Abstract
Soil salinity is a significant threat to sustainable agricultural production worldwide. Plants must adjust their developmental and physiological processes to cope with salt stress. Although the capacity for adaptation ultimately depends on the genome, the exceptional versatility in gene regulation provided by the spliceosome-mediated alternative splicing (AS) is essential in these adaptive processes. However, the functions of the spliceosome in plant stress responses are poorly understood. Here, we report the in-depth characterization of a U1 spliceosomal protein, AtU1A, in controlling AS of pre-mRNAs under salt stress and salt stress tolerance in Arabidopsis thaliana. The atu1a mutant was hypersensitive to salt stress and accumulated more reactive oxygen species (ROS) than the wild-type under salt stress. RNA-seq analysis revealed that AtU1A regulates AS of many genes, presumably through modulating recognition of 5′ splice sites. We showed that AtU1A is associated with the pre-mRNA of the ROS detoxification-related gene ACO1 and is necessary for the regulation of ACO1 AS. ACO1 is important for salt tolerance because ectopic expression of ACO1 in the atu1a mutant can partially rescue its salt hypersensitive phenotype. Our findings highlight the critical role of AtU1A as a regulator of pre-mRNA processing and salt tolerance in plants.
INTRODUCTION
Given the sessile nature of plants, their survival under adverse environmental conditions requires that they rapidly perceive environmental cues and convert the cues into adaptive metabolic and developmental changes (1–3). One such environmental cue, high soil salinity, greatly limits plant growth and development and the quality and productivity of agricultural crops worldwide. Understanding how plants perceive and respond to salt stress is critical for improving plant resistance to salt stress through rational breeding and genetic engineering strategies.
High levels of salts including chlorides of sodium, calcium and magnesium cause soil sodicity, alkalinity and other soil problems (4). High soil salinity harms plants because of the toxicity of Na+ and other ions. To deal with high salinity, plant cells have evolved mechanisms to regulate ion influx and efflux at the plasma membrane and to sequester salts in vacuoles in order to maintain ion homeostasis in the cell (2). Salinity and many other stresses also cause plants to accumulate reactive oxygen species (ROS) including superoxide (O2.−) and hydroxyl (OH.) free radicals, hydrogen peroxide (H2O2) and free singlet oxygen (5,6). To maintain redox homeostasis, plants have developed enzymatic strategies (involving superoxide dismutase, peroxidase and catalase) and non-enzymatic strategies (involving antioxidants and some secondary metabolites) to detoxify excessive ROS (5). In addition to induce these cellular responses, salt stress can induce the expression of many stress-responsive genes, including those encoding transcription factors and protein kinases (7,8). Most studies on stress-regulated gene expression in plants have focused on the regulation of gene expression at the transcriptional level; in comparison, the regulation of salt-induced gene expression at the post-transcriptional level is much less known (9).
Post-transcriptional regulation of gene expression includes pre-mRNA splicing, capping and polyadenylation, mRNA transport, mRNA stability and translation of the functional mRNA (10), although some of these processes occur co-transcriptionally. Among these processes, pre-mRNA splicing, which is an essential step between transcription and translation of most eukaryotic mRNAs, is mediated by a dynamic macromolecular complex termed the spliceosome (11,12). The major spliceosome consists of five small nuclear ribonucleoprotein particles (snRNPs), namely U1, U2, U4, U5 and U6, and hundreds of non-snRNP proteins, which include serine- and arginine-rich proteins (SR proteins). Genome-wide studies have demonstrated that some of these spliceosome components are involved in responses to various abiotic stresses, including high soil salinity, temperature stress and high light irradiation (13–17). Overexpression of certain spliceosome or other splicing factors can increase plant tolerance to salt and other stresses (14,18). Thus, accurate and efficient regulation of these spliceosome components is necessary for gene function and may increase plant adaption to salt and other stresses (19).
In the present study, we characterize a novel mutant with reduced salt stress tolerance and show that the mutation disrupts the function of a gene encoding the Arabidopsis U1 snRNP-specific protein (AtU1A). Our study reveals that AtU1A is a component of a spliceosome complex in Arabidopsis and plays a pivotal role in pre-mRNA splicing of the oxidative stress-related gene ACO1 through direct pre-mRNA binding. The atu1a mutation alters genome-wide splicing patterns, especially in response to salt stress. Overexpression of AtU1A in Arabidopsis increases salt tolerance and the expression of many stress-related genes. Our study reveals a critical role for the AtU1A spliceosomal protein in regulating alternative splicing (AS) and in determining salt tolerance.
MATERIALS AND METHODS
Plant materials and growth conditions
Arabidopsis thaliana seedlings on Murashige and Skoog (MS) medium agar plates (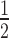 MS salts, 2% sucrose, 1.2% agar, pH 5.7) were routinely grown under continuous white light (∼75 μmol m−2 s−1) at 23 ± 1°C. Soil-grown plants were grown at 23 ± 1°C with a 16-h-light/8-h-dark photoperiod. Seeds of the T-DNA insertion line of AtU1A (SALK_074230) and other homozygous T-DNA mutants described in this study were obtained from the Arabidopsis Biological Resource Center at the Ohio State University (Columbus, OH, USA).
MS salts, 2% sucrose, 1.2% agar, pH 5.7) were routinely grown under continuous white light (∼75 μmol m−2 s−1) at 23 ± 1°C. Soil-grown plants were grown at 23 ± 1°C with a 16-h-light/8-h-dark photoperiod. Seeds of the T-DNA insertion line of AtU1A (SALK_074230) and other homozygous T-DNA mutants described in this study were obtained from the Arabidopsis Biological Resource Center at the Ohio State University (Columbus, OH, USA).
Physiological assays
For a seed germination assay, seeds of Col-0 and the atu1a mutant were sown horizontally on a half-strength MS medium with or without NaCl. In each experiment, at least 100 seeds per genotype were stratified at 4°C for 3 days, and radicle emergence was used as an indication of seed germination.
For a salt sensitivity assay on plates, Col-0, the atu1a mutant, and the atu1a complementation lines (U1Apro:U1A in atu1a #1 to #4) were grown vertically on MS medium with 1.2% agar under continuous light for 4 days at 23°C. Seedlings were transferred to MS medium with or without NaCl. Seedlings were photographed and fresh weights were measured at the indicated times.
For a salt sensitivity assay in soil, Col-0, atu1a and the AtU1A overexpression lines (OX1 and OX2) were grown in soil under a long-day photoperiod (16 h light/8 h dark) for 2 weeks. Subsequently, the soil was irrigated with 300 mM NaCl every 2 day, and plants were allowed to grow for an additional 1 to 3 weeks before they were photographed and chlorophyll content was analyzed.
For an oxidative stress assay on plates, Col-0 and atu1a mutants were grown vertically on MS medium with 1.2% agar under continuous light for 4 days at 23°C. Seedlings were transferred to MS medium with or without oxidative reagent (methyl viologen [MV) or H2O2). Seedlings were photographed and fresh weights were measured 14 days after the transfer.
For a chlorophyll content analysis, the leaves of soil-grown Col-0 and atu1a mutant seedlings were harvested, weighed and placed in 1.5-ml Eppendorf tubes. The leaves were ground with steel beads and extracted with 80% acetone. After they were incubated in the dark for 2 h at room temperature, the samples were centrifuged at 16 000 × g for 10 min and the supernatants were transferred to new tubes. Absorption at 645 and 663 nm was determined with a spectrophotometer. The concentration of chlorophyll was calculated as previously described (20).
Plasmid construction for gene complementation
To conduct a gene complementation of the atu1a mutant, we amplified a 3623-bp fragment of the AtU1A genomic sequence with the U1Apro attB1 and U1Ag attB2 primers (Supplementary Table S3) and cloned the fragment into the pDONR207 vector by BP recombination reaction (Gateway Cloning, Thermo Fisher Scientific). The resulting entry clone (pDONR207-AtU1Ag) was then introduced into the Gateway destination vector pGWB501 to yield the complementation clone by LR recombination reaction (Gateway Cloning, Thermo Fisher Scientific). The pGWB501-AtU1A genomic construct was introduced into the atu1a mutant through Agrobacterium-mediated plant transformation (21). T3 transgenic lines were confirmed by reverse transcriptase-polymerase chain reaction (RT-PCR) and were used for salt sensitivity assays.
qRT-PCR and RT-PCR analyses
We used Trizol reagent (Thermo Fisher Scientific) to extract total RNA from 12-day-old seedlings grown on MS medium under a 16-h-light/8-h-dark photoperiod. Total RNA was treated with RNase-free DNase I to remove potential genomic DNA contaminations. A 5-μg quantity of RNA was used for reverse transcription with M-MLV reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. The synthesized cDNA mixture was used as a template for quantitative RT-PCR (qRT-PCR) or RT-PCR analysis with the primers listed in Supplementary Table S3. UBQ10 was used as an internal control.
Analysis of AtU1A subcellular localization and AtU1A promoter activity
For subcellular localization, the AtU1A coding region or AtU1A genomic sequence without the stop codon was amplified and then cloned into the pDONR207 vector by BP recombination reaction. The resulting entry vector was then recombined with the destination vector pGWB505 or pGWB504 through LR recombination reaction to fuse the target protein to the N terminus of green fluorescent protein (GFP). For a promoter β-glucuronidase (GUS) assay, a 1895-bp genomic sequence 5′-upstream of the gene was amplified and then cloned into the pDONR207 vector by BP recombination reaction. The resulting entry vector was then recombined with the destination vector pGWB533 by LR recombination reaction. The constructs were transformed into Col-0 or the atu1a mutant, and transgenic T3 lines or T2 lines were used for GFP observation or GUS staining, respectively.
