhou N., Germann M.W., van de Sande J.H., Pattabiraman N., Vogel H.J.. Solution structure of the parallel-stranded hairpin d(T8.ltbbrac..rtbbrac.C4A8) as determined by two-dimensional NMR. Biochemistry. 1993; 32:646–656. [DOI] [PubMed] [Google Scholar]
- 36. Parvathy V.R., Bhaumik S.R., Chary K.V., Govil G., Liu K., Howard F.B., Miles H.T.. NMR structure of a parallel-stranded DNA duplex at atomic resolution. Nucleic Acids Res. 2002; 30:1500–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang X.-L., Sugiyama H., Ikeda S., Saito I., Wang A.H.J.. Structural studies of a stable parallel-stranded DNA duplex incorporating isoguanine:cytosine and isocytosine:guanine basepairs by nuclear magnetic resonance spectroscopy. Biophys. J. 1998; 75:1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deleavey G.F., Watts J.K., Alain T., Robert F., Kalota A., Aishwarya V., Pelletier J., Gewirtz A.M., Sonenberg N., Damha M.J.. Synergistic effects between analogs of DNA and RNA improve the potency of siRNA-mediated gene silencing. Nucleic Acids Res. 2010; 38:4547–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Manoharan M., Akinc A., Pandey R.K., Qin J., Hadwiger P., John M., Mills K., Charisse K., Maier M.A., Nechev L. et al. Unique gene-silencing and structural properties of 2′-fluoro-modified siRNAs. Angew. Chem. Int. Ed. 2011; 50:2284–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ui-Tei K., Naito Y., Zenno S., Nishi K., Yamato K., Takahashi F., Juni A., Saigo K.. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008; 36:2136–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Anzahaee M.Y., Deleavey G.F., Le P.U., Fakhoury J., Petrecca K., Damha M.J.. Arabinonucleic acids: 2′-stereoisomeric modulators of siRNA activity. Nucleic Acid Ther. 2014; 24:336–343. [DOI] [PubMed] [Google Scholar]
- 42. Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T.. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001; 411:494–498. [DOI] [PubMed] [Google Scholar]
- 43. Phan A.T., Mergny J.-L.. Human telomeric DNA: G-quadruplex, i-motif and Watson–Crick double helix. Nucleic Acids Res. 2002; 30:4618–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lima W.F., Prakash T.P., Murray H.M., Kinberger G.A., Li W., Chappell A.E., Li C.S., Murray S.F., Gaus H., Seth P.P. et al. Single-stranded siRNAs activate RNAi in animals. Cell. 150:883–894. [DOI] [PubMed] [Google Scholar]
- 45. Yu D., Pendergraff H., Liu J., Kordasiewicz H.B., Cleveland D.W., Swayze E.E., Lima W.F., Crooke S.T., Prakash T.P., Corey D.R.. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant Huntingtin expression. Cell. 2012; 150:895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wilson J.A., Zhang C., Huys A., Richardson C.D.. Human Ago2 is required for efficient microRNA 122 regulation of hepatitis C virus RNA accumulation and translation. J. Virol. 2011; 85:2342–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Elbashir S.M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T.. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001; 20:6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Parrish S., Fleenor J., Xu S., Mello C., Fire A.. Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol. Cell. 2000; 6:1077–1087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.