Abstract
Four different types (α4, α′2, (αβ)2 and ϵ2) of RNA-splicing endonucleases (EndAs) for RNA processing are known to exist in the Archaea. Only the (αβ)2 and ϵ2 types can cleave non-canonical introns in precursor (pre)-tRNA. Both enzyme types possess an insert associated with a specific loop, allowing broad substrate specificity in the catalytic α units. Here, the hyperthermophilic euryarchaeon Methanopyrus kandleri (MKA) was predicted to harbor an (αβ)2-type EndA lacking the specific loop. To characterize MKA EndA enzymatic activity, we constructed a fusion protein derived from MKA α and β subunits (fMKA EndA). In vitro assessment demonstrated complete removal of the canonical bulge-helix-bulge (BHB) intron structure from MKA pre-tRNAAsn. However, removal of the relaxed BHB structure in MKA pre-tRNAGlu was inefficient compared to crenarchaeal (αβ)2 EndA, and the ability to process the relaxed intron within mini-helix RNA was not detected. fMKA EndA X-ray structure revealed a shape similar to that of other EndA types, with no specific loop. Mapping of EndA types and their specific loops and the tRNA gene diversity among various Archaea suggest that MKA EndA is evolutionarily related to other (αβ)2-type EndAs found in the Thaumarchaeota, Crenarchaeota and Aigarchaeota but uniquely represents constrained substrate specificity.
INTRODUCTION
Transfer RNAs (tRNAs) are essential molecules for all living organisms and play a fundamental role in decoding genetic information through the codon–anticodon base pairing between a messenger RNA (mRNA) and a tRNA molecule charged with the amino acid corresponding to the triplet sequences within ribosomes. The tRNA needs to be matured for the proper functioning of protein synthesis. In addition to the general translation event, tRNA-related fragments, so-called tRFs, have also been found to have roles in cell proliferation, regulation of gene expression, modulation of the DNA damage response, and tumor suppression (1).
During tRNA maturation, a splicing event occurs in the precursor (pre)-tRNA to remove the intron sequences. Either autocatalytic or enzymatic tRNA splicing mechanisms are adopted across the three domains of life. Group I self-splicing introns, which are auto-catalytically removed with an external guanosine-5′-triphosphate (GTP), are found in pre-tRNA from some bacteria and higher eukaryote plastids (2,3). In contrast, introns of archaeal pre-tRNA are enzymatically removed by an RNA-splicing endonuclease (EndA) (4), and then, the two halves of the tRNA exon are connected by a GTP-dependent 3′–5′ RNA ligase (RtcB), an enzyme requiring manganese (5–9). Archaeal EndA is classified into four types: homotetramer (α4), heterotetramer ((αβ)2), and two different homodimers (α′2 and ϵ2), according to their subunit component and subdomain arrangement (10–18). The structural and functional features of archaeal EndAs have been well characterized; the overall structures show a common shape of rectangular parallelepiped, and the catalytic triad (histidine, tyrosine and lysine), along with two arginine or arginine and tryptophan residues responsible for substrate recognition, are well conserved (11,13). The enzymatic tRNA splicing mechanism in the Archaea can also be found in eukaryotes, with the conserved structural and functional similarity between their EndA and RtcB enzymes suggesting their common origin (11,19,20). Archaeal and eukaryotic EndAs can recognize and remove introns, forming a structure known as the bulge–helix–bulge (BHB) motif, which consists of two bulges (3 nt) separated by one helix (4 nt) located at the intron–exon boundary of pre-tRNAs (11,21). The majority of the canonical BHB motifs are found in the anticodon loop between position 37 and 38 (37/38) of archaeal pre-tRNA, although in some cases, this motif is observed in other RNA species such as pre-mRNA (22–24) and pre-ribosomal RNA (rRNA) and requires removal for their maturation (25,26). The relaxed forms of the BHB motif, known as the non-canonical introns (HBh′ and hBH), have a disrupted 5′- or 3′-bulge, and in an extreme case, one of the bulges is missing to form a relaxed bulge-helix-loop (BHL) motif. It has been shown that only the (αβ)2 and ϵ2 types of archaeal EndAs can remove these non-canonical introns with the help of a structurally inserted ‘specific loop,’ in which the conserved lysine residue plays an important role in acquiring broad substrate specificity, i.e. the capability of cleaving introns located outside the anticodon loop, with the relaxed BHB motif lacking one of the two bulges (13,16,27–29). In comparison, eukaryotic EndA requires a mature domain of pre-tRNA for the removal of non-canonical introns (11,30).
It has been hypothesized that the α4 type, which encodes a single catalytic subunit, constitutes the ancestral form of the EndA protein, and that subsequent subfunctionalization through gene duplication and fusion led to the emergence of the other three types (α′2, (αβ)2 and ϵ2) (10). While the EndAs found in Euryarchaeota are mostly α4 and α′2 types (10), the hyperthermophilic euryarchaeon Methanopyrus kandleri (MKA) genome has the α and β subunit genes encoding the EndA resembling the (αβ)2 type (31,32). The (αβ)2 type is widely conserved within the TACK superphylum (33), including Thaumarchaeota, Crenarchaeota, and Aigarchaeota, with the exception of Korarchaeota, but is also found in the genome of Nanoarchaeum equitans, belonging to the Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota, and Nanohaloarchaea (DPANN) superphylum (34). The evolutionary origin of the ϵ2 type remains elusive because it has only been discovered from Candidatus Micrarchaeum acidiphilum (Archaeal Richmond Mine acidophilic nano-organism (ARMAN)-1 and ARMAN-2), also belonging to DPANN (12,26). Structurally, EndAs found in the TACK superphylum have acquired a specific loop in the N-terminus of the catalytic α subunit, known as a Crenarchaea-specific loop (CSL), capable of cleaving the non-canonical intron from pre-tRNA. Notably, the ARMAN-2 ϵ2 EndA also harbors an ARMAN-2-type specific loop (ASL) at the same structural location, and in both cases, a conserved lysine residue is responsible for providing the broad substrate specificity (13,28). Organisms with (αβ)2 and ϵ2 EndA possess higher numbers of disrupted tRNA genes with features including multiple intron-containing tRNAs (35), split and tri-split tRNAs in which the tRNA fragments are encoded by two or three genes (36,37), and permuted tRNAs in which the 5′ and 3′ halves of the coding sequences are inverted (38). Whereas the precise mechanism of intron insertion and disruption remains unclear, it has been proposed that acquisition of the specific loop in the EndA catalytic subunit, leading to the recognition of relaxed RNA intron motifs, allowed the overall disruption of the tRNA gene structure (13,39,40). To assess the above evolutionary scenario, we performed functional and structural studies of the MKA EndA, given that it stands as the only known (αβ)2 type that lacks the specific loop. The placement of M. kandleri in the archaeal phylogenetic tree remains debated owing to the high rate of gene loss, capture, and recombination (41). Thus, bioinformatics analysis was also applied to fully capture the correlation between tRNA gene diversity and EndA subfunctionalization through changes in unit/subunit architectures and the acquisition of specific loops.
MATERIALS AND METHODS
Protein expression and purification
The M. kandleri genomic DNA (BRC number, JGD7495) (32) was provided by Riken BRC through the National Bio-Resource project of MEXT, Japan. The genes encoding the α and β subunits of MKA EndA were individually cloned into the NdeI and BamHI sites of the vector pET-15 b (Novagen), which contains the coding sequence for a thrombin-cleavable N-terminal His6 tag, using the primers MKA_aF, MKA_aR, MKA_bF and MKA_bR, and designated as plasmids pETA and pETB. In pETB, the sequences of the thrombin recognition site were removed by inverse PCR using the primers Del_F and Del_R, resulting in pETB1. To obtain a plasmid expressing the fusion gene of β and α subunits of MKA EndA, the α subunit gene (including the nine coding sequences at its 5′ terminus, which encode three glycine residues functioning as the linker of the fusion gene) was amplified by PCR with pETA as the template DNA, using the primers GlyF and GlyR. Then, the PCR product was inserted into the site between the sequences of the β subunit gene and the stop codon in pETB1 using the In-Fusion HD Cloning Kit (Clontech), resulting in the plasmid pFMKAEndA. The primers used for the construction of the plasmid harboring the β/α subunit fusion gene are listed in Supplementary Table S1. The pFMKAEndA plasmid was transformed into Escherichia coli BL21 (DE3) Rossetta2 (Novagen), and the transformants were grown in LB medium supplemented with 100 μg/ml ampicillin at 37°C. When the cell density reached OD600 = −0.8, isopropylthio-β-galactoside was added to a final concentration of 0.5 mM, and cells were grown for an additional 24 h at 20°C. Cells were then harvested by centrifugation (4320 × g at 20°C for 20 min). Twenty-three grams of the cells was suspended in 40 ml buffer A (50 mM Tris–HCl [pH 8.0], 400 mM KCl, 5 mM imidazole, and 5% glycerol) supplemented with Halt protease inhibitor single-use cocktail (Thermo Scientific) and then disrupted using an ultrasonic disruptor (model VCX-500; Sonics & Materials. Inc.). To remove a fraction of E. coli proteins by heat denaturation, lysed cells were incubated at 70°C for 20 min and pelleted by centrifugation (38 900 × g at 4°C for 20 min). The supernatant was loaded onto a Ni-NTA Superflow column (Qiagen) equilibrated with buffer A, and the enzyme was eluted using buffer A containing 500 mM imidazole. The eluted fractions were collected and loaded onto a HiTrap Heparin-Sepharose column (GE Healthcare) pre-equilibrated with buffer B (20 mM Tris–HCl [pH 7.6], 20 mM KCl, 10 mM 2-mercaptoethanol, and 5% glycerol). The bound protein was eluted with a linear gradient of buffer B containing 50 mM of 1.5 M KCl and then pooled. Ammonium sulfate was further added to a final concentration of 65% saturation. The precipitate was collected by centrifugation (38 900 × g at 4°C for 20 min) and dissolved with 300 μl of the buffer (10 mM Tris–HCl [pH 7.6]). The concentration of recombinant fMKA EndA was ∼7.0 mg/ml. The purity of concentrated fMKA EndA was confirmed by SDS-PAGE (Supplementary Figure S2). Mutant genes (Y295A, H303A and K334) were generated using the QuickChange site-directed mutagenesis kit (Stratagene), and the mutations were verified by DNA sequencing. Mutant proteins were expressed and purified in the same manner as for the wild-type protein. Recombinant Archaeoglobus fulgidus (AFU) and Aeropyrum pernix (APE) EndAs were prepared as reported previously (28).
