Abstract
Histone deacetylase inhibitors (HDACIs) are known to alter gene expression by both up- and down-regulation of protein-coding genes in normal and cancer cells. However, the exact regulatory mechanisms of action remain uncharacterized. Here we investigated genome wide dose-dependent epigenetic and transcriptome changes in response to HDACI largazole in a transformed and a non-transformed cell line. Exposure to low nanomolar largazole concentrations (<GI50) predominantly resulted in upregulation of gene transcripts whereas higher largazole doses (≥GI50) triggered a general decrease in mRNA accumulation. Largazole induces elevation of histone H3 acetylation at Lys-9 and Lys-27 along many gene bodies but does not correlate with up- or down-regulation of the associated transcripts. A higher dose of largazole results in more RNA polymerase II pausing at the promoters of actively transcribed genes and cell death. The most prevalent changes associated with transcriptional regulation occur at distal enhancer elements. Largazole promotes H3K27 acetylation at a subset of poised enhancers and unexpectedly, we also found active enhancers that become decommissioned in a dose and cell type-dependent manner. In particular, largazole decreases RNA polymerase II accumulation at super-enhancers (SEs) and preferentially suppresses SE-driven transcripts that are associated with oncogenic activities in transformed cells.
INTRODUCTION
Reversible lysine acetylation is important for homeostatic regulation of many cellular processes. The lysine residues in the N-terminal tail of histones are tightly regulated by acetylation and deacetylation modifications catalyzed by enzymes known as histone acetyltransferases (HATs) and histone deacetylases (HDACs) respectively (1). It has been long recognized that HDACs are predominantly involved in transcriptional repression as loss of histone lysine acetylation, a hallmark of transcriptionally active chromatin, decreases chromatin accessibility. HDACs often exist as the catalytic module of chromatin remodeling machineries, including CoREST, NuRD, Sin3 and N-CoR (2). These molecular complexes target specific genomic regions through sequence specific interactions mediated by non-histone proteins such as transcription factors, methyl binding proteins (MBDs), or other epigenetic modifier enzymes such as DNA and histone methyltransferases (DNMTs or HMTs).
Deregulation of histone post-translational modifications have been observed in human tumors. Indeed, transcription of tumor suppressor genes is frequently silenced in tumor cells due the hyper- or aberrant activity of HDACs (3). Accordingly, HDACIs are used clinically for the treatment of a subset of hematologic tumors (4). There are 18 HDAC enzymes in the human genome, belonging to four distinct classes (4). Class I, II and IV enzymes contain a zinc (Zn2+) ion in their catalytic site and are inhibited by pan-HDACIs such as Vorinostat, Belinostat, or Panobinostat (4).
Aberrant recruitment of HDACs, as seen in cells with chromosomal translocations or mutations in certain transcription factors, contribute to development of tumors (3). Hence, HDACIs are used to de-repress silenced genes in cancer treatment (4). The therapeutic benefits of HDAC inhibition are thought to be associated with their chromatin remodeling activities and the resulting transcriptional reprogramming changes. However, exactly what type of chromatin remodeling activities or resulting chromatin mark changes are responsible for HDACI-regulated gene expression are still not fully understood. As expected, previous transcriptome analysis in the presence of HDACIs revealed the drastic up-regulation of a large number of genes (5–7). Surprisingly, transcription of many genes is also repressed by HDACI exposure (5–7). The opposing functions of HDACIs on transcription are difficult to reconcile. Genome-wide HDAC localization analyses indicate that HDACs are associated strongly with actively transcribed genes in human cells (8). Divergent activities of HDACIs on transcription could be a result of deacetylation activity towards different classes of targets. For example, non-histone substrates, including certain transcription factors, are activated when deacetylated (9–11). A recent study suggests that HDACIs target the transcription elongation complex and cause redistribution of other elongation factors across the genome (12).
Despite the clinical efficacy of HDAC inhibition for certain tumors, it has been generally challenging to understand the disparate activities of HDAC inhibitors in vitro and in vivo. As numerous biological activities of HDAC inhibitors are dose-dependent, it is imperative to characterize dose-dependent changes at molecular and genome-wide levels. The importance of HDACIs as anticancer therapies warrants more in-depth understanding of their dose-dependent activities in transcriptional regulation.
Largazole, a marine natural product discovered in cyanobacteria, is a highly potent Class I, Class IIb, and Class IV selective HDACI and displays selective killing of tumor cells (13–15). Largazole offers a unique tool to address the mechanism of HDAC inhibition in cancer biology due to its selectivity, superb potency, and minimal off-target activities. Parsing out various mechanisms underlying largazole-induced transcription activation and repression could offer fundamental insights critical for developing superior HDACIs with better clinical efficacy and low toxicity. To this end, we conducted comprehensive analysis of the specificity and molecular mechanisms of action for largazole in both transformed and non-transformed cell lines. We show that largazole selectively inhibits class I and class IIb HDAC enzymes at a subnanomolar range and causes cytostatic responses in a variety of tumor cell lines. We performed genome wide studies to identify histone marks and gene signatures whose dose-responsive changes, upon exposure to increasing concentrations of largazole, closely match the GI50 curve of the cytostatic response. Our data indicates that largazole induces profound dose-dependent changes in H3K9ac, H3K27ac, H3K4me1, H3K4me2 and perturbs the association of RNAPII with enhancers, promoters, and gene bodies. Low doses of largazole exposure resulted mostly in the up-regulation of gene transcripts whereas mid to high doses lead to more pronounced transcriptional suppression. We observe a correlation between the effect of largazole on cellular proliferation and transcriptional suppression with its effects on enhancer elements. Thus, our results reveal that largazole causes remodeling of numerous enhancer elements by modulating H3K27ac and retooling the enhancer atlas in a dose-dependent manner.
MATERIALS AND METHODS
Cell culture and largazole treatment
Cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (Sigma), 1% penicillin streptomycin, and 1% GlutaMAX (Invitrogen) and maintained at 37°C and 5% CO2. Prior to treatment, HCT116 and RPE cells were grown in complete media and passaged for three consecutive days. Cells were treated with the indicated largazole concentration or equivalent amount of vehicle (DMSO) at 70% confluency and harvested after 16 hours for all ChIP-seq experiments as well as for immunoblotting assays. Synthesis of largazole has been described previously (15).
RNA extraction and library preparation
Total RNA was extracted from 16 h treated HCT116 cells using TRIzol reagent (Life Technologies) according to the manufacturer's protocol. The concentration of each sample was quantified using the Qubit™ 3.0 Fluorometer (Thermo Fisher), and integrity was measured on an Agilent Bioanalyzer 2100 (Agilent Technologies). The Illumina TruSeq RNA Sample Prepartaion kit (Illumina) was used to generate the RNA sequencing libraries. Briefly, mRNA was purified from 2.5 ug total RNA from each sample, fragmented, and converted to double-stranded cDNA with the use of modified oligo(dT) primers. Sequencing barcodes were ligated to the cDNA fragments, and the resulting fragments were amplified using PCR. The final lengths of oligos from each library were validated on an Agilent Bioanalyzer 2100.
Sequencing
Libraries were quantified using the Qubit™ 3.0 Fluorometer and sequenced at the Next-Generation Sequencing Facility at the University of Colorado BioFrontiers Institute and University of Colorado Anschutz Medical Campus. All sequencing libraries were multiplexed and sequenced on an Illumina HiSeq 2000 sequencing system (Illumina).
RNA mapping and normalization
Reads were trimmed to a final length of 43 bp and mapped to human genome 18 (RefSeq) using Bowtie version 2.02.0 and TopHat version 2.0.6 (16). After mapping, alignment files were processed using SAMtools version 0.1.18.0 (17). Using Cuffdiff version 2.1.1 we counted the total number of sequencing reads that aligned to each putative gene model in the human genome. To determine which genes were differentially expressed, we used the R package DESeq version 1.30.0 (18).
Immunoblotting, antibodies and signal quantification
Western blots were carried out using standard protocols. Briefly, HCT116 and RPE cells were grown, treated and harvested as previously mentioned. Nuclear protein lysates were separated by SDS-PAGE and transferred to GVS nitrocellulose 0.22 μm membranes. Blots were probed with primary antibodies, followed by peroxidase-conjugated secondary antibodies (GE Healthcare Life). Signal for all immunoblots was acquired using the ImageQuant LAS 4000 biomolecular imager (GE Healthcare LS) with an average exposure of 30 s. Antibodies used are as follows: H3K9ac (abcam, cat. # ab4441); H3K27ac (abcam, cat. # ab4729); H3K4me1 (abcam, cat. # ab8895); H3K4me2 (abcam, cat. # ab7766); H3K4me3 (abcam, cat. # ab8580); total H3 (abcam, cat #1791).
Chromatin immunoprecipitation
HCT116 and RPE cells were treated with largazole or vehicle for 16 h and cross-linked with 1% formaldehyde for 15 min at room temperature (25°C). Cells were washed two times with PBS and membranes ruptured in hypotonic buffer (50 mM NaCl, 1% NP-40 alternative, 2 mM EDTA, 10 mM Tris, 1 mM DTT, 2 mM EDTA, 1X protease inhibitor cocktail (Roche # 04693124001). The cell nuclei were recovered by centrifugation and resuspended in lysate buffer (150 mM NaCl, 0.5% Triton X-100, 2 mM EDTA, 0.1% SDS, 20 mM Tris, 1 mM DTT, 1X protease inhibitor cocktail). Resuspended samples were sonicated for 25 cycles (30 s ‘on’ at high level and 30s ‘off’ per cycle) using a Bioruptor (Diagenode; Denville, NJ, USA) and spun for 10 min at 16 000 × g in a microcentrifuge. Samples were incubated for 5 h at 4°C with 5–20 μg of antibodies and 20 μl of 50% slurry with protein A beads (Millipore; Billerica, MA, USA). The immunoprecipitated chromatin was then recovered and DNA purified using phenol chloroform extraction. Sequencing libraries were prepared using an Illumina ChIP-Seq DNA Sample Prep Kit (cat. # IP-102-1001), with a starting sample varying from 2 to 20 ng of DNA isolated from the immunoprecipitation step. Antibodies used are as follows: RNAPII (Santa Cruz sc-899 lot # K0111); H3K9ac (abcam, cat. # ab4441); H3K27ac (abcam, cat. # ab4729); H3K4me1 (abcam, cat. # ab8895); H3K4me2 (abcam, cat. # ab7766).
ChIP-seq mapping and normalization
ChIP-seq datasets were aligned using Bowtie mapping software version 0.12.7 (19). To maintain the same read length across all experiments, 1 × 150 bp ChIP-seq raw datasets (fastq files) were trimmed to 50 bp using FASTX-toolkit (version 0.0.13.2) (http://hannonlab.cshl.edu/fastx_toolkit/) or Trimmomatic (version 0.36) (20). All reads were mapped to the hg18 reference human genome with a number of base pairs mismatch not >2 (96% sequence match). We used SAMtools version 0.1.19 (21) to generate a sorted pileup format of the aligned reads. Reads were then extended from the 3′–end to a final length of 150 bp. For each experiment, genome coverage bed graph files were generated using BEDTools2 version 2.25.0 (22) and then normalized by multiplying the read density times 100 and dividing by the total number of mapped reads. Normalized bed graph files were subsequently converted to bigwig files and uploaded to UCSC Genome Browser for visualization. We downloaded ChIP-seq data for p300, MLL4, and the corresponding input from HCT116 cells previously published (23), from the GEO database accession number GSE1176. In addition, we also acquired published GRO-seq data for HCT116 cells from the GEO database accession number GSE53964 (24). Raw ChIP-seq data for p300, MLL4, as well as the GRO-seq data were processed in the same manner as mentioned above.
Identification of ChIP-seq signal
H3K4me1, H3K4me2, RNAPII, p300 and MLL4
With the exception of H3K9ac and –K27ac, signal analyses for all ChIP-seq datasets experiments were performed using MACS2 version 2.1.0.20150731 (25) under default settings and a P-value cutoff of 1e–05. We used –broad -g hs –keep-dup = auto -p 1e-5 -m 10 200 –bw 200 and selected broad peak calls.
