Abstract
When a stop codon is at the 80S ribosomal A site, there are six nucleotides (+4 to +9) downstream that are inferred to be occupying the mRNA channel. We examined the influence of these downstream nucleotides on translation termination success or failure in mammalian cells at the three stop codons. The expected hierarchy in the intrinsic fidelity of the stop codons (UAA>UAG>>UGA) was observed, with highly influential effects on termination readthrough mediated by nucleotides at position +4 and position +8. A more complex influence was observed from the nucleotides at positions +5 and +6. The weakest termination contexts were most affected by increases or decreases in the concentration of the decoding release factor (eRF1), indicating that eRF1 binding to these signals was rate-limiting. When termination efficiency was significantly reduced by cognate suppressor tRNAs, the observed influence of downstream nucleotides was maintained. There was a positive correlation between experimentally measured signal strength and frequency of the signal in eukaryotic genomes, particularly in Saccharomyces cerevisiae and Drosophila melanogaster. We propose that termination efficiency is not only influenced by interrogation of the stop signal directly by the release factor, but also by downstream ribosomal interactions with the mRNA nucleotides in the entry channel.
INTRODUCTION
Termination of protein synthesis involves the decoding of a stop signal through an interaction between mRNA and proteins (release factors, RFs) and occurs in response to the translocation of a termination signal sequence, containing a stop codon (UAA, UAG or UGA) into the A site of the ribosome (1). In eukaryotes, a single eRF1 recognizes all three stop codons on the ribosome. When a stop signal is decoded by a release factor, it facilitates the hydrolytic release of the nascent polypeptide chain from the peptidyl-tRNA bound at the P site within the peptidyl-transferase center. However, in some rare cases, the stop codons can be decoded as sense codons by near-cognate tRNAs, resulting in continued translation and ‘readthrough’ of the stop codon (2,3).
Crosslinking studies revealed interactions between the stop codon and the conserved NIKS, YxCxxF and GTS motifs of domain 1 of eRF1 (4,5). Cryo-electron microscopy structures of mammalian ribosomal complexes containing eRF1 in the A site (6–8) have revealed that direct interactions with the NIKS sequence, the lysine of which can be hydroxylated (9), impose a requirement for uridine in the first (+1) position of the stop codon. Remarkably, eRF1 binding induces an unusual conformation in which four mRNA nucleotides—the stop codon and one additional 3′ nucleotide—occupy the A site, stabilised by interactions with eRF1 and the ribosome (6–8). This explains the early observation that a minimum of four bases is required for a functional termination signal in an in vitro assay (10), and later that the identity of the nucleotide immediately downstream of a stop codon is highly influential in determining the efficiency of the signal (11). Interactions with the YxCxxF motif and E55 of eRF1 with the +2 and +3 bases provide a basis for stop codon discrimination (6) with the GTS motif closing the binding pocket formed between the stop codon and eRF1 (7). These interactions suggest a complex three-dimensional pattern of stop codon recognition, as has also been shown in the high-resolution structures of the prokaryotic ribosome release factor complexes, and this is quite different from the ‘anticodon-type’ interaction seen with sense codons and tRNAs.
Changes to domain 1 of eRF1, originally deduced to be the site of stop codon recognition (12–15), can cause profound alterations in the stop codon recognition profile. For example, swapping this domain from Tetrahymena eRF1 (UGA recognition only) to yeast eRF1 resulted in a hybrid eRF1 that could direct termination only at UGA stop codons in vitro (16). Hatin and co-workers (17) showed in Saccharomyces cerevisiae that mutations in domains 1 and 3 of eRF1 had differential effects on anti-suppressor activity depending on the stop codon analysed, reinforcing the conclusion that specific regions of the eRF1 protein can influence decoding depending on the identity of the stop codon.
The consequences of stop codons being recognised directly by a protein release factor rather than a tRNA, as in polypeptide elongation, means that the signal for translation termination could extend beyond the three nucleotides specified in the genetic code (18). Earlier studies of gene sequences in eukaryotes (19,20) and studies in yeast (21,22), plants (23) and mammals (24,25), revealed a bias in the occurrence of nucleotides 5′ and 3′ of stop codons. These findings led to the proposal that the RF recognised a tetra-nucleotide sequence containing limited redundancy (U1 A/G2 A/G3 A/G/T/C4) and not simply each of the three tri-nucleotide stop codons (26) that were tested experimentally (11).
Translation termination signal readthrough at a limited set of selected eukaryotic sequences has been investigated experimentally, and indeed the nucleotide sequences both 5′ and 3′ of the stop codon can modulate this event (27–29). Atkins and co-workers’ pioneering studies showed the importance of the 3′ context (30,31). Rousset and co-workers screened oligonucleotide libraries degenerate in the six positions 3′ of the stop codon for their ability to promote >5% readthrough in an ADE2 gene yeast suppressible marker assay (32). Nineteen unique sequences were identified, and they showed bias in the first, second, third and sixth positions downstream of the stop codon. The conclusion was that the consensus sequence, [STOP CODON] C1A2(A/G)3N4(U/C/G)5A6 conferred inefficient termination in yeast (32).
Cryo-EM and structural studies of the 80S eukaryotic ribosome complex have shown that ∼30 nucleotides of the mRNA interact with the ribosome at any one time during translation (33,34). During translation, the mRNA is wrapped in a groove that circles the 40S subunit, and nucleotides downstream from the A site codon pass through a tunnel, possibly guaranteeing directional movement (34). During translation termination, when a stop codon is in the A site and eRF1 is bound, the mRNA assumes a compact configuration that accommodates four nucleotides in the A site instead of three (6). This allows for the two nucleotides beyond the stop codon (+4 and +5) to base stack with 18S rRNA bases G626 and C1698 respectively (6–8). The nucleotides beyond these are in contact with a layer of rRNA and protein extending to the solvent (34).
The nucleotides upstream of the stop codon can also modulate translational termination (28,29,35). The P site tRNA provides a face to which the decoding factor (in this case eRF1) in the A site must be able to accommodate (1), and indeed there is evidence that some codons are highly avoided immediately before the stop codon (36). A number of additional protein factors have also been implicated in translation termination (37–40). The ribosome recycling factor Rli1/ABCE1 modulates the efficiency of translation termination by an unknown mechanism (41), and the translation (re) initiation factor eIF3 enhances readthrough specifically at weak termination contexts, perhaps by facilitating the incorporation of near-cognate tRNAs (42). A DEAD-box RNA helicase enhances termination at the point of stop codon recognition by eRF1 (43,44).
In our current study, we employed a systematic approach to examine specifically the influence of the six nucleotides within the mRNA channel following the stop codon (+4 to +9) on translation termination. All 192 possible combinations of the bases in the three nucleotides directly downstream of each stop codon (+4 to +6), together with a selected set of the next three positions (+7 to +9), were tested for efficiency of termination in cultured mammalian cells. We also assessed if an alteration in the cellular concentrations of eRF1 and competing cognate suppressor tRNAs could affect the efficiency of termination at selected stop codon contexts. In addition, we determined whether the genomic frequency of particular stop codon contexts correlated with their experimental termination efficiency.
MATERIALS AND METHODS
Plasmids
The bi-cistronic pGL3-hRLuc plasmid was generated by subcloning of the codon optimized hRLuc gene (phRL-SV40, Promega) into the dual luciferase reporter plasmid pGL3-NCO (45). In addition, a novel SalI restriction site was introduced into the multiple cloning sites (MCS) of the plasmid. In turn, the test plasmids were generated by subcloning a 23-nucleotide fragment from the pMAL-RF2 series of constructs into the EcoRI and SalI sites of the pGL3-hRluc MCS (46). The pGL3-hRLuc test plasmids were designed so that all 64 possible combinations of the three nucleotides 3′ of the internal stop codon (+4 to +6) were constructed. Three series of pGL3-hRLuc plasmids were developed which varied according to the stop codon in the test window (UAA NNN, UGA NNN and UAG NNN). The invariant six nucleotides upstream of the stop codon, which can, in turn, contribute to translation termination, were UAUCUU. All clones were initially screened for the presence or absence of the introduced sequence by restriction digestion, and then the identity of each insert was confirmed by sequencing. Two independent clones were isolated for each of the 64 UAANNN, 64 UAGNNN and 64 UGANNN constructs, resulting in a total of 384 clones.
For the eRF1 overexpression plasmid, the eRF1 gene coding sequence was amplified by PCR from an existing construct, heRF1 (47), and cloned into the pcDNA3.1 (+) vector (Invitrogen) to give the plasmid pcDNA-eRF1. The eRF1 insert was verified by sequencing.
For eRF1 knockdown, two siRNA target sequences were obtained from (48): si90, which targets nucleotides 90–108 of the eRF1 mRNA (5′-UGGCACCAGCAUGAUAUCA-3′), and si1186 (modified from si1187), which targets nucleotides 1186–1204 (5′-UCACAAGAAGGGUCUCAGU-3′). These were expressed as small hairpin RNAs (shRNAs) from the pSilencer 3.0-H1 vector (Ambion) as reported previously (47).
The ptRNA series of serine suppressor tRNAs, one for each stop codon (ptRNAoc—ochre UAA, ptRNAam—amber UAG and ptRNAop—opal UGA) were kindly gifted by Dr Olivier Jean-Jean, Institut de Biologie Paris Seine, Paris, France (49).
Cell culture
COS-7 and HEK 293T cells (American Type Culture Collection) were maintained in Dulbecco's Modified Eagle Medium (Invitrogen) supplemented with 4 mM l-glutamine, 1.5 g/l sodium bicarbonate, 4.5 g/l glucose and adjusted to contain 10% fetal calf serum (Gibco) at 37°C and 5% CO2. Cell lines were maintained according to standard techniques. Cells were grown to 70–80% confluence, seeded into 24-well plates at a density of 40 000 cells per well and allowed to grow to a confluence of 50–60% (∼24 h) before transfection. Typically, 0.25 μg of pGL3s-hRLuc plasmid DNA in a 6:1 ratio with FuGENE® 6 Transfection Reagent (Roche) was used for each transfection. Optimisation of siRNAs and eRF1 over-expression by immunoblot blot and real-time PCR utilised a titration of plasmid DNA ranging from 0.25 to 1.0 μg (siRNAs) or 0.25 to 1.5 μg (pcDNA-eRF1) in a 3:1 ratio with FuGENE® 6. In co-expression experiments, 1 μg pcDNA-eRF1, 1 μg pSilencer shRNA or 0.2 μg suppressor tRNA was cotransfected with 0.25 μg pGL3s-hRLuc in a 3:1 ratio with FuGENE® 6. Transfected cells were incubated at 37°C and 5% CO2 for 48 h for pcDNA-eRF1 and suppressor tRNAs or 120 h for siRNAs, and then washed twice with 200 μl phosphate-buffered saline before lysis. Cells were lysed by addition of 100 μl of 1 × Passive Lysis Buffer (Promega) for immunoblot blot and luciferase assays, or 200 μl of TRIzol® Reagent (Invitrogen) for quantitative PCR and northern blot analysis. Cell lysates were vortexed and centrifuged at 12 500 × g for 1 min before being assayed for luciferase activity.