Intron-cleavage assay by the splicing endonuclease
The transcript of M. kandleri pre-tRNAAsn (GUU) was prepared using T7 RNA polymerase as described in our previous report (42). For the preparation of the M. kandleri pre-tRNAGlu (UUC) transcript, the plasmid harboring its coding gene was constructed using the Mighty TA-cloning kit (TaKaRa). The pre-tRNAGlu gene was amplified by PCR from the plasmid DNA template and then transcribed using T7 RNA polymerase. The mini-helix RNA oligo forming the hBH motif was purchased from Hokkaido System Science, Japan. Splicing reactions were performed as follows: 0.45 nmol EndA was mixed with 4.5 nmol transcripts in 225 μl buffer (50 mM Tris–HCl [pH 7.6], 5 mM MgCl2, 6 mM 2-mercaptoethanol, and 50 mM KCl) and incubated at 70°C. Aliquots (50 μl) were taken at 1, 5, 10 and 15 min, and the cleaved RNA fragments were extracted using phenol/chloroform/isoamyl alcohol solution and analyzed using 15% PAGE/7 M urea. The gel was stained with 0.05% toluidine blue. To identify the cleavage sites, we further performed northern blot analysis (Supplementary Figures S3 and S4). After the cleavage reaction, the RNA fragments were separated by 15% PAGE/7 M urea and then transferred to a Hybond-N+ membrane (GE Healthcare) by electro blotting, and fixed by UV254 nm irradiation. Northern hybridization was performed with the hybridization buffer (GE Healthcare) using a 5′-32P-labeled DNA probe at 52°C overnight. Nucleotide sequences of the DNA probes used in this study are shown in Supplementary Figures S3 and S4. The hybridized bands were monitored using a FLA-2000 Typhoon laser scanner (GE Healthcare).
Crystallization of fMKA EndA protein
The concentrated solution of ∼7 mg/ml recombinant fMKA EndA was used for crystallization. Initial crystallization trials were set up in VDX48 plates with sealant (Hampton Research) using the hanging-drop vapor diffusion method and commercial crystallization screens from Hampton Research (Index, Salt RX, and Crystal screen). The drop solution was equilibrated against 200 μl of reservoir solution at 20°C. A few single crystals of fMKA EndA appeared after 4 days with Index screen reagent #18 (i.e. 1.4 M sodium/potassium phosphate pH 6.9). The single crystal featured a rectangular shape with a full size of 200 × 100 × 100 μm. For the experimental phase determination by the single-wavelength anomalous dispersion (SAD) method, the crystal was soaked in mother liquor supplemented with 4.0 mM F2OsCl6 at 20°C for 6 h and then soaked in mother liquor at 20°C for 1 h. Cryoprotection of the native and Os-induced crystals was achieved by stepwise transfer to the respective artificial mother liquor containing 25% ethylene glycol. The crystals were then flash-frozen in liquid nitrogen.
Data collection and structure determination
X-ray diffraction data sets from native crystals (λ = 1.0000) and SAD data sets from Os-induced crystals (λ = 1.13986) were collected at 100 K on the BL26B2 beamline at the SPring-8 synchrotron radiation facility (Hyogo, Japan). All data sets were processed, merged, and scaled using the HKL2000 program (43). Using the deduced Os-SAD data set, all 10 Os positions were identified and refined in the orthorhombic space group I222, and the initial phase was calculated by using AutoSol in PHENIX (44), followed by automated model building using RESOLVE (45). The resulting map and partial model were used for manually building the model using COOT (46). The model was further refined by using PHENIX (44). Using the native data set and the refined model as a search coordinate, the structure of the fMKA EndA was determined by molecular replacement using the Phaser-MR in PHENIX (44). The model was further manually built with COOT (46) and refined with PHENIX (44). The structure of fMKA EndA was refined to a Rwork/Rfree of 15.8%/19.6% at a 1.53 Å resolution (Table 1). The space group of the crystal belonged to I222, where one fMKA EndA molecule is present in an asymmetric unit. The final model contained residues 14–180 and 190–359, 292 water molecules, and two PO42− molecules. The final model of the fMKA EndA structure was further checked using PROCHECK (47), showing the quality of the refined model. Ramachandran plots (%) of the fMKA EndA structure are tabulated in Table 1. The structure factor and coordinates have been deposited in the Protein Data Bank (PDB code 5X89). All structural figures were generated using PyMOL (DeLano Scientific). The amino acid sequence alignment was generated using ClustalW 1.83 (48) and ESPript (49) programs.
Table 1. Data collection and refinement statistics.
| fMKA EndA | fMKA EndA | |
|---|---|---|
| Os-derivative | ||
| Data collection | ||
| Space group | I222 | I222 |
| Cell dimensions | ||
| a, b, c (Å) | 77.674, 93.059, 126.109 | 77.528, 93.495, 126.206 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50 to 1.53 (1.56–1.53) | 50 to 1.89 (1.92–1.89) |
| R merge a | 4.7 (23.9) | 5.7(17.7) |
| I / σI | 52.0 (8.7) | 108.3 (31.3) |
| Completeness (%) | 100(100) | 100(100) |
| Redundancy | 9.1 (9.0) | 29.4 (29.4) |
| Refinement | ||
| Resolution (Å) | 38.85 to 1.53 | |
| No. reflections | 69,045 | |
| R work b/ Rfreec | 15.8/19.6 | |
| No. atoms | 3,101 | |
| Protein | 2,799 | |
| Water | 292 | |
| No. PO42- | 2 | |
| Avg. B-factors (Å2) | 17.9 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | |
| Bond angles (°) | 1.051 | |
| Ramachandran plot (%) | ||
| Most favored | 95.9 | |
| Additional allowed | 3.8 | |
| Generously allowed | 0.3 | |
| Disallowed | 0.0 | |
The value in the parentheses is for the highest resolution shell.
a R merge = ΣΣj|<I(h)> – I(h)j|/ΣΣj|<I(h)>|, where <I(h)> is the mean intensity of symmetry-equivalent reflections.
b R work = Σ (IIFp(obs) – Fp(calc)II)/ΣIFp(obs)I.
c R free = R factor for a selected subset (10%) of reflections that was not included in earlier refinement calculations.
tRNA gene prediction
tRNA gene information was obtained and updated for 46 archaeal species based on previous publications (12,38). In addition, we predicted tRNA genes from four recently discovered DPANN superphylum members (Candidatus Pacearchaeota archaeon RBG_13_33_26, Candidatus Woesearchaeota archaeon RBG_13_36_6, Nanohaloarchaea archaeon SG9 and Candidatus Mancarchaeum acidiphilum Mia14), four Asgard superphylum members (Candidatus Odinarchaeota archaeon LCB_4, Candidatus Lokiarchaeota archaeon CR_4, Candidatus Thorarchaeota archaeon SMTZ1–83 and Heimdallarchaeota archaeon AB_125) and one Stygia superclass, Hadesarchaea archaeon YNP_N21, using tRNAscan-SE 2.0 with a setting of source ‘Archaeal’ and search mode ‘Default’ (50). tRNA gene candidates predicted with irregular loop or stem size were further subjected to BHB search using SPLITS, with the given parameters –p ‘0.5’ –f ‘’−5’ –h ‘4’ (51) to predict correct intron sequences.
Phylogenetic analysis of EndA proteins
We obtained conserved C-terminal domain sequences of 81 EndA protein units/subunits corresponding to the species used for the tRNA gene prediction. We excluded structural α′ unit sequences belonging to the archaeal orders Thermoplasmatales, Halobacteriales, and Archaeoglobales, due to the deletion of a sequence found in the C-terminal domain. Sequences were aligned using Seaview version 4 (52), and conserved core residues (45 amino acids in length) were used as the input for computing the maximum-likelihood tree using the PhyML v3.1 program (53) with default parameters of LG evolutionary model, aLRT for branch support, and the NNI tree search algorithm. The output data tree was visualized by iToL (54).