H3K9ac and H3K27ac
The FStitch algorithm (26) was used to identify genomic regions enriched with H3K9ac and –K27ac signal from HCT116 ChIP-seq experiments. In order to acquire uniform FStitch signal calls across experiments targeting the same acetylated lysine, we determined the minimal number of unique reads found in datasets for H3K9ac as well as in those for H3K27ac (Supplementary Figure S1D). Based on these numbers, we randomly subsample 12 844 004 unique reads from all H3K9ac ChIP-seq experiments and 9 122 018 unique reads from all nine H3K27ac ChIP-seq datasets. For H3K9ac ChIP-seq data analyses, we used 20 genomic regions from untreated HCT116 H3K9ac ChIP-seq data as FStitch-training genomic locations (Supplementary Table S3). In a similar manner, we used 19 genomic regions from H3K27ac under basal experimental conditions as FStitch training parameters (Supplementary Table S4). Segmentation analysis for all ChIP-seq experiments targeting the same lysine on histone H3 were conducted using the output parameters gathered from the training sessions. The same analysis was performed on the input experiment and any resulting signal was subtracted from all ChIP-seqs. Signal analysis for H3K27ac ChIP-seq experiments gathered from RPE cells was performed using SICER (version 1.1) (27) with flags ‘hg18 1 200 150 0.74 600 0.01’.
Defining promoter proximal start sites and positive association with RNAPII
To select for genes bound by RNAPII at transcription start sites, we defined proximal gene promoter regions as 100 bp ± from annotated TSSs using the January 2016 UCSC RefSeq gene assembly (hg18). RNAPII ChIP-seq signal was determined using MACS2 (version 2.1.0.20150731) narrow peak analysis based on default settings and a P value cutoff equal to 1e–05. Using merged peak-signals of fragments within a 1 kb range resulting from MACS2 analysis, we identified proximal gene promoter regions positively associated with RNAPII in untreated cells. Because many annotated genes contain multiple isoforms associated with a single TSS, we selected for the longest annotated gene versions and for genes which bodies did not overlap with other genes. From this list, we excluded genes which associated TSSs were within 2 kb from neighboring genes, genes which annotated lengths are smaller than 3 kb, genes that contained intragenic enhancer elements (based on H3K27ac and H3K4me1 co-occupancy), as well as genes that displayed multiple internal TSSs occupied by RNAPII. Using this method, we identified 2352 genes in HCT116 and 1,644 in RPE cells that were bound by RNAPII at the corresponding TSS and deemed suitable for pausing index assessment.
Pausing index calculation
Calculations were performed as in (28). For the selected genes, we defined promoter regions from –30 to +300 relative to the TSS and the gene body extending from +300 bp to the end of the gene annotation. RNAPII accumulation at promoters and gene bodies was determined using unique mapped reads from RNAPII ChIP-seq experiments of untreated, and largazole treated cells with 75 and 300 nM concentrations. Read density for promoter and gene body windows were calculated by dividing the number of unique reads by the total base pairs associated with each specified window. Pausing index was assigned to each gene from the ratio between RNAPII density in the promoter region to that of the gene body.
De novo motif analysis
For de novo motif discovery, we used MEME (29). Analysis were performed with a search window of 800 and 500 bp flanking the center enhancer elements associated with the defined cluster. The reported E-value is the output of the MEME de novo motif finding algorithm. To identify related transcription factors, each identified motif was input to TOMTOM version 4.11.1 (16) using motif database JASPAR DNA CORE (2016) or HUMAN DNA HOCOMOCO (v10). We also report the E-value describing the certainty of the match between the identified de novo motif and the database position weight matrices. The images were prepared using Adobe Illustrator CS6 or Photoshop CS6.
Identification of conventional enhancer elements
We first determined H3K27ac ChIP-seq signal (FStitch or SICER), as mentioned above, resulting from unstimulated and largazole treated cells. We performed fragment intersect analyses to extract genomic regions with overlapping H3K27ac and H3K4me1 accumulation for both cell lines. To further define the boundaries of enhancer regions in HCT116 cells, we trimmed the co-occupied regions using MACS2 broad peak calls gathered from H3K4me2, RNAPII, and MACS2 narrow peaks from p300 ChIP-seq data gathered from unstimulated HCT116 cells (23). We then eliminated all genomic regions which coordinates overlapped with annotated transcription start sites based on the January 2016 UCSC RefSeq gene assembly (hg18). This led to identification of 41 017 and 28 299 putative enhancer elements in unstimulated HCT116 and RPE cells, respectively.
Identification of super-enhancers
Genomic locations and associated genes for super-enhancers in HCT116 cells were extracted from the dbSUPER database (http://bioinfo.au.tsinghua.edu.cn/dbsuper) (30). The effects of largazole on RNAPII occupancy along SEs for both HCT116 and RPE cells were determined using ROSE (https://bitbucket.org/young_computation/rose) (31). We used an exclusion zone of 5 kb (‘-t 2500’) flanking the transcription start site and the default stitching size of 12.5 kb. RNAPII peaks as determined by MACS2 were used as input constituent enhancers.
K–means clustering of H3K27ac signal along enhancer regions
K–means clustering was performed on the enhancer data set referred as ‘isolated enhancers’ (n = 12 172) from HCT116 cells. These elements are characterized by a single H3K27ac peak, co-occupied by H3K4me1, centered along a 20 kb genomic region, either under basal cellular conditions (n = 8667) or resulting from stimulation with 300 nM largazole treatment (n = 3505). To this end, we used H3K27ac FStitch calls from all nine ChIP-seq experiments along ±1 kb distance centered on overlapping peak regions (H3K27ac, H3K4me1 (MACS2 BP) and H3K4me2 (MACS2 BP), RNAPII (MACS2 BP) or p300 (MACS2 NP) when present). Two filters were applied on the analyzed enhancer list. First, the K-means clusters were selected based on two general H3K27ac signal trends, decreasing or increasing under largazole treatments. Second, an additional filtered was applied on these clusters based on RNAPII normalized read density patterns; selecting enhancer regions with both decreasing H3K27ac ChIP-seq signal in concomitance with an overall decreasing in RNAPII occupancy (based on DMSO, 75 nM and 300 nM data) or enhancer elements with increasing H3K27ac ChIP-seq signal accompanied by the systematic increase of RNAPII binding.
Cell viability assay
Cell viability for HCT116 cells, treated for 48 h with the indicated largazole concentration or unstimulated (DMSO), was measured using the crystal violet staining method. In short, treated cells were gently washed once with phosphate buffer saline (PBS) and fixed for 20 mins at room temperature with 4% paraformaldehyde under constant rocking. After a single wash with PBS, fixed cells were stained with 0.5% crystal violet (Sigma) in 20% methanol at room temperature for 10 mins. Cells were then thoroughly washed with water and left overnight to dry. Last, 150 μl of developing solution (4:1:1 mix of methanol, ethanol, and water) was added to each well and absorbance was measured at λ = 560 nM.
Flow cytometry analysis
HCT116 and RPE cells (1 × 106) were treated with vehicle (DMSO) or the indicated dose of largazole for 25 h. For each cell population analyzed, we washed with ice-cold PBS, treated with trypsin solution, and fixed in cold 70% ethanol overnight. Fixed cells were then washed with ice cold PBS, and incubated in 0.25 mg/ml or RNase (Sigma) for 1 h at 37°C. Before analysis, cells were stained with 10 ug/ml of propidium iodide (PI) (Sigma) at 4°C for 1 h. Analysis was performed using a FACSAccuri flow cytometer (Becton-Dickinson). Data obtained from the cell cycle distribution were analyzed using FlowJo version 10.1 (Tree Star). Gaussian distributions and S-phase polynomial were assigned to each cell population using the Watson pragmatic model. Starting from samples treated with 9.4 nM largazole dose and above, we specified the range of G1 and G2 peaks in order to gather percentage of cells in each cell cycle phase.
RESULTS
Dose-dependent cytostatic activity of largazole
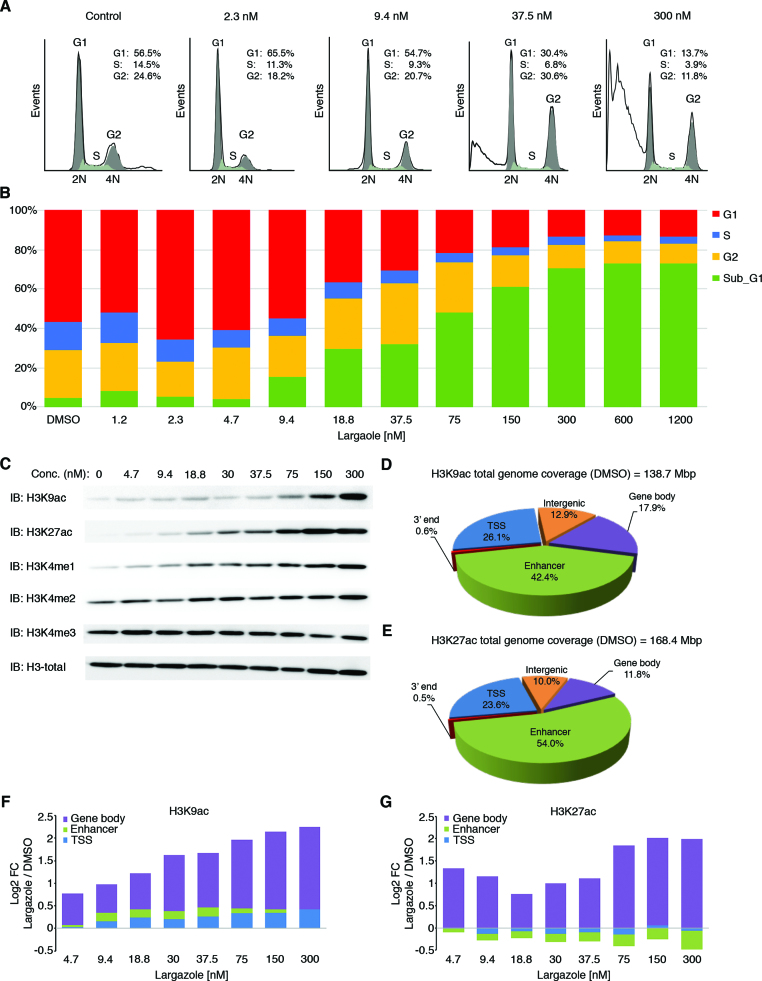
As a HDAC inhibitor, the natural product largazole selectively inhibits class I, class IIb, and to a lesser extent class IV HDACs but spares class IIa HDACs (Supplementary Table S1). Largazole has a broad spectrum tumor inhibitory activity toward most of the NCI 60 cell lines, with an average GI50 of ∼10 nM (Supplementary Table S2). We found that colorectal cell lines are particularly sensitive to largazole and because HCT116 cells have been consistently investigated by genome-wide sequencing analyses, we chose this cell line for all follow-up studies. Cell cycle analysis of HCT116 cells by flow cytometry revealed that largazole stimulation for 25 h alters cell cycle progression and leads to significant cell death (Figure 1A and B). Specifically, exposure of HCT116 cells to low concentrations of largazole induces cell cycle arrest at G1 (∼2 nM) and G2/M (∼37 nM) phases; however, these effects systematically dissipate at higher largazole concentrations. Significantly, largazole caused a dramatic increase of sub-G1 phase (apoptotic) cells in a dose-dependent manner that becomes apparent at ∼18 nM and plateaus at ∼300 nM. These results confirm that largazole inhibits proliferation and induces significant cell death of HCT116 cells at low nanomolar concentrations (GI50 = ∼34 nM) and further demonstrate that its effect on cell cycle profile is dose dependent.
Figure 1.