Luciferase assays
The Renilla reniformis luciferase (RLuc) assay was performed using the conditions described (50) with modifications (51). The Photinus pyralis luciferase (Luc+) assay was performed using the conditions stated (52). Luciferase activities of whole cell lysate were determined in duplicate by adding 50 μl of either Assay Buffer Renilla Luciferase or Assay Buffer Photinus Luciferase to 1–10 μl of sample at 25°C. Luminesence was measured immediately after the addition of assay buffer for 30 s on a POLARstar OPTIMA luminometer (BMG Labtechnologies, Germany), or with an AutoLumat LB953 (EG&G Berthold, Germany). Relative light unit (RLU) counts, normalized for the background were used to calculate the Luc+:RLuc ratio for each sample. The Luc+:RLuc ratio was used to determine the rate of stop codon readthrough of the test construct by comparing it to the Luc+:RLuc ratio of a control that contained a near-cognate codon in place of the stop codon, which resulted in 100% readthrough. The absolute rate of readthrough was calculated by determining the ratio of Luc+ to RLuc activity from the test samples relative to the ratio of Luc+ to RLuc from the control samples as described by Grentzmann et al. (53).
Total RNA extraction
Total RNA was isolated from cultured cells using TRIzol® Reagent (Invitrogen). TRIzol® Reagent was added directly to aspirated cells (1 ml per 10 cm2). The resulting lysate was phase separated with chloroform (0.2 ml per 1 ml TRIzol®) and total RNA precipitated with isopropanol (0.5 ml per 1 ml TRIzol®). The RNA was washed twice with 75% ethanol (1 ml per 1 ml TRIzol®) and resuspended in DEPC treated ddH2O by incubating at 60°C for 10 min. The resuspended RNA was assessed for quality (260/280 nm absorbance ratio) and integrity (formaldehyde agarose gel electrophoresis).
Quantitative PCR
Total RNA (10 μg) was incubated with 2 U DNase I (Roche) for 30 min at 37°C to remove any contaminating DNA. First strand cDNA was synthesised from 1 μg of total RNA using 200 U SuperScript™ III Reverse Transcriptase (Invitrogen) and 250 ng random p(dN)6 primers (Roche) following the manufacturer's instructions. Levels of eRF1 mRNA were measured in an Applied Biosystems 7500 Fast Real-Time PCR System using ABsolute™ QPCR Low ROX Mix (ABgene) and a TaqMan® Gene Expression Assay (ETF1 (eRF1) Gene Assay ID: Hs01107358_g1; GAPDH Gene Assay ID: Hs99999905_m1 Applied Biosystems). 18S rRNA was measured as a reference for relative quantification using TaqMan® Ribosomal Control Reagents (Applied Biosystems).
Northern analysis
Polyacrylamide gel electrophoresis was used to separate RNA species to allow for detection of suppressor tRNAs and 5.8S rRNA by northern blot analysis. Total RNA (15 μg) was fractionated on a vertical 8% (w/v) polyacrylamide gel (Acryl:Bis 38:2) containing 8 M urea. Gels were equilibrated before loading with pre-running buffer in 1 × TBE, 50 V/cm for 2 h. Samples were fractionated by electrophoresis at 25 V/cm for 2 h and transferred to Hybond-Ntrade membrane (Amersham) using the Panthertrade Semi-Dry Electroblotting system (Owl Separation Systems, USA). The transfer was performed at 14 V maximum, 46 Amp for 1 h. The RNA was fixed to the membrane by UV light exposure for 5 min. Hybridization with 5′ end-labelled [γ-32P]dATP oligonucleotide probes (5 pmol) was performed overnight at 60°C in Otago Hybridization Buffer (4 × SSPE, 0.1% sodium pyrophosphate, 0.5 mg/ml heparin, 0.5% (w/v) SDS) before washing in SSPE and exposure to X-ray film. Hybridization signals were quantified with a GS-800 Calibrated Densitometer. Degenerate oligonucleotide probes were designed to detect the suppressor tRNAs (5′-tttrragtccatcgcc-3′). Suppressor tRNA signal was normalised to total 5.8S rRNA (5′-cgaagtgtcgatgatcaat-3′).
Immunoblot analysis
Total protein concentration of the cell lysates was determined by the BCA assay (54). A total of 25 μg of lysate was separated by 12.5% (w/v) SDS-PAGE and transferred to Protran® nitrocellulose transfer membrane (Whatman). Membranes were blocked for 1 h at 4°C in TBS (40 mM Tris–HCl, 150 mM NaCl, 0.05% (v/v) Tween-20, pH 7.6) with 5% (w/v) milk powder, then washed three times with TBS. Membranes were incubated overnight at 4°C in TBS with 5% (w/v) BSA and rabbit anti-eRF1 polyclonal antibody (1:10 000). The anti-eRF1 polyclonal antibody was kindly gifted by Dr Adam Geballe, Fred Hutchinson Cancer Research Center, Seattle, USA. Membranes were washed three times with TBS and incubated at room temperature for 1 h in TBS with 2% (w/v) milk powder and mouse anti-β-actin monoclonal antibody (1:5000) (Sigma). After washing three times with TBS, membranes were incubated at room temperature for 1 h in TBS with 5% (w/v) milk powder and goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:5000) (Sigma). After a further three washes with TBS, ECLtrade Western Blotting Detection Reagents (Amersham Biosciences) were added according to the manufacturer's instructions, and the blot exposed to X-ray film. The level of eRF1 over-expression or knockdown was estimated relative to untransfected controls using a Molecular Imager FX (Bio-Rad).
Bioinformatics
The heat map was generated with the heatmap.2 function of the gplots package in R. Microsoft Excel or Prism 7 (Graphpad Software, Inc) were used for basic data handling and graphing.
Human termination sequences were obtained from the TransTerm database, using RefSeq 77 coordinates (55). For Drosophila melanogaster (assembly BDGP release 6) and S. cerevisiae (assembly sacCer3) sequences [open reading frames (ORFs) with downstream extensions] predicted by AUGUSTUS (56) were obtained from the UCSC genome table browser (57). Gene sequences were excluded if they did not contain a valid stop codon or were mitochondrial, and genes that were not unique were collapsed into a single instance. This resulted in 5 467 stop signals to be analysed for S. cerevisiae, 12 172 for D. melanogaster, and 19 549 for Homo sapiens. A visual representation of nucleotide bias at downstream positions was generated with the BLogo programme, normalised to background frequencies of the +4 to +9 nucleotides for each species (58). The bias in the occurrence of each nucleotide was determined using the Fisher's exact test. To determine the random number of expected occurrences of each hexanucleotide stop signal, the nucleotides in positions +4 to +9 downstream of each stop codon instance were shuffled 100 times, the resulting hexanucleotide stop signals (which included the original stop codon) were counted, and each count divided by 100 using Python. The sign of the frequency ratio was taken from the sign of the observed – expected value for each hexanucleotide stop codon. This approach is similar to that used by Williams et al. (22). The Spearman rank correlation coefficient was calculated with R.
RESULTS
Cis effects of nucleotides in the mRNA channel downstream of the stop codon
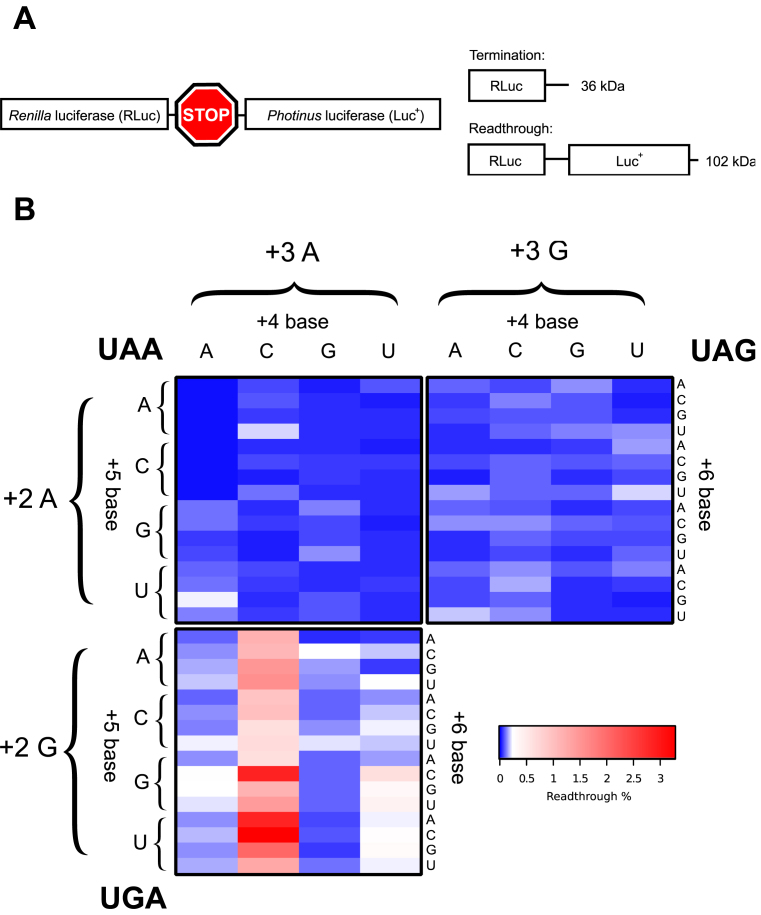
We initially focused on the first three nucleotides (+4 to +6) within the mRNA channel downstream of the stop codon (+1 to +3) positioned at the ribosomal A site. We determined how all permutations (192) for the three stop codon-containing sequences +1UGANNN+6, +1UAANNN+6 and +1UAGNNN+6 affected faithful translation termination. Within this region positions +4 to +5 are known to base stack with 18S rRNA within the mRNA channel (8), possibly affecting termination efficiency. Relative termination success was assayed in cultured mammalian cells using a bi-cistronic reporter construct pGL3s-hRLuc that was developed for this purpose (Materials and Methods and Figure 1A). Upon translation of the reporter construct two possible proteins are generated: either a 36 kDa RLuc product when termination occurs as expected, or, a 102 kDa RLuc–Luc+ readthrough protein when termination fails because a near-cognate tRNA decodes the stop codon (Figure 1A). The activity of the two different luciferase enzymes can then be measured to estimate the extent of stop codon readthrough. Rates of readthrough were normalised against a control reporter construct where the stop codon was substituted with an appropriate sense codon, which results in translation of the full-length dual luciferase fusion protein (100% readthrough). The specific enzymatic activity of RLuc is not affected by the C-terminal extension of Luc+ (45,47). In Figure 1B a heat map shows the readthrough efficiency for each stop codon in separate panels with all permutations of the three downstream nucleotides (+4 to +6). Each stop codon had its own pattern of efficiencies dependent upon the specific nucleotides in the +4 to +6 positions. In most contexts termination failed in less than 0.2% of translational events. The exception were signals of the +1UGACNN+6 series, for which readthrough approached 3% in some contexts. The way termination efficiency was modulated for each stop codon by downstream nucleotides +4 to+6 is analysed and described in detail below.