RESULTS
tRNA intron cleavage by the M. kandleri RNA-splicing endonuclease
According to the genomic tRNA database (GtRNAdb), 35 tRNA genes are predicted from the M. kandleri genome, among which seven genes are disrupted by intron insertion (55). RNA-Seq analysis has previously identified the matured form of M. kandleri tRNAs (56), indicating that these pre-tRNA species are likely processed by the M. kandleri RNA-splicing endonuclease (MKA EndA). The majority of the introns are located at the canonical position between 37 and 38 (37/38) in the anticodon loop of pre-tRNAAsn (GUU), except the single relaxed HBh′ motif located at the position between 20b and 21 (20b/21) in the D-loop of pre-tRNAGlu (UUC), which is a multiple intron-containing tRNA also harboring a canonical intron (Figure 1). To verify the removal of the two introns found in pre-tRNAGlu (UUC), we first attempted to co-express the α and β subunits of MKA EndA in an E. coli expression system. However, recombinant proteins were not obtained, presumably owing to the formation of a non-active homodimer, as reported in the case of the (αβ)2-type nanoarchaeal EndA (17). Hence, we expressed each subunit separately, and the purified proteins were mixed in the presence of buffer solution with 6 M urea, allowing the partial unfolding of proteins for the promotion of heterodimer (αβ)2 formation. After the dialysis, very weak intron cleavage activity was observed, indicating that most of the subunits remained in an inactive state (data not shown).
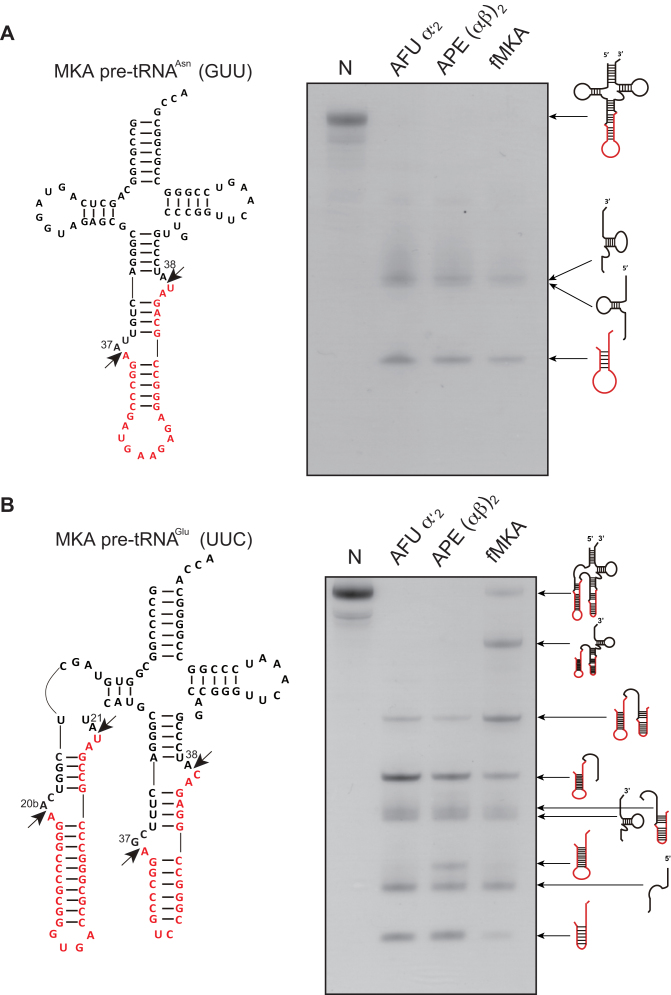
Figure 1.
Splicing activity and specificity of AFU EndA, APE EndA and fMKA EndA. (A) Left, predicted secondary structure of MKA pre-tRNAAsn (GUU) containing the intron with the BHB motif at the canonical position 37/38 (red). The two arrows show the cleavage sites. Right, splicing reaction of the MKA pre-tRNAAsn (GUU) at 70°C for 5 min. (B) Left, predicted secondary structure of MKA pre-tRNAGlu (UUC) containing double introns consisting of a non-canonical HBh′ intron at 20b/21 and a canonical intron at 37/38 (red). The two arrows show the cleavage sites. Right, splicing reaction of the MKA pre-tRNAGlu (UUC) at 70°C for 5 min. Reaction mixtures were separated on 15% polyacrylamide/7 M urea gels. In each gel, ‘N’ (first lane) indicates the control (no enzyme). The cleavage products are shown using schematic models at the right-hand side of the gel.
To overcome this problem, we further constructed a recombinant fusion-MKA EndA (fMKA EndA) protein of α and β subunits to form an α–β fusion dimer resembling the native heterotetrameric structure. The enzymatic activity of fMKA was compared with those of α′2-type Euryarchaeota AFU EndA and (αβ)2-type Crenarchaeota APE EndA. Using M. kandleri pre-tRNAAsn with a canonical intron as a substrate, we confirmed that all three types of EndAs were capable of removing the canonical intron from M. kandleri pre-tRNAAsn in 5 min at 70°C. However, under the same condition, clear differences were observed for the cleavage pattern of pre-tRNAGlu (UUC) harboring both canonical and non-canonical introns. Whereas the (αβ)2-type APE EndA could splice both the strict canonical BHB and relaxed non-canonical HBh′ intron located in the D-loop, AFU EndA and fMKA EndA failed to show the efficient removal of the non-canonical intron, instead leaving a sizable portion of the pre-tRNA 5′ fragment, suggesting that cleavage mostly occurred at the bulge of the HBh′ motif (Figure 1B). We have previously identified that the removal of the non-canonical intron by APE EndA is achieved by the acquisition of the CSL, which confers the ability to recognize both the strict and relaxed intronic forms (28).
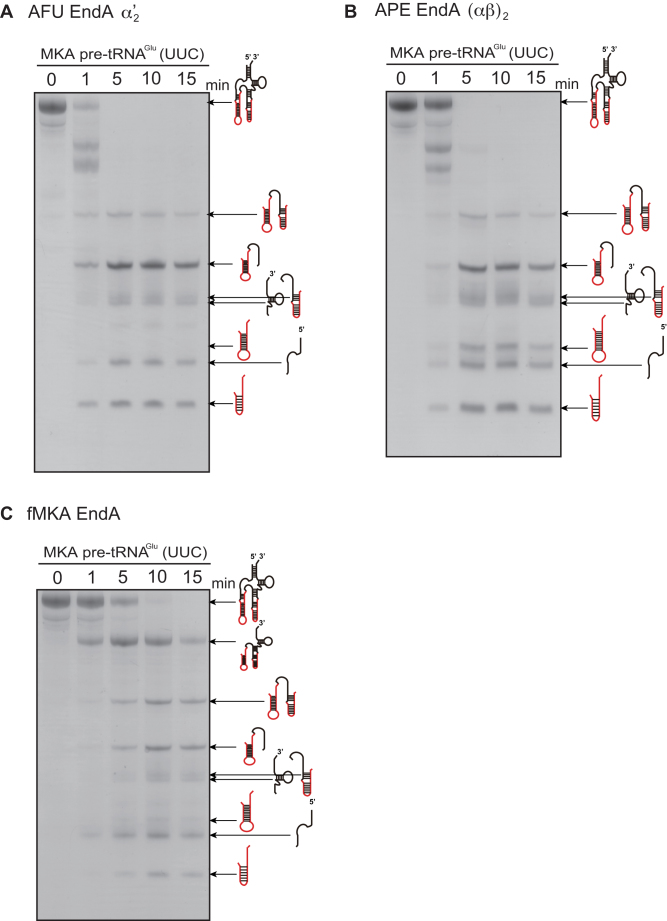
To further characterize the splicing pattern and efficiency of intron cleavage by the three EndAs, we performed a time-sensitive intron cleavage assay using the same pre-tRNAGlu (UUC) (Figure 2). The AFU and APE EndAs exhibited cleavage of the canonical intron within 1 min (Figures 2A and 2B), whereas fMKA EndA required 5 min to obtain the same splicing pattern (Figure 2C). Thus, the rate of intron processing in the fMKA EndA is slower than that in the AFU and APE EndAs under the given experimental conditions. In addition, non-canonical intron cleavage by APE EndA was obtained after 1 min (Figure 2B), whereas a trace of the cleaved free non-canonical intron became visible for fMKA EndA only after 10 min (Figure 2C), and no clear evidence was seen for AFU EndA (Figure 2A). Northern blot analysis was further conducted to determine the relative positions of the processed tRNA fragments (Supplementary Figure S4), although the shortest fragment between position A21 and G37 was not detected because the fragment could not be obtained by phenol/chloroform/isoamyl alcohol and ethanol extraction after the splicing reaction. From the splicing patterns of the three different EndAs, even after 20 min of incubation, it appeared that the non-canonical intron and canonical intron both remained to be processed. Among the remaining partially cleaved pre-tRNA fragments, the most prominent was the fragment corresponding to the non-canonical intron, followed by the sequence from the D-stem (Figure 2). Collectively, these results indicate that the complete removal of the non-canonical intron requires additional effort, and thus, removal of the canonical intron before the non-canonical intron likely occurs in the case of all three EndAs.
Figure 2.
Time-dependent intron cleavage activity of the MKA pre-tRNAGlu (UUC) by AFU EndA (A), APE EndA (B) and fMKA EndA (C). Reaction mixtures were separated on 15% polyacrylamide/7 M urea gels. In each gel, ‘N’ (first lane) indicates the control (no enzyme). The cleavage products are shown using schematic models at the right-hand side of the gel.