Dose-dependent cellular and histone acetylation responses to largazole treatment in HCT116 cells. (A) Quantitative analysis of the cell cycle progression by propidium iodide staining using flow cytometry in HCT116 cells treated with the indicted largazole concentration for 25 h. (B) Histogram showing the percentages of cells in G1 (red), S (blue), and G2 (yellow) phases of the cell cycle as well as subG1 fraction (green). (C) Dose-dependent global changes in indicated histone marks upon largazole exposure for 16 h as determined by immunoblotting with antibodies against each histone mark. Total histone H3 was used as a loading control. (D and E) Changes in H3K9ac and H3K27ac induced by largazole according to genomic territories. Pie charts illustrate the distribution of H3K9ac and H3K27ac signals (as determined by FStitch) from ChIP-seq experiments in vehicle (DMSO) treated HCT116 cells. Genomic territories are divided by gene bodies (purple), enhancer regions (green), TSS (blue), intergenic locations (orange) and 3′ ends (red). (F and G) The log2 fold change ratio for increasing H3K9ac and H3K27ac in each genomic territory with various doses of largazole (nM) exposure.
Histone marks undergo dose-dependent changes in response to largazole treatment
Histone H3 acetylation has been used as a reliable pharmacodynamic mark for HDACI on-target activity. To determine largazole's effects on acetylation of specific lysine residues, we analyzed dose-dependent accumulation of acetylated lysine 9 and 27 on histone H3 (H3K9/27ac). It is important to note that largazole-induced cell death occurs after 17 h exposure based on time and dose dependent GI50 measurement. At the 16 h time point, there is no significant cell death (data not shown). For most of the genome-wide studies in this report, we collected cells at 16 h post treatment. As expected, both H3K9ac and H3K27ac signals elevated significantly over the range of largazole doses used but the EC50 of the two marks appear to be different (Figure 1C). Moreover, three H3 lysine methylation marks were also analyzed. While there was a noticeable increase in global H3K4me1, no significant changes in H3K4me2 or H3K4me3 were observed (Figure 1C). Because hyperacetylation of histones is linked to gene activation, we hypothesized that genomic location-specific changes in histone acetylation may account for the altered regulation of the largazole responsive genes. To test this hypothesis, we employed chromatin immunoprecipitation coupled with massive parallel sequencing (ChIP-seq) using antibodies targeting H3K9ac and H3K27ac in HCT116 cells treated with increasing concentrations of largazole.
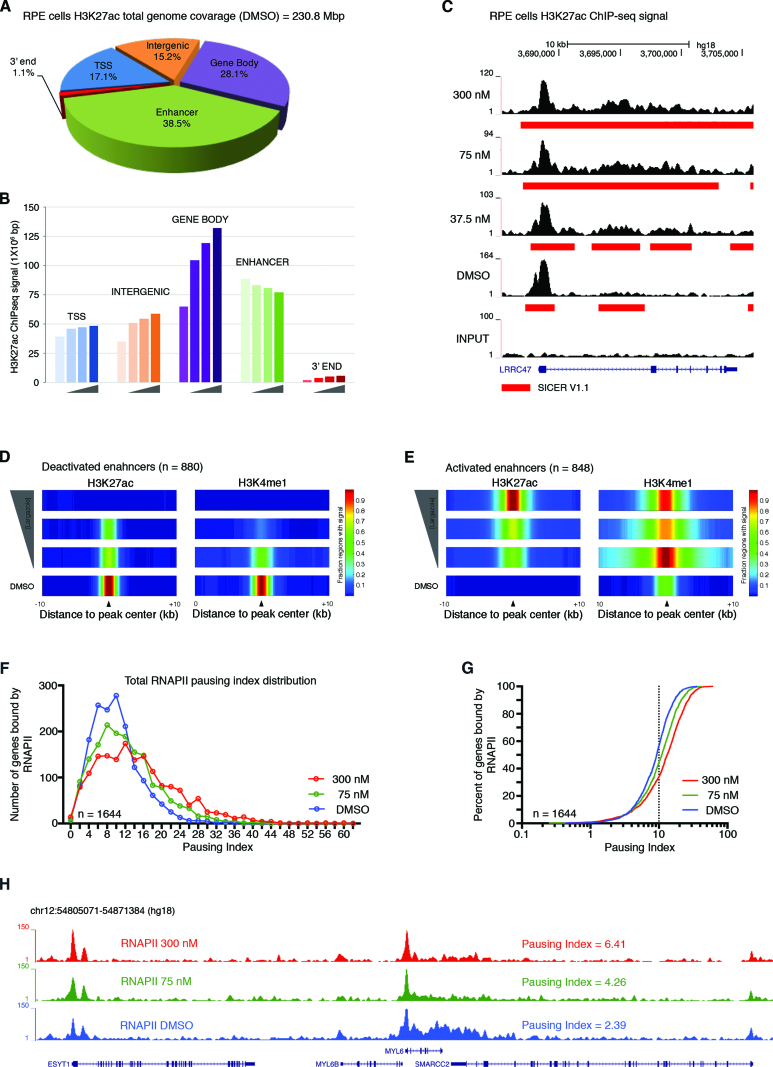
Because largazole confers an extensive degree of acetylation over genomic regions that can extend for several kilobases (Supplementary Figure S1A), we used Fast Read Stitcher (FStitch) under default signal threshold, to identify broad regions of enrichment over a wide range of signal strength (26). FStitch exhibited a detection range superior to MACS2 (32) for low-profile acetylation signal found in cells treated with high largazole concentrations. Over 96% of the genomic space identified by FStitch in H3K27ac ChIP-seq data from cells treated with 300nM largazole was not detected by MACS2 (Supplementary Figure S1B). To gain a better understanding of the signal distribution of H3K9/27ac, we divided the human genome into five territories based on the RefSeq hg18 gene alignment from the UCSC Genome Browser (25): proximal promoter (±2 kb from the TSS), gene region (–2 kb from TSS to end of annotated gene), 3′ end (end of annotated gene to 2 kb downstream), enhancer elements (±2 kb from the determined center), and intergenic regions. In unstimulated cells, ∼5% of the genome was associated with H3K9ac and –K27ac ChIP-seq signal, with both marks heavily enriched at enhancer elements (54.0% for H3K27ac and 42.4% for H3K9ac) and transcription start sites (23.6% for H3K27ac and 26.1% for H3K9ac) (Figure 1D and E).
Increasing doses of largazole (nM) exposure shows a general trend of gradual H3K9ac enrichment for all genomic territories analyzed, with the gene body regions displaying the highest elevation (expanding purple region of the log2 ratio of largazole/vehicle) (Figure 1F and Supplementary Figure S1C). In contrast, H3K27ac enrichment is mostly restricted to gene body regions (Figure 1G and Supplementary Figure S1C). Unexpectedly, a notable reduction in H3K27ac signal was seen in the enhancer elements as largazole concentration increased (Figure 1G, green). Collectively, our data indicates that H3K9ac and H3K27ac undergo similar dose-dependent changes along gene body regions but opposite trends at enhancer locations in response to largazole.
Dose-dependent spreading of H3K9ac and –K27ac at specific regions of the genome upon largazole exposure
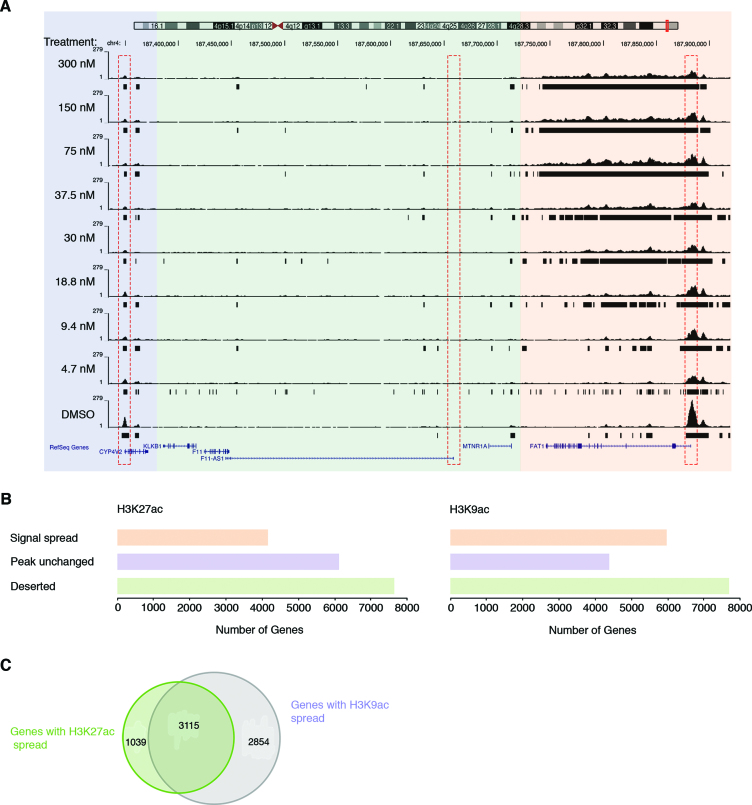
Since H3K9ac and –K27ac are functionally associated with proximal promoters and the most drastic acetylation changes occurred at protein-coding regions, we further examined genes that were occupied by H3K9ac and –K27ac under basal conditions. A total of 10 356 unique genes with a minimal length of 3 kb exhibited H3K9ac at their transcription start site (TSS) and 10 272 genes were positive for H3K27ac signal. As shown in Figure 2A, we found distinct gene patterns associated with H3K9 and –K27 acetylation changes. One class of genes displays a TSS highly occupied by acetylated H3K27 or –K9 at basal state, and the signal spreads in a dose-dependent manner into the transcribed region upon largazole treatment (Figure 2A, orange region; box on the right). The second class of genes displays a moderate amount of histone acetylation at the TSS with DMSO treatment and the signal remains anchored to the promoter throughout all largazole doses (Figure 2A, blue region; box on the left). Lastly, we found over 7600 genes that are not associated with H3K9/27ac and remain acetylation free under any dose of largazole stimulation (Figure 2A, green region; dash box in the middle). Using a cutoff of a two-fold spread in histone acetylation signal from untreated (DMSO) versus 300 nM largazole, we found 4154 unique genes exhibiting H3K27 hyperacetylation and 5969 genes with higher levels of acetylated H3K9 (Figure 2B and C). The relative number of genes without significant acetylation (acetylation ‘deserted’ genes) is very similar for both histone marks. Thus, largazole-induced dose-dependent changes in histone acetylation are both acetylation mark specific and restricted to defined genomic regions.
Figure 2.
Distinct H3K9 and H3K27 hyperacetylation responsive patterns upon largazole treatment. (A) A representative genomic snapshot of H3K27ac peaks illustrating different responses of gene bodies to newly acetylated histones. The signal initiates from the TSS (red dotted rectangle) of the FAT1 gene (orange panel) and spreads into the coding region or as in the case of CYP4V2 (purple panel), the preexisting acetylated TSS remains unmodified throughout all largazole dose treatments. Genes that do not show H3K9 or –K27 acetylation at the TSS (green panel) under basal conditions do not associate with the two histone marks as a result of largazole treatment. (B) Number of gene regions associated with the three response categories for H3K27ac (left) and H3K9ac (right). (C) Venn diagram showing the number of genes that exhibit new association with H3K27ac (green), H3K9ac (purple), and those that display both acetylation marks.