Figure 1.
(A) Schematic of the pGL3s-hRLuc construct. Upon translation of the dual luciferase mRNA two protein products were produced, a 36 kDa termination product (RLuc only) and a 102 kDa readthrough product (RLuc:Luc+ fusion). (B) Heat map of mean measured readthrough efficiency for 192 constructs (all possible permutations of the +4 to +6 positions for each stop codon). Top left quadrant, UAA; top right quadrant, UAG; bottom left quadrant, UGA. ‘+2’ and ‘+3’ refer to the identity of the second and third bases in the stop codon; ‘+4’, ‘+5’ and ‘+6’ refer to the three nucleotides immediately 3′ to the stop codon. Data are available in Supplementary Table S1.
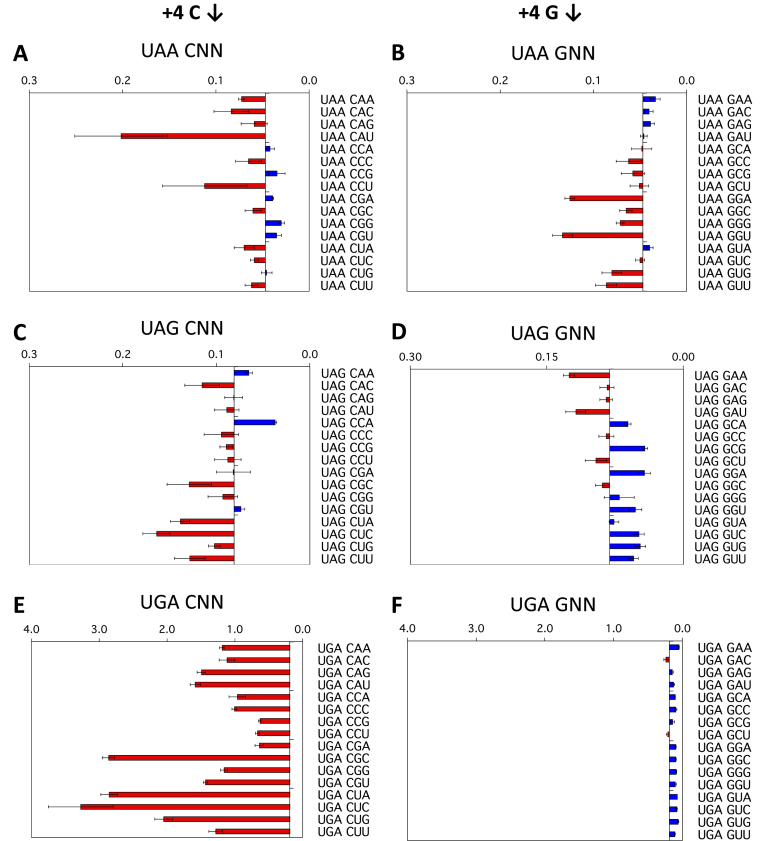
To illustrate the effects of the +4 to +6 positions, termination efficiencies normalised to the readthrough control are shown in Figure 2A–F for a representative strong (G) and a representative weak (C) fourth (+4) base context, that is +1UAA(C/G)NN+6, +1UAG(C/G)NN+6 and +1UGA(C/G)NN+6. Termination success or failure is influenced by the identities of the first (+4), second (+5) and third position (+6) downstream of the stop codon. To aid interpretation, in each panel the results are graphed relative to the median termination efficiency for the 16 constructs within each group. Dark blue bars represent higher efficiency stop signals (lower levels of stop codon readthrough) than the median. By contrast, light red bars represent lower efficiency stop signals with higher levels of termination failure than the median.
Figure 2.
Termination efficiencies of the +1UAA(C/G)NN+6, +1UAG(C/G)NN+6 and +1UGA(C/G)NN+6 signals (+4 position). Termination efficiencies of the +1UAA(C/G)NN+6 (A, B), +1UAG(C/G)NN+6 (C, D) and +1UGA(C/G)NN+6 (E, F) sequence contexts measured in COS-7 cultured cells. Readthrough (%) was calculated by comparison to near-cognate control constructs (CAG/UAA, UAU/UAG, and UGG/UGA) and plotted on the x-axis. Termination efficiencies are presented according to the identity of the base immediately following the termination codon (+4). Results are graphed relative to the median termination efficiency for the series (0.047% UAA, 0.081% UAG and 0.190% UGA). Red bars represent levels of readthrough greater than the median (left) and blue bars represent levels of readthrough less than the median (right). Experiments were performed in triplicate with two independently isolated constructs for each signal context. Mean values are presented ± SEM.
The +4 position
Decoding failure at the UGA stop codon was high (median 0.19%, range 0.10–3.27%) compared to the UAA and UAG sequences (Figure 1B, Supplementary Figure S1, Table S1). Despite the relative abundance of UGA stop signals in eukaryotic genes (20), they are distinct from UAA and UAG in being more sensitive to changes in the stop codon contexts. The identity of the base following the UGA stop codon (+4) had a profound effect on termination success or failure. The most dramatic example of this occurred with +4C in this position when there was up to six-fold higher termination signal failure above the median value (Figure 2E). The other pyrimidine base, +4U, also provided a relatively poor termination context (Supplementary Figure S1F), but not as marked as +4C. By contrast, +4G in this position (Figure 2F) and as well the other purine +4A (Supplementary Figure S1E) promoted efficient termination at UGA, indicating how important purines are as the base following UGA to promote successful termination. Two mammalian cell lines, COS-7 (Figure 2A–F) and the human cell line HEK293T (Supplementary Figure S2) were used as the host cells, and similar results were obtained for the relative efficiencies of the 24 stop signal contexts tested, with only one exception—UAAGUC showed a readthrough of 0.05% in COS-7 cells compared with 0.17% in HEK293T, but these values were both within the low range of 0.015% to 0.228% for the UAA series of termination signal contexts.
By contrast to the UGA series, the UAA series of constructs showed a four-fold lower median readthrough (0.049%), and as well, a much lower range (0.015–0.228%) compared to UGA. In the UAA series, the identity of the +4 nucleotide had less impact on termination. Nevertheless, the identity of the nucleotide that followed the UAA (+4) still modulated the event in a sequence-specific manner. In the cell culture assay, +4C or +4G nucleotides (Figure 2A, B) were the least effective for termination, allowing a higher rate of readthrough than the other two nucleotides. Consistent with these data, it has previously been shown that the +4 nucleotide is essential for active termination with UAA termination signals in an in vitro assay, with the UAAC oligonucleotide a particularly poor termination signal (11).
The range of termination decoding failures at the UAG stop codon contexts (0.028–0.203% readthrough) was similar to UAA, although the median was higher (0.081% compared with 0.047% for UAA). The downstream nucleotides, however, affected termination at UAG signals differently to UAA signals. As with UAA when +4C was present as the nucleotide following the stop codon the rate of UAG readthrough was higher than the mean for the series in almost all contexts (Figure 2A, C). In contrast to UAA but similarly to UGA, +4G and +4A in this position in UAG contexts had lower readthrough (Figure 2D, and Supplementary Figure S1C), but in most cases, this was dependent on the +5 base that followed, with +5U improving and +5A reducing termination success respectively.
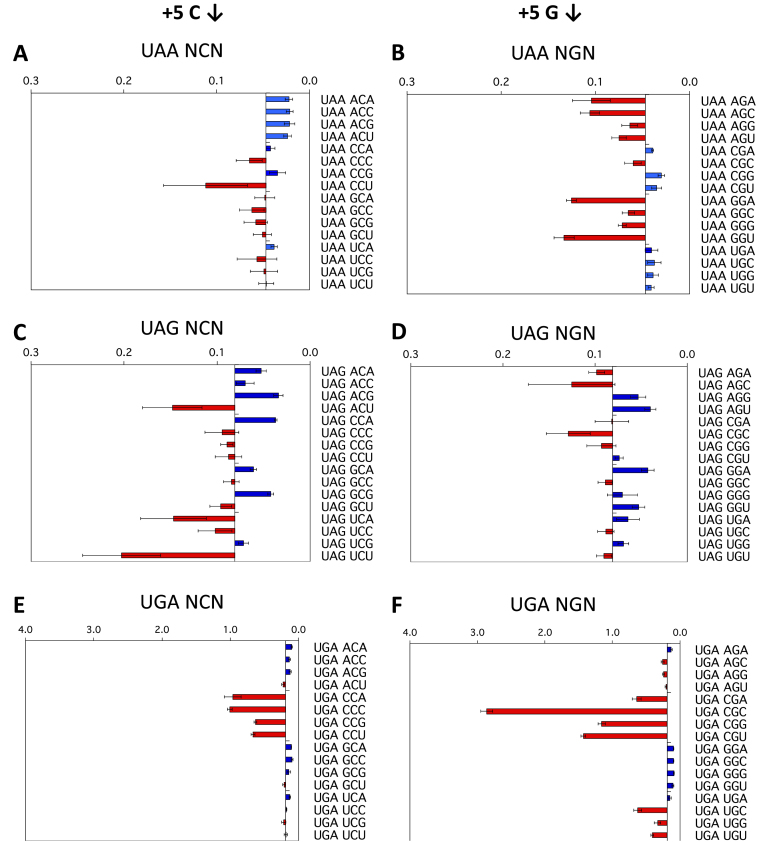
The +5 and +6 positions
As all 64 permutations for each of the three stop codon-containing sequences were assessed, we were able to examine the effect of nucleotides within the +5 to +6 region of the termination signal on termination success or failure. With UGA contexts, the identities of both the +5 and +6 nucleotides were major determinants of increased or decreased readthrough (Figure 3E, F, Supplementary Figure S3E, F). An example of this effect was that +5C but not 5+G improved termination with +4 nucleotides other than C (Figure 3E, F). With the higher observed readthrough of the UGA stop codons, the +6 nucleotide showed a marked effect on termination in some cases; +6C sometimes weakened an already poor context for termination (e.g. UGA CGC) (Figure 3F), indicating that nucleotides even well beyond the triplet stop codon could modulate the rate of stop codon failure and be an important influence on translation termination. This also implies that the constrained binding surface formed between the UGA stop codon and eRF1 (6) was not providing equivalent interaction energy compared to UAA and UAG and that the downstream nucleotides were able to stabilise the UGA/eRF1 interaction directly or indirectly.
Figure 3.