Yeast two- or three-hybrid assays
To create the AtU1A bait plasmid, we cloned the AtU1A cDNA into the pGBKT7 vector. Yeast transformation was performed according to the manufacturer's instructions (Clontech, CA, USA). To detect the interaction between AtU1A and other spliceosome protein (AtU1-70K and AtU1C), we amplified Arabidopsis U1-70K and U1C cDNAs from Col-0 plants and cloned them into the pDONR207 vector by BP recombination reaction. The entry vector pDONR207-AtU1A, pDONR207-AtU1-70K CDS and pDONR207-AtU1C CDS were recombined into the destination vector pGADT7 by LR recombination reactions to yield the yeast expression constructs. These constructs were then co-transformed into yeast stain Y2H gold with the corresponding full-length partner (AtU1A in pGBKT7) and were plated on -tryptophan/-leucine (-TL, for transformation control) and –leucine/-tryptophan/-histidine (-L-T-H, for selection) media. To detect the interaction between AtU1A and pre-mRNA (U1 snRNA and ACO1 5S), we cloned the Arabidopsis pre-mRNAs of U1 snRNA and ACO1 5S into the plllA/MS2-2 vector. These constructs were then co-transformed into the yeast strain L40-coat and plated on media that that contained 2 mM 3-aminotriazole but lacked leucine and uracil. The empty plllA/MS2-2 vector and pAD-AtU1A plasmid were co-transformed into the yeast strain L40-coat and used as negative controls. To detect the interaction between AtU1 snRNA and U2B'', we cloned the Arabidopsis U2B'' into the pGAD vector. The plllA-AtU1 snRNA and pAD-AtU2B'' plasmids were then co-transformed into the yeast strain L40-coat and plated on media that contained 2 mM 3-aminotriazole but lacked leucine and uracil. The empty pGAD vector and plllA-AtU1 snRNA were co-transformed into the yeast strain L40-coat and used as negative controls. The plllA/IRE-MS2 vector and pAD-IRP plasmid were co-transformed into the yeast strain L40-coat and used as positive controls.
Bimolecular fluorescence complementation (BiFC) assays in N. benthamiana
The entry vectors pDONR207-AtU1A CDS, pDONR207-AtU1-70K CDS and pDONR207-AtU1C CDS were recombined into the destination vector pEarlygate201YN or pEarlygate202YC by LR recombination reaction to yield the corresponding expression constructs. The resulting constructs were transformed into Agrobacterium tumefaciens strain GV3101, and the transformed Agrobacteria were grown overnight at 28°C in LB medium. After they were harvested by centrifugation, the bacteria were re-suspended and incubated in induction medium (10 mM MES, pH 5.6, 10 mM MgCl2 and 150 μM acetosyringone; Sigma) for 2 h. The bacteria were then resuspended in the induction medium to a final concentration of OD600 = 0.5 before inoculation. The bacterial suspensions were infiltrated into young but fully expanded Nicotiana benthamiana leaves. After infiltration, plants were immediately covered with plastic bags and kept at 23°C for 48–72 h before they were examined with confocal microscopy.
Poly-A RNA in situ hybridization
Poly-A RNA in situ hybridization was performed as described before with minor modifications (22). In brief, 7-day-old seedlings grown in MS plates treated or not treated with salt (150 mM NaCl for 3 h) were immersed in 10 ml of fixation cocktail containing a mixture of 50% fixation buffer (120 mM NaCl, 7 mM Na2HPO4, 3 mM NaH2PO4, 2.7 mM KCl, 0.1% Tween 20, 80 mM EGTA, 5% formaldehyde and 10% Dimethyl Sulphoxide (DMSO)) and 50% heptane. The samples were gently agitated for 30 min at room temperature. After dehydration twice for 5 min each time in 100% methanol and three times for 5 min each time in 100% ethanol, the samples were incubated for 30 min in ethanol:xylene (50:50). After they were washed twice for 5 min each time in 100% ethanol and twice for 5 min each time in 100% methanol, the samples were first incubated for 5 min each time in methanol:fixation buffer without formaldehyde (50:50) before they were incubated for 30 min in fixation buffer with formaldehyde. After being rinsed twice for 5 min each time with fixation buffer without formaldehyde and once for 5 min with 1 ml of PerfectHyb Plus Hybridization Buffer (Sigma-Aldrich; H-7033), the samples were treated with 1 ml of the PerfectHyb Plus Hybridization Buffer and were pre-hybridized for 1 h at 50°C. After pre-hybridization, 5 pmol of 5′-labeled (Alex-488; Invitrogen) oligo d(T) was added and the samples were hybridized overnight at 50°C in darkness. The samples were then washed once for 60 min in 2 × Saline Sodium Citrate (SSC) (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate), and 0.1% sodium dodecyl sulphate (SDS) at 50°C and once for 20 min in 0.2 × SSC, and 0.1% SDS at 50°C in darkness. The samples were then immediately examined using a Leica confocal laser-scanning microscope with a 488-nm excitation laser. Each experiment was repeated at least three times, and similar results were obtained.
RNA immunoprecipitation assays
A 3-g quantity of fresh leaves per sample from seedlings at the rosette stage was harvested, and incubated in 1% formaldehyde in vacuum for 15 min to crosslink protein–RNA. The crosslinking was stopped by adding 125 mM glycine and incubating for 5 min. Leaves were ground and resuspended in lysis buffer (100 mM KCl, 10 mM HEPES, 5 mM ethylenediaminetetraacetic acid (EDTA), 10% glycerol, 0.1% NP-40, 200 U/ml RNasin, 2 mM Ribonucleoside Vanadyl Complexes, 2 mM Phenylmethanesulfonyl fluoride (PMSF) and 10 μl/ml protease inhibitor [PI, Sigma P9599]). The lysed supernatant was sonicated to produce fragments of 300–500 bp, which were then treated with DNase to remove DNA before immunoprecipitation. RNA-IP was then performed using anti-GFP antibody (catalog number ab290, Abcam) coupled with Dynabeads Protein G (catalog number 10003D, Invitrogen). After the preparation was washed five times, the protein on the crosslinked complex was digested by proteinase K, and the RNA was extracted with phenol–chloroform–isoamyl alcohol. Following reverse transcription, qRT-PCR was performed with the UBC30 gene as an internal control.
RNA-seq analysis and validation
The 12-day-old seedlings of Col-0 and the atu1a mutant grown on MS medium were transferred to MS medium containing 0 or 150 mM NaCl and allowed to grow for an additional 3 h. Total RNA was extracted with Trizol reagent (Invitrogen) from these above seedlings and was treated with Turbo RNase-free DNase I (Thermo Fisher Scientific) to remove genomic DNA. Polyadenylated RNAs were isolated using the Oligotex mRNA Midi Kit (70042, Qiagen Inc., Valencia, CA, USA). The RNA-seq libraries generated from Col-0 and the atu1a mutant plants under normal and salt stress condition (there are two biological replicates per genotype) were constructed using the Illumina Whole Transcriptome Analysis Kit following the standard protocol (Illumina, HiSeq system) and were sequenced on the HiSeq 2000 platform to generate high-quality pair-end reads of 101 nt. To identify differential AS events corresponding to all five basic types of AS patterns (23), we subjected the RNA-seq data to analysis by rMATS v3.2.5 (http://rnaseq-mats.sourceforge.net). Statistical parameters for significant splicing change were defined as FDR < 0.05 and P-value < 0.05. The selected AS and intron retention (IR) events were validated by RT-PCR using a set of primers (See Supplementary Table S3) that were designed based on each AS event. The sequencing data of RNA sequencing (RNA-seq) experiments are available in the SRA database (Accession number: SRS1498194).
Nucleotide frequencies around novel and known splicing sites were determined and visually displayed using sequence logos (24).
RESULTS
The atu1a mutant is hypersensitive to salt stress
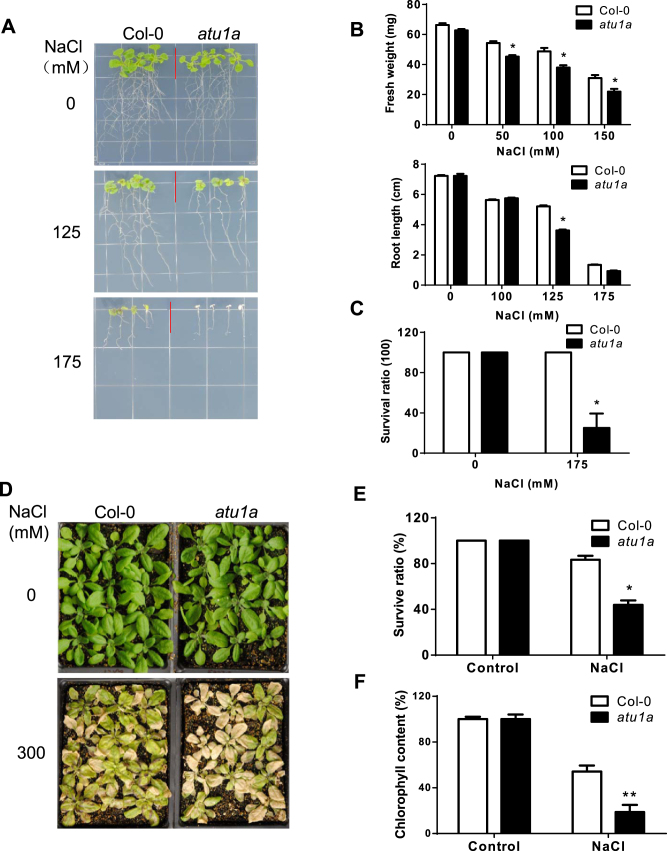
We previously reported a genome-wide analysis of AS of pre-mRNAs under salt stress in Arabidopsis (15). Further studies suggested that AS in plants might be involved in the regulation of salt tolerance (14,25). To find new components of AS pathways involved in salt stress responses, we performed a genetic screen with our collection of homozygous Arabidopsis T-DNA insertion mutants. From this analysis, we isolated a mutant, atu1a, with reduced salt tolerance (Figure 1). When 4-day-old wild-type (ecotype Col-0) and atu1a seedlings were transferred to MS medium containing 125 mM NaCl, the growth of the primary root and shoot was mildly reduced for wild-type seedlings, but was greatly reduced for atu1a seedlings (Figure 1A and B). When seedlings were grown on 175 mM NaCl, most of the atu1a mutant seedlings died while the wild-type seedlings remained green (Figure 1A and Supplementary Figure S1A). We further examined the phenotypes of wild-type and the atu1a mutant during seed germination and cotyledon greening under salt stress. In the absence of salt stress, germination rates did not significantly differ between the wild-type and the atu1a mutant (Supplementary Figure S1C). Although germination of both atu1a mutant and wild-type seeds was delayed under salt stress, the germination rate of the atu1a mutant seeds was substantially lower (Supplementary Figure S1C). Furthermore, inhibition of cotyledon greening by salt was greater in the atu1a mutant than in the wild-type (Supplementary Figure S1D). We then determined the salt tolerance of soil-grown plants. When treated with 300 mM NaCl for 2 weeks, about 60% of atu1a mutant plants died while only 20% of wild-type plants died (Figure 1D and E). Chlorophyll content of salt-stressed plants was also lower for the atu1a mutant than for the wild-type (Figure 1F), which was consistent with the increased salt damage in the mutant. To determine whether the salt-sensitive phenotype of the atu1a mutant is specific to Na+, we transferred 4-day-old atu1a mutant plants to MS medium containing different concentrations of LiCl, KCl or sorbitol. We found that atu1a mutant plants were more sensitive than wild-type plants to LiCl but not to KCl (Supplementary Figure S1A and B). However, the response of atu1a mutant plants to sorbitol, which induces general osmotic stress, was similar to that of wild-type plants (Supplementary Figure S1A and B). Thus, the atu1a mutant is specifically sensitive to Na+ and Li+, but not to K+ or Cl−.