X-ray structure of fMKA EndA
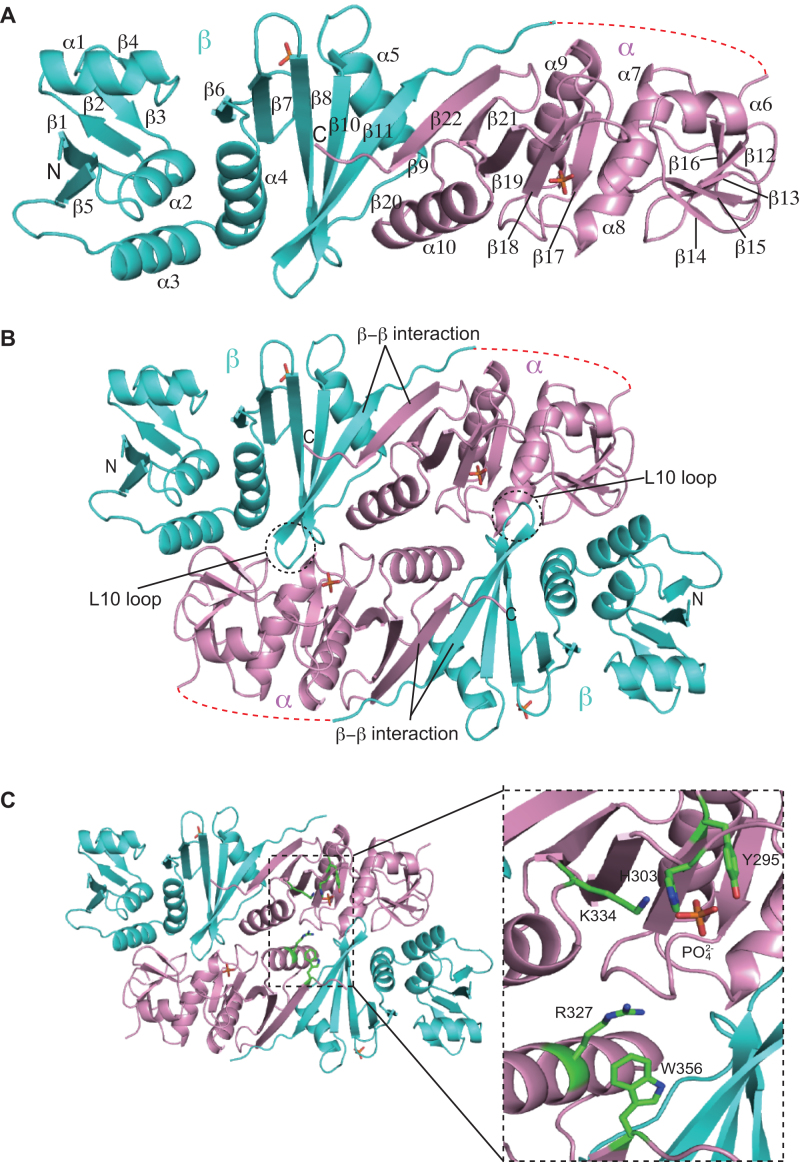
The X-ray structure of the α–β subunit fusion protein fMKA EndA was determined at a 1.53 Å resolution using SAD (Figure 3A). Crystallographic statistics are tabulated in Table 1. One molecule exists in each asymmetric unit of the fMKA EndA crystal. The N-terminal β unit (residues 14–178) is connected to the C-terminal α unit (residues 190–359) through the linker region (residues 179–189). The larger part of the linker region (residues 181–189, dotted red line in Figure 3A) and the N-terminal 6 × His tag plus the protease site (residues 1–13) was disordered. Two phosphate ions are observed at each of the β and α units. The N-terminal β unit consists of five α-helices (α1–α5) and eleven β strands forming mixed anti-parallel and parallel β sheets (β1–β5 and β6–β11). The overall structure of the C-terminal α unit is similar to the N-terminal β unit with mixed anti-parallel and parallel β sheets (β12–β16 and β17–β22) surrounded by the five α-helices (α6–α10). Figure 3B shows a homodimer structure of fMKA EndA, presumably resembling the native (αβ)2 heterotetramer. In particular, the overall shape of the homodimer is rectangular parallelepiped, which is similar to the shape of other active EndA structures (Supplementary Figure S5). The dimer formation of fMKA EndA is achieved by the interaction of a negatively charged L10 loop within the β unit with the positively charged pocket of the α unit (dotted circle in Figure 3B). D167 in the L10 loop forms a salt bridge with K287 at a distance of 2.75 Å. Similarly, interaction between the α and β subunit is maintained by a β−β strand hydrophobic interaction (β11–β22). Both the electrostatic interaction and hydrophobic interaction are well conserved in all four types of EndA structures to facilitate intra/inter-unit assembly (Supplementary Figures S5 and S6).
Figure 3.
X-ray crystal structure of fMKA EndA. (A) Ribbon diagram of the monomeric structure in the asymmetric unit of the fMKA crystal. The α and β units are colored magenta and cyan, respectively. The N- and C-terminal ends are labeled as N and C, respectively. The secondary structures of the α helix and β strand are labeled (in order) as α and β, respectively. The disordered linker region connecting the β and α subunits is depicted by red dotted lines. Two phosphate ions are shown by the stick model (orange). (B) Ribbon diagram of the overall structure of the functional homodimeric complex. The β–β interaction responsible for inter/intra-unit formation, and the L10 loop and pocket responsible for dimer/tetramer formation are highlighted. (C) Close-up view of the active and substrate recognition sites. Three residues (Y295, H303 and K334) and two residues (R327 and W356) are expected to be involved in catalysis and substrate recognition, respectively. These residues are shown by a stick model (green).
Putative residues responsible for catalysis and substrate recognition
RNA cleavage is known to be conducted by the catalytic triad (tyrosine, histidine, and lysine) and the two substrate recognition residues (two arginines or arginine and tryptophan) that are well conserved among archaeal EndAs (11,57). In fMKA EndA, we found candidate residues for the catalytic triad (Y295, H303 and K334) and two substrate recognition residues (R327 and W356) in the α unit responsible for recognition of the bulge structure in the BHB motif (Figure 3C). Notably, a single phosphate ion is bound to the catalytic pocket near the catalytic triad, thereby mimicking a phosphate group of the substrate RNA.
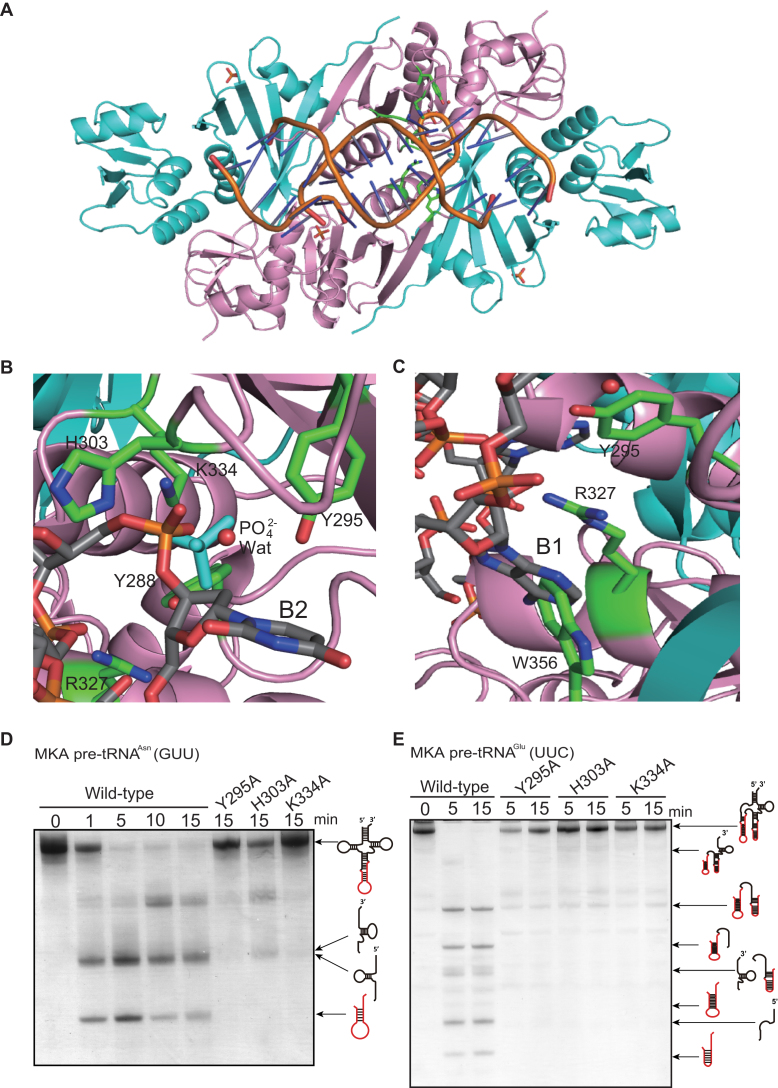
To further explore the possibility that the candidate residues are involved in intron cleavage and substrate RNA recognition, we created a docking model of fMKA EndA with its substrate RNA harboring the BHB motif, on the basis of the previously reported AFU-EndA-RNA complex structure (57) (Figure 4A). Given the identical structure between fMKA EndA and AFU EndA (Supplementary Figures S5A and S6), the model clearly represents that the phosphate group from the second RNA bulge (B2) is found at the similar location with the phosphate ion bound near the catalytic triad (Figure 4B). The tyrosine Y295 residue is located adjacent to the 2′-hydroxyl of the ribose of B2, suggesting that this residue plays an important role in the deprotonation of the 2′-nucleophilic oxygen during the cleavage. H303 and K334 residues both form a hydrogen bond with the phosphate ion bound to the α unit of fMKA EndA (Supplementary Figure S7, Nζ K334-O2 PO42− [2.8 Å] and Nδ H303-O2 PO42− [2.8 Å]), indicating that H303 is an acid catalytic residue responsible for donating a proton to the 5′-leaving group of B2, and that K334 contributes to the overall stabilization of the acid/base catalytic reaction carried out by H303 and Y295, consistent with the catalytic mechanism proposed for other EndAs (11,57). The adenine base in the first bulge (B1) is known to be flipped out from the RNA strand owing to the cation–π interaction caused by two adjacent arginine residues in α2- and α4-type EndAs, or by arginine and tryptophan residues in the case of (αβ)2- and ϵ2-type EndAs (11,13). Here, R327 and W356 residing near the adenine base appear to interact through cation–π interactions (Figure 4C).