Termination efficiencies of the +1UAAN(C/G)N+6, +1UAGN(C/G)N+6 and +1UGAN(C/G)N+6 signals (+5 and +6 positions). Termination efficiencies of the +1UAAN(C/G)N+6 (A, B), +1UAGN(C/G)N+6 (C, D) and +1UGAN(C/G)N+6 (E, F) sequence contexts measured in COS-7 cultured cells. Readthrough (%) was calculated and results graphed on the x-axis relative to the median termination efficiency for the series as in Figure 2. Termination efficiencies are presented according to the identity of the second base following the termination codon (+5). Presentation format, numbers of replicates and error bars are as in Figure 2.
When the data from the +1UAANNN+6 series of constructs were ordered according to the identity of the second (+5) or third (+6) nucleotides from the stop codon, the pattern indicated that, even with this high-efficiency termination codon, the downstream nucleotides affected the efficiency of stop signal decoding (Figure 3A and B, Supplementary Figure S3A, B). This was primarily evident in the UAAANN context, in which a +5 A or C enhanced and a +5 G or U suppressed termination. The selected series shown in Figure 3A, B comparing +5C and +5G, illustrates the complex variations observed. With +5G (and with +5U [Supplementary Figure S3B]) termination success was generally lower at the UAA stop codon when challenged by near-cognate tRNA competition compared with +5C in that position (Figure 3A) (and with +5A [Supplementary Figure S3A]). These data show that each nucleotide in the mRNA channel downstream of the stop codon is not isolated from the influence of neighbouring nucleotides.
The effects of the +5 and +6 positions on UAG readthrough were distinctively different from that of UAA signals, implying that recognition of each of these codons by eRF1 is unique (see Figure 1B). With the UAGNNN contexts (Figure 3C, D, Supplementary Figure S3C, D), it was generally observed that if the sequence contained two different pyrimidines (C/U) or two different purines (A/G) in positions +4 and +5, termination context became poorer (e.g. +4UCN+6), but not with the same pyrimidine or purine pairs (e.g. +4CCN+6 see Figure 3C).
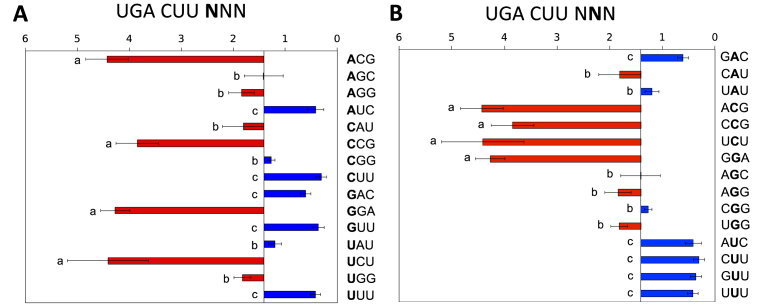
The +7 to +9 positions
How far does the effect on termination extend downstream of the stop codon? We investigated whether the influence on termination extended out to the nucleotides protected by the outer regions of the mRNA channel (34,59–61), where interaction with ribosomal proteins rather than rRNA is more likely (34,62).
A series of contexts of the weak stop signal +1UGACUU+6 varying in the following three nucleotides +7 to +9 were tested. A highly significant effect on termination success was observed (Figure 4A), with readthrough varying 20-fold from 0.3 to 4.4%. This substantial effect is consistent with that observed previously in yeast when +7UUA+9 was changed to +7GGA+9 downstream of +1UAGCAA+6, and with mutational analyses of the readthrough in the tobacco mosaic virus (30,32). The nature of the nucleotide in the +7 and +8 positions was influential in modulating the relative strength of the termination signal but the identity of the nucleotide in the +8 position imprinted its own pattern on the termination signal effectiveness (Figure 4B). In this position C was unable to rescue a poor signal, U could strengthen the signal strongly, and A or G had more modest effects (Figure 4B). We deduced that the +9 position was not highly influential in the limited range of selected contexts tested.
Figure 4.
Termination efficiencies of the +1UGACUUNNN+9 signal (+7 to +9 positions). (A) Termination efficiencies of representative high and low efficiency +1UGACUUNNN+9 sequence contexts measured in COS-7 cultured cells. Readthrough (%) was calculated by comparison to a near-cognate control construct (UGG). Termination efficiencies are presented according to the identity of the fourth base following the termination codon (+7). Results are graphed relative to the median termination efficiency for the series (1.41%). Presentation format, numbers of replicates and error bars are as in Figure 2. Means not sharing the same letter are significantly different (Tukey HSD, P < 0.01). (B) The same data as in (A) but ordered according to the identity of the +8 nucleotide.
How does measured strength of the termination signal correlate with frequency of occurrence?
A number of features influence the sequence of nucleotides immediately downstream of stop codons: the presence of in-frame downstream stop codons, the amino acid sequence of readthrough products, local RNA structure, or the products of ribosome reinitiation close to the termination site (20,22,63,64), while neutral mutation bias relative to local GC content is also influential at stop signals (25). Indeed, previous studies evaluating eukaryote genomes have identified bias in nucleotide usage both 5′ and 3′ to the stop codon (20,26,45), correlated with expression of both the mRNA and protein (25,45,65), and a higher degree of non-randomness is observed in known readthrough elements compared to efficient terminators (31).
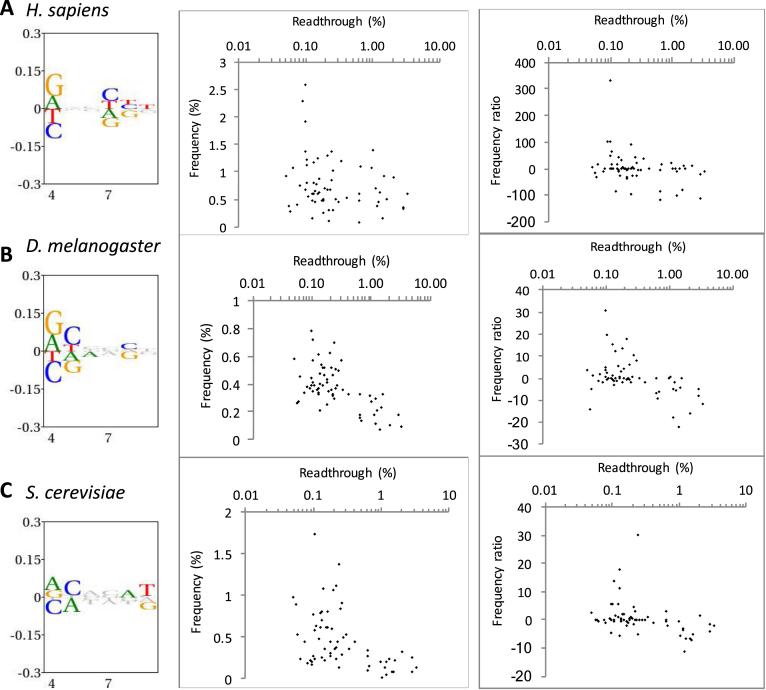
To assess how the bias observed around the stop codon correlated with our measured readthrough percentages we assessed the abundance of all 192 permutations for the three stop codon-containing sequences +1UGANNN+6, +1UAANNN+6 and +1UAGNNN+6 in the genome of H. sapiens, S. cerevisiae, and D. melanogaster (Supplementary Figure S4, Table S2). In the genomes investigated there were both significant positive and negative biases in the occurrence of the hexa-nucleotide sequences for the three stop codon contexts (Figure 5, left panels). As expected, the greatest bias was observed in the +4 position, where purines were overrepresented. Consistent with our measured readthrough capacity, nucleotide bias was less evident further downstream of the stop codon although, notably, a bias was clearly observed at +7 to +9 which is consistent with Figure 4 and previous investigations (31,32).
Figure 5.
Hexanucleotide bias and stop codon readthrough in the genomes of three eukaryotic species. Left panels show information content and bias through the +4 to +9 positions downstream of all stop codons in each organism. The height of all symbols at any given position specifies the information content, and over-and under-represented nucleotides are assigned positive and negative values, respectively. Middle and right panels show the correlation between measured readthrough and hexanucleotide stop codons beginning with UGA. In the middle panel, frequency among all analysed genes is shown. In the right panels, readthrough is plotted against a frequency ratio, (observed − expected)2/expected (see Methods for details) to adjust for local nucleotide bias. (A) H. sapiens (9482 analysed sequences). (B) D. melanogaster (2280 sequences). (C) S. cerevisiae (1598 sequences).
Among all hexanucleotide termination signals, in S. cerevisiae and D. melanogaster, in particular, there was an inverse correlation between the level of measured readthrough and the frequency of stop signals starting with UGA in the genome, as assessed by the Spearman rank correlation test. That is, higher-fidelity signals were more frequently found (Figure 5 and Table 1). This was not evident in H. sapiens, perhaps because of the very high proportion of relatively weak UGA stop codons in higher eukaryotes (48.9% of H. sapiens in our analysis; Supplementary Table S2 and Figure S4A).
Table 1. Spearman's rank coefficient analysis of UGA stop signal occurrence in three genomes versus measured readthrough in COS-7 cells.
| Frequency | Frequency ratio | |||
|---|---|---|---|---|
| Rho | P-value | Rho | P-value | |
| H. sapiens | –0.2191056 | n.s. | –0.2925824 | P < 0.05 |
| D. melanogaster | –0.5418644 | P < 0.001 | –0.4180909 | P < 0.001 |
| S. cerevisiae | –0.4270498 | P < 0.001 | –0.4505952 | P < 0.001 |
n.s., not significant.
UGA has the highest capacity for readthrough and therefore is most likely to be under selection for efficient termination. When UGA stop signals were evaluated separately, an inverse correlation between stop signal strength and frequency was observed in D. melanogaster and S. cerevisiae (Figure 5B and C, middle panels). A correlation between poorer signal strength and lower frequency of occurrence was less evident in H. sapiens, (Supplementary Figure S4A and Table S2) (25). Nonetheless, it has previously been reported that a subset of highly expressed human genes—for example, those encoding ribosomal proteins—are enriched for efficient UAA stop signals (25,65).
This inverse correlation between signal strength and frequency of occurrence largely reflected the local bias in nucleotide usage in this region of the genome. When the observed frequencies of each hexanucleotide stop signal were corrected for local nucleotide usage by shuffling (see Methods) the observed correlation was weaker or disappeared. However, there still was an inverse correlation between signal strength and occurrence in S. cerevisiae and D. melanogaster at UGA stop codons after correction (Table 1 and Figure 5B and C, right panels). The observed bias in these nucleotides downstream of the stop codon within the mRNA entry channel may reflect a selection for sequences that improved stop signal fidelity (Supplementary Figure S4B, C).