Figure 1.
Salt-hypersensitive phenotype of the atu1a mutant. (A) Growth of wild-type (ecotype Col-0) and atu1a seedlings with response to salt. The 4-day-old seedlings grown on MS medium were transferred to MS medium containing different levels of NaCl and were grown for additional 10 days. Representative plants were photographed. (B) The fresh weights (upper panel) and root elongation (lower panel) of plants shown in (A). (C) Survival rates of wild-type and atu1a seedlings under 175 mM NaCl treatment. (D) Salt tolerance of soil-grown wild-type and atu1a plants. The 14-day-old seedlings were treated with 300 mM NaCl solution and were then grown for additional 10 days. Representative plants were photographed. (E and F) Survival (shown as a percentage of survival on normal MS medium,) and chlorophyll content of plants shown in (D). Error bars represent SD (n = 16 in [B and C], and 48 in [E and F]). Asterisks indicate significant differences (*P < 0.05, **P < 0.01) as determined by a two-tailed paired Student's t-test.
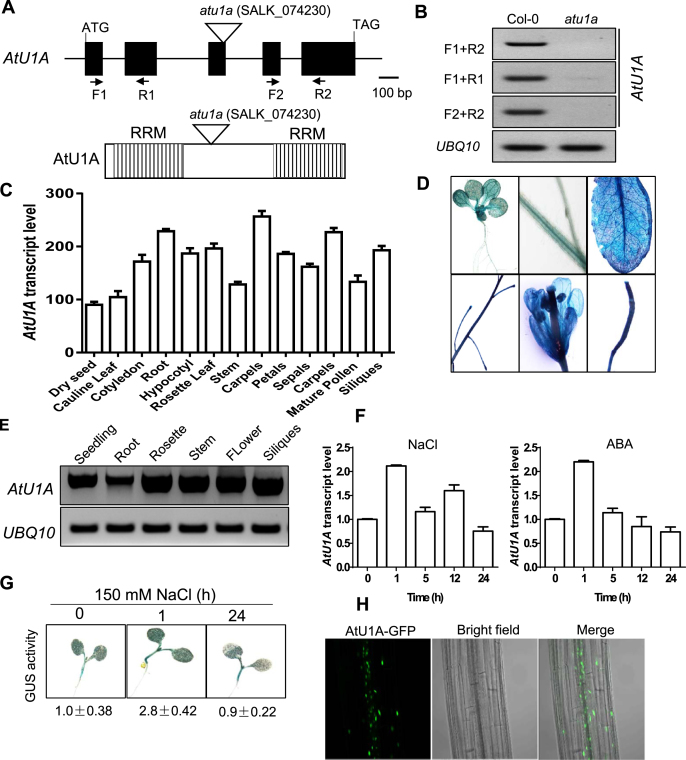
The atu1a/SALK_074230 mutant carries a T-DNA insertion in the At2g47580 (AtU1A) gene, and diagnostic PCR using a gene-specific primer and the T-DNA left border primer confirmed that this T-DNA insertion is homozygous (Supplementary Figure S2A and B). RT-PCR analysis revealed that AtU1A expression is abolished in the atu1a mutant (Figure 2A and B). The AtU1A protein, one of the U1 snRNP-specific proteins, contains two evolutionarily conserved RNA recognition motifs (RRMs) involved in the biosynthesis of cellular RNA (26). The human N-terminal RRM (amino acids [aa] 2–95) of U1A is necessary and sufficient for binding to the stem-loop2 sequence of U1 snRNA, while the C-terminal RRM (aa 202–283) of the human U1A does not seem to have any affinity for RNA (27–29). To further test whether shorter AtU1A transcripts are still present in the mutant as T-DNA insertion site was located in the bridge between two domains of RRMs of AtU1A (Figure 2A). The AtU1A transcript is severely disrupted in the mutant when N-terminal primers are used (F1 + R1, Figure 2A and B), and is abolished in the mutant when C-terminal primers are used (F2 + R2, Figure 2A and B).
Figure 2.
Expression profiles of AtU1A and its protein subcellular localization. (A) Structure of the AtU1A gene. Exons and introns are illustrated with black boxes and black lines, respectively. The triangle indicates the T-DNA insertion. Positions of primers used for RT-PCR analysis in (B) are included. Shown also are the two RRM in AtU1A. (B) RT-PCR analysis of AtU1A expression in the atu1a mutant. (C) Expression of AtU1A in wild-type plants at different developmental stages as indicated by publicly available gene expression data (30). (D) AtU1Apro:GUS reporter gene expression. T2 transgenic plants expressing GUS under the control of the AtU1A promoter were stained with X-Gluc and and examined and imaged with a microscope. (E) RT-PCR analysis of AtU1A expression level in different tissues of wild-type plants. (F) Transcript level of AtU1A in response to salt and ABA treatments. (G) AtU1Apro:GUS reporter gene expression in response to salt. The 7-day-old T3 transgenic plants were treated or not treated with 150 mM NaCl for the indicated times and were then stained with X-Gluc and examined and imaged with a microscope. GUS activities were determined by measuring fluorescence with a Tecan 200 fluorometer using 360 and 465 nm as excitation and emission wavelengths, respectively. The GUS activity under control conditions was set to 1. (H) Subcellular localization of AtU1A. The 7-day-old transgenic plants harboring the AtU1A-GFP construct were examined with a confocal microscope. UBQ10 was used as an internal control in (B and E) and (F). Error bars represent SD (n = 3 in [C] and [F]).
To confirm that the T-DNA insertion in AtU1A was responsible for the salt stress sensitive phenotype of the atu1a mutant, we transformed a genomic DNA fragment containing the entire AtU1A gene (with a ∼1.9-kb promoter region, the AtU1A coding region, and 185-bp fragment downstream of the translation stop codon) into the atu1a mutant. Thirty-two independent transgenic lines were obtained, and four T3 lines were randomly chosen to test for salt stress sensitivity. The mutant seedlings remained sensitive to salt stress, but the four transgenic lines did not (Supplementary Figure S3). These data demonstrated that the T-DNA insertion mutation in AtU1A is responsible for the atu1a mutant phenotypes.
Expression pattern of AtU1A and subcellular localization of AtU1A
To further determine the function of AtU1A in planta, we investigated its expression patterns. AtU1A is expressed at a relatively high level throughout the plant life cycle as revealed by publicly available gene expression data (Figure 2C) (30). In addition, GUS activity with transgenic plants harboring AtU1A promoter-driven GUS (AtU1Apro::GUS) and RT-PCR analyses confirmed AtU1A expression profiles in different plant tissues (Figure 2D and E). We then examined the expression of AtU1A in response to salt stress or abscisic acid (ABA). Total RNA was extracted from wild-type seedlings treated with 150 mM NaCl or 50 μM ABA for various periods of time. We found that the transcript levels of AtU1A were up-regulated by salt or ABA (Figure 2F). Consistent with the qRT-PCR results, transgenic plants harboring AtU1A promoter-driven GUS showed a slight upregulation in GUS activity in response to salt stress (Figure 2F). To determine the subcellular localization of AtU1A in vivo, we transiently expressed AtU1A fused in frame with the GFP at the C-terminus (AtU1A-GFP) in Arabidopsis protoplasts. Consistent with observations from a previous study (31), the AtU1A protein was localized in both the nucleus and cytoplasm in Arabidopsis protoplasts (Supplementary Figure S2C). We next obtained several independent transgenic plants that stably express AtU1A-GFP. The AtU1A-GFP signals in these plants were mainly detected in the nuclei of root cells, and only very weak signals were detected in the cytoplasm (Figure 2H).
