Abstract
Recent studies have reported the emerging role of microRNAs (miRNAs) in human cancers. We systematically characterized miRNA expression and editing in the human brain, which displays the highest number of A-to-I RNA editing sites among human tissues, and in de novo glioblastoma brain cancer. We identified 299 miRNAs altered in their expression and 24 miRNAs differently edited in human brain compared to glioblastoma tissues. We focused on the editing site within the miR-589–3p seed. MiR-589–3p is a unique miRNA almost fully edited (∼100%) in normal brain and with a consistent editing decrease in glioblastoma. The edited version of miR-589–3p inhibits glioblastoma cell proliferation, migration and invasion, while the unedited version boosts cell proliferation and motility/invasion, thus being a potential cancer-promoting factor. We demonstrated that the editing of this miRNA is mediated by ADAR2, and retargets miR-589–3p from the tumor-suppressor PCDH9 to ADAM12, which codes for the metalloproteinase 12 promoting glioblastoma invasion. Overall, our study dissects the role of a unique brain-specific editing site within miR-589–3p, with important anticancer features, and highlights the importance of RNA editing as an essential player not only for diversifying the genomic message but also for correcting not-tolerable/critical genomic coding sites.
INTRODUCTION
Post-transcriptional modifications, such as RNA editing, are emerging as important players in RNA/protein diversification, microRNA (miRNA) modulation, cell differentiation and are determinants of several human diseases (1,2). A-to-I RNA editing, the most abundant type of RNA editing in mammals, is a tightly controlled molecular process that changes nucleotide sequences of targeted double-stranded RNAs (dsRNAs) by the deamination of the canonical Adenosine (A) nucleic acid base to the specialized RNA-base Inosine (I). A-to-I editing occurs in both coding and non-coding RNA molecules in all human tissues and targets adenosines at different levels diversifying and increasing RNA/protein isoforms (3). In mammals, this essential molecular mechanism is mediated by the Adenosine Deaminases Acting on dsRNA (ADAR) enzymes (4,5). Mammals have three ADAR genes: ADAR1 (also known as ADAR), ADAR2 (also known as ADARB1) and ADAR3 (also known as ADARB2) sharing similar domain structures with RNA-binding domains, which bind the dsRNA substrates, and the carboxy-terminal catalytic deaminase domain (6–8). Inosine has different biochemical properties compared to Adenosine and it is recognized as Guanosine by splicing and translational machineries. For these properties, Inosine can alter splicing, mRNA sequence, protein–RNA and miRNA–RNA interactions (9,10). Indeed, editing within miRNA precursors can interfere with miR-maturation (leading to modulation of miRNA expression) and/or can alter miR-seed sequence, generating novel miRNAs (edi-miRs), that can cause miRNA differential targeting (11). Mammalian brain shows the highest level of A-to-I editing (12) and one of the most-studied recoding editing site occurs in the brain within the glutamate receptor 2 (GRIA2) that codes for a subunit of the α-amino-3-hydroxy-5-methylisoxasole-4propionate (AMPA) channel. GRIA2 is the only transcript to be ∼100% edited at the recoding Q/R site, which is then considered as a unique editing site. This editing, mediated by ADAR2, is responsible for the Ca2+ permeability of the AMPA channel (4) and alone can rescue the lethal neurological phenotype observed in Adar2-null mice (4). Consistently with the essential role played in brain by Adar2 in vivo, accumulating evidence testified the importance of this enzyme in many human brain diseases, including cancer such as glioblastoma (13–17). Glioblastoma (GBM or astrocytoma grade IV) is the most aggressive, infiltrative and lethal brain tumor in humans (18). The most common form of glioblastoma is de novo glioblastoma, which starts from the beginning as a highly fast-growing and aggressive mass and does not evolve from lower-grade astrocytoma (secondary glioblastoma). Glioblastomas are currently incurable, due to their resistance to conventional therapies and their invasive nature. One of the biggest challenges in the treatment of GBM is the presence of highly invasive tumor cells that disseminate into the normal brain parenchyma(19,20).
Despite recent efforts in searching novel edited transcripts in both brain and glioblastoma (21,22), little has been done to identify and dissect the role of key-editing site(s) important for de novo glioblastoma progression and invasion.
Herein, by deeply exploring miRNA expression and A-to-I editing, we report that (i) novel differently edited and expressed miRNAs were identified in brain cortex compared to de novo glioblastomas; (ii) editing (∼100%) within miR-589–3p seed sequence is mediated by ADAR2; (iii) edi-miR-589–3p inhibits glioblastoma cell proliferation, migration and invasion; (iv) editing within miR-589–3p retargets the miRNA from the protocadherin PCDH9 to the metalloprotease ADAM12, which is involved in glioblastoma cell invasion.
MATERIALS AND METHODS
Cell culture and transfection
The well-characterized human glioblastoma cell lines T98G, U87-MG and A172 were obtained from American Type Culture Collection (ATCC) and routinely maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Gibco-Life Technologies), 100 U/ml penicillin and 100 μg/ml streptomycin, at 37°C in 5% CO2.
The BLOCK-iT™ inducible Pol II miR RNAi Expression Vector Kit EmGFP (Invitrogen-Life Technologies) and EGFP-ADAR2-vector (16) were used to obtain glioblastoma cell lines silenced or overexpressing ADAR2 respectively. Specific primers to silence ADAR2 (selected by Block-iT RNAi Designer) and the scramble primers (provided by the kit) were cloned in the pcDNA™6.2-GW/EmGFP-miR vector, according to the manufacturer’s instructions. A172 cells were transfected with Lipofectamine 2000 (Invitrogen-Life Technologies). For stable clones, the cells were maintained under blasticidin selection (20 μg/ml). Immediately after transfection, cells were selected on a Fluorescent Activated Cell Sorting (FACS) according to their Green Fluorescent Protein (GFP) expression units (MIF = mean index fluorescent).
Human brain tissues
We used total RNA of human adult brains from a pool of four individuals (636530, Clontech) and human fetal brain (pool of 21 female and male) (636526, Clontech).
RNA isolation
The small (up to 200 nt) and total RNA fractions were isolated using miRVana™ miRNA Isolation Kit (Ambion-Life Technologies) and TRIzol reagent (Invitrogen-Life Technologies), respectively. Both procedures were performed according to the manufacturer's recommendations. RNA concentration and purity (A260/A280 nm ratio) were evaluated using a NanoDrop ND-2000 (Thermo Scientific). RNA quality was assessed by gel electrophoresis or by an Agilent 2100 Bioanalyzer microfluidics-based platform (Agilent Technologies).
De novo systematic screening of microRNA editing and expression
In house miRNA library was generated by the Illumina’s TruSeq Small RNA Sample Prep Kit, according to the manufacturer’s protocol. The mature miRNA libraries were sequenced with bar-codes and run to the Illumina HiSeq2000 instrument following the manufacturer's protocol. All reads were filtered and trimmed as in (22,23) and editing events identified as previously described (22,23). The sequencing data have been submitted to the NCBI Sequence Read Archive (SRA) under accession number SRP125247.
A similar bioinformatics procedure was applied for the de novo screening of editing sites within miRNAs isolated from all the tissue samples analyzed in the present study [adult cerebral cortex, de novo glioblastoma - ERP012317 (24) and lower grade gliomas (The Cancer Genome Atlas -TCGA-https://tcga-data.nci.nih.gov/tcga/)]. Specifically, for the ERP012317 runs the pre-processing step has been conducted by using Cutadapt (https://cutadapt.readthedocs.io/en/stable/) and FASTQ Quality Filter from the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Then, REDItoolDnaRna version 1.1 (25) (Q 30 and minimum 10 reads coverage cutoff in the edited position) was used to call positions for de novo and already known editing sites (23,26,27). Two-sided Mann-Whitney U test (calculated with the Python library SciPy) with P-value correction for multiple tests (False Discovery Rate, FDR 5%) was applied and only the positions detected in at least two replicates were considered ‘informative.’
MiRNA expression profiles of cell lines (in house reads) were calculated by normalizing the samples and statistically significant differences were identified as described previously (22,23,28). Briefly, the expression profiles were normalized using a variation of the Trimmed Mean of M-values normalization method. Fold-changes between counts in samples A and B were calculated using the formula log2[(A + 1)/(B + 1)], in order to avoid problems associated with zero values, and only miRNAs covered by at least 10 reads (on average) were considered.
For TCGA Glioblastoma and Lower Grade Glioma (ac n. phs000178.v9.p8), Genotype-Tissue Expression (GTEx) Brain cortex (ac n. phs000424.v7.p2) and six different human tissues [SRA code SRP058632, ac n. phs000870.v1.p1, (12)] datasets, we downloaded reads after authorization from the database of Genotypes and Phenotypes (dbGaP). For miRNAs, raw counts were generated using featureCounts 1.5.1 (29), we considered only samples covered by at least 10 reads (on average) and fold-changes were calculated using DESeq2, Bioconductor version 3.4, P-value < 0.05. The rlog transformation method was used to convert counts to regularized log2 values showed as a heatmap (generated with R heatmap.2 function of gplots package).
For mRNA expression, normal brains (132 cerebral cortex from GTEx project) and primary untreated glioblastomas (145 samples from the TCGA) RNA-Seq data, derived from unstranded libraries of polyA enriched RNA (2 × 76 bp), were converted in standard .fastq and mapped to GRCh37 human genome by HISAT2 version 2.0.4. Transcriptome quantification and differential expression test were performed using Cuffquant and CuffDiff2 software.
Real-Time (qRT-PCR)
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed to validate the expression of specific mature miRNAs, using pre-designed stem-loop primers (TaqMan MiRNA Assay, Applied Biosystems-Life Technologies). cDNA was synthesized from 10 ng of total RNA using TaqMan miR Reverse Transcriptase Kit (Applied Biosystems-Life Technologies) according to the manufacturer's instructions. For mRNA, 1 μg of total RNA (pre-treated with DNase I) was used to generate cDNA by the ImProm-II Reverse Transcription System (Promega) using random hexamer primers according to the manufacturer’s instructions.