Potential factors influencing the rate of stop codon failure
How important is the eRF1 stop signal interaction for the efficiencies of different stop codon contexts? The cellular concentration of the release factor eRF1 was manipulated in the assay system to determine whether it was a key determinant of the patterns of readthrough observed, through its interaction with the stop codon, the +4 nucleotide and even perhaps a more extended element. An eRF1 over-expression plasmid (pcDNA-eRF1) and one of two shRNA pSilencer plasmids were used to achieve either an increased or decreased concentration of eRF1 in the cells as described previously (47). Consistent with our previous studies, cells transfected with the pcDNA-eRF1 plasmid exhibited a 4-fold increase in eRF1 protein levels, and conversely, cells transfected with the pSilencer-si90 or pSilencer-si1186 plasmid exhibited a 50% decrease in the level of eRF1 mRNA and protein (47,48).
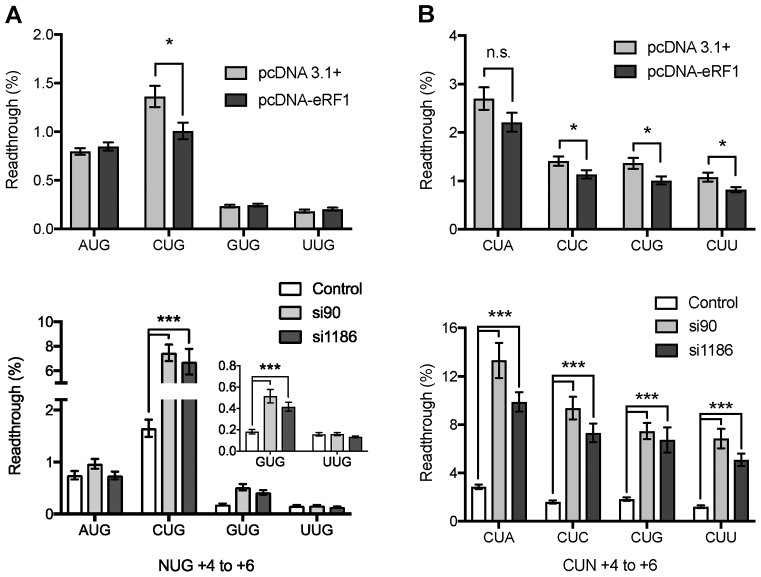
Initially, to determine the importance of cellular eRF1 concentration on stop codon readthrough four UGA contexts were chosen for a study that varied in the identity of the +4 nucleotide (Figure 6A). Readthrough at the poor termination sequence element UGACUG was significantly reduced in cells expressing additional eRF1 compared with non-supplemented cells (Figure 6A upper panel). By contrast, the previously identified strong termination contexts UGAAUG, UGAGUG and UGAUUG did not show any significant decrease in their low rate of readthrough with high concentrations of eRF1 (Figure 6A, upper panel). These results indicate that eRF1 binding might be rate limiting but only at certain termination contexts, and as a consequence near-cognate tRNAs would be more competitive for decoding the stop codon as sense in these cases. Higher concentrations of release factor counter this effect.
Figure 6.
Effect of regulation of eRF1 expression on termination efficiency. Termination efficiencies of +1UGANUG+6 (A) and +1UGACUN+6 (B) sequence contexts in HEK 293T cultured cells co-transfected with either the control plasmid pcDNA or the pcDNA-eRF1 over-expression plasmid (eRF1) (upper panels), or the shRNAs pSilencer negative control (NC), pSilencer-si90 or pSilencer-si1186 (lower panels). Readthrough (%) was calculated by comparison to a near-cognate control construct (UGG). GUG result is expanded in inset in left lower panel. Experiments were performed in triplicate with 12 replicates for each condition, and 24 replicates for pSilencer NC. Mean values are presented. Error bars are SEM. *P < 0.05, ***P < 0.01, n.s., not significant. Data are available in Supplementary Table S3.
Co-transfection of either the pSilencer-si90 or pSilencer-si1186 siRNA plasmid with the test vector resulted in a large increase in the rate of stop codon readthrough at the weak UGACUG context, as well as a smaller yet still significant increase at the stronger UGAGUG context (Figure 6A, lower panel inset). No change was observed when A or U was present in the +4 position (Figure 6A, lower panel). This implies that lowering intracellular levels of eRF1 by about 50% causes it to become rate limiting for termination at a weak context, compared to efficient stop signals. At stronger contexts, perhaps modulation of the levels of additional termination factors such as eRF3 would be required as well. Taken together the eRF1 overexpression and shRNA experiments highlight that sequence elements can vary in their ability to respond to changing eRF1 concentrations with regard to termination efficiency.
To further determine the importance of the termination context on the eRF1 binding to the termination signal four additional sequences that varied at the +6 base were assessed for stop codon readthrough at different concentrations of eRF1 (Figure 6B). Both weak UGACUN and strong UGAGUN sequence elements were assessed to determine the response to changes in the cellular levels of eRF1. Co-transfection of the previously identified weak termination sequence context UGACUN and the pcDNA-eRF1 plasmid resulted in significant improvement in termination at this stop signal in most contexts (Figure 6B, upper panel). The UGACUA context was, however, not responsive to enhanced levels of the cellular levels of eRF1. For the previously identified strong context, UGAGUN readthrough was below detection before supplementation with higher concentrations of eRF1 (data not shown).
Reducing the cellular levels of eRF1 by siRNAs resulted in a significant increase in the readthrough of all the UGACUN containing stop contexts assayed (Figure 6B, lower panel). The greatest response to the decrease in eRF1 levels was exhibited by UGACUA—the weakest signal—increasing readthrough by 9% above non-supplemented controls. The strong UGAGUN contexts exhibited a small but significant increase in the rate of stop codon readthrough under conditions of reduced eRF1 (data not shown). These data indicate that, in general, weak termination signals are particularly sensitive to modulation of eRF1 levels, although there are marked context-specific effects.
In-trans competition for decoding the termination codon by cognate suppressor tRNAs
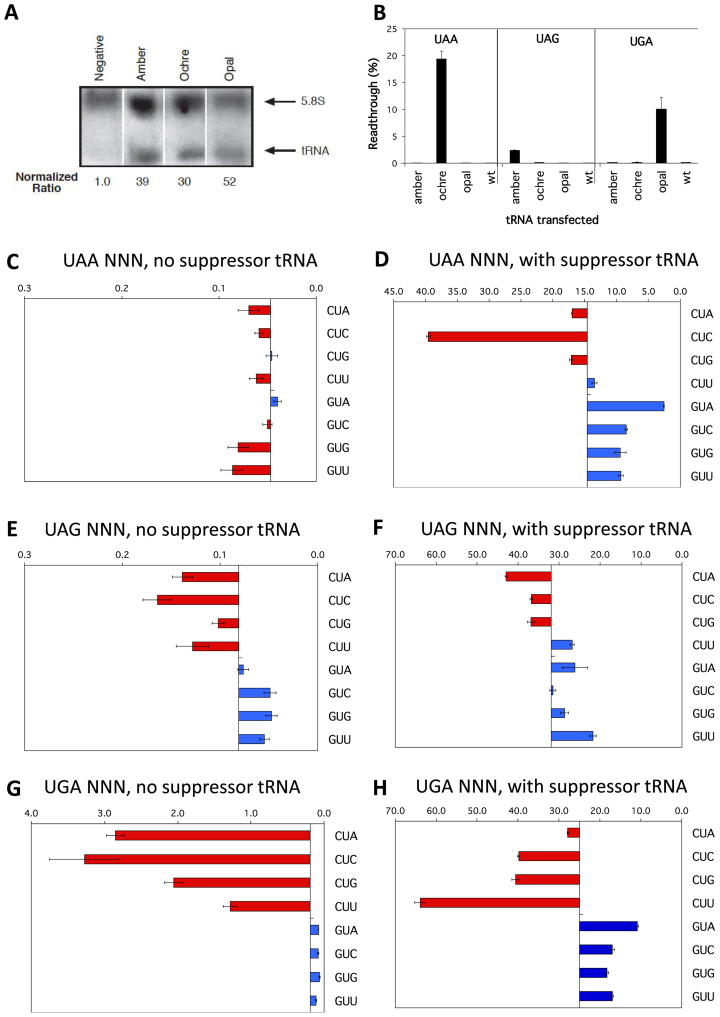
Despite the observed effects of modulating eRF1 (Figure 6), the range in the percentages of readthrough of termination signals observed in Figures 2–4 could also reflect a unique enhancement or weakening of the competitiveness in decoding the stop signal by each competing near-cognate tRNA, rather than a reduction in the binding of eRF1 to the stop signal. We, therefore, tested the competitiveness of near-cognate tRNA binding utilising three cognate suppressor tRNAs (sup-tRNAs). The sup-tRNAs possess the same structural backbone as the near cognate tRNAs, having been derived from a serine–tRNA and they compete directly with eRF1 for binding to the stop codon context. Each novel sup-tRNAs is specific for a stop codon (ptRNAoc, ochre UAA; ptRNAam, amber UAG; and ptRNAop, opal UGA) have been documented to show functional specificity in their suppression of these stop codons in HEK293T cells (49,66). The levels of expression of the ptRNAs were comparable in transfected cells as shown by Northern blot, normalised to 5.8S rRNA (Figure 7A and B). The oligonucleotide probe used for Northern detection produced a low signal in non-transfected control cells—likely detecting the endogenous serine tRNA from which the suppressor tRNAs were derived (Figure 7A).
Figure 7.
Competition by cognate tRNAs at termination signals. (A) Northern blot probed for sup-tRNA (lower arrow) and 5.8S rRNA (upper arrow) transcripts. Expression levels of sup-tRNA relative to 5.8S rRNA are indicated. The sup-tRNA probe was able to detect ptRNA amber, ptRNA ochre and ptRNA opal as it was complementary to the anticodon loop of the tRNA, but degenerative for the specific anticodon sequence. (B) Termination efficiencies of the +1UAAAGA+6, +1UAGAGA+6 and +1UGAAGA+6 sequence contexts measured in COS-7 cultured cells co-transfected with cognate sup-tRNAs ptRNAoc (ochre UAA), ptRNAam (amber UAG), or ptRNAop (opal UGA); or with the parent control ptRNAser (wild-type tRNAser). Readthrough (%) was calculated by comparison to near-cognate control constructs (CAG/UAA, UAU/UAG, and UGG/UGA) and plotted on the x-axis. Presentation and replicates are as in Figure 2. (C-H) Termination efficiencies of selected UAA (C, D), UAG (E, F) and UGA (G, H) sequence contexts were measured in COS-7 cultured cells co-transfected with cognate suppressor tRNAs (right panels) or without (left panels). UAA sequence contexts were co-transfected with ptRNAoc (ochre UAA), UAG sequence contexts were co-transfected with ptRNAamber (amber UAG), and UGA sequence contexts were co-transfected with ptRNAop (opal UGA). Readthrough (%) was calculated as before. Results are graphed relative to the median termination efficiency for the series (without and with suppressors respectively: 0.047% and 14.61% UAA, 0.081% and 31.94% UAG, 0.190% and 25.0% UGA). Presentation, replicates and error bars were as before.