AtU1A is a U1 snRNP-specific protein in planta
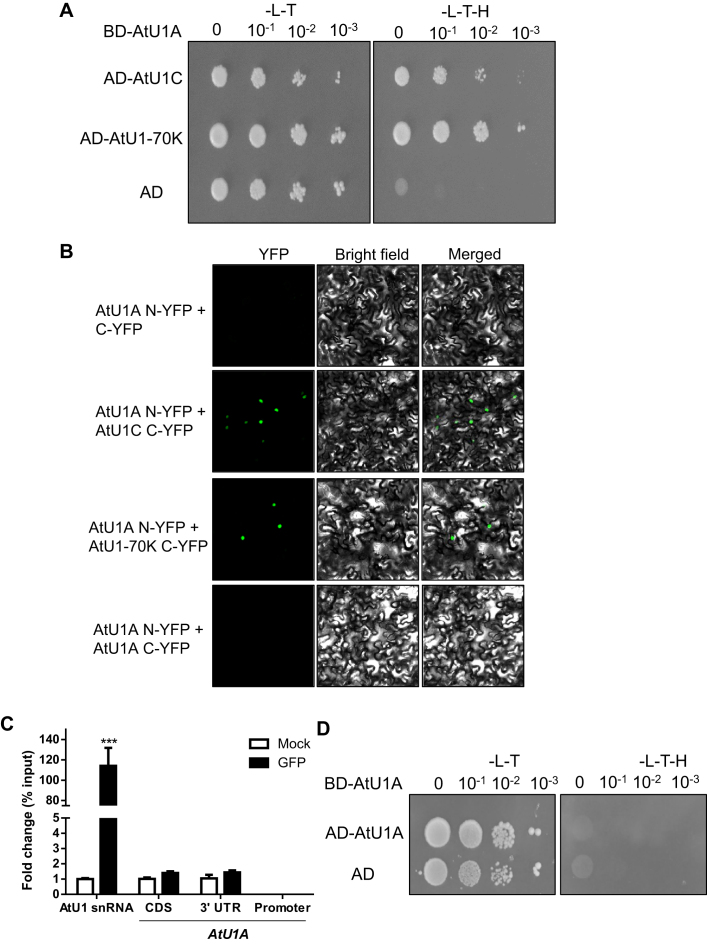
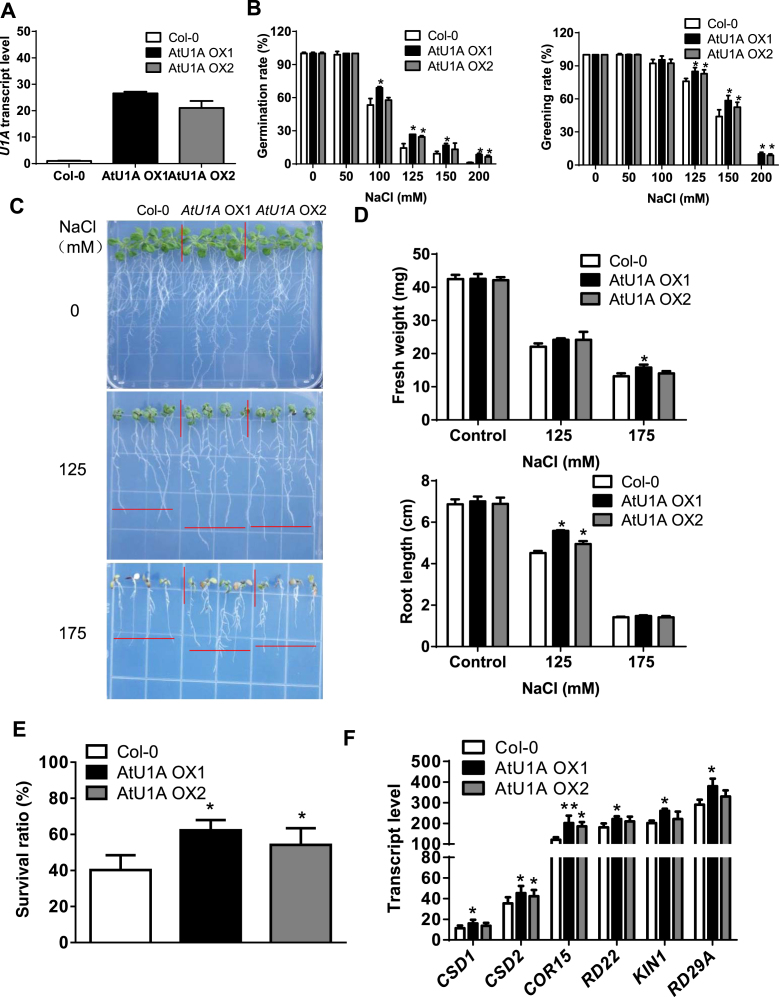
Molecular phylogenetic analysis of AtU1A and its plant and animal orthologs by the maximum likelihood method revealed that AtU1A is closely related to the human U1A and that its orthologs in other plant species are present (Supplementary Figure S4A). It is well documented that the human U1 snRNP complex comprises of three U1-specific proteins (U1A, U1-70K and U1C) and seven ‘Smith antigen’ (Sm) proteins (Sm B/B’, Sm C, Sm D1, Sm D2, Sm D3, Sm F and Sm G), and is required for spliceosome assembly, pre-mRNA splicing and AS by precisely recognizing the 5′ splice sites (32). To determine whether AtU1A in this study is a U1 snRNP-specific protein in planta, we performed a yeast two-hybrid assay. Interestingly, AtU1A interacted with both AtU1-70K and AtU1C in yeast (Figure 3A). To confirm whether these interactions occur in vivo, we performed bimolecular fluorescence complementation (BiFC) assays in tobacco leaves by Agrobacterium infiltration. Consistent with the yeast two-hybrid results, AtU1A interacted with both AtU1-70K and AtU1C in vivo (Figure 3B). These results indicate that AtU1A is coupled with AtU1-70K and AtU1C and that these three proteins form a stable sub-complex of the U1 snRNP in planta. The AtU1A protein plays a critical role in the identification of 5′ splice site by the U1 snRNP through base-paring interactions of the U1 snRNA. We therefore performed RNA-IP analysis to determine whether AtU1A can directly bind to U1 snRNA in Arabidopsis. We took advantage of the AtU1Apro:U1A-GFP transgenic line in the atu1a mutant and applied antibody against GFP to immunoprecipitate AtU1A-GFP. We found that AtU1A could directly bind to U1 snRNA in vivo (Figure 3C). Researchers previously reported that the U1A protein is able to autoregulate its own production by binding to and inhibiting the polyadenylation of its own pre-mRNA in vertebrates (33). The 3′ untranslated region of U1A pre-mRNA contains a 50-nt polyadenylation-inhibitory element RNA that is required for the cooperative binding between two molecules of U1A protein (34). Further study demonstrated that 14 residues of the U1A protein are necessary for homodimerization, RNA binding and inhibition of polyadenylation (35). To determine whether AtU1A protein can dimerize, we used the yeast two-hybrid system to detect protein–protein interaction. Unexpectedly, the AtU1A protein did not homodimerize in yeast, i.e. the affinity between two molecules of the protein was not greater than the affinity for one molecule of the protein with the empty vector (Figure 3D). We confirmed that AtU1A does not homodimerize in vivo with a BiFC assay (Figure 3B). Because two molecules of AtU1A protein do not homodimerize, we reasoned that the plant AtU1Amight not have an additional role in determining the polyadenylation. The RNA-IP analysis showed that AtU1A cannot bind to its own pre-mRNA located in 3′ untranslated region (Figure 3C). This observation is consistent with a previous study that found that the AtU1A proteins do not bind to their own RNAs in vitro (36).
Figure 3.
AtU1A is a U1 snRNP-specific protein in planta. (A) AtU1A interacts with two other U1 snRNP-specific proteins, AtU1-70K and AtU1C in yeast. The pGBK (AtU1A) or pGAD (AtU1-70K and AtU1C) plasmids were co-transformed into yeast and plated on –leucine/-tryptophan/-histidine (-L-T-H) medium. The Empty vector pGAD and AtU1A with pGBK were co-transformed into yeast and used as a negative control. (B) BiFC assays of interactions between AtU1A and AtU1-70K, AtU1A and AtU1C. C-YFP is an empty vector. The different combinations of plasmids were transformed into tobacco epidermal cells and the YFP signals were detected with a confocal microscope. (C) RNA-IP analysis of the association of AtU1A protein with AtU1 snRNA or the pre-mRNA of AtU1A with diverse fragments. (D) AtU1A does not interact with itself in yeast. The BD-AtU1A in pGBK and AD-AtU1A in pGAD plasmids were co-transformed into yeast and plated on –leucine/-tryptophan/-histidine (-L-T-H) medium. The empty vector pGAD and AtU1A in pGBK were co-transformed into yeast and served as a negative control. Asterisks indicate significant differences (***P < 0.001) as determined by a two-tailed paired Student's t-test.
In addition to the U1 snRNA and the common Sm proteins, the human (and most likely the plant) U1 snRNP complex contains three specific proteins: U1-70K, U1 and U1C. We then determined whether transcript levels of other Arabidopsis U1 snRNP proteins as well as AtU1 snRNA were changed in the atu1a mutant. Compared with transcript levels in wild-type plants, the transcript levels of AtU1-70K and AtU1C were not changed in the atu1a mutant under normal or salt stress conditions. However, the transcript level of AtU1 snRNA was significantly reduced in the atu1a mutant under salt stress but not under normal growth conditions (Supplementary Figure S4B).