Largazole induces dose-dependent changes in RNA transcript levels irrespective of spreading of histone acetylation marks
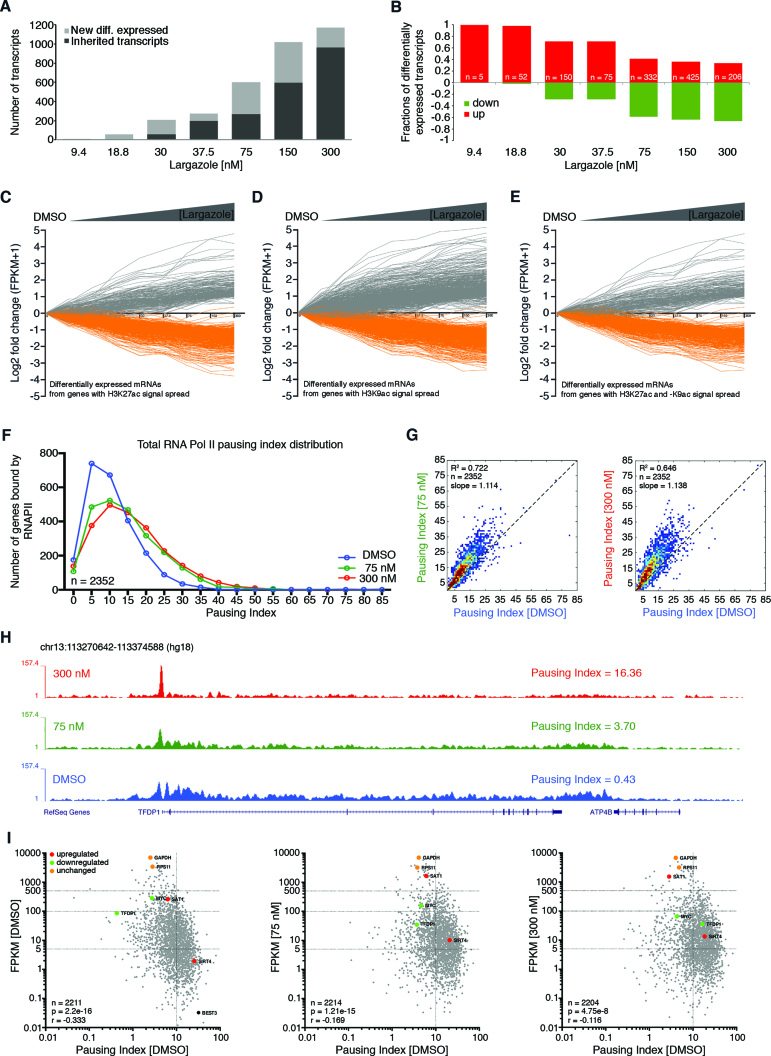
Since a subset of genes exhibited dose-dependent H3K9ac and H3K27ac signal spreading, we wondered if these patterns are predictive of changes in gene expression. We harvested poly(A) RNA from HCT116 cells treated for 16 h with increasing concentrations of largazole and performed RNA-seq analysis. Only transcripts that exhibit dose-dependent up- or down-regulation based on DESeq analysis with an adjusted P-value cutoff of <0.1 were selected for further analysis (18). Dose-dependent transcripts were identified through this approach. A striking pattern emerged when differentially expressed transcripts were plotted against largazole dose. Whereas a higher dose of largazole resulted in more significantly deregulated genes (Figure 3A), increasing dosage led more genes to be down-regulated than up-regulated (Figure 3B). This event is unlikely due to the cell loss associated with nonspecific toxic effects of largazole as a more polarized gene expression pattern is observed with no significant decrease in cell numbers at this endpoint. Thus, low dose largazole exposure triggers selective gene activation and higher dose of largazole is associated with wide spread transcriptional suppression.
Figure 3.
Increasing dose of largazole causes more gene suppression than activation and hyperacetylation of gene body territories does not predict higher mRNA accumulation. (A) Total number of genes differentially expressed at each largazole dose treatment showing newly differentially expressed in grey and transcripts that were inherited from a lower dose in black. (B) Differentially expressed transcripts unique to each largazole dose treatment based on DESeq analysis with an adjusted P-value cutoff of <0.1. Transcripts are shown as a fraction of total elements per dose where those that are upregulated (red) are plotted above the zero line and below those that are downregulated (green). DREM version 2.0 software was used to visualize dynamic transcript changes as a function of largazole dose with a minimal absolute expression fold change of 2. Differentially expressed mRNAs from genes with only (C) H3K27 hyperacetylation, (D) H3K9 hyperacetylation, or (E) those displaying an increase association with both histone marks. (F) Effects of largazole on RNAPII pausing index. Histograms depicting the calculated PI distribution of a group of genes treated with DMSO (blue), 75 nM largazole (green), and 300 nM largazole (red). (G) Contour plots showing Pearson correlation analysis between the calculated pausing indexes under two different largazole treatments. (H) Screen shot of the TFDP1 loci showing total RNAPII ChIP-seq signal from HCT116 cells treated with vehicle (blue), largazole at 75 nM (green) and largazole at 300 nM (red) showing the calculated pausing index. (I) Correlation between pausing index and relative transcript levels (FPKMs). Pearson correlation method was implemented using the ln(PI) and ln(FPKM) values from ∼2200 transcribable gene regions (grey). The input gene list was generated from gene bodies with a minimum length of 3 kb and that were bound by total RNAPII at the transcription start site, as determined from MACS2 narrow peak calling signal. Overlapping gene regions and genes containing intergenic enhancers were excluded from the analysis. Three categories of transcriptionally regulated genes are shown as representative elements of the data; downregulated (green), upregulated (red), and not changing gene transcripts (orange).
To examine the effect of hyperacetylation of gene bodies on transcription, we plotted mRNA levels from all nine treatments for the set of genes that showed significant acetylation spread. Surprisingly, hyperacetylated genes with H3K9ac or –K27ac showed no consistent behavior in expression changes, as we found both up- and down-regulated transcripts (based on an arbitrary 2-fold change cutoff) as well as a relatively large set of genes with no significant changes in mRNA levels (Figure 3C and D). It is worth noting that the change in transcript levels associated with H3K9ac occurs at a lower largazole dose than the observed changes in H3K27ac spread (Figure 3D).
To explore a possible synergy between H3K9ac and –K27ac with respect to transcriptional effects, we also looked at the mRNA expression levels of 3115 genes that developed enrichment for both histone marks (Figure 2C). Similar to mRNA expression patterns from genes hyperacetylated at H3K9 or –K27, mRNAs from genes whose coding regions exhibited spread of both acetylation marks showed both events of up- and down-regulation (Figure 3E). Taken together, our findings show that the spectacular elevation and spreading of H3K9ac or H3K27ac upon largazole exposure appears to be insufficient to predict the direction of change (i.e. up or down) in transcript levels with largazole exposure.
Largazole induces RNAPII pausing at a subset of genes
Because previous reports suggest HDACI target the transcription elongation complex (12), we sought to examine the genome wide effects of largazole-induced hyperacetylation on RNAPII occupancy along gene bodies. To this end, we conducted ChIP-seq experiments targeting total RNAPII in HCT116 cells treated with DMSO and those treated with either 75 and 300 nM largazole. We used the ‘pausing index’ (PI) (28) as the measurement to determine the extent of RNAPII pausing in a representative set of genes (refer to methods and materials for selection criteria). Pausing index was calculated by dividing the RNAPII ChIP-seq unique read density in the proximal promoter region by that in the gene body. We observed that RNAPII PIs increased systematically upon largazole treatment for most of the ∼2300 genes analyzed. As shown in Figure 3F, most genes exhibit an increased PI when comparing 75 nM largazole treated to DMSO treated cells. Treatment with 300 nM largazole further increased PIs relative to 75 nM treatment, but this increase was not as pronounced as that between DMSO and 75 nM largazole-treated cells. Correlation comparisons of pausing indices from the three cellular conditions further illustrate that PIs increase with largazole dosage (refer to slopes in Figure 3G). In addition, examination of specific gene loci suggests that the relative distribution of RNAPII along genes is different across largazole-dose treatments. For example, the PI for the region coding of transcription factor DP1 (TFDP1) in DMSO treated cells is 0.43 and the index increased to 3.70 in cells treated with 75 nM largazole (Figure 3H, blue and green). In this case, depletion of RNAPII signal throughout the gene body is the main contributing factor to the increase of PI at the TFDP1 locus. However, at higher largazole dose treatment (300 nM), the TFDP1 gene region displays a PI of 16.36 that mainly reflects the vast accumulation of RNAPII restricted to the proximal promoter (Figure 3H, red).
We then examined the influence of RNAPII pausing on the relative accumulation of transcripts associated with affected genes. In untreated cells, we found a modest but strongly supported (P = 2.2e–16) negative correlation (r = –0.333) between RNAPII pausing indices and relative accumulation of mRNAs (FPKM values in RNA-seq) (Figure 3I, left). We observed that highly expressed genes, such as TFDP1 and MYC (FPKMs > 80), are associated with relatively low PIs (0.46 and 2.74, respectively), whereas silent genes or those with low levels of expression (FPKMs < 1) such as BEST3 illustrate PIs greater than 20 (Figure 3I, left). Analyses of data from cells treated with either 75 nM and 300 nM concentrations of largazole showed a marginal decrease in the correlation between PIs and FPKM values, however a general unidirectional trend of some genes became evident when looking at the changes in both PIs and FPKM values from downregulated genes (Figure 3, middle and right). Increase in pausing indices is clearly the most dominant pattern observed among the analyzed gene regions; however, we also see a small number of genes with unchanged PIs and relatively constant FPKM values (GAPDH and RPS11) as well as genes that became less paused and more transcriptionally active (SAT1 and SIRT4). Thus for most genes analyzed, largazole specifically interrupts RNAPII occupancy downstream of proximal promoters and this event most likely affects the transition from initiation to elongation or the elongation steps of RNA synthesis and plausibly contributes to down-regulation of gene expression at higher dose of largazole.
Genes with promoter-paused RNAPII are more resistant to H3K27 hyperacetylation by largazole
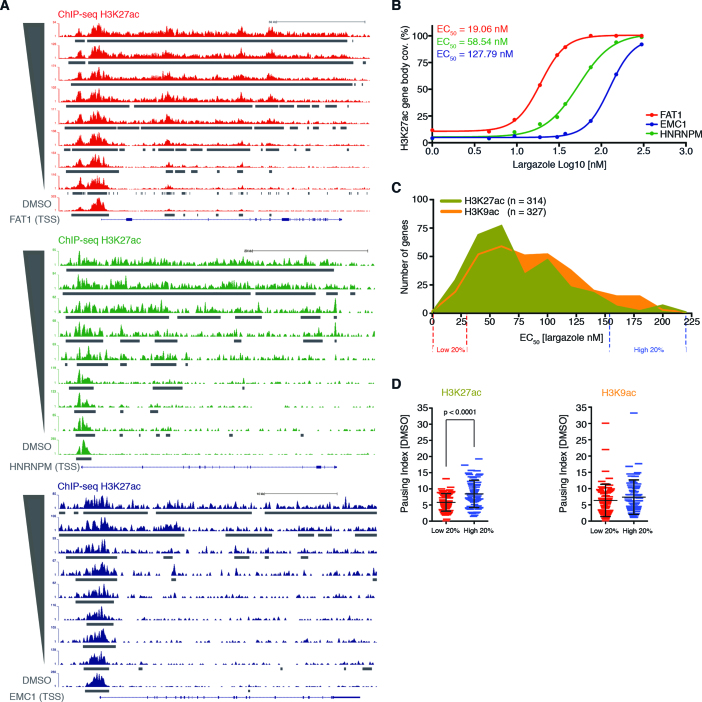
Class I HDACs preferentially occupy promoters of active genes and positively correlate with transcription levels (8). Accordingly, it is expected that highly expressed (low paused) genes should be more sensitive to largazole. This should be reflected in our data by the accumulation of newly acetylated H3K9 and –K27 signal along highly expressed genes in cells treated with relatively low concentrations of largazole. Among the genes displaying hyperacetylation of histone H3K9 and –K27, we noticed clear differences in dose-specific signal spread. For instance, the transcribed region of the protocadherin gene FAT1, displays a substantial association with H3K27ac in cells treated with 18.75 nM largazole and the signal reaches complete gene body coverage in cells stimulated with 75 nM (Figure 4A, top). We also found gene regions, such as the HNRNPM locus that do not associate with significant H3K27ac levels until ∼30 nM largazole treatments (Figure 4A, middle). Finally, there is a group of genes that are more resistant to hyperacetylation changes. For example, in the EMC1 gene region newly acetylated histones are only detected in cells treated with largazole concentrations at or above 75 nM (Figure 4A, bottom).