The small endogenous nuclear RNA U6 (RNU6B), GAPDH and/or β-actin were used as controls for normalization of mature miRNAs and mRNAs, respectively. The relative amount of each substrate was calculated by the 2−ΔΔCT method (30). Expression levels were represented as relative fold increase compared to the control sample, which was arbitrarily set to 1. All qRT-PCR reactions were performed in duplicates and repeated at least twice from independent RT-PCRs and P- values were calculated (two-sided t-test). All the primers were supplied by Applied Biosystems: ADAR2, ID Hs00953730_m1; ADAR1, ID Hs00241666_m1; ADAM12, ID Hs01106101_m1; GAPDH, ID Hs99999905_m1; β-actin, ID Hs99999903_m1; RNU6B, ID 001093. All primers for the miRNAs used in the studies were supplied by Applied Biosystems: miR-222–5p ID002276; miR-21–3p ID002438; miR-25–3p ID477994; miR-30a-5p ID000417; miR-135b-5p ID478582; miR-589–3p ID001543; miR-155–5p ID002623.
Immunoblotting
Total protein extracts were isolated with RIPA lysis buffer in the presence of a protease inhibitor mixture and phosphatase inhibitor cocktail (Sigma-Aldrich). Protein extracts were quantified with the BCA Protein Assay Kit (Pierce). Equal amounts of total cellular lysates (30 μg) were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membrane, analyzed by immunoblotting with the appropriate antibodies and then revealed by ECL (GE Healthcare). The antibodies used in this study were: anti-ADAR2 (1:200, Sigma), anti-β-actin (1:5000, Santa Cruz Biotechnology) and anti-α-tubulin (1:10 000, Abcam), anti-PCDH9 (1:500, Abcam) and anti-ADAM12 (1:500 Santa Cruz Biotechnology). The protein-specific signals were quantified by densitometric analysis using ImageJ v1.47 software.
Proliferation
Cells were seeded in dishes and live cells (Trypan blue dye exclusion) were determined daily, from day 1 to day 4. The assay was repeated at least three times in duplicate. For statistical analysis, we used the two-sided t-test.
MicroRNA mimic/antagomir transfection
miRIDIAN miRNA mimics (Dharmacon-GE Healthcare) miR-589–3p (5′-ucagaacaaaugccgguucccaga-3′) or edited miR-589–3p (5′-ucagagcaaaugccgguucccaga-3′) were transfected using Oligofectamine (Invitrogen-Life Technologies), according to the manufacturer’s instructions. A total of 100 nM miRIDIAN miRNA mimics (Dharmacon-GE Healthcare) miR-589–3p or edited miR-589–3p plus 100 nM miRIDIAN miRNA hsa-miR-589–3p hairpin inhibitor (Dharmacon-GE Healthcare) were transfected using Oligofectamine (Invitrogen-Life Technologies), according to the manufacturer’s instructions.
Migration assay
For evaluation of in vitro cell motility, a monolayer-wounding assay and a transwell assay were performed. For the wounding assay, cells were allowed to form a monolayer on a culture dish surface and, when they were approaching 100% cell confluence, a wound was made by scratching the monolayer with a pipette tip. After the scratching, the cells were incubated in a 5% CO2 incubator at 37°C for a further 24 h. Photographs (10× magnification using Leica DMi8 microscope) of the wound were taken at various time points. Three independent series of experiments were performed. The wound area was measured by the program Image J software (NIH, Bethesda, MD, USA). The percentage of wound closure was estimated by the following equation: wound closure % = [1 − (wound area at Tt/wound area at T0)] x 100%. With T0 the time immediately after wound and Tt the time 12 h post wound.
Transwell inserts with a 8 μm pore size in 24-well plates (Corning, Life Sciences) were used for migration assays. 1 × 104 cells were added to the upper chamber in 0.2 ml serum-free medium; the bottom chamber contained medium with 10% of Fetal Bovine Serum (FBS) which acted as cell attractant. After 24 h incubation, cells that reached the underside of the filter were stained with Diff-Quik staining set (Medion Diagnostics) and counted based on five field digital images taken randomly at 10× magnification. Three independent experiments were performed. For statistical analysis, we used the two-sided t-test.
Invasion assay
Transwell insert with 8 μm pore size in 24-well plates (Corning, Life Sciences) were used for invasion assays and coated with 50 μl of matrigel (Corning Life Sciences, Sigma-Aldrich) for the in vitro cancer cell invasion assays. In both cases 1.5 × 104 cells were added to the upper chamber in 0.2 ml serum-free medium; the bottom chamber contained medium with 10% FBS which acted as cell attractant. After 24 h incubation, cells that reached the underside of the filter were stained with Diff-Quik staining set (Medion Diagnostics) and counted based on five field digital images taken randomly at 4× and 10× magnification using Olimpus IX71. Three independent experiments were performed. For statistical analysis, we used the two-sided t-test.
Gelatin zymography analysis
Conditioned media from A172 and U87-MG cells, respectively transfected with miR-589–3p mimic and miR-589–3p edited mimic, were used for the detection of MMP-9 by gelatin zymography analysis after 48 h of culture. Samples were applied without reduction on a 12% polyacrylamide gel containing 0.1% gelatin. After electrophoresis, gels were washed twice with 2% Triton-X for 1 h at room temperature to remove SDS. Gels were then incubated at 37°C overnight in 50 mM Tris–HCl, 10 mM CaCl2, 100 mM NaCl2, 2% Triton-X at pH 7.6 and stained with 0.5% Coomassie blue R25, destained and scanned. Quantification of MMP-9 was achieved after image capture and by computerized image analysis using image J software. The secretion of MMP-9 in mock cells was taken as 1. Three independent experiments were performed.
HEK editing assay
Approximately 7 × 105 HEK 293T cells at 60% confluence were transfected with 3 μg of either EGFP-ADAR2 or EGFP-ADAR1 using Lipofectamine 2000 (Invitrogen), following the manufacturer’s instructions. Forty-eight hours post-transfection, total RNA was extracted from the cells using TRIzol reagent and reverse transcribed using random primers. Editing at specific sites was tested by PCR of the specific targets. Direct sequencing was performed, and editing was calculated as previously described (31). Pri-miR-589 primers: fw 5′-gagcagggccatggagaag-3′, rev 5′-tttcctggcacaggtgctg-3′.
RNA editing analysis by Sanger sequencing
For editing analysis, RNA samples were pre-treated with DNase I and cDNAs were generated by Superscript II Reverse Transcriptase (Invitrogen-Life Technologies) using random hexamer primers or transcript-specific oligonucleotides, according to the manufacturer's instructions. The cDNAs were utilized for PCR reactions by using the Expand high-fidelity Plus PCR System (Roche) (16). The specific PCR products were gel purified (Qiaquick, Qiagen). Direct sequencing (ABI 3500 Genetic Analyzer, Applied Biosystems-Life Technologies) was performed and editing levels were calculated as previously described (31).
Luciferase assay
The 3′UTR portion of ADAM12, containing the putative binding site for the edi-miR-589–3p, was amplified using the primers Fw:5′-agaattccacccacaccgcctatatta-3′ and Rev:5′-aaagcggccgctctggtaccataagcacagc-3′. PCR product was then digested (with EcoR1 and Not1) and ligated into the pMirTarget vector for luciferase assay (Origene). In a 96-well plate, a total of 1 × 104 cells/well were transfected with 0.25 μg of the pmiR target vector containing ADAM12 3′UTR together with 50 nM of negative control mimic, 50nM of miR-589–3p mimic or 50 nM edi-mir 589–3p mimic respectively. For each transfection, 25 ng of Renilla luciferase reporter construct (pRL- CMV, Promega) was also transfected. After 48h the luciferase activity was assayed using a Dual-Glo Luciferase assay system (Promega) according to the manufacturer’s instructions. Luciferase luminescence was measured with EnSpire® multimode reader (Perkin Elmer). The ratio of firefly luciferase activity to the Renilla luciferase activity was used to normalize any difference in transfection efficiency among samples. All luciferase analyses were performed in triplicates. For statistical analysis we used the two-sided t-test.
The experimental methods comply with the Helsinki declaration.