Mutation of AtU1A leads to genome-wide splicing defects under salt stress
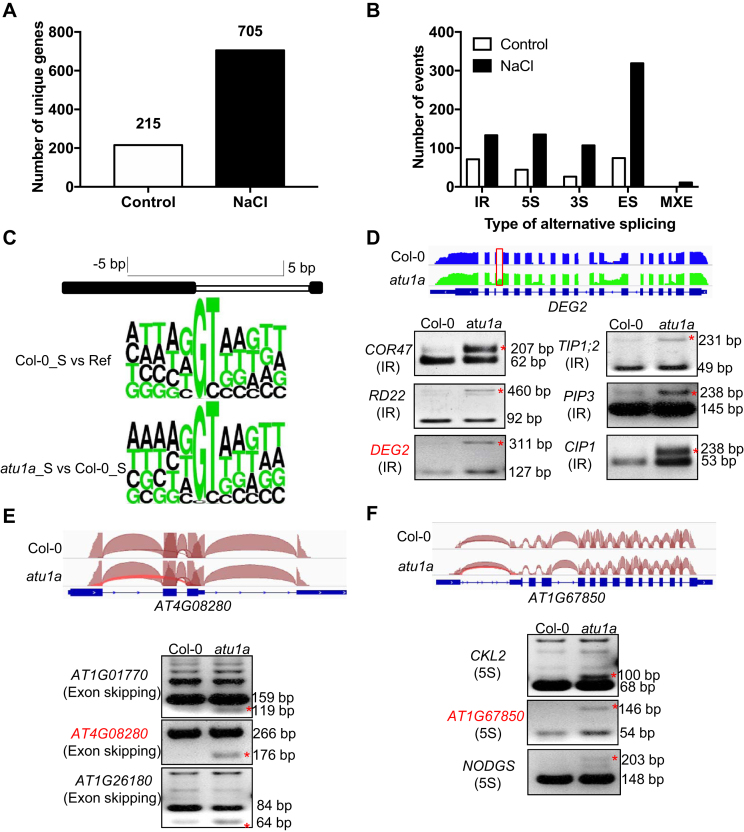
U1 snRNPs play an essential role in defining the 5′ splicing site through base-paring interactions of the 5′ end of the U1 snRNA in eukaryotes. To determine the role of AtU1A in pre-mRNA splicing, we first performed RNA-seq using the Illumina Hi-seq platform in order to examine the global defects in pre-mRNA splicing in the atu1a mutant. We detected aberrant splicing events in only 215 unique genes in the atu1a mutant plants under normal growth conditions (Figure 4A). Our previous studies showed that splicing defects could be enhanced by salt stress in the hos5 (25) or sad1/lsm5 mutant (14). We therefore determined whether salt stress affects pre-mRNA splicing in the atu1a mutant. We generated 65.59 and 52.59 million reads with an average length of 101 bp from the wild-type and atu1a, respectively. Almost 90% of these reads could be unambiguously aligned to the TAIR10 reference genome sequence (fragments per kilo base of exons per million mapped reads- [FPKM] > 0.85). Comparison of the mapped reads against the gene mode (version TAIR10) revealed that ∼95% of the reads were mapped to the exon regions, whereas only ∼3% were mapped to the intergenic regions. Plotting the coverage of reads along each transcribed unit revealed a uniform distribution, with no obvious 3′/5′ bias, which indicates that the cDNA library was of high quality. Interestingly, we detected aberrant splicing events in 705 unique genes in the atu1a mutant under salt stress (Figure 4A). These splicing events belong to five categories: IR, alternative 5′ or 3′ splice site (5S or 3S), exon skipping (ES) and mutually exclusive exon (Figure 4B; Supplementary Tables S1 and 2). Previous studies showed that U1 snRNP-specific proteins are required for 5′ splice site recognitions, and consensus sequences at the 5S are important for accurate splicing of pre-mRNAs in plants (10,37). Our previous studies revealed that the majority of splice sites are conformed to consensus sequences under salt stress (14,15). In the current study, however, the frequency distribution of the nucleotides at 5S from the aberrant splicing events in the atu1a mutant differed from these consensus sequences, with an obvious decrease in the frequency distribution of the base G at +1 positions of the alternative 5′ splice sites (Figure 4C). This suggests that, as is the case in other eukaryotes, the AtU1A protein in Arabidopsis functions in alternative 5′ splice site recognition (37,38).
Figure 4.
Comparison of global AS events between the wild-type and the atu1a mutant under salt stress. (A) Summary of genes whose transcripts were abnormally spliced in atu1a as determined by RNA-seq experiments. (B) Genes with defects in different types of AS patterns in atu1a as determined by RNA-seq experiments. (C) The frequency distribution of nucleotides at consensus 5′ alternative splice sites. Sequence logos illustrate consensus sequences for 5′ splice sites in the wild-type (top logos) and atu1a mutant (bottom logos) under salt stress. Ref is defined as the Arabidopsis reference genome of TAIR10. (D–F) Representative AS events visualized by the Integrative Genomics Viewer browser (IGV) and validation of AS by RT-PCR analysis between the wild-type and atu1a mutant under salt stress (150 mM NaCl for 3 h). For the IGV visualization, the exon–intron structure of each gene is displayed at the bottom of each panel. The arcs generated by IGV browser indicate splice junction reads that support the junctions. The IR, exon-skipping and 5′ alternative sites are shown in (D), (E) and (F), respectively. These events are highlighted by asterisks.
To validate the differential AS data revealed in the RNA-seq experiments, we selected and assessed representative splicing events of IR, 5S and ES by using RT-PCR analysis with primers flanking these events in the salt-treated atu1a mutant. We found that the aberrant splicing events were readily detected in the salt-treated atu1a mutant, whereas they were not detected or only weakly detected in the wild-type plants (Figure 4E and F; Supplementary Figure S5). When we attempted to detect above selected aberrant splicing events under normal growth conditions, no obvious difference was evident between the wild-type and the atu1a mutant plants (Supplementary Figure S6). These results indicate that AtU1A is mainly involved in pre-mRNA splicing in the presence of but not in the absence of salt stress.
Gene ontology analysis of the genes with aberrant 5′ splice site recognition in the atu1a mutant revealed a striking enrichment in the response-to-abiotic-stress categories (Supplementary Figure S7A). We also analyzed these genes with Genevestigator and found that they are closely associated with the response to salt stress (Supplementary Figure S7B). These results suggest that AtU1A plays critical roles in regulating splicing of stress-responsive genes under salt stress.
Mutation of AtU1A impairs mRNA export under salt stress
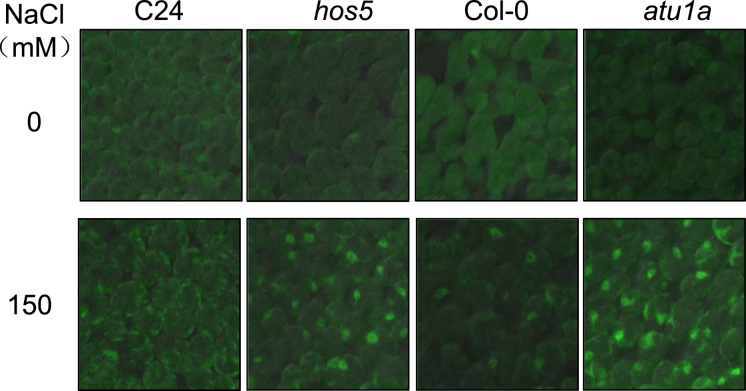
In view of the connections between splicing and mRNA export, we examined mRNA accumulation in the nuclei of the atu1a mutant. Leaf samples from seedlings with or without NaCl treatment were fixed by formaldehyde and hybridized with an oligo(dT) probe labeled with Alexa Fluor 488 (polyA RNA). In agreement with the differential AS results in the atu1a mutant, Fluor 488 signal strength did not significantly differ between the wild-type and the atu1a mutant under normal growth conditions. However, accumulation of polyA RNA was increased in the wild-type and atu1a mutant under salt stress, and the accumulation was greater in the atu1a mutant (Figure 5). The defective mRNA export phenotypes of hos5 versus C24 (background of the hos5 mutant) in the presence and absence of salt stress indicated that our experimental conditions were similar to those reported previously (25). Together, these data suggest that AtU1A is involved in mRNA export under salt stress.
Figure 5.
Impairment of mRNA export in the atu1a mutant under salt stress. The 7-day-old seedlings of C24, hos5, Col-0 and atu1a grown on MS medium were transferred to MS medium containing 0 or 150 mM NaCl and allowed to grow for an additional 5 h. PolyA in situ hybridization analysis was performed and polyA signals were detected with a confocal microscope.
The atu1a mutant over-accumulates ROS under salt stress
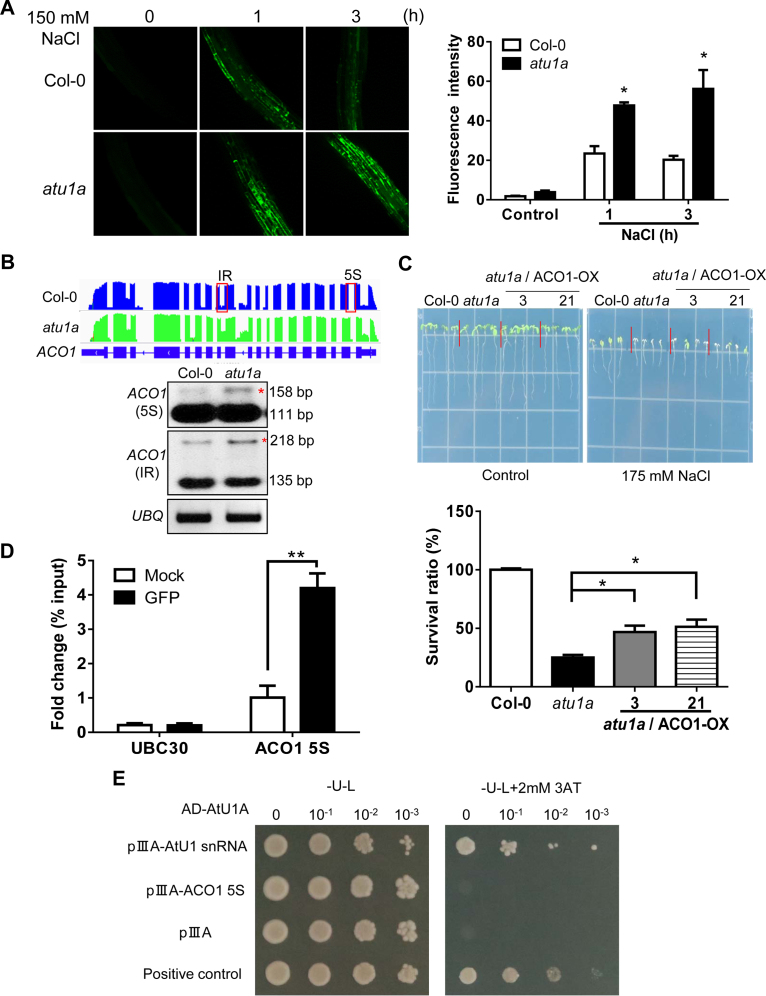
Salt stress can cause over-production of ROS and thereby affects salt stress tolerance in plants. We determined whether the increased salt sensitivity of the atu1a mutant is due to over-accumulation of ROS. The fluorescent dye 5-(and 6)-chloromethyl-2′7′-dichlorodihydrofluo resceindiacetate acetyl ester (CM-H2DCFDA) was used to visualize and quantify total ROS in the roots. Under normal growth conditions, ROS fluorescence signals did not significantly differ in the atu1a mutant versus the wild-type (Figure 6A). In response to 150 mM NaCl, however, ROS levels were higher in the atu1a mutant plants than the wild-type plants and the ROS levels also remained high for a longer period in the mutant than in the wild-type plants (Figure 6A). These results suggested that the increased salt sensitivity in the atu1a mutant might be caused by over-accumulation of ROS. We further tested the sensitivity of atu1a mutants to two exogenous chemicals, H2O2 or MV, that can lead to an increase in the generation of toxic superoxide free radicals in plants. We found that primary root growth was more sensitive to H2O2 (1 mM) or MV (0.2 μM) for of the atu1a mutant than for the wild-type (Supplementary Figure S8A and B).