Figure 4.
Correlation between histone acetylation signal spread sensitivity along gene bodies and pausing index. (A) Snap shots from UCSC Genome Browser showing H3K27ac ChIP-seq signal over three different gene regions illustrate different dose-dependent acetylation of H3K27; low dose (red), mid dose (green), and high dose responders (blue). (B) EC50 values calculated by dose response plots of the normalized H3K27ac gene body coverage (FStitch signal) for the three genes in (A). (C) Histograms showing the distribution of EC50 values, for both H3K27ac (green) and –K9ac (orange), associated with a set of selected gene regions. (D) Student's unpaired t-test analysis between the calculated RNAPII pausing indexes from genes with the lowest (20%) and highest (20%) EC50 values determined from the histogram shown in (C). Individual genes with associated PI are plotted and the mean and standard deviation are shown with black horizontal lines.
To elucidate the sensitivity of each gene to largazole-induced acetylation changes we determined the largazole concentration necessary to induce a half-maximal acetylation response (EC50) in genes displaying 50% or greater H3K9ac or –K27ac signal coverage over the annotated gene lengths. Using FStitch calls, we calculated the total acetylation signal for both H3K9ac and –K27ac along the gene bodies for each of the nine corresponding ChIP-seq experiments and used the Sigmoidal Dose Response Search algorithm (SDRS) with a P = 0.05 cutoff (33). This approach allowed us to effectively assign each gene a largazole dosage sensitivity for both H3K9ac and –K27ac changes (Figure 4B). Distribution analysis of EC50 values from the two histone marks revealed a similar overall range of largazole dose sensitivity (∼8 nM to ∼210 nM) (Figure 4C). However, within the population of gene regions associated with H3K27ac, we observed a higher number of genes responsive to low concentrations of largazole as compared to H3K9ac.
To explore a possible correlation between the basal pause state of genes and sensitivity to largazole-induced acetylation changes, we compared the PIs from the 20% of genes most sensitive to largazole (lowest EC50) to that of the 20% of genes exhibiting the most resistance to acetylation changes (highest EC50) (Figure 4D). We found that gene bodies with low H3K27ac EC50 scores (more sensitive) are significantly less paused under basal conditions, when compared to the pausing indices of genes most resistant to H3K27ac association (least sensitive) (Figure 4D, left). In contrast, a similar analysis revealed that the pausing state of RNAPII from genes in untreated cells has no statistically significant influence on the dose-dependent changes of H3K9ac (Figure 4D, right). Overall, our data shows that dose-dependent changes in H3K9ac and H3K27ac by largazole have distinct dose-response behaviors. Genes that are highly expressed with low RNAPII pausing prior to treatment are more sensitive to low dose H3K27 hyperacetylation whereas H3K9 acetylation dose-dependent changes do not seem to be influenced by pausing state.
Largazole induces major changes in the landscapes of histone marks in distal regulatory elements
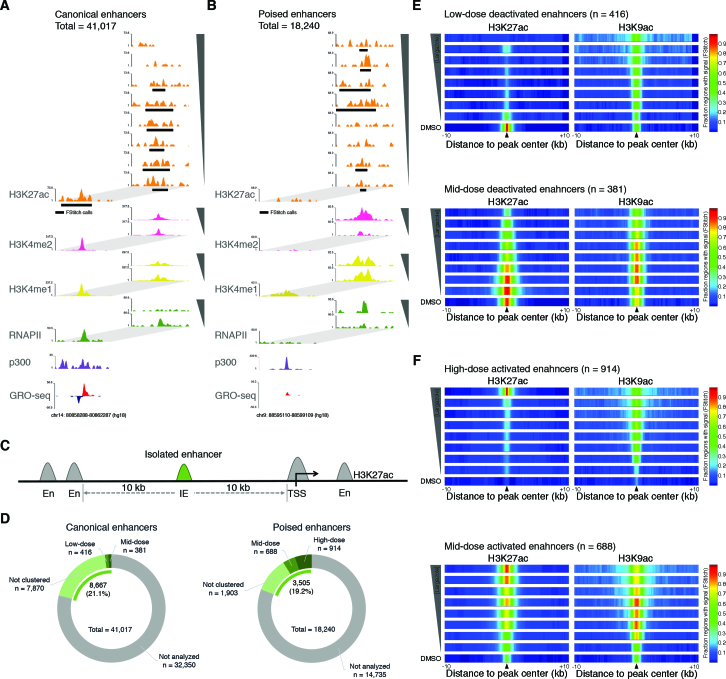
Although H3K9ac and H3K27ac undergo a broad dose-dependent increase in the gene body region, we observe dramatic differences between the two marks at enhancer regions. Specifically, we observe loss of H3K27ac but not H3K9ac with higher doses of largazole (Figure 1F and G). We wondered whether the increase in RNAPII pausing observed with higher concentrations of largazole may be the result of remodeling of enhancer elements. To this end, we measured enhancer associated histone acetylation (H3K4me1 and H3K4me2 (34,35)) and RNAPII binding as a function of largazole dose in treated cells. We performed H3K4me1 and H3K4me2 ChIP-seq of crosslinked nuclear extracts obtained following either vehicle (DMSO), 75 nM, or 300 nM largazole treatment of HCT116 cells. To help identify active enhancer regions, we used published GRO-seq (24) as well as ChIP-seq data for MLL4 and p300 (23) in untreated HCT116 cells. We then searched for genomic regions containing overlapping H3K27ac and H3K4me1 peaks (as determined by FStitch and MACS2, respectively) that were not superimposed over annotated transcription start sites. We identified 41,017 inter- and intragenic enhancer locations co-occupied by both H3K27ac and H3K4me1 prior to largazole treatment (Figure 5A). We refer to these enhancers as ‘canonical enhancers’. When post-largazole treatment data was examined, another class of enhancer elements we refer to as ‘poised enhancers’ became apparent. These enhancers are characterized by the dramatic dose-dependent increase of H3K27ac, H3K4me2, and RNAPII occupancy and are marked by H3K4me1 prior to largazole stimulation (Figure 5B). These regions are frequently occupied by MLL4, display unusually high levels of p300, and produce low amounts of eRNA in the basal cellular state. We identified 18,240 poised enhancer elements in HCT116.
Figure 5.
Dose dependent largazole effects on the epigenetic features of distal enhancer elements. (A and B) Screen shots from Genome Browser (UCSC) showing ChIP-seq and associated signal determined by FStitch (black rectangles) from HCT116 cells targeting H3K27ac (orange) starting with untreated cells (DMSO) at the bottom and followed by eight increasing largazole dose treatments on top (4.7–300 nM). ChIP-seq signal accumulation for p300 (purple) (23), total RNAPII (green), H3K4me1 (yellow), and H3K4me2 (pink) is shown for untreated HCT116 cells and for those treated with either 75 nM or 300 nM largazole concentrations (insets to the right). GRO-seq data from unstimulated HCT116 cells illustrate the presence of nascent transcripts resulting from the plus (red) and negative strand (blue) (24). (C) and (D) Schematic diagram shown illustrates the features used to identify isolated enhancers (IE) for genomic regions displaying both H3K27ac and H3K4me1 signal (determined by FStitch and MACS2 respectively). Only enhancers elements (green) located with a minimal distance of ±10 kb from neighboring H3K27ac/H3K4me1 locations from canonical (n = 8667) and poised (n = 3505) enhancers were used for further cluster analyses. (E and F) Largazole induces both the decommission and activation of transcriptional enhancers in a dose dependent manner. Shown are the fraction of IE regions with H3K27ac (left) and H3K9ac (right) signal (FStitch calls) along a ±10 kb distance centered on overlapping peak regions. Peak center locations are indicated by black triangles. Nine ChIP-seq experiments are illustrated with vehicle (DMSO) at the bottom and followed by increasing doses of largazole treatments to a maximum of 300 nM at the top. The fraction of IE elements with significant signal (FStitch) for each histone acetylation marks is illustrated by the heat-color scale: all regions (red); half of regions (green); no regions with signal detected (dark blue).
Many enhancer elements span large regions likely containing multiple nucleosomes, which makes it difficult to analyze dose responsive changes in H3K27ac. To minimize the number of false-positive deactivated and activated enhancers, we focused on a subset of isolated enhancer regions marked with a single, centered H3K27ac peak in a 20 kb genomic window for further analysis (Figure 5C). From the originally identified ∼41 000 putative enhancers in untreated cells, we selected 8,667 isolated active enhancers that met the above criteria (Figure 5D, left). Similarly, we selected 3505 isolated enhancers from an initial 18,240 identified poised elements (Figure 5D, right). To examine epigenetic modifications of enhancer elements as a function of dose, we quantified H3K27ac signal coverage (FStitch) over ±1.5 kb of the enhancer regions in data from each largazole treatment. K-means clustering of the 8667 isolated active enhancers and subsequent filtering for decreased RNAPII accumulation, revealed the presence of 797 largazole-inactivated regulatory elements. An example of a largazole-deactivated enhancer is seen in the hnRNPU locus (Supplementary Figure S2A), a gene that is required for the association of Xist RNA with the Xi and accumulation of H3K27me3 to ensure X chromosome inactivation (36). While the gene body of hnRNPU undergoes largazole-dependent H3K27ac increase and expansion, the upstream enhancer region (identified by p300, RNAPII, MLL4, H3K4me1 and H3K4me2 marks) exhibits dose-dependent decline in H3K27ac, H3K4me1 and H3K4me2 signal. Moreover, the hnRNPU mRNA levels show dose-dependent inhibition, which correlates with the loss of H3K27ac at its enhancer (Supplementary Figure S2B).
We further segregated the deactivated enhancers into low-dose (416 elements) and mid-dose (381 elements) affected subsets (Figure 5D, left). The low dose deactivated enhancer cluster displays a high H3K27ac and low H3K9ac signature at the basal state (Figure 5E, top). Low dose largazole treatments erase H3K27ac while the H3K9ac signal retains a low profile. Interestingly, the H3K9ac boundaries associated with these genomic regions undergo a significant expansion with increasing largazole dosage. Consistent with deactivation of these enhancers, dose-dependent reduction of H3K4me1 and RNAPII association were observed and loss of H3K4me2 only occurred at high dose exposure (Supplementary Figure S3D). The mid-dose deactivated cluster exhibits gradual loss of H3K27ac and a bell-shaped response in H3K9ac changes with increasing largazole exposure (Figure 5E, bottom).In this cluster of enhancers, H3K4me1 association shows a dose-dependent decline while H3K4me2 is unchanged (Supplementary Figure S3C). Hence, largazole treatment causes a significant number of enhancers to be deactivated with the characteristic loss of H3K27ac.
A similar analysis on the selected poised enhancers yielded mid-dose (688 elements) and high-dose (914 elements) activated subsets. The high dose cluster exhibited a largazole dose-dependent enrichment of H3K27ac and to a lesser degree it also accrued H3K9ac signal (Figure 5F, top). There is a significant RNAPII association and only a slight increase in H3K4me1 in this group. In contrast, H3K4me2 signal was barely detectable with DMSO and 75 nM largazole but elevated drastically upon treatment with 300 nM largazole which correlates with the dose-dependent rise of H3K27ac (Supplementary Figure S3B). The mid-dose induced cluster showed a gradual increase in H3K27ac and RNAPII but a fluctuating H3K9ac level as largazole dose was increased (Figure 5F, bottom and Supplementary Figure S3A). H3K4me2 was unchanged and H3K4me1 displayed a slight decrease at 300 nM largazole. A striking feature associated with the activation of the poised enhancers is that under basal conditions they tend to display high p300 occupancy yet minimal or absent H3K27ac signal (Supplementary Figure S3F). The presence of H3K4me1 and absence of H3K27ac in untreated HCT116 cells suggest that this subset of enhancers is in a poised state but likely primed for prompt activation. It is interesting to note that certain sequence motifs such as the binding site for transcription factor AP-1 (activator protein-1) are enriched in these epigenetically remodeled enhancer regions (Supplementary Figure S3A–C). Furthermore, the expression of AP-1 is upregulated by largazole (Supplementary Figure S3E), suggesting that AP-1 may play a role in enhancer remodeling with HDACs. Our results suggest that HDACs are probably actively involved in maintaining the poised state and largazole inhibition of histone deacetylases tip the balance in favor of H3K27 acetylation. Taken together, our findings show that largazole acts through inhibition of HDAC targets both in the deactivation and activation of distinct classes of enhancer elements that can be discerned by their dose sensitivity.