To confirm the stop codon specificity of each sup-tRNA, cells were co-transfected with the ptRNAs and a test vector, pGL3s-hRLuc (containing stop codons UAA or UAG or UGA). In the test vector, the downstream Luc+ gene (Figure 1A) has two in-frame stop codons spaced one codon apart (i.e., UAA UUC UAG AGU) that allow termination in the presence of any specific sup-tRNA. As shown in Figure 7B the absolute value of readthrough for each stop codon context increased many fold with the cognate sup-tRNA in direct competition with eRF1 for the codon and was specific for the particular cognate ptRNA species with no overlap in suppression exhibited by any of the three tRNAs. The degree of competition to decode the stop codon against eRF1 varied among the three different ptRNA suppressors (UAA ∼3%, UGA ∼10% and UAG 20%) and was independent of the concentration of the sup-tRNA (Figure 7B). Cells co-transfected with wild-type tRNASer exhibited no increase in decoding competition with eRF1 (Figure 7B, wt).
Next, we examined whether the previously observed patterns of termination efficiencies at different contexts of each stop codon changed with the addition of competing sup-tRNA (suppressor tRNA versus endogenous near-cognate tRNA). Representative termination sequence contexts (‘weak’ +4CUN+6 and ‘strong’ +4GUN+6) were tested with the ptRNA series of plasmids (Figure 7D, F and H) and compared to the previous results (Figure 7C, E and G). For UAG and UGA stop codons the pattern of readthrough at the different contexts was largely consistent with that observed without suppressor tRNAs (i.e. when competition is dependent on endogenous near-cognate tRNAs only) (Figure 7E and G versus F and H). The UAG and UGA weaker stop contexts (+4CUN+6) were more susceptible to readthrough by the specific suppressor tRNA, consistent with less competitive termination decoding with these contexts. This implies that the downstream nucleotides of the mRNA enclosed in the channel mediate their effect on termination efficiency with UGA and UAG primarily not by enhancing or depressing decoding from competing tRNAs, but rather more directly by affecting the binding and/or activity of eRF1. The pattern of readthrough in the high fidelity UAA contexts, by contrast, did change when sup-tRNA was expressed (Figure 7C versus D). Perhaps sup-tRNA expression here simply allows sub-hierarchies of stop codon contexts to be revealed through the greater competition for decoding UAA.
DISCUSSION
Stop signal: a sequence element with multifaceted contributions to efficiency
A number of studies have provided evidence that nucleotides downstream of the eukaryotic translational stop codon can affect the efficiency of translation termination (21,28,30–32). While nucleotides close to the codon could have a direct effect by interacting with the decoding release factor, eRF1, as shown with the +4 base (6,7) it has been puzzling how nucleotides quite distant could have such a direct effect. In the current study, we have investigated the influences of those nucleotides within the mRNA channel of the ribosome, when a stop codon is in the A site of the decoding centre, during translation of that codon.
Cryo-EM studies of the eukaryotic translational termination complex with eRF1 have shown that the +4 and +5 nucleotides of the stop codon context base stack with 18S rRNA bases G626 and C1698 respectively (6–8). Stacking would be more stable if a purine were present in the +4 position (G626) (6), and a purine in the +5 position (C1698) (8) increasing the stability of the decoding complex. Additionally, hydroxylation of a proline residue in the ribosomal protein uS12, which contacts the mRNA backbone near the +4 nucleotide, modulates readthrough efficiency in a direction dependent on the base in the +4 position, implying a special contribution of this nucleotide to the termination process (67). Consistent with this we found that the position following the A site stop codon (+4) has a particularly strong influence on the efficiency and fidelity of the decoding event. Additionally, the next two positions (+5 and +6) are also significantly influential in a complex manner, both from the perspective of the identity of the nucleotide in each position and through combinations of nucleotides from +4 to +6. This indicates that the interaction between the mRNA and the ribosomal components, like the rRNA nucleotides that line the channel, influence the translation decoding rate. RNase footprinting studies indicate that the length of mRNA protected within the eukaryotic ribosome is approximately 28–31 nt, depending on digestion conditions (60,61,68,69). This suggests that the mRNA channel in the eukaryotic ribosome protects a very similar number of nucleotides to that of the bacterial ribosome (59,70). Consistent with this we found that even in the outer reaches of the channel near to the entry point for the mRNA, the +8 nucleotide position can have strong effects to improve the efficiency of a poorly performing +1UGA+3 stop codon as an efficient termination signal. Although we conducted only limited studies of sequence variations for the +7 to +9 distant positions, the results suggested the +8 position was dominant among this trio, with +7 and +9 not particularly influential in establishing an effector pattern (Figure 4). A similar complex pattern was observed in the readthrough element of tobacco mosaic virus. Notably, however, the +9 nucleotide has been observed to have a significant effect on readthrough depending upon the upstream context, indicating that caution is required when generalising these results to all stop signals (30,31).
Both bioinformatics (Figure 5) and previous in vitro and in vivo experimental studies have implied the existence of a eukaryotic translation termination element that encompasses nucleotides both upstream and downstream of the stop codon (20,22,35,45,71). Parts of such an indicative element could have quite different influences on the efficiency of termination depending on where they are positioned. For example, the upstream sequence through its coding potential determines which amino acids are present in the PTC and exit tunnel as well as which tRNAs are favoured in the E and P sites preceding a stop codon. A nascent peptide designated MTO1 within the Arabidopsis thaliana CGS1 coding region causes ribosomes to stall in response to S-adenosyl-l-methionine during elongation (72). Expression of the human cytomegalovirus (CMV) gp48 gene is reduced by translation of its uORF2 nascent peptide (73), which causes ribosomes to stall at the uORF2 termination codon (74). These studies reveal the paradoxical potential for interactions between a nascent peptide and eRF1 to obstruct the translation termination cascade.
Eukaryotic eRF3 strongly stimulates peptide release by eRF1 (75) through its GTPase activity indicating a role for class-2 release factors in eukaryotic termination. It is of note that binding of eRF1–eRF3–GTP to pre-termination complexes (pre-TCs) also induces a 2-nt forwardshift of their toeprint (75), caused by the compaction of the mRNA in the termination complex (6,76). Therefore, it is possible that sequences particularly amenable to such contraction would be favoured, suggesting another mechanism of how the extended termination signal could operate in vivo.
It has emerged that the evolutionarily distinct bacterial and eukaryotic decoding release factors interact with the stop codon through different parts of their structures, including the major recognition loop and helix a5 region for bacterial RFs (77,78) and three distinctly separate peptide regions from the N-domain of eRF1 that surround the stop codon on the ribosome (5,6). The exquisite geometry required for productive interaction and recognition of the stop codons highlights the importance of the correct orientation of the bases with respect to the amino acids of the eRF1. There is strong evidence for a significant conformational change in eRF1 on binding to eRF3 in solution or alone on the ribosome, causing eRF1 to adopt a bent conformation that resembles a tRNA (5–8,79,80). This dynamic structural change highlights how each individual factor/nucleotide interaction might be highly sensitive to perturbation from the distal nucleotides. As the more distal downstream bases in the mRNA can make interactions with ribosomal moieties in the channel they may distort the positioning of the upstream RNA bases affecting individual factor/nucleotide interaction required for decoding.
The overall conformation of the ribosome in the pre-termination state resembles the post-translocation state ribosome, with peptidyl-tRNA in the P site, which is locked in the nonratcheted conformation (7). In this conformation, there is a ∼8.5° rotation of the small subunit compared to the ratcheted state, reminiscent of the range of rotation for bacterial ribosomes (70,81). Therefore, during translation termination, with sense codons in the P and E sites, there is a closure of the ribosomal mRNA channel that must trigger a network of interactions between the mRNA and the ribosome. This could explain why the downstream nucleotides distant from the stop codon can influence termination so markedly. In the model generated from the cryo-electron microscopy structure of a termination complex reported by Shao et al. (8) continuous density around the +7 base is observed which is perhaps indicative of direct contact between the ribosome and mRNA in the entrance channel.
The translation termination elements of UAA and UAG
UAA and UAG stop codons have a very high degree of termination fidelity and competitiveness against near-cognate events. It is tempting to dismiss the concept of a signal element with these codons since they are less markedly influenced by the downstream nucleotides. The oligonucleotide UAAC is, however, several-fold less efficient than UAAG at facilitating termination in a cell-free in vitro termination assay where there is no competition from tRNAs to decode the signal and the UAA codon itself is inactive (65). In our current study in cultured cells, the identity of the +4 nucleotide also has a significant effect on fidelity, so evidence for a four-base stop signal is compelling even with these high strength signals.
The patterns of stop failure were different between UAA and UAG signals, possibly resulting from the differences in UAG stop codon recognition by eRF1 (utilising E55) from that of UAA (6). There was, by contrast, a good correlation in the patterns for particular signals where data are available between yeast and our two mammalian cell lines, consistent with the high conservation of eRF1 among eukaryotes (82). The twelve sequences investigated by Bonetti et al. (21) to examine the effect of the +4 base on readthrough in yeast were included in our examined sequences, and comparison between the two studies reveals a comparable pattern of termination failure rates for the UAGNAU and UAANAU signals. However, there is a difference in signal readthrough in the UAGNAU context, with our study identifying higher readthrough with the UAGUAU context than Bonetti et al., who reported higher readthrough with UAGCAU (21). These differences may reflect the reported influence of the last C terminal amino acid and mRNA sequence 5′ of the stop codon on termination efficiencies (28,29), as the 5′ context varied significantly between the two studies.
The translation termination element of UGA
Accurate decoding of the UGA stop codon was strongly dependent upon the three downstream nucleotides. The most marked example is with C in the +4 position immediately following the stop codon, which results in termination failure up to six-fold above the median value for the 64 contexts examined (Figure 1B). These results correlate with structural studies that show stacking between the +4 nucleotide and residue G626 of 18S rRNA would be more stable with purines in this position (6). This nucleotide stacking might increase termination efficiency through stabilising the interaction with eRF1 and the mRNA to contribute significant interaction energy. This observation correlates well with bioinformatic analyses that have found that the termination sequences TGAC and TGAT are under-represented in eukaryotic genomes (45). Stop codon recognition has been proposed to occur in two stages, an initial recognition, followed by a tightening of the interaction after GTP hydrolysis (83). The second step has been proposed to occur more slowly for UGA codons, and this would allow a greater chance of a near-cognate decoding event to compete. Additionally, as UGA stop codons make unique interactions with eRF1 (6), it is possible that these accommodations also affect the rate of decoding. It is significant that higher concentrations of eRF1 and presumably higher probability of occupancy of the A site enhanced the chance of a productive termination event at weaker UGA contexts (84).
The amelioration of nonsense mutations in inherited diseases that promote premature translational termination has been of continuing clinical interest. Stop codon failure at premature termination codons, influenced by context, may highlight sites at which the nonsense mutation may be more amenable to readthrough manipulation. Also, since premature termination triggers nonsense-mediated mRNA decay (NMD) (85); an extended termination signal may be important within the cell for translational machinery to distinguish between mature and premature stop codons (39).