RESULTS
MicroRNA differential expression signature in brain cortex and de novo glioblastoma tissues
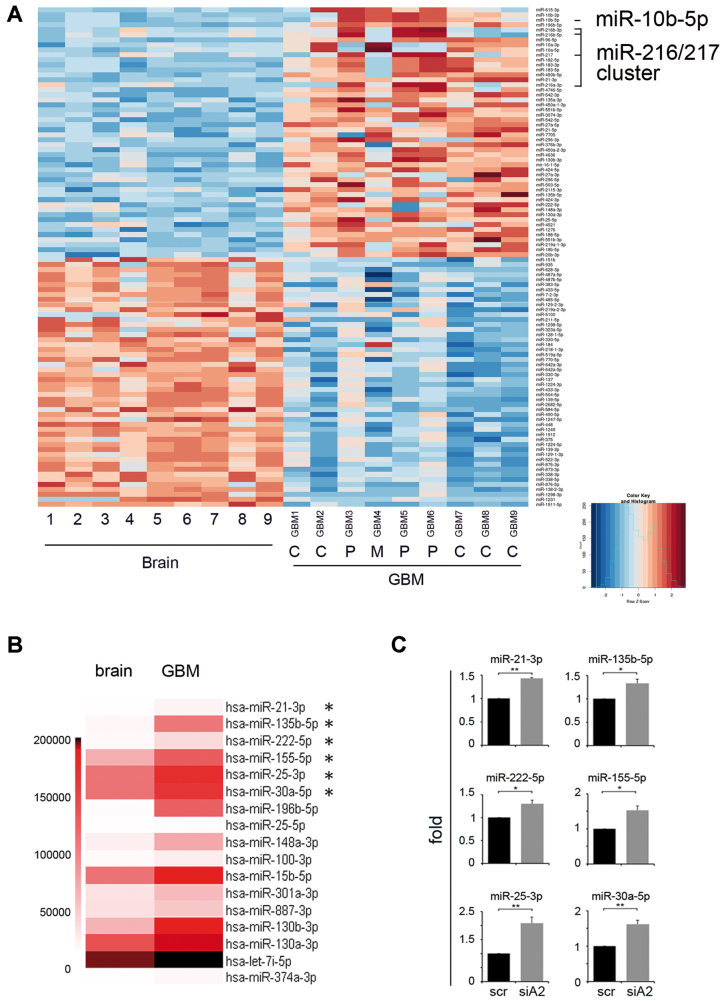
First we performed a comprehensive miRNA expression profile of miRNA-seq runs comparing nine de novo glioblastoma samples [three Proneural (P), one Mesenchymal (M) and five Classical (C) subtypes as Verhaak classification (32)] and nine adult brain cortex samples (24). Our analysis showed that miRomes of normal and cancer tissues are deeply different, with 299 modulated miRNAs (DESeq2 corrected P < 0.05) (Figure 1A and Supplementary Table S1). We have found that a specific miRNA involved in glioma growth (33), miR-10b-5p, was overexpressed in de novo glioblastoma tissues, with a strong upregulation mainly in GBM belonging to the Proneural subtype (Figure 1 and Supplementary Table S1). Similarly, miR-10b-3p was found over-expressed in glioblastomas compared to normal tissues. Our analysis also showed, for the first time, that a specific miR-cluster (miR-216a/217) is overexpressed in glioblastoma tissues (Figure 1 and Supplementary Table S1). This cluster was associated with drug resistance and recurrence in liver cancer (34). Among several known onco-miRs found overexpressed in GBM, there were well-known onco-miRs (such as miR-21, miR-148a/b, miR-221, miR-96), while other miRNAs known to act as tumor suppressors in glioblastoma (such as miR-485–5p, miR-338–3p, miR-31, miR-218) were found significantly downregulated (Supplementary Table S1).
Figure 1.
Differentially expressed miRNAs in de novo glioblastoma and normal brain tissues. (A) Heatmap shows differentially expressed miRNAs (FDR < 0.05) of glioblastoma tissues and normal brain cortex tissues. Top 50 most upregulated and top 50 most downregulated miRNAs are shown. Statistical analysis was performed using DESeq2. The matrix of Z-scores representing rlog normalized counts was calculated and scaled by row. Negative values (blue) indicate miRNAs under expressed in glioblastoma and positive values (red) indicate miRNAs over expressed in glioblastoma. (B) Heatmap showing the expression levels of miRNAs found significantly overexpressed in GBM (means) versus brain cortex (means) (FDR < 0.05) (Supplementary Figure S1) and also found overexpressed in siADAR2 versus scr glioblastoma cells (see Supplementary Table S2). (C) qRT-PCR of common miRNAs (asterisks in B) validated in siADAR2 (siA2) versus control (scr) glioblastoma cells. Mean ± s.e.m. (n = 2), **P < 0.01, *P < 0.05 (two-sided t-test). Expression levels were normalized to RNU6B levels and shown as relative fold increase compared to the scramble.
As A-to-I RNA editing can deeply modulate miRNA expression, by altering the RNA structure of the miRNA precursor (23,35), we wondered whether loss of ADAR2, a major deaminase involved in brain and poorly expressed/active in glioblastoma (16,21,36), was able to modulate miRNAs important in GBM. Therefore, we silenced ADAR2 in glioblastoma cells (A172) (Supplementary Figure S2) and then we performed a de novo miR-expression screening of siADAR2 glioblastoma cells and controls by using a next-generation sequencing (NGS) approach. We found that, downregulating ADAR2, 43 miRNAs were upregulated while none among the downregulated miRNAs passed our screening. The 36 most overexpressed miRNAs (siADAR2 + scr ≥200 reads) following ADAR2 silencing are shown in Supplementary Table S2. Among them, miR-143–3p and miR-155–5p are both reported as miRNAs involved in promoting glioma aggressiveness (37,38). Several (15/36) miRNAs upregulated in siADAR2 cancer cells (Supplementary Table S2, miR shown in bold), and also overexpressed in tissues from de novo glioblastoma patients (Figure 1B), were then validated in siADAR2 A172 cells, by qRT-PCR (Figure 1C and data not shown).
Overall, our data identified miRNAs aberrantly expressed in de novo glioblastoma never reported before and indicated that loss of ADAR2 specifically upregulates a definite group of miRNAs, most of which are overexpressed in glioblastoma and involved in the onset and progression of this aggressive cancer.
Shaping the microRNA editing signature in glioblastoma and normal brain
A-to-I RNA editing can modulate miRNA expression and also change the sequence of mature miRNAs, with possible consequences on gene-retargeting (39).
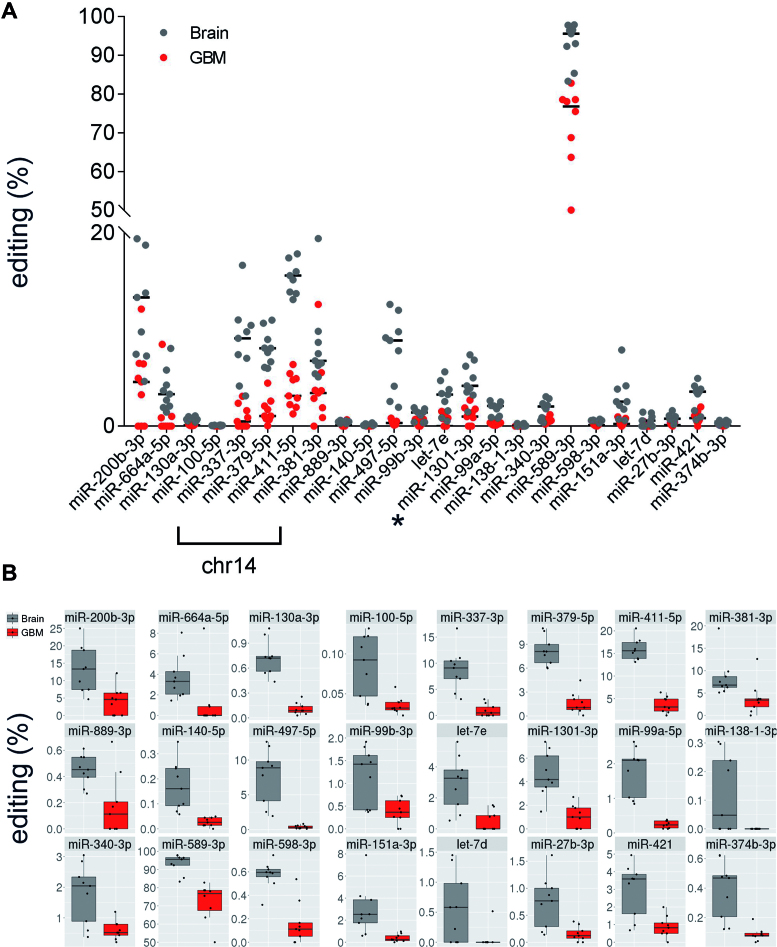
To identify mature miRNAs undergoing editing, herein indicated as edi-miRs, we systematically analyzed the miRNA editing profiles of 9 brain cortex and 9 de novo glioblastoma tissues (24). MiRNA NGS data were analyzed with a specific algorithm for miR-editing detection (22). We identified 24 miRNAs differentially edited in normal cortex and glioblastoma samples (Figure 2, Table 1 and Supplementary Table S3). All miRNAs showed an overall decreased editing signature in glioblastoma samples compared to brain with a particular enrichment of miRNAs located on Chromosome 14 (Figure 2 and Table 1). We report that several (15/24) miRNAs, differently edited in normal brain versus glioblastoma, underwent editing within their seed sequences (Table 1) and, among them, we identified a miRNA with a novel edited site, miR-1301–3p, edited at position +5 of the mature miRNA (Figure 2 and Table 1). We also showed that editing within mature miRNAs occurred generally at low level (average <5%). However, miR-589–3p, also edited within its seed sequence, came out as the miRNA with the highest editing (median 95.59%) in normal brain tissues and with a consistent editing decrease in glioblastoma samples compared with brain cortex (18.81% editing Δ median, P = 0.0006, q = 0.0055) (Figure 2 and Table 1).
Figure 2.
MiRNA editing fingerprint in glioblastoma and brain cortex tissues. (A) Differently edited miRNAs in de novo glioblastomas (red dots) and brain tissues (gray dots) are shown (P < 0.05 calculated with two-sided Mann–Whitney U test followed by Benjamini–Hochberg multiple testing correction) (see also Table 1). Dots represent the editing level (%) for each sample (either normal and GBM). We found that several of the miRNAs reported are transcribed from genes on chromosome 14. Asterisk indicates the novel edi-miR-1301–3p. Medians of editing values are shown as black bars. (B) Editing levels distribution details of sites shown in (A).