Largazole perturbs super-enhancers (SEs) and preferentially suppresses SE-associated transcripts
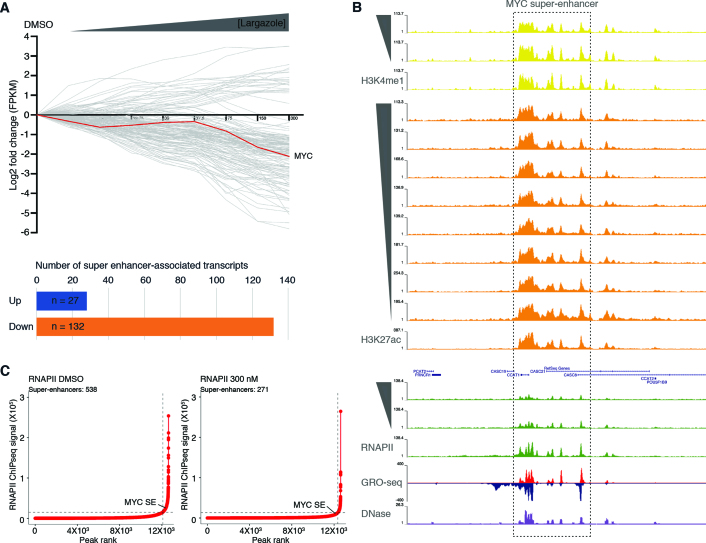
Initially identified as large clusters of transcription factor binding sites (37,38), super-enhancers have been shown to be involved in driving expression of genes that play prominent roles in cell proliferation and differentiation (39). Perturbations of super-enhancer function are frequently associated with human diseases including tumorigenesis. Since we showed that largazole suppresses expression of some genes in a dose dependent manner, we wondered whether largazole targets transcripts known to be regulated by super-enhancers. We analyzed RNA-seq data from HCT116 cells exposed to increasing doses of largazole. Among 387 super-enhancer associated genes previously identified in HCT116 cells (30), only 285 of these have significant expression (FPKM values > 5) and were selected for further analysis. We found that in this subset of genes, 132 transcripts are down-regulated (≥2-fold), in a dose-dependent manner (Figure 6A). In contrast, only 27 are up-regulated under the same parameters. No significant changes were observed in the levels of the remaining 126 transcripts. We noted that among the super-enhancer associated genes, the c-Myc oncogene, which is frequently up regulated in tumor cells, is suppressed by high dose largazole treatment (Figure 6A). Furthermore, inspection of the corresponding c-Myc super-enhancer region revealed a reduction of RNAPII and H3K4me1 ChIP-seq signals upon largazole treatment (Figure 6B). These observations suggest that largazole perturbs super-enhancer function and preferentially suppresses super enhancer-associated transcripts.
Figure 6.
Dose dependent largazole-induced depletion of RNAPII occupancy at most individual elements within super-enhancer regions. (A) Graph illustrates the log2 fold change in mRNA accumulation from HCT116 cells treated for 16 h in the absence (DMSO) or presence of increasing concentrations of largazole (4.7–300 nM). Each grey line depicts the trend of an individual gene transcript, with upregulated mRNAs (log2 > 1) shown above the black horizontal line and below those downregulated (log2 < –1). Distribution of total number of SE-associated transcripts differentially expressed under largazole. (B) Screenshot of Genome Browser (UCSC) along the MYC super enhancer (SE #34241) (30) showing normalized ChIP-seq data in untreated (DMSO) or increasing largazole dose treatment of HCT116 cells targeting H3K4me1, H3K27ac and RNAPII. GRO-seq signal (red and blue) from (24) and the associated DNAse I hypersensitivity peak clusters from ENCODE under basal conditions (www.encodeproject.org). (C) Delineation of super-enhancers based on RNAPII occupancy in untreated and largazole exposed HCT116 cells using the ROSE algorithm (31,40).
Super-enhancers consist of a collection of many individual enhancers. To determine the effect of largazole on the super-enhancers previously delineated in HCT116 cells (30), we characterized the largazole response of the individual enhancer regions that make up SEs. Reduction of RNAPII signal was the most dramatic perturbation observed on the c-Myc SE. Therefore, we utilized RNAPII occupancy trends resulting from largazole treatments and identified distinct responsive patterns (Supplementary Figure S4A). In cells treated with largazole, 65.7% (1008 out of 1534) of individual RNAPII peak-regions showed decreased densities of RNAPII signal when compared to untreated cells (patterns a, b and c). We found 18% of individual enhancers with no change in RNAPII occupancy (pattern d) and only ∼9% displayed a gained in RNAPII occupancy (patterns e, f and g). The remaining 7% of single peaks did not cluster. All individual enhancers analyzed displayed a general loss of H3K4me1 in response to largazole (Supplementary Figure S4A). To further evaluate the largazole-dependent depletion of RNAPII on super-enhancers in an unbiased approach, we determined the number of SEs in vehicle (DMSO) and largazole treated cells using RNAPII ChIP-seq peak-enrichment ranking method (31,40) to separate super-enhancers from conventional enhancers. Under basal cellular conditions we identified 538 super-enhancers characterized with high levels of RNAPII signal and these number is reduced by half to 271 elements in cells under high largazole treatment (Figure 6C). Unlike conventional enhancers that become inactivated by largazole, we detected no discernable change in the boundaries of H3K27 acetylation along super enhancers under largazole treatment (Supplementary Figure S4B, black centroids). Taking together, largazole promotes a dose dependent depletion of RNAPII and H3K4me1 levels at most individual peaks within super-enhancer regions but spares histone H3K27 acetylation marks.
Largazole-induced genome response in transformed and non-transformed cell lines
Largazole is known to have differential growth inhibitory activity between transformed and non-transformed cells (13,41). We sought to investigate the divergent and convergent genome wide responses to largazole between HCT116 and RPE cells, a nontumorigenic retinal pigmented epithelial cell line immortalized by telomerase overexpression. As seen with HCT116, largazole treatment leads similar dose-dependent increases in H3 acetylation measured by immunoblotting in RPE cells (Supplementary Figure S5A). Cell cycle analysis with RPE shows that higher doses of largazole result in more G2/M cells and a reduction in G1 and S phase cells, suggesting G2/M arrest in response to largazole treatment (Supplementary Figure S5B). However, the frequency of sub G1 cells is lower in largazole treated RPE cells than in HCT116. Consistent with less cell death, RPE cells are less sensitive to largazole in the growth inhibition assay (GI50 = 86 nM). Next, we performed ChIP-seq studies in RPE cells exposed to 0, 37.5, 75 and 300 nM largazole for 16 h with antibodies against H3K27ac and H3K4me1 and 0, 75 and 300 nM targeting RNAPII. Vehicle treated RPE cells have higher H3K27ac signal enrichment along gene body regions (∼65 Mb versus ∼20 Mb) in comparison to HCT116 (Figure 7A vs. Figure 1E). Similar H3K27ac signal was detected at enhancer elements (∼90 Mb) and TSS defined regions (∼40 Mb) for both cell lines. With increasing doses of largazole, there is an increase in H3K27ac signal along gene bodies and decrease at enhancer locations in RPE cells resembling the effect on HCT116 (Figure 7B versus Figure 1G). Examples of H3K27ac signal spreading are shown in Figure 7C and Figure 4. One notable difference between RPE and HCT116 cells, however, is the number of enhancers defined by the histone marks H3K27ac and H3K4me1, and occupied by RNAPII. RPE cells have fewer enhancer elements in comparison to HCT116 (1332 versus 8045) (Supplementary Figure S5C). Nevertheless, dose dependent deactivation and activation of enhancers seen in HCT116 also occurs in RPE with increasing exposure to largazole (Figure 7D and E). Finally, we investigated the extent of RNAPII promoter pausing in a representative set of genes in RPE cells (n = 1644). As seen with HCT116, higher doses of largazole exposure led to more RNAPII pausing, ascertained by the pausing indices of the genes analyzed (Figure 7F–H). Therefore, largazole treatment induces shared global genome-responses in transformed and non-transformed cells through spreading of H3K27ac signal, loss of H3K27ac at enhancer elements, and increase promoter pausing of RNAPII.
Figure 7.
Dose-dependent largazole effects on epigenetic features in RPE cells. (A) Distribution of H3K27 acetylation by type of genomic region. Pie chart illustrates the distribution of H3K27ac signals (as determined by SICER) from ChIP-seq experiments using RPE cells exposed to vehicle (DMSO) for 16 h. (B) Histogram showing H3K27ac ChIP-seq signal coverage in million base pair windows along the indicated genomic territory in RPE cells untreated (DMSO) or treated with 35.7, 75 or 300 nM largazole. Genomic territories are divided by gene bodies (purple), enhancer regions (green), TSS (blue), intergenic locations (orange) and 3′ ends (red). (C) Snapshot from UCSC Genome Browser showing H3K27ac ChIP-seq signal spread (red rectangle) into the coding region of the LRRC47 gene. (D and E) Largazole stimulates the decommission and activation of canonical enhancers in RPE cells. Shown are the fraction of enhancer regions with H3K27ac (left) and H3K4me1 (right) signal (SICER and MACS2 calls, respectively) along a ±10 kb distance centered on overlapping peak regions. Four ChIP-seq experiments are illustrated with vehicle (DMSO) at the bottom and followed by 37.5, 75 and 300 nM at the top. (F–H) Largazole promotes RNAPII pausing at proximal promoters of genes in RPE cells. Histogram and cumulative index plot depicting the calculated PI distribution of a group of genes treated with DMSO (blue), 75 nM largazole (green) and 300 nM largazole (red). Screen shot of the MYL6 loci showing total RNAPII ChIP-seq signal from RPE cells treated with vehicle (blue), largazole at 75 nM (green) and largazole at 300 nM (red) showing the calculated pausing index.