Figure 6.
The atu1a mutant plants accumulate more ROS than wild-type plants under salt and have abnormally AS transcripts of ACO1. (A) Representative images of ROS production (determined by staining with the fluorescent dye H2DCF-DA) in root tips and root elongation zones after treatment with 150 mM NaCl for the indicated times. Quantification of ROS production based on fluorescence pixel intensity (right panel). Error bars indicate SD (n = 30 [number of seedlings]). (B) ACO1 is mis-spliced in the atu1a plants under salt stress. Shown are results from RNA-seq (graphic display in the upper panel) and RT-PCR analysis (lower panel). (C) Growth of transgenic lines overexpressing ACO1 in the atu1a mutant in response to salt (upper panel) and survival rates of these plants under salt stress (lower panel). (D) RNA-IP analysis of the association of AtU1A with the pre-mRNA of ACO1 5S, and UBC30. (E) Direct selection for RNA–protein interaction. The pAD-AtU1Aand plllA/MS2-2 (AtU1 snRNA or pre-mRNA of ACO1 5S) plasmids were co-transformed into L40-coat yeast and plated on a medium lacking leucine and uracil and containing 2 mM 3-aminotriazole. The empty vector plllA/MS2-2 and pAD-AtU1A were co-transformed into yeast and used as a negative control. The plllA/IRE-MS2 vector and pAD-IRP were co-transformed into yeast and used as a positive control. Error bars indicate SD (n = 16 in [C], 3 in [D]). Asterisks indicate significant differences (*P < 0.05, **P < 0.01) as determined by a two-tailed paired Student's t-test.
Because AtU1A is one of the U1 snRNP-specific proteins involved in alternative pre-mRNA splicing in other eukaryotes (39), we reasoned that ROS-related genes might be mis-spliced under salt stress. Notably, we found that the CSD1 gene in the atu1a mutant has an IR event and that the ACO1 gene in the atu1a mutant has two types of splicing defects including IR and alternative 5′ splice selection (Figure 6B and Supplementary Figure S9A). CSD1 encodes a cytosolic copper/zinc superoxide dismutase that can detoxify superoxide radicals (40). ACO1 encodes an aconitase that can catalyze the conversion of citrate to isocitrate through a cis-aconitate intermediate and possibly functions in the tricarboxylic acid cycle. ACO1 protein can specifically bind to the 5′-UTR of CSD2 (it encodes a cytosolic copper/zinc superoxide dismutase that can detoxify superoxide radicals) transcript and affect CSD2 transcript level, and thus it may contribute to the response to oxidative stress (41). To determine whether these ROS detoxification-related enzymes encoding genes are critical for the salt stress-sensitivity phenotype of the atu1a mutant, we generated transgenic plants that overexpress ACO1 (ACO1 OX) or CSD1 (CSD1 OX) in the atu1a mutant background (Figure 6C; Supplementary Figure S9B and C). We found that overexpression of ACO1 in the atu1a mutant could partially restore the salt sensitive phenotype of the atu1a mutant (Figure 6C and Supplementary Figure S9E), while overexpression of CSD1 in atu1a only slightly rescued its salt-sensitive phenotype (Supplementary Figure S9B and C). These results indicate that in addition to ACO1 and CSD1, aberrant splicing patterns in other AtU1A target genes are contributing to the increased salt sensitivity of the atu1a mutant. Because splicing of pre-mRNAs requires recruitment of pre-mRNAs to the spliceosome, we determined whether the pre-mRNAs of CSD1 or ACO1 associate with AtU1A in planta. ACO1 pre-mRNAs, but not the pre-mRNAs of CSD1, were detected in the immunoprecipitation samples from the AtU1Apro:AtU1A-GFP transgenic line (Figure 6D and Supplementary Figure S9D). To clarify the function of AtU1A acting in isolation or as a part of a U1 snRNP complex, we used a yeast three-hybrid system in order to detect RNA–protein interactions in vitro (42). We found that AtU1A protein can bind to the AtU1 snRNA but not to the 5′S of ACO1 pre-mRNA in yeast (Figure 6E). The results support a role for the AtU1A-containing spliceosome in recognizing the 5S of intron in planta. These results also suggest that AtU1A uses more than one pathways to control salt-responsive splicing. Previous study found that U1A and U2B'' are functionally redundant in worms (43). To clarify whether the plant orthologs of U1A and U2B'' are functionally redundant in planta, we carried out additional yeast three-hybrid experiments. Consistent with a previous study (44), we showed that the plant U2B'' cannot bind to the U1 snRNA in vitro suggesting that they may not have redundant function in plant (Supplementary Figure S10). Overall, these results suggest that splicing defects in ROS detoxification-related genes contribute to the overall increased salt-sensitive phenotype of the atu1a plants.
Overexpression of AtU1A increases plant salt tolerance
Because AtU1A is required for salt stress tolerance and salt stress-responsive gene expression, we generated Arabidopsis transgenic plants expressing AtU1A under the control of the 35S promoter to investigate whether overexpression of AtU1A could improve plant performance under salt stress (Figure 7A). We examined the phenotypes of these AtU1A overexpression lines during seed germination and cotyledon greening under salt stress. Under normal growth conditions, seed germination rates of the wild-type and AtU1A overexpression lines did not significantly differ. Under salt stress, however, seed germination rates were higher for AtU1A overexpression lines for the wild-type (Figure 7B). Furthermore, cotyledon greening was less inhibited by salt stress for the seedlings of AtU1A overexpression lines than for the wild-type seedlings (Figure 7B). On MS medium, shoot and root growth were less inhibited by salt stress for the AtU1A-overexpressing plants than for the wild-type plants (Figures 7C and D). To determine salt tolerance in soil, we treated 2-week-old soil-grown plants with 300 mM NaCl for additional 14 days. Although the salt stress treatment reduced the survival of both kinds of plants, survival was much higher for the AtU1A overexpression plants than for the wild-type plants (Figure 7C and E). To determine whether transcript levels of genes related to salt or oxidative stress were changed by overexpression of AtU1A, we used qRT-PCR to quantify the expression of CSD1, CSD2, COR15, RD22, KIN1 and RD29A. We found that the transcript levels of these stress-related genes were moderately higher in the AtU1A overexpression lines than in the wild-type plants under salt stress (Figure 7F). Taken together, these results indicate that overexpression of AtU1A can increase plant tolerance to salt stress.
Figure 7.
Overexpression of AtU1A confers salt tolerance. (A) Transcript level of AtU1A in two AtU1A overexpression lines. (B) Seed germination and seedling greening of wild-type and two AtU1A overexpression plants under salt stress. (C) Growth of the wild-type (Col-0) and two AtU1A overexpression lines in response to salt stress. The 4-day-old seedlings grown on MS medium were transferred to MS medium containing different levels of NaCl and were allowed to grow for additional 10 days. Representative plants were photographed. (D) Fresh weights (upper panel) and root elongation (lower panel) of plants shown in (C). (E) Survival (shown as a percentage of survival on normal medium) ratio of soil-grown wild-type and two AtU1A overexpression plants under salt stress. The 14-day-old seedlings grown in soil were irrigated with 300 mM NaCl and allowed to grow for additional 10 days. (F) Transcript levels of ROS detoxification- or stress-related genes in wild-type and two AtU1A overexpression plants under salt stress. UBQ10 was used as an internal control in [A] and [F]. Error bars represent SD (n = 3 in [A and F], 100 [number of seeds or seedlings] in B, 16 in D, 48 in E). Asterisks indicate significant differences (*P < 0.05, **P < 0.01) as determined by a two-tailed paired Student's t-test.
DISCUSSION
In this study, we identified a knock-out mutant of AtU1A that was more sensitive to salt stress than the wild-type, and we found that the increased salt sensitivity in atu1a mutant could be completely rescued by re-introducing the wild-type AtU1A gene into the mutant. Previous studies showed that U1A primarily functions as a component of the U1 snRNP complex, which is required for splicing of pre-mRNAs in mammals and yeast (26,31). Our study also revealed that AtU1A is one of the U1 snRNP-specific proteins in Arabidopsis that is closely associated with other two U1 snRNP-specific particles, AtU1-70K and AtU1C. Mutation of AtU1A led to a defect in the AS of a portion of pre-mRNAs encoded by the Arabidopsis genome, and this defect in splicing of pre-mRNAs was much more abundant under salt stress than under normal growth conditions. In response to salt stress, the atu1a mutant accumulated more ROS than the wild-type suggesting that AtU1A is required for suppress ROS levels. We found that AtU1A is required for salt tolerance because it regulates the splicing of pre-mRNAs of ROS-related genes (ACO1 and CSD1) in Arabidopsis. The AtU1A is able to binding to the pre-mRNA of ACO1 through the U1 snRNA but this binding is not evident for the pre-mRNA of CSD1. The importance of AtU1A protein in salt stress responses was supported by the increased salt tolerance of AtU1A overexpression lines in Arabidopsis.