Table 1. Editing events in miRNAs in brain and glioblastoma tissues.
| Chromosome | Position | Strand | miRNA | pre-miRNA | miRNA | BRAIN median | GBM median | Δ medians | P-value | q-value |
|---|---|---|---|---|---|---|---|---|---|---|
| chr7 | 5 535 483 | − | hsa-miR-589–3p | 66 | 6 | 95.59 | 76.78 | 18.81 | 0.0006 | 0.0055 |
| chr14 | 101 489 681 | + | hsa-miR-411–5p | 20 | 5 | 15.6 | 3.14 | 12.45 | 0.0004 | 0.0041 |
| chr1 | 1 102 544 | + | hsa-miR-200b-3p | 61 | 5 | 13.33 | 4.59 | 8.75 | 0.0035 | 0.0167 |
| chr14 | 101 340 895 | + | hsa-miR-337–3p | 66 | 6 | 9.09 | 0.49 | 8.6 | 0.0004 | 0.0041 |
| chr17 | 6 921 317 | − | hsa-miR-497–5p | 25 | 2 | 8.88 | 0.34 | 8.54 | 0.0004 | 0.0041 |
| chr14 | 101 488 412 | + | hsa-miR-379–5p | 10 | 5 | 8.08 | 1.05 | 7.03 | 0.0004 | 0.0041 |
| chr14 | 101 512 308 | + | hsa-miR-381–3p | 52 | 4 | 6.78 | 3.44 | 3.34 | 0.0081 | 0.0297 |
| chr1 | 220 373 944 | − | hsa-miR-664a-5p | 18 | 8 | 3.31 | 0 | 3.31 | 0.0053 | 0.0232 |
| chr19 | 52 196 095 | + | hsa-let-7e | 57 | 5 | 3.27 | 0 | 3.27 | 0.0024 | 0.013 |
| chr2 | 25 551 539 | − | hsa-miR-1301–3p | 52 | 5 | 4.18 | 1 | 3.18 | 0.0011 | 0.0069 |
| chrX | 73 438 236 | − | hsa-miR-421 | 61 | 14 | 3.59 | 0.83 | 2.76 | 0.0062 | 0.0241 |
| chr8 | 141 742 704 | − | hsa-miR-151a-3p | 49 | 3 | 2.53 | 0.24 | 2.28 | 0.0015 | 0.0086 |
| chr21 | 17 911 421 | + | hsa-miR-99a-5p | 13 | 1 | 2.1 | 0.23 | 1.87 | 0.0004 | 0.0041 |
| chr5 | 179 442 328 | − | hsa-miR-340–3p | 70 | 13 | 2.04 | 0.52 | 1.53 | 0.0134 | 0.0444 |
| chr19 | 52 195 911 | + | hsa-miR-99b | 47 | 3 | 1.42 | 0.37 | 1.06 | 0.0134 | 0.0444 |
| chr9 | 97 847 792 | + | hsa-miR-27b-3p | 66 | 6 | 0.77 | 0.12 | 0.64 | 0.0062 | 0.0241 |
| chr11 | 57 408 726 | + | hsa-miR-130a-3p | 56 | 2 | 0.72 | 0.09 | 0.63 | 0.0004 | 0.0041 |
| chr9 | 96 941 181 | + | hsa-let-7d | 66 | 5 | 0.58 | 0 | 0.58 | 0.0159 | 0.0483 |
| chr8 | 10 892 751 | − | hsa-miR-598–3p | 62 | 2 | 0.6 | 0.11 | 0.48 | 0.0011 | 0.0069 |
| chrX | 73 438 433 | − | hsa-miR-374b-3p | 21 | 11 | 0.47 | 0.08 | 0.39 | 0.0008 | 0.0062 |
| chr14 | 101 514 299 | + | hsa-miR-889–3p | 62 | 14 | 0.45 | 0.11 | 0.34 | 0.0165 | 0.0483 |
| chr16 | 69 967 021 | + | hsa-miR-140–5p | 38 | 16 | 0.16 | 0.03 | 0.13 | 0.0004 | 0.0041 |
| chr11 | 122 023 004 | − | hsa-miR-100–5p | 13 | 1 | 0.09 | 0.03 | 0.06 | 0.0036 | 0.0167 |
| chr3 | 44 155 777 | + | hsa-miR-138–1-3p | 74 | 12 | 0.05 | 0 | 0.05 | 0.014 | 0.0444 |
Editing levels are shown as percentages. Chromosome coordinates refer to human genome assembly hg19. Only sites with q-value ≤ 0.05 (Benjamini–Hochberg corrected P-value) are shown.
Next, siADAR2 and control (scr) glioblastoma cells (Supplementary Figure S2) were analyzed for possible editing events within miRNAs. We identified only a few miRNAs being differently edited in glioblastoma cells: miR-196a2–3p site +9 (10.9%), miR-589–3p site +6 (10.4%) and miR-561–3p site +9 (44.9%). These editing levels resulted further reduced in siADAR2 cells (with ≥50% ADAR2 decrease): 0% for miR-196a2–3p, 6.5% for miR-589–3p and 35% for miR-561–3p. Among them, the editing in miR-561–5p and miR-196a2–3p occurs outside the seed sequence. Of note, edi-miR-196a2–3p is novel and identified herein for the first time.
We report that only few miRNAs (miR-1301–3p, miR-340–3p, miR-889–3p and miR-138–1-3p) were both edited and expressed to a lower level in glioblastoma tissues compared to brain cortex; by contrast, two miRNAs (miR-497–5p and miR-130a-3p) were edited to a low extent but over-expressed in glioblastoma compared to normal brain.
Crossing data from glioblastoma tissues and glioblastoma siADAR2 cells, we observed that only miR-589–3p was found in both screenings and consistently less edited in glioblastoma tissue samples than controls (18.81% editing Δ median, P = 0.0006, q = 0.0055) (Table 1).
Overall, our data demonstrated that in de novo glioblastomas editing was decreased within a specific subset of miRNAs and that this editing mainly occurs within seed sequences; among the modulated miRNAs, the edi-miR-589–3p shows the strongest drop in editing level in glioblastoma tissues compared to brain cortex and was also less edited in siADAR2 glioblastoma cells.
A-to-I editing within miR-589-3p seed is mediated by ADAR2
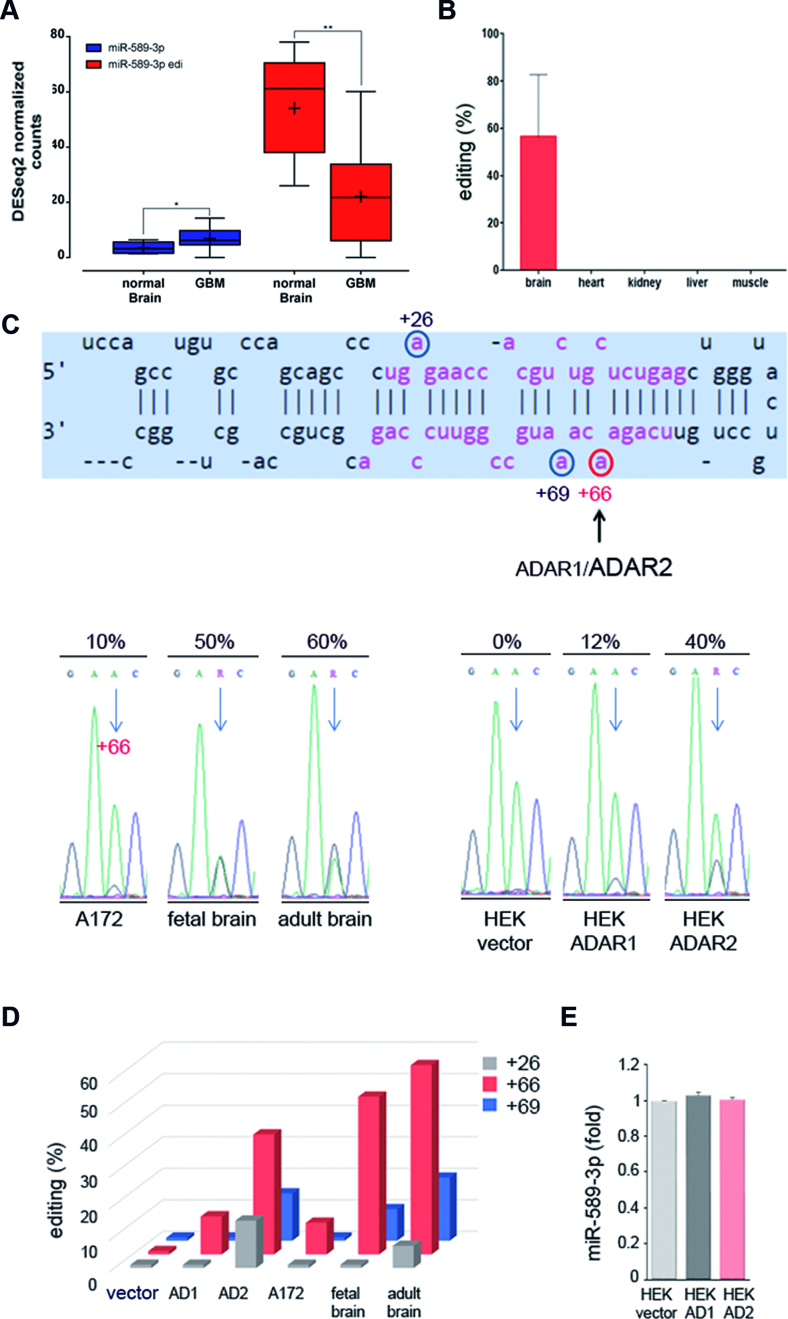
Next, we studied the role of the edi-miR-589–3p in glioblastoma. We first interrogated deep-sequence miR-data (24) in order to detect the expression levels of the unedited and the edited miR-589–3p in normal brain and in de novo glioblastoma. We found that edi-miR-589–3p is highly expressed in normal brain with a decreased expression in GBMs, while the unedited miRNA was found extremely low expressed in both tissues (Figure 3A). Furthermore, we interrogated available NGS-datasets (12) for human normal tissues in order to find this editing event within miR-589–3p (Chr7, position 5535483) in brain, heart, kidney, liver and muscle. Despite the few samples analyzed, we found that editing at this position (position +6 of miR-589–3p) was observed exclusively in brain, while no editing was observed in the other tissues analyzed (Figure 3B). Overall, miR-589–3p is highly expressed only in brain and almost only as edited-miR (edi-miR-589–3p).