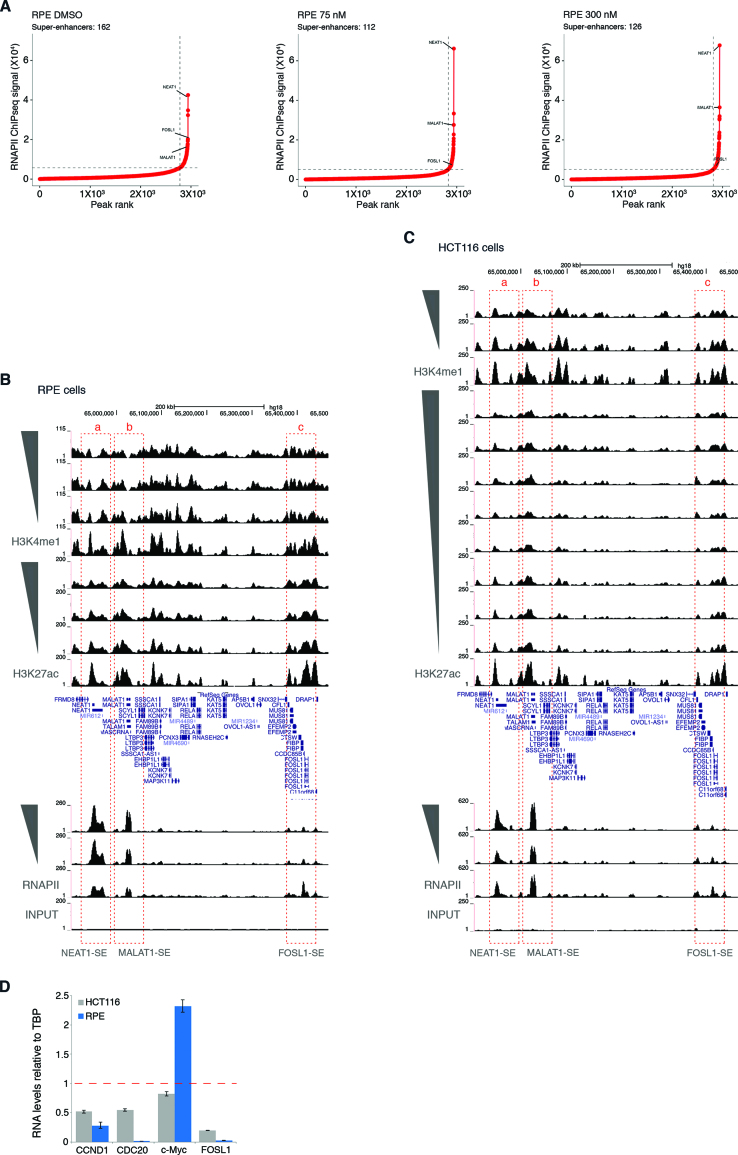
Differential super-enhancer responses to largazole in transformed and non-transformed cell lines
HCT116 and RPE cells differ widely in the number of active super-enhancers. In vehicle treated RPE cells, only 162 elements possess the hallmarks of SEs compared to 538 in HCT116 cells (Supplementary Figure S5C). In contrast to SEs in HCT116, fewer SEs exhibit significant changes in RNAPII occupancy in response to increasing doses of largazole in RPE cells (Figure 8A versus Figure 6C). Two super-enhancers associated with the non-coding RNAs NEAT1 and MALAT1 appear to be more active with largazole exposure (Figure 8B). Conversely, the SE encompassing the FOSL1 locus shows dose-dependent decrease in activity based on RNAPII occupancy (Figure 8B). FOSL1 is a member of the Fos gene family that can dimerize with components of the JUN family of proteins to form AP-1 transcription factor complexes. Unlike other Jun/Fos genes which are up-regulated by largazole (Supplementary Figure S3E), expression of FOSL1 is suppressed in both HCT116 and RPE cells. Previously, FOSL1 was identified as a gene involved in controlling G1/S phase transition by upregulating CCND1 (42). Depletion and gene expression studies have revealed that FOSL1 is an oncogene and its elevated expression is essential for KRAS-driven lung and pancreatic cancer by regulating cell motility and invasion as well as mitotic progression (43–45). Interestingly, the FOSL1-associated SE is deactivated in both RPE and HCT116 cells (Figure 8B and C). To independently validate changes in gene expression of SE associated transcripts in HCT116 and RPE cells in response to largazole treatment, quantitative real-time RT-PCR analysis was performed with selected SE-driven transcripts (Figure 8D). In agreement with RNA-seq and ChIP-seq results, FOSL1, CCND1, CDC20 are down-regulated by largazole in both cell lines while c-Myc is suppressed in HCT116 but not RPE cells. Collectively, our results indicate that largazole preferentially targets super-enhancers in transformed cells and suppresses oncogenes that fuel cellular transformation.
Figure 8.
Largazole spares most super-enhancers in RPE cells. (A) Delineation of super-enhancers in RPE cells based on RNAPII occupancy in untreated and largazole exposed HCT116 cells using the ROSE algorithm. Screenshot of Genome Browser (UCSC) encompassing three super-enhancers near the locus of the (a) NEAT1, (b) MALAT1 lncRNAs, and the (c) FOSL1 gene in (B) RPE and (C) HCT116 cells. (D) Real time quantitative PCR analysis of mRNA levels in HCT116 and RPE cells exposed for 16 h to 75 nM largazole. Data are represented as mean ± S.E.M. (n = 2).
DISCUSSION
HDAC inhibitors are known to induce extensive transcriptome changes in tumor and normal cells and gene regulation is thought to contribute at least in part to their effects on cell proliferation and death. Therapeutically effective HDAC inhibitors target multiple HDAC enzymes. Using largazole as a prototypical HDACI, we show that largazole induces dose-dependent changes in transcriptome, histone marks, and cell death. Low dose largazole induces mostly transcriptional activation while high dose causes mostly transcriptional repression. Our results reveal that largazole differentially regulates acetylation of H3K9 and –K27 in a dose-dependent manner. The most notable effect of largazole is in the enhancer regions where largazole treatment promotes poised enhancers to become fully active with increased H3K27ac. Notably, at higher doses of largazole, we observe the loss of H3K27ac at enhancers, a subset of which are super enhancers. This loss leads to preferential suppression of super-enhancer associated transcripts, which is associated with cell death response. While largazole has similar effects on hyperacetylation of histones and transcription pausing in both transformed and non-transformed cell lines, it has more profound and polarizing effects on enhancers and super-enhancers in transformed cell lines. Collectively these results uncover novel mechanisms of action for largazole and most likely other HDAC inhibitors in their suppression of gene expression and tumor suppression.
The prevailing notion about HDACI transcriptional regulation is that hyperacetylation of histones positively regulate gene expression. For example, in Drosophila cells, HDAC inhibition by both TSA and SAHA contribute to histone H3 acetylation at promoters and downstream regions. This event stimulates both transcription initiation and elongation (46). More recent studies from human cells suggest that TSA or SAHA induce a decrease of transcription along gene bodies without affecting nascent transcript production at the corresponding promoters (12). This effect has been attributed to failure of NELF eviction at promoters and loss of eRNA synthesis at some enhancers (12). Our dose dependent analysis of the effect of largazole on transcription may provide a new insight into transcriptional activation and repression by a HDAC inhibitor. Low doses of largazole (9.4 and 18.8 nM) mostly induce transcriptional activation but as the dose of largazole treatment increases the fraction of up-regulated transcripts decreases (Figure 3B). At 300 nM, more genes are repressed than activated. Therefore, transcriptional reprogramming by HDAC inhibition is dose-dependent. In agreement with the previous observations (7,12), we saw an increase in RNAPII pausing which may be responsible for elevated transcriptional repression (47). Increase in RNAPII pausing could be the result of a defect in the formation of the preinitiation complex or perturbations of the transcription elongation process (48). To our surprise, we discovered that a subset of active enhancers lose H3K27ac upon largazole exposure in a dose-dependent manner. It remains a challenge to unequivocally link a particular enhancer to a specific target gene as long distance enhancer-promoter communication is ubiquitous in eukaryotic cells. Since H3K9ac and H3K27ac elevation and spreading in the gene bodies do not correlate well with observed transcription changes (activation or repression) but H3K27ac loss at enhancers does correlate with target gene repression, we propose that deactivation of enhancers is likely to be part of the underlying mechanism of largazole induced transcriptional repression. As to the function of HDACs in gene bodies, our data is more in line with previous findings that histones are deacetylated co-transcriptionally to suppress histone eviction and spurious intragenic transcription (49,50) rather than to regulate the rate of transcription.
Loss of H3K27 acetylation at enhancer elements with higher doses of largazole is rather counterintuitive. One possibility is that H3K27ac histones were replaced by non-modified histones or through wholesale nucleosome replacement or loss. While we cannot entirely rule out this possibility, we consider this event less likely since the H3K9ac mark was retained in many cases despite the loss of H3K27ac. Another factor that may contribute to loss of H3K27ac is suppression of the enzymes that make H3K27ac. CBP and p300 are known writers for H3K27ac (51). While there is little change in p300 RNA levels across our experiments, CBP expression shows dose-dependent reduction up to ∼40% at the highest dose of largazole treatment (Supplementary Figure S5D). For H3K9ac writers, the level of KAT2A (GCN5) mRNA is reduced by ∼75% while no significant expression of KAT2B (PCAF) is seen in HCT116 cells. Thus, despite the general increase in histone acetylation particularly along gene bodies upon largazole treatment, loss of H3K27ac at certain enhancers could be the result of decreased CBP levels or recruitment when it becomes limiting. It will be interesting to test whether CBP and p300 have non-overlapping control of largazole sensitive enhancers. Finally, loss of H3K27ac at enhancers could also be a result of aberrant recruitment of another class of lysine deacetylases to the enhancer regions. It has been shown previously that sirtuins also possess histone deacetylase activity. For instance, in rat cardiomyocytes Sirt6 interacts with a c-Jun homodimer, is recruited to proximal promoters, and inhibits the transcription activation of insulin-like growth factor (IGF) signaling-related genes by deacetylating histone H3 at lysine 9 (52). Redundant activity of HDACs and sirtuins could lead to the disappearance of H3K27ac at the enhancers. Further experiments are necessary to distinguish these possibilities.
Our global analysis of cis-regulatory elements highlights the differences in the number of active enhancers and super-enhancers in HCT116 from RPE cells. This is not very surprising as tumor cells are dedifferentiated and generally more transcriptionally active. Previous studies suggest super-enhancers are bound by terminal transcription factors of signaling pathways and super-enhancer alterations are frequently found in tumor cells in response to oncogenic signaling (39,40). Targeting super-enhancer associated factors such as BRD4 and CDK7 has emerged as a promising cancer treatment strategy (40,53). Our genome-wide analysis points to super-enhancers from tumor cells as distinctly sensitive to perturbation by largazole. Of particular interest is the fact that SE-driven genes are predominantly down-regulated by largazole. Genes that have been shown to be critical for oncogenic signaling such as c-Myc and FOSL1 are particularly sensitive to such down-regulation. This result suggests that largazole may suppress oncogenic signaling through down-regulation of SE-driven genes, which may explain why HDAC inhibition has anti-tumor activity.
Accumulating evidence now supports the notion that there are active, poised and latent enhancers in the genome which are defined by distinct histone marks (54,55). Poised enhancers bear the features of H3K4me1 or H3K4me2 histone marks, absent or low acetylation of H3K27 and minimal association with RNAPII. Largazole-induced increase in H3K27ac at poised enhancers suggests that these enhancers are normally maintained by a steady-state active deacetylation and acetylation cycle. This interpretation is in agreement with a model in which both HATs and HDACs occupy the poised enhancer region but overall maintain a repressed state (56). Establishment of this repressed or poised state could be a result of association of these enhancers with sequence-specific transcriptional repressors, which are known to recruit HDACs (57) or the HDAC containing Nucleosome Remodeling and Deacetylation (NuRD) complex (58). Once HDACs are inhibited by largazole, the balance shifts toward acetylation of H3K9/27, critical for subsequent enhancer activation. Several lines of evidence support this hypothesis. In macrophages, the NCoR1/HDAC3 complex is recruited to promoter regions bearing AP-1 binding sites where it modulates transcription factor accessibility likely by mediating deacetylation of histone tails required for transcription activity (59). These observations imply that HDAC complexes could be recruited to particular enhancer elements for active histone deacetylation and repression of specific gene transcripts. Indeed, in CD4+ T cells, class I HDACs (HDAC2 and 3) and several HATs (p300, CBP, PCAF, MOF and Tip60) can be found bound to the same intergenic regions at high frequency, suggesting dynamic histone acetylation remodeling at these locations (8). Interestingly, under basal cellular conditions we found high levels of p300 ChIP-seq signal at intergenic regions that become activated enhancers under largazole stimulation. However, these genomic locations display minimal H3K27ac signal. Given largazole has superb potency against class I HDAC enzymes, we speculate that this class of HDACs is involved in maintaining poised enhancers in the repressive state.
Dose-dependent transcriptome changes correlate with the biological responses of cancer cells. It has been shown previously that a low dose of largazole induces cell growth arrest at the G1 phase of the cell cycle while a high dose of largazole causes G2/M arrest and apoptosis (60). Based on our results, it is tempting to speculate that transcriptional activation at low largazole doses may contribute to cell cycle arrest at G1 and that the profound transcriptional repression observed upon treatment with a high largazole dose is linked to G2 arrest and apoptosis. Since most therapeutic drugs are administered just below the maximum dose tolerance (MTD), HDACI-induced transcriptional repression is probably both relevant to their therapeutic benefits and their undesirable toxicity. Recent clinical success in treatment of ER positive breast cancer with Cdk4/6 inhibitor Palbociclib (Pfizer) sparks renewed interest in developing inhibitors that block G1 to S transition and promote cell differentiation (61). One implication of our study is that low doses of HDACI could also be an effective yet unexplored treatment strategy, especially in developing combination therapies.