An increasing number of highly efficient ‘programmed’ readthrough elements have been described in eukaryotes, sometimes with a defined biological function for the extended, ‘read-through’ polypeptides (86–89). For example, in Drosophila spp. it has recently been reported that an evolutionarily recent point mutation causing a premature termination codon can facilitate readthrough approaching 100% efficiency, resulting in the translation of a functional protein from an apparent pseudogene. Although the extended sequence that allows this to occur is not understood, the effect is independent of the identity of the stop codon but strongly dependent upon the +4 nucleotide (90). Furthermore, a case report of a patient with a predicted premature termination codon but with a weak termination context—+1UGACUA+6, among the weakest observed in our library—that can facilitate readthrough to mitigate the expected fatal disease phenotype has been reported (35). Although the nucleotides directly upstream and downstream of the described endogenous readthrough-prone termination contexts are frequently described to affect termination, in many instances an extended signal, often containing a structured element, is required to mediate efficient readthrough (91).
The biological function of the translation termination sequence element
A key question is whether the influence of the downstream sequence on the efficiency of stop codon decoding, as we have shown, is simply variation within an allowable physiological window, or whether there is a broader significance for the overall biology of the eukaryotic cell. In prokaryotes, where transcription and translation are spatially linked, efficient termination signals might convey a selective advantage to the organism and hence be positively selected over time. Indeed, in Escherichia coli the most highly expressed genes have the most rapidly decoded stop signals (predominantly UAAU) (92). What happens in eukaryotes, where transcription and translation are separated spatially? One key feature may lie with the role the translation termination sequence plays in the proposed mRNA recycling loop with eRF1 and eRF3 (93). More efficient sequence elements will transit eRF1 and eRF3 more rapidly, expediting the formation of the recycling loop and maintaining mRNA stability, increasing the rate of translation and the amount of protein expressed per transcript.
Do relatively inefficient termination signals also have a specific function? The ability of specific sequences to promote rare recoding events like stop codon readthrough (e.g. the histidine decarboxylase gene), selenocysteine incorporation at a UGAC (e.g. the glutathione peroxidase gene) and frameshifting also at a UGAC (e.g. the antizyme gene) allow for expression of a small alternative transcriptome (reviewed in (94)). Also, human genes that contain inefficient termination signals have common biological functions, such as cell cycling and anti-apoptosis (95). This is an intriguing observation considering that cell cycling proteins interact with eRF3 (96), over-expression of eRF1 and eRF3 in some cell lines causes apoptosis (Cridge, unpublished), and overexpression of eRF3/GSPT1 has been reported in intestinal type gastric tumors (97). Is there a hidden code in the nucleotides surrounding the stop codon that our study has highlighted, providing a subtle layer of regulation for the expression of a gene or co-ordinated pathways of genes, only the most obvious of which have so far been revealed?
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Pat Flawn, Tina Edgar, Andrew Gray and Dr Peter Stockwell, at the University of Otago, for their support of this work.
Dedication: A.G.C. wishes to dedicate this paper to the memory of Pat (Parvati) Flawn, whose encouragement and enthusiasm helped to inspire the work reported here.
Author contributions: A.G.C. conducted the majority of the experimental work and made a significant contribution to the preparation of the manuscript. C.C.M. was also a major contributor, making significant contributions to the bioinformatics analyses and the presentation of the manuscript. S.F.M. carried out some key experimental studies. W.P.T. planned and supervised the study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Health Research Council of New Zealand [01/317 to W.P.T]; Marsden Fund of New Zealand [02-UOO-901 to W.P.T and doctoral scholarship to A.G.C]. Funding for open access charge: Biochemistry Department, University of Otago.
Conflict of interest statement. None declared.
REFERENCES
- 1. Poole E.S., Young D.J., Askarian-Amiri M.E., Scarlett D.J., Tate W.P.. Accommodating the bacterial decoding release factor as an alien protein among the RNAs at the active site of the ribosome. Cell Res. 2007; 17:591–607. [DOI] [PubMed] [Google Scholar]
- 2. Roy B., Leszyk J.D., Mangus D.A., Jacobson A.. Nonsense suppression by near-cognate tRNAs employs alternative base pairing at codon positions 1 and 3. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:3038–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blanchet S., Cornu D., Argentini M., Namy O.. New insights into the incorporation of natural suppressor tRNAs at stop codons in Saccharomyces cerevisiae. Nucleic Acids Res. 2014; 42:10061–10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chavatte L., Seit-Nebi A., Dubovaya V., Favre A.. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 2002; 21:5302–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bulygin K.N., Khairulina Y.S., Kolosov P.M., Ven’yaminova A.G., Graifer D.M., Vorobjev Y.N., Frolova L.Y., Kisselev L.L., Karpova G.G.. Three distinct peptides from the N domain of translation termination factor eRF1 surround stop codon in the ribosome. RNA. 2010; 16:1902–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown A., Shao S., Murray J., Hegde R.S., Ramakrishnan V.. Structural basis for stop codon recognition in eukaryotes. Nature. 2015; 524:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Matheisl S., Berninghausen O., Becker T., Beckmann R.. Structure of a human translation termination complex. Nucleic Acids Res. 2015; 43:8615–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shao S., Murray J., Brown A., Taunton J., Ramakrishnan V., Hegde R.S.. Decoding mammalian ribosome-mRNA states by translational GTPase complexes. Cell. 2016; 167:1229–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feng T., Yamamoto A., Wilkins S.E., Sokolova E., Yates L.A., Munzel M., Singh P., Hopkinson R.J., Fischer R., Cockman M.E. et al. . Optimal translational termination requires C4 lysyl hydroxylation of eRF1. Mol. Cell. 2014; 53:645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Konecki D.S., Aune K.C., Tate W., Caskey C.T.. Characterization of reticulocyte release factor. J. Biol. Chem. 1977; 252:4514–4520. [PubMed] [Google Scholar]
- 11. McCaughan K.K., Brown C.M., Dalphin M.E., Berry M.J., Tate W.P.. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:5431–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song H., Mugnier P., Das A.K., Webb H.M., Evans D.R., Tuite M.F., Hemmings B.A., Barford D.. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000; 100:311–321. [DOI] [PubMed] [Google Scholar]
- 13. Bertram G., Bell H.A., Ritchie D.W., Fullerton G., Stansfield I.. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000; 6:1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frolova L., Seit-Nebi A., Kisselev L.. Highly conserved NIKS tetrapeptide is functionally essential in eukaryotic translation termination factor eRF1. RNA. 2002; 8:129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Seit-Nebi A., Frolova L., Kisselev L.. Conversion of omnipotent translation termination factor eRF1 into ciliate-like UGA-only unipotent eRF1. EMBO Rep. 2002; 3:881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ito K., Frolova L., Seit-Nebi A., Karamyshev A., Kisselev L., Nakamura Y.. Omnipotent decoding potential resides in eukaryotic translation termination factor eRF1 of variant-code organisms and is modulated by the interactions of amino acid sequences within domain 1. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:8494–8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hatin I., Fabret C., Rousset J.P., Namy O.. Molecular dissection of translation termination mechanism identifies two new critical regions in eRF1. Nucleic Acids Res. 2009; 37:1789–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tate W.P., Poole E.S., Dalphin M.E., Major L.L., Crawford D.J., Mannering S.A.. The translational stop signal: codon with a context, or extended factor recognition element. Biochimie. 1996; 78:945–952. [DOI] [PubMed] [Google Scholar]
- 19. Cavener D.R., Ray S.C.. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991; 19:3185–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun J., Chen M., Xu J., Luo J.. Relationships among stop codon usage bias, its context, isochores, and gene expression level in various eukaryotes. J. Mol. Evol. 2005; 61:437–444. [DOI] [PubMed] [Google Scholar]
- 21. Bonetti B., Fu L., Moon J., Bedwell D.M.. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 1995; 251:334–345. [DOI] [PubMed] [Google Scholar]
- 22. Williams I., Richardson J., Starkey A., Stansfield I.. Genome-wide prediction of stop codon readthrough during translation in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2004; 32:6605–6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Angenon G., Van Montagu M., Depicker A.. Analysis of the stop codon context in plant nuclear genes. FEBS Lett. 1990; 271:144–146. [DOI] [PubMed] [Google Scholar]
- 24. Arkov A.L., Korolev S.V., Kisselev L.L.. 5′ contexts of Escherichia coli and human termination codons are similar. Nucleic Acids Res. 1995; 23:4712–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Trotta E. Selective forces and mutational biases drive stop codon usage in the human genome: a comparison with sense codon usage. BMC Genomics. 2016; 17:366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brown C.M., Stockwell P.A., Trotman C.N., Tate W.P.. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990; 18:6339–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mottagui-Tabar S., Tuite M.F., Isaksson L.A.. The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur. J. Biochem. 1998; 257:249–254. [DOI] [PubMed] [Google Scholar]
- 28. Cassan M., Rousset J.P.. UAG readthrough in mammalian cells: effect of upstream and downstream stop codon contexts reveal different signals. BMC Mol. Biol. 2001; 2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tork S., Hatin I., Rousset J.P., Fabret C.. The major 5′ determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res. 2004; 32:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Skuzeski J.M., Nichols L.M., Gesteland R.F., Atkins J.F.. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol. 1991; 218:365–373. [DOI] [PubMed] [Google Scholar]
- 31. Harrell L., Melcher U., Atkins J.F.. Predominance of six different hexanucleotide recoding signals 3′ of read-through stop codons. Nucleic Acids Res. 2002; 30:2011–2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Namy O., Hatin I., Rousset J.P.. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2001; 2:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spahn C.M., Beckmann R., Eswar N., Penczek P.A., Sali A., Blobel G., Frank J.. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell. 2001; 107:373–386. [DOI] [PubMed] [Google Scholar]
- 34. Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M.. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011; 334:1524–1529. [DOI] [PubMed] [Google Scholar]
- 35. Pacho F., Zambruno G., Calabresi V., Kiritsi D., Schneider H.. Efficiency of translation termination in humans is highly dependent upon nucleotides in the neighbourhood of a (premature) termination codon. J. Med. Genet. 2011; 48:640–644. [DOI] [PubMed] [Google Scholar]
- 36. Boycheva S.S., Bachvarov B.I., Berzal-Heranz A., Ivanov I.G.. Effect of 3′ terminal codon pairs with different frequency of occurrence on the expression of cat gene in Escherichia coli. Curr. Microbiol. 2004; 48:97–101. [DOI] [PubMed] [Google Scholar]