Figure 3.
miR-589–3p is highly edited within its seed in the brain. (A) MiR-589–3p and edi-miR-589–3p expression in glioblastoma tissues (nine samples) and normal brain cortex (nine samples), **P < 0.01, *P < 0.05 (two-sided t-test). The expression of the two forms inferred from normalized counts of reads supporting only the genomically transcribed (uned-miR) and the edited version (edi-miR) were shown respectively. (B) Editing (%) of miR-589–3p (+6 site) as calculated in different normal human tissues (two/three samples per tissue were analyzed); mean ± s.e.m. (C) Sequence structure of pri-miR-589 with the edited positions identified by circles, in red the position +6 of the miR-589–3p. Below, chromatograms showing the editing (as A/G double peaks) at nucleotide +66 of the pri-miR-589 (corresponding to nucleotide +6 of the mature miR-589–3p) in glioblastoma cells (A172), fetal brain, adult brain and HEK293T cells transfected with either empty vector, ADAR1 or ADAR2. After transfection (48 h), editing within pri-miR-589 was analyzed. (D) Editing values (%) at three sites (nucleotides +26, +66 and +69 of the pri-miR) as found in fetal brain, adult brain, glioblastoma cell line A172 and in transfected HEK293T cells. (E) qRT-PCR (48h p.t.) of miR-589–3p in HEK293T cells transfected with empty vector, ADAR1 (AD1) or ADAR2 (AD2).
We have already reported that miR-589–3p editing decreased upon ADAR2 silencing in A172 glioblastoma cells (see previous paragraph). In order to demonstrate that ADAR2 edits miR-589–3p, we used an ADAR-overexpressing system in HEK293T cells and specifically amplified and sequenced the endogenous pri-miR-589 48h after ADAR1, ADAR2 or empty vector transfection. In parallel, we amplified the pri-miR-589 from human adult brain cortex and human fetal brain. We found that position +66 of the pri-miR-589–3p (corresponding to position +6 of the mature miR-589–3p, within the seed sequence) is edited ∼ 40% by ADAR2 and only ≤10% by ADAR1 (Figure 3C and D; Supplementary Figure S3). Of note, this site was found edited at 50% in fetal brain and >50% in adult brain, while in A172 glioblastoma cells it was edited at low level (∼10%) (Figure 3C and D; Supplementary Figure S3). In total, we identified three editing sites within pri-miR-589, one editing site within miR-589–5p (+26 of the pri-miR) and two editing sites within miR-589–3p (+66 and +69 of the pri-miR) with the highest editing level found at position +66 of the pri-miR-589 (Figure 3 and Supplementary Figure S3). No differences in mature miRNA expression levels were observed in HEK293T cells after transfection with either ADAR1 or ADAR2 (Figure 3E).
To further confirm that ADAR2 edits miR-589–3p at position +6 in glioblastoma cells, we also sequenced the pri-miR-589 from total RNA isolated from two glioblastoma cell lines (A172 and U87-MG) in which we overexpressed or silenced ADAR2. We showed that position +66 of the pri-miR-589 is modulated by ADAR2 in glioblastoma cells (Supplementary Figure S4).
Overall, we showed that miR-589–3p is almost fully edited in the brain and that this editing is ADAR2-mediated; additionally, we showed that editing within miR-589–3p progressively increases from fetal to adult brain, while is lost in glioblastoma cells/tissues.
A-to-I editing within miR-589-3p seed inhibits glioblastoma cell aggressiveness
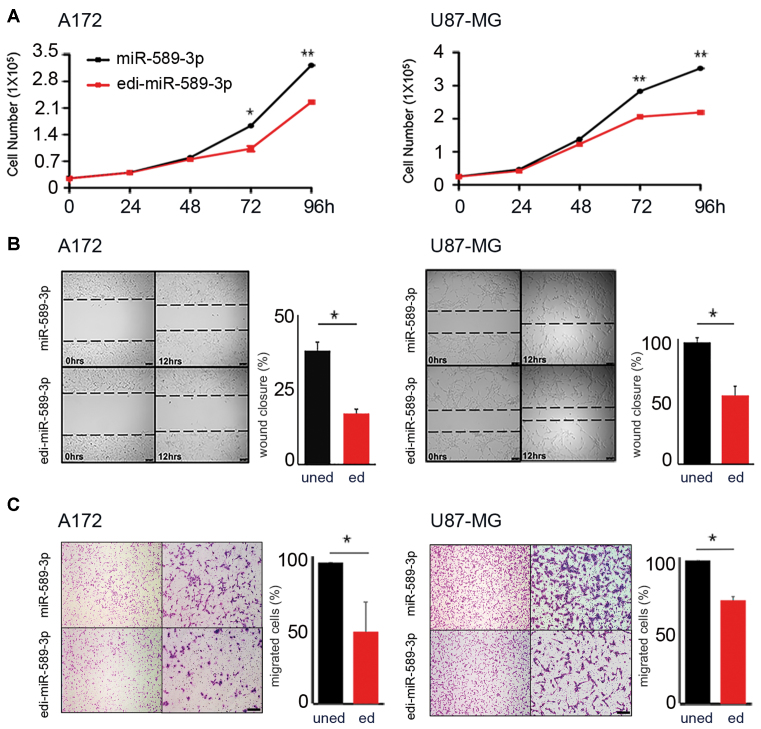
Next, we tested whether the editing decrease within miR-589–3p in glioblastoma has any impact on cancer cells. We overexpressed equal amounts (Supplementary Figure S5) of either miR-589–3p mimic or its edited version (carrying a G at position +6 of the seed sequence) edi-miR-589–3p mimic, in several glioblastoma cell lines (A172, U87-MG and T98G) and we tested them for cell proliferation, migration and invasion. We found that the edi-miR-589–3p significantly decreased cell proliferation in all the cell lines tested (Figure 4A and Supplementary Figure S6). Similarly, when we tested the cells for their ability to migrate, within 24 h post-transfection (a time in which these cells do not duplicate), we observed that edi-miR-589–3p significantly reduced cell motility compared to the unedited-miR, as tested using both scratch and transwell assays (Figure 4B and C; Supplementary Figure S6).
Figure 4.
Edi-miR-589–3p inhibits both proliferation and migration of glioblastoma cells. (A) A172 (2.5 × 104) and U87-MG (2.5 × 104) cells were transiently transfected with miR-589–3p mimic or edi-miR-589–3p mimic and cell proliferation (Trypan blue exclusion) monitored over days. Mean ± standard deviation (n = 3), *P < 0.05, **P < 0.01 (two-sided t-test). (B) 1 × 105 A172 and U87-MG cells, transiently transfected with miR-589–3p (uned) or edi-miR-589–3p (ed) mimic, were seeded to confluence for scratch assay. Wound closure was monitored after 12 h (see representative pictures, scale bar 100 μm). Quantitation of wound closure is reported, mean ± standard deviation (n = 3), *P < 0.05 (two-sided t-test). (C) Transwell migration assay was performed 48 h p.t. Representative photographs of migrated cells are shown (4× and 10× magnifications). Cells were visualized by Diff Quick staining and counted 24 h post seeding. Scale bar 400 μm. Histograms show the migration ability relative to the uned-mimic (100%) (fold increase ± standard deviation, n = 3) of cells that passed through the 8 μm filter pore, *P < 0.05 (two-sided t-test).
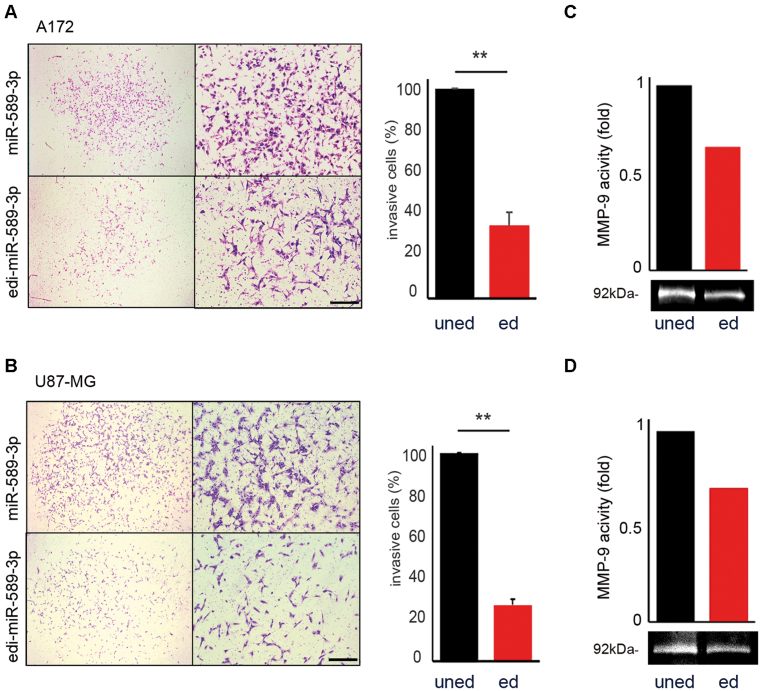
To assess the effects of the unedi- and edi-miR-589–3p on the invasion ability of glioblastoma cells, we used a Transwell Chamber in which the top side of 8 μm pore membrane was coated with matrigel. Identical GBM cells number (A172 and U87-MG), transfected with either the unedi- or edi-miR-589–3p mimics, were placed on the top of the chamber and 24 h later, we observed that cells transfected with miR-589–3p invaded significantly more compared with those transfected with the edited mimic in both cell lines (Figure 5A and B).
Figure 5.
Edi-miR-589–3p inhibits glioblastoma invasiveness. (A) A172 and (B) U87-MG cells (1.5 × 104) transfected with either miR-589–3p mimic (uned) or edi-miR-589–3p mimic (ed) were seeded, 24 h post-transfection, over matrigel-coated inserts in serum-free DMEM with a below chamber containing medium supplemented with FBS. Twenty-four hours after seeding the invasive cells were visualized by Diff Quick staining and counted. Representative photographs of the invasive cells are shown (4× and 10× magnifications). Histograms show the invasive ability relative to the uned-mimic (100%). Scale bar 400 μm, **P < 0.01 (two-sided t-test). (C and D) Representative Gelatin-Zymography assays (MMP-9) of A172 and U87-MG cells transfected with uned- and edi-miR-589–3p mimic were shown (n = 3), histograms (upper image) show densitometric quantification of the corresponding MMP-9 digestion activity (bottom image).