Figure 4.
Docking model of the fMKA EndA-RNA complex (A). The model was created on the basis of the AFU EndA-RNA complex (57). The α and β units are colored as in Figure 3A. The phosphate backbones of double-stranded RNA are colored orange, and the base components are colored gray. (B) The candidates for the three catalytic residues (Y295, H303, and K334) are shown by a green stick model in the active site. The phosphate ion (PO42−) and water (Wat) are depicted as stick (cyan) and ball (red) models, respectively. The uracil in the second bulge (B2) of the BHB motif is shown by a stick model (gray). (C) The candidates for the two residues (R327 and W356) responsible for substrate recognition are shown by a green stick model. The adenosine in the first bulge (B1) of the BHB motif appears to be in a stacking coordination with two amino acid residues, R327 and W356. (D) Intron cleavage activity of MKA pre-tRNAAsn (GUU) by wild-type fMKA EndA and three Ala-substituted mutants (Y295A, H303A, and K334A). (E) Intron cleavage activity of MKA pre-tRNAGlu (UUC) by wild-type fMKA EndA and three Ala-substituted mutants (Y295A, H303A, and K334A). Reaction mixtures were separated on 15% polyacrylamide/7 M urea gels. The cleavage products are shown using schematic models at the right-hand side of the gel.
To examine the relevance of these three residues (Y295, H303 and K334) in catalysis, we constructed three fMKA EndA mutants, with alanine substituting for each residue (Y295A, H303A and K334A), and carried out the intron cleavage assay for comparison with the wild type. Notably, all three mutants failed to process pre-tRNAAsn (GUU) with a single canonical intron (Figure 4D), as well as pre-tRNAGlu (GUU) with multiple introns (Figure 4E), strongly indicating that the residues Y294, H303 and K334 are in fact responsible for the RNA cleavage in fMKA EndA.
Substrate specificity of the specific loop lacking fMKA EndA
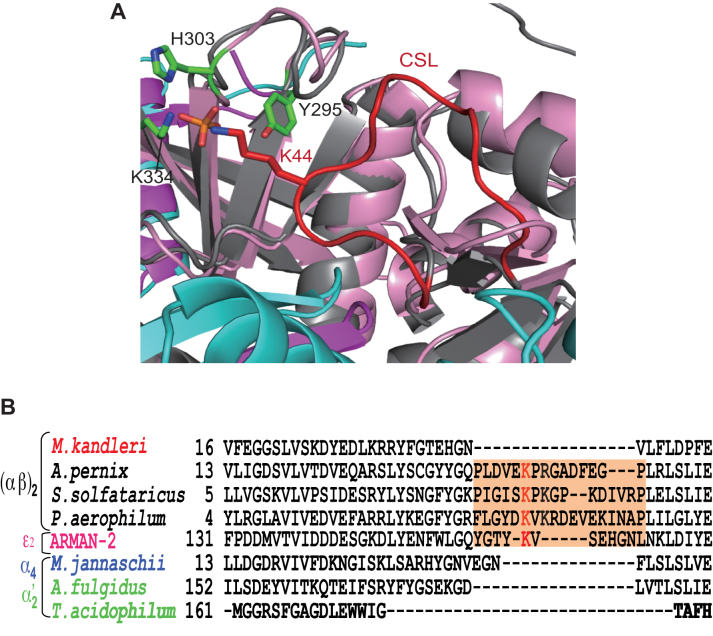
Native fMKA EndA should be able to remove both the canonical and non-canonical introns, otherwise M. kandleri would not survive. However, the efficiency of non-canonical intron cleavage was very low compared to that by the (αβ)2-type APE EndA (Figure 2). This feature brings into question whether fMKA EndA possesses the CSL or the ASL, the key loop structures conserved among (αβ)2-type and ϵ2-type EndAs for providing broad substrate specificity (13,28). The superimposition of the fMKA EndA and APE EndA protein structure around the catalytic pocket revealed that, although their overall structures clearly overlapped, the specific loop was absent in fMKA EndA (Figure 5A). Furthermore, protein sequence alignment clearly indicated that M. kandleri EndA does not harbor a specific loop in the α subunit (Figure 5B).
Figure 5.
Structural and sequence positioning of specific loops responsible for broad substrate specificity. (A) Superimposition diagram of Cα atoms of the α subunit (gray) in APE EndA onto that of the α unit (magenta) in fMKA EndA. The three catalytic residues (Y295, H303, and K334) are shown by a green stick model. The conserved K44 residue in the CSL (red) is depicted as a red stick model. (B) Amino acid sequence alignment of α subunits around the CSL region (highlighted in orange). Full names of the archaeal species are as follows: Methanopyrus kandleri, Aeropyrum pernix, Sulfolobus solfataricus, Pyrobaculum aerophilum, Candidatus Micrarchaeum acidiphilum (ARMAN-2), Methanocaldococcus jannaschii, Archaeoglobus fulgidus and Thermoplasma acidophilum. The key Lys residues responsible for broad substrate specificity are shown in red.
To confirm that fMKA EndAs do not have the same level of broad substrate specificity compared to other specific loop-harboring (αβ)2-type EndAs, we further performed two additional assays. Considering the hyperthermophilic nature of M. kandleri, which has an optimal growth temperature of 98°C (58), we performed high-temperature in vitro cleavage assays of pre-tRNAAsn (GUU) and pre-tRNAGlu (UUC) at 80°C and 90°C (Supplementary Figure S8). However, under both conditions, the splicing patterns of the introns of all three EndAs (AFU, APE and fMKA) remained unchanged compared to that with the assay performed at 70°C (Figure 2), indicating that fMKA indeed remains functional near the optimal growth temperature, but temperature change does not induce a change in substrate recognition or enzymatic activity to solve the inefficiency of HBh′ intron cleavage.
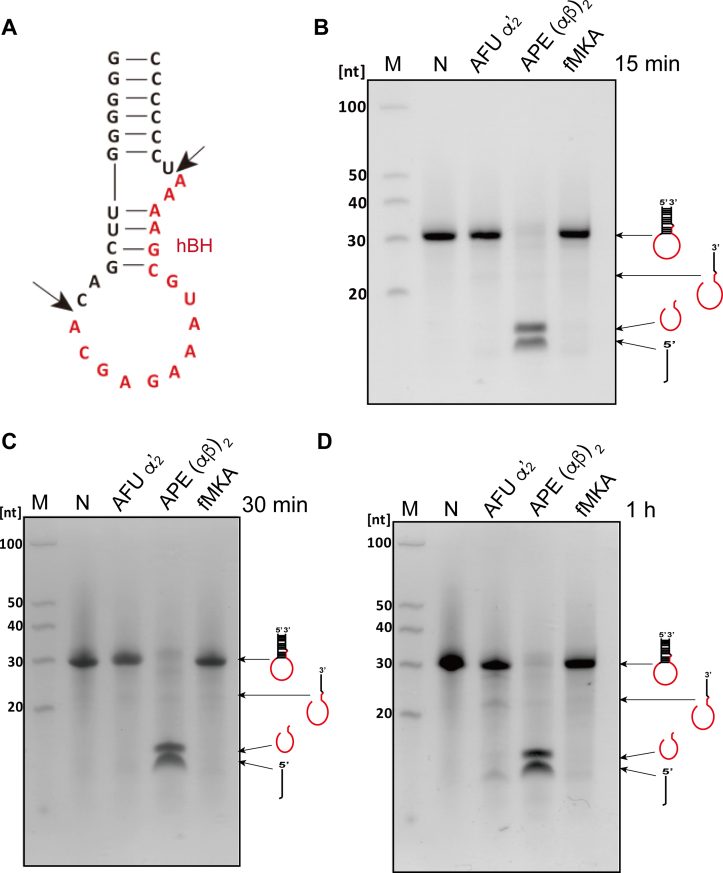
Second, we used mini-helix RNA, forming a non-canonical hBH structure, as a substrate known to be only processed by EndA (Figure 6A). Within 15 min, APE EndA could completely process the non-canonical hBH intron, whereas no obvious AFU EndA or fMKA EndA activity was detected even after an hour of incubation (Figure 6B–D). The shortest 3′-end fragment between positions U29 and G36 removed by APE EndA was not detected probably because of the loss of fragment by phenol/chloroform/isoamyl alcohol and ethanol extraction. Collectively, these results demonstrate that fMKA EndA does not possess broad substrate specificity, but rather harbors limited substrate specificity, owing to the lack of a specific loop.