- 37. Bolger T.A., Folkmann A.W., Tran E.J., Wente S.R.. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008; 134:624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rispal D., Henri J., van Tilbeurgh H., Graille M., Seraphin B.. Structural and functional analysis of Nro1/Ett1: a protein involved in translation termination in S. cerevisiae and in O2-mediated gene control in S. pombe. RNA. 2011; 17:1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Amrani N., Ganesan R., Kervestin S., Mangus D.A., Ghosh S., Jacobson A.. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004; 432:112–118. [DOI] [PubMed] [Google Scholar]
- 40. Henri J., Rispal D., Bayart E., van Tilbeurgh H., Seraphin B., Graille M.. Structural and functional insights into Saccharomyces cerevisiae Tpa1, a putative prolylhydroxylase influencing translation termination and transcription. J. Biol. Chem. 2010; 285:30767–30778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Khoshnevis S., Gross T., Rotte C., Baierlein C., Ficner R., Krebber H.. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 2010; 11:214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Beznoskova P., Wagner S., Jansen M.E., von der Haar T., Valasek L.S.. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res. 2015; 43:5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gross T., Siepmann A., Sturm D., Windgassen M., Scarcelli J.J., Seedorf M., Cole C.N., Krebber H.. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007; 315:646–649. [DOI] [PubMed] [Google Scholar]
- 44. Mikhailova T., Shuvalova E., Ivanov A., Susorov D., Shuvalov A., Kolosov P.M., Alkalaeva E.. RNA helicase DDX19 stabilizes ribosomal elongation and termination complexes. Nucleic Acids Res. 2017; 45:1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cridge A.G., Major L.L., Mahagaonkar A.A., Poole E.S., Isaksson L.A., Tate W.P.. Comparison of characteristics and function of translation termination signals between and within prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2006; 34:1959–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tate W.P., Mansell J.B., Mannering S.A., Irvine J.H., Major L.L., Wilson D.N.. UGA: a dual signal for ‘stop’ and for recoding in protein synthesis. Biochemistry (Moscow). 1999; 64:1342–1353. [PubMed] [Google Scholar]
- 47. Mathew S.F., Crowe-McAuliffe C., Graves R., Cardno T.S., McKinney C., Poole E.S., Tate W.P.. The highly conserved codon following the slippery sequence supports −1 frameshift efficiency at the HIV-1 frameshift site. PLoS One. 2015; 10:e0122176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Carnes J., Jacobson M., Leinwand L., Yarus M.. Stop codon suppression via inhibition of eRF1 expression. RNA. 2003; 9:648–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Le Goff X., Philippe M., Jean-Jean O.. Overexpression of human release factor 1 alone has an antisuppressor effect in human cells. Mol. Cell. Biol. 1997; 17:3164–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Matthews J.C., Hori K., Cormier M.J.. Purification and properties of Renilla reniformis luciferase. Biochemistry. 1977; 16:85–91. [DOI] [PubMed] [Google Scholar]
- 51. Srikantha T., Klapach A., Lorenz W.W., Tsai L.K., Laughlin L.A., Gorman J.A., Soll D.R.. The sea pansy Renilla reniformis luciferase serves as a sensitive bioluminescent reporter for differential gene expression in Candida albicans. J. Bacteriol. 1996; 178:121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tanguay R.L., Gallie D.R.. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol. Cell. Biol. 1996; 16:146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grentzmann G., Ingram J.A., Kelly P.J., Gesteland R.F., Atkins J.F.. A dual-luciferase reporter system for studying recoding signals. RNA. 1998; 4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 54. Smith P.K., Krohn R.I., Hermanson G.T., Mallia A.K., Gartner F.H., Provenzano M.D., Fujimoto E.K., Goeke N.M., Olson B.J., Klenk D.C.. Measurement of protein using bicinchoninic acid. Annu. Rev. Biochem. 1985; 150:76–85. [DOI] [PubMed] [Google Scholar]
- 55. Jacobs G.H., Chen A., Stevens S.G., Stockwell P.A., Black M.A., Tate W.P., Brown C.M.. Transterm: a database to aid the analysis of regulatory sequences in mRNAs. Nucleic Acids Res. 2009; 37:D72–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Keller O., Kollmar M., Stanke M., Waack S.. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics. 2011; 27:757–763. [DOI] [PubMed] [Google Scholar]
- 57. Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J. et al. . The UCSC Genome Browser Database. Nucleic Acids Res. 2003; 31:51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li W., Yang B., Liang S., Wang Y., Whiteley C., Cao Y., Wang X.. BLogo: a tool for visualization of bias in biological sequences. Bioinformatics. 2008; 24:2254–2255. [DOI] [PubMed] [Google Scholar]
- 59. Steitz J.A. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969; 224:957–964. [DOI] [PubMed] [Google Scholar]
- 60. Wolin S.L., Walter P.. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988; 7:3559–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Anger A.M., Armache J.P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., Beckmann R.. Structures of the human and Drosophila 80S ribosome. Nature. 2013; 497:80–85. [DOI] [PubMed] [Google Scholar]
- 63. Young D.J., Guydosh N.R., Zhang F., Hinnebusch A.G., Green R.. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3′UTRs in vivo. Cell. 2015; 162:872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jungreis I., Lin M.F., Spokony R., Chan C.S., Negre N., Victorsen A., White K.P., Kellis M.. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 2011; 21:2096–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McCaughan K.K., Brown C.M., Dalphin M.E., Berry M.J., Tate W.P.. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:5431–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Capone J.P., Sharp P.A., RajBhandary U.L.. Amber, ochre and opal suppressor tRNA genes derived from a human serine tRNA gene. EMBO J. 1985; 4:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Loenarz C., Sekirnik R., Thalhammer A., Ge W., Spivakovsky E., Mackeen M.M., McDonough M.A., Cockman M.E., Kessler B.M., Ratcliffe P.J. et al. . Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:4019–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Namy O., Moran S.J., Stuart D.I., Gilbert R.J., Brierley I.. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006; 441:244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S.. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012; 7:1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jenner L., Demeshkina N., Yusupova G., Yusupov M.. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat. Struct. Mol. Biol. 2010; 17:1072–1078. [DOI] [PubMed] [Google Scholar]
- 71. Shabalina S.A., Ogurtsov A.Y., Rogozin I.B., Koonin E.V., Lipman D.J.. Comparative analysis of orthologous eukaryotic mRNAs: potential hidden functional signals. Nucleic Acids Res. 2004; 32:1774–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Onouchi H., Nagami Y., Haraguchi Y., Nakamoto M., Nishimura Y., Sakurai R., Nagao N., Kawasaki D., Kadokura Y., Naito S.. Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis. Genes Dev. 2005; 19:1799–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Janzen D.M., Frolova L., Geballe A.P.. Inhibition of translation termination mediated by an interaction of eukaryotic release factor 1 with a nascent peptidyl-tRNA. Mol. Cell. Biol. 2002; 22:8562–8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cao J., Geballe A.P.. Inhibition of nascent-peptide release at translation termination. Mol. Cell. Biol. 1996; 16:7109–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Alkalaeva E.Z., Pisarev A.V., Frolova L.Y., Kisselev L.L., Pestova T.V.. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006; 125:1125–1136. [DOI] [PubMed] [Google Scholar]
- 76. Ingolia N.T., Lareau L.F., Weissman J.S.. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011; 147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Petry S., Brodersen D.E., Murphy F.V.t., Dunham C.M., Selmer M., Tarry M.J., Kelley A.C., Ramakrishnan V.. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005; 123:1255–1266. [DOI] [PubMed] [Google Scholar]
- 78. Young D.J., Edgar C.D., Poole E.S., Tate W.P.. The codon specificity of eubacterial release factors is determined by the sequence and size of the recognition loop. RNA. 2010; 16:1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cheng Z., Saito K., Pisarev A.V., Wada M., Pisareva V.P., Pestova T.V., Gajda M., Round A., Kong C., Lim M. et al. . Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 2009; 23:1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Preis A., Heuer A., Barrio-Garcia C., Hauser A., Eyler D.E., Berninghausen O., Green R., Becker T., Beckmann R.. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep. 2014; 8:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Svidritskiy E., Brilot A.F., Koh C.S., Grigorieff N., Korostelev A.A.. Structures of yeast 80S ribosome-tRNA complexes in the rotated and non-rotated conformations. Structure. 2014; 22:1210–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Inagaki Y., Blouin C., Doolittle W.F., Roger A.J.. Convergence and constraint in eukaryotic release factor 1 (eRF1) domain 1: the evolution of stop codon specificity. Nucleic Acids Res. 2002; 30:532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Fan-Minogue H., Du M., Pisarev A.V., Kallmeyer A.K., Salas-Marco J., Keeling K.M., Thompson S.R., Pestova T.V., Bedwell D.M.. Distinct eRF3 requirements suggest alternate eRF1 conformations mediate peptide release during eukaryotic translation termination. Mol. Cell. 2008; 30:599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Salas-Marco J., Bedwell D.M.. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell Biol. 2004; 24:7769–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jacobson A., Peltz S.W.. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 1996; 65:693–739. [DOI] [PubMed] [Google Scholar]
- 86. Schueren F., Lingner T., George R., Hofhuis J., Dickel C., Gartner J., Thoms S.. Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals. Elife. 2014; 3:e03640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Loughran G., Chou M.Y., Ivanov I.P., Jungreis I., Kellis M., Kiran A.M., Baranov P.V., Atkins J.F.. Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 2014; 42:8928–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S.. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013; 2:e01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Stiebler A.C., Freitag J., Schink K.O., Stehlik T., Tillmann B.A., Ast J., Bolker M.. Ribosomal readthrough at a short UGA stop codon context triggers dual localization of metabolic enzymes in fungi and animals. PLoS Genet. 2014; 10:e1004685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Prieto-Godino L.L., Rytz R., Bargeton B., Abuin L., Arguello J.R., Peraro M.D., Benton R.. Olfactory receptor pseudo-pseudogenes. Nature. 2016; 539:93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Firth A.E., Wills N.M., Gesteland R.F., Atkins J.F.. Stimulation of stop codon readthrough: frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res. 2011; 39:6679–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Poole E.S., Brown C.M., Tate W.P.. The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J. 1995; 14:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Uchida N., Hoshino S., Imataka H., Sonenberg N., Katada T.. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in cap/poly(A)-dependent translation. J. Biol. Chem. 2002; 277:50286–50292. [DOI] [PubMed] [Google Scholar]
- 94. Namy O., Rousset J.P., Napthine S., Brierley I.. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell. 2004; 13:157–168. [DOI] [PubMed] [Google Scholar]
- 95. Cridge A.G. Characterisation of the eukaryotic termination element. PhD thesis. 2004; University of Otago. [Google Scholar]
- 96. Basu J., Williams B.C., Li Z., Williams E.V., Goldberg M.L.. Depletion of a Drosophila homolog of yeast Sup35p disrupts spindle assembly, chromosome segregation, and cytokinesis during male meiosis. Cytoskeleton. 1998; 39:286–302. [DOI] [PubMed] [Google Scholar]
- 97. Malta-Vacas J., Aires C., Costa P., Conde A.R., Ramos S., Martins A.P., Monteiro C., Brito M.. Differential expression of the eukaryotic release factor 3 (eRF3/GSPT1) according to gastric cancer histological types. J. Clin. Pathol. 2005; 58:621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.