In summary, our genome-wide dose-response analysis of transcriptome and histone signatures revealed new target specificity of largazole in transcriptional reprogramming. Our studies provide a more mechanistic explanation of the effect of HDACI on gene expression. Future studies focusing on dynamic changes of histone signatures and more comprehensive profiling of histone marks should provide more insights into remodeling of enhancer landscapes and their link to therapeutic responses in vivo and ultimately uncover predictive biomarkers. Such studies will be critical for developing more effective clinical use of HDAC inhibitors in cancer therapies and expanding their clinical potential beyond limited hematological malignancy.
AVAILABILITY
The genomic datasets from this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) database under accession number GSE101708.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Zhong Wang, Dr Bifeng Gao and Eric Gunther for technical assistance and Dr Andrew Phillips for providing largazole. We also want to thank Dr Dylan Taatjes for critical reading of the manuscript and advice from; Drs Natalie Ahn, James Goodrich, Tom Cech and members of Dowell and Liu laboratories for discussion and suggestions. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute and National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health [CA107098, AR068254 to X.L.]; National Cancer Institute [CA107098S1 to G.S.]; Bioscience Discovery Evaluation Grant Program [14BGF-28] from the State of Colorado; National Institute of General Medical Sciences [T32GM08759 to E.B.]; ImageXpress MicroXL was supported by the National Center for Research Resources [S10 RR026680]; FACSAria was supported by the National Institutes of Health [S10OD021601]; high-memory supercomputer was supported by National Institutes of Health [1S10OD012300-01]. Funding for open access charge: NIH/NIAMS [R01AR068254].
Conflict of interest statement. X.L. and the University of Colorado-Boulder have a financial interest in development of HDAC inhibitors for therapeutics and own equity in OnKure Inc. X.L. is one of the scientific founders of OnKure that has licensed proprietary HDAC inhibitors different from largazole from the University of Colorado-Boulder. OnKure has neither involvement in experimental design nor funding of this study.
REFERENCES
- 1. Strahl B.D., Allis C.D.. The language of covalent histone modifications. Nature. 2000; 403:41–45. [DOI] [PubMed] [Google Scholar]
- 2. Yang X.-J., Seto E.. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell. Biol. 2008; 9:206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Minucci S., Pelicci P.G.. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006; 6:38–51. [DOI] [PubMed] [Google Scholar]
- 4. Marks P.A. Histone deacetylase inhibitors: a chemical genetics approach to understanding cellular functions. Biochim. Biophys. Acta. 2010; 1799:717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rada-Iglesias A., Enroth S., Ameur A., Koch C.M., Clelland G.K., Respuela-Alonso P., Wilcox S., Dovey O.M., Ellis P.D., Langford C.F. et al. . Butyrate mediates decrease of histone acetylation centered on transcription start sites and down-regulation of associated genes. Genome Res. 2007; 17:708–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rafehi H., Balcerczyk A., Lunke S., Kaspi A., Ziemann M., Harikrishnan K.N., Okabe J., Khurana I., Ooi J., Khan A.W. et al. . Vascular histone deacetylation by pharmacological HDAC inhibition. Genome Res. 2014; 24:1271–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim Y.J., Greer C.B., Cecchini K.R., Harris L.N., Tuck D.P., Kim T.H.. HDAC inhibitors induce transcriptional repression of high copy number genes in breast cancer through elongation blockade. Oncogene. 2013; 32:2828–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Z., Zang C., Cui K., Schones D.E., Barski A., Peng W., Zhao K.. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009; 138:1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen G., Fernandez J., Mische S., Courey A.J.. A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes Dev. 1999; 13:2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wolf D., Rodova M., Miska E.A., Calvet J.P., Kouzarides T.. Acetylation of β-catenin by CREB-binding protein (CBP). J. Biol. Chem. 2002; 277:25562–25567. [DOI] [PubMed] [Google Scholar]
- 11. Xu M., Nie L., Kim S.H., Sun X.H.. STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBP?. EMBO J. 2003; 22:893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Greer C.B., Tanaka Y., Kim Y.J., Xie P., Zhang M.Q., Park I.H., Kim T.H.. Histone Deacetylases Positively Regulate Transcription through the Elongation Machinery. Cell Rep. 2015; 13:1444–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taori K., Paul V.J., Luesch H.. Structure and activity of largazole, a potent antiproliferative agent from the Floridian marine cyanobacterium Symploca sp. J. Am. Chem. Soc. 2008; 130:1806–1807. [DOI] [PubMed] [Google Scholar]
- 14. Bowers A., West N., Taunton J., Schreiber S.L., Bradner J.E., Williams R.M.. Total synthesis and biological mode of action of largazole: a potent class I histone deacetylase inhibitor. J. Am. Chem. Soc. 2008; 130:11219–11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nasveschuk C.G., Ungermannova D., Liu X., Phillips A.J.. A concise total synthesis of largazole, solution structure, and some preliminary structure activity relationships. Org. Lett. 2008; 10:3595–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S.. Quantifying similarity between motifs. Genome Biol. 2007; 8:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frietze S., Wang R., Yao L., Tak Y., Ye Z., Gaddis M., Witt H., Farnham P.J., Jin V.X.. Cell type-specific binding patterns reveal that TCF7L2 can be tethered to the genome by association with GATA3. Genome Biol. 2012; 13:R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Simon A., Wolfgang H.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu D., Gao X., Morgan M.A., Herz H.-M., Smith E.R., Shilatifard A.. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell Biol. 2013; 33:4745–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Allen M.A., Andrysik Z., Dengler V.L., Mellert H.S., Guarnieri A., Freeman J.A., Sullivan K.D., Galbraith M.D., Luo X., Lee Kraus W. et al. . Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife. 2014; 2014:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler A.D.. The Human Genome Browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Azofeifa J., Allen M., Lladser M., Dowell R.. An annotation agnostic algorithm for detecting nascent RNA transcripts in GRO-seq. IEEE/ACM Trans. Comput. Biol. Bioinf. 2016; 14:1070–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zang C., Schones D.E., Zeng C., Cui K., Zhao K., Peng W.. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009; 25:1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A.. C-Myc regulates transcriptional pause release. Cell. 2010; 141:432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Khan A., Zhang X.. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res. 2016; 44:D164–D171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nussbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ji R.R., Siemers N.O., Lei M., Schweizer L., Bruccoleri R.E.. SDRS—an algorithm for analyzing large-scale dose-response data. Bioinformatics. 2011; 27:2921–2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. He H.H., Meyer C.A., Shin H., Bailey S.T., Wei G., Wang Q., Zhang Y., Xu K., Ni M., Lupien M. et al. . Nucleosome dynamics define transcriptional enhancers. Nat. Genet. 2010; 42:343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaikkonen M.U., Spann N.J., Heinz S., Romanoski C.E., Allison K.A., Stender J.D., Chun H.B., Tough D.F., Prinjha R.K., Benner C. et al. . Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell. 2013; 51:310–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hasegawa Y., Brockdorff N., Kawano S., Tsutui K., Tsutui K., Nakagawa S.. The matrix protein hnRNP U is required for chromosomal localization of xist RNA. Dev. Cell. 2010; 19:469–476. [DOI] [PubMed] [Google Scholar]
- 37. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parker S.C.J., Stitzel M.L., Taylor D.L., Orozco J.M., Erdos M.R., Akiyama J.A., van Bueren K.L., Chines P.S., Narisu N., Black B.L. et al. . Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. PNAS. 2013; 110:17921–17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hnisz D., Schuijers J., Lin C.Y., Weintraub A.S., Abraham B.J., Lee T.I., Bradner J.E., Young R.A.. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol. Cell. 2015; 58:362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lovén J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A.. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013; 153:320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nasveschuk C.G., Ungermannova D., Liu X., Phillips A.J.. A concise total synthesis of largazole, solution structure, and some preliminary structure activity relationships. Org. Lett. 2008; 10:3595–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mechta F., Lallemand D., Pfarr C.M., Yaniv M.. Transformation by ras modifies AP1 composition and activity. Oncogene. 1997; 14:837–847. [DOI] [PubMed] [Google Scholar]
- 43. Vial E., Sahai E., Marshall C.J.. ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell. 2003; 4:67–79. [DOI] [PubMed] [Google Scholar]
- 44. Doehn U., Hauge C., Frank S.R., Jensen C.J., Duda K., Nielsen J. V, Cohen M.S., Johansen J. V, Winther B.R., Lund L.R. et al. . RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells. Mol. Cell. 2009; 35:511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vallejo A., Perurena N., Guruceaga E., Mazur P.K., Martinez-Canarias S., Zandueta C., Valencia K., Arricibita A., Gwinn D., Sayles L.C. et al. . An integrative approach unveils FOSL1 as an oncogene vulnerability in KRAS-driven lung and pancreatic cancer. Nat. Commun. 2017; 8:14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao Y., Lu J., Sun H., Chen X., Huang W., Tao D., Huang B.. Histone acetylation regulates both transcription initiation and elongation of hsp22 gene in Drosophila. Biochem. Biophys. Res. Commun. 2005; 326:811–816. [DOI] [PubMed] [Google Scholar]
- 47. Muse G.W., Gilchrist D.A., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K.. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007; 39:1507–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adelman K., Lis J.T.. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 2012; 13:720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P. et al. . Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005; 123:581–592. [DOI] [PubMed] [Google Scholar]
- 50. Govind C.K., Qiu H., Ginsburg D.S., Ruan C., Hofmeyer K., Hu C., Swaminathan V., Workman J.L., Li B., Hinnebusch A.G.. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol. Cell. 2010; 39:234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jin Q., Yu L.-R., Wang L., Zhang Z., Kasper L.H., Lee J.-E., Wang C., Brindle P.K., Dent S.Y.R., Ge K.. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011; 30:249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sundaresan N.R., Vasudevan P., Zhong L., Kim G., Samant S., Parekh V., Pillai V.B., Ravindra P. V, Gupta M., Jeevanandam V. et al. . The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat. Med. 2012; 18:1643–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kwiatkowski N., Zhang T., Rahl P.B., Abraham B.J., Reddy J., Ficarro S.B., Dastur A., Amzallag A., Ramaswamy S., Tesar B. et al. . Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014; 511:616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Heinz S., Romanoski C.E., Benner C., Glass C.K.. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015; 16:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shlyueva D., Stampfel G., Stark A.. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014; 15:272–286. [DOI] [PubMed] [Google Scholar]
- 56. Whyte W.A., Bilodeau S., Orlando D.A., Hoke H.A., Frampton G.M., Foster C.T., Cowley S.M., Young R.A.. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012; 482:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Koenecke N., Johnston J., He Q., Meier S., Zeitlinger J.. Drosophila poised enhancers are generated during tissue patterning with the help of repression. Genome Res. 2017; 27:64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Reynolds N., Latos P., Hynes-Allen A., Loos R., Leaford D., O’Shaughnessy A., Mosaku O., Signolet J., Brennecke P., Kalkan T. et al. . NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012; 10:583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ogawa S., Lozach J., Jepsen K., Sawka-Verhelle D., Perissi V., Sasik R., Rose D.W., Johnson R.S., Rosenfeld M.G., Glass C.K.. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. PNAS. 2004; 101:14461–14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu Y., Salvador L.A., Byeon S., Ying Y., Kwan J.C., Law B.K., Hong J., Luesch H.. Anticolon cancer activity of largazole, a marine-derived tunable histone deacetylase inhibitor. J. Pharmacol. Exp. Ther. 2010; 335:351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Turner N.C., Ro J., André F., Loi S., Verma S., Iwata H., Harbeck N., Loibl S., Huang Bartlett C., Zhang K. et al. . Palbociclib in hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 2015; 373:209–219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.