Figure 6.
RNA cleavage assay using the non-canonical hBH motif. (A) The predicted secondary structure of the substrate mini-helix RNA, mimicking a tRNA intron with an hBH motif located at the non-canonical position 59/60 in ARMAN-2 pre-tRNACys (GCA) (12). Intron cleavage activity of the mini-helix RNA by AFU EndA, APE EndA and fMKA EndA at 70°C for 15 min (B), at 70°C for 30 min (C), and at 70°C for 1 h. (D) Reaction mixtures were separated on 15% polyacrylamide/7 M urea gels. In each gel, ‘M’ (first lane) and ‘N’ (second lane) indicate the molecular markers of fragmented RNA and control (no enzyme), respectively. The cleavage products are shown using schematic models at the right-hand side of the gel.
Coevolution of tRNA genes and broad substrate specificity
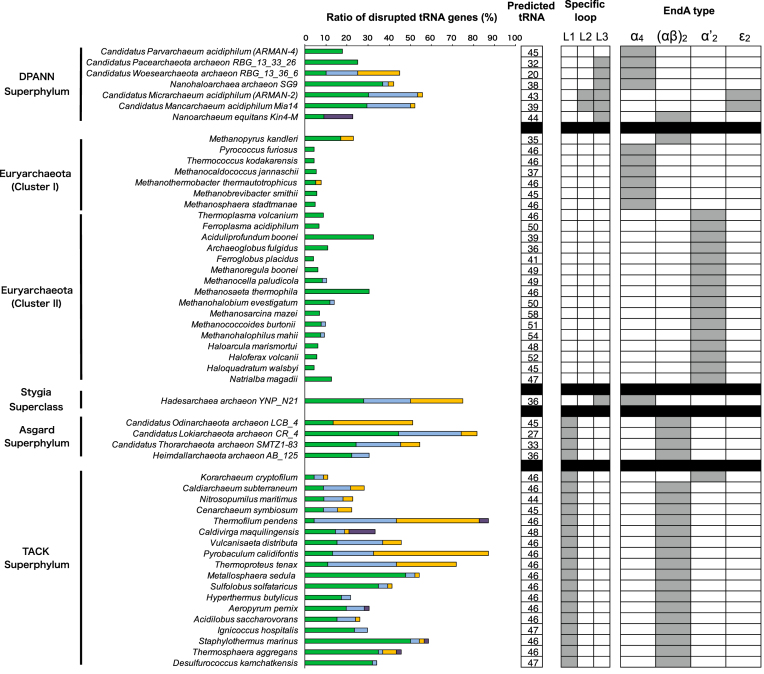
M. kandleri EndA remains the only known (αβ)2-type heterotetramer that lacks the specific loop. Recognition of diverse relaxed BHB motifs is likely an acquired feature specifically associated with the loop insertion event. Hence, to provide an updated view on the coevolution of EndA and tRNA genes, we mapped the EndA type, specific loop type, and tRNA gene diversity for 46 archaeal species, including the recently characterized DPANN and Asgard superphyla (Figure 7). For clarification, we reannotated the three distinct insertion sequences CSL, ASL, and Nanoarchaea/ARMAN-2 specific loop as L1, L2 and L3, respectively. The L1 loop is conserved throughout the TACK and Asgard superphyla but is not found in DPANN. In comparison, the L2 loop is known to confer broad substrate specificity in ARMAN-2 EndA, and is specifically found in homologous ϵ2-type EndAs from Candidatus Mancarchaeum acidiphilum Mia14, a recently sequenced archaeon related to ARMAN-2 (59). The L3 loop is found among diverse DPANN lineages, except for Candidatus Parvarchaeum acidiphilum (ARMAN-4). Previously, the α4 type was recognized to strictly cleave only the BHB motif; however, we found growing evidence of L3 harboring α4-type EndAs in multiple DPANN lineages, as well as in the newly proposed Stygia superclass Hadesarchaea (previously known as the South-African Gold Mine Miscellaneous Euryarchaeal Group), to be associated with a marked increase in tRNA gene disruption. Therefore, subunit type seems to have the least contribution towards substrate recognition, which has also been shown in a previous study (16). L1 and L2 reside at the N-terminal insertion site of the EndA protein sequence near the histidine residue of the catalytic triad, whereas the L3 loop is located in the C-terminus region, indicating a convergent evolution of EndAs toward acquiring broad substrate specificity (13,17,27–28). In particular, archaeal species with EndA inserts clearly represent a trend of increased tRNA gene diversity, unrelated to their EndA types. In addition, the inability to predict the complete set of tRNAs in some of the newly sequenced DPANN and Asgard lineages indicated the presence of unidentified disrupted tRNA genes, warranting in-depth sequence searches and manual curation.
Figure 7.
Divergence of EndA and tRNA genes in Archaea. tRNA genes are predicted for a total of 46 archaeal species that belong to five major groups: DPANN, Euryarchaeota, Stygia, Asgard and TACK. Classification of Clusters I and II, the Euryarchaeota and Stygia superclass, is as proposed by Adam et al. (62). Four types of EndAs (α4, α′2, (αβ)2 and ϵ2) and the relative ratio of the four types of disrupted tRNA genes are indicated: single intron located at the canonical position 37/38 (blue), single intron located at the non-canonical position (light blue), multiple introns (orange), split introns, and permuted introns (purple). The boxed number indicates the total number of predicted tRNA genes for each species. The insertion of the three major EndA-specific loops, formally known as CSL (L1), ASL (L2) and Nanoarchaea/ARMAN-2-specific loop (L3), are indicated.
DISCUSSION
Our study demonstrated an active recombinant fusion protein, M. kandleri EndA, which could remove the canonical tRNA precursor intron but processed the non-canonical HBh′ intron in a less efficient manner than the (αβ)2-type APE EndA with a specific loop could (Figure 2 and Supplementary Figure S8). The determined X-ray crystal structure of fMKA EndA revealed a rectangular parallelepiped formation, which is in good alignment with the structures from other previously elucidated EndA types (Supplementary Figure S5). The presence of mature tRNA sequences obtained through M. kandleri RNA-Seq indicates that the HBh′ intron located in the D-arms of isoacceptor pre-tRNAGlu (UUC) are indeed processed for maturation (56). The inefficiency of HBh′ intron cleavage could be the consequence of several possibilities combined, such as improper folding of the pre-tRNA during the cleavage assay at high temperature, a unique C-to-U editing at position C8 (60), and diverse RNA modifications observed in M. kandleri tRNAs (61). The major contribution of U8 is its base-pairing nature with A14 to stabilize the tertiary structure of mature tRNA. We speculate that having the intron located at position 20b/21 within the D-loop of pre-tRNAGlu (UUC) will most likely disrupt the formation of the tRNA tertiary structure, and therefore, U8 should have a minimal effect on the folding of the non-canonical HBh′ motif, as well as the overall splicing activity of EndA. However, we have previously shown that the APE pre-tRNAThr (UGU) harboring the U8-A14 base pair with the intron located within the D-loop can be spliced efficiently by the APE EndA at 70°C (28), indicating the possibility that the C-to-U editing is not a limiting step for efficient intron splicing in the D-loop of M. kandleri pre-tRNAGlu (UUC). Furthermore, we showed that the fMKA protein does consistently operate under a wide range of temperature conditions (70–90°C), while other nucleotide editing/modifications need to be taken into account to fully understand the underlying mechanism.
Interestingly, the L1 loop appears to compensate for such irregular arrangement of the tertiary core structure and/or lack of nucleotide modification and allow EndA to recognize diverse relaxed BHB motifs under a wide range of temperature conditions, as seen for APE EndA in our experiment. Other possible reasons, such as the contribution of tRNA-binding proteins or polyamines, remain to be investigated. Collectively, the evidence of mature tRNAGlu (UUC) from M. kandleri RNA-Seq (56) and the observations from the intron cleavage assay and the crystal structure, together support the concept that native MKA EndA is capable of processing the non-canonical HBh′ intron but not the hBH mini-helix, suggesting this heterotetrameric enzyme to be unique in having a constrained range of substrate specificity. The lack of broad substrate specificity can be explained by the missing L1 loop at the N-terminus of the α catalytic subunit. Our bioinformatics analysis revealed that Asgard Archaea harbor the (αβ)2-type EndA, with an L1 insert previously known to be specific to the TACK superphylum (40); this strongly implies that the L1 insertion event has most likely occurred at the root of Asgard and TACK superphyla, after the MKA EndA α subunit diverged (red arrow, Supplementary Figure S9).