Splicing of pre-mRNAs is a key step in the regulation of gene expression, transcriptome plasticity and proteome diversity in eukaryotes (45,46). Splicing of introns requires the recognition and subsequent cleavage at the 5′ donor and 3′ acceptor sites in genes as mediated by the spliceosome (11,47). The spliceosome consists of five snRNPs and over 200 additional proteins (39). The core particles of the U1, U2, U4 and U5 snRNPs are formed by Sm proteins, whereas the U6 snRNP contains the related LSM2 to LSM8 proteins. The U1 snRNP particles are involved in the first step of spliceosome formation, in which it binds to the 5′ splice site of the pre-mRNAs. In addition to the U1 snRNA and the common Sm proteins, the U1 snRNP particles contain three specific proteins: U1-70K, U1A and U1C. U1-70K and U1C strongly rely on each other to ensure correct 5′ splice site recognition because the stable incorporation of U1C into the U1 snRNP requires the presence of U1-70K, and the presence of U1C is necessary for the interaction of U1-70K with SRSF1 (ASD/SF2), which triggers U1 snRNP binding to the 5′ splice site (48). Our study reveals that AtU1A interacts with AtU1-70K and AtU1C suggesting their interdependent role in the formation of the U1 snRNP (Figure 3). As a component of the U1 snRNP, U1A has two functions in the snRNP biogenesis pathway (33,49). First, U1A binds directly to stem-loop 2 of the U1 snRNA and such binding is required for pre-mRNA splicing (26–28). Consistent with the previous reports (31,36), we found that AtU1A can directly bind to AtU1 snRNA both in vitro and in vivo (Figures 3D and 6E), and global RNA-seq study revealed a novel role of AtU1A during AS, primarily during 5′ splice site recognition under salt stress (Figure 4). The nucleotide sequences of this novel 5′ splice-site recognition diverge from consensus sequences, indicating that AtU1A defines correct 5′ splice recognition sites of pre-mRNAs in plants (Figure 4C). Moreover, a rare nucleotide C at +1 positions of the alternative 5′ splice sites observed in the atu1a mutant may at least partially explain the differentially spliced transcripts in the mutant plants. A second function of U1A is that it modulates polyadenylation by autoregulating its expression by blocking polyadenylation of its own mRNA in the 3′ untranslated region. Tandem-affinity purification of U1A identified a large collection of pre-mRNA and RNA processing factors, as well as the transcriptional machinery (50–53), suggesting a role of U1A in coupling 3′ processing and splicing. However, plant U1A could not bind to its own mRNA (Figure 3D) suggesting that plant U1A is not involved in polyadenylation (36). Thus, it is possible that snRNP-free AtU1A protein does not tightly regulate polyadenylation or that such regulation is achieved by a different mechanism in plants.
Because regulation of gene expression is important for the adaptation and survival of plants under environment stress conditions, it has been the focus of many previous studies (1,54–57). Recent genome-wide studies have revealed that pre-mRNA splicing is affected by development and stress treatment (13,15,58), and that activities of spliceosome-like proteins, such as pre-mRNA splicing as mediated by LSM5 and SKIP, are important for plant stress tolerance (14,59). A defect in the homolog of the LSM5 protein, SAD1/LSM5, leads to an increased sensitivity to drought and ABA (60). The sad1/lsm5 mutant also has reduced levels of U6 snRNA and increased levels of unspliced pre-mRNAs, suggesting that SAD1/LSM5 may contribute to U6 stability in pre-mRNA splicing. Similarly, the lsm4 mutant is hypersensitive to salt and ABA and shows mis-splicing of some stress-related genes (61). The STABILIZED1 (STA1) protein, which is similar to the human U5 small ribonucleoprotein-associated 102-kD protein, is required for the response to cold stress in Arabidopsis (62). A component of the U4/U6 snRNP, RDM16, which encodes a pre-mRNA-splicing factor 3 and is involved in pre-mRNA splicing in planta. Mutation of RDM16 resulted in hypersensitivity to salt stress and ABA, and transcriptome analysis revealed a novel role of RDM16 in the RNA-directed DNA methylation pathway (63). RBM25 is a novel splicing factor that modulates the response to ABA during and after seed germination by recognizing the pre-mRNAs of HAB1 and many other genes (64,65). Mutants defective in the subunits of the cap-binding complex, At-CBP20/80, which interact with the m7G cap of pre-mRNAs, showed splicing defects for genes involved in proline and sugar metabolism (66). Mutation in a spliceosomal protein, PRPF31, reduces tolerance to low temperature. AtPRP31, which regulates the formation of the U4/U6.U5 snRNP, is required for splicing of cold-induced genes, especially under cold stress (67,68).
Our study indicates that AtU1A protein is responsible for recognizing the 5′ splice site in the initial step of pre-mRNA splicing, and that mutation in AtU1A results in defective splicing of many stress-responsive genes. Moreover, overexpression of AtU1A increases plant tolerance to salt stress, indicating that AtU1A and the spliced genes that it regulates are essential for salt tolerance (Figure 8). Database searches revealed that close orthologs of AtU1A are present in many other plant species (Supplementary Figure S4). These findings suggest that the spliceosome complex is conserved across plant species. Perhaps these homologs have functions similar to those of AtU1A in response to salt stress. Because overexpression of AtU1A increases salt tolerance in Arabidopsis, manipulation of AtU1A (or its close orthologs) levels in corps may also increase crop salt tolerance.
Figure 8.
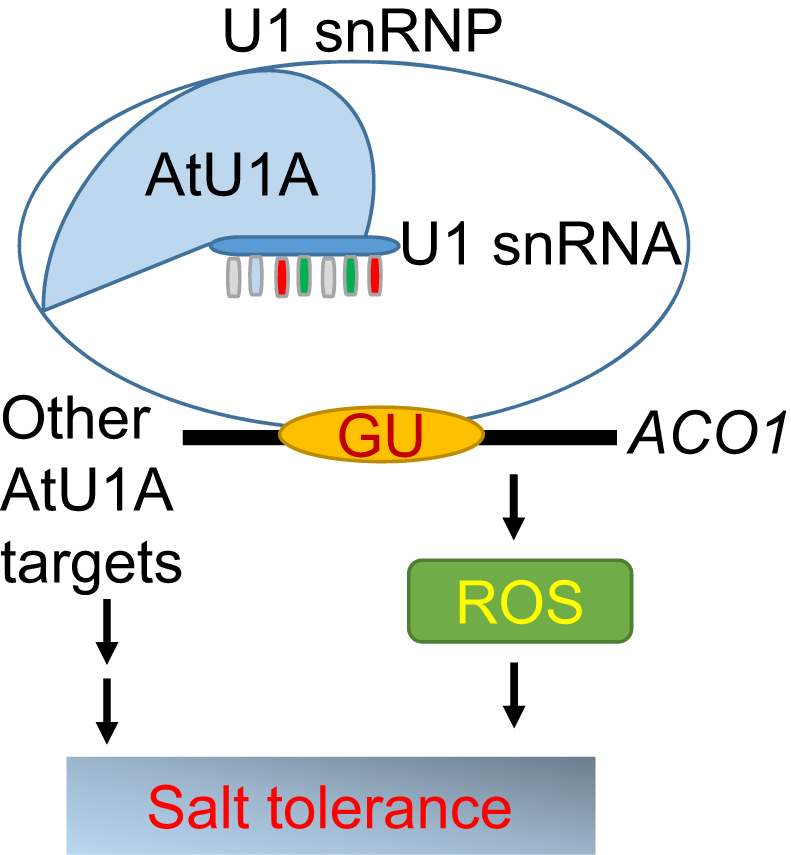
A model of AtU1A function in pre-mRNA splicing and salt stress tolerance. This schematic illustration shows how the 5′ splice site in the Arabidopsis aconitase (ACO1) pre-mRNA can be sequentially recognized by the U1 snRNA that is associated with the U1 snRNP-specific protein AtU1A. Splicing of ACO1 pre-mRNA is important for maintaining proper ROS levels in the cell and for contributing to salt tolerance. Other genes whose pre-mRNA splicing are under the control of AtU1A may also contribute to salt tolerance.
DATA AVAILABILITY
The sequencing data of RNA-seq experiments is available in the SRA database (Accession number: SRS1498194).
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the ABRC for providing the T-DNA insertion lines. We thank Prof. Marvin Wickens (University of Wisconsin-Madison) for providing the yeast three-hybrid system.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31670250 to Z.W.]; Natural Science Foundation of Hainan Province [20163041 to Z.W.]; Hainan University Startup Fund [KYQD1562 to Z.W.]; YNTC [YNTC-2016YN22 to H.X.]; KAUST Faculty Baseline Funds [#BAS/1/1007-01-01 to L.M.X.]; National Key Technology Support Program [2015BAD01B02 to Y.H.L., X.Y.J.]; National Science Foundation [MCB0950242 to J.H.Z.]. Funding for open access charge: Hainan University Startup Fund [KYQD1562].
Conflict of interest statement. None declared.