Highly aggressive and invasive glioblastoma cells move through extracellular matrix (ECM) into neighbouring brain parenchyma in a process involving ECM degradation and proteolysis. Two gelatinases, MMP-2 (gelatinase A) and MMP-9 (gelatinase B) are unique among MMPs for preferentially degrading type IV collagen, a major component of cerebrovascular basement membranes and a major structural barrier for tumor cell invasion (40). Then, we explored the possibility that loss of editing within miR-589–3p may enhance the metalloprotease activity of these MMPs in glioblastoma cells. We analyzed glioblastoma cells (A172 and U87-MG), transfected with equal amount of unedi- and edi-miR-589–3p mimic, by using a gelatin-zymography assay (40). Our data show that MMP-9 activity was clearly inhibited by edi-miR-589–3p, while it was enhanced by the miR-589–3p (Figure 5C and D). Similar findings, but with a mild effect, were also obtained with MMP-2 (data not shown).
In summary, we demonstrated that editing within miR-589–3p at +6 site restricts glioblastoma cell proliferation, migration, invasion and the ability to digest ECM, while the unedited miR-589–3p promotes glioblastoma progression and invasion.
Editing within miRNA-589-3p progressively decreases with the increased astrocytoma malignancy grades and targets a different set of RNAs
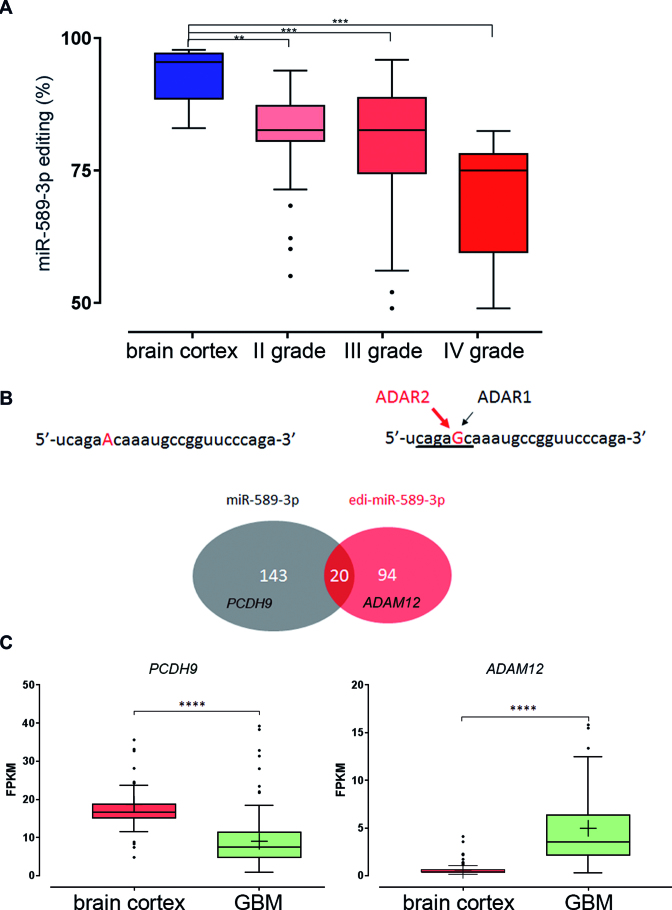
Next, we measured editing level of miR-589–3p (+6 site) in normal brain and astrocytoma samples of increasing malignancy grades (grades I–IV). By screening multiple miR-datasets (NGS) (TCGA, https://tcga-data.nci.nih.gov/tcga/) (24) we found that editing at this site progressively decreased from normal brain to astrocytoma grade II, and grade III being lowest in glioblastoma samples (Figure 6A).
Figure 6.
Editing within miR-589–3p progressively decreases with astrocytomas grade of malignancy and re-targets the edi-miR589–3p to a smaller and different set of transcripts compared to the miR-589–3p. (A) Box-plot showing distribution of editing values (%) within miR-589–3p seed (position +6) as analyzed in normal brain, astrocytomas grade II, astrocytomas III and astrocytomas IV (or glioblastoma) (NGS runs), medians are shown, **P < 0.01, ***P < 0.001 (two-sided Mann–Whitney U test). (B) Sequences of the miR-589–3p with the Adenosine undergoing editing in red and the edi-miR-589–3p with the Guanosine in red. Below a cartoon showing the number of the predicted target genes of the unedited miR-589–3p (in gray, among them PCDH9) and of the edi-miR-589–3p (in red, among them ADAM12). Only 20 target genes were found in common. (C) Expression levels of PCDH9 and ADAM12 (FPKM) in de novo glioblastoma (145 samples) and normal brain (132 samples), ****P < 0.0001 (P-values calculated by Cuffdiff2).
Considering the impact of this specific editing event, we focused on possible target genes modulated by the unedited and the edited version of miR-589–3p, by interrogating available software [miRDB (41) and TargetScan (42)]. By crossing results from both software and picking the common targets, we found that while the unedited miR-589–3p may target 143 genes, the edi-miR-589–3p could affect a different and smaller set of genes (94 target genes), with only 20 potential target genes in common (Figure 6B and Supplementary Table S4). Among the RNA–target genes exclusively modulated by edi-miR-589–3p, there were several involved in cell proliferation and migration/invasion (Supplementary Table S4).
Protocadherin 9 (PCDH9), a tumor suppressor gene associated to glioma progression (43), came out as a specific RNA target gene controlled by the miR-589–3p. The Disintegrin and Metalloproteinase 12 (ADAM12) gene transcript, over-expressed in various cancers (44,45), resulted to be a specific candidate for the edi-miR-589–3p (Figure 6B). Then, we analyzed the expression levels of both PCDH9 and ADAM12 in normal brain cortex and de novo glioblastoma tissues by interrogating 145 GBMs (TCGA dataset) and 132 normal brain tissue samples (GTEx database). We found that, while PCDH9 is downregulated, ADAM12 is upregulated in glioblastoma tissues compared with normal brain tissues (Figure 6C).
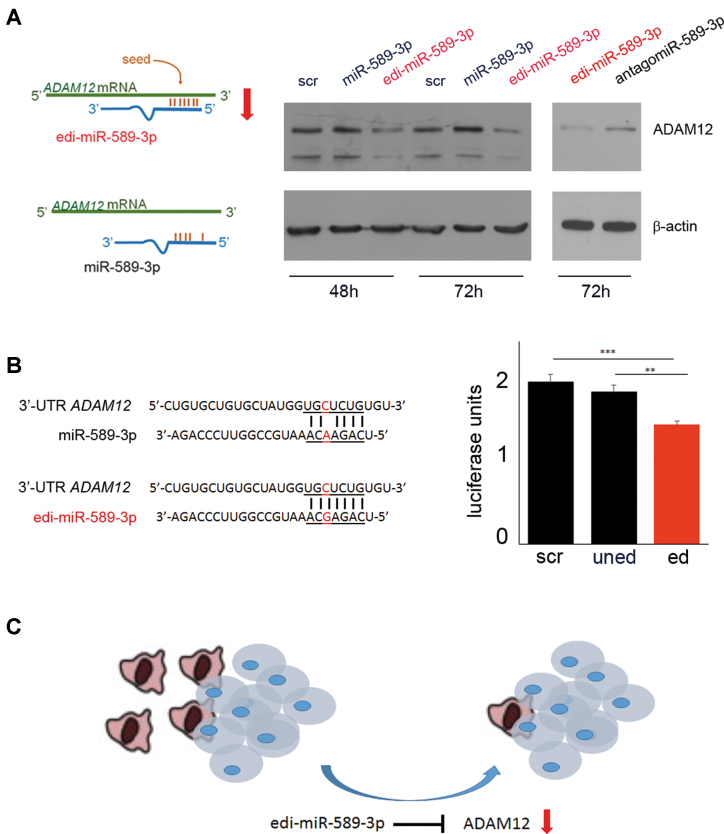
To validate PCDH9 and ADAM12 as unedi- and edi-miR-589–3p target genes respectively, we transfected glioblastoma cells with either unedited or edited-miR-589–3p mimic and tested PCDH9 and ADAM12 levels. We demonstrated that while miR-589–3p silenced PCDH9 48h post-transfection (Supplementary Figure S7), edi-miR-589–3p specifically silenced ADAM12 48 and 72 h post-transfection (Figure 7A and Supplementary Figure S8). We showed that the ability of edi-miR-589–3p to silence ADAM12 is specific as it was lost when we quenched the overexpressed edi-miR-589–3p with the specific inhibitor (antagomir) (Figure 7A). Luciferase assay was also performed using HEK293T cells. The reporter construct containing the ADAM12 3′UTR was co-transfected with either the unedited or the edited miR-589–3p mimic. Specific binding to ADAM12 3′UTR was observed only with the edited version of miR-589–3p, revealing a specific contribution of I:C/G:C base pairs for hybridization of the edited-miR to its target (Figure 7B).
Figure 7.
Editing retargets miR-589–3p from PCDH9 to ADAM12. (A) Cartoon summarizing the binding between ADAM12 3′-UTR with the edi-miR-589–3p but not with miR-589–3p. On the right, western blotting analysis showing the reduction of ADAM12 protein level only with the edi-miR-589–3p mimic and not with the miR-589–3p mimic (100 nM) in U87-MG cells 48 and 72 h post-transfection. AntagomiR (100 nM) experiments were also performed showing that the specific binding of edi-miR-589–3p was lost if edi-miR-589–3p was co-transfected with the corresponding inhibitor (antagomiR). (B) Nucleotides details of Luc-ADAM12 3′-UTR and the uned- or edi-miR-589–3p. On the right, histogram showing the results of the luciferase assay (n = 4) **P < 0.01, ***P < 0.001 (two-sided t-test). (C) Picture resuming the molecular pathway showing edi-miR-589–3p as a potent anti-glioblastoma molecule able to block glioblastoma invasive cells.
Finally, in order to recapitulate what observed in glioblastoma tissues (with editing reduction from ∼100 to ∼80%, see Table 1), we co-transfected U87-MG glioblastoma cells with the two miRNAs mixed to different ratios (edited/unedited) and we measured the invasion ability of the cells. We show that a little decrease of edi-miR-589–3p (of about 20%) already enhanced the invasive cell behaviour that further progresses with the edi-miR reduction (Supplementary Figure S9).
Collectively, our data demonstrated that both unedited and edited miR-589–3p are critical partners with opposite effects in glioblastoma cells. Specifically, we showed that the fully edited miR-589–3p, highly expressed in normal brain, keep silenced the metalloprotease ADAM12, while the unedited miR-589–3p released the control over ADAM12, increasing glioblastoma cell invasiveness (Figure 7C).
DISCUSSION
The identification of genes involved in glioblastoma progression and ability to switch from a stationary condition to a diffuse mobile cell state, with a tendency to spread into the brain, is of high interest for cancer research (46). Herein, we wondered whether ADAR enzymes are involved in glioma aggressiveness/invasiveness and we aimed to identify the ADAR target genes involved.
We focused on identifying edited target genes involved in a highly malignant cell-reprogramming switch, with a particular attention to miRNAs. MiRNAs play multiple and crucial roles in cell proliferation, migration and invasion (47,48) and are also attractive molecules as new targets for potential therapeutic interventions in several cancers, including glioblastoma (49). A-to-I RNA editing, mediated by ADAR enzymes, affects miRNA-controlled pathways by interfering with miRNA maturation steps as well as with miRNA targeting (39). Indeed, editing within miRNA seed sequence (the miRNA–RNA recognition site) may alter miRNA target specificity, thus re-orchestrating miRNA-mediated gene silencing (11,22). However, despite the fact that most edited RNAs are concentrated in mammalian brain (12), only one example of edi-miRNA (miR-376a*), affecting glioblastoma, has been reported (50).
Interestingly, editing within miR-376* is mediated by ADAR2 and, when decreased, can promote glioblastoma invasion (50). Of note, miR-376a* is edited at low level and results difficult to detect from NGS data (51). Indeed, also in the present study, only few samples showed editing within this miRNA and no statistical significant differences were observed between normal brain and GBMs (Mann–Whitney U test, Benjamini–Hochberg corrected P value < 0.05) (Supplementary Table S3).
Overall, it is conceivable that in vivo multiple edi-miRs (such as the edi-miR-589–3p, miR-376a* and other edi-miRs) can contribute together to restrict glioma aggressiveness with a possible synergistic action.
Herein, we compared the miRNA signature (expression and editing) of de novo glioblastoma and normal brain tissues showing that a characteristic miRNA-profile distinguishes cancer from normal tissues. Cancer tissues are enriched in onco-miRNAs, such as miR-10b and the ancient chordate-specific miRNA cluster miR-216/217 (52), found overexpressed in glioblastoma tissues (mainly in the Proneural subtype) and recently connected to the induction of epithelial-mesenchymal transition in liver cancer (34).
The editing signature of miRNAs is also a characteristic feature distinguishing glioblastoma from normal brain tissue with a generalized and significantly decreased editing occurring in 24 miRNAs, some of which (∼20%) mapped to Chromosome 14 (miR-337, miR-379, miR-411, miR-381 and miR-889, Figure 2 and Table 1). All these miRs are organized in three independent miR-clusters. Specifically, miR-379 and miR-411 belong to the miR-379/411 cluster (14q32) that contributes to drug resistance in malignant pleural mesothelioma (53). Another miR-cluster, comprising 19 miRNAs, includes the miR-381, the miR-889 and the already mentioned miR-376a*. It is intriguing that significant editing decrease in glioblastoma occurs in miR-clusters mapping to the same chromosomal region (14q32). Of note, loss of heterozygosity (LOH) involving 14q31.3–14q32.1 was reported in ∼22% of glioblastomas (54), indicating that genes acting as tumor suppressors are located in this specific chromosome portion.
Among the significantly modulated edi-miRs, here we report a novel editing event within miR-1301–3p (in its seed sequence). We found that most (15/24) of the editing events altered the miR-seed sequence (Table 1), suggesting that editing is not a random event, but occurs at essential-specific sites. Among the significantly modulated edi-miRs, miR-589–3p, originally identified in our laboratory (22,23), showed the highest editing level among miRNAs in the brain (median 95.59%), with also the highest drop in editing proceeding from normal brain to glioblastoma (Δ median 18.81%, P = 0.0006). We showed that this distinctive miR-589–3p is edited and expressed almost exclusively as edited-miR in the brain compared to other human tissues, and editing progressively decreased in astrocytomas with increasing grade of malignancy, being significantly edited to a low extent in astrocytoma grade IV or glioblastoma (Figure 6).
We demonstrated that ADAR2 is the principal editase involved in editing pri-miR-589–3p at position +66 (position +6 of the mature miR-589–3p); additionally, we also identified other two ADAR2-medited editing sites at position +26 and +69. Remarkably, miR-589–3p +6 site is peculiar as miR-site displaying ∼100% editing and, for this reason, it reminds to the well-known, almost fully-edited, GRIA2 Q/R site (4). Indeed, both sites are ADAR2-/brain-specific and loss of editing at these positions promotes glioblastoma cell invasion [Figures 4 and 5 and (55)].
We showed that editing at +6 of miR-589–3p is sufficient to significantly decrease both cell proliferation and migration in several glioblastoma cell lines (A172, T98G and U87-MG), while the unedited miR-589–3p promoted cell proliferation and accelerated cell migration. We demonstrated that the unedited miR-589–3p promoted cell invasion and increased the metalloproteinase 9 (MMP-9) activity; on the contrary, the edited miR-589–3p constrained cell invasion and decreased MMP-9 activity.
To investigate whether this single A → I/G base change within miR-589–3p affects the selection of target genes, we took advantage of available computational algorithms, [miRDB (41) and TargetScan (42)], to predict uned-/edi-miRNA-target specific interactions. Among possible target genes controlled by edi-miR-589–3p, we concentrated on the Disintegrin and Metalloproteinase 12 (ADAM12) that is a novel and important protease promoting glioblastoma aggressiveness (44,56). We demonstrated that, while miR-589–3p silences PCDH9 mRNA, the edited miR-589–3p (edi-miR-589–3p) specifically targets ADAM12 and inhibits cell migration/invasion. Of note, edi-miR-589–3p constrained not only cell migration/invasion but also cell proliferation. Recently ADAM12 has been also connected with glioblastoma cell proliferation through its shedding activity over HB-EGF (56).
Since miR-589–3p increases cell proliferation and migration/invasion, it appears reasonable why, in normal brain, we found a high editing level (almost 100%) of this miRNA, as it might prevent abnormal cell proliferation/migration/invasion. It is intriguing that this key miR-site (+6 miR-589–3p) is controlled post-transcriptionally by RNA editing machinery. We hypothesize that editing, acting as an ‘on/off system’, may be necessary to quickly respond to different stimuli (for example during brain development, memory process, cell plasticity or brain injury). Indeed, we found that editing within miR-589–3p increased from fetal to adult brain. At the same time, glioblastoma cells may take advantage of this editing machinery by decreasing editing within miR-589–3p to increase cell aggressiveness/spreading when necessary.
This study demonstrates that the edi-miR-589–3p could be an alternative more specific and effective approach to control simultaneously abnormal cell proliferation/migration and, most importantly, invasion/spreading of glioblastoma cells. Altogether, our data highlight the importance of RNA editing as a promising field of research for brain and brain cancer.
DATA AVAILABILITY
The sequencing data have been submitted to the NCBI Sequence Read Archive (SRA) under accession number SRP125247.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Lucia Ricci-Vitiani for providing ADAM12 antibody, Federica Galeano, Lorenzo Cucina and Nicolò Mangraviti for their scientific support in some experiments. We also thank Graziano Pesole and IBBE CNR for providing computational facilities.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Associazione Italiana Ricerca sul Cancro (AIRC) IG Grants [13202, 17615 to A.G.]; AIRC 5 × 1000 [9962 to F.L.]. Funding for open access charge: AIRC Grants.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kornblihtt A.R., Schor I.E., Allo M., Dujardin G., Petrillo E., Munoz M.J.. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013; 14:153–165. [DOI] [PubMed] [Google Scholar]
- 2. Scholzova E., Malik R., Sevcik J., Kleibl Z.. RNA regulation and cancer development. Cancer Lett. 2007; 246:12–23. [DOI] [PubMed] [Google Scholar]
- 3. Gallo A., Locatelli F.. ADARs: allies or enemies? The importance of A-to-I RNA editing in human disease: from cancer to HIV-1. Biol. Rev. Camb. Phil. Soc. 2012; 87:95–110. [DOI] [PubMed] [Google Scholar]
- 4. Higuchi M., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H.. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000; 406:78–81. [DOI] [PubMed] [Google Scholar]
- 5. Wang Q., Khillan J., Gadue P., Nishikura K.. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000; 290:1765–1768. [DOI] [PubMed] [Google Scholar]
- 6. Keegan L.P., Gallo A., O’Connell M.A.. Development. Survival is impossible without an editor. Science. 2000; 290:1707–1709. [DOI] [PubMed] [Google Scholar]
- 7. Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002; 71:817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gallo A., Locatelli F.. ADARs: allies or enemies? The importance of A-to-I RNA editing in human disease: from cancer to HIV-1. Biological reviews of the Cambridge Philos. Society. 2011; 87:95–110. [DOI] [PubMed] [Google Scholar]
- 9. Tomaselli S., Locatelli F., Gallo A.. The RNA editing enzymes ADARs: mechanism of action and human disease. Cell Tissue Res. 2014; 356:527–532. [DOI] [PubMed] [Google Scholar]
- 10. Rueter S.M., Dawson T.R., Emeson R.B.. Regulation of alternative splicing by RNA editing. Nature. 1999; 399:75–80. [DOI] [PubMed] [Google Scholar]
- 11. Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K.. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007; 315:1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Picardi E., Manzari C., Mastropasqua F., Aiello I., D’Erchia A.M., Pesole G.. Profiling RNA editing in human tissues: towards the inosinome Atlas. Sci. Rep. 2015; 5:14941–14957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kitaura H., Sonoda M., Teramoto S., Shirozu H., Shimizu H., Kimura T., Masuda H., Ito Y., Takahashi H., Kwak S. et al. Ca2+ -permeable AMPA receptors associated with epileptogenesis of hypothalamic hamartoma. Epilepsia. 2017; 58:e59–e63. [DOI] [PubMed] [Google Scholar]
- 14. Gaisler-Salomon I., Kravitz E., Feiler Y., Safran M., Biegon A., Amariglio N., Rechavi G.. Hippocampus-specific deficiency in RNA editing of GluA2 in Alzheimer's disease. Neurobiol. Aging. 2014; 35:1785–1791. [DOI] [PubMed] [Google Scholar]
- 15. Tomaselli S., Galeano F., Massimi L., Di Rocco C., Lauriola L., Mastronuzzi A., Locatelli F., Gallo A.. ADAR2 editing activity in newly diagnosed versus relapsed pediatric high-grade astrocytomas. BMC Cancer. 2013; 13:255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Galeano F., Rossetti C., Tomaselli S., Cifaldi L., Lezzerini M., Pezzullo M., Boldrini R., Massimi L., Di Rocco C.M., Locatelli F. et al. ADAR2-editing activity inhibits glioblastoma growth through the modulation of the CDC14B/Skp2/p21/p27 axis. Oncogene. 2013; 32:998–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peng P.L., Zhong X., Tu W., Soundarapandian M.M., Molner P., Zhu D., Lau L., Liu S., Liu F., Lu Y.. ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron. 2006; 49:719–733. [DOI] [PubMed] [Google Scholar]
- 18. Dolecek T.A., Propp J.M., Stroup N.E., Kruchko C.. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro-oncology. 2012; 14(Suppl. 5):v1–v49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D., Misra A., Nigro J.M., Colman H., Soroceanu L. et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006; 9:157–173. [DOI] [PubMed] [Google Scholar]
- 20. van den Boom J., Wolter M., Kuick R., Misek D.E., Youkilis A.S., Wechsler D.S., Sommer C., Reifenberger G., Hanash S.M.. Characterization of gene expression profiles associated with glioma progression using oligonucleotide-based microarray analysis and real-time reverse transcription-polymerase chain reaction. Am. J. Pathol. 2003; 163:1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maas S., Patt S., Schrey M., Rich A.. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. 2001; 98:14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alon S., Mor E., Vigneault F., Church G.M., Locatelli F., Galeano F., Gallo A., Shomron N., Eisenberg E.. Systematic identification of edited microRNAs in the human brain. Genome Res. 2012; 22:1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tomaselli S., Galeano F., Alon S., Raho S., Galardi S., Polito V.A., Presutti C., Vincenti S., Eisenberg E., Locatelli F. et al. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol. 2015; 16:5–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamoun A., Idbaih A., Dehais C., Elarouci N., Carpentier C., Letouze E., Colin C., Mokhtari K., Jouvet A., Uro-Coste E. et al. Integrated multi-omics analysis of oligodendroglial tumours identifies three subgroups of 1p/19q co-deleted gliomas. Nat. Commun. 2016; 7:11263–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Picardi E., Pesole G.. REDItools: high-throughput RNA editing detection made easy. Bioinformatics. 2013; 29:1813–1814. [DOI] [PubMed] [Google Scholar]
- 26. Gong J., Wu Y., Zhang X., Liao Y., Sibanda V.L., Liu W., Guo A.Y.. Comprehensive analysis of human small RNA sequencing data provides insights into expression profiles and miRNA editing. RNA Biol. 2014; 11:1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nigita G., Acunzo M., Romano G., Veneziano D., Lagana A., Vitiello M., Wernicke D., Ferro A., Croce C.M.. microRNA editing in seed region aligns with cellular changes in hypoxic conditions. Nucleic Acids Res. 2016; 44:6298–6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alon S., Vigneault F., Eminaga S., Christodoulou D.C., Seidman J.G., Church G.M., Eisenberg E.. Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res. 2011; 21:1506–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 30. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 31. Larder B.A., Kohli A., Kellam P., Kemp S.D., Kronick M., Henfrey R.D.. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature. 1993; 365:671–673. [DOI] [PubMed] [Google Scholar]
- 32. Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010; 17:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gabriely G., Yi M., Narayan R.S., Niers J.M., Wurdinger T., Imitola J., Ligon K.L., Kesari S., Esau C., Stephens R.M. et al. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011; 71:3563–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xia H., Ooi L.L., Hui K.M.. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 2013; 58:629–641. [DOI] [PubMed] [Google Scholar]
- 35. Kawahara Y., Zinshteyn B., Chendrimada T.P., Shiekhattar R., Nishikura K.. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Reports. 2007; 8:763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cenci C., Barzotti R., Galeano F., Corbelli S., Rota R., Massimi L., Di Rocco C., O’Connell M.A., Gallo A.. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J. Biol. Chem. 2008; 283:7251–7260. [DOI] [PubMed] [Google Scholar]
- 37. Koo S., Martin G.S., Schulz K.J., Ronck M., Toussaint L.G.. Serial selection for invasiveness increases expression of miR-143/miR-145 in glioblastoma cell lines. BMC Cancer. 2012; 12:143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu S., Yin F., Zhang J., Wicha M.S., Chang A.E., Fan W., Chen L., Fan M., Li Q.. Regulatory roles of miRNA in the human neural stem cell transformation to glioma stem cells. J. Cell Biochem. 2014; 115:1368–1380. [DOI] [PubMed] [Google Scholar]
- 39. Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016; 17:83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Toth M., Fridman R.. Assessment of gelatinases (MMP-2 and MMP-9) by gelatin zymography. Methods Mol. Med. 2001; 57:163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wong N., Wang X.. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015; 43:D146–D152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lewis B.P., Burge C.B., Bartel D.P.. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005; 120:15–20. [DOI] [PubMed] [Google Scholar]
- 43. Wang C., Yu G., Liu J., Wang J., Zhang Y., Zhang X., Zhou Z., Huang Z.. Downregulation of PCDH9 predicts prognosis for patients with glioma. J. Clin. Neurosci. 2012; 19:541–545. [DOI] [PubMed] [Google Scholar]
- 44. Lulli V., Buccarelli M., Martini M., Signore M., Biffoni M., Giannetti S., Morgante L., Marziali G., Ilari R., Pagliuca A. et al. miR-135b suppresses tumorigenesis in glioblastoma stem-like cells impairing proliferation, migration and self-renewal. Oncotarget. 2015; 6:37241–37256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Iba K., Albrechtsen R., Gilpin B., Frohlich C., Loechel F., Zolkiewska A., Ishiguro K., Kojima T., Liu W., Langford J.K. et al. The cysteine-rich domain of human ADAM 12 supports cell adhesion through syndecans and triggers signaling events that lead to beta1 integrin-dependent cell spreading. J. Cell Biol. 2000; 149:1143–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hoelzinger D.B., Mariani L., Weis J., Woyke T., Berens T.J., McDonough W.S., Sloan A., Coons S.W., Berens M.E.. Gene expression profile of glioblastoma multiforme invasive phenotype points to new therapeutic targets. Neoplasia. 2005; 7:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee Y.S., Dutta A.. MicroRNAs: small but potent oncogenes or tumor suppressors. Curr. Opin. Investig. Drugs. 2006; 7:560–564. [PubMed] [Google Scholar]
- 48. Esquela-Kerscher A., Slack F.J.. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 2006; 6:259–269. [DOI] [PubMed] [Google Scholar]
- 49. Dalmay T. MicroRNAs and cancer. J. Intern. Med. 2008; 263:366–375. [DOI] [PubMed] [Google Scholar]
- 50. Choudhury Y., Tay F.C., Lam D.H., Sandanaraj E., Tang C., Ang B.T., Wang S.. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J. Clin. Investig. 2012; 122:4059–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paul D., Sinha A.N., Ray A., Lal M., Nayak S., Sharma A., Mehani B., Mukherjee D., Laddha S.V., Suri A. et al. A-to-I editing in human miRNAs is enriched in seed sequence, influenced by sequence contexts and significantly hypoedited in glioblastoma multiforme. Sci. Rep. 2017; 7:2466–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Candiani S. Focus on miRNAs evolution: a perspective from amphioxus. Brief. Funct. Genomics. 2012; 11:107–117. [DOI] [PubMed] [Google Scholar]
- 53. Yamamoto K., Seike M., Takeuchi S., Soeno C., Miyanaga A., Noro R., Minegishi Y., Kubota K., Gemma A.. MiR-379/411 cluster regulates IL-18 and contributes to drug resistance in malignant pleural mesothelioma. Oncol. Rep. 2014; 32:2365–2372. [DOI] [PubMed] [Google Scholar]
- 54. Dichamp C., Taillibert S., Aguirre-Cruz L., Lejeune J., Marie Y., Kujas M., Delattre J.Y., Hoang-Xuan K., Sanson M.. Loss of 14q chromosome in oligodendroglial and astrocytic tumors. J. Neuro-oncol. 2004; 67:281–285. [DOI] [PubMed] [Google Scholar]
- 55. Ishiuchi S., Tsuzuki K., Yoshida Y., Yamada N., Hagimura N., Okado H., Miwa A., Kurihara H., Nakazato Y., Tamura M. et al. Blockage of Ca(2+)-permeable AMPA receptors suppresses migration and induces apoptosis in human glioblastoma cells. Nat. Med. 2002; 8:971–978. [DOI] [PubMed] [Google Scholar]
- 56. Kodama T., Ikeda E., Okada A., Ohtsuka T., Shimoda M., Shiomi T., Yoshida K., Nakada M., Ohuchi E., Okada Y.. ADAM12 is selectively overexpressed in human glioblastomas and is associated with glioblastoma cell proliferation and shedding of heparin-binding epidermal growth factor. Am. J. Pathol. 2004; 165:1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data have been submitted to the NCBI Sequence Read Archive (SRA) under accession number SRP125247.