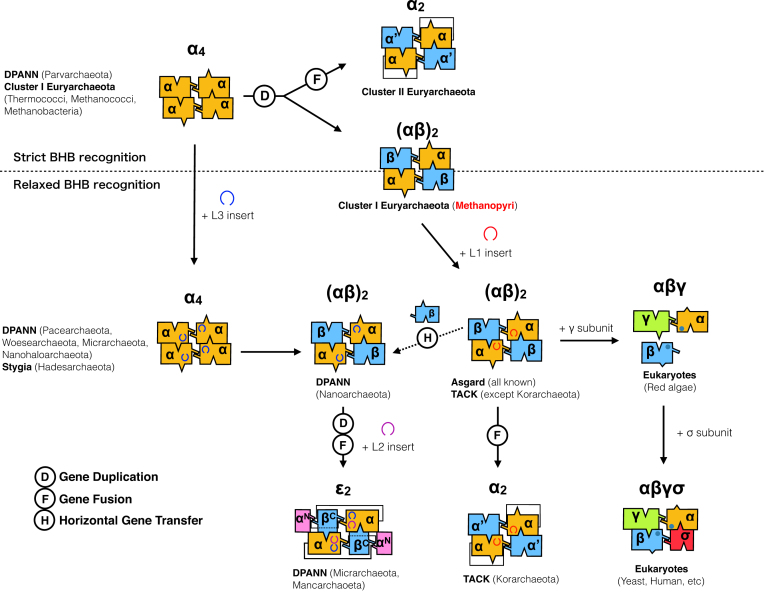
On the basis of these observations, we provide the latest view of the possible evolutionary scenario of archaeal EndAs (Figure 8). Transition from α4 to α′2 type is consistent with the most recent perspective on archaeal phylogeny, in which the new root falls within Euryarchaeota as opposed to the traditional root separating Euryarchaeota and the TACK superphylum (62). The first cluster (Cluster I) includes the subset of euryarchaeal species such as those in class Methanococci, Thermococci, Methanobacteria, and Methanopyri, as well as Stygia, Asgard and TACK superclasses/superphyla, harboring α4- and (αβ)2-type EndAs. The remaining Euryarchaeota are reclassified as Cluster II, exclusively encoding the α′2-type EndA. The irregular α′2-type EndA found in Korarchaea can be explained by lineage-specific gene fusion, given the presence of the L1 loop within its EndA α subunit (12,26).
Figure 8.
Possible evolutionary pathways of EndA proteins. Box representation of the four archaeal (α4, α′2, (αβ)2, ϵ2,) and two eukaryotic (αβγ, αβγδ) EndA types are shown with unit/subunit structure color coordination. A dotted line separates the EndA based on their predicted functional capability (strict versus relaxed BHB recognition) derived from the tRNA gene types (Figure 7). Possible gene diversification events such as gene duplication, gene fusion, and horizontal gene transfer are indicated as circled D, F and H, respectively.
The origin of the (αβ)2-type EndA is more complicated, given its presence in three distinct archaeal groups (Methanopyri, Nanoarchaeota and Asgard/TACK). However, the EndA phylogenetic tree supports the evolutionary relationships of MKA EndA α and β subunits with those of TACK/Asgard EndAs but not with N. equitans EndA subunits (Supplementary Figure S9). The origins of DPANN EndA could be non-monophyletic owing to their scattered distribution and weak branch support of α and β subunits in the phylogenetic tree. The diversity of DPANN EndA architecture (α4, (αβ)2 and ϵ2) further implies the occurrence of active gene duplication, fusion, and horizontal transfer, which is in good correspondence with their symbiotic nature and high rates of genome evolution (63). However, the missing α′2 type and their consistency in harboring the L3 loop place the origins of the DPANN EndA catalytic α subunit near the α4-type class I Euryarchaeota EndA.
The origin of eukaryotic EndA poses yet another issue to be solved, although the heterotetrameric form (αβγδ), consisting of two catalytic (Sen2 and Sen34) and two accessory (Sen15 and Sen54) subunits, has been studied in model eukaryotes such as Saccharomyces cerevisiae, Arabidopsis thaliana and Homo sapiens (64). An evolutionary relationship between the archaeal α subunit and Sen2 and Sen34 exists, with approximately 50 amino acid residues being highly conserved (4). However, this region does not include the two major insertion sites (L1–L3) seen in archaeal EndA. Notably, the primitive eukaryotic red alga Cyanidioschyzon merolae is known to harbor disrupted tRNA genes, including permuted tRNA and multiple intron-containing tRNAs, a feature also found in Archaea (65,66). In C. merolae, three EndA subunits (cmSen2p, cmSen34p and cmSen54p) are predicted to be responsible for processing these pre-tRNAs through relaxed BHB recognition, and different modes of subunit interactions might occur in vivo (67). Nevertheless, future structural analysis of C. merolae EndA is expected to reveal commonalities and evolutionary relationships between archaeal and eukaryotic EndAs.
In summary, analysis of the structure and function of an (αβ)2-type MKA EndA lacking a specific loop insert has provided a proof-of-concept regarding the relationships among subunit architecture, insertion, and broad substrate specificity of the EndA enzyme. MKA EndA stands as a pivotal enzyme during the subfunctionalization history of EndA, likely representing the ancestral form of the (αβ)2 type lacking the specific loop, albeit harboring a constrained range of substrate specificity. Herein, our experimental results, along with the bioinformatics analysis, strengthen the view that specific loop insertions (L1, L2 and L3) might be the major factors determining the efficient cleavage of diverse relaxed BHB motifs.
DATA AVAILABILITY
The crystal structure factor and coordinate have been deposited in the Protein Data Bank (PDB code 5X89).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the staff members of the beamline facility at SPring-8 (Hyogo, Japan) for their technical support during data collection. The synchrotron radiation experiments were performed at BL26B1, BL26B2, BL38B1 and BL41XU in the SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (Proposal No. 2012B1916, 2013A1110, 2014B1221 and 2015A1047).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
JSPS KAKENHI [JP15K06975 to A.H.]. Funding for open access charge: JSPS KAKENHI [JP15K06975].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kumar P., Kuscu C., Dutta A.. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem. Sci. 2016; 41:679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu M., Kathe S., Goodrich-Blair H., Nierzwicki-Bauer S., Shub D.. Bacterial origin of a chloroplast intron: conserved self-splicing group I introns in cyanobacteria. Science. 1990; 250:1566–1570. [DOI] [PubMed] [Google Scholar]
- 3. Haugen P., Simon D.M., Bhattacharya D.. The natural history of group I introns. Trends Genet. 2005; 21:111–119. [DOI] [PubMed] [Google Scholar]
- 4. Abelson J., Trotta C., Li H.. tRNA splicing. J. Biol. Chem. 1998; 273:12685–12688. [DOI] [PubMed] [Google Scholar]
- 5. Tanaka N., Meineke B., Shuman S.. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J. Biol. Chem. 2011; 286:30253–30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tanaka N., Shuman S.. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J. Biol. Chem. 2011; 286:7727–7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tanaka N., Chakravarty A.K., Maughan B., Shuman S.. Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-Phosphate/5′-hydroxyl ligation reactions. J. Biol. Chem. 2011; 286:43134–43143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Englert M., Sheppard K., Aslanian A., Yates J.R., Söll D.. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:1290–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Englert M., Xia S., Okada C., Nakamura A., Tanavde V., Yao M., Eom S.H., Konigsberg W.H., Söll D., Wang J.. Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3′-terminal phosphate and 5′-OH. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:15235–15240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tocchini-Valentini G., Fruscoloni P., Tocchini-Valentini G.. Structure, function, and evolution of the tRNA endonucleases of Archaea: an example of subfunctionalization. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:8933–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calvin K., Li H.. RNA-splicing endonuclease structure and function. Cell. Mol. Life Sci. 2008; 65:1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fujishima K., Sugahara J., Miller C.S., Baker B.J., Di Giulio M., Takesue K., Sato A., Tomita M., Banfield J.F., Kanai A.. A novel three-unit tRNA splicing endonuclease found in ultrasmall Archaea possesses broad substrate specificity. Nucleic Acids Res. 2011; 39:9695–9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hirata A., Fujishima K., Yamagami R., Kawamura T., Banfield J.F., Kanai A., Hori H.. X-ray structure of the fourth type of archaeal tRNA splicing endonuclease: insights into the evolution of a novel three-unit composition and a unique loop involved in broad substrate specificity. Nucleic Acids Res. 2012; 40:10554–10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Calvin K., Hall M., Xu F., Xue S., Li H.. Structural characterization of the catalytic subunit of a novel RNA splicing endonuclease. J. Mol. Biol. 2005; 353:952–960. [DOI] [PubMed] [Google Scholar]
- 15. Li H., Trotta C., Abelson J.. Crystal structure and evolution of a transfer RNA splicing enzyme. Science. 1998; 280:279–284. [DOI] [PubMed] [Google Scholar]
- 16. Yoshinari S., Shiba T., Inaoka D., Itoh T., Kurisu G., Harada S., Kita K., Watanabe Y.. Functional importance of crenarchaea-specific extra-loop revealed by an X-ray structure of a heterotetrameric crenarchaeal splicing endonuclease. Nucleic Acids Res. 2009; 37:4787–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mitchell M., Xue S., Erdman R., Randau L., Söll D., Li H.. Crystal structure and assembly of the functional Nanoarchaeum equitans tRNA splicing endonuclease. Nucleic Acids Res. 2009; 37:5793–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim Y., Mizutani K., Rhee K., Nam K., Lee W., Lee E., Kim E., Park S., Hwang K.. Structural and mutational analysis of tRNA intron-splicing endonuclease from Thermoplasma acidophilum DSM 1728: catalytic mechanism of tRNA intron-splicing endonucleases. J. Bacteriol. 2007; 189:8339–8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Popow J., Englert M., Weitzer S., Schleiffer A., Mierzwa B., Mechtler K., Trowitzsch S., Will C.L., Lührmann R., Söll D. et al. . HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011; 331:760–764. [DOI] [PubMed] [Google Scholar]