REFERENCES
- 1. Zhu J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003; 6:441–445. [DOI] [PubMed] [Google Scholar]
- 2. Chinnusamy V., Zhu J., Zhu J.K.. Salt stress signaling and mechanisms of plant salt tolerance. Genet. Eng. (N Y). 2006; 27:141–177. [DOI] [PubMed] [Google Scholar]
- 3. Tang X., Mu X., Shao H., Wang H., Brestic M.. Global plant-responding mechanisms to salt stress: physiological and molecular levels and implications in biotechnology. Crit. Rev. Biotechnol. 2015; 35:425–437. [DOI] [PubMed] [Google Scholar]
- 4. Hasegawa P.M., Bressan R.A., Zhu J.K., Bohnert H.J.. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000; 51:463–499. [DOI] [PubMed] [Google Scholar]
- 5. Apel K., Hirt H.. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004; 55:373–399. [DOI] [PubMed] [Google Scholar]
- 6. Golldack D., Li C., Mohan H., Probst N.. Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 2014; 5:151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kreps J.A., Wu Y., Chang H.-S., Zhu T., Wang X., Harper J.F.. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002; 130:2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zeller G., Henz S.R., Widmer C.K., Sachsenberg T., Ratsch G., Weigel D., Laubinger S.. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J. 2009; 58:1068–1082. [DOI] [PubMed] [Google Scholar]
- 9. Feng J., Li J., Gao Z., Lu Y., Yu J., Zheng Q., Yan S., Zhang W., He H., Ma L. et al. SKIP confers osmotic tolerance during salt stress by controlling alternative gene splicing in Arabidopsis. Mol. Plant. 2015; 8:1038–1052. [DOI] [PubMed] [Google Scholar]
- 10. Reddy A.S. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007; 58:267–294. [DOI] [PubMed] [Google Scholar]
- 11. Lee Y., Rio D.C.. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 2015; 84:291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kornblihtt A.R., Schor I.E., Allo M., Dujardin G., Petrillo E., Munoz M.J.. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013; 14:153–165. [DOI] [PubMed] [Google Scholar]
- 13. Staiger D., Brown J.W.S.. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell. 2013; 25:3640–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cui P., Zhang S., Ding F., Ali S., Xiong L.. Dynamic regulation of genome-wide pre-mRNA splicing and stress tolerance by the Sm-like protein LSm5 in Arabidopsis. Genome Biol. 2014; 15:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ding F., Cui P., Wang Z., Zhang S., Ali S., Xiong L.. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics. 2014; 15:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ben Chaabane S., Liu R., Chinnusamy V., Kwon Y., Park J.H., Kim S.Y., Zhu J.K., Yang S.W., Lee B.H.. STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res. 2013; 41:1984–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deng X., Gu L., Liu C., Lu T., Lu F., Lu Z., Cui P., Pei Y., Wang B., Hu S. et al. Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:19114–19119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guan Q., Wu J., Zhang Y., Jiang C., Liu R., Chai C., Zhu J.. A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell. 2013; 25:342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cui P., Xiong L.. Environmental stress and pre-mRNA splicing. Mol. Plant. 2015; 8:1302–1303. [DOI] [PubMed] [Google Scholar]
- 20. Lichtenthaler H., Wellburn A.. Determinations of total carotenoids and chlorophyll a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983; 11:591–592. [Google Scholar]
- 21. Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H.. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006; 1:641–646. [DOI] [PubMed] [Google Scholar]
- 22. Gong Z., Dong C.H., Lee H., Zhu J., Xiong L., Gong D., Stevenson B., Zhu J.K.. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell. 2005; 17:256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shen S., Park J.W., Huang J., Dittmar K.A., Lu Z.X., Zhou Q., Carstens R.P., Xing Y.. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res. 2012; 40:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thomsen M.C., Nielsen M.. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 2012; 40:W281–W287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen T., Cui P., Chen H., Ali S., Zhang S., Xiong L.. A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet. 2013; 9:e1003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scherly D., Boelens W., van Venrooij W.J., Dathan N.A., Hamm J., Mattaj I.W.. Identification of the RNA binding segment of human U1 A protein and definition of its binding site on U1 snRNA. EMBO J. 1989; 8:4163–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lutz-Freyermuth C., Keene J.D.. The U1 RNA-binding site of the U1 small nuclear ribonucleoprotein (snRNP)-associated A protein suggests a similarity with U2 snRNPs. Mol. Cell. Biol. 1989; 9:2975–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lutz-Freyermuth C., Query C.C., Keene J.D.. Quantitative determination that one of two potential RNA-binding domains of the A protein component of the U1 small nuclear ribonucleoprotein complex binds with high affinity to stem-loop II of U1 RNA. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:6393–6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Law M.J., Lee D.S., Lee C.S., Anglim P.P., Haworth I.S., Laird-Offringa I.A.. The role of the C-terminal helix of U1A protein in the interaction with U1hpII RNA. Nucleic Acids Res. 2013; 41:7092–7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J.U.. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005; 37:501–506. [DOI] [PubMed] [Google Scholar]
- 31. Lorkovic Z.J., Barta A.. Role of Cajal bodies and nucleolus in the maturation of the U1 snRNP in Arabidopsis. PLoS One. 2008; 3:e3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ast G. How did alternative splicing evolve. Nat. Rev. Genet. 2004; 5:773–782. [DOI] [PubMed] [Google Scholar]
- 33. Boelens W.C., Jansen E.J., van Venrooij W.J., Stripecke R., Mattaj I.W., Gunderson S.I.. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell. 1993; 72:881–892. [DOI] [PubMed] [Google Scholar]
- 34. van Gelder C.W., Gunderson S.I., Jansen E.J., Boelens W.C., Polycarpou-Schwarz M., Mattaj I.W., van Venrooij W.J.. A complex secondary structure in U1A pre-mRNA that binds two molecules of U1A protein is required for regulation of polyadenylation. EMBO J. 1993; 12:5191–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klein Gunnewiek J.M.T., Hussein R.I., van Aarssen Y., Palacios D., de Jong R., van Venrooij W.J., Gunderson S.I.. Fourteen residues of the U1 snRNP-specific U1A protein are required for homodimerization, cooperative RNA binding, and inhibition of polyadenylation. Mol. Cell. Biol. 2000; 20:2209–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simpson G.G., Clark G.P., Rothnie H.M., Boelens W., van Venrooij W., Brown J.W.. Molecular characterization of the spliceosomal proteins U1A and U2B" from higher plants. EMBO J. 1995; 14:4540–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu Y., Maroney P.A., Denker J.A., Zhang X.H.F., Dybkov O., Lührmann R., Jankowsky E., Chasin L.A., Nilsen T.W.. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell. 2008; 135:1224–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Puig O., Gottschalk A., Fabrizio P., Seraphin B.. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev. 1999; 13:569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wahl M.C., Will C.L., Luhrmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009; 136:701–718. [DOI] [PubMed] [Google Scholar]
- 40. Guan Q., Lu X., Zeng H., Zhang Y., Zhu J.. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013; 74:840–851. [DOI] [PubMed] [Google Scholar]
- 41. Moeder W., Del Pozo O., Navarre D.A., Martin G.B., Klessig D.F.. Aconitase plays a role in regulating resistance to oxidative stress and cell death in Arabidopsis and Nicotiana benthamiana. Plant Mol. Biol. 2007; 63:273–287. [DOI] [PubMed] [Google Scholar]
- 42. SenGupta D.J., Zhang B., Kraemer B., Pochart P., Fields S., Wickens M.. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saldi T., Wilusz C., MacMorris M., Blumenthal T.. Functional redundancy of worm spliceosomal proteins U1A and U2B''. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:9753–9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Terzi L.C., Simpson G.G.. Arabidopsis RNA immunoprecipitation. Plant J. 2009; 59:163–168. [DOI] [PubMed] [Google Scholar]
- 45. Jurica M.S., Moore M.J.. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003; 12:5–14. [DOI] [PubMed] [Google Scholar]
- 46. Nilsen T.W., Graveley B.R.. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010; 463:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Seraphin B., Rosbash M.. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989; 59:349–358. [DOI] [PubMed] [Google Scholar]
- 48. Du H., Rosbash M.. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature. 2002; 419:86–90. [DOI] [PubMed] [Google Scholar]
- 49. Gunderson S.I., Beyer K., Martin G., Keller W., Boelens W.C., Mattaj L.W.. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994; 76:531–541. [DOI] [PubMed] [Google Scholar]
- 50. Brody Y., Neufeld N., Bieberstein N., Causse S.Z., Bohnlein E.M., Neugebauer K.M., Darzacq X., Shav-Tal Y.. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 2011; 9:e1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Das R., Yu J., Zhang Z., Gygi M.P., Krainer A.R., Gygi S.P., Reed R.. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol. Cell. 2007; 26:867–881. [DOI] [PubMed] [Google Scholar]
- 52. Lewis J.D., Izaurralde E., Jarmolowski A., McGuigan C., Mattaj I.W.. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996; 10:1683–1698. [DOI] [PubMed] [Google Scholar]
- 53. Lutz C.S., Murthy K.G., Schek N., O’Connor J.P., Manley J.L., Alwine J.C.. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996; 10:325–337. [DOI] [PubMed] [Google Scholar]
- 54. Kawasaki S., Borchert C., Deyholos M., Wang H., Brazille S., Kawai K., Galbraith D., Bohnert H.J.. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell. 2001; 13:889–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhu J.-K. Plant salt tolerance. Trends Plant Sci. 2001; 6:66–71. [DOI] [PubMed] [Google Scholar]
- 56. Julkowska M.M., Testerink C.. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 2015; 20:586–594. [DOI] [PubMed] [Google Scholar]
- 57. Guan Q., Wu J., Yue X., Zhang Y., Zhu J.. A nuclear calcium-sensing pathway is critical for gene regulation and salt stress tolerance in Arabidopsis. PLoS Genet. 2013; 9:e1003755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Filichkin S.A., Priest H.D., Givan S.A., Shen R., Bryant D.W., Fox S.E., Wong W.K., Mockler T.C.. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010; 20:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Feng J., Li J., Gao Z., Lu Y., Yu J., Zheng Q., Yan S., Zhang W., He H., Ma L. et al. SKIP confers osmotic tolerance during salt stress by controlling alternative gene splicing in Arabidopsis. Mol. Plant. 2015; 8:1038–1052. [DOI] [PubMed] [Google Scholar]
- 60. Xiong L., Gong Z., Rock C.D., Subramanian S., Guo Y., Xu W., Galbraith D., Zhu J.K.. Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell. 2001; 1:771–781. [DOI] [PubMed] [Google Scholar]
- 61. Zhang Z., Zhang S., Zhang Y., Wang X., Li D., Li Q., Yue M., Li Q., Zhang Y.E., Xu Y. et al. Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell. 2011; 23:396–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lee B.H., Kapoor A., Zhu J., Zhu J.K.. STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell. 2006; 18:1736–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Huang C.F., Miki D., Tang K., Zhou H.R., Zheng Z., Chen W., Ma Z.Y., Yang L., Zhang H., Liu R. et al. A pre-mRNA-splicing factor is required for RNA-directed DNA methylation in Arabidopsis. PLoS Genet. 2013; 9:e1003779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang Z., Ji H., Yuan B., Wang S., Su C., Yao B., Zhao H., Li X.. ABA signalling is fine-tuned by antagonistic HAB1 variants. Nat. Commun. 2015; 6:8138. [DOI] [PubMed] [Google Scholar]
- 65. Zhan X., Qian B., Cao F., Wu W., Yang L., Guan Q., Gu X., Wang P., Okusolubo T.A., Dunn S.L. et al. An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat. Commun. 2015; 6:8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Raczynska K.D., Simpson C.G., Ciesiolka A., Szewc L., Lewandowska D., McNicol J., Szweykowska-Kulinska Z., Brown J.W., Jarmolowski A.. Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2010; 38:265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Du J.L., Zhang S.W., Huang H.W., Cai T., Li L., Chen S., He X.J.. The splicing factor PRP31 is involved in transcriptional gene silencing and stress response in Arabidopsis. Mol. Plant. 2015; 8:1053–1068. [DOI] [PubMed] [Google Scholar]
- 68. Schaffert N., Hossbach M., Heintzmann R., Achsel T., Luhrmann R.. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 2004; 23:3000–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data of RNA-seq experiments is available in the SRA database (Accession number: SRS1498194).