- 20. Popow J., Schleiffer A., Martinez J.. Diversity and roles of (t)RNA ligases. Cell. Mol. Life Sci. 2012; 69:2657–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marck C., Grosjean H.. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: evolutionary implications. RNA. 2003; 9:1516–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Watanabe Y., Yokobori S., Inaba T., Yamagishi A., Oshima T., Kawarabayasi Y., Kikuchi H., Kita K.. Introns in protein-coding genes in Archaea. FEBS Lett. 2002; 510:27–30. [DOI] [PubMed] [Google Scholar]
- 23. Yoshinari S., Itoh T., Hallam S.J., DeLong E.F., Yokobori S., Yamagishi A., Oshima T., Kita K., Watanabe Y.. Archaeal pre-mRNA splicing: a connection to hetero-oligomeric splicing endonuclease. Biochem. Biophys. Res. Commun. 2006; 346:1024–1032. [DOI] [PubMed] [Google Scholar]
- 24. Yokobori S., Itoh T., Yoshinari S., Nomura N., Sako Y., Yamagishi A., Oshima T., Kita K., Watanabe Y.. Gain and loss of an intron in a protein-coding gene in Archaea: the case of an archaeal RNA pseudouridine synthase gene. BMC Evol. Biol. 2009; 9:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kjems J., Garrett R.A.. Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell. 1988; 54:693–703. [DOI] [PubMed] [Google Scholar]
- 26. Baker B.J., Tyson G.W., Webb R.I., Flanagan J., Hugenholtz P., Allen E.E., Banfield J.F.. Lineages of acidophilic archaea revealed by community genomic analysis. Science. 2006; 314:1933–1935. [DOI] [PubMed] [Google Scholar]
- 27. Okuda M., Shiba T., Inaoka D.K., Kita K., Kurisu G., Mineki S., Harada S., Watanabe Y., Yoshinari S.. A conserved lysine residue in the Crenarchaea-specific loop is important for the Crenarchaeal splicing endonuclease activity. J. Mol. Biol. 2011; 405:92–104. [DOI] [PubMed] [Google Scholar]
- 28. Hirata A., Kitajima T., Hori H.. Cleavage of intron from the standard or non-standard position of the precursor tRNA by the splicing endonuclease of Aeropyrum pernix, a hyper-thermophilic Crenarchaeon, involves a novel RNA recognition site in the Crenarchaea specific loop. Nucleic Acids Res. 2011; 39:9376–9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Randau L., Calvin K., Hall M., Yuan J., Podar M., Li H., Söll D.. The heteromeric Nanoarchaeum equitans splicing endonuclease cleaves noncanonical bulge-helix-bulge motifs of joined tRNA halves. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:17934–17939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reyes V., Abelson J.. Substrate recognition and splice site determination in yeast tRNA splicing. Cell. 1988; 55:719–730. [DOI] [PubMed] [Google Scholar]
- 31. Burggraf S., Stetter K.O., Rouviere P., Woese C.R.. Methanopyrus kandleri: an archaeal methanogen unrelated to all other known methanogens. Syst. Appl. Microbiol. 1991; 14:346–351. [DOI] [PubMed] [Google Scholar]
- 32. Slesarev A.I., Mezhevaya K.V., Makarova K.S., Polushin N.N., Shcherbinina O.V., Shakhova V.V., Belova G.I., Aravind L., Natale D.A., Rogozin I.B. et al. . The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:4644–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guy L., Ettema T.J.. The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 2011; 19:580–587. [DOI] [PubMed] [Google Scholar]
- 34. Rinke C., Schwientek P., Sczyrba A., Ivanova N.N., Anderson I.J., Cheng J.F., Darling A., Malfatti S., Swan B.K., Gies E.A. et al. . Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013; 499:431–437. [DOI] [PubMed] [Google Scholar]
- 35. Sugahara J., Kikuta K., Fujishima K., Yachie N., Tomita M., Kanai A.. Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order thermoproteales. Mol. Biol. Evol. 2008; 25:2709–2716. [DOI] [PubMed] [Google Scholar]
- 36. Randau L., Münch R., Hohn M., Jahn D., Söll D.. Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature. 2005; 433:537–541. [DOI] [PubMed] [Google Scholar]
- 37. Fujishima K., Sugahara J., Kikuta K., Hirano R., Sato A., Tomita M., Kanai A.. Tri-split tRNA is a transfer RNA made from 3 transcripts that provides insight into the evolution of fragmented tRNAs in archaea. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:2683–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chan P.P., Cozen A.E., Lowe T.M.. Discovery of permuted and recently split transfer RNAs in Archaea. Genome Biol. 2011; 12:R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tocchini-Valentini G., Fruscoloni P., Tocchini-Valentini G.. Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15418–15422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fujishima K., Kanai A.. tRNA gene diversity in the three domains of life. Front. Genet. 2014; 5:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brochier C., Forterre P., Gribaldo S.. Archaeal phylogeny based on proteins of the transcription and translation machineries: tackling the Methanopyrus kandleri paradox. Genome Biol. 2004; 5:R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hori H., Yamazaki N., Matsumoto T., Watanabe Y., Ueda T., Nishikawa K., Kumagai I., Watanabe K.. Substrate recognition of tRNA (Guanosine-2′-)-methyltransferase from Thermus thermophilus HB27. J. Biol. Chem. 1998; 273:25721–25727. [DOI] [PubMed] [Google Scholar]
- 43. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 44. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Terwilliger T.C. Automated side-chain model building and sequence assignment by template matching. Acta Crystallogr. D Biol. Crystallogr. 2003; 59:45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 47. Laskowski R., MacArthur M., Moss D., Thoronton J.. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1992; 26:283–291. [Google Scholar]
- 48. Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D.. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003; 31:3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gouet P., Robert X., Courcelle E.. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003; 31:3320–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lowe T.M., Eddy S.R.. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997; 25:955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sugahara J., Yachie N., Sekine Y., Soma A., Matsui M., Tomita M., Kanai A.. SPLITS: a new program for predicting split and intron-containing tRNA genes at the genome level. In Silico Biol. 2006; 6:411–418. [PubMed] [Google Scholar]
- 52. Gouy M., Guindon S., Gascuel O.. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010; 27:221–224. [DOI] [PubMed] [Google Scholar]
- 53. Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O.. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010; 59:307–321. [DOI] [PubMed] [Google Scholar]
- 54. Letunic I., Bork P.. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016; 44:W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chan P.P., Lowe T.M.. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009; 37:D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Su A.A., Tripp V., Randau L.. RNA-Seq analyses reveal the order of tRNA processing events and the maturation of C/D box and CRISPR RNAs in the hyperthermophile Methanopyrus kandleri. Nucleic Acids Res. 2013; 41:6250–6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xue S., Calvin K., Li H.. RNA recognition and cleavage by a splicing endonuclease. Science. 2006; 312:906–910. [DOI] [PubMed] [Google Scholar]
- 58. Kurr M., Huber R., König H., Jaasch W.H., Fricke H., Trincone A., Kristjansson K.J., Stetter O.K.. Methanopyrus kandleri, gen. and sp. nov. represents a novel group of hyperthermophilic methanogens, growing at 110°C. Arch. Microbiol. 1991; 156:239–247. [Google Scholar]
- 59. Golyshina O.V., Toshchakov S.V., Makarova K.S., Gavrilov S.N., Korzhenkov A.A., La Cono V., Arcadi E., Nechitaylo T.Y., Ferrer M., Kublanov I.V. et al. . ‘ARMAN’ archaea depend on association with euryarchaeal host in culture and in situ. Nat. Commun. 2017; 8:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Randau L., Stanley B.J., Kohlway A., Mechta S., Xiong Y., Söll D.. A cytidine deaminase edits C to U in transfer RNAs in Archaea. Science. 2009; 324:657–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sauerwald A., Sitaramaiah D., McCloskey J.A., Söll D., Crain P.F.. N6-Acetyladenosine: a new modified nucleoside from Methanopyrus kandleri tRNA. FEBS Lett. 2005; 579:2807–2810. [DOI] [PubMed] [Google Scholar]
- 62. Adam P.S., Borrel G., Brochier-Armanet C., Gribaldo S.. The growing tree of Archaea: new perspectives on their diversity, evolution and ecology. ISME J. 2017; 11:2407–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Williams T.A., Szöllősi G.J., Spang A., Foster P.G., Heaps S.E., Boussau B., Ettema T.J.G., Embley T.M.. Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E4602–E4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Trotta C., Miao F., Arn E., Stevens S., Ho C., Rauhut R., Abelson J.. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell. 1997; 89:849–858. [DOI] [PubMed] [Google Scholar]
- 65. Maruyama S., Sugahara J., Kanai A., Nozaki H.. Permuted tRNA genes in the nuclear and nucleomorph genomes of photosynthetic eukaryotes. Mol. Biol. Evol. 2010; 27:1070–1076. [DOI] [PubMed] [Google Scholar]
- 66. Soma A., Onodera A., Sugahara J., Kanai A., Yachie N., Tomita M., Kawamura F., Sekine Y.. Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science. 2007; 318:450–453. [DOI] [PubMed] [Google Scholar]
- 67. Soma A., Sugahara J., Onodera A., Yachie N., Kanai A., Watanabe S., Yoshikawa H., Ohnuma M., Kuroiwa H., Kuroiwa T. et al. . Identification of highly-disrupted tRNA genes in nuclear genome of the red alga, Cyanidioschyzon merolae 10D. Sci. Rep. 2013; 3:2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystal structure factor and coordinate have been deposited in the Protein Data Bank (PDB code 